Abstract
Bacterial vaginosis (BV) is the most common cause of vaginal discharge. It is associated with an increased risk of preterm delivery, pelvic inflammatory disease, and an increased risk of acquisition of sexually transmitted infections including human immunodeficiency virus (HIV). The epidemiology of BV supports sexual transmission. However, its etiology remains unknown. At the center of the debate is whether BV is caused by a primary pathogen or a polymicrobial consortium of microorganisms that are sexually transmitted. We previously published a conceptual model hypothesizing that BV is initiated by sexual transmission of Gardnerella vaginalis. Critics of this model have iterated that G. vaginalis is found in virginal women and in sexually active women with a normal vaginal microbiota. In addition, colonization does not always lead to BV. However, recent advances in BV pathogenesis research have determined the existence of 13 different species within the genus Gardnerella. It may be that healthy women are colonized by nonpathogenic Gardnerella species, whereas virulent strains are involved in BV development. Based on our results from a recent prospective study, in addition to an extensive literature review, we present an updated conceptual model for the pathogenesis of BV that centers on the roles of virulent strains of G. vaginalis, as well as Prevotella bivia and Atopobium vaginae.
Keywords: Bacterial vaginosis, biofilm, Gardnerella vaginalis, Prevotella bivia, Atopobium vaginae
This article provides an updated conceptual model for the pathogenesis of bacterial vaginosis, centering on the roles of virulent strains of Gardnerella vaginalis, as well as Prevotella bivia and Atopobium vaginae.
Bacterial vaginosis (BV), the most common cause of vaginal discharge [1], is a vaginal dysbiosis characterized by loss of lactic acid–producing lactobacilli and proliferation of facultative and strict anaerobes [2]. It is associated with multiple adverse outcomes, including an increased risk of preterm birth, as well as acquisition of human immunodeficiency virus (HIV) and other sexually transmitted pathogens [2]. BV is diagnosed clinically, using the Amsel criteria, and microbiologically, using the Nugent score [2]. Epidemiological data strongly suggest that BV is a sexually transmitted infection (STI) with an incubation period of around 4 days, similar to that of other bacterial causes of STIs, such as Neisseria gonorrhoeae [3]. The most significant risk factor for incident BV is a new sex partner [4], whereas that for recurrent BV is a regular sex partner [5].
Despite >60 years of research, the etiology of BV remains unknown. Gardnerella vaginalis, present in 95%–100% of BV cases [6], was originally thought to be the primary BV pathogen [7]. In vitro, it is more virulent (ie, it has a greater ability to harm a host) than other BV-associated bacteria, adhering in larger aggregates to vaginal epithelial cells and exhibiting greater cytotoxic activity through production of a pore-forming toxin (vaginolysin) [8–10]. However, G. vaginalis has been found in virginal women [11] and in sexually active women with a normal vaginal microbiota [12]. In addition, colonization with G. vaginalis does not always lead to BV [13], suggesting that G. vaginalis alone may be necessary but not sufficient for BV development.
Recent advances in BV pathogenesis research have suggested distinct roles for G. vaginalis subgroups [14]. Earlier genomic studies of G. vaginalis revealed that it consists of 4 nonrecombining groups/clades of organisms with distinct gene pools and genomic properties that may confer distinct ecological or pathological properties [15]. The presence of multiple G. vaginalis clades has since been found to have a positive association with BV (ie, clades 1 and 2), whereas the presence of a single clade (ie, clade 4) has been negatively linked with BV [16]. More recently, Vaneechoutte et al analyzed 81 full-genome sequences of G. vaginalis by performing digital DNA-DNA hybridization and measuring the average nucleotide identity and demonstrated the existence of at least 13 different species within the genus Gardnerella [17]. While these Gardnerella species may be closely related genetically, only a few may be involved in disease (ie, BV) [17]. These new genomic data put into perspective previous criticisms regarding G. vaginalis colonization in sexually active women with normal vaginal microbiota as evidence against its central role in BV etiology [13]. It may be that healthy women are colonized by Gardnerella species with low virulence potential and that other virulent strains are involved in BV development. Future research should include study of potential sexual transmission of the different Gardnerella strains.
Recent data now suggest a potentially important synergistic relationship between G. vaginalis, Prevotella bivia, and Atopobium vaginae in BV pathogenesis [18, 19]. Gilbert et al recently showed in a mouse model that coinfection with G. vaginalis and P. bivia recapitulates several features of BV [19]. In a prospective study of women who have sex with women, we found that the mean relative abundance of P. bivia and G. vaginalis became sequentially higher 4 days (for P. bivia) and 3 days (for G. vaginalis) prior to the development of incident BV, compared with findings for women who maintained a normal vaginal microbiota; the mean relative abundance of A. vaginae became significantly higher on the day of incident BV in this study [18]. P. bivia has been found in high concentrations in women with BV [20] and in women with preterm birth [21]. A. vaginae is highly specific for BV and rarely occurs in the absence of G. vaginalis [22].
To put these new data into context, we performed an updated literature review on BV pathogenesis research, focusing on more-recent data on G. vaginalis [8, 14, 16, 17, 23, 24], as well as interactions among G. vaginalis, P. bivia, and A. vaginae [18, 19, 25–27]. We revised our prior hypothetical model [28], which mainly centered on G. vaginalis, and present an updated conceptual model (Figure 1) that includes these additional BV-associated bacteria as potential key pathogens in the development of BV.
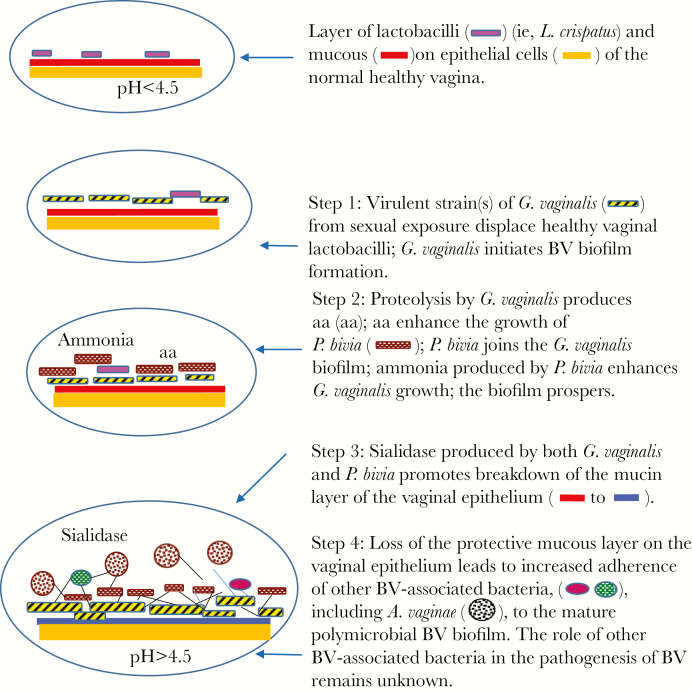
Figure 1.
Updated conceptual model for the pathogenesis of bacterial vaginosis (BV). This model is a schematic representation and is not to scale. aa, amino acids; A. vaginae, Atopobium vaginae; G. vaginalis, Gardnerella vaginalis; L. crispatus, Lactobacillus crispatus; P. bivia, Prevotella bivia.
PROPOSED STEPS IN BV DEVELOPMENT
Sexual Transmission of Virulent Strain(s) of G. vaginalis Displace Vaginal Lactobacilli and Initiate BV Biofilm Formation on the Vaginal Epithelium
A notable feature of BV is the appearance of a polymicrobial biofilm on vaginal epithelial cells. Presence of this biofilm creates a favorable environment for anaerobic bacteria, owing to the presence of an oxygen gradient within the biofilm [29]. The BV biofilm has been found to contain abundant G. vaginalis, fewer A. vaginae, Lactobacillus species, and other bacterial species [30]. Desquamation of vaginal epithelial cells coated with BV biofilm results in clue cells, which can be seen on wet-mount analysis of vaginal fluid specimens. Polymicrobial biofilms, such as the BV biofilm, incorporate secondary bacteria after an initial colonizer species adheres to the surface; a synergetic relationship between these species allows the bacterial biofilm to prosper and mature [31].
Initial adhesion to the vaginal epithelium is a crucial step in BV development [30] and the first step in BV biofilm formation [32]. We have previously proposed that BV development is triggered by sexual transmission of G. vaginalis, which displaces healthy vaginal lactobacilli, such as L. crispatus, and initiates BV biofilm formation on the vaginal epithelium [28]. As a microaerophilic and facultative anaerobe, G. vaginalis, unlike other strict BV-associated bacteria, can tolerate the high redox potential of a lactobacillus-dominated vaginal microbiota [28]. Similar to facultative anaerobes involved in dental plaque biofilms [33], virulent strain(s) of G. vaginalis, perhaps at a certain threshold [34], may lower the redox potential in the vaginal microbiota, leading to a marked decrease in lactobacilli and an increase in other strict anaerobic BV-associated bacteria (eg, P. bivia and A. vaginae) [28]. These other BV-associated bacteria, acquired from maternal and environmental sources, may normally be present in very low concentrations [35]. Non-BV strains of G. vaginalis are less likely to displace vaginal lactobacilli from monolayers of HeLa cells in vitro than strains of G. vaginalis from women with BV [23].
During lactobacillus displacement, G. vaginalis adheres to vaginal epithelial cells in large numbers and triggers BV biofilm growth [9, 23]. In a study by Patterson et al, strains of G. vaginalis obtained from women with BV were found to form significantly thicker biofilms on tissue-culture–treated polystyrene, compared with other common BV-associated bacteria (P. bivia, A. vaginae, Mobiluncus mulieris, Fusobacterium nucleatum, Veillonella species, Peptostreptococcus species, and Peptoniphilus species) [9]. Correspondingly, in a study of 30 BV-associated bacteria, using an in vitro biofilm formation model, Alves et al also found that BV strains of G. vaginalis had the greatest propensity to form biofilms [8]. Interestingly, in this study, many of the other BV-associated bacteria also had a tendency to grow as biofilms, particularly Mycoplasma hominis, Staphylococcus hominis, Brevibacterium mcbrellneri, and Enterococcus faecalis. However, when tested for initial adhesion to HeLa cells, these species had a significantly lower ability to adhere than G. vaginalis. This suggests that, when in contact with human cell lines, smaller numbers of BV-associated bacteria are not sufficient to induce biofilm formation.
G. vaginalis biofilms tolerate H2O2 and lactic acid (substances produced by vaginal lactobacilli that reduce the vaginal pH to <4.5 and prevent colonization by pathogenic anaerobes) better than planktonic (free-floating) cells [36]. This protective mode of growth allows bacteria embedded in the BV biofilm to survive in harsh environments. Castro et al have recently shown that G. vaginalis biofilm cells alter their gene expression profiles significantly, which may contribute to antimicrobial resistance and BV biofilm persistence, promoting the recurring and chronic nature of BV [24].
Synergy Between Virulent Strain(s) of G. vaginalis and P. bivia Occurs on the Vaginal Epithelium
After virulent strain(s) of G. vaginalis displace vaginal lactobacilli, adhere to the vaginal epithelium in high numbers, and initiate BV biofilm formation, we propose that key interactions between G. vaginalis and P. bivia promote the next step in BV development. It is during this time that P. bivia joins G. vaginalis in the lower layers of the BV biofilm. P. bivia is a common constituent of the human vaginal microbiota during BV [37], as are other Prevotella species, and has been found in infected amniotic fluid specimens, as well as in uterine and placental tissue specimens from women undergoing preterm birth and other pregnancy complications [38]. G. vaginalis and P. bivia have a well-established symbiotic relationship in vitro [39], and interactions between these bacteria may be the next key step in BV pathogenesis. Proteolysis by G. vaginalis produces amino acids [40] that enhance P. bivia growth [39], and ammonia produced by P. bivia stimulates G. vaginalis growth in vitro [39]. It has also been shown that P. bivia enhances biofilm formation initiated by G. vaginalis [26]. If this mutualistic relationship exists in vivo, it could account for the presence of both of these bacteria in high concentrations during BV [27].
While no in vivo studies have examined the incorporation of P. bivia in the BV biofilm along with G. vaginalis, we have found that P. bivia coaggregates with G. vaginalis and can incorporate multispecies BV biofilms in vitro [26]. Future research should explore whether P. bivia incorporates into the BV biofilm in vivo. To determine the presence of P. bivia in the BV biofilm in vivo, a specific probe for P. bivia, such as a peptide nucleic acid probe, would need to be developed.
Vaginal Sialidase, Produced by BV-Associated Bacteria, Including G. vaginalis and P. bivia, Promotes Breakdown of the Mucin Layer on the Vaginal Epithelium
We next propose that vaginal sialidase, produced by several BV-associated bacteria, promotes breakdown of the protective mucus layer on the vaginal epithelium and leads to enhanced susceptibility to ascending infection in the female genital tract. Vaginal sialidase is highly correlated with BV [41] and preterm birth [42] and has been implicated in multiple aspects of host-pathogen interactions, including mucosal barrier degradation (which likely contributes to the characteristically thin consistency of vaginal fluid during BV), bacterial attachment, and release of carbon sources to facilitate bacterial growth [19]. Sialidase activity also has immunomodulatory consequences for receptors on host cells.
G. vaginalis and P. bivia are 2 BV-associated bacteria that produce sialidase in vivo [41]. Other Prevotella species (Prevotella oralis and Prevotella loescheii) and B. fragilis also produced sialidase in this study, but in much smaller quantities. In contrast, other BV-associated bacteria, including Mobiluncus species, Peptostreptococcus species, and M. hominis, did not produce detectable sialidase.
Gilbert et al have recently shown in their mouse model that both G. vaginalis and P. bivia produce measurable levels of vaginal sialidase, with G. vaginalis producing higher levels than P. bivia [19]. In this model, G. vaginalis but not P. bivia also induced vaginal epithelial cell exfoliation. Furthermore, G. vaginalis enhanced the invasive potential of P. bivia and facilitated its ascension into mouse uteri. Interestingly, neither G. vaginalis nor P. bivia induced a purulent inflammatory response. The authors concluded that this study provides strong evidence that G. vaginalis and P. bivia are both direct contributors to the pathogenesis of BV, as well as its associated symptoms and outcomes [19]. However, they did note several limitations to their study, which included the use of single strains of G. vaginalis and P. bivia, as well as the inherent limitations of murine models in BV pathogenesis research (ie, mice do not have dominant Lactobacillus microbiomes in the vagina as do many women; thus, the mouse model may not accurately reflect all aspects of human vaginal physiology that accompany a shift away from a lactobacillus-dominant vaginal microbiota). Nevertheless, the findings by Gilbert et al provide an important framework for advancing BV pathogenesis research, particularly that involving G. vaginalis and P. bivia, as well as other BV-associated bacteria.
Loss of the Protective Mucous Layer on the Vaginal Epithelium Leads to Increased Adherence of Other BV-Associated Bacteria, Including A. vaginae, to the Mature Polymicrobial BV Biofilm
We next propose that loss of the protective mucous layer on the vaginal epithelium leads to increased adherence of other BV-associated bacteria, including A. vaginae [43], and to the formation of a mature, polymicrobial BV biofilm. There is considerable evidence supporting the involvement of A. vaginae in the pathogenesis of BV. As previously mentioned, A. vaginae is highly specific for BV and rarely occurs in the absence of G. vaginalis [22]. A. vaginae has been found in BV biofilms in vivo, although in smaller numbers than G. vaginalis [30]. Similar to G. vaginalis, A. vaginae has also been found in a dispersed form that is nonadherent [25]. In a study of 120 women participating in a clinical trial in Rwanda, A. vaginae and G. vaginalis were visualized by fluorescence in situ hybridization in 54.1% and 82.0% of vaginal specimens containing BV biofilms, respectively [25]. A. vaginae was accompanied by G. vaginalis in 99.5% of the samples in this study. The odds of having a Nugent score >4 were increased for vaginal samples with dispersed G. vaginalis and/or A. vaginae present (odds ratio [OR], 4.5; 95% confidence interval [CI], 2–10.3). The odds of having a high Nugent score were even higher when a combination of adherent G. vaginalis and dispersed A. vaginae were present (OR, 75.6; 95% CI, 13.3–429.5) and highest when both bacteria were found in the BV biofilm (OR, 119; 95% CI, 39.9–360.8). Only rarely (ie, in only 2 of the vaginal samples in the Rwanda study [25]) has A. vaginae been identified in BV biofilms in the absence of G. vaginalis. Women with G. vaginalis and A. vaginae have higher rates of recurrent BV than women with G. vaginalis alone [22]. In addition, it has been demonstrated that a combination of a G. vaginalis DNA level ≥109 copies/mL and an A. vaginae DNA level ≥108 copies/mL is the best diagnostic definition of BV [44].
A. vaginae has been found to stimulate a stronger immune response from vaginal epithelial cells than G. vaginalis, leading to localized cytokine and β-defensin production [45]. Thus, it may be a potent component of the host response to BV. This host immune response may contribute to the symptoms (ie, vaginal discharge and odor) of BV, as well as to the adverse outcomes (ie, preterm birth and pelvic inflammatory disease), as increased vaginal levels of inflammatory cytokines and neutrophils are predictive of both [46]. Vaginal odor in particular is also linked to increase levels of vaginal biogenic amines, including putrescine, cadaverine, and trimethylamine, which are produced by multiple BV-associated bacteria and may mitigate the acidic barrier that favors vaginal lactobacilli [47].
The presence of A. vaginae in the BV biofilm may also have an impact on BV treatment outcomes [25]. A. vaginae is highly resistant to metronidazole but susceptible to clindamycin (3 of 3 clinical isolates tested in one study) [48]. In a study of 0.75% topical metronidazole gel applied by women once daily for 5 days, a high pretreatment concentration of A. vaginae was associated with partial or complete BV treatment failure at 4 weeks after treatment [49]. However, this study was small, and quantitative polymerase chain reaction analysis of other BV-associated bacteria before and after treatment was not performed. If A. vaginae is integral to the pathogenesis of BV, one would expect superior cure rates by using clindamycin as compared to metronidazole. However, metronidazole and clindamycin were found to have similar short-term efficacy in a review of BV clinical trials [50]. It is important to note that the quality of trials included in that review varied widely, which could have biased comparisons of efficacy between drugs. Although the authors included a sensitivity analysis in their study, they acknowledged that making direct comparisons of treatment efficacy between metronidazole and clindamycin was difficult. In addition, A. vaginae is not present in all women with recurrent BV [22]. Thus, the role of A. vaginae in BV pathogenesis remains controversial and should be further investigated.
Discussion
Understanding the etiology of BV is essential for improvements in diagnosis, treatment, and prevention of this extremely common clinical condition. We have proposed an updated conceptual model highlighting the potential key roles of virulent strains of G. vaginalis, P. bivia, and A. vaginae in BV pathogenesis. We propose that this model of BV pathogenesis is the same regardless of how BV is diagnosed (eg, by the Amsel criteria, Nugent score, or molecular diagnostic assay) or whether patients perceive themselves to be symptomatic (of note, patients’ perception of their vaginal symptoms varies greatly and does not necessarily correlate with signs of BV [3]). Nevertheless, it is essential to note that other BV-associated bacteria may play a role in BV pathogenesis but remain less well characterized and are not included in our current model; this should be an active area of research moving forward. Along these lines, the ecological interactions between G. vaginalis and 15 other BV-associated bacteria were recently analyzed by Castro et al, using a dual-species biofilm model [26]. Interestingly, this study revealed distinct biofilm structures between each bacterial consortium, leading to at least 3 unique dual-species biofilm morphotypes. A. vaginae, Corynebacterium tuscaniense, Mobiluncus mulieris, P. bivia, and Streptococcus anginosus were the bacterial species that coaggregated the most with G. vaginalis in vitro (coaggregation is a virulence factor and a common mechanism for survival of bacteria in nature). However, surprisingly, transcriptomic findings indicated that Enterococcus faecalis and Actinomyces neuii had the highest impact on enhancing key G. vaginalis genes associated with BV development, namely vly (which encodes the pore-forming toxin vaginolysin), sld (which encodes sialidase), and HMPREF0424_0821 (which is thought to be associated with biofilm formation metabolism). Interestingly, P. bivia also enhanced vly, while A. vaginae enhanced HMPREF0424_0821. The authors suggested that perhaps not all BV-associated bacteria contribute to the enhancement of BV pathogenesis by influencing G. vaginalis virulence. Nevertheless, these results suggest that “social networking” between G. vaginalis and an array of BV-associated bacteria can occur. Additional research focusing on these complex interactions between bacteria during BV is needed.
If G. vaginalis, P. bivia, and A. vaginae are found to be central to the pathogenesis of BV, future studies should focus on the design of molecular diagnostic assays targeting all 3 of these bacteria. Additional research should also focus on interventions that modify or block the synergistic relationship between these bacteria, particularly the synergistic relationship between G. vaginalis and P. bivia. In addition, clinical trials evaluating the treatment of P. bivia or A. vaginae colonization in women without BV should be conducted. Of note, we are currently conducting a clinical trial of treatment of G. vaginalis colonization with amoxicillin among women without BV (clinical trials registration NCT03211156 [J. R. S., principal investigator]), with results forthcoming.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant K23AI106957 to C. A. M.).
Potential conflicts of interest. C. A. M. is a consultant for Lupin Pharmaceuticals, BioFire Diagnostics, and Cepheid and has also received honoraria from Roche Diagnostics. J. R. S. has been a consultant for and received research support from Hologic/GenProbe, BD Diagnostics, Cepheid, Quidel, Symbiomix, StarPharma, Lupin Pharmaceuticals, and Toltec. All other authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Koumans EH, Sternberg M, Bruce C, et al. The prevalence of bacterial vaginosis in the United States, 2001–2004; associations with symptoms, sexual behaviors, and reproductive health. Sex Transm Dis 2007; 34:864–9. [DOI] [PubMed] [Google Scholar]
- 2. Hillier SL, Marrazzo J, Holmes KK. Bacterial vaginosis. In: Holmes KK, Sparling PF, Mardh PA, et al. , eds. Sexually transmitted diseases. 4th ed. New York: McGraw-Hill, 2008:737–68. [Google Scholar]
- 3. Muzny CA, Lensing SY, Aaron KJ, Schwebke JR. Incubation period and risk factors support sexual transmission of bacterial vaginosis in women who have sex with women. Sex Transm Infect 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vodstrcil LA, Walker SM, Hocking JS, et al. Incident bacterial vaginosis (BV) in women who have sex with women is associated with behaviors that suggest sexual transmission of BV. Clin Infect Dis 2015; 60:1042–53. [DOI] [PubMed] [Google Scholar]
- 5. Bradshaw CS, Morton AN, Hocking J, et al. High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence. J Infect Dis 2006; 193:1478–86. [DOI] [PubMed] [Google Scholar]
- 6. Hill GB. The microbiology of bacterial vaginosis. Am J Obstet Gynecol 1993; 169:450–4. [DOI] [PubMed] [Google Scholar]
- 7. Gardner HL, Dukes CD. Haemophilus vaginalis vaginitis: a newly defined specific infection previously classified non-specific vaginitis. Am J Obstet Gynecol 1955; 69:962–76. [PubMed] [Google Scholar]
- 8. Alves P, Castro J, Sousa C, Cereija TB, Cerca N. Gardnerella vaginalis outcompetes 29 other bacterial species isolated from patients with bacterial vaginosis, using in an in vitro biofilm formation model. J Infect Dis 2014; 210:593–6. [DOI] [PubMed] [Google Scholar]
- 9. Patterson JL, Stull-Lane A, Girerd PH, Jefferson KK. Analysis of adherence, biofilm formation and cytotoxicity suggests a greater virulence potential of Gardnerella vaginalis relative to other bacterial-vaginosis-associated anaerobes. Microbiology 2010; 156:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gelber SE, Aguilar JL, Lewis KL, Ratner AJ. Functional and phylogenetic characterization of Vaginolysin, the human-specific cytolysin from Gardnerella vaginalis. J Bacteriol 2008; 190:3896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bump RC, Buesching WJ 3rd. Bacterial vaginosis in virginal and sexually active adolescent females: evidence against exclusive sexual transmission. Am J Obstet Gynecol 1988; 158:935–9. [DOI] [PubMed] [Google Scholar]
- 12. Aroutcheva AA, Simoes JA, Behbakht K, Faro S. Gardnerella vaginalis isolated from patients with bacterial vaginosis and from patients with healthy vaginal ecosystems. Clin Infect Dis 2001; 33:1022–7. [DOI] [PubMed] [Google Scholar]
- 13. Hickey RJ, Forney LJ. Gardnerella vaginalis does not always cause bacterial vaginosis. J Infect Dis 2014; 210:1682–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schellenberg JJ, Paramel Jayaprakash T, Withana Gamage N, Patterson MH, Vaneechoutte M, Hill JE. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS One 2016; 11:e0146510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed A, Earl J, Retchless A, et al. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol 2012; 194:3922–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janulaitiene M, Paliulyte V, Grinceviciene S, et al. Prevalence and distribution of Gardnerella vaginalis subgroups in women with and without bacterial vaginosis. BMC Infect Dis 2017; 17:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaneechoutte M, Guschin A, Van Simaey L, Gansemans Y, Van Nieuwerburgh F, Cools P. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int J Syst Evol Microbiol 2019; 69:679–87. [DOI] [PubMed] [Google Scholar]
- 18. Muzny CA, Blanchard E, Taylor CM, et al. Identification of key bacteria involved in the induction of incident bacterial vaginosis: a prospective study. J Infect Dis 2018; 218:966–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilbert NM, Lewis WG, Li G, et al. Gardnerella vaginalis and Prevotella bivia trigger distinct and overlapping phenotypes in a mouse model of bacterial vaginosis. J Infect Dis 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol 2010; 48:1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krohn MA, Hillier SL, Lee ML, Rabe LK, Eschenbach DA. Vaginal Bacteroides species are associated with an increased rate of preterm delivery among women in preterm labor. J Infect Dis 1991; 164:88–93. [DOI] [PubMed] [Google Scholar]
- 22. Bradshaw CS, Tabrizi SN, Fairley CK, Morton AN, Rudland E, Garland SM. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J Infect Dis 2006; 194:828–36. [DOI] [PubMed] [Google Scholar]
- 23. Castro J, Alves P, Sousa C, et al. Using an in-vitro biofilm model to assess the virulence potential of bacterial vaginosis or non-bacterial vaginosis Gardnerella vaginalis isolates. Sci Rep 2015; 5:11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castro J, França A, Bradwell KR, Serrano MG, Jefferson KK, Cerca N. Comparative transcriptomic analysis of Gardnerella vaginalis biofilms vs. planktonic cultures using RNA-seq. NPJ Biofilms Microbiomes 2017; 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hardy L, Jespers V, Abdellati S, et al. A fruitful alliance: the synergy between Atopobium vaginae and Gardnerella vaginalis in bacterial vaginosis-associated biofilm. Sex Transm Infect 2016; 92:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castro J, Machado D, Cerca N. Unveiling the role of Gardnerella vaginalis in polymicrobial Bacterial Vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J 2019; 13:1306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Santiago GL, Tency I, Verstraelen H, et al. Longitudinal qPCR study of the dynamics of L. crispatus, L. iners, A. vaginae, (sialidase positive) G. vaginalis, and P. bivia in the vagina. PLoS One 2012; 7:e45281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwebke JR, Muzny CA, Josey WE. Role of Gardnerella vaginalis in the pathogenesis of bacterial vaginosis: a conceptual model. J Infect Dis 2014; 210:338–43. [DOI] [PubMed] [Google Scholar]
- 29. Beebout CJ, Eberly AR, Werby SH, et al. Respiratory heterogeneity shapes biofilm formation and host colonization in uropathogenic Escherichia coli. MBio 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swidsinski A, Mendling W, Loening-Baucke V, et al. Adherent biofilms in bacterial vaginosis. Obstet Gynecol 2005; 106:1013–23. [DOI] [PubMed] [Google Scholar]
- 31. Machado A, Cerca N. Influence of biofilm formation by gardnerella vaginalis and other anaerobes on bacterial vaginosis. J Infect Dis 2015; 212:1856–61. [DOI] [PubMed] [Google Scholar]
- 32. Joo HS, Otto M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol 2012; 19:1503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruby J, Goldner M. Nature of symbiosis in oral disease. J Dent Res 2007; 86:8–11. [DOI] [PubMed] [Google Scholar]
- 34. Roy S, Sharma M, Ayyagari A, Malhotra S. A quantitative microbiological study of bacterial vaginosis. Indian J Med Res 1994; 100:172–6. [PubMed] [Google Scholar]
- 35. Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. Microbes on the human vaginal epithelium. Proc Natl Acad Sci U S A 2005; 102:7952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patterson JL, Girerd PH, Karjane NW, Jefferson KK. Effect of biofilm phenotype on resistance of Gardnerella vaginalis to hydrogen peroxide and lactic acid. Am J Obstet Gynecol 2007;197:170 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 2005; 353:1899–911. [DOI] [PubMed] [Google Scholar]
- 38. Hecht JL, Onderdonk A, Delaney M, et al. ; ELGAN Study Investigators Characterization of chorioamnionitis in 2nd-trimester C-section placentas and correlation with microorganism recovery from subamniotic tissues. Pediatr Dev Pathol 2008; 11:15–22. [DOI] [PubMed] [Google Scholar]
- 39. Pybus V, Onderdonk AB. Evidence for a commensal, symbiotic relationship between Gardnerella vaginalis and Prevotella bivia involving ammonia: potential significance for bacterial vaginosis. J Infect Dis 1997; 175:406–13. [DOI] [PubMed] [Google Scholar]
- 40. Chen KC, Forsyth PS, Buchanan TM, Holmes KK. Amine content of vaginal fluid from untreated and treated patients with nonspecific vaginitis. J Clin Invest 1979; 63:828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Briselden AM, Moncla BJ, Stevens CE, Hillier SL. Sialidases (neuraminidases) in bacterial vaginosis and bacterial vaginosis-associated microflora. J Clin Microbiol 1992; 30:663–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cauci S, Culhane JF. High sialidase levels increase preterm birth risk among women who are bacterial vaginosis-positive in early gestation. Am J Obstet Gynecol 2011; 204:142 e1–9. [DOI] [PubMed] [Google Scholar]
- 43. Schauer R. Chemistry, metabolism, and biological functions of sialic acids. Adv Carbohydr Chem Biochem 1982; 40:131–234. [DOI] [PubMed] [Google Scholar]
- 44. Menard JP, Fenollar F, Henry M, Bretelle F, Raoult D. Molecular quantification of Gardnerella vaginalis and Atopobium vaginae loads to predict bacterial vaginosis. Clin Infect Dis 2008; 47:33–43. [DOI] [PubMed] [Google Scholar]
- 45. Libby EK, Pascal KE, Mordechai E, Adelson ME, Trama JP. Atopobium vaginae triggers an innate immune response in an in vitro model of bacterial vaginosis. Microbes Infect 2008; 10:439–46. [DOI] [PubMed] [Google Scholar]
- 46. Ramsey PS, Lyon MD, Goepfert AR, et al. Use of vaginal polymorphonuclear to epithelial cell ratios for the prediction of preterm birth. Obstet Gynecol 2005; 105:139–44. [DOI] [PubMed] [Google Scholar]
- 47. Nelson TM, Borgogna JL, Brotman RM, Ravel J, Walk ST, Yeoman CJ. Vaginal biogenic amines: biomarkers of bacterial vaginosis or precursors to vaginal dysbiosis? Front Physiol 2015; 6:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferris MJ, Masztal A, Aldridge KE, Fortenberry JD, Fidel PL Jr, Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis 2004; 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ferris MJ, Norori J, Zozaya-Hinchliffe M, Martin DH. Cultivation-independent analysis of changes in bacterial vaginosis flora following metronidazole treatment. J Clin Microbiol 2007; 45:1016–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koumans EH, Markowitz LE, Hogan V; CDC BV Working Group Indications for therapy and treatment recommendations for bacterial vaginosis in nonpregnant and pregnant women: a synthesis of data. Clin Infect Dis 2002; 35:S152–72. [DOI] [PubMed] [Google Scholar]