Abstract
Dental caries is the most prevalent chronic condition worldwide. Early detection can significantly improve treatment outcomes and reduce the need for invasive procedures. Recently, near-infrared transillumination (TI) imaging has been shown to be effective for the detection of early stage lesions. In this work, we present a deep learning model for the automated detection and localization of dental lesions in TI images. Our method is based on a convolutional neural network (CNN) trained on a semantic segmentation task. We use various strategies to mitigate issues related to training data scarcity, class imbalance, and overfitting. With only 185 training samples, our model achieved an overall mean intersection-over-union (IOU) score of 72.7% on a 5-class segmentation task and specifically an IOU score of 49.5% and 49.0% for proximal and occlusal carious lesions, respectively. In addition, we constructed a simplified task, in which regions of interest were evaluated for the binary presence or absence of carious lesions. For this task, our model achieved an area under the receiver operating characteristic curve of 83.6% and 85.6% for occlusal and proximal lesions, respectively. Our work demonstrates that a deep learning approach for the analysis of dental images holds promise for increasing the speed and accuracy of caries detection, supporting the diagnoses of dental practitioners, and improving patient outcomes.
Keywords: caries detection/diagnosis/prevention, digital imaging/radiology, artificial intelligence, dental informatics/bioinformatics, informatics, oral diagnosis
Introduction
Dental caries is the most prevalent chronic disease globally. Even with recent declines in the rates of large cavitated lesions, early lesions are present in most of the population (Kassebaum et al. 2015). Conventional caries detection methods that rely on visual inspection and the use of a dental probe are effective for large, clearly visible caries and for those partially obscured but accessible with a handheld mirror (Fejerskov and Kidd 2009). On the other hand, dental radiography is used for detecting hidden or inaccessible lesions. However, it is known that early detection of dental lesions, which is an important determinant of treatment outcomes, would benefit from the introduction of new tools (Schwendicke et al. 2015).
To this end, near-infrared transillumination (TI), a nonionizing imaging technology that leverages differences in scattering and absorption of near-infrared light depending on the degree of tooth mineralization, promises to fill the gap. In vitro and in vivo studies investigating near-infrared TI for the detection of early caries have yielded encouraging results (Fried et al. 2005; Simon, Darling et al. 2016; Simon, Lucas et al. 2016; Litzenburger et al. 2018).
A commercially available system, DIAGNOcam (KaVo), uses this technology to provide grayscale images with useful information about early enamel caries and dentin caries (Söchtig et al. 2014; Abdelaziz and Krejci 2015; Kühnisch et al. 2016; Abdelaziz et al. 2018). The interpretation of these images, as for any diagnostic imaging technology, is limited by interrater disagreement. For this reason, there has been increasing interest in computer-assisted image analysis to support dental procedures (Behere and Lele 2011; Tracy et al. 2011).
In recent years, deep learning models based on convolutional neural networks (CNNs) have been successfully deployed for computer vision applications. These models have achieved high accuracy, to the extent that they now represent the state of the art for a wide range of applications, including image classification (Chollet 2017), object detection (Redmon and Farhadi 2017), and semantic segmentation (Chen et al. 2018). Increased interest in deep learning models has also led to their adoption for medical imaging (Litjens et al. 2017).
In particular, a first deep learning model for semantic segmentation was introduced in Long et al. (2015). A subsequent model, SegNet (Badrinarayanan et al. 2017), offers improvements through the adoption of a symmetric autoencoder architecture. Another model, U-Net (Ronneberger et al. 2015a), takes inspiration from autoencoders but also introduces skip connections between corresponding layers in the encoding and decoding path, yielding further improvements in accuracy. An even more recent model, DeepLab (Chen et al. 2018), uses a deep encoder with residual connections and dilated convolution, generating more accurate multiscale predictions.
Previous efforts have attempted to apply machine learning techniques to dental disease diagnostics. A distinction should be made between models for image classification (Ali et al. 2016; Imangaliyev et al. 2016; Prajapati et al. 2017; Lee et al. 2018) and models for semantic segmentation (Ronneberger et al. 2015b; Rana et al. 2017; Srivastava et al. 2017). Classification models predict a categorical label for an entire image. These labels are typically binary (e.g., healthy/diseased), although multiclass schemes are also used (Prajapati et al. 2017). Classification-based models are simpler and represent the majority of the literature, but they are more limited in scope as they merely predict a single value for the image without delineating the spatial extent of the disease. On the other hand, segmentation-based models are able to provide a pixelwise classification of the input image. Such models usually employ pixelwise binary classification (Rana et al. 2017; Srivastava et al. 2017), although Ronneberger et al. (2015b) implemented a more sophisticated model capable of distinguishing between background, types of dental tissue, caries, and dental restorations. However, the accuracy of this segmentation model was poor, especially for carious tissue (Wang et al. 2016).
Since X-ray radiography is the most common dental imaging modality in clinical practice, most studies of automated caries detection pertain to X-ray images. However, machine learning techniques have also been applied to other oral diseases and a variety of imaging techniques, including intraoral fluorescence imaging for detecting inflammation on gingival surfaces (Rana et al. 2017) and quantitative light-induced fluorescence imaging for plaque classification (Imangaliyev et al. 2016).
To summarize, we observed that all of the aforementioned studies suffered from 1 or more of the following shortcomings: solving a simplified problem (classification instead of segmentation), requiring hundreds to thousands of samples for training, or having poor prediction accuracy. Finally, while machine learning methods have been applied to X-ray data sets, to date they have not been tested on near-infrared TI images.
In this work, we train a CNN-based model for the detection and classification of dental lesions images obtained with near-infrared TI imaging. We investigate various techniques to overcome issues related to the scarcity of training data, and we test the performance of our model on 2 tasks: pixelwise segmentation into tissue classes and binary labeling (healthy/diseased) of regions of interest.
Materials and Methods
Inputs and Segmentation Labels
Our data set consists of 217 grayscale images of upper and lower molars and premolars, obtained with the DIAGNOcam system. Images were acquired at the Geneva university dental clinics, by graduate and postgraduate dental students. Images included in this study were acquired between 2013 and 2018. The project was reviewed by the Geneva Swissethics committee (CCER_Req-2017-00361). Each image is roughly centered on the occlusal surface of a tooth. To speed up the training of our model, we downsampled each image from 480 × 640 pixels to 256 × 320 pixels. We collected reference segmentation maps of our images from dental experts with clinical experience using the DIAGNOcam. Five possible segmentation classes were considered: background (B), enamel (E), dentin (D), proximal caries (PC), and occlusal caries (OC). Images containing dental restorations were discarded, as near-infrared TI imaging is mainly targeted at primary lesions, due to the impaired transmission of light through the bulk of the thick filling material. We included 2 distinct classes for enamel and dentin in our segmentation maps, as the identification of these tissues is essential for the evaluation of the extension and severity of the lesion.
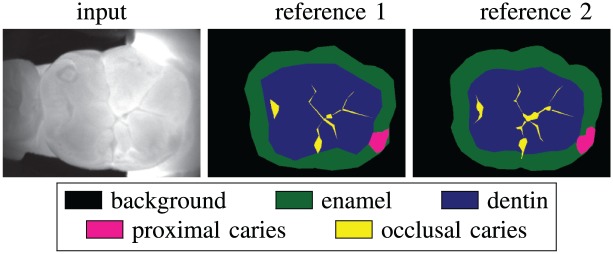
It is important to emphasize that manual labeling by experts provides a reference that is necessary for training and evaluating the model but does not necessarily represent ground truth. Histological staining of dental sections is the gold standard for evaluating the degree and extent of decay, but for our data set, it would neither be practically feasible nor readily provide a pixelwise accurate segmentation map. While interrater agreement for dental diagnostics is generally within an acceptable range for clinical practice (Litzenburger et al. 2018), some degree of discrepancy is inevitable, as we observed in the expert-drawn labels for our semantic segmentation task. For example, Figure 1 shows an image with segmentation labels drawn by 2 experts. Although they appear qualitatively similar, their mean intersection-over-union (mIOU) (see Materials and Methods) is 74.2%, and for the occlusal lesion, the intersection-over-union (IOU) score is as low as 42.5%. This observation demonstrates the importance of a second opinion for a reliable and accurate diagnosis. Indeed, our model could be integrated into a medical device to support dental diagnoses. On the other hand, this example argues that pixelwise metrics alone may not supply a complete description of the similarity between 2 labels, whether they are drawn by dentists or by algorithms. For this reason, we also introduced a high-level binary labeling scheme that was derived from the segmentation labels.
Figure 1.
Example of grayscale image of a molar obtained with DIAGNOcam and reference segmentation labels as drawn by 2 dentists. The mean intersection-over-union (mIOU) across all classes is 74.2%, while per-class scores are IOU = 97.0% (background), IOU = 78.4% (enamel), IOU = 85.7% (dentin), IOU = 67.2% (proximal caries), IOU = 42.5% (occlusal caries).
Binary Labels for Regions of Interest
In the auxiliary binary labeling scheme referred to above, we divided each tooth into 3 regions of interest (ROIs): mesial proximal surface, occlusal surface, and distal proximal surface. For each ROI, we set a binary label (healthy/diseased) equal to 1 if at least 1 pixel in that ROI was labeled as carious (PC for the proximal surfaces, OC for the occlusal surface) in the segmentation map and 0 otherwise.
This approach allows a high-level evaluation of segmentation labels. For example, the labels shown in Figure 1 have slight disparities that cause significant downstream differences in pixelwise metrics. However, binary ROI labeling predicts a healthy mesial proximal surface, carious occlusal surface, and carious distal proximal surface for both images.
While this approach provides useful information regarding the overall agreement of 2 segmentation maps, the localization of lesions is lost. Thus, pixelwise analysis is still necessary to provide a complete overview of disease progression. In Results, we present and evaluate the results of our model using both the original pixelwise segmentation labels and the derived ROI binary labels.
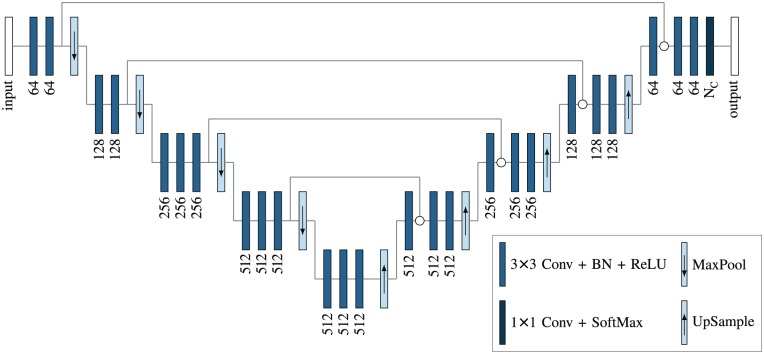
Neural Network Architecture
We compared the results obtained using the different approaches proposed in the machine learning literature for semantic segmentation to design an optimal architecture for our neural network. The architecture that we selected for this study is shown in Figure 2. It comprises a symmetric autoencoder with skip connections similar to U-Net (Ronneberger et al. 2015a) and an encoding path inspired by the structure of the VGG16 classifier (Simonyan and Zisserman 2015).
Figure 2.
Neural network architecture used in this work. Nc is the number of segmentation classes. In our work, Nc = 5.
Improving Model Training
A typical tactic for deep learning applications with limited training data is to use pretrained weights for the encoding path. To this end, we tested the possibility of replacing the encoding path of our architecture with a VGG16 model pretrained on the ImageNet data set.
Another common performance-enhancing strategy is data augmentation, wherein the size and diversity of the data set are increased by applying various transformations to the original images. We implemented a data augmentation step in our training pipeline using the following transformations: flip, zoom, rotation, translation, contrast, and brightness.
We also considered several approaches to prevent overfitting. U-Net (Ronneberger et al. 2015a) uses dropout regularization, but we achieved better performance by instead applying batch normalization before each rectified linear unit (ReLU) activation, as shown in Figure 2.
Finally, CNNs for semantic segmentation are typically trained by minimizing categorical cross-entropy loss with an optimization method based on mini-batch stochastic gradient descent (Ronneberger et al. 2015a). This approach gives equal weight to all classes, which is not suitable for unbalanced data sets. This is indeed our case: 49.4% of pixels represent background, while only 0.7% of pixels are occlusal lesions. One approach for addressing the imbalance is to modify the loss function by introducing weights that are inversely proportional to the pixel distribution of each class. In this study, we tested the effects of both inversely proportional weights and weights inversely proportional to the square root of the pixel distribution, the latter yielding better performance. To our knowledge, this is the first time that this weighting scheme has been used.
Statistical Analysis and Evaluation Metrics
Selecting a task-appropriate performance metric for a model is critical, as outcomes tend to be highly sensitive to this choice. We employ the IOU metric, also known as the Jaccard index. For each class k, this metric computes the ratio between the number of pixels that are correctly predicted as k, divided by the number of pixels that belong to class k in either the reference label or in the predicted label. This quantity is often summarized by the mean of all IOU values in each class (mIOU). The output of our model is a vector of Nc = 5 elements for each pixel, representing a probabilistic prediction that the pixel belongs to each class. To compute the IOU, we transformed this output into a segmentation map by taking the class with the highest predicted probability for each pixel.
To evaluate our model on the higher-level task of binary classification of proximal and occlusal regions of interest, we used the receiver operating characteristic (ROC) curve, which plots the true-positive rate against the false-positive rate as a function of varying discrimination threshold. The curve can be summarized by the area under curve (AUC) statistic. For each ROI, we defined the true value to be 1 if at least 1 pixel is labeled as carious and the predicted value as the highest predicted probability over all pixels in that region for each respective lesion class.
To test the model’s predictive performance and assess its generalization capacity, we used a Monte Carlo cross-validation scheme with 20 stratified splits, ensuring that the training and validation data sets had the same ratio of upper and lower molars and premolars. At each iteration, we trained our model on 185 samples and validated it on the remaining 32 samples.
Results
Comparing the performance achieved by our architecture (Fig. 2) with deeper neural networks (e.g., DeepLab), we observed that the latter tended to overfit our data set, generating worse predictions in the validation phase, even with the inclusion of residual connections. We also found that dilated convolution gave no improvements in performance. In all cases, using an encoding path that was pretrained on the ImageNet data set provided no benefit.
The Table summarizes the validation mIOU scores (averaged across the 20 splits) obtained using different strategies. The results indicate that the baseline model gave a mIOU of 63.1%. Introducing data augmentation and batch normalization provided a significant improvement in accuracy, bringing the mIOU up to 72.0%. Finally, a weighted loss further improved the mIOU score. In particular, weights inversely proportional to the square root of the per-class pixel counts gave better results than the linear weights commonly used for unbalanced data sets.
Table.
Summary of Validation Scores Achieved on the Semantic Segmentation Task Using Different Approaches to Train the Network.
| Data Augmentation | Batch Normalization | Loss Weights | mIOU, % |
|---|---|---|---|
| 63.1 | |||
| ✓ | 70.7 | ||
| ✓ | ✓ | 72.0 | |
| ✓ | ✓ | ✓—linear | 72.3 |
| ✓ | ✓ | ✓—square root | 72.7 |
mIOU, mean intersection-over-union.
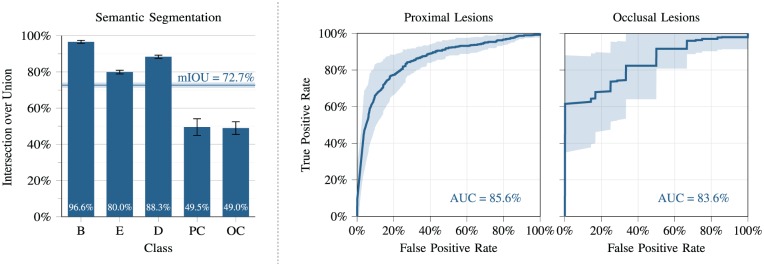
Figure 3 offers a more detailed analysis of the results obtained using the best version of our model, scoring an overall mIOU = 72.7%. The IOU histogram shows that our model achieved excellent performance on the background (IOU = 96.6%), enamel (IOU = 80.0%), and dentin (IOU = 88.3%) classes. For proximal and occlusal carious regions, the IOU scores are 49.5% and 49.0%, respectively. More specifically, we observed that 33.0% and 35.5% of pixels labeled as proximal and occlusal lesions in the reference were instead predicted as enamel and dentin, respectively.
Figure 3.
Quantitative analysis of the results achieved by our model for the semantic segmentation task (left) and for the binary region-of-interest classification task (right).
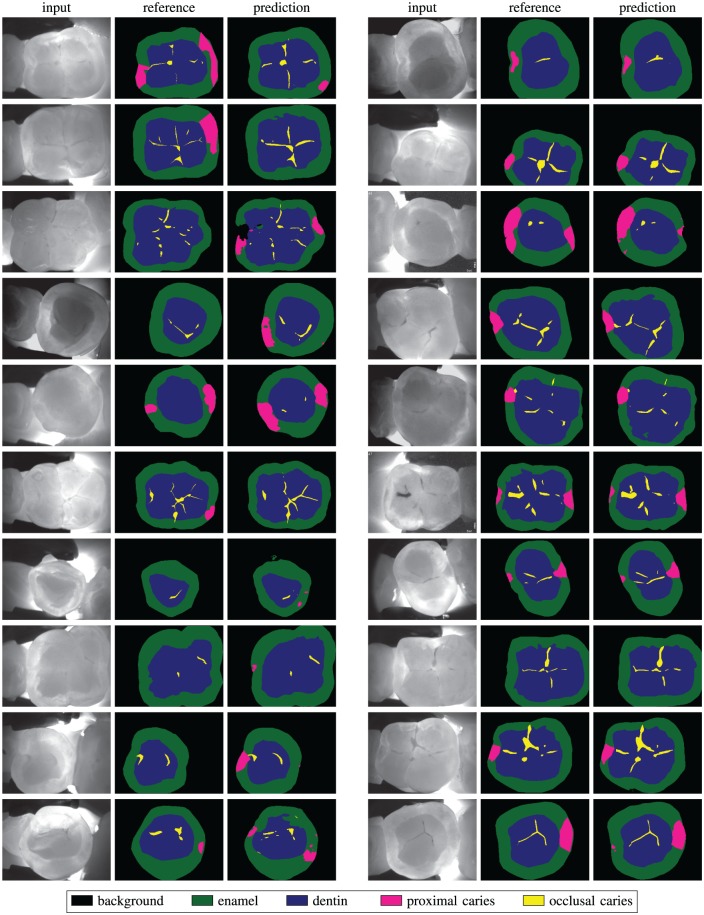
Figure 4 depicts selected results obtained on validation data for a qualitative overview of our model’s predictions. On the right-hand side, we see samples where the correspondence between the reference and the model’s predictions is both numerically high and visually salient. On the left-hand side, we see samples where references and predictions diverge.
Figure 4.
Examples of segmentation results on the validation set. Left: samples where predictions of our model and reference segmentation show discrepancies. Right: samples where agreement between our model and the reference is high.
As for the analysis of our results in terms of a high-level binary classification of regions of interest, in Figure 3, we see that the predicted and reference labels largely agree in both proximal and occlusal regions (AUC = 85.6% and AUC = 83.6%, respectively).
Discussion
A pixelwise comparison of our model’s predictions with reference segmentations drawn by experts shows an excellent agreement for background, enamel, and dentin classes (which all have an IOU score above 80%). For carious lesions, our model gives a lower IOU, but this can be in part explained by the observation that the precise boundaries of lesions are difficult to trace, especially for narrow occlusal lesions. As we noted in Figure 1, the IOU metric is sensitive to small discrepancies, and even similar-looking segmentation maps labeled by different experts can have low IOU scores for classes with few pixels.
A qualitative analysis of our predicted segmentation (Fig. 4) suggests that the model can effectively learn to provide predictions that reproduce those of a trained dental expert, given sufficient training data. In most of the model’s failure cases depicted in Figure 4, the deviation from the reference is eclipsed by possible disagreement among experts on the interpretation of the presence or exact extension of a lesion. In a few cases, however, we see physically unrealistic segmentation artifacts; most of these artifacts are produced in overexposed or underexposed areas, so a straightforward way to improve these results would be to collect more training data. Another possibility would be to use a conditional generative adversarial network (cGAN) (Isola et al. 2017), which allows the network to learn its own loss function and automatically penalize unrealistic artifacts.
The analysis of our predictions in terms of binary classification of regions of interest (Fig. 3) highlights how, even if our model’s predicted segmentation maps differ from the reference maps in the exact location and extent of lesions, there can be substantial agreement on the presence or absence of lesions in the regions of interest of the dental scan.
Finally, we note that in the ROC curve for occlusal lesions, a high true-positive rate was measured at every discrimination threshold. This is due to an important class imbalance, as only about 10.1% of our images are free of lesions on the occlusal surface. This is representative of clinical reality, as most individuals are likely to develop occlusal lesions in the course of their life (Carvalho et al. 2016).
Overall, the performance of our model represents a dramatic improvement over the previous effort of Ronneberger et al. (2015b) that achieved IOU = 7.3% and IOU = 7.8% (on 2 different validation data sets), using a similar approach on X-ray images (Wang et al. 2016). This improvement is chiefly accounted for by 2 observations. First, we trained and validated our model on near-infrared TI images, while Ronneberger et al. (2015b) worked with a different data set comprising X-ray images. Second, as shown in the Table, we employed various techniques to overcome the scarcity of training data, which significantly improved the generalization accuracy of our model.
As previously mentioned, accuracy did not improve with deeper architectures. In fact, very deep networks are either trained on large data sets or leverage pretrained weights. With only 185 training samples, our data set was insufficient to train a deep network from scratch. In addition, the poor results obtained with a pretrained encoder are expected, since transfer learning does not improve performance if the original data and pretraining data are sufficiently dissimilar (Yosinski et al. 2014). This was true in our case, since the large databases (e.g., ImageNet) where the weights are pretrained consist of images unrelated to our application.
Our model still exhibits several shortcomings. We occasionally observed physically unrealistic labeling artifacts, especially in overexposed or underexposed regions. Moreover, our data set was limited both in terms of size and in terms of the certainty of the ground truth labels. Data augmentation is known to play an important role in computer vision applications as it helps with the scarcity of training data by reducing overfitting and improving the model’s generalization performance (Krizhevsky et al. 2012), although this approach is limited and collecting new diverse training samples would be required to further improve the performance.
For the future, we acknowledge different ways in which our work could be extended. First, the size of the data set could be increased and its quality enhanced through the inclusion of histological exams and consensus-based labels provided by a panel of experts. Second, a cGAN-inspired approach could reduce labeling artifacts. Finally, our model could benefit from varied inputs such as images centered on the proximal surfaces or from using video inputs instead of static images to allow for real-time, augmented reality predictions.
In this way, we envision future iterations of the algorithmic foundation described here being integrated into tools with real clinical applicability, facilitating the chronic monitoring and diagnosis of dental disease.
Conclusion
Computer-assisted analysis tools could support dentists by providing high-throughput diagnostic assistance. In this work, we demonstrated the use of an automatic deep learning approach for semantic segmentation of dental scans obtained using the DIAGNOcam system. Our results suggest that a similar approach could be used for the automated interpretation of DIAGNOcam scans to facilitate dental diagnoses.
Author Contributions
F. Casalegno, T. Newton, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; R. Daher, M. Abdelaziz, A. Lodi-Rizzini, I. Krejci, contributed to conception, data acquisition, and interpretation, critically revised the manuscript; F. Schürmann, H. Markram, contributed to conception and data analysis, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Footnotes
This study was supported by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology, and by internal funds of the Division of Cariology and Endodontology, Clinique universitaire de médecine dentaire, Université de Genève.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abdelaziz M, Krejci I. 2015. DIAGNOcam: a Near Infrared Digital Imaging Transillumination (NIDIT) technology. Int J Esthet Dent. 10(1):158–165. [PubMed] [Google Scholar]
- Abdelaziz M, Krejci I, Perneger T, Feilzer A, Vazquez L. 2018. Near infrared transillumination compared with radiography to detect and monitor proximal caries: a clinical retrospective study. J Dent. 70:40–45. [DOI] [PubMed] [Google Scholar]
- Ali RB, Ejbali R, Zaied M. 2016. Detection and classification of dental caries in X-ray images using deep neural networks. In: International Conference on Software Engineering Advances (ICSEA). p. 236. [Google Scholar]
- Badrinarayanan V, Kendall A, Cipolla R. 2017. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 39(12):2481–2495. [DOI] [PubMed] [Google Scholar]
- Behere RR, Lele SM. 2011. Reliability of Logicon caries detector in the detection and depth assessment of dental caries: an in-vitro study. Indian J Dent Res. 22(2):362. [DOI] [PubMed] [Google Scholar]
- Carvalho JC, Dige I, Machiulskiene V, Qvist V, Bakhshandeh A, Fatturi-Parolo C, Maltz M. 2016. Occlusal caries: biological approach for its diagnosis and management. Caries Res. 50(6):527–542. [DOI] [PubMed] [Google Scholar]
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. 2018. Deeplab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 40(4):834–848. [DOI] [PubMed] [Google Scholar]
- Chollet F. 2017. Xception: Deep learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition IEEE p. 1251–1258. [Google Scholar]
- Fejerskov O, Kidd E. 2009. Dental caries: the disease and its clinical management. Hoboken, NJ: John Wiley. [Google Scholar]
- Fried D, Featherstone JD, Darling CL, Jones RS, Ngaotheppitak P, Buhler CM. 2005. Early caries imaging and monitoring with near-infrared light. Dent Clin North Am. 49(4):771–793, vi. [DOI] [PubMed] [Google Scholar]
- Imangaliyev S, van der Veen MH, Volgenant CM, Keijser BJ, Crielaard W, Levin E. 2016. Deep learning for classification of dental plaque images. In: International Workshop on Machine Learning, Optimization, and Big Data. Cham, Switzerland: Springer; p. 407–410. [Google Scholar]
- Isola P, Zhu JY, Zhou T, Efros AA. 2017. Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition IEEE p. 1125–1134. [Google Scholar]
- Kassebaum NJ, Bernabé E, Dahiya M, Bhandari B, Murray CJ, Marcenes W. 2015. Global burden of untreated caries: a systematic review and metaregression. J Dent Res. 94(5):650–658. [DOI] [PubMed] [Google Scholar]
- Krizhevsky A, Sutskever I, Hinton GE. 2017. ImageNet classification with deep convolutional neural networks. Commun ACM. 60(6):84–90. [Google Scholar]
- Kühnisch J, Söchtig F, Pitchika V, Laubender R, Neuhaus KW, Lussi A, Hickel R. 2016. In vivo validation of near-infrared light transillumination for interproximal dentin caries detection. Clin Oral Investig. 20(4):821–829. [DOI] [PubMed] [Google Scholar]
- Lee JH, Kim DH, Jeong SN, Choi SH. 2018. Detection and diagnosis of dental caries using a deep learning-based convolutional neural network algorithm. J Dent. 77:106–111. [DOI] [PubMed] [Google Scholar]
- Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. 2017. A survey on deep learning in medical image analysis. Med Image Anal. 42:60–88. [DOI] [PubMed] [Google Scholar]
- Litzenburger F, Heck K, Pitchika V, Neuhaus KW, Jost FN, Hickel R, Jablonski-Momeni A, Welk A, Lederer A, Kühnisch J. 2018. Inter- and intraexaminer reliability of bitewing radiography and near-infrared light transillumination for proximal caries detection and assessment. Dentomaxillofac Radiol. 47(3):20170292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Shelhamer E, Darrell T. 2015. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition IEEE p. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Prajapati SA, Nagaraj R, Mitra S. 2017. Classification of dental diseases using CNN and transfer learning. In: 5th International Symposium on Computational and Business Intelligence (ISCBI) IEEE p. 70–74. [Google Scholar]
- Rana A, Yauney G, Wong LC, Gupta O, Muftu A, Shah P. 2017. Automated segmentation of gingival diseases from oral images. In: Proceedings of the IEEE Conference on Healthcare Innovations and Point of Care Technologies (HI-POCT) IEEE p. 144–147. [Google Scholar]
- Redmon J, Farhadi A. 2017. YOLO9000: better, faster, stronger. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition IEEE p. 7263–7271. [Google Scholar]
- Ronneberger O, Fischer P, Brox T. 2015. a. U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention Cham, Switzerland: Springer; p. 234–241. [Google Scholar]
- Ronneberger O, Fischer P, Brox T. 2015. b. Dental X-ray image segmentation using a U-shaped Deep Convolutional network. ISBI. http://www-o.ntust.edu.tw/~cweiwang/ISBI2015/challenge2/isbi2015_Ronneberger.pdf (accessed 16 May 2019).
- Schwendicke F, Tzschoppe M, Paris S. 2015. Radiographic caries detection: a systematic review and meta-analysis. J Dent. 43(8):924–933. [DOI] [PubMed] [Google Scholar]
- Simon JC, Darling CL, Fried D. 2016. A system for simultaneous near-infrared reflectance and transillumination imaging of occlusal carious lesions. Proc SPIE Int Soc Opt Eng. 2016:9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JC, Lucas SA, Staninec M, Tom H, Chan KH, Darling CL, Cozin MJ, Lee RC, Fried D. 2016. Near-IR transillumination and reflectance imaging at 1,300 nm and 1,500-1,700 nm for in vivo caries detection. Lasers Surg Med. 48(9):828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A. 2015. Very deep convolutional networks for large-scale image recognition. In: Proceedings of the International Conference on Learning Representations (ICLR) New Orleans, LA: ICLR. [Google Scholar]
- Söchtig F, Hickel R, Kühnisch J. 2014. Caries detection and diagnostics with near-infrared light transillumination: clinical experiences. Quintessence Int. 45(6):531–538. [DOI] [PubMed] [Google Scholar]
- Srivastava MM, Kumar P, Pradhan L, Varadarajan S. 2017. Detection of tooth caries in bitewing radiographs using deep learning. arXiv preprint arXiv:1711.07312. https://arxiv.org/abs/1711.07312 (accessed 16 May 2019).
- Tracy KD, Dykstra BA, Gakenheimer DC, Scheetz JP, Lacina S, Scarfe WC, Farman AG. 2011. Utility and effectiveness of computer-aided diagnosis of dental caries. Gen Dent. 59(2):136–144. [PubMed] [Google Scholar]
- Wang CW, Huang CT, Lee JH, Li CH, Chang SW, Siao MJ, Lai TM, Ibragimov B, Vrtovec T, Ronneberger Oet al. 2016. A benchmark for comparison of dental radiography analysis algorithms. Med Image Anal. 31:63–76. [DOI] [PubMed] [Google Scholar]
- Yosinski J, Clune J, Bengio Y, Lipson H. 2014. How transferable are features in deep neural networks? In: Advances in neural information processing system p. 3320–3328. [Google Scholar]