Abstract
Background
We evaluated use of phylogenetic methods to predict the direction of human immunodeficiency virus (HIV) transmission.
Methods
For 33 pairs of HIV-infected patients (hereafter, “index patients”) and their partners who acquired genetically linked HIV infection during the study, samples were collected from partners and index patients close to the time when the partner seroconverted (hereafter, “SC samples”); for 31 pairs, samples collected from the index patient at an earlier time point (hereafter, “early index samples”) were also available. Phylogenies were inferred using env next-generation sequences (1 tree per pair/subtype). The direction of transmission (DoT) predicted from each tree was classified as correct or incorrect on the basis of which sequences (those from the index patient or the partner) were closest to the root. DoT was also assessed using maximum parsimony to infer ancestral node states for 100 bootstrap trees.
Results
DoT was predicted correctly for both single-pair and subtype-specific trees in 22 pairs (67%) by using SC samples and in 23 pairs (74%) by using early index samples. DoT was predicted incorrectly for 4 pairs (15%) by using SC or early index samples. In the bootstrap analysis, DoT was predicted correctly for 18 pairs (55%) by using SC samples and for 24 pairs (73%) by using early index samples. DoT was predicted incorrectly for 7 pairs (21%) by using SC samples and for 4 pairs (13%) by using early index samples.
Conclusions
Phylogenetic methods based solely on the tree topology of HIV env sequences, particularly without consideration of phylogenetic uncertainty, may be insufficient for determining DoT.
Keywords: Networks, epidemiology, viral dynamics
We evaluated the accuracy of phylogenetic methods to predict the direction of human immunodeficiency virus transmission in 33 partner–index patient pairs. Phylogenetic methods that fail to incorporate phylogenetic uncertainty may provide useful information for population-level analyses but are insufficient in legal contexts.
The rapid evolutionary rate of human immunodeficiency virus (HIV) can be used to identify transmission groups based on the genetic similarity of HIV [1]. HIV network studies often seek to identify genetically linked infections, determine when transmission occurred, and infer the likely source of infection. Such studies have provided information about social, community, and global HIV transmission networks [2–6] and informed the design of HIV prevention interventions and interpretation of HIV prevention studies [3, 4, 7]. Phylogenetic analysis of HIV has also been used in court cases to determine the genetic linkage and direction of transmission; however, a great deal of caution is needed when interpreting results of phylogenetic analyses in legal settings [8–12]. Results can be significantly influenced by methodological factors, including the model, the sequencing method, the genetic distance threshold, the time since infection, and the methods to address ambiguous nucleotides in sequence alignments [13–16].
Transmission clusters of HIV infections are typically defined using genetic distance measures alone [6, 14] or in conjunction with branch support values [17, 18]. It is possible to confirm genetic linkage if appropriate local controls are included in the analysis and if extensive contact tracing is performed; however, it is often impossible to rule out the possibility that additional individuals with genetically linked infection remain unsampled [11]. In legal settings, analysis of the genetic linkage of HIV between 2 persons should include as many sequences as possible from the local outbreak [11]. However, there are no clear guidelines on the number or relatedness of the reference sequences necessary for accurate determination of the direction of HIV transmission between 2 individuals.
HIV genetic diversity is often assumed to correlate with time since infection [6, 19, 20]. More-sophisticated models that incorporate time-sampled sequences can account for variation in the evolutionary rate and more accurately predict the timing of transmission events [21, 22]. These molecular clock methods are appropriate for small data sets (eg, consensus sequences from cross-sectional population surveys or clonal sequences from a few potentially linked cases [3, 4, 23, 24]). However, inferring the timing of HIV transmission events is complicated by the preferential transmission of ancestral viruses [25] and differences in intrahost and interhost evolutionary rates [26]. Transmission models that take these factors into account may provide greater accuracy [25].
The direction of transmission is difficult to assign by using phylogenetic methods since many factors may confound the analysis, including variable viral population size, heterogeneous evolutionary rates, ongoing reinfection between long-term partners, unidentified additional partners, drug-resistant mutations creating parallel evolution, transmission of multiple and/or recombinant variants, lack of phylogeny branch support, an inadequate number of sequences and/or time points from the potential donor/recipient, and insufficient sequences from other infected individuals from the local outbreak (ie, the “background” sequences) [11, 27, 28]. However, tree topologies may provide some information [9, 29]. Two informative characteristics of phylogenetic trees are the ancestral node placement and the topological pattern (eg, monophyly, polyphyly, and paraphyly; see Methods) [9, 30]. The concordance between topological pattern and direction of transmission were substantiated in retrospective analyses of 2 court cases [9], in simulated data sets [31], and, most recently, in documented transmission pairs [30].
Here, we evaluated the accuracy of phylogenetic methods to predict the direction of transmission in 33 pairs of index patients and their partners (hereafter, “index-partner pairs”) from the HIV Prevention Trials Network (HPTN) 052 clinical trial [32–34]; index patients were infected with HIV at study enrollment, and their partners acquired genetically linked infection during the trial. The analysis was performed using HIV env sequences obtained with next-generation sequencing (NGS). This data set was ideally suited for this study, since the 33 index-partner pairs were previously shown to have genetically linked infections and since direction of transmission was known for all pairs.
HIV sequences were analyzed using different sample sets and phylogenetic methods. All analyses were performed using partner samples collected near the time of the partner’s seroconversion (hereafter, “SC samples”). Two different sets of samples from index patients were compared to those from partners: (1) a sample collected close to the time of the partner’s seroconversion and (2) samples collected at an earlier study visit (hereafter, “early index samples”). In the first method, maximum likelihood (ML) trees were inferred using sequences from each index-partner pair, as well as sequences of the same HIV subtype from all index-partner pairs. The direction of transmission was predicted by determining which sequences (those from the index patient or those from the partner) were closer to the root of the tree, based on topological patterns. The second method used maximum parsimony to infer the state of the ancestral node in 100 bootstrap replicates for each index-partner pair.
METHODS
Study Cohort and HIV Sequences
HIV sequence data was obtained from samples collected in the HPTN 052 clinical trial [34]. This trial enrolled HIV-serodiscordant pairs and assessed the impact of early antiretroviral treatment (ART) on HIV transmission. A full description of the study protocol and institutional review board oversight is available in the original publication [34]. Genetic linkage of most index-partner pairs was based on phylogenetic and Bayesian analysis of HIV pol sequences obtained by bulk Sanger sequencing; in selected cases, linkage was confirmed by neighbor-joining tree analysis of NGS, using the 454-Roche Biotechnology platform [32, 33]. This study only included pairs in which both the index patient and their partner were infected with a single HIV strain.
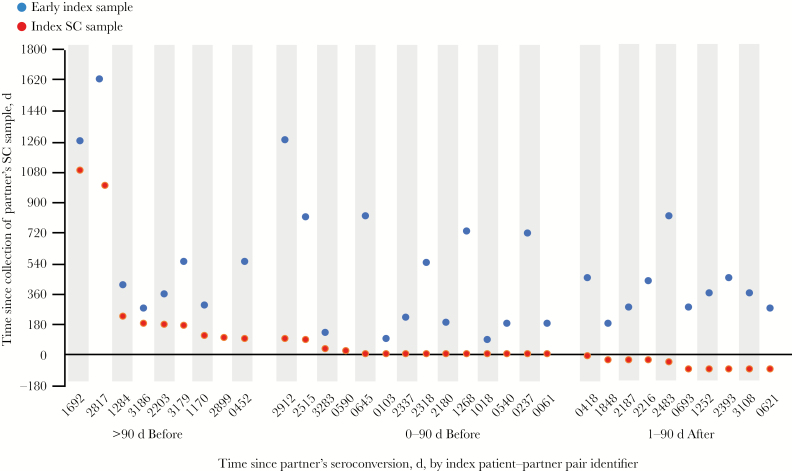
Samples were obtained from partners at the visit when seroconversion was documented or at the next study visit (hereafter, “SC samples”; median time of collection, 91 days after the last visit during which an HIV-negative test result was obtained [range, 84–588 days); partners were not followed in the HPTN 052 trial after HIV infection was confirmed. Paired SC samples were collected from index patients at the visit closest to the visit when their partner’s seroconversion was documented (for 9 index patients, their SC sample was collected >90 days before seroconversion detection in the partner; for 14, their SC sample was collected 0–90 days before seroconversion detection; and for 10, their SC sample was collected 1–90 days after seroconversion detection; Figure 1). For 31 index-partner pairs, additional samples collected earlier from index patients (hereafter, “index samples”; median time of collection, 362 days before collection of the index patients’ SC samples [range, 84–1174 days]) were also available and were analyzed separately.
Figure 1.
Time between collection of samples from human immunodeficiency virus (HIV)–infected index patients and their partners who acquired genetically linked HIV infection during the study. Samples were collected from partners and index patients close to the time when the partner seroconverted (hereafter, “SC samples”); for 31 pairs, samples collected from the index patient at an earlier time point (hereafter, “early index samples”) were also available. Positive values indicate that the SC or early index sample was collected before the partner sample; negative values indicate that the index patient’s SC or early index sample was collected after the partner’s SC sample. The identifier for each index patient and partner pair is shown on the x-axis. Pairs were created on the basis of the timing of collection of SC samples from partners and index patients. Two pairs did not have an early index sample available for analysis.
A total of 450 336 NGS-derived reads from env (nucleotides 7941–8264 relative to HXB2) were obtained from plasma samples from the 33 index-partner pairs [34, 35]. From these reads, 9051 consensus sequences (hereafter, “sequences”) were generated using GS Amplicon Variant Analyzer, version 2.5 (Roche); each sequence represented a cluster of ≥10 individual reads. Each sample had an average of 91 sequences representing 4503 reads. Sequence alignments were manually edited by codon, using AliView [36], and frameshift insertions were removed. Sequences were subtyped using REGA (available at: http://dbpartners.stanford.edu:8080/RegaSubtyping/stanford-hiv/typingtool). Reference sequences were obtained from the Los Alamos HIV Database (available at: https://www.hiv.lanl.gov/).
Single-Tree Method
For each index-partner pair, separate sequence alignments were constructed from 2 sample sets: (1) index patient and partner SC samples (the SC/SC sample set) and (2) early index samples and partner SC samples (the early/SC sample set). In addition, sequences from all pairs of the same subtype (subtype A1, 2 pairs; AE, 1; B, 3; and C, 27) were combined using the SC/SC sample set and the early/SC sample set (pairs with subtypes A1 and AE were combined, resulting in 6 analyses, 2 for each subtype). ML trees were inferred for all alignments by use of the HKY model of nucleotide substitution with gamma-distributed among-site variation, using PhyML [37] in the Geneious software package (available at: http://www.geneious.com) and RaxML v8.2.9 [38].
The direction of transmission for each assessment was independently scored by 2 investigators as described elsewhere [31], and discrepancies in scoring were reconciled by a third party. Three topological patterns were assessed: (1) a monophyletic pattern for both subjects (all sequences from a given participant shared a common ancestor that excluded sequences from any other subject), (2) a paraphyletic/monophyletic pattern (the monophyletic clade from one subject shared a common ancestor with some but not all of the sequences from the other subject), and (3) a paraphyletic/polyphyletic pattern (a mixed clade containing all sequences from one subject shared a common ancestor with some but not all of the sequences from the other subject; Supplementary Figure 1). The direction of transmission was scored as “correct” if index patient sequences were paraphyletic and partner sequences were monophyletic/polyphyletic and as “incorrect” if partner sequences were paraphyletic and index patient sequences were monophyletic/polyphyletic. If sequences from both the index patient and partner were monophyletic, the direction of transmission was scored as “equivocal.”
Bootstrapping Method
For each of the 33 pairs, separate sequence alignments were constructed for the SC/SC sample set and the early/SC sample set. All alignments also included a reference set consisting of a single random sequence from each of the other index-partner pairs and the HXB2 sequence for rooting. One hundred bootstrap phylogenies of each alignment were generated with RAxML v8.2.9 [38]. For each phylogeny, Phyloscanner v1.6.4 was used to infer the ancestral state of each of the internal nodes of the tree, using a modified maximum parsimony procedure [39]. Ancestral states were classified as an “index” state, a “partner” state, or an “unsampled” state representing either a third party or an unclear ancestry.
For each of the 100 trees generated for an index-partner pair, we identified the earliest node(s) in the tree (ie, the node that had no ancestral nodes with a sampled state). The state of this node (ie, index, partner, or unsampled) was considered to represent the transmitting subject for that tree.
If there was no such node (ie, separate clades from each subject with no implied ancestry), then the tree was labeled as (1) “equivocal” if there were no tips from the reference set descended from the most recent common ancestor node of both patients or (2) “unlinked” if there was at least 1 tip (Supplemental Figure 2).
For each index-partner pair, the direction of transmission was assigned as (1) “correct” if the state of the earliest node was classified as index in at least twice as many trees as those where it was classified as partner, (2) “incorrect” if the state of the earliest node was classified as partner in twice as many trees as those where it was classified as index, (3) “unlinked” if at least 1 tip from the reference set was descended from the earliest node in more than half of the trees, and (4) “indeterminate” for all other cases.
RESULTS
Transmission Direction Predicted Using the Single-Tree Method
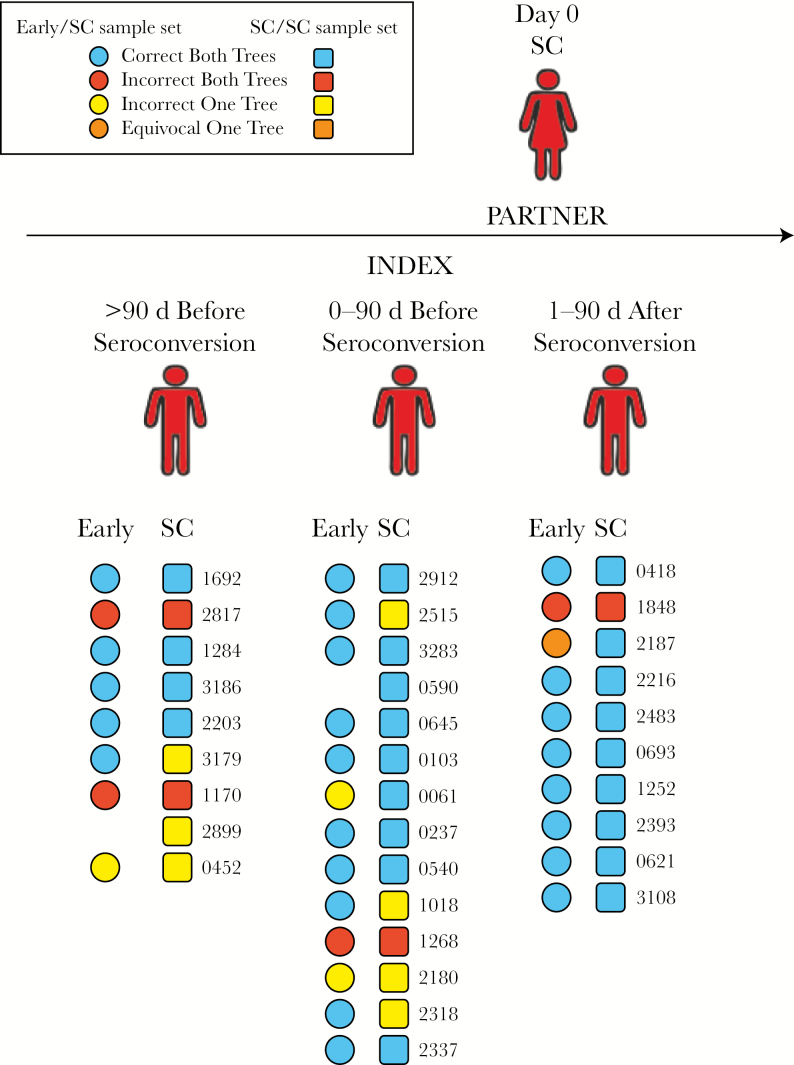
For each of the 33 index-partner pairs, we first evaluated the predicted direction of transmission, using the single-tree method. Two trees were evaluated for each index-partner pair: individual (only sequences from that pair plus subtype reference sequences) and subtype specific (all sequences of the same subtype combined). The analysis was first performed using the SC/SC sample set. The predicted direction of transmission was correct in both trees for 22 pairs (67%) and incorrect in both trees for 4 pairs (12%). Trees were discordant for the remaining 7 pairs (Figure 2 and Supplementary Table 1).
Figure 2.
Predicted direction of human immunodeficiency virus (HIV) transmission, using the single-tree method. Samples were collected from HIV-infected index patients and their partners close to the time when the partner seroconverted (hereafter, “SC samples”); for 31 pairs, samples collected from the index patient at an earlier time point (hereafter, “early index samples”) were also available. Each square or circle represents 1 index patient and partner pair; pairs were created on the basis of the timing of collection of SC samples from partners and index patients. Squares show data obtained for the SC/SC sample set, comprising index patient and partner SC samples; circles show data obtained for the early/SC sample set, comprising early index samples and partner SC samples. The identifier for each index patient and partner pair is shown to the right of the corresponding square. Colors of the squares and circles correspond to the direction of transmission predicted from individual pair trees and subtype-specific trees.
The analyses described above were next performed using the early/SC sample set, available for 31 pairs (Figure 2). The predicted direction of transmission was correct for both trees for 23 pairs (74%), incorrect for both trees for 4 pairs (13%), and discordant and/or equivocal for 4 pairs.
We then compared results obtained using the SC/SC and early/SC sample sets to determine whether the timing of sample collection influenced the prediction of the direction of transmission. This analysis was performed for the 31 index-partner pairs that had results from an early index sample. Nineteen pairs (61%) had the correct direction of transmission predicted in both trees for both sample sets. Four pairs (13%) had the incorrect direction predicted for both trees for both sample sets. Two pairs had the incorrect direction predicted in 1 tree for both sample sets. The remaining 6 pairs had discordant predictions for the 2 sample sets. Incorrect and/or equivocal predictions did not appear to be correlated with the time between collection of the SC sample from the index patient and the corresponding SC sample from the partner.
Transmission Direction Predicted Using the Bootstrapping Method
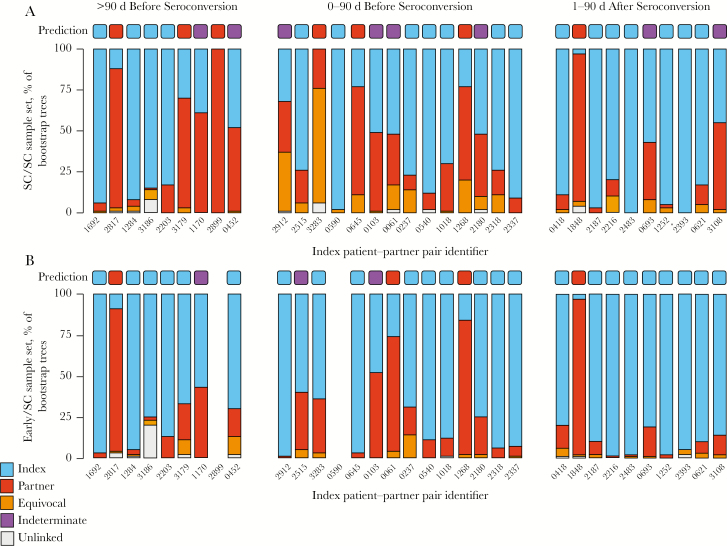
We next evaluated the accuracy of predictions of the direction of transmission, using replicate bootstrap trees (ie, bootstrap support for the predicted direction, using the modified maximum parsimony approach implemented in Phyloscanner). For the SC/SC sample set, the direction of transmission was predicted correctly for 18 pairs (55%), predicted incorrectly for 7 pairs (21%), and indeterminate for 8 pairs (24%; Figure 3A and Supplementary Table 1). For the 31 pairs with the early/SC sample set, the direction of transmission was predicted correctly for 24 pairs (73%), predicted incorrectly for 4 pairs (13%), and indeterminate for 3 pairs (12%; Figure 3B). Sixteen pairs (52%) had the correct direction predicted for both sample sets, and 3 pairs (10%) had the incorrect direction predicted for both sample sets. The predicted direction of transmission for the remaining 12 pairs was either indeterminate or inconsistent between sample sets.
Figure 3.
Predicted direction of human immunodeficiency virus (HIV) transmission, using the bootstrap method (inferred ancestral state of 100 bootstrap trees). Samples were collected from partners and index patients close to the time when the partner seroconverted (hereafter, “SC samples”); for 31 pairs, samples collected from the index patient at an earlier time point (hereafter, “early index samples”) were also available. Each bar shows the percentage of trees with different predicted ancestral states for 100 bootstrap trees, colored according to the key. The identifier for each index patient and partner pair is shown below each bar. Pairs were created on the basis of the timing of collection of SC samples from partners and index patients. A, Trees inferred using the SC/SC sample set, comprising index patient and partner SC samples. B, Trees inferred using the early/SC sample set, comprising early index samples and partner SC samples.
Comparison of Predictions From the Single-Tree and Bootstrap Methods
In general, results from the 2 approaches (ie, single trees vs bootstrapped trees) were consistent. For the SC/SC sample set, both the bootstrap method and the single-tree method for both individual and subtype-specific trees predicted the correct direction of transmission for 15 pairs (45%) and the incorrect direction for 3 pairs (9%; Figure 3 and Supplementary Table 1). The bootstrap method predicted an indeterminate direction for 5 pairs (0103, 2912, 0061, 0693, and 3108) that were correctly assessed using the single-tree method for both trees and for 1 pair (1170) that was incorrectly assessed using the single-tree method for both trees. The bootstrap analysis also predicted the incorrect direction in 2 pairs (0645 and 3283) that were correctly assessed for both trees by using the single-tree method. For one of these pairs (0645), approximately 25% of the bootstrap trees predicted the correct direction of transmission; for the other pair (3283), none of the bootstrap trees predicted the correct direction of transmission. Of note, the single trees for this pair (3283) showed that only 1 index sequence was basal to the whole clade; the remaining index sequences clustered together elsewhere.
For the 7 pairs where results from both single trees were inconsistent, the bootstrap method predicted a correct direction in 3 pairs (2515, 1018, and 2318), predicted an incorrect direction in 2 pairs (3179 and 2899), and had indeterminate results for 2 pairs (2180, 0452, and 3108).
For the early/SC sample set, both the bootstrap method and the single-tree method for both individual and subtype-specific trees predicted the correct direction of transmission for 21 pairs (68%) and the incorrect direction for 3 pairs (10%; Figure 3 and Supplementary Table 1). The bootstrap method predicted an indeterminate direction for 2 pairs (0103 and 2515) that were correctly assessed using the single-tree method for both trees and for 1 pair (1170) that was incorrectly assessed using the single-tree method for both trees. For the 4 pairs with inconsistent or equivocal single trees, the bootstrap analysis predicted a correct direction of transmission for 3 pairs (0452, 2180, and 2187) and an incorrect direction for 1 pair (0061). Taken together, these results suggest that using a single tree may overestimate the number of cases that are correctly classified for the direction of transmission.
DISCUSSION
We evaluated the accuracy of using tree topology to predict the direction of HIV transmission in 33 index-partner pairs with genetically linked HIV infection and a known direction of transmission. We compared different phylogenetic methods (the single-tree method and the bootstrap method), different sampling strategies (individual index-partner pairs and subtype-specific analysis), and different sample sets (samples collected from index patients near the time of partner seroconversion or earlier).
The direction of transmission was predicted correctly for both individual and subtype-specific trees in 67% of index-partner pairs from the SC/SC sample set. Similarly, direction of transmission was predicted correctly for both trees in 74% of index-partner pairs from the early/SC sample set. The direction of transmission was predicted correctly for only 61% of the index-partner pairs for both trees and both sample sets. It is concerning that the direction of transmission was predicted incorrectly in 13% of index-partner pairs for both trees and sample sets. In some cases, conflicting results were obtained for the 2 tree types; this suggests that the choice and/or number of background sequences may be an important factor in topological reconstruction.
The proportion of cases in this study where the direction of transmission was predicted correctly was lower than that reported in previous studies that used a similar method of predicting the direction of transmission by using topological patterns [30, 31]. However, low branch support could produce an incorrect result by chance placement of one or a few sequences. To address this, we compared results obtained with the single-tree method to results obtained using a maximum parsimony–based method to infer the state of the ancestral node for 100 bootstrap replicates for each pair. In this analysis, the direction of transmission was correctly predicted for only 18 index-partner pairs (55%) by using the SC/SC sample set and for 24 pairs (73%) by using the early/SC sample set. Only 16 pairs (52%) had the correct direction predicted by using both sample sets. The lower percentage of correct predictions by the bootstrap method demonstrates the potential of stochasticity to skew inferences and suggests that using only a single tree may overestimate confidence in determining the correct direction of transmission. Additional metrics (eg, the viral genetic diversity of host virus vs recipient virus) could potentially provide additional information that could enhance phylogenetic methods; however, this avenue has yet to be explored fully.
While both the single-tree and the bootstrap methods predicted the correct direction of transmission in more trees by using the early/SC sample sets as compared to the SC/SC sample sets, in general there was no clear trend between the predicted direction and the timing of index samples relative to the partner’s sample. Because partners were not followed in the trial after infection was confirmed, we were not able to evaluate the performance of the methods for predicting the direction of transmission when partner samples were collected from individuals with longer-term infections.
It is possible that some other factor specific to the HPTN 052 trial could have influenced our results. While most pairs (n = 27) studied in this report were infected with HIV subtype C, both correct and incorrect predictions were found for pairs of 3 different subtypes (A1, B, and C), which suggests that subtype is not a major factor influencing the accuracy of the methods used. Differences in rates of evolution and population growth of the virus may be a factor [27], which could result from antiretroviral therapy (although only 1 of 64 samples from index patients were collected after the index patient started antiretroviral therapy).
Other factors that may have influenced the accuracy of these methods include the sequence length and genomic location of the env sequences analyzed. While diversity of the env region likely enhanced the phylogenetic signal, selection bias during sample preparation might have resulted in more-frequent variants being preferentially amplified. HIV env is also subject to within-host selection pressure, which may have resulted in homoplasies caused by convergent evolution (ie, identical but independent changes) and/or lost variation; both factors could have potentially masked true transmission patterns. Additionally, recombination during amplification/sequencing could have also resulted in homoplasies. We are currently investigating the accuracy of these methods for predicting the direction of transmission, using full HIV genome sequences (using methods similar to those described by Wymant et al [39]).
The findings here are particularly important because data from phylogenetic analyses have been used as evidence in the criminal and civil justice systems in cases of suspected HIV transmission [11]. Since the repercussions of incorrect conclusions are potentially severe in legal settings, considerable effort has been invested in assessing the appropriateness and accuracy of phylogenetic methods used to assess the genetic linkage of HIV strains and the timing and direction of HIV transmission [11]. It is widely acknowledged that current methods are best used for excluding potential persons as the source of infections, and/or for assessing the duration of HIV infections, rather than for determining the direction of transmission (eg, between a plaintiff and the person suspected of being the source of the plaintiff’s infection). Our results strongly indicate that methods to determine the direction of HIV transmission that are based solely on the tree topology of HIV env sequences, particularly without consideration of phylogenetic uncertainty, should be considered insufficient for forensic or legal applications, especially in settings where additional epidemiological information is unavailable. However, these methods may provide useful insights in the context of population-level analyses (eg, to identify factors associated with increased transmission risk).
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the HPTN 052 team, for their dedication, commitment, and efforts; the participants in the HPTN 052 trial, for their invaluable contributions; the laboratory staff at Johns Hopkins University, the Rocky Mountain Laboratories, and the HPTN 052 study sites, for assistance with sample and data management; and David Nolan, for his assistance with interpretation of data.
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported by the HIV Prevention Trials Network (HPTN; sponsored by the National Institute of Allergy and Infectious Diseases [NIAID], the National Institute on Drug Abuse, the National Institute of Mental Health, and the Office of AIDS Research, National Institutes of Health [NIH], Department of Health and Human Services, under grants UM1AI068613 [to the HPTN Network Laboratory, S. H. E., principal investigator], UM1AI068617 [to the HPTN Statistical and Data Management Center, Deborah Donnell, principal investigator), and UM1AI068619 (to the HPTN Core and Operations Center, Wafaa El-Sadr, principal investigator]); the Division of Intramural Research, NIAID, NIH; and the National Cancer Institute, NIH (contract HHSN261200800001E).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: 22nd International AIDS Conference, Amsterdam, the Netherlands, 23–27 July 2018.
References
- 1. Hassan AS, Pybus OG, Sanders EJ, Albert J, Esbjörnsson J. Defining HIV-1 transmission clusters based on sequence data. AIDS 2017; 31:1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wertheim JO, Kosakovsky Pond SL, Forgione LA, et al. . Social and genetic networks of HIV-1 transmission in New York City. PLoS Pathog 2017; 13:e1006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT; UK HIV Drug Resistance Collaboration Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 2011; 204:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ; UK HIV Drug Resistance Collaboration Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog 2009; 5:e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wertheim JO, Oster AM, Hernandez AL, Saduvala N, Bañez Ocfemia MC, Hall HI. The international dimension of the U.S. HIV transmission network and onward transmission of HIV recently imported into the United States. AIDS Res Hum Retroviruses 2016; 32:1046–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wertheim JO, Leigh Brown AJ, Hepler NL, et al. . The global transmission network of HIV-1. J Infect Dis 2014; 209:304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wertheim JO, Kosakovsky Pond SL, Little SJ, De Gruttola V. Using HIV transmission networks to investigate community effects in HIV prevention trials. PLoS One 2011; 6:e27775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leitner T, Albert J. Reconstruction of HIV-1 transmission chains for forensic purposes AIDS Review 2000; 2:241–51. [Google Scholar]
- 9. Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A 2010; 107:21242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernard EJ, Azad Y, Vandamme AM, Weait M, Geretti AM. HIV forensics: pitfalls and acceptable standards in the use of phylogenetic analysis as evidence in criminal investigations of HIV transmission. HIV Med 2007; 8:382–7. [DOI] [PubMed] [Google Scholar]
- 11. Abecasis AB, Pingarilho M, Vandamme AM. Phylogenetic analysis as a forensic tool in HIV transmission investigations. AIDS 2018; 32:543–54. [DOI] [PubMed] [Google Scholar]
- 12. Abecasis AB, Geretti AM, Albert J, Power L, Weait M, Vandamme AM. Science in court: the myth of HIV fingerprinting. Lancet Infect Dis 2011; 11:78–9. [DOI] [PubMed] [Google Scholar]
- 13. Rose R, Lamers SL, Dollar JJ, et al. . Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res Hum Retroviruses 2017; 33:211–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (Transmission Cluster Engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018; 35:1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poon A. Impacts and shortcomings of genetic clustering methods for infectious disease outbreaks. Virus Evol 2016; 2:vew031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le Vu S, Ratmann O, Delpech V, et al. . Comparison of cluster-based and source-attribution methods for estimating transmission risk using large HIV sequence databases. Epidemics 2018; 23:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prosperi MC, Ciccozzi M, Fanti I, et al. ; ARCA collaborative group A novel methodology for large-scale phylogeny partition. Nat Commun 2011; 2:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. ; UK HIV Drug Resistance Database Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puller V, Neher R, Albert J. Estimating time of HIV-1 infection from next-generation sequence diversity. PLoS Comput Biol 2017; 13:e1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kouyos RD, von Wyl V, Yerly S, et al. ; Swiss HIV Cohort Study Ambiguous nucleotide calls from population-based sequencing of HIV-1 are a marker for viral diversity and the age of infection. Clin Infect Dis 2011; 52:532–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol 2006; 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drummond AJ, Pybus OG, Rambaut A, Forsberg R, Rodrigo AG. Measurably evolving populations. Trends in Ecology and Evolution 2003; 18:481–8. [Google Scholar]
- 23. Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med 2008; 5:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jacka B, Applegate T, Poon AF, et al. . Transmission of hepatitis C virus infection among younger and older people who inject drugs in Vancouver, Canada. J Hepatol 2016; 64:1247–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vrancken B, Rambaut A, Suchard MA, et al. . The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Comput Biol 2014; 10:e1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev 2006; 8:125–40. [PubMed] [Google Scholar]
- 27. Romero-Severson EO, Bulla I, Hengartner N, et al. . Donor-recipient identification in para- and poly-phyletic trees under alternative HIV-1 transmission hypotheses using approximate bayesian computation. Genetics 2017; 207:1089–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemey P, Derdelinckx I, Rambaut A, et al. . Molecular footprint of drug-selective pressure in a human immunodeficiency virus transmission chain. J Virol 2005; 79:11981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Siljic M, Salemovic D, Cirkovic V, et al. . Forensic application of phylogenetic analyses–exploration of suspected HIV-1 transmission case. Forensic Sci Int Genet 2017; 27:100–5. [DOI] [PubMed] [Google Scholar]
- 30. Leitner T, Romero-Severson E. Phylogenetic patterns recover known HIV epidemiological relationships and reveal common transmission of multiple variants. Nat Microbiol 2018; 3:983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero-Severson EO, Bulla I, Leitner T. Phylogenetically resolving epidemiologic linkage. Proc Natl Acad Sci U S A 2016; 113:2690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eshleman SH, Hudelson SE, Redd AD, et al. . Treatment as prevention: characterization of partner infections in the HIV prevention trials network 052 Trial. J Acquir Immune Defic Syndr 2017; 74:112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eshleman SH, Hudelson SE, Redd AD, et al. . Analysis of genetic linkage of HIV from couples enrolled in the HIV prevention trials network 052 trial. J Infect Dis 2011; 204:1918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cohen MS, Chen YQ, McCauley M, et al. . Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med 2011; 365:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cohen MS, Chen YQ, McCauley M, et al. . Antiretroviral therapy for the prevention of HIV-1 transmission. N Engl J Med 2016; 375:830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014; 30:3276–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 2009; 537:113–37. [DOI] [PubMed] [Google Scholar]
- 38. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. .Wymant C, Hall M, Ratmann O, et al. . PHYLOSCANNER: inferring transmission from within- and between-host pathogen genetic diversity. Mol Biol Evol 2017. doi: 10.1093/molbev/msx304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.