Abstract
Summary
Large scale technologies produce massive amounts of experimental data that need to be investigated. To improve their biological interpretation we have developed ClueGO, a Cytoscape App that selects representative Gene Onology terms and pathways for one or multiple lists of genes/proteins and visualizes them into functionally organized networks. Because of its reliability, userfriendliness and support of many species ClueGO gained a large community of users. To further allow scientists programmatic access to ClueGO with R, Python, JavaScript etc., we implemented the cyREST API into ClueGO. In this article we describe this novel, complementary way of accessing ClueGO via REST, and provide R and Phyton examples to demonstrate how ClueGO workflows can be integrated into bioinformatic analysis pipelines.
Availability and implementation
ClueGO is available in the Cytoscape App Store (http://apps.cytoscape.org/apps/cluego).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
High-throughput technologies produce large amounts of experimental data that need to be investigated to gain insights into biological processes. Systemic approaches for data integration, analysis and visualization were developed to reflect not only individual biological components, but also their interactions in pathways and networks. Software tools that perform such type of analyses in an automatic way are more and more needed.
Cytoscape (Shannon et al., 2003) is a major computational platform to visualize and analyze networks. Cytoscape Automation (https://github.com/cytoscape/cytoscape-automation) enables scientific workflows written in many languages and scales Cytoscape to large datasets and pipelines. The Cytoscape programmatic interface for this is CyREST (Ono et al., 2015).
We have contributed to the Cytoscape App collection (Saito et al., 2012) with ClueGO (Bindea et al., 2009) and CluePedia (Bindea et al., 2013) apps that are broadly used by the scientific community to enhance the interpretation of biological data (Mlecnik et al., 2018). Within ClueGO, representative gene ontology (GO) terms (Ashburner et al., 2000) as well as KEGG (Kanehisa et al., 2002), WikiPathways (Pico et al., 2008) and Reactome (Croft et al., 2011) pathways are integrated into a functionally organized network. Furthermore, ClueGO can compare the biological role of several lists of genes/proteins.
To perform multiple analyses in complex workflows can be time consuming and prone to errors. We have thus enabled the cyREST Application Programming Interface (API) in ClueGO, to allow scientists programmatic access to functional analyses. We describe here this new functionality of the ClueGO App.
2 ClueGO functional analysis via cyREST
The Cytoscape App Manager (http://cytoscape.org/) allows the automatic download of the latest version of ClueGO from the App Store (Lotia et al., 2013). The yFiles Layout Algorithms App should be installed as well. ClueGO is written in Java programing language implementing the OSGi interface of Cytoscape. Starting with version 2.5.0 ClueGO implements the cyREST core plugin API and provides programmatic access to its functionality (Fig. 1A). ClueGO features REST enabled can be explored in the cyREST API Swagger (Fig. 1B), accessible via the Cytoscape menu, Help → Automation → CyREST API.
Fig. 1.
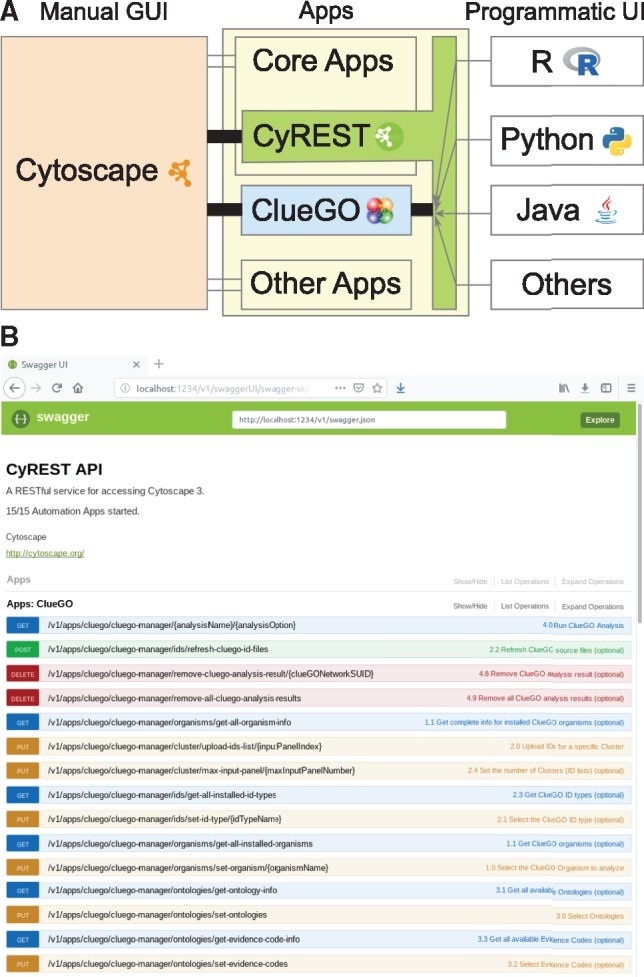
(A) ClueGO can be accessed through the Cytoscape Graphical User Interface (GUI) and the cyREST Programmatic User Interface (UI). (B) ClueGO functions REST enabled in the CyREST API Swagger
ClueGO can be hence accessed with both the graphical user interface of Cytoscape as well as programmatical through cyREST (Fig. 1 and Supplementary Material). These two ways of performing analyses complement each other and answer to different types of user requests.
2.1 ClueGO analysis steps
The four main steps of a typical ClueGO analysis and additional optional REST enabled features are shown in Supplementary Table S1. The ClueGO-REST enabled functions are accessed by a URL that is built up by the host address (e.g. localhost), the port (e.g. 1234) and the feature requested by the user. The HTTP request return types are then encoded as JSON, tab delimited text or binary data.
The selection of an organism is the first step. Human and mouse data sources are included by default in ClueGO and more than 200 other organisms are available for download. The organism to analyze has to be set e.g. human:/v1/apps/cluego/cluego-manager/organisms/set-organism/‘Homo Sapiens’.
The second step requires the upload of one or several lists of genes/proteins to analyze. ClueGO automatically recognizes multiple identifier types based on information from NCBI (NCBI Resource Coordinators, 2018), UniProtKB (The UniProt Consortium, 2017) and Ensembl (Aken et al., 2017) databases. Different colors and shapes are automatically attributed to the clusters, to visualize them on the network.
In the third step representative GO terms and pathways are selected using predefined filters based on the number of associated genes found from the uploaded list, their percentage from the total number of genes of the term or the GO tree level. Additionally, GO terms can be selected based on particular evidence codes of the gene-term associations. The significance of the pathways and their similarity in terms of associated genes are automatically mapped on the network and illustrated in different interchangeable visual styles.
After running the enrichment analysis through the last Step 4 the functionally grouped network with the terms connected and grouped based on kappa score is created (Supplementary Fig. S1). The kappa score is calculated by taking into account how many genes are shared among two terms and is also used to define the functional groups of terms and pathways. The results of the statistical analysis, the network as well as other graphical representations of the results can be downloaded through cyREST functions. The analysis and visualization can be customized. Large networks can be refined by applying the fusion of similar terms or by visualizing only significant pathways (optional steps).
Debug messages appear during the workflow if mandatory analysis steps are skipped, if the term selection is too restrictive/permissive or when other exceptions occur.
2.2 Use cases
R and Python ClueGO analysis examples with one or two lists of genes are provided as Supplementary Material. B and NK cell genes (Critchley-Thorne et al., 2007; Edgar et al., 2002) were analysed, and results are illustrated as a network of pathways showing either functional groups (Supplementary Fig. S1) or the origin of the genes in the two lists (Supplementary Fig. S2). All available ClueGO-REST endpoints are illustrated, including optional features.
3 Summary
In this article, we describe the REST enabled ClueGO functionality and show how scientists can integrate ClueGO with other Cytoscape apps and non-Cytoscape libraries to perform functional analyses in a programmatic way. Detailed information on ClueGO can be found at http://www.ici.upmc.fr/cluego.
Supplementary Material
Acknowledgements
The authors thank the Cytoscape team for their great work and support.
Funding
This work was supported by grants from INSERM (‘Integrative Cancer Immunology’, ‘Cancer et Environnement’, ‘Heterogeneity of Colorectal and Liver Cancer’ (HETCOLI), the National Cancer Institute of France (INCa), Canceropole Ile de France, MedImmune, AstraZeneca, the Transcan ERAnet European Project [Grant number TRANS201401218], La Ligue contre le Cancer, the Qatar National Research Fund (QNRF), Cancer research for personalized medicine (CARPEM), Paris Alliance of Cancer Research Institutes (PACRI) and LabEx Immuno-Oncology.
Conflict of Interest: none declared.
References
- Aken B.L. et al. (2017) Ensembl 2017. Nucleic Acids Res., 45, D635–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. et al. (2000) Gene ontology: tool for the unification of biology The Gene Ontology Consortium. Nat. Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G. et al. (2009) ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25, 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G. et al. (2013) CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics, 29, 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley-Thorne R.J. et al. (2007) Down-regulation of the interferon signaling pathway in T lymphocytes from patients with metastatic melanoma. PLoS Med., 4, e176.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D. et al. (2011) Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res., 39, D691–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. et al. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res., 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. et al. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotia S. et al. (2013) Cytoscape app store. Bioinformatics, 29, 1350–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlecnik B. et al. (2018) Comprehensive functional analysis of large lists of genes and proteins. J. Proteomics, 171, 2–10. [DOI] [PubMed] [Google Scholar]
- NCBI Resource Coordinators (2018) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 46, D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K. et al. (2015) CyREST: turbocharging Cytoscape access for external tools via a RESTful API. F1000Res., 4, 478.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pico A. et al. (2008) WikiPathways: pathway editing for the people. PLoS Biol., 6, e184.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R. et al. (2012) A travel guide to Cytoscape plugins. Nat. Methods, 9, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res., 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res., 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


