Abstract
Background
Autologous saphenous vein is the most common choice for coronary artery bypass grafting. This study was conducted to identify and characterize differentially expressed genes (DEGs) induced by overexpressing DEPTOR in human saphenous vein endothelial cells (hsVECs) that might play roles in restenosis.
Material/Methods
hsVECs isolated from the saphenous veins were transfected with DEPTOR overexpression vector and analyzed for mTOR expression. RNA was prepared from the cells and sequenced using high-throughput sequencing technology (RNA-Seq). The DEGs were analyzed based on enrichment scores in GO terms and KEGG pathways.
Results
The cells had typical hsVEC morphology and characteristics based on the HE staining and immunohistochemical and immunofluorescence assays. The expression of mTOR increased, and 102 genes were upregulated, and 409 genes were downregulated after DEPTOR overexpression. KEGG analysis showed that the DEGs were mainly enriched in 20 signal pathways, such as Focal adhesion and ECM-receptor interaction pathways. The DEGs were enriched in GO terms such as integrin binding and glycosaminoglycan binding. For cellular components, GO analysis revealed that the DEGs were enriched in main axon, plasma membrane part, cell junction, and proteinaceous extracellular matrix. DEGs included many cytokines, such as bone morphogenetic protein-7, interleukin-8, interleukin-1β, and inhibin, which have important effects on vascular growth and inflammation.
Conclusions
The overexpression of DEPTOR in hsVECs results in DEGs that are involved in cell proliferation and differentiation, intercellular junction, and extracellular matrix receptor. These findings may provide valuable molecular information for improving venous permeability through manipulation of DEPTOR and related mTOR pathways.
MeSH Keywords: Coronary Artery Bypass, Coronary Restenosis, Gene Expression
Background
Coronary artery bypass grafting (CABG) is a typical application of arterial bypass grafting, and is mainly used to treat ischemic heart diseases [1]. Autologous saphenous vein is the most common choice for CABG [2]. However, restenosis rates at 1 year and 10 years after CABG have been reported to be 15% and 50%, respectively [3]. Rapamycin is an inhibitor of mammalian target of rapamycin (mTOR). It inhibits cell proliferation and is used to prevent restenosis[4–6]. mTOR has 2 structurally and functionally different complexes – mTORC1 and mTORC2 – and the latter is relatively insensitive to rapamycin [7,8]. DEPTOR (domain-containing mTOR-interacting protein) is another common component of mTORC1 and m TORC2. DEPTOR also directly interacts with mTOR. It has been reported that the overexpression of DEPTOR downregulates the activity of mTORC1 and mTORC2 [9,10]. In addition, studies have reported that DEPTOR regulates the synthesis of fat [11]. Therefore, DEPTOR has great value in improving blood flow in the human saphenous vein. To better explore the molecular roles of DEPTOR, high-throughput sequencing technique (RNA-Seq) was used to identify and characterize differentially expressed genes (DEGs) induced by DEPTOR. These findings may provide valuable molecular information and clues for improving venous permeability through manipulation of DEPTOR and related mTOR pathways.
Material and Methods
Tissues and reagents
Human saphenous veins abandoned in CABG were obtained from the Surgery Department of the First Affiliated Hospital of Nanchang University. pcDNA3.1 was used to construct a DEPTOR expression vector pcDNA-DEPTOR. DMEM/F-12 (1: 1) (cat. no. 1861453) and Opti-MEM (cat. no. 331985-062) were obtained from GIBICO, USA. Lipofectamine 3000 (cat. no. 18882752) was purchased from Invitrogen, USA. Human bFGF (cat. no. L10402031) and human EGF (cat. no. N10504031) were obtained from Cyagen, USA. Mouse monoclonal antibody against GAPDH (1/2000, cat. no. TA-08), goat anti-mouse IgG (1/2000, cat. no. ZB-2305) and anti-rabbit IgG (1/2000, cat. no. ZB-2301) were purchased from Zsbio, Beijing, China. Rabbit polyclonal antibody against mTOR (1/2000, cat. no. ab2732) was purchased from Abcam, USA. Rabbit antibodies against CD31 (1/2000, cat. no. BA2966), CD34 (1/2000, cat. no. PB0031) and vWF (1/2000, cat. no. bs-20428R) were purchased from BOSTER, Beijing, China. Conjugated goat anti-rabbit IgG Cy3 (1/2000, cat. no. CW0159S) was purchased from CWBIO, Wuhan, China. Rabbit anti-factor VII (1/1000, cat. no. bs-2974R) and HRP labeled anti-rabbit IgG (1/2000, cat. no. SV0002) were obtained from Bioss, Beijing, China.
Cell isolation and culture
hsVECs were isolated from the abandoned saphenous veins as described previously [12]. Six segments of 1 cm end segments of saphenous vein were taken from each of 3 patients and used together in the study. Briefly, a syringe with a lavage needle was inserted into one end of the vein. The venous cavity was washed repeatedly with PBS. Then, the washed vein was injected with collagenase II (1 g/L) and incubated in a CO2 incubator at 37°C for 15 min. The digest was collected and centrifuged at 4°C for 5 min. The cells were suspended and cultured in DMEM/F12 medium with 20% FBS, streptomycin and penicillin mixture (cat. no. 1400, Solarbio, USA), hEGF(10μg/L), 1% insulin-transferrin-selenium (ITS) (cat. no. 41400-045, Gibco, USA), and hbFGF (3ng/mL) in a 5% CO2 incubator at 37°C.
HE staining
The tissue samples were fixed with formalin, embedded in paraffin, and sectioned into 4-μm-thick slices. The sections were baked, dewaxed, rehydrated, and stained with hematoxylin solution for 3 min. After differentiation with hydrochloric acid ethanol solution for 15 s, the slides were stained again for 15 s with eosin. The slides were viewed under a microscope after being sealed with neutral resin.
Immunohistochemistry
The cells were incubated with antibodies against factor VIII for 2 h at room temperature followed by incubation with appropriate secondary antibodies for 1 h, and mounted with ProLong antifade reagent (Invitrogen). Diaminobenzidine (DAB) and hematoxylin chromogen (Dako, Glostrup, Denmark) method was used. The sections were subsequently examined by light microscopy.
Immunofluorescence
The cells were immobilized with 4% paraformaldehyde for 15 min, cleared with 0.5%Triton X-100 at room temperature for 20 min, and blocked with 5% BSA at 37°C for 30 min. The cells were stained with antibodies against CD31, CD34, or vWF at 4°C overnight. Cy3 antibody was added drop-wise to cells and incubated at 37°C for 30 min. DAPI was used to stain the nuclei, and the slides were viewed under a fluorescence microscope. The red fluorescence was from the proteins of interest, while the blue fluorescence was from the nucleus.
Transfection
hsVECs were grown to 90% confluence and transfected with pcDNA- DEPTOR using Lipofectamine 3000 according to the manufacturer’s instructions. pcDNA 3.1 was used as negative control. Transfected cells were cultured in DMEM/F12 medium with 20% FBS for 24 h and harvested for transfection efficiency assays.
Construction of Illumina sequencing library
Total RNA was isolated from the cells using the TRIzol reagent (Invitrogen). The concentration and purity of the RNA were detected using Nanodrop 2000 and the RNA integrity was checked using agarose gel electrophoresis. We used the Ribo-Zero Magnetic Kit and RNase R (EpiCenter, USA) to remove rRNA and linear RNA. The purified RNA was used to construct a paired-end library using the TruSeq™ Stranded Total RNA Library Prep Kit (Illumina, USA) for sequencing on the Hiseq4000 platform according to the supplier’s instructions.
Sequence analysis
SepPrep and Sickkle software were used to test the quality of the reads. The reads were then aligned with the human reference genome [13] using Bowtie to generate the gene lists. Sequences from cells transfected with pcDNA- DEPTOR or transfected with pcDNA 3.1 were compared to generate differentially expressed genes (DEGs).
Enrichment analysis of DEGs
Goatools was used to enrich GO function (molecular function, cell components, biological processes) [14–16] and KOBAS was used to enrich KEGG Pway and annotate KEGG functions [17,18].
Statistical analysis
Data were expressed as means ±s.d. and were analyzed using the statistical analysis software SPSS 19.0. Student’s t-test was performed to compare the difference between groups and P<0.05 were considered as statistically significant.
Results
Characterization of primary hsVEC culture
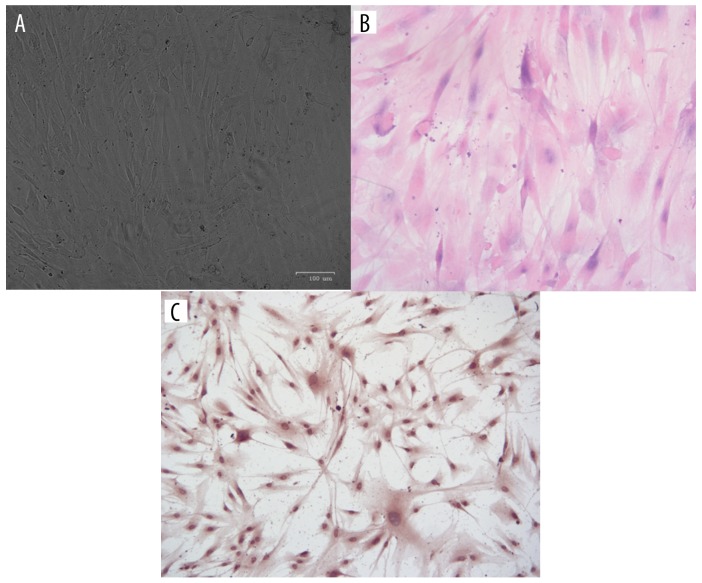
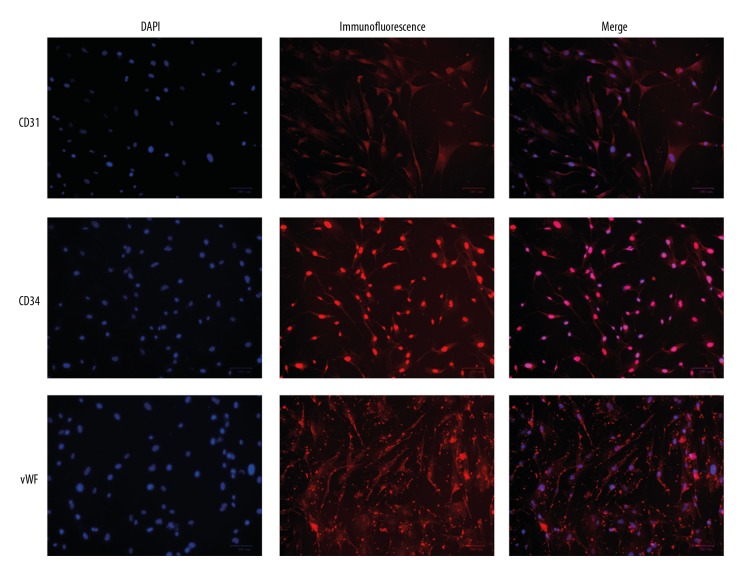
The morphology of primary hsVEC culture is shown in Figure 1A. HE staining and immunohistochemical staining of endothelial factor VIII (Figure 1B, 1C) showed that the culture had typical features of hsVECs. In addition, endothelial markers were detected in the cytoplasm (CD31), nuclei (CD34), and the junctions between the cells (vWF) in the cultured cells (Figure 2), further demonstrating that they are hsVECs.
Figure 1.
Culture and characterization of hsVEC. (A) Morphology of primary hsVEC culture. (B, C). HE staining and immunohistochemical staining of endothelial factor VIII.
Figure 2.
Immunofluorescence assays of endothelial markers CD31, CD34, and vWF in cultured hsVECs.
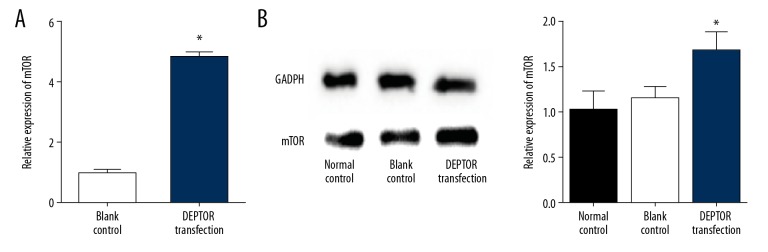
DEPTOR increased after transfection
When the cells were transfected with pcDNA- DEPTOR, the mRNA and protein levels of DETOR were significantly upregulated (p<0.05, Figure 3).
Figure 3.
Expression of mTOR mRNA and protein in hsVECs transfected with pcDNA- DEPTOR. (A) mRNA level. (B) Left panel: representative Western blots, right panel: relative protein levels. * Denotes significantly difference vs. control.
Differentially expressed genes following DEPTOR overexpression
To obtain the gene expression profiles following DEPTOR overexpression, RNA samples were isolated from hsVECs transfected with and without DEPTOR expression cassette. The samples were sequenced on an Illumina HiSeq 4000 platform. Quality control on the raw reads was performed using SepPrep and Sickkle. Analysis showed that there were 102 upregulated and 409 downregulated genes in DEPTOR-transfected cells as compared with empty vector-transfected cells (Figure 4, Supplementary Table 1, [Supplemantary/raw data available from the corresponding author on request]).
Figure 4.
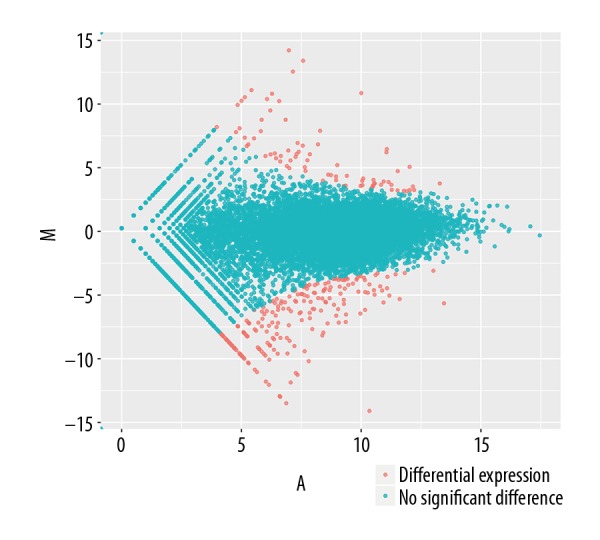
MA-plots showing fold change in expression generated by comparing DEPTOR overexpression vs. empty vector. The plots were obtained using DEGseq package, where M (Y-axis) represents the intensity ratio log2 (fold change), and A (X-axis) represents the average intensity.
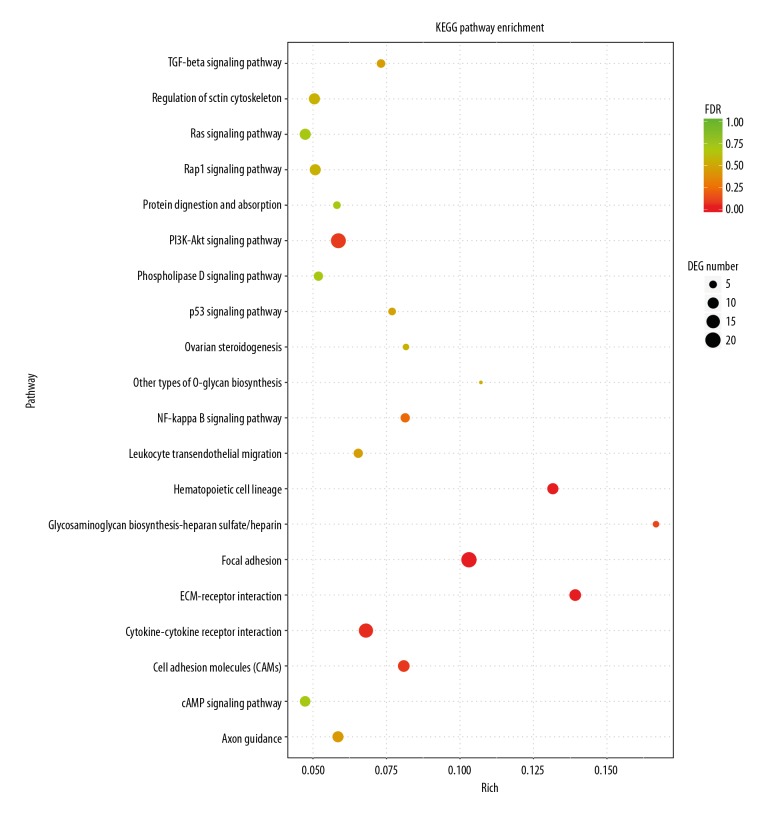
KEGG pathway analysis of DEGs
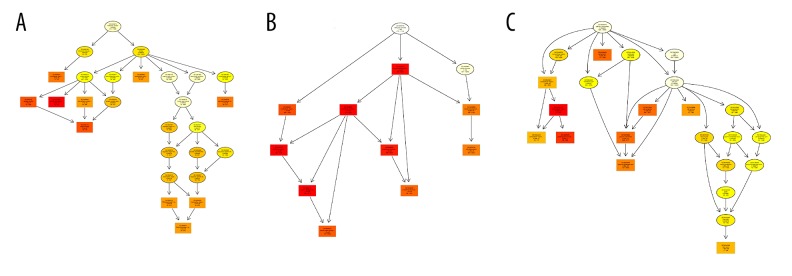
KEGG pathway analysis showed that the differentially expressed genes were mainly concentrated in 20 signaling pathways, such as Focal adhesion, ECM-receptor interaction, Cytokine-cytokine receptor interaction, and PI3K-Akt signaling pathway (Figure 5).
Figure 5.
KEGG pathway analysis of DEGs identified in in hsVECs. The ordinate is the KEGG Pathway entry; the abscissa is the ratio of the number of differentially enriched genes to the number of genes annotated on the pathway. The size of the dot in the graph represents the number of differentially enriched genes and the number of genes annotated on the pathway. The color represents the significance of the pathway P. The smaller the P value is, the more significant the enrichment is.
GO analysis of DEGs
GO enrichment analysis showed that in molecular functions, the DEGs were concentrated in integrin binding, glycosaminoglycan binding, RNA polymerase II regulatory region sequence-specific DNA binding, and extracellular matrix structural constituent (Figure 6A). In biological process, enriched GO terms included cell differentiation, anatomical structure morphogenesis, and system development (Figure 6B). For cellular components, GO analysis revealed that differentially expressed genes were enriched in GO terms such as main axon, plasma membrane part, cell junction, and proteinaceous extracellular matrix (Figure 6C).
Figure 6.
GO term analysis of DEGs identified in in hsVECs. (A) Molecular functions. (B) Biological process. (C) Cellular components.
Discussion
CABG uses autologous veins to bridge arteries, and the vein segment actually acts as an artery after the operation, which is thus subjected to different hemodynamic conditions and increased partial pressure of oxygen after the surgery. In order to adapt to this change, the wall of the vein will gradually become thicker and harder and the diameter expands [19,20]. It has been reported that if the vein was grafted back to the venous environment 2 weeks after CABG, there are restorative changes in the vein [21]. In this dynamic and reversible process, the role of endothelial cells needs to be defined.
The main reason for the gradual hardening of the vein wall is the remodeling of matrix proteins [22]. These proteins are in the signal pathways identified in this study, such as Focal adhesion, Cell adhesion molecules, ECM-receptor interaction in KEGG enrichment and integrin binding, glycosaminoglycan binding, and extracellular matrix structural component in GO enrichment. In the Focal adhesion pathway, 20 DEGs were enriched; among them, 7 were upregulated and 13 were downregulated. In the Cell adhesion molecules pathway, 11 DEGs were enriched; among them, 3 were upregulated and 8 were downregulated. There were 11 common DEGs in the Focal adhesion and ECM-receptor interaction pathways; therefore, all DEGs in the ECM-receptor interaction pathway are also DEGs in the Focal adhesion pathway. On the other hand, there was only 1 common DEG (integrin α4, ENSG00000115232) between the Focal adhesion and Cell adhesion molecules pathways.
DEPTOR is a component of mTORC1 and mTORC2, and mTOR regulates cell proliferation mainly through downstream PI3K-Akt pathways [23]. In our work, these pathways were enriched with 19 DEGs; among them, 5 were upregulated and 14 were downregulated after DEPTOR overexpression. In addition, DEGs were identified in GO terms RNA polymerase II regulatory region sequence-specific DNA binding, suggesting that it has a regulatory role in gene expression. Cytokine-cytokine receptor interaction, TGF-beta signaling pathway, and NF-kappa B signaling pathway are also significant signaling pathways that are associated with inflammatory response [24,25]. We found that 3 upregulated and 14 downregulated DEGs were enriched in the Cytokine-cytokine receptor interaction pathway; 1 upregulated and 5 downregulated DEGs were enriched in the TGF-beta signaling pathway; and 3 upregulated and 4 downregulated DEGs were enriched in the NF-kappa B signaling pathway. The Cytokine-cytokine receptor interaction and GF-beta signaling pathways had the same 4 DEGs: ENSG00000139269 (inhibin β E chain), ENSG00000101144 (bone morphogenetic protein 7), ENSG00000122641 (inhibin β A chain), and ENSG00000163083 (inhibin β B chain). These DEGs were all downregulated. The Cytokine-cytokine receptor interaction and NF-kappa B signaling pathways also had the same 4 DEGs ENSG00000101017 (CD40, upregulated) and ENSG00000125538 (interleukin-1 β), ENSG00000169429 (Interleukin-8) and ENSG00000111321 (LTBR, all downregulated). Furthermore, more than half of the DEGs in the TGF-beta signaling pathway and NF-kappa B signaling pathway were also found in the Cytokine-cytokine receptor interaction pathway. The enrichment of DEGs in these pathways suggest that DEPTOR may have a regulatory role in inflammatory response and release of cytokines in hsVEC.
It is worth mentioning that GO Term and KEGG signaling pathways of adipogenesis are not enriched, although DEPTOR is related to fat synthesis in hsVECs, suggesting that overexpression of DEPTOR does not change the expression patterns of genes related to adipogenesis.
Since more than 200 DEGs have been identified, these genes would provide candidates for functional analysis and lay a foundation for functional molecular and biological analysis.
Conclusions
Differentially expressed genes induced by overexpressing DEPTOR in hsVECs are involved in cell proliferation and differentiation, intercellular junction, and extracellular matrix receptor. These genes should be further explored for their biological roles and may provide molecular cues to improve venous permeability through manipulation of DEPTOR and related mTOR pathways.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (grants no. 81760078, 81260055, 30960380), the Youth Fund for Science and Technology, Jiangxi Department of Education (grant no. 2018.1-2020.12), and the Fund for Science and Technology, Jiangxi Health and Family Planning Commission (grant no. 20873999)
Conflicts of interests
None.
References
- 1.Hillis LD, Smith PK, Anderson JL, et al. 2011 ACCF/AHA Guideline for Coronary Artery Bypass Graft Surgery: executive summary: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation. 2011;124:2610–42. doi: 10.1161/CIR.0b013e31823b5fee. [DOI] [PubMed] [Google Scholar]
- 2.Bhasin M, Huang Z, Pradhan-Nabzdyk L, et al. Temporal network based analysis of cell specific vein graft transcriptome defines key pathways and hub genes in implantation injury. PLoS One. 2012;7:e39123. doi: 10.1371/journal.pone.0039123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Head SJ, Borgermann J, Osnabrugge RL, et al. Coronary artery bypass grafting: Part 2 – optimizing outcomes and future prospects. Eur Heart J. 2013;34:2873–86. doi: 10.1093/eurheartj/eht284. [DOI] [PubMed] [Google Scholar]
- 4.Sousa JE, Costa MA, Abizaid A, et al. Lack of neointimal proliferation after implantation of sirolimus-coated stents in human coronary arteries: A quantitative coronary angiography and three-dimensional intravascular ultrasound study. Circulation. 2001;103:192–95. doi: 10.1161/01.cir.103.2.192. [DOI] [PubMed] [Google Scholar]
- 5.Morice MC, Serruys PW, Sousa JE, et al. RAVEL Study Group. Randomized Study with the Sirolimus-Coated Bx Velocity Balloon-Expandable Stent in the Treatment of Patients with de Novo Native Coronary Artery Lesions: A randomized comparison of a sirolimus-eluting stent with a standard stent for coronary revascularization. N Engl J Med. 2002;346:1773–80. doi: 10.1056/NEJMoa012843. [DOI] [PubMed] [Google Scholar]
- 6.Hausleiter J, Kastrati A, Mehilli J, et al. OSIRIS Investigators. Randomized, double-blind, placebo-controlled trial of oral sirolimus for restenosis prevention in patients with in-stent restenosis: The Oral Sirolimus to Inhibit Recurrent In-stent Stenosis (OSIRIS) trial. Circulation. 2004;110:790–95. doi: 10.1161/01.CIR.0000138935.17503.35. [DOI] [PubMed] [Google Scholar]
- 7.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–86. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caron A, Baraboi ED, Laplante M, Richard D. DEP domain-containing mTOR-interacting protein in the rat brain: Distribution of expression and potential implication. J Comp Neurol. 2015;523:93–107. doi: 10.1002/cne.23668. [DOI] [PubMed] [Google Scholar]
- 11.Laplante M, Horvat S, Festuccia WT, et al. DEPTOR cell-autonomously promotes adipogenesis, and its expression is associated with obesity. Cell Metab. 2012;16:202–12. doi: 10.1016/j.cmet.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Liu N. [In vitro culture of human saphenous vein endothelial cells]. Chinese Journal of Cell Biology. 2005;27:474–76. [in Chinese] [Google Scholar]
- 13.Szabo L, Morey R, Palpant NJ, et al. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, Wang X, Bowers JE, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–54. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westfall PH, Young SS. p Value adjustments for multiple tests in multivariate binomial models. Journal of the American Statistical Association. 1989;84:780–86. [Google Scholar]
- 16.Aickin M, Gensler H. Adjusting for multiple testing when reporting research results: the Bonferroni vs. Holm methods. Am J Public Health. 1996;86:726–28. doi: 10.2105/ajph.86.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoav B, Yosef H. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- 18.Xie C, Mao X, Huang J, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conte MS. Molecular engineering of vein bypass grafts. J Vasc Surg. 2007;45(Suppl A):A74–81. doi: 10.1016/j.jvs.2007.02.031. [DOI] [PubMed] [Google Scholar]
- 20.Wong AP, Nili N, Jackson ZS, et al. Expansive remodeling in venous bypass grafts: novel implications for vein graft disease. Atherosclerosis. 2008;196:580–89. doi: 10.1016/j.atherosclerosis.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 21.Min SK, Kenagy RD, Clowes AW. Induction of vascular atrophy as a novel approach to treating restenosis. A review. J Vasc Surg. 2008;47:662–70. doi: 10.1016/j.jvs.2007.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owens CD, Wake N, Jacot JG, et al. Early biomechanical changes in lower extremity vein grafts – distinct temporal phases of remodeling and wall stiffness. J Vasc Surg. 2006;44:740–46. doi: 10.1016/j.jvs.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Nemazanyy I, Espeillac C, Pende M, Panasyuk G. Role of PI3K, mTOR and Akt2 signalling in hepatic tumorigenesis via the control of PKM2 expression. Biochem Soc Trans. 2013;41:917–22. doi: 10.1042/BST20130034. [DOI] [PubMed] [Google Scholar]
- 24.Ashcroft GS. Bidirectional regulation of macrophage function by TGF-beta. Microbes Infect. 1999;1:1275–82. doi: 10.1016/s1286-4579(99)00257-9. [DOI] [PubMed] [Google Scholar]
- 25.Hargrave BY, Tiangco DA, Lattanzio FA, Beebe SJ. Cocaine, not morphine, causes the generation of reactive oxygen species and activation of NF-kappaB in transiently cotransfected heart cells. Cardiovasc Toxicol. 2003;3:141–51. doi: 10.1385/ct:3:2:141. [DOI] [PubMed] [Google Scholar]