Abstract
Background
PAS domain containing repressor 1 (PASD1), the cancer-testis antigen (CTA), has been reported to be aberrantly expressed in various cancer tissues and cancer cell lines; however, normal PASD1 expression can be detected in normal tissue, excluding testicular tissue. Moreover, PASD1 is reported to be abnormally expressed in various malignant tumors. However, it remains unclear whether PASD1 participates in tumorigenesis of glioma.
Material/Methods
PASD1 expression was detected by immunohistochemistry in 155 glioma tissue specimens in this study. Furthermore, the relationship of PASD1 expression with clinicopathological features in glioma cases was statistically analyzed. In addition, PASD1 was knocked down by small interference RNA (shRNA) in glioma cell line (LN229), so as to assess the potential to use it as the target for treating glioma.
Results
Our findings suggested that PASD1 expression in glioma patients was extremely upregulated compared with that in normal tissue samples and cell lines. Moreover, PASD1 expression was found to be markedly correlated with gender, The World Health Organization grade and p53 expression; in addition, high PASD1 expression indicated poor prognosis for glioma patients. Additionally, downregulation of PASD1 inhibited the proliferation ability of cells and resulted in cell arrest at the G2/M phase, which was achieved through accelerating apoptosis. Furthermore, our results indicated that PASD1 downregulation could upregulate some apoptosis-modulating proteins at the same time it downregulated some cycle-regulating proteins.
Conclusions
Taken together, our findings demonstrated that PASD1, an oncogene, can potentially serve as an independent prognostic factor for glioma patients.
MeSH Keywords: Apoptosis, Cell Cycle, Glioma
Background
Glioma, the most frequently-seen intracranial tumor in the world, is characterized by invasive growth and active angiogenesis [1]. Unfortunately, despite the surgical resection, radiotherapy, and chemotherapy, the median survival time among the glioma population is only 16 to 19 months, while the 2-year survival rate is approximately 25% to 30%, even though excision, radiotherapy, and chemotherapy may be available [2,3]. Thus, there is an urgently need to further investigate effective diagnosis and treatment methods, considering the poor prognosis of glioma.
As a type of protein antigen, the normal expression of cancer-testis antigen (CTA) can only be detected in the reproductive cells of adult testis, but abnormal expression can be detected in a variety of human cancers [4]. PAS domain containing repressor 1 (PASD1), one of the CTAs known as CT63 or CT64 (https://www.ncbi.nlm.nih.gov/gene/139135), has been newly identified as a CTA and has been reported to be abnormally expressed in a variety of malignancies [5]. PASD1, a type of CT-X antigen, has been localized in chromosome Xq28, but it does not belong to the multi-gene family. PASD1 expression in glioma tissues as well as normal adjacent tissues was evaluated in this study to examine its prognostic and clinicopathological value. In addition, the role of PASD1 in the pathogenesis of glioma was also explored, and its effects on the glioma cell line LN229 in vivo was evaluated. Our findings demonstrated that PASD1 might work as a new indicator of the poor prognosis for glioma patients, which might thereby serve as a potential diagnostic and therapeutic marker in glioma.
Material and Methods
Tissue specimens
We collected 155 tissue samples from the First Hospital Affiliated to Zhengzhou University from May 2016 to May 2018 (Table 1). Meanwhile, normal brain tissue samples were provided from the Department of Histology and Embryology of Zhengzhou University and served as the controls. Tumor staging was assessed according to the 2007 World Health Organization (WHO) classification of nervous system tumors. Inform consent was obtained from patients to use tissue specimens. This work was authorized by the Research Ethics Committee of our hospital. All cases were treatment-naive prior to surgery, and all tissue specimens were immersed into liquid nitrogen promptly for RNA isolation.
Table 1.
Correlation between the PASD1 and clinical characteristics of glioma patients.
| Characteristics | Positive/total (%) | Positive/total (%) | χ2 | P | |
|---|---|---|---|---|---|
| High | Low | ||||
| Gender | |||||
| Male | 106/155 | 79 | 27 | 8.388 | 0.004** |
| Female | 49/155 | 25 | 24 | ||
| Age (years) | |||||
| <38 | 43/155 | 25 | 18 | 2.163 | 0.141 |
| ≥38 | 112/155 | 79 | 33 | ||
| Tumor size (cm) | |||||
| <5 | 78/155 | 56 | 22 | 1.570 | 0.21 |
| ≥5 | 77/155 | 48 | 29 | ||
| WHO grade# | |||||
| I–II | 59/155 | 33 | 26 | 5.378 | 0.02* |
| III–IV | 96/155 | 71 | 25 | ||
| Ki-67 (%) | |||||
| <10 | 86/155 | 53 | 33 | 2.617 | 0.106 |
| ≥10 | 69/155 | 51 | 18 | ||
| p53 (%) | |||||
| <10 | 71/155 | 36 | 35 | 15.946 | 0.000*** |
| ≥10 | 84/155 | 68 | 16 | ||
| KPS score | |||||
| <70 | 66/155 | 42 | 24 | 0.623 | 0.430 |
| ≥70 | 89/155 | 62 | 27 | ||
Classified by the 2007 WHO (World Health Organization) Classification of Tumors of the Nervous System;
statistically significant (P<0.05);
statistically significant (P<0.01);
statistically significant (P<0.00).
KPS – Karnofsky performance scale.
Immunohistochemistry (IHC)
Immunostaining reagents for polyclonal anti-human PASD1 antibody were obtained from Abcam (ab 224362). Continuous sections of 4 μm thickness were prepared from formalin-fixed, paraffin embedded tissue. Subsequently, the sections were heated in citrate buffer (pH 6.0) for high-temperature antigen retrieval. After endogenous peroxidase was inactivated by 3% hydrogen peroxide, the sections were immunostained with anti-PASD1 polyclonal antibody (1: 200 dilution) or pre-immune serum (negative control) overnight at 4°C. Then the treated sections were recovery at room temperature and incubated with biotinylated second antibodies (ZSGB-BIO, China). Lastly, immunoreactivity was visualized with 3, 3′-diaminobenzidine (DAB) (Maixin Biotechnology, China) followed by hematoxylin counterstain.
Cell lines and cell transfection
There were 4 glioma cell lines (U251, SHG-44, A172, and LN229), together with 1 human glial (HEB), cell line collected from the Type Culture Collection Cell Bank, of the Chinese Academy of Science Committee (Shanghai, China). Subsequently, cells were cultivated in DMEM (GIBCO-BRL, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Gibco) in an incubator at 37°C and 5% CO2. For stabile knockdown of PASD1 in glioma cells, the shRNA sequence that specifically targeted PASD1 (sh-PASD1: 5′-CCAAGAAACAACAGAAACA-3′) had been cloned into the lentiviral plasmid pLKO.1 puro (Addgene, Cambridge, MA, USA). Additionally, a scrambled shRNA sequence targeting none of the known gene (NC, 5′-CAUCGCAUCGUGUAATTTCTG-3′) had also been processed in parallel as the negative control. Besides, pcDNA3.1 (control) and pcDNA3.1/PASD1 (specifically overexpress PASD1) was brought from Applied Biological Materials (ABM, Canada). Cells were transfected using the Lipofectamine 2000 kit (Thermo Fisher Scientific) in strict accordance with the manufacturer protocols.
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the collected tissues, which was then separated with the TRIzol reagent (Invitrogen, Grand Island, CA, USA). Subsequently, the isolated RNA would be reversely transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Japan) following manufacturer instructions. Afterwards, the SYBR Prime Script RT-PCR Kit (Takara, Japan) would be employed for qRT-PCR according to manufacturer instruction. All primers used in this study had been presented as follows: PASD1 primers, 5′-CTGTTAAGGTGGTGGCATTG-3′ (forward), and 5′-GGAGGCTCATACTGGCTGAT-3′ (reverse); GAPDH primers, 5′-CTCGCTTCGGCAGCACA-3′ (forward), and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). All experiments were carried out 3 times, and relative expression was calculated according to the 2−ΔΔCt method [6] and standardized based on GAPDH mRNA expression.
Western blot analysis of the isolating proteins
After being separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), cell protein lysates were first transferred onto polyvinylidene fluoride membranes (Roche), followed by incubation with specific rabbit anti-human antibodies (Abcam, Shanghai), including cyclin B1 (ab32053, 1: 3000 dilution), p-CDK1 (ab18, 1: 3000 dilution), cleaved caspase-3 (ab32042, 1: 5000 dilution), and cleaved caspase-9 (ab2324, 1: 5000 dilution). Subsequently, cells were stored at 4°C overnight, and incubated with the secondary anti-rabbit antibodies (A32732, 1: 1000 dilution, Thermo Fisher Scientific, American), where the ECL chromogenic substrate had been applied in quantification through densitometry (Quantity One software; Bio-Rad, Hercules, CA, USA).
Cell proliferation assay
Cell viability was assessed using the Cell Counting Kit 8 (CCK-8) kit (Dojindo Laboratories, Kumamoto, Japan). Afterwards, cells were seeded in a 96-well plate at a density of 1×104 cells/well. After 24 hours of culture, the corresponding sh-PASD1 and NC were transfected and cultivated in the normal mediums. Later, the CCK-8 solution was added at 0, 24, 48 and 72 hours, and relative cell number was evaluated at the wavelength of 450 nm. All assays were performed 3 times.
Colony forming assay
After trypsin digestion, an appropriate number of transfected cells were seeded into the culture dish and cultivated in DMED supplemented with 10% FBS for 2 weeks. Afterwards, cells were fixed with paraformaldehyde and stained with crystal violet (Beyotime, Haimen, China). Subsequently, cells were air-dried, and photos were used to count the visible colonies.
Distribution of cell cycle, as well as analysis of cell apoptosis
The sh-PASD1- or NC-transfected cells were seeded in 6-cm dishes, harvested after reaching 70% convergence, and washed with cold phosphate-buffered saline (PBS). To assess the cell cycle, cells were initially fixed into cold ethanol (70%) overnight at −20°C, washed with PBS, stained with PI/RNase Staining Buffer (BD Pharmingen, USA), and incubated at room temperature for 30 minutes in dark prior to analysis on the BD FACSVerse. For apoptosis, cells were washed with cold PBS, resuspended in the binding solution, stained with Annexin V-FITC (Beckman Coulter), and PI/RNase staining buffer, and incubated at room temperature for 30 minutes in dark.
Statistical analysis
SPSS 22.0 (SPSS Inc., Chicago, IL, USA) was employed for all statistical analyses. The Student’s t-test was adopted for inter-group pairwise comparisons, while one-way analysis of variance (ANOVA) or chi-square test was utilized for multiple group comparisons. Moreover, Spearman’s correlation analysis was employed to detect the correlation of PASD1 expression in glioma tissues with that in cells. Besides, survival was analyzed by the Kaplan-Meier method. Multivariate analysis of prognostic parameters in glioma patients was analyzed, and meanwhile, the multi-factor Cox proportional hazards model was used to analyze the prognosis-related factors among glioma patients. A difference of P<0.05 was deemed to show statistical significance.
Results
PASD1 was elevated in glioma
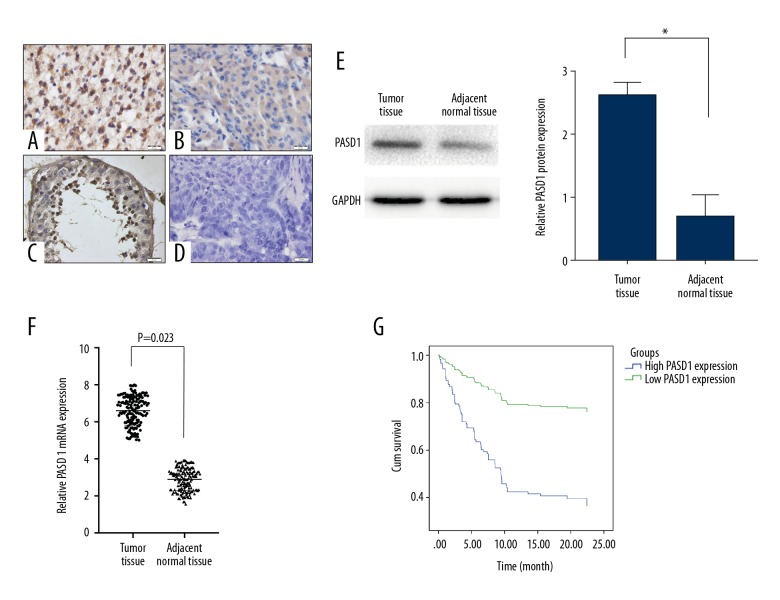
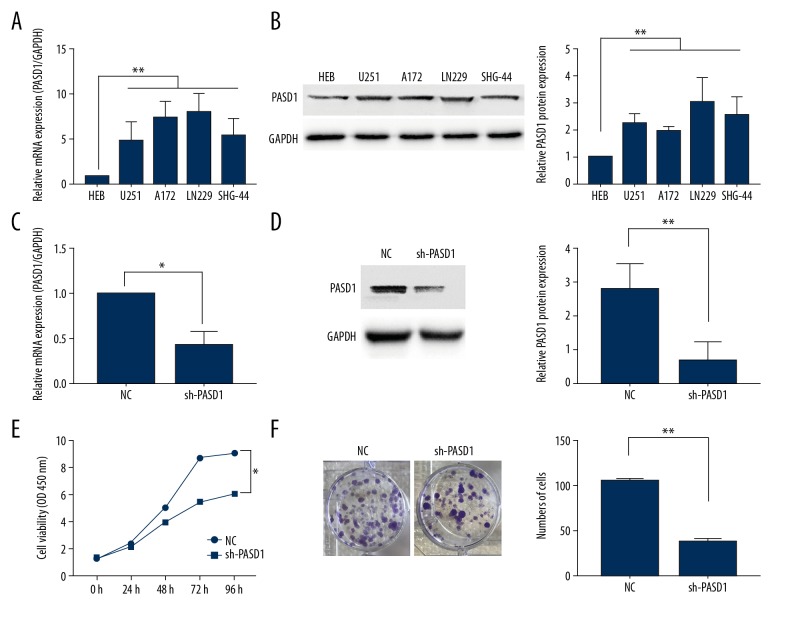
To explore the role of PASD1 in glioma, PASD1 expression in glioma as well as normal adjacent tissue specimens was detected among the 155 glioma samples through immunohistochemical (IHC) staining, along with PASD1 specific antibody testing. As was seen in our results, positive staining was mainly observed in cytoplasm as well as nucleus (Figure 1A–1C) with pre-immune serum as negative control (Figure 1D). Moreover, the intensity of staining indicated that high and low PASD1 protein expression levels were 67.10% (104 out of 155) and 32.90% (51 out of 155), respectively (Table 1). At the same time, 5 normal cerebral tissues were detected, which suggested that PASD1 was rarely detected in normal cerebral specimens (Figure 1C). In addition, relative PASD1 levels in glioma specimens were detected through western blot and qRT-PCR, and were shown to be markedly upregulated relative to that in normal cerebral specimens (P<0.001) (Figure 1E, 1F). Furthermore, PASD1 was also over-expressed in glioma cell lines compared with that in human glial (HEB) cell line (P<0.05) whether in protein level or mRNA level (Figure 2A, 2B). Therefore, the LN229 cell line with the highest PASD1 expression was selected to downregulate PASD1 through infection with the PASD1-specific sh-PASD1 (sh-NC was used as the control) (Figure 2C, 2D).
Figure 1.
PASD1 is upregulated in glioma tissues and cell lines. (A–D) Typical immunohistochemical staining of PASD1 protein in glioma tissues (A, B), normal brain tissue (C) and paired glioma tissues probed with pre-immune serum as negative control (D). (E, F) Relative expression of PASD1 protein and mRNA in 155 pairs of glioma tissues and adjacent normal brain tissues by western blot assay (E) and qRT-PCR (F). (G) The overall survival in glioma patients presented by Kaplan-Meier curves. P<0.05. Bar, 20 μm. Data are presented as mean ± standard error based on at least 3 independent experiments.
Figure 2.
PASD1 promotes cell proliferation. (A, B) qRT-PCR assay (A) and western blot assay (B) were performed to examine the expression of PASD1 in 5 glioma cells and 1 normal human glial (HEB) cell line. (C, D) qRT-PCR assay (C) and western blot assay (D) were performed to examine the expression of PASD1 in LN229 cell line transfection with PASD1-specific sh-PASD1 (shRNA as control). (E, F) CCK-8 assay (E) and cell colony assay (F) of cell viability. The viability of sh-PASD1 group was significantly decreased compared to control at 24 hours. The columns show the mean for 3 separate experiments; bars, standard deviation. * P<0.05, ** P<0.01.
Higher PASD1 expression was related to the poorer prognosis among glioma patients
PASD1 was over-expressed in glioma; therefore, it was proposed that PASD1 expression might be associated with the pathogenesis of glioma. As shown in Table 1, PASD1 expression was detected to be markedly correlated with gender (P=0.004), WHO grade (P=0.02) as well as p53% (P=0.000), but not with age of patient (P=0.141), tumor size (P=0.21), Ki-67 (P=0.106) or KPS score (P=0.430). Moreover, high PASD1 expression could be used as an independent factor to predict the prognosis of glioma patients (Table 2, P=0.000). Likewise, Figure 1G showed that high PASD1 expression was associated with the dismal prognosis for glioma patients (P<0.05). Taken together, these findings indicated the potential of PASD1 as a detrimental prognosis factor in glioma.
Table 2.
Cox proportional Hazards model was used to analyze the effect of PASD1 expression on the survival of glioma patients.
| Variable | Category | HR (95% CI) | P value |
|---|---|---|---|
| Gender | Male | 0.986 (0.562–1.641) | 0.957 |
| Female | |||
| Age (years) | <38 | 0.737 (0.407–1.334) | 0.313 |
| ≥38 | |||
| Tumor size (cm) | <5 | 0.919 (0.570–1.479) | 0.727 |
| ≥5 | |||
| WHO grade | I–II | 0.768 (0.456–1.292) | 0.320 |
| III–IV | |||
| Ki-67 expression | <10 | 0.902 (0.563–1.444) | 0.667 |
| ≥10 | |||
| p53 expression | <10 | 1.034 (0.628–1.703) | 0.895 |
| ≥10 | |||
| KPS score | <70 | 0.851 (0.526–1.377) | 0.511 |
| ≥70 | |||
| PASD1 expression | High | 3.696 (1.893–7.218) | 0.000*** |
| Low |
Univariate analysis was performed using the log-rank test. Multivariate analysis was performed using the Cox proportional hazards model. HR – Hazard ratio; 95%CI – 95 percent confidence interval for relative risk.
Statistically significant (P<0.05);
statistically significant (P<0.01);
statistically significant (P<0.00).
PASD1 promoted cell proliferation through inhibiting apoptosis in vitro
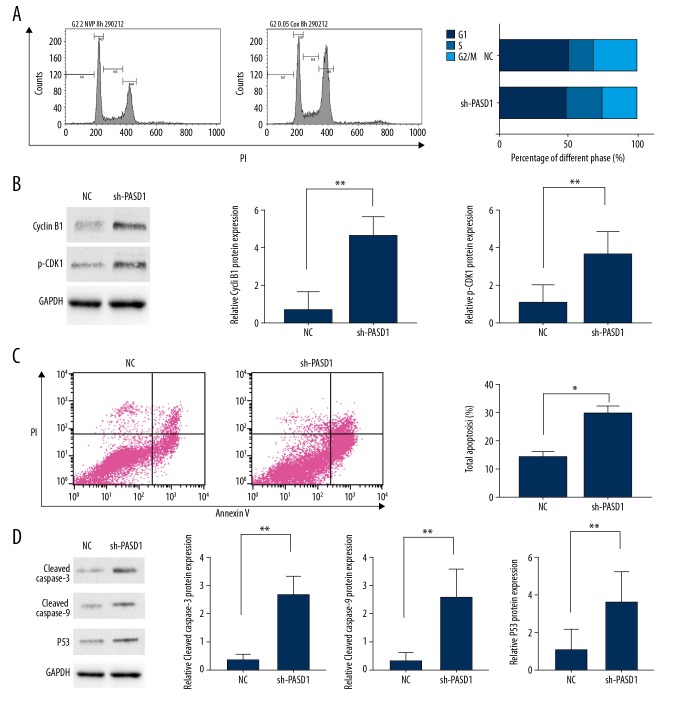
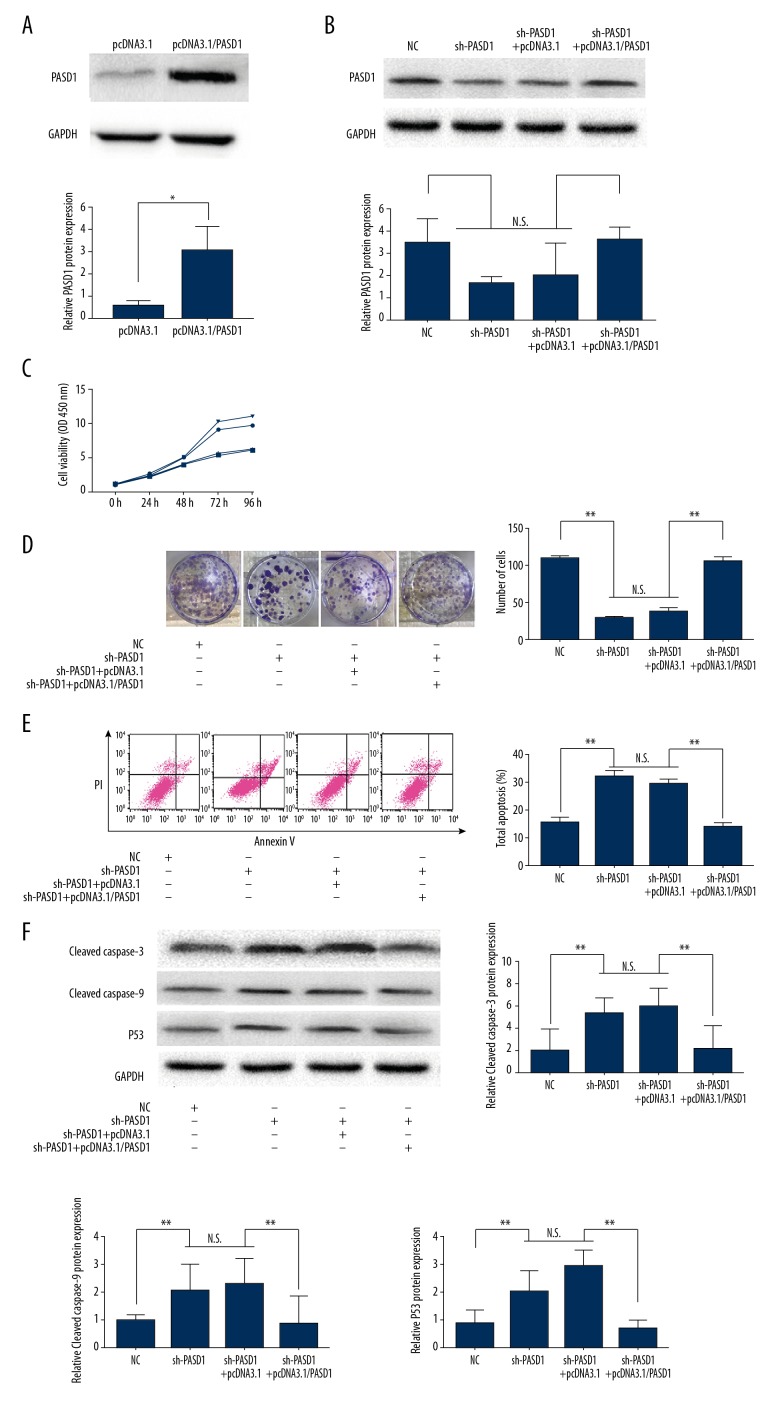
Results of the CCK-8 assay revealed that transfection with sh-PASD1 could remarkably reduce the proliferation rate of LN229 compared with that in cells transfected with sh-RNA (P<0.01, Figure 2E). Similarly, results of colony forming assay indicated that PASD1 downregulation could lead to decreased proliferation relative to that in the control group (P<0.05; Figure 2F). Disturbance of the cell cycle could result in persistent cell proliferation [7]; as a result, flow cytometry could be utilized to detect whether PASD1 could impair the cell cycle. Our results showed that, PASD1 knockdown could dramatically reduce the number of cells at S phase, along with increased number of cells at G2/M phase relative to those in the control group (P<0.05, Figure 3A). Moreover, cell counts at G0/G1 phase displayed a statistically significant difference. Meanwhile, western blot assay was performed to further verify the influence of PASD1 on the proliferation of glioma. As shown in Figure 3B, cyclin B1 expression was evidently upregulated in sh-PASD1 transfected cells (P<0.05). Meanwhile, cyclin-dependent kinase 1 (CDK1) expression was markedly downregulated compared with that in the control group (P<0.05). Moreover, flow cytometry was performed to further verify whether PASD1 could regulate cell proliferation through interfering cell apoptosis. As presented in Figure 4A, downregulating PASD1 in the LN229 cell lines accelerated cell apoptosis (P<0.01; Figure 3C). Therefore, the protein expression levels of cleaved caspase-3 as well as cleaved caspase-9 were evaluated; in the meantime, the levels of apoptosis markers belonging to the caspase family, together with p53, a well-known pro-apoptotic protein, were also detected. As presented in Figure 3D, compared with the control group, downregulating PASD1 could upregulate the levels of cleaved caspase-3, cleaved caspase-9, and p53 (P<0.01; Figure 3D). Therefore, PASD1 downregulation in glioma cells would inhibit the proliferation ability of cells and result in cell arrest at G2/M phase by accelerating apoptosis. Then, rescue assays were applied to further illuminate that the function of PASD1 in glioma. Satisfactory transfection efficiency of PASD1-overexpression was detected in the A172 cell line (P<0.05; Figure 4A). After co-transfected with sh-PASD1 and pcDNA3.1/PASD1 in the LN229 cell line (Figure 4B), CCK8 assays and colony formation assays were performed. As revealed in Figure 4C and 4D, the reducing of proliferation induced by downregulation of PASD1 was restored by co-transfecting pcDNA3.1/PASD1 in the LN229 cell line. Additionally, as shown in Figure 4E, co-transfection with pcDNA3.1/PASD1 recovered the enhancing of apoptosis induced by PASD1 upregulation. Similarly, up-expression of cleaved caspase-3, cleaved caspase-9, and p53 were reduced by co-transfection with pcDNA3.1/PASD1 (P <0.05; Figure 4F). These data indicated that PASD1 promoted cell proliferation through inhibiting apoptosis in vitro.
Figure 3.
Flow cytometric analysis of cell cycle and cell apoptosis. (A) Flow cytometry to detect cell cycle. Cell counts in different phases suggest that downregulation of PASD1 would result in cell arrest at G2/M phase. (B) Results of western blot assay on cyclin B1 as well as CDK1 expression. (C) Flow cytometric analysis of cell apoptosis and the percentage of total apoptosis cells showed that downregulation of PASD1 accelerated apoptosis. (D) Western blot analysis of cleaved caspase-3, cleaved caspase-9 and p53 levels. The columns show the mean for 3 separate experiments; bars, standard deviation. * P<0.05, ** P<0.01
Figure 4.
Recue assay. (A, B) Efficiency of transfected pcDNA3.1/PASD1 in A172 (A) and co-transfected sh-PASD1 and pcDNA3.1/PASD1 in LN229 (B) were detected by western blot assay. (C, D) CCK-8 assay (C) and cell colony assay (D) of cell viability. (E) Flow cytometric analysis of cell apoptosis and the percentage of total apoptosis cells. (F) Western blot analysis of cleaved caspase-3, cleaved caspase-9 and p53 levels. The columns show the mean for 3 separate experiments; bars, standard deviation. * P<0.05, ** P<0.01
Discussion
Nowadays, an increasing number of studies have revealed the relationship between cancer-associated cancer-testis antigen (CTAs) and the tumorigenesis of various cancers. Moreover, many CTAs have been identified to play a vital role in glioma. Li et al. revealed that high OY-TES-1 expression could be detected in glioma tissues; besides, the anti-OY-TES-1 antibodies existed in the serum of 5 out of 36 glioma patients (14%), which could not be detected in the serum samples from 107 healthy donors [6]. Saito et al. demonstrated that KIF20A was a tumor-associated antigen, which was involved in the growth and survival of glioma cells, suggesting that KIF20A was an onco-antigen as well as a candidate novel immunotherapeutic target for glioma [7]. On the other hand, Deng et al. declared that multiple CTAs, such as ADAMTS1, ADAM23, SPANXA1, SPANXB1/2, IL13RA2, VCY, and VCX3A, had been upregulated in pediatric glioma, which was correlated with pediatric gliomagenesis [8]. Therefore, CTAs can potentially serve as prognostic factors and diagnostic biomarkers for glioma. However, the molecular mechanism by which PASD1 affects glioma cells remains largely unknown.
PASD1 was originally identified as a CTA in diffuse large B cell lymphoma [8], and was discovered to be expressed in many other human cancers, including various hematological malignancies such as acute myeloid leukemia [9], Burkitt’s lymphoma [9], Hodgkin’s lymphoma [10], and multiple myeloma [10], as well as solid tumors like melanoma [11], lung cancer [12], head and neck cancer [11], cervical cancer [13], and colorectal carcinoma [14]. PASD1 has become a promising immunotherapeutic target for many cancers, which can be ascribed to its restricted expression and good immunogenicity [14]. Recently, PASD1 was reported to positively regulate IL-6–STAT3 signaling, which facilitates tumor growth [14]. In our experiment, high PASD1 expression was also be detected in glioma tissues compared with that in adjacent tissues. Additionally, PASD1 expression was found to be closely related to gender, WHO grade, and p53 (%), and the over-expression of PASD1 indicated the poor prognosis for glioma patients. These data demonstrated the potential of PASD1 as a novel indicator for diagnosis and poor prognosis in glioma.
Moreover, it was also found that knockdown of PASD1 could result in reduced proliferation of glioma cells in vivo, which was achieved through inducing cell arrest at G2/M phase; meanwhile, differences in the cell counts at G0/G1 phase displayed no statistical significance. To further verify flow cytometric (FCM) results, the protein expression levels of cyclin B1 and p-CDK1 were detected after cell infection. As is well-known, cell cycle progression is modulated through numerous factors, including CDKs, as well as the inhibitors of CDKs [15]. Among these, cyclin B1 is associated with CDK1 and is expressed at the G2 phase; however, the cyclin B1/CDK1 complexes remains inactive until late in the G2 phase when their activation is required to initiate the mitosis [16]. Therefore, cyclin B1 and p-CDK1 were selected in our study for detection. Our experiments suggested that cyclin B1 expression was increased in glioma cells in the presence of downregulated PASD1, whereas p-CDK1 was downregulated. Thereby, it was proposed that PASD1 might be an upstream modulator of both cyclin B1 and p-CDK1. Obviously, the imbalanced apoptosis and proliferation usually result in the progression of tumor growth. Consequently, the role of PASD1 in the apoptosis of glioma cells would also be detected, which would suggest a significant increase compared with the control group in the presence of the downregulated PASD1. The FCM results were consistent with upregulated p53 expression as well as cleaved caspase-3, an apoptosis marker belonging to the caspase family. It has been shown that p53, a well-known typical transcription factor, exerts a vital part in numerous cell processes, like aging, cell cycle, cell growth, as well as cell apoptosis, which is achieved by regulating various downstream target genes, like p21 as well as Bcl-2 [17]. It is thus expected that downregulated PASD1 would upregulate cleaved caspase-3, cleaved caspase-9, and p53 expression, which suggests the role of PASD1 in stimulating glioma progression. However, it remains unclear whether Bcl-2 [18] and Bax [19], and other caspase family members, are also involved in apoptosis induced by sh-PASD1, which should be further investigated.
Conclusions
This study revealed that PASD1 expression was remarkably over-expressed in glioma. Moreover, PASD1 might serve as an oncogene, which can indicate poor survival of glioma patients. In the meantime, it was demonstrated in this study that knockout of PASD1 could facilitate cell apoptosis in vitro. Likewise, downregulating the expression of PASD1 could inhibit cell proliferation and induce the arrest of cell cycle in vivo. These results demonstrate that PASD1 might potentially serve as a novel target for the prognosis and the future treatment of glioma.
Footnotes
Source of support: Departmental sources
References
- 1.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–46. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neurooncology. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salmaninejad A, Zamani MR, Pourvahedi M, et al. Cancer/testis antigens: Expression, regulation, tumor invasion, and use in immunotherapy of cancers. Immunol Invest. 2016;45:619–40. doi: 10.1080/08820139.2016.1197241. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, Ohta S, Kawakami Y, et al. Functional analysis of KIF20A, a potential immunotherapeutic target for glioma. J Neurooncol. 2017;132:63–74. doi: 10.1007/s11060-016-2360-1. [DOI] [PubMed] [Google Scholar]
- 5.Ait-Tahar K, Liggins AP, Collins GP, et al. CD4-positive T-helper cell responses to the PASD1 protein in patients with diffuse large B-cell lymphoma. Haematologica. 2011;96:78–86. doi: 10.3324/haematol.2010.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 7.Cheng T, Xu Y. Effects of enhancer of zeste homolog 2 (EZH2) expression on brain glioma cell proliferation and tumorigenesis. Med Sci Monit. 2018;24:7249–55. doi: 10.12659/MSM.909814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ait-Tahar K, Liggins AP, Collins GP, et al. Cytolytic T-cell response to the PASD1 cancer testis antigen in patients with diffuse large B-cell lymphoma. Br J Haematol. 2009;146:396–407. doi: 10.1111/j.1365-2141.2009.07761.x. [DOI] [PubMed] [Google Scholar]
- 9.Hardwick N, Buchan S, Ingram W, et al. An analogue peptide from the Cancer/Testis antigen PASD1 induces CD8+ T cell responses against naturally processed peptide. Cancer Immunity. 2013;13:16. [PMC free article] [PubMed] [Google Scholar]
- 10.Joseph-Pietras D, Gao Y, Zojer N, et al. DNA vaccines to target the cancer testis antigen PASD1 in human multiple myeloma. Leukemia. 2010;24:1951–59. doi: 10.1038/leu.2010.196. [DOI] [PubMed] [Google Scholar]
- 11.Stamell EF, Wolchok JD, Gnjatic S, et al. The abscopal effect associated with a systemic anti-melanoma immune response. Int J Radiat Oncol Biol Phys. 2013;85:293–95. doi: 10.1016/j.ijrobp.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prins GS, Hu WY, Xie L, et al. Evaluation of bisphenol A (BPA) exposures on prostate stem cell homeostasis and prostate cancer risk in the NCTR-Sprague-Dawley rat: an NIEHS/FDA CLARITY-BPA Consortium Study. Environ Health Perspect. 2018;126:117001. doi: 10.1289/EHP3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan G, Brooks SE, Mills KI, Guinn BA. Infrequent expression of the cancer-testis antigen, PASD1, in ovarian cancer. Biomark Cancer. 2015;7:31–38. doi: 10.4137/BIC.S28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu ZS, Zhang HX, Zhang YL, et al. PASD1 promotes STAT3 activity and tumor growth by inhibiting TC45-mediated dephosphorylation of STAT3 in the nucleus. J Mol Cell Biol. 2016;8:221–31. doi: 10.1093/jmcb/mjw005. [DOI] [PubMed] [Google Scholar]
- 15.Lim S, Kaldis P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development. 2013;140:3079–93. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z, Fan M, Candas D, et al. Cyclin B1/CDK1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell. 2014;29:217–32. doi: 10.1016/j.devcel.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469:325–46. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 18.Edlich F. BCL-2 proteins and apoptosis: Recent insights and unknowns. Biochem Biophys Res Commun. 2018;500:26–34. doi: 10.1016/j.bbrc.2017.06.190. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Ding Y, Ye N, et al. Direct activation of Bax protein for cancer therapy. Med Res Rev. 2016;36:313–41. doi: 10.1002/med.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]