Abstract
The AAA+ ATPase spastin remodels microtubule arrays through severing, and its mutation is the most common cause of hereditary spastic paraplegias (HSP). Polyglutamylation of the tubulin C-terminal tail recruits spastin to microtubules and modulates severing activity. Here, we present a ~3.2 Å resolution cryo-EM structure of the Drosophila melanogaster spastin hexamer with a polyglutamate peptide bound in its central pore. Two electropositive loops arranged in a double-helical staircase coordinate the substrate sidechains. The structure reveals how concurrent nucleotide and substrate binding organizes the conserved spastin pore loops into an ordered network that is allosterically coupled to oligomerization, and suggests how tubulin tail engagement activates spastin for microtubule disassembly. This allosteric coupling may apply generally in organizing AAA+ protein translocases into their active conformations. We show that this allosteric network is essential for severing and is a hotspot for HSP mutations.
Spastin is a microtubule-severing AAA ATPase (ATPases Associated with diverse cellular Activities) whose function is important in basic cell biological processes ranging from neurogenesis and axonal maintenance to nuclear envelope breakdown, vesicle trafficking, mitosis, and cytokinesis1. Spastin is recruited to endosomes and the midbody through its interaction with ESCRTIII and is thought to participate in the coordinated remodeling of membranes and microtubules1. Spastin ATPase activity is stimulated by microtubules2–4 and microtubule severing activity requires adenosine triphosphate (ATP) hydrolysis2,5–7. Spastin mutations are responsible for ~50% of hereditary spastic paraplegias (HSPs), a large and clinically diverse family of neurodegenerative disorders8. In the pure form of the disease, HSP patients typically exhibit axonal degeneration in motor axons of the corticospinal tract, leading to progressive lower limb spasticity and weakness8–10. Studies in model organisms11 as well as patient derived induced pluripotent stem cells12 have shown that neurons are especially sensitive to spastin gene dosage. Cellular and biochemical studies have shown that many of the disease associated mutations examined impair microtubule severing and ATP hydrolysis5–7,13. However, the etiology of spastin induced HSP is still poorly understood.
Spastin has a modular structure comprising a three-helix bundle microtubule-interacting and trafficking (MIT) domain, a poorly-conserved linker region, and an AAA+ ATPase domain. This domain architecture is shared among all three known microtubule severing enzymes - spastin, katanin, and fidgetin1. The ATPase domain is structurally homologous to that of other members of the AAA+ protein family and consists of an α/β nucleotide-binding domain (NBD) and a four-helix bundle domain (HBD). Both spastin and katanin differ from other AAA+ ATPase enzymes in that they contain two additional helices within the NBD that are essential for severing6,14. The AAA+ ATPase domains hexamerize in a nucleotide-dependent manner and the tubulin substrate lowers the critical concentration for oligomerization15,16. Consistent with this, high-affinity binding of spastin to microtubules is highly cooperative and requires ATP4. The substrate-dependent oligomerization is likely important for the specificity and timing of action of these AAA+ ATPases in the cell, as unregulated spastin and katanin microtubule-severing activity is highly deleterious17–19. The majority of disease associated mutations of the SPAST gene are found in the ATPase domain of spastin.
Microtubule severing by spastin requires the β-tubulin C-terminal tail, an intrinsically disordered element that decorates the microtubule surface. While the α-tubulin tail contributes to binding, it is not required for severing activity20. It was proposed that microtubule severing involves the hexamerization of spastin protomers around the ~20 residue, negatively charged tubulin tail, and the subsequent pulling of the tubulin subunit out the microtubule lattice using ATP-driven conformational changes in three pore loops that help translocate the substrate through the central pore of the hexameric ATPase6,21. However, there has been no direct demonstration of the tubulin tail engagement by the spastin pore nor any atomistic structural information on the spastin hexamer or its interaction with the tubulin tail substrate. Spastin’s interaction with the microtubule is strongly enhanced by polyglutamylation of the β-tubulin tail20, a post-translational modification that involves the addition of multiple glutamate chains of variable lengths; unmodified microtubules are not effectively severed at in vivo spastin concentrations20,22. As many as 21 glutamates have been detected on tubulin tails in vivo23,24. Polyglutamylation is highly abundant in neurons where spastin activity is required for neurite extension as well as axonal maintenance and regeneration11.
Here, we present the cryo-EM structure of the spastin hexamer in complex with a polyglutamate peptide at ~3.2 Å resolution. The structure reveals an asymmetric hexamer with the AAA domains arranged in a split lock-washer conformation. Two conserved pore loops from the six protomers form a double-helical staircase gripping the odd and even glutamates of the substrate peptide, respectively. Residues in this double helix are positively charged and neutralize the electronegative polyglutamate peptide, consistent with the regulation of microtubule severing by polyglutamylation of tubulin tails. The substrate binding pore loops are allosterically coupled to the ATP binding site as well as the oligomerization interfaces, providing a structural explanation for the tubulin substrate binding-induced spastin activation. The majority of the residues in this allosteric network are mutated in HSP patients, underscoring their importance to spastin function. Our comprehensive structural analysis of all reported HSP-associated spastin missense mutations in its AAA core provides a framework for understanding spastin molecular dysfunction.
RESULTS
Spastin forms a hexameric spiral
Unlike many AAA+ ATPases, spastin exists as a monomer or dimer in the absence of nucleotide or bound to ADP, and only assembles into hexamers upon ATP-binding3,6 This hexamer is labile and falls apart at lower concentrations6. In order to stabilize the hexameric state for structural studies by cryo-electron microscopy (cryo-EM) we used a commonly used mutation in the Walker B motif of the ATPase domain (E583Q), which retains ATP binding but prevents ATP hydrolysis6,13. Analytical ultracentrifugation (AUC) shows that this construct assembles into hexamers in the presence of ATP at concentrations as low as 6 μM (Supplementary Fig. 1a). Initial structure determination yielded a reconstruction of the spastin hexamer limited to ~3.8 Å resolution showing cryo-EM density of adventitious peptide binding. As polyglutamylation enhances substrate binding20 and spastin has been shown to interact with polyglutamate peptides, we incubated our spastin preparation with polyglutamate (Methods). This increased the proportion of intact hexamers and allowed structural determination of the spastin-peptide complex to ~3.2 Å resolution (Table 1, Fig. 1, Supplementary Figs. 2 and 3). Poly-glutamate activates spastin ATPase with a maximal activation of ~six-fold above the basal level (Supplementary Fig. 1b), comparable to the activation reported with microtubules2. Analytical centrifugation with a fluorescently labeled poly-glutamate peptide shows that it comigrates with the spastin hexamer (Supplementary Fig. 1c). The choice of polyglutamate also overcomes the experimental issue of placing enzymes in a specific register with respect to the tubulin tail sequence which is overrepresented in polyglutamates, but also contains other amino acid residues.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| SpastinE583Q (PDB 6P07) (EMDB 20226) | |
|---|---|
| Data collection and Processing | |
| Microscope | Thermo Fischer Talos Arctica |
| Camera | Gatan K2 Summit DED |
| Magnification (nominal) | 36,000x |
| Magnification (calibrated) | 43478.3x |
| Voltage (kV) | 200 |
| Total electron exposure (e−/Å2) | 52 |
| Exposure rate (e-/pixel/sec) | 5.6 |
| Defocus range (μm) | −1.0 to −2.0 |
| Pixel size (Å) | 1.15 |
| Micrographs collected (no.) | 2,534 |
| Micrographs used (no.) | 2,534 |
| Total extracted particles (no.) | 2,736,865 |
| Refined particles (no.) | 1,259,553 |
| Final particles (no.) | 488,385 |
| Symmetry imposed | C1 |
| Resolution (global) | |
| FSC 0.5 (unmasked/masked) | 7.0/3.6 |
| FSC 0.143 (unmasked/masked) | 4.3/3.2 |
| Resolution Range (local) | 3 – 5 |
| Model composition | |
| Nonhydrogen atoms | 14,089 |
| Protein residues | 1804 |
| Ligands | 12 |
| Refinement | |
| Initial model used (PDB code) | 3B9P |
| Average FSC | 3.2 |
| B factors (Å2) | |
| Protein residues | 46.8 |
| Ligands | 50.5 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.14 |
| Validation | |
| MolProbity score | 1.46 |
| Clashscore | 3.05 |
| Poor rotamers (%) | 0.0 |
| C-beta deviations | 0 |
| Mean per-residue Cα RMSD (Å) | 0.64 |
| Per-residue Cα RMSD range (Å) | 0.03–5.66 |
| Ramachandran plot | |
| Favored (%) | 94.65% |
| Allowed (%) | 5.35% |
| Disallowed (%) | 0.00% |
| EMRinger score84 | 3.00 |
| CaBLAM outliers | 3.35% |
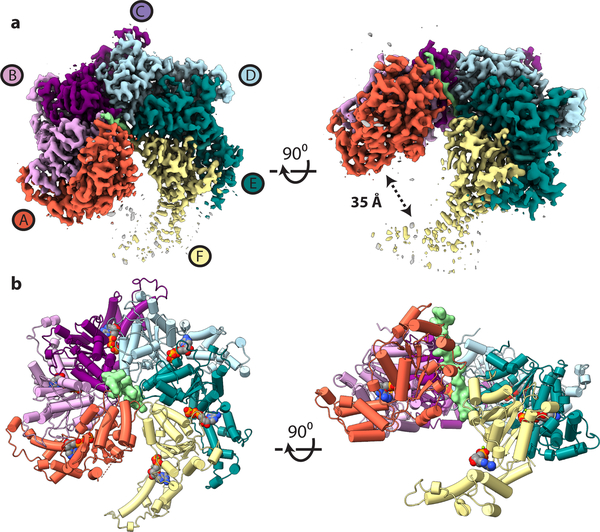
Figure 1. The architecture of the substrate-bound spastin hexamer in a split lock-washer conformation.
(a) Top (Left) and side view (Right) of the EM map of the spastin hexamer with individual protomers colored separately. Substrate density is colored in light green. The ~35 Å gap between protomer A and F is apparent in the sideview. (b) Top (Left) and side view (Right) of the atomic model in ribbon representation. Nucleotides are depicted as space-filling models and the polyglutamate peptide substrate is shown in surface representation.
The overall architecture of the spastin hexamer resembles that of a split lock-washer, as previously observed for other homohexameric ATPases25,26, including the related severing enzyme katanin14 (Fig. 1a). The individual protomers assemble into a right-handed spiral, with a ~6 Å rise for each protomer and a ~35 Å gap between the topmost and lowest protomers. The large (NBD) subdomain of the ATPase forms the central pore of the hexamer, with the HBD directed peripherally and in contact with the NBD of the neighboring protomer. The cryo-EM density corresponding to the “gating” protomers A and F is lower in resolution (particularly the HBD of protomer F) than that of the central four protomers, indicative of increased flexibility (Supplementary Fig. 3b). The MIT (which is dispensable for microtubule severing activity13) and most of the linker are not observed in the reconstruction, and were likely averaged out during image processing due to their conformational variability, as seen for katanin14. However, the EM map was of sufficient quality to model 98.7% of the residues in the hexameric ATPase domains (Table 1; Fig. 1b, Supplementary Fig. 4a).
Our reconstruction contains interpretable cryo-EM density features within each of the six nucleotide binding pockets. With the exception of the gating protomer F, all protomers contain a clearly defined ATP molecule with a magnesium ion coordinated between the β- and γ-phosphates and T530 of the phosphate binding loop (Supplementary Fig. 4b). R640 and R641 from the neighboring protomer complete the ATP binding site for all protomers except for the gating protomer F. The Arginine finger residue R641 is in a catalytically competent conformation and within H-bonding distance from the γ-phosphate (Supplementary Fig. 4b). The nucleotide binding pocket of protomer F is not as well resolved as in the other protomers (Supplementary Fig. 4b), and the cryo-EM density could correspond to either ADP or ATP. The binding pocket of protomer F is exposed to solution, presumably flexible, and the lack of clear density for the γ-phosphate could arise from either incomplete coordination of the ATP or to background hydrolysis of the ATP. However, comparison of the overall fold of this F protomer to the ATP-bound protomers (Supplementary Fig. 5a) reveals that their structures are highly similar (0.695 Å Cα-RMSD). In contrast, comparing the structures of the apo spastin monomer (PDB 3B9P)6 with the ATP-bound protomers in our structure (Supplementary Fig. 5b) shows extensive conformational rearrangement of the NBD, as well as a ~9° rotation of the HBD relative to the NBD, consistent with nucleotide binding being the main driver of the conformational changes required for hexamerization of the ATPase protomers. Notably, three conserved and functionally important loops in the AAA domain are disordered or poorly ordered in the nucleotide-free structures of spastin6,27, but are well-ordered in our hexamer structure and line the central pore, where they are involved in substrate binding and inter-protomer contacts. Linker residues 455–463 preceding α1 also become ordered in the hexamer and are stabilized through interactions with helix α1 from the neighboring protomer (Supplementary Fig. 6a). Interestingly, the structure of this linker region is distinct from the “fishhook” motif found in katanin, which decorates the N-terminal side of the central pore and also participates in protomer-protomer contacts14. Helix α1 is unique to severing enzymes, and mutations in this helix impair severing6. The linker connecting helices α1 and α2 moves in concert with the closure of the NBD and HBD around the nucleotide, establishing stabilizing interactions with the α1-β4 loop of the neighboring lower protomer. The latter loop is disordered in the apo structure6. On the opposite face of the hexamer, the C-terminus of the AAA domain (res. 753–757), which is disordered in prior crystal structures6,27, is engaged in intimate contacts with the α10- α11 loop of the neighboring protomer (Supplementary Fig. 6b), and together with helix α11 forms a belt around the hexamer as in katanin14. Invariant Y753 at the end of helix α11 is part of the interface with the α11- α12 linker of the neighboring protomer (Supplementary Fig. 6c), and its mutation severely impairs severing6.
A double-helical staircase of pore loops coordinates the substrate
Density for the polyglutamate peptide is clearly visible at the center of the spastin hexamer (Fig. 2). While the added polyglutamate peptide ranges in length from 10 to 37 amino acids, only the 15 residues that undergo stabilizing interactions with the spastin AAA core are clearly visible (Fig. 2a, b and Fig. 3a). We were unable to assign the polarity of the substrate peptide in the cryo-EM density with certainty at this resolution, as both polarities resulted in acceptable fits to the map (Supplementary Fig. 7) and neither orientation resulted in an increase in stabilizing interactions with the spastin hexamer. We modeled the polarity of the peptide with the N-terminus of the peptide engaged to the uppermost protomer (protomer A), and the C-terminus closest to the lowest protomer (protomer F). We note that structural and functional studies have so far been unable to unambiguously assign the polarity of the substrate peptide for the meiotic class of AAA ATPases to which spastin and katanin belong, and the current assigned polarity has been inferred from functional studies and analogy with other AAA+ ATPases, including VPS428, HSP10429 and the proteasome30. Interestingly, a recent study of the AAA ATPase VAT shows that it is able to translocate peptides in either direction31. It is possible that the polarity of the peptide through the spastin pore is not dictated by pore interactions alone and also results from the engagement of other interfaces with the microtubule substrate.
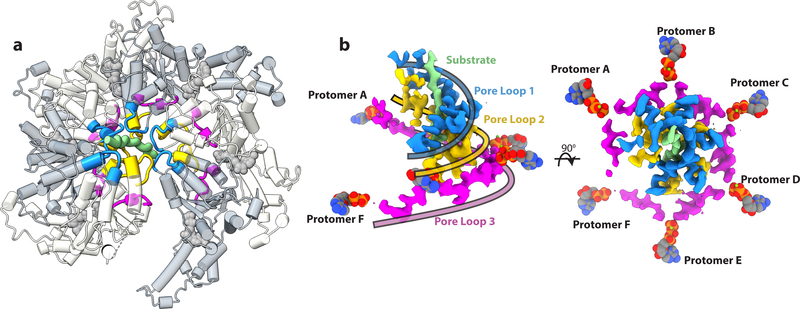
Figure 2. A double-helical staircase of pore loops surrounds the substrate and is coupled by a third pore loop spiral to the ATP binding site.
(a) The three solvent exposed pore loops highlighted on the spastin hexamer structure. Pore loop 1 is colored in blue, pore loop 2 in gold, and pore loop 3 in magenta. (b) Side (Left) and top (Right) view of the EM density of the triple spiral staircase generated by pore loops 1, 2 and 3. Pore loops 1 and pore loops 2 form a tight double spiral that engages to the peptide substrate shown in light green. Pore loops 3 form a shallow spiral between pore loops 1–2 and the nucleotide. Ribbons colored as their corresponding pore loops are overlaid on the EM map to highlight the trajectory of the three pore loop spirals.
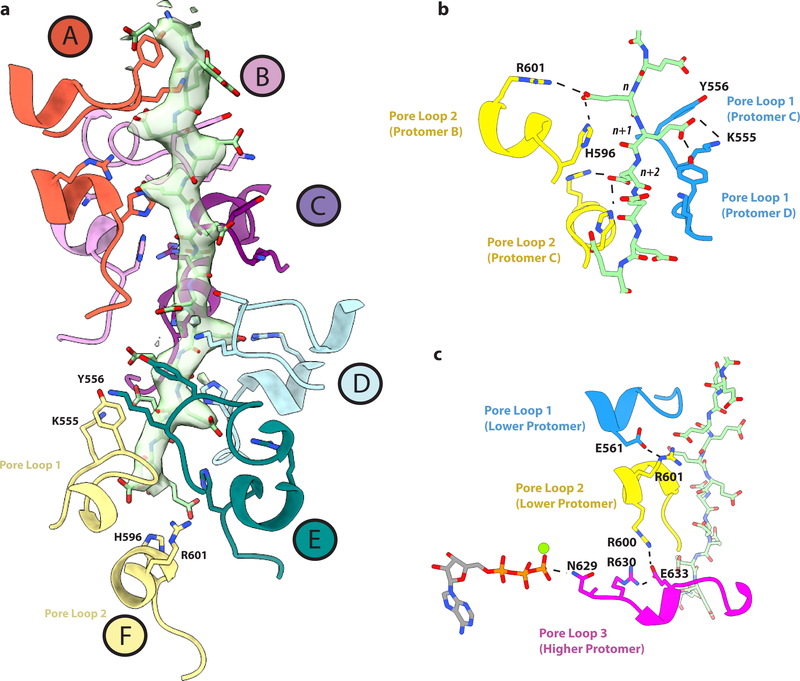
Figure 3. A pore loop network couples nucleotide binding to substrate engagement and oligomerization.
(a) Pore loops 1 and 2 interacting with the polyglutamate substrate. Substrate density is shown as a transparent surface with the polyglutamate model in light green. Pore loops are colored according to their assigned protomer, as in Figure 1. Residues that interact directly with the substrate (K555, Y556, H596, R601) are shown in stick representation. (b) Interactions between the substrate and conserved residues in pore loops 1 and 2. (c) A network of interactions between charged residues connects the peptide substrate to the nucleotide. Hydrogen bonds are shown as dashed lines. All interactions depicted have a measured distance of less than 4 Å.
Consistent with the cryo-EM structures of other AAA+ ATPases, pore loop 1 (555–562), which is characterized as having a conserved aromatic residue important for substrate translocation32, interacts directly with the bound substrate (Fig. 2b and Figs. 3a, b)28,30,33–35. Conserved Y556 intercalates between n and n+2 residues of the substrate, sandwiching the glutamate sidechain and forming a hydrogen bond with one of the carboxyl oxygens (Fig. 3a, b). Consistent with its role in substrate engagement, its mutation abolishes severing while retaining full ATPase activity (Fig. 4). This intercalating organization between pore loops and substrate has been previously observed in other ATPases28,29,33 and likely serves as a general mode of substrate interaction for AAA+ ATPases.
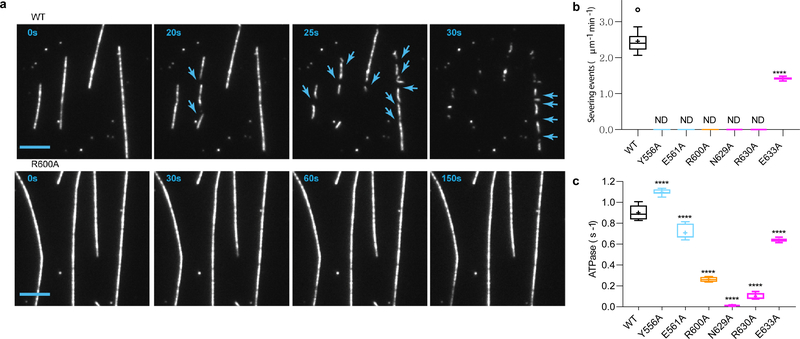
Figure 4. Mutations in the three pore loops impair ATPase and microtubule severing.
(a) Microtubule severing assays of wild-type spastin and pore loop mutant R600A. Severing events are highlighted with blue arrows. Scale bar, 5 μm. (b) Tukey plots of severing rates of structure-based spastin mutants, n = 30 microtubules from 2, 4, 4, 4, 4, 4 and 3 chambers for wild-type, Y556A, E561A, R600A, E629A, R630A, E633A mutants (c) and ATPase rates of structure-based spastin mutants, n = 24, 16, 12, 8, 8, 7, 8 for wild-type, Y556A, E561A, R600A, E629A, R630A, E633A mutants. Plus denotes mean, outliers shown as circles; ****, p < 0.0001, determined by the Mann-Whitney test. ND, not detectable. Pore loop 1, 2 and 3 mutants shown in cyan, orange and magenta, respectively.
Together with invariant K555, which forms a salt bridge with the glutamate of the n+1 substrate residue, Y556 constitutes the first of two pore-loop spiral staircases that coordinate the substrate. The sidechain of pore loop 1 residue K555 is also likely stabilized by a hydrogen bond with conserved E462 in helix α1, the secondary structure element unique to severing enzymes6,14. VPS4, the other member of the meiotic clade of AAA ATPases, has a short β-strand at this location36. Mutation of residues in α1 impair severing without a notable effect on ATPase activity6. Invariant H596 and R601 of pore loop 2 (594–601) form electrostatic interactions with every other peptide sidechain substrate (n, n+2), generating a second spiral staircase (Fig. 2b and Fig. 3b). Mutation of either of these residues inactivates severing and impairs ATPase activity6. Together, pore loops 1 and 2 enclose the poly-Glu substrate with a double helix of positively charged residues (Fig. 2b and Fig. 3b), neutralizing the highly negative charge of the peptide and providing substrate specificity for either the poly-Glu chains on the tubulin tails, or the β-tubulin tails themselves, which contain a high density of negatively charged residues (8–10 glutamates in addition to 2–3 aspartates)37. This double coordination by both pore loop 1 and 2 is distinct from VPS4, whose pore loop 2 does not directly contact its polypeptide substrate28. Participation of pore loop 2 in substrate translocation has been previously inferred from crosslinking studies of ClpA38, where mutagenesis of certain residues within the loop inhibits substrate degradation but not substrate binding. Additionally, crystallographic studies of HSP104 show pore loop 2 interacting with an adjacent N’ domain molecule, acting as a “substrate mimic”39.
An allosteric network mutated in HSP couples substrate binding to oligomerization
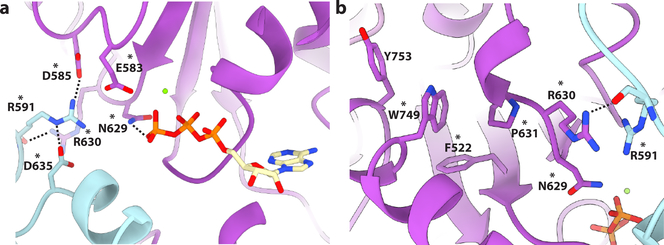
A third highly conserved pore loop in the AAA domain (629–637) runs nearly orthogonal to pore loop 2, and does not contact the substrate peptide directly, but functions as a structural bridge linking the substrate-interacting pore loops to the nucleotide-binding pocket (Fig. 3c). Invariant E561 in α4, which immediately follows pore loop 1, is within proximity of H-bonding interactions, with invariant R601 from pore loop 2 of the same protomer, potentially allowing direct coordination between these two pore loops. Pore loop 2 is disordered in crystal structures of the apo spastin monomer6,27 (Supplementary Fig. 5b), and the fact that we observe direct interactions between R601 and substrate strongly suggests that the E561-R601 interaction depends on the presence of substrate. Consistent with this role, mutation of E561 abolishes severing, while still retaining ATPase activity (Fig. 4b,c). R601 is also within H-bonding distance to S599 of the lower neighboring protomer, thus possibly linking substrate binding to oligomerization (Supplementary Fig. 6d). Both S599 and R601 are mutated in HSP (Supplementary Table 1) and the R601L disease mutation impairs ATPase and abolishes microtubule severing6. The organization of these pore loops gives rise to an interaction network wherein invariant R600 of pore loop 2 forms a salt bridge with invariant E633 in pore loop 3 of the next, lower protomer, which in turn interacts with invariant R630 which H-bonds with the backbone of R591 in loop 2 of the upper protomer (Fig. 3c). Invariant N629 in pore loop 3 is within H-bonding distance from the γ-phosphate of the ATP of the same protomer and thus can potentially contribute to nucleotide sensing in addition to the traditional sensor residues. Consistent with this, mutation of N629 abolishes both ATPase activity and severing (Fig. 4b,c). Together with invariant D585, N629 is also within H-bonding distance of invariant R591 of the neighboring lower protomer, which in turn is within hydrogen bonding distance to the catalytic glutamate in the Walker B motif (E583) of the higher protomer (Fig. 5a). The R591 backbone is also stabilized through interaction with the R630 sidechain from the lower protomer (Figs. 5b). Thus, R591 is at the center of an allosteric network that can couple the substrate binding loops to ATP hydrolysis and oligomerization. Moreover, the conformation of pore loop 3 containing R630 is coupled through a network of hydrophobic residues to helix α11 involved in oligomerization interactions between HBD domains (Fig. 5b and Supplementary Fig. 6b). Mutation of Y753 at this oligomerization interface severely impairs severing6. Strikingly, all residues in this extensive network are mutated in HSP patients, highlighting their importance for spastin function (Fig. 5 and Supplementary Table 1). Thus, our structure sheds light on how ATP and peptide substrate binding establish a network of intra- and inter-protomer interactions that stabilize the spastin hexamer. Since spastin intracellular concentrations are below the critical concentration for hexamer assembly6,22, we speculate that this pore loop network primes spastin for assembly around the tubulin tails for effective microtubule severing.
Figure 5. An allosteric network mutated in HSP couples substrate binding to oligomerization and ATP hydrolysis.
(a) Interaction network centered on R591. Residues colored plum and cyan for protomers B and C, respectively. Residues mutated in HSP are indicated with an asterix. All interactions depicted have a measured distance of less than 4 Å. (b) W749 makes van der Walls contacts with several hydrophobic residues mutated in HSP and connects pore loop 3 to the C-terminal helix α11 involved in oligomerization. Residues are colored purple and cyan for protomers C and D, respectively. Residues mutated in HSP are indicated with an asterix. All interactions depicted have a measured distance of less than 4 Å.
Effect of HSP mutations on spastin activity
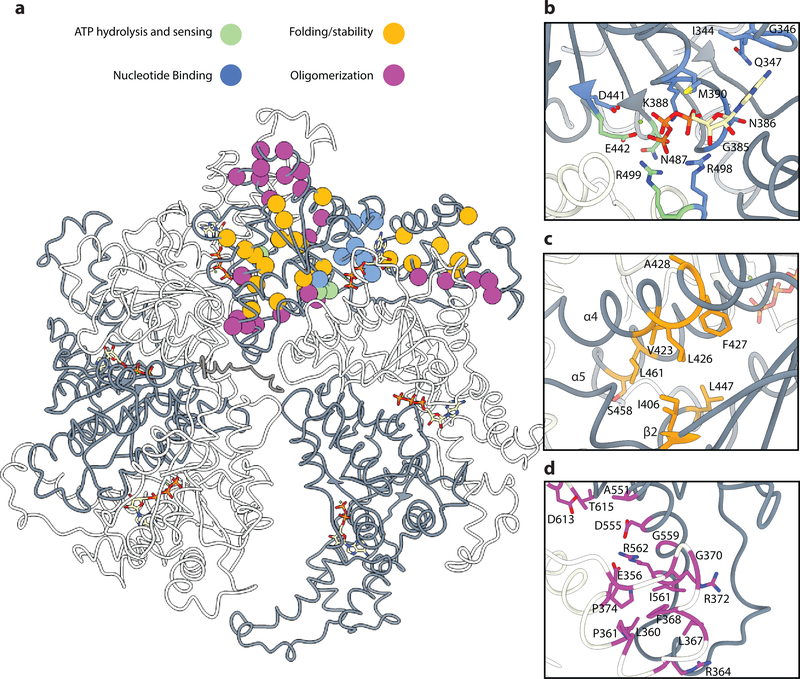
There are over 200 HSP-associated spastin mutations reported to date and 30% of them are missense mutations10. The majority of spastin missense mutations linked to HSP are located within the ATPase domain10, and several missense disease mutations examined have been shown to inactivate or severely impair microtubule severing activity in biochemical and cellular studies5–7,10,13,40. Our structure provides a mechanistic understanding of how HSP mutations in the ATPase domain lead to loss of spastin function. Based on the high sequence identity and known structural conservation between drosophila and human spastin6,27, we generated a homology model of the human spastin hexamer (Fig. 6a) that provides complete coverage of the highly-conserved AAA domain (amino acids 89–388). Structural analysis of all known missense spastin mutations identified to date in the AAA core6,41–61 suggests that most impair oligomerization, nucleotide binding, or disrupt the folding or stability of the AAA domain’s structure to variable degrees (Supplementary Table 1). Several mutations map directly to the nucleotide binding pocket (Fig. 6b) and impair nucleotide binding (I344K, G385W, N386K, K388R, M390V, R498S) and thus hexamerization. One mutation interferes with magnesium coordination (D441G). Two disease mutations affect ATP hydrolysis and sensing of the γ-phosphate (E442A or K, and R499C). E442 is part of the Walker B motif and participates in catalysis in all AAA ATPases, while R499 serves as the Arg finger. The Arg finger coordinates the γ-phosphate and electrostatically stabilizes the transition state after ATP hydrolysis. N487D (N629 in our Drosophila cryo-EM structure) is likely involved in both sensing the γ-phosphate and oligomerization interactions (Figs. 6b and 5a)
Figure 6. Structure-based insight into the mechanism of action of human spastin HSP disease mutations.
(a) Homology model of the human spastin hexamer depicting the location of missense mutations identified in HSP. Protomers are colored in alternating grey and white around the hexamer. Mutations are depicted as spheres at their Cα position and colored according to their proposed mechanism of action: interference with nucleotide binding (blue), defects in ATP hydrolysis and γ-phosphate sensing (green), disruption of interactions important for folding or conferring stability to the protomer (orange), and disruption of oligomerization interfaces (magenta). (b) Close-up view of the nucleotide pocket, showing a large number of mutations that likely disrupt nucleotide binding or impair ATP hydrolysis and sensing. (c) Close-up view of hydrophobic interfaces between α4, α5 and β2 highlighting potentially destabilizing HSP mutations. (d) Close-up view of the NBD-HBD interaction interface between protomers showing a high-density of HSP mutations.
Other HSP mutations are likely deleterious because they disrupt the overall fold and stability of the AAA domain structure (Fig. 6c) (I406V, V423L, L426V, F427C, A428P, L447V, S458R, and L461P among others). A large number of disease mutations map to protomer-protomer interfaces and likely interfere with oligomerization (Fig. 6d) (E356G, L360P, P361L/S, R364M, L367S, F368L/S, G370R, R372G, C448Y, A551P/Y, D555G/N, E356V, G559D, I561G, R562Q, D613H, T615I). Consistent with this, cellular studies have shown that the C448Y mutant is unable to associate with wild-type spastin and inhibit microtubule severing62. Thus, the preponderance of HSP mutations likely impact oligomerization interfaces or the overall stability of spastin and may lead to spastin haploinsufficiency. However, there is a large phenotypic variability in spastin-linked HSP and more recent work has shown that some spastin disease mutations can also have a toxic gain-of-function that is further exacerbated by spastin haploinsufficiency63. We note that spastin mutants that are deficient in oligomerization could still bind microtubules through a stretch of residues in the linker that connects the AAA and MIT3,6,7,13,64 domains and thus have a toxic gain of function on the microtubule cytoskeleton. Interestingly, mutations in the Arg finger R499 have been associated with early onset HSP41,52,53,59,65–68, possibly suggesting a dominant negative effect of this mutation. The Walker B residue E442 is the other strong candidate for a dominant negative mechanism, however not enough phenotypic data are currently available for this HSP mutation. Our structure will serve as a platform for the analysis of HSP mutations and the correlation between genotype and molecular phenotype.
Discussion
Recent cryo-EM studies of AAA+ ATPase protein translocases have shown how a conserved mechanistic infrastructure can be generally utilized for effective nucleotide-driven protein translocation, while also highlighting the allosteric diversity of this family of ATPases. Even within the meiotic clade of AAA+ ATPases, we observe differences in structural motifs such as the substrate-interacting pore loops. For example, while spastin surrounds its substrate through a network of interactions that utilizes both pore-loops, the closely related family member VPS4 primarily interacts with the translocating substrate via an aromatic-containing pore loop 1, without direct interactions from pore loop 2. Such differences may evolve to accommodate substrate specificity or to modulate the force exerted upon a targeted polypeptide.
Given its structural homology to the C-terminal AAA domain of spastin, it is possible that the katanin ATPase establishes a nucleotide-substrate interaction network that operates under a homologous allosteric mechanism. However, in contrast to previous structural studies of katanin, we only observed the split locker-washer conformation of spastin, and were unable to identify a closed-ring conformer within our datasets as has been observed for other AAA ATPases, including the closely related meiotic clade members katanin14 and VPS428. One possible explanation for this could be a higher ATP-binding affinity for spastin versus katanin, which could prevent the dissociation of nucleotide in protomer F under our experimental conditions, a requirement for adoption of the closed-ring conformation previously observed for katanin14 and VPS428. We speculate that the split locker-washer conformation of spastin observed here represents a pre-hydrolysis state where all six protomers are engaged with the substrate peptide.
In summary, our structure of spastin elucidates for the first time the details of substrate engagement by a microtubule severing enzyme, and reveals how three conserved pore loops in spastin establish a conduit that links substrate binding with oligomerization and ATP hydrolysis. Our comprehensive analysis of human HSP mutations reveals the extensive coverage of disease mutations on the spastin hexamer and how they would impact all major facets of hexamer function: ATP binding, hydrolysis, and oligomerization.
Online Methods
Protein expression and purification
Full-length Drosophila melanogaster spastin6 with a Walker B mutation (E583Q) was expressed in Escherichia coli BL21DE3 cells as a N-terminal glutathione S-transferase (GST) fusion and purified by affinity chromatography, followed by Prescission protease cleavage of the GST69. After cleavage with Prescission protease, spastin was further purified on a HiTrapQ chromatography column (subtractive step) followed by a MonoS column (GE Healthcare). Spastin eluted as two peaks that were collected separately. The purer spastin peak was concentrated to ~ 5mg/ml and injected on a Superdex 200 size exclusion column (GE Healthcare) in 20 mM HEPES pH 7.5, 300 mM KCl, 10 mM MgCl2, 5 mM DTT, 5 % glycerol. The sample was concentrated to 10 mg/ml, flash-frozen in liquid nitrogen and stored at −80°C after supplementing the glycerol concentration to 15%. For ATPase and severing assays, the gel filtration step was omitted and the protein was concentrated to 5mg/ml and buffer exchanged into 20 mM HEPES pH 7.5, 300 mM KCl, 10mM MgCl2, 5 mM DTT, 15% glycerol. Single use aliquots were flash frozen in liquid nitrogen and stored at −80°C. All structure-based point mutants were generated using Quickchange mutagenesis and subjected to the same purification protocol as the wild-type protein.
Analytical ultracentrifugation
AUC experiments were conducted in a ProteomeLab XL-I analytical ultracentrifuge (Beckman Coulter) using absorption optics. The spastin Walker B mutant was subjected to size exclusion chromatography in 20 mM HEPES, 300 mM KCl, 10 mM MgCl2 and 1 mM TCEP. The eluted protein was used for preparation of AUC samples in the same buffer supplemented with 0.1 mM ATP. The sample was loaded into a 12 mm cell, placed in a AN-Ti rotor and equilibrated thermally in the ultracentrifuge. After thermal equilibrium was reached at rest at 10°C, the rotor was accelerated to 45,000 rpm and intensity scans at 280 nm were started immediately and collected until no further sedimentation boundary movement was observed. Data were analyzed in terms of continuous c(s) distributions using the SEDFIT program70. All accepted fits had the Root Mean Square Deviations (RMSD) less than 0.008. Partial specific volume and buffer parameters were calculated using the Sednterp (http://sednterp.unh.edu/). Sedimentation coefficients distributions were corrected to standard conditions at 20°C in water, s20,w. For the detection of spastin and peptide co-sedimentation, a VGSEEEEEEEEEE peptide was synthesized and purified by Biosynthesis and then labeled with Atto488 on its N-terminus using Atto488-NHS ester (Sigma- Aldrich #41698) according the manufacturer’s protocol. After labeling, peptides were RP-HPLC purified. Experiments were performed for peptide (16.7 μM) alone and spastin (10 μM for the monomer or 1.67 μM for the hexamer) and excess peptide (16.7 μM). The samples were placed in the four-hole AN-Ti rotor and equilibrated thermally in the ultracentrifuge. After thermal equilibrium was reached at rest at 10°C, rotor was accelerated to 50,000 rpm and the intensity scans at 280 nm and 483 nM were started immediately and collected until no further sedimentation boundary movement was observed. Data were analyzed in terms of continuous c(s) distributions using the SEDFIT program. All accepted fits had the Root Mean Square Deviations (RMSD) less than 0.008. Partial specific volume and buffer parameters were calculated using the Sednterp (http://sednterp.unh.edu/). Sedimentation coefficients distributions were corrected to standard conditions at 20°C in water, s20,w. Modeling the 9.9s peak as a monodisperse species gives statistically identical solutions, indicating that the 9.9s peak is not a mixture of oligomers71.
Sample preparation for electron microscopy
1.2 mg ml−1 of spastinE583Q was incubated on ice for ~20 min in 20 mM HEPEs buffer (pH 7.5) containing 300 mM KCl, 10 mM MgCl2, 5 mM DTT, 1 mM ATP and 0.05 % LMNG detergent with and without 0.22 μM 1,500 – 5,500 mol wt polyglutamate peptide (Sigma Aldrich). 2.5μl of sample was applied to a UltraAuFoil R1.2/1.3 300-mesh grid (Electron Microscopy Services) freshly plasma-cleaned using a Gatan Solarus (75% argon/2% oxygen atmosphere, 15 W for 7 s). Grids were manually blotted with filter paper (Whatman No.1) for ~4 s in a 4º C cold room prior to plunge freezing in liquid ethane cooled by liquid nitrogen.
Electron microscopy data acquisition
All cryo-EM data were acquired using the Leginon automated data-acquisition program72. All image pre-processing (frame alignment, CTF estimation, particle picking) were performed in real-time using the Appion image-processing pipeline during data collection73.Images were collected on a Thermo Fischer Talos Arctica operating at 200 keV, equipped with a Gatan K2 Summit DED, at a nominal magnification of 36,000x corresponding to a physical pixel size of 1.15 Å/pixel. 2,534 movies (48 frames/movie) were collected using an exposure time of 12 s with an exposure rate of 5.6 e−/pixel/s, resulting in a total dose of 52 e−/Å2 (1.08 e−/frame) with a nominal defocus range from −1.0 to −2.0 μm.
Image processing
A total of 564 images were used for automated particle picking using Difference of Gaussians (DoG) picker to yield 92,387 particles73. CTFFIND4 was used for CTF estimation prior to extracting particles. Particles were Fourier-binned 4×4 and subjected to reference-free 2D classification using multivariate statistical analysis (MSA) and multi-reference alignment (MRA) in the Appion pipeline. The best 3 classes representing end-on, tilted and side views were selected for template-based particle picking using FindEM resulting in 2,736,856 particle picks. The ~2.7 million particles picks were extracted unbinned, and subjected to two rounds of reference-free 2D classification in cryoSPARC74 to remove non-particles and poorly aligning particles in the data (Supplementary Fig. 3). Particles from exemplary classes (strong secondary structural features present in the 2D classes) comprised a subset of 1,259,553 particles. This subset then went on to reference-free ab initio model generation in cryoSPARC, resulting in an initial 3D model. Homogeneous refinement resulted in a map with a reported resolution of ~3.4 Å. Some disordered density above the topmost protomer was observed, which we attributed to an off-register misalignment of a subset of particles relative to the majority register. 3D classification without alignment in RELION75 using a tau-fudge value of 20 was successful in identifying the misaligned hexamers, which constituted 12% of the input particles. One class, which comprised 22% of the input particles, did not display the previously observed disordered density and was better ordered compared to the other 5 classes. 3D autorefinement in Relion with limited local and global search angles resulted in a ~4.2 Å map. We then used this map as a reference map for a heterogeneous refinement into 2 classes in cryoSPARC, using the original 1,259,553 particles that were kept after 2D classification. One class was significantly better ordered, and was selected for an additional round of heterogenous refinement with 2 classes. One class was selected, based on visual inspection for quality of the reconstruction, for homogenous refinement resulting in a sharpened map of ~3.2 Å from 488,385 particles. This final reconstruction includes only 18% of the total particles initially selected from the micrographs. We attribute this to false particle-picks, damaged particles from sample freezing, inaccurate defocus values, non-substrate-bound particles, and sample heterogeneity. Notably, throughout the image processing of spastin, we did not observe any evidence of an additional conformational state other than the split lock-washer conformation, even at low resolution.
An additional data set of the spastin hexamer without the addition of the polyglutamate substrate resulted in a ~3.8 Å reconstruction with an unknown density within the central pore that resembled an unfolded polypeptide substrate (Supplementary Fig. 2). Similar densities have been observed for numerous AAA+ ATPase protein translocases25,28,33. Notably, of the ~300,000 particles selected from the images, only ~30,000 particles contributed to this final reconstruction. We were unable to identify the peptide species through mass spectrometry analysis due to its low abundance. We speculate that the unknown density is either due to a single low-abundant peptide species, or an average of many low-abundant peptides of differing sequences bound to spastin hexamers. Due to the presence of substrate density in the central pore of the hexamer, we speculated that substrate binding may have a stabilizing effect on the overall organization of the complex, and that a small minority of selected particles contained residual substrate that may have been retained through the protein purification. Indeed, pre-incubation our spastin with polyglutamate peptide, known to bind to spastin and inhibit microtubule severing20, increased the percentage of well-ordered hexamers observed in our dataset by ~2-fold and resulted in a substantial improvement in resolution to ~3.2 Å of the substrate-bound spastin hexamer, which allowed atomic model building (Table 1 and above).
Atomic model building and refinement
A crystal structure of the Drosophila melanogaster spastin monomer was used as a starting point6 (PDB 3B9P) for modeling. Using Chimera76, all loops and coils were deleted, and secondary structural elements were docked into the EM map. After one round of real-space refinement in Phenix77, Coot78 was used to rebuild coils and improve main chain and side chains. The rebuilt model was then used as the initial input for a multi-model-generating pipeline79, which allowed for accessing model quality. 200 models were generated in Rosetta using the refined map and model, and the top 10 scoring models were selected for further model refinement using Phenix, using a per-residue Cα-RMSD. Regions with poor model convergence were remodeled and refined. The Molprobity80 server (http://molprobity.biochem.duke.edu/) and PDB validation Service server (https://validate-rcsb-1.wwpdb.org/) were used to identify problem regions for subsequent correction in Coot. Residues 502–516 and 613–620 of protomer A are missing in the final model. The polyglutamate peptide was built de novo in Coot.
ATP hydrolysis assays
Basal ATP hydrolysis rates were measured at room temperature using the EnzChec Phosphate Assay Kit (Thermo Fisher Scientific) in 50 mM PIPES pH 8.8, 50 mM KCl, 1 mM EGTA, 2 mM MgCl2 and 1 mM DTT for 50 nM spastin and point mutants. Initial rates were calculated from the linear portion of the reaction curve after addition of 2 mM ATP. ATPase rates were corrected by subtraction of the measured release of phosphate in the absence of ATP. Poly-glutamate stimulated ATP hydrolysis rates were measured at room temperature in 20 mM HEPES pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM DTT and 2 mM ATP at 200 nM spastin and poly-glutamate concentrations ranging from 0 to 100 μM. Stock solutions(100mM) of poly-glutamate (0.75 – 5.0 kDa) was made in water and adjusted to pH 7.5 with potassium hydroxide. Ten-fold serial dilutions were made in water and added to the ATPase reaction, before addition of ATP. Initial rates were calculated from the linear portion of the reaction curve. ATPase rates were corrected by subtraction of the measured release of phosphate in the absence of ATP and poly-glutamate.
Microtubule severing assays
Microtubules were polymerized from 2 mg/ml bovine cycled brain tubulin (PurSolutions), 4% tetramethyl rodamine labeled and 1% biotin-labeled porchine brain tubulin (Cytoskeleton). Microtubules were double-cycled with the slow-hydrolyzable GTP analog guanylyl (α,β)-methylene diphosphonate (GMPCPP). The first polymerization was 1 hour and the second polymerization overnight81. The microtubules were spun down at 126,000 xg using a TLA100 rotor (Beckman Coulter), re-suspended in warm BRB80 (80 mM K-PIPES pH 6.8, 1 mM MgCl2, 1 mM EGTA) and stored at 37°C. Chambers for TIRF microscopy were assembled as described previously69. Microtubules were immobilized in the chamber using 2-mg/ml NeutrAvidin (Thermo Fisher Scientific) and imaged by TIRF microscopy in severing buffer containing BRB80, 2-mg/ml casein, 9.1 mM 2-mercaptoethanol, 2.5% glycerol, 50 mM KCl, 2.5 mM MgCl2, 1 mM ATP, 1% Pluronic F127 (Life Technologies), and oxygen scavengers69. Severing reactions were carried out using 20nM full-length wild-type and mutants of Drosophila melanogaster spastin. Severing was scored as previously described20. Images were acquired with a Nikon Ti-E microscope equipped with a 60X 1.49 NA oil objective and a TI-TIRF adapter (Nikon) at 1Hz with a 50ms exposure time. Excitation was provided by a 561nm laser (Agilent MLC) set to 20 mW before being coupled to the microscope by an optical fiber. The final pixel size for images was 90 nm. Image acquisition was carried out using Micro-Manager82. Image analysis was carried out in Fiji. Prism (Graphpad Inc.) was used for graphing and statistical analysis.
Quantification and data analysis
N numbers are reported for all experiments in figure legends. Data in Fig. 4b, c were subjected to a Mann-Whitney statistical test.
Homology modeling
A human spastin hexamer homology model was generated using the online server SWISS-MODEL83 (https://swissmodel.expasy.org/). Residues 228–617 of the M1 isoform (uniprot: Q9UBP0) were used as a targeting sequence and our 3.2 Å atomic model of the D. melanogaster spastin was used as a template.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
All data used in this study is available from the corresponding authors upon reasonable request. Electron microscopy map and the top scoring model of 5 atomic models obtained from an EM multi-model pipeline have been deposited at the Electron Microscopy Data Bank and Protein Data Bank under accession numbers EMDB: 20226 and PDB: 6P07
Supplementary Material
Acknowledgements
We thank J.C. Ducom at The Scripps Research Institute High Performance Computing for computational support, and B. Anderson at The Scripps Research Institute electron microscopy facility for microscope support. We thank M. Herzik and A. Hernandes for help with atomic modeling, E. Szczesna for help with microtubule severing assays, G. Piszcek from the Biophysics Core of the National Heart, Lung and Blood Institute for help with AUC experiments, S. Chowdhury, C. Puchades, and M. Wu for helpful discussion. C.R.S. is supported by a National Science Foundation predoctoral fellowship. G.C.L. is supported as a Searle Scholar, a Pew Scholar, an Amgen Young Investigator, and by the National Institutes of Health (NIH) DP2EB020402. Computational analyses of EM data were performed using shared instrumentation funded by NIH S10OD021634 to G.C.L. A.R.M. is supported by the intramural programs of the National Institute of Neurological Disorders and Stroke (NINDS) and the National Heart, Lung and Blood Institute (NHLBI).
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Mcnally FJ & Roll-Mecak A Microtubule-severing enzymes: From cellular functions to molecular mechanism. 1–13 (2018). doi: 10.1083/jcb.201612104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salinas S et al. Human spastin has multiple microtubule-related functions. J. Neurochem 95, 1411–1420 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Eckert T, Le DT Van, Link S, Friedmann L & Woehlke G Spastin’s Microtubule-Binding Properties and Comparison to Katanin. PLoS One 7, 1–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen M & Wang C The nucleotide cycle of spastin correlates with its microtubule-binding properties. FEBS J. 280, 3868–3877 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG & Lauring BP Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol 168, 599–606 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roll-Mecak A & Vale RD Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature 451, 363–367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roll-Mecak A & Vale RD The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol 15, 650–655 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hazan J et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet 23, 296–303 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Blackstone C, O’Kane CJ & Reid E Hereditary spastic paraplegias: Membrane traffic and the motor pathway. Nat. Rev. Neurosci 12, 31–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solowska JM & Baas PW Hereditary spastic paraplegia SPG4: What is known and not known about the disease. Brain 138, 2471–2484 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone MC et al. Normal Spastin Gene Dosage Is Specifically Required for Axon Regeneration. Cell Rep. 2, 1340–1350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havlicek S et al. Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients’ neurons. Hum. Mol. Genet 23, 2527–2541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White SR, Evans KJ, Lary J, Cole JL & Lauring B Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol 176, 995–1005 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zehr E et al. Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat. Struct. Mol. Biol 24, 717–725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman JJ & Vale RD Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science (80-. ). 286, 782–785 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Eckert T et al. Subunit Interactions and Cooperativity in the Microtubule-severing AAA ATPase Spastin. 287, 26278–26290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummings CM, Bentley CA, Perdue SA, Baas PW & Singer JD The Cul3/Klhdc5 E3 ligase regulates p60/katanin and is required for normal mitosis in mammalian cells. J. Biol. Chem 284, 11663–11675 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu C, Srayko M & Mains PE The Caenorhabditis elegans Microtubule-severing Complex MEI-1/MEI-2 Katanin Interacts Differently with Two Superficially Redundant β-Tubulin Isotypes. Mol. Biol. Cell (2004). doi: 10.1091/mbc.e03-06-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherwood NT, Sun Q, Xue M, Zhang B & Zinn K Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valenstein ML & Roll-Mecak A Graded Control of Microtubule Severing by Tubulin Glutamylation. Cell 164, 911–921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vemu A et al. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science (80-. ). 361, eaau1504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itzhak DN, Tyanova S, Cox J & Borner GHH Global, quantitative and dynamic mapping of protein subcellular localization. Elife 5, 1–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geimer S, Teltenkötter A, Plessmann U, Weber K & Lechtreck KF Purification and characterization of basal apparatuses from a flagellate green alga. Cell Motil. Cytoskeleton 37, 72–85 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Schneider A, Plessmann U, Felleisen R & Weber K Posttranslational modifications of trichomonad tubulins; identification of multiple glutamylation sites. FEBS Lett. 429, 399–402 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Abid Ali F et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat. Commun 7, 10708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skordalakes E & Berger JM Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Taylor JL, White SR, Lauring B & Kull FJ Crystal structure of the human spastin AAA domain. J. Struct. Biol 179, 133–137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han H, Monroe N, Sundquist WI, Shen PS & Hill CP The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife 6, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gates SN et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science (80-. ). 357, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De la Peña AH, Goodall EA, Gates SN, Lander GC & Martin A Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science (80-. ). 362, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augustyniak R & Kay LE Cotranslocational processing of the protein substrate calmodulin by an AAA+ unfoldase occurs via unfolding and refolding intermediates. Proc. Natl. Acad. Sci 115, E4786–E4795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlieker C et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11, 607–615 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Puchades C et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science (80-. ). 358, eaao0464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ripstein ZA, Huang R, Augustyniak R, Kay LE & Rubinstein JL Structure of a AAA+ unfoldase in the process of unfolding substrate. Elife 6, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alfieri C, Chang L & Barford D Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature 559, 274–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scott A et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 24, 3658–3669 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roll-Mecak A Intrinsically disordered tubulin tails: Complex tuners of microtubule functions? Semin. Cell Dev. Biol 37, 11–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW & Horwich AL Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–1041 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Lee J et al. Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase. Biosci. Rep 37, BSR20171399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charvin D et al. Mutations of SPG4 are responsible for a loss of function of spastin, an abundant neuronal protein localized in the nucleus. Hum. Mol. Genet 12, 71–78 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Mészárosová AU et al. SPAST mutation spectrum and familial occurrence among Czech patients with pure hereditary spastic paraplegia. J. Hum. Genet 61, 845–850 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Ishiura H et al. Molecular epidemiology and clinical spectrum of hereditary spastic paraplegia in the Japanese population based on comprehensive mutational analyses. J. Hum. Genet 59, 163–172 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Depienne C et al. Spastin mutations are frequent in sporadic spastic paraparesis and their spectrum is different from that observed in familial cases. J. Med. Genet 43, 259–265 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang BS et al. Clinical features of hereditary spastic paraplegia with thin corpus callosum: Report of 5 Chinese cases. Chin. Med. J. (Engl) (2004). [PubMed] [Google Scholar]
- 45.Hentati A et al. Novel mutations in spastin gene and absence of correlation with age at onset of symptoms. Neurology 55, 1388–90 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Meijer IA, Hand CK, Cossette P, Figlewicz DA & Rouleau GA Spectrum of SPG4 mutations in a large collection of North American families with hereditary spastic paraplegia. Arch. Neurol 281–286 (2002). doi: 10.1001/archneur.59.2.281 [DOI] [PubMed] [Google Scholar]
- 47.Patrono C et al. Autosomal dominant hereditary spastic paraplegia: DHPLC-based mutation analysis of SPG4 reveals eleven novel mutations. Hum. Mutat 25, 506 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Balicza P et al. Genetic background of the hereditary spastic paraplegia phenotypes in Hungary - An analysis of 58 probands. J. Neurol. Sci. 364, 116–121 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Bürger J et al. Hereditary spastic paraplegia caused by mutations in the SPG4 gene. Eur. J. Hum. Genet (2000). doi: 10.1038/sj.ejhg.5200528 [DOI] [PubMed] [Google Scholar]
- 50.Elert-Dobkowska E et al. Molecular spectrum of the SPAST, ATL1 and REEP1 gene mutations associated with the most common hereditary spastic paraplegias in a group of Polish patients. J. Neurol. Sci 359, 35–39 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Lu X et al. Genetic analysis of SPG4 and SPG3A genes in a cohort of Chinese patients with hereditary spastic paraplegia. J. Neurol. Sci 347, 368–371 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Park H et al. Mutational spectrum of the SPAST and ATL1 genes in Korean patients with hereditary spastic paraplegia. J. Neurol. Sci 357, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Polymeris AA et al. A series of Greek children with pure hereditary spastic paraplegia: clinical features and genetic findings. J. Neurol 263, 1604–1611 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Aulitzky A et al. A complex form of hereditary spastic paraplegia in three siblings due to somatic mosaicism for a novel SPAST mutation in the mother. J. Neurol. Sci 347, 352–355 (2014). [DOI] [PubMed] [Google Scholar]
- 55.McDermott CJ et al. Clinical features of hereditary spastic paraplegia due to spastin mutation. Neurology (2006). doi: 10.1212/01.wnl.0000223315.62404.00 [DOI] [PubMed] [Google Scholar]
- 56.Yabe I, Sasaki H & Tashiro K Spastin gene mutation in Japanese with hereditary spastic paraplegia. J. Med … 39, 14–15 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong EL et al. Clinical spectrum and genetic landscape for hereditary spastic paraplegias in China. Mol. Neurodegener 13, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y et al. A diagnostic gene chip for hereditary spastic paraplegias. Brain Res. Bull 97, 112–118 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Shoukier M et al. Expansion of mutation spectrum, determination of mutation cluster regions and predictive structural classification of SPAST mutations in hereditary spastic paraplegia. Eur. J. Hum. Genet 17, 187–194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fonknechten N et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 9, 637–644 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Proukakis C, Moore D, Labrum R, Wood NW & Houlden H Detection of novel mutations and review of published data suggests that hereditary spastic paraplegia caused by spastin (SPAST) mutations is found more often in males. J. Neurol. Sci 306, 62–65 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Solowska JM et al. Pathogenic Mutation of Spastin Has Gain-of-Function Effects on Microtubule Dynamics. J. Neurosci 34, 1856–1867 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qiang L et al. Hereditary spastic paraplegia: gain-of-function mechanisms revealed by new transgenic mouse. Hum. Mol. Genet 28, 1136–1152 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Errico A, Ballabio A & Rugarli EI Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum. Mol. Genet 11, 153–163 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Crippa F et al. Eight novel mutations in SPG4 in a large sample of patients with hereditary spastic paraplegia. Arch. Neurol. (2006). doi: 10.1001/archneur.63.5.750 [DOI] [PubMed] [Google Scholar]
- 66.França MC et al. SPG4-related hereditary spastic paraplegia: Frequency and mutation spectrum in Brazil. Clin. Genet. 86, 194–196 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Kim T-H et al. Mutation analysis of SPAST, ATL1, and REEP1 in Korean Patients with Hereditary Spastic Paraplegia. J. Clin. Neurol 10, 257–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gillespie MK, Humphreys P, McMillan HJ & Boycott KM Association of Early-Onset Spasticity and Risk for Cognitive Impairment With Mutations at Amino Acid 499 in SPAST. J. Child Neurol (2018). doi: 10.1177/0883073818756680 [DOI] [PubMed] [Google Scholar]
Methods References
- 69.Ziolkowska N & Roll-Mecak A In Vitro Microtubule Severing Assays. Neurochem. Int 37, 399–400 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Schuck P Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown PH, Balbo A & Schuck P Using prior knowlede in the determination of macromolecular size-disrtributions by analytical ultracentrifugation. Biomacromolecules 8, 2011–2024 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carragher B et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol 132, 33–45 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Voss NR, Yoshioka CK, Radermacher M, Potter CS & Carragher B DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol 166, 205–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Scheres SHW RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pettersen EF et al. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 77.Adams PD et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr (2010). doi: 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P & Cowtan K Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr (2004). doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 79.Herzik MA, Fraser JS & Lander GC A Multi-model Approach to Assessing Local and Global Cryo-EM Map Quality. Structure (2018). doi: 10.1016/j.str.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen VB et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr (2010). doi: 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gell C et al. Purification of tubulin from porcine brain. Methods Mol. Biol (2011). doi: 10.1007/978-1-61779-252-6_2 [DOI] [PubMed] [Google Scholar]
- 82.Edelstein AD et al. Advance methods of microscope control using microManager software. 1, 1–18 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waterhouse A et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barad BA et al. EMRinger: Side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cardone G, Heymann JB & Steven AC One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol 184, 226–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng EC, Pettersen EF, Couch GS, Huang CC & Ferrin TE Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics 7, 1–10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study is available from the corresponding authors upon reasonable request. Electron microscopy map and the top scoring model of 5 atomic models obtained from an EM multi-model pipeline have been deposited at the Electron Microscopy Data Bank and Protein Data Bank under accession numbers EMDB: 20226 and PDB: 6P07