Summary
CENP-A is a centromere-specific histone H3 variant that epigenetically determines centromere identity, but how CENP-A is deposited at the centromere remains obscure. We previously reported that CENP-A K124 ubiquitylation, mediated by the CUL4A-RBX1-COPS8 complex, is essential for CENP-A deposition at the centromere. However, a recent report stated that CENP-A K124R mutants show no defects in centromere localization and cell viability. In the present study, we found that EYFP tagging induces additional ubiquitylation of EYFP-CENP-A K124R, which allows the mutant protein to bind to HJURP. Using a previously developed conditional CENP-A knockout system and our CENP-A K124R knock-in mutant created by the CRISPR-Cas9 system, we show that the Flag-tagged or untagged CENP-A K124R mutant is lethal. This lethality is rescued by monoubiquitin fusion, indicating that CENP-A ubiquitylation is essential for viability.
eTOC
Nikura et al. find that EYFP tagging induces additional ubiquitylation of CENP-A K124R, which allows the mutant EYFP-CENP-A K124R protein to bind HJURP. Flag-tagged or untagged CENP-A K124R mutants are lethal, but can be rescued by a monoubiquitin fusion. These results support the idea that CENP-A ubiquitylation is essential for viability.
Introduction
CENP-A is a centromere-specific histone H3 variant required for kinetochore assembly to ensure proper chromosome segregation (Fukagawa and Earnshaw, 2014). In most eukaryotes, centromere identity relies not on DNA sequence but on the presence of CENP-A–containing nucleosomes, which are formed with the canonical histones H2A, H2B, and H4 at the active centromeres. CENP-A nucleosomes bind to 171-bp α-satellite DNA and are required for active centromeres to recruit a constitutive centromere-associated network (CCAN). The kinetochore proteins, together with the spindle checkpoint, orchestrate the kinetochore-microtubule attachment and regulate cycle progression.
Because of its singular role in centromere formation, CENP-A is proposed as the epigenetic mark of the centromere. After DNA replication, centromeric nucleosomes with existing CENP-A are distributed to the replicated chromatids, and newly synthesized CENP-A is deposited at the centromere in the G1 phase in humans (Jansen et al., 2007). This regulation is crucial for proper centromere inheritance and function. Therefore, determining the molecular mechanism of the recruitment of CENP-A to the centromere is an important biological issue.
The assembly of new centromeric nucleosomes requires the Holliday junction recognition protein (HJURP), which is a CENP-A–specific chromatin assembly factor (Dunleavy et al., 2009; Foltz et al., 2009). HJURP and CENP-A are recruited during the early G1 phase.
CENP-A undergoes a variety of posttranslational modifications (PTMs) including phosphorylation, ubiquitylation, methylation, and acetylation on its amino terminus and histone-fold domain. However, understanding of CENP-A PTMs is limited compared with those of canonical histones, which directly or indirectly regulate their function (Srivastava and Foltz, 2018).
We have previously shown that CENP-A deposition at the centromere requires ubiquitylation of lysine 124 (K124) that is mediated by the CUL4A-RBX1-COPS8 E3 ligase (Niikura et al., 2015). Introduction of the K124R mutation of CENP-A reduces interaction with HJURP and abrogates the centromeric localization of CENP-A, and addition of mono-ubiquitin at the C terminus of CENP-A K124R restores its centromeric localization and the interaction with HJURP, suggesting that signaling ubiquitylation is important for CENP-A deposition at the centromere (Niikura et al., 2015).
In contrast to our findings, another report stated that the CENP-A K124R mutants showed no defects in centromere localization and cell viability (Fachinetti et al., 2017). However, there are substantive issues with the experiments that yielded these results. We published our response describing the potential problems with their results and conclusions (Niikura et al., 2017). A major caveat is that they used a fusion protein much larger than CENP-A: in the RPE-1 CENP-A−/F knockout system, the enhanced yellow fluorescent protein (EYFP) is approximately 30 kDa, and endogenous CENP-A is about 16 kDa. One possibility is that the tagging of a large protein may induce ubiquitylation at an amino acid other than K124 in the CENP-A K124R mutant protein, and this ubiquitylation at another site could suppress the mutant phenotype. Therefore, we hypothesized that the presence of a large fusion protein induces ubiquitylation at a different lysine in the CENP-A K124R mutant protein.
Results
We re-created the system described by Fachinetti et al. (2017): EYFP-CENP-A was expressed from the pBabe-EYFP retrovirus vector in diploid human (RPE-1) cells carrying one disrupted and one “floxed” CENP-A allele (CENP-A−/F), the latter of which could be inactivated by Cre recombinase (Fachinetti et al., 2017). CENP-A rescue gene constructs encoded for EYFP fused to the N terminus of wild-type CENP-A or CENP-A K124R, and these constructs were stably expressed upon retroviral integration. The remaining “floxed” CENP-A allele was then inactivated by Cre recombinase. Endogenous CENP-A was not detectable 7 days after induction of Cre recombinase (data not shown). EYFP-CENP-A K124R and EYFP-CENP-A WT localized to the centromere 7 days after inactivation of the remaining endogenous CENP-A allele (data not shown). Cell viability was examined by colony formation 14 days after inactivation of the remaining endogenous CENP-A allele. The EYFP-CENP-A K124R mutant yielded a comparable number of “rescued” colonies that became visible 14 days after inactivation of the remaining endogenous CENP-A gene (data not shown). Therefore, we reproduced the results obtained by Fachinetti et al. (Fachinetti et al., 2017).
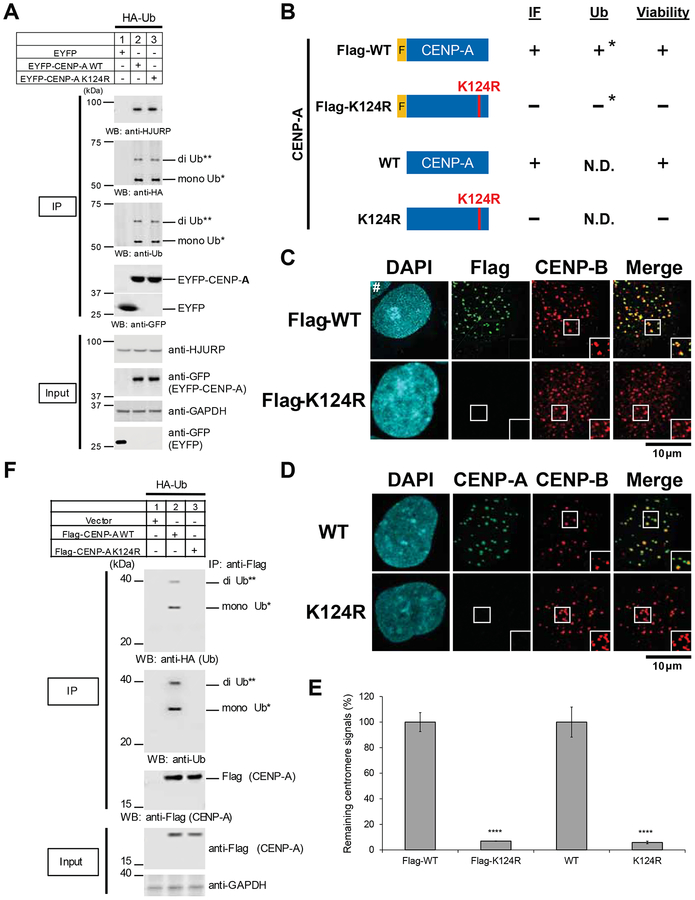
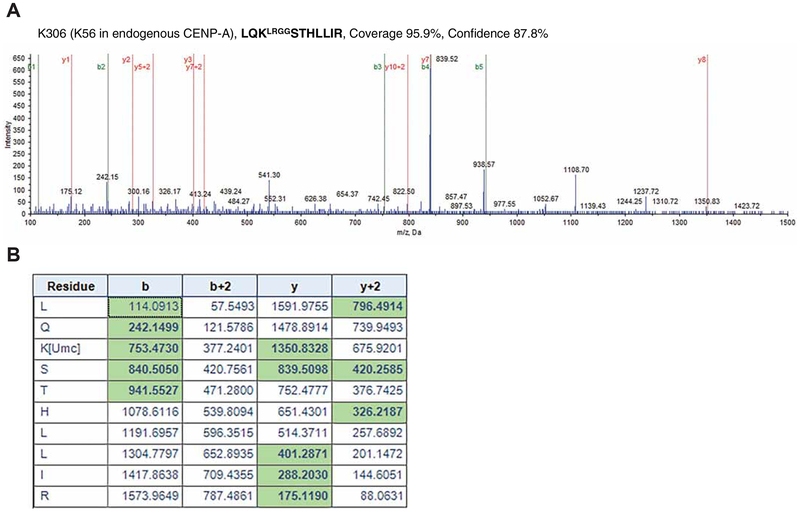
Although the initial results were comparable, we found that EYFP-CENP-A WT and the K124R mutant are ubiquitylated and bind to HJURP (Figure 1A). Our data suggest that EYFP tagging induces ubiquitylation at a lysine other than K124 in EYFP-CENP-A K124R, and this ubiquitylation allows EYFP-CENP-A K124R to bind to HJURP. Our immunoprecipitation mass spectrometry analysis revealed that lysine 306 (K306) in the EYFP-CENP-A K124R mutant is ubiquitylated in CENP-A−/F cells (Figures 2A and 2B). This site corresponds to lysine 56 (K56) in CENP-A. These data indicate that EYFP tagging induced ubiquitylation at a different lysine than K124 in CENP-A.
Figure 1. EYFP-K124R mutant is ubiquitylated and binds to HJURP.
(A) The EYFP-CENP-A K124 mutant is ubiquitylated and interacts with HJURP. In vivo ubiquitylation assay (see CENP-A In Vivo Ubiquitylation Assay in STAR METHODS). CENP-A−/F RPE-1 cells were transfected with the indicated constructs (STAR METHODS; pQCXIP-EYFP-CENP-A). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) using anti-GFP rabbit polyclonal antibody (KEY RESOURCES TABLE, ANTI #76) were detected by Western blot analysis using the indicated antibodies. Putative di-Ub-EYFP-CENP-A (**) and putative mono-Ub-EYFP-CENP-A (*) are indicated.
(B) Scheme for representation of the different Flag-tagged CENP-A at the N terminus (top) and untagged CENP-A (bottom) constructs. Red letters highlight the amino acid substitution of K124R used in the indicated construct (left). Results of assays are indicated (right). *Ubiquitylation loss of CENP-A K124R-Flag (tagged at the C-terminus) compared to WT was also previously shown (Niikura et al., 2015).
(C) Flag-CENP-A K124R mutants delocalize from centromeres. CENP-A−/F RPE-1 cells were cotransfected with indicated constructs, cultured 14 days under blasticidin S selection, and infected with retro-Cre virus infection (which mimics culture conditions shown in Figure 3A, top). Cells were collected and immunostained (see Immunofluorescence in STAR METHODS) at 4 days after Cre infection. DAPI (cyan), Flag (green), and endogenous CENP-B (red), which served as a centromere location control, were visualized. Hash (#) indicates other interphase cells just outside the inset. Scale bar, 10 μm.
(D) Untagged CENP-A K124R mutant delocalizes from centromeres. CENP-A−/F RPE-1 cells were cotransfected with indicated constructs, cultured 14 days under blasticidin S selection, and infected with retro-Cre virus infection (which mimics culture conditions shown in Figure 3A, bottom). Cells were collected and immunostained (see Immunofluorescence in STAR METHODS) 4 days after Cre infection. DAPI (cyan), CENP-A (green), and endogenous CENP-B (red), which served as a centromere location control, were visualized. Scale bar, 10 μm.
(E) Flag signals (left) or CENP-A signals (right) at centromeres shown in (C) and (D) were quantified (see Immunofluorescence and Image Acquisition, Processing, and Quantification in STAR METHODS). Signals were normalized to those of each WT, and the mean percentages (±SEM) are shown. ****p < 0.0001 compared to each WT (Student’s t test).
(F) The CENP-A K124R mutation abrogates ubiquitylation of Flag-tagged CENP-A in vivo (see CENP-A In Vivo Ubiquitylation Assay in STAR METHODS). Cells were transfected with the indicated constructs (STAR METHODS; pcDNA3-Flag-CENP-A). Proteins in 5% of the total cell lysates (Input) and immunoprecipitates (IP) were detected by Western blot analysis using the indicated antibodies. Putative di-Ub-Flag-CENP-A (**) and putative mono-Ub-Flag-CENP-A (*) are indicated.
Figure 2. Representative image: evidence of fragmentation in ubiquitylated EYFP-CENP-A K124R peptides.
(A) Lysine 306 (K306) in EYFP-CENP-A K124R is ubiquitylated in RPE-1 CENP-A−/F cells. This site corresponds to lysine 56 (K56) in CENP-A. Collision-induced dissociation analysis (STAR METHODS; Mass Spectrometry) of the LQKLRGGSTHLLIR peptide is displayed (coverage 95.9%, confidence 87.8%). The b (green) and y (red) ions in the right spectra detected during fragmentation are highlighted with green in the table of (B). The b-3 ion (m/z 753.4730) and y-8 ion (m/z 1350.8328) confirm the modification of K306 by the LRGG motif (incomplete cleavage).
(B) The b (green) and y (red) ions in the spectra of (A) are highlighted in the table. The LQKLRGGSTHLLIR peptide shown in (A) is indicated as LQK[Umc]STHLLIR in the table.
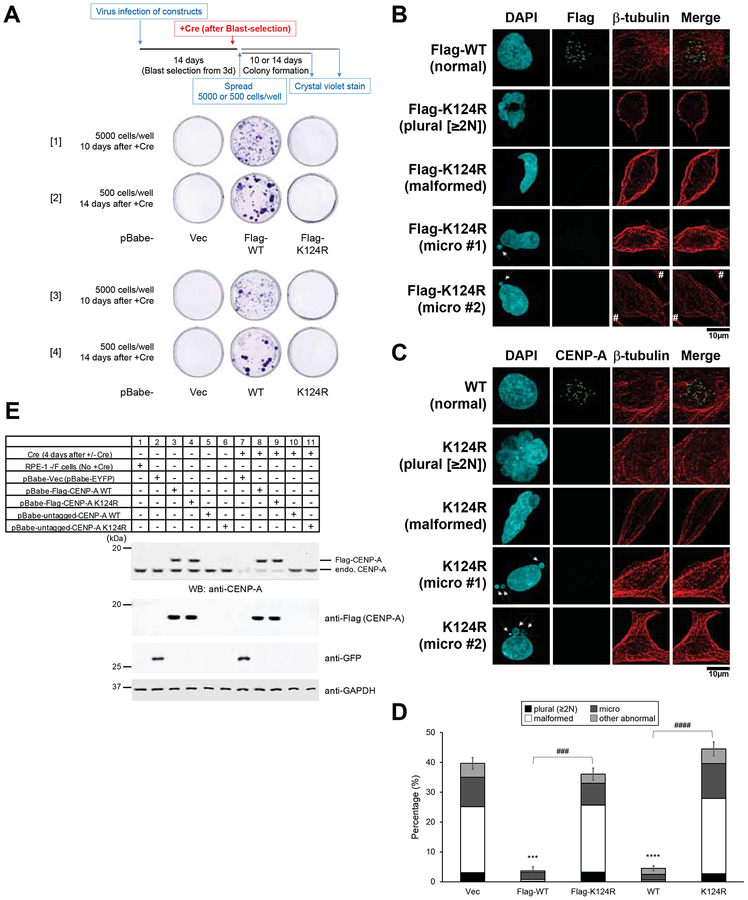
Therefore, we made pBabe-Flag-CENP-A WT and K124R mutant constructs (Figure 1B, top). These proteins were expressed at a level similar to the endogenous level of CENP-A expression (Figure 3E). As shown previously (Niikura et al., 2015), Flag-CENP-A K124R did not localize to the centromere (Figures 1C and 1E, left), and Flag-CENP-A K124R was not ubiquitylated (Figure 1F). We have previously shown that Flag-CENP-A K124R does not interact with HJURP efficiently (Niikura et al., 2015). As expected, pBabe-Flag-CENP-A WT but not the pBabe-Flag-CENP-A K124R mutant rescued the lethality of CENP-A−/F cells induced by Cre expression (Figures 3A, [1] and [2]). Consistent with this result, CENP-A−/− cells that expressed the Flag-CENP-A K124R mutant exhibited substantial numbers of abnormal nuclei 4 days after Cre infection (Figures 3B and 3D, left).
Figure 3. Flag-tagged and untagged CENP-A-K124R mutants are lethal.
(A) Representative images from the colony outgrowth assay as shown in schematic (top) of different conditions ([1] and [2] of Flag-CENP-A; [3] and [4] of untagged CENP-A). Experiments conducted under other conditions yielded similar results (data not shown). pBabe-Vec: pBabe-EYFP.
(B) Abnormal nuclei were revealed by DAPI staining in the Flag-CENP-A K124R transfectants at a higher frequency than in WT. CENP-A−/F RPE-1 cells were cotransfected with indicated constructs, cultured 14 days under blasticidin S selection, and infected with retro-Cre virus infection (which mimics the culture conditions shown in Figure 3A, [1] and [2]). Cells were collected and immunostained (see Immunofluorescence in STAR METHODS) at 4 days after Cre infection. DAPI (blue), Flag (green), and endogenous -tubulin (red) were used to show the cytoskeletal structure. Hash (#) indicates signals from other interphase cells just outside the inset. Arrows indicate micronuclei. Plural, fragmented, and aggregated nuclei consisting of 2 or more nuclei-like fragments including binuclei; malformed, malformed nuclei; micronuclei (arrow); normal, normally shaped nuclei. Scale bar, 10 μm.
(C) Abnormal nuclei were observed by DAPI stain in the untagged-CENP-A K124R transfectants more frequently than in WT. CENP-A−/F RPE-1 cells were cotransfected with the indicated constructs, cultured 14 days under blasticidin S selection, and infected with retro-Cre virus infection (which mimics culture conditions shown in Figure 3A, [3] and [4]). Cells were collected and immunostained (see Immunofluorescence in STAR METHODS) 4 days after Cre infection. DAPI (blue), CENP-A (green), and endogenous -tubulin (red) were used to show the cytoskeletal structure. Arrows indicate micronuclei. Plural, fragmented and aggregated nuclei consisting of 2 or more nuclei-like fragments including binuclei; malformed, malformed nuclei; micronuclei (arrow); normal or normaly shaped nuclei. Scale bar, 10 μm.
(D) Histogram summarizing the abnormal nuclei shown in panels (B) and (C). More than 200 interphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SEM) are shown based on the summed population: plural + malformed + micro + other abnormal. Other types of abnormal nuclei previously reported (Niikura et al., 2015) (e.g., chromosome bridges), or nuclei of damaged cells or dead cells are presumably due to transfection or other treatments (images not shown). ****p < 0.0001 and ***p < 0.001 when compared with the vector control (Vec: pBabe-EYFP) (Student’s t test). ####p < 0.0001 and ###p < 0.001 when WT and K124R were compared (Student’s t test).
(E) Confirmation of CENP-A KO by Western blot analysis. Immunoblots were performed to determine the level of expression of the indicated rescue constructs (ca. 15–20 kDa), and the loss of endogenous CENP-A protein (ca. 16 kDa) was confirmed in the CENP-A−/− cell lines collected 4 days after Cre infection. GAPDH protein was used as a loading control.
To avoid the effects of tagging, we generated pBabe-untagged CENP-A WT and K124R mutant constructs (Figure 1B, bottom). These proteins were also expressed at a level similar to the endogenous level of CENP-A expression (Figure 3E). Untagged CENP-A K124R did not localize to the centromere (Figures 1D and 1E, right). As before, the pBabe-untagged CENP-A WT but not the pBabe-untagged CENP-A K124R mutant rescued the lethality of CENP-A−/F cells induced by Cre expression (Figures 3A, [3] and [4]). The CENP-A−/− cells that expressed the untagged CENP-A-K124R mutant consistently exhibited substantial numbers of abnormal nuclei (Figures 3C and 3D, right).
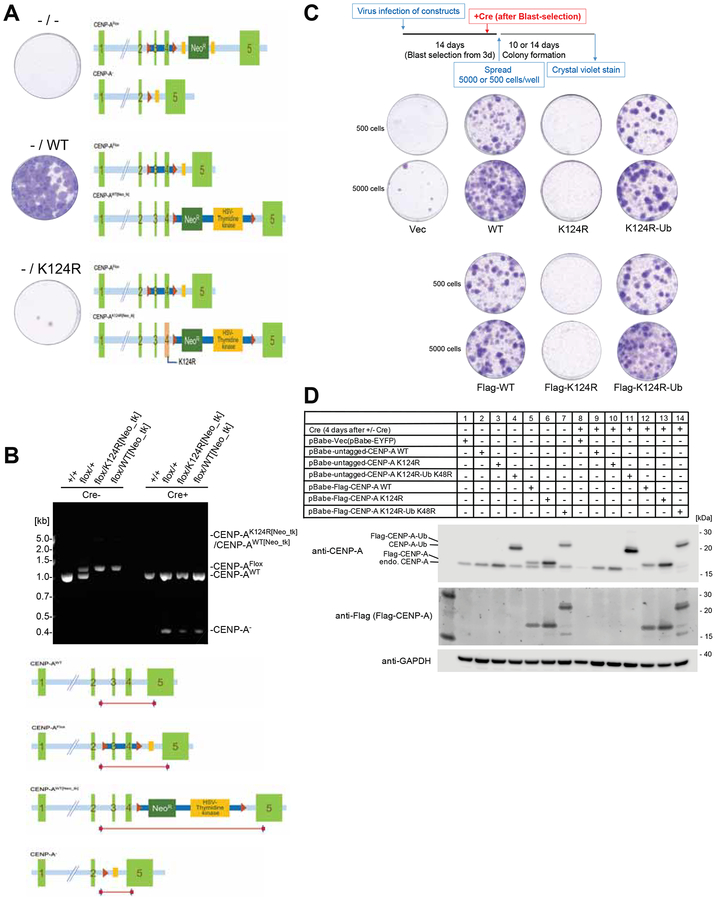
We also constructed CENP-A K124R knock-in cells carrying one CENP-A K124R allele and one “floxed” CENP-A WT allele using the CRISPR-Cas9 system and performed colony outgrowth assays (Figures 4A and 4B). In these knock-in cells, the “floxed” CENP-A allele was excised after Cre-induction (Figure 4B). The CENP-A−/K124R cells showed severe lethality compared with the CENP-A−/WT cells (Figures 4A and 4B). These results indicate that the K124R mutation is lethal, suggesting that K124 ubiquitylation is important for cell viability.
Figure 4. The hemizygous CENP-A K124R knock-in mutant created by the CRISPR-Cas9 system is lethal.
(A) The hemizygous K124R mutant is lethal to cells. Representative images from the colony outgrowth assay for cells harboring the modified CENP-A genomic locus shown as schematic illustration. Positions of the CENP-A exons, loxP sites, FRT sites, and the neomycin-resistant gene-thymidine kinase gene cassette are indicated. Images of colonies visualized with crystal violet were taken 14 days after infection with Cre expressing retrovirus vector.
(B) CENP-A genotypes validated by genomic PCR. Genomic structures of CENP-Aflox/K124R[Neo_tk] cells and its wildtype counterpart CENP-Aflox/WT[Neo_tk] cells were confirmed by genomic PCR using the specific primer sets. Genomic DNA was isolated from Wild type (+/+), Parental strain (flox/+), CENP-Aflox/K124R[Neo_tk] (flox/K124R[Neo_tk]) and CENP-Aflox/WT[Neo_tk] (flox/WT[Neo_tk]) before (Cre-) and 4 days after infection with Cre expressing retrovirus (Cre+). The schematic illustration indicates the positions of PCR products.
(C) Viability of Flag-tagged/untagged CENP-AK124R mutants is recovered by mono-ubiquitin fusion. Representative images from the colony outgrowth assay as shown in schematic (top) of different conditions (See STAR METHODS). Vec: pBabe-EYFP; WT: pBabe-CENP-A; K124R: pBabe-CENP-A K124R; K124R-Ub: pBabe-CENP-A K124R-Ub K48R; Flag-WT: pBabe-Flag-CENP-A; Flag-K124R: pBabe-Flag-CENP-A K124R; Flag-K124R-Ub: pBabe-Flag-CENP-A K124R-Ub K48R.
(D) Confirmation of expression levels of the indicated rescue constructs and endogenous CENP-A protein (ca. 16 kDa) before and 4 days after Cre infection. GAPDH protein was used as a loading control.
In addition, Bui et al. reported that K124 is acetylated and methylated in vivo (Bui et al., 2017). They used K124A (unacetylated) and K124Q (acetylation mimic) mutants for their analyses and found no defects in centromere localization. However, they used GFP-tagged proteins for the localization analysis. We found that GFP tagging also induces ubiquitylation at another lysine in GFP-CENP-A K124A and K124Q mutants (data not shown). Thus, our finding that CENP-A K124R is lethal does not necessarily mean that K124 ubiquitylation is essential. However, the addition of ubiquitin to the CENP-A-K124R mutant rescues the defect of this mutant in centromere localization (Niikura et al., 2015). Since the lethality of CENP-A K124R may be due to the lack of centromere localization, it is likely that CENP-A K124R is lethal because CENP-A K124R is not ubiquitylated. To confirm this idea, we tested the effects of adding ubiquitin to the CENP-A K124R. Monoubiquitin fusion rescued the lethality of K124R mutants (Figures 4C and 4D), indicating that CENP-A ubiquitylation is essential for viability.
Discussion
We tested various hypotheses to explain the discrepancy between the results of Fachinetti et al. (2017) (Fachinetti et al., 2017) and our findings; in particular, why they observed no phenotypes of the CENP-A K124R mutant. Our experiments revealed that EYFP tagging induced ubiquitylation at a lysine other than K124 in EYFP-CENP-A K124R. CENP-A has 6 lysins in addition to K124, and EYFP and the linker region have 20 lysins. Our in vivo ubiquitylation assay showed indistinguishable ubiquitination patterns between EYFP-CENP-A-WT and K124R, although EYFP alone shows no ubiquitination pattern (Figure 1A). Our immunoprecipitation mass spectrometry analysis showed that lysine 306 (K306) in the EYFP-CENP-A K124R mutant is ubiquitylated in vivo (Figure 2). This site corresponds to lysine 56 (K56) in CENP-A. These data suggest that once EYFP-is tagged to a K124R mutant, another ubiquitylation occurs at a different site than K124. Fachinetti et al. also used SNAP tags, and they found that SNAP-CENP-A K124R showed no defects in centromere deposition. Because the SNAP tag (20 kDa) is also a larger tag than CENP-A (approximately 16 kDa) and has 10 lysins, SNAP-CENP-A K124R, presumably, is ubiquitylated at a site different than K124.
In addition to the Flag tag, we tested the hemagglutinin (HA) tag in our studies. Like Flag-CENP-A K124R, HA-CENP-A K124R did not localize properly to the centromere (data not shown). We then sought to determine whether studies using EYFP or Flag/HA better depicted events in wild-type cells. We tested untagged CENP-A K124R to characterize its effect on cell viability. Flag-tagged CENP-A K124R behaves similarly to untagged CENP-A K124R regarding localization, cell viability, and terminal phenotypes (Figures 1 and 3).
Our current work suggests a caveat in the use of GFP/EYFP as a tool to analyze the function of a protein, and still supports that the CENP-A ubiquitylation is indispensable to cell viability.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Katsumi Kitagawa (kitagawak@uthscsa.edu). There are restrictions to the availability of cell lines and recombinant DNA (plasmids) due to material transfer agreement (MTA) at the Greehey Children’s Cancer Research Institute, UT Health Science Center San Antonio.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Culture and Transfection
CENP-A−/F RPE-1 cells (female) and CENP-AF/+ RPE-1 cells (female) (KEY RESOURCES TABLE) were cultured in DMEM and F12 medium (1:1 dilution ratio; Hyclone) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 100 units penicillin/100 μg streptomycin (Gibco). HeLa cells (female) and 293T cells (female) (KEY RESOURCES TABLE) were cultured in high-glucose Dulbecco’s modified Eagle’s (DMEM) medium (BioWhittaker) with 10% fetal bovine serum (FBS) (Gibco) and 100 units penicillin/100 μg streptomycin (Gibco). Cells were grown at 37°C in an atmosphere of 5% CO2 in a humidified incubator.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| antibodies | ||
| Mouse monoclonal anti-Flag | Sigma-Aldrich | F3165 |
| Rabbit polyclonal anti-Flag | Sigma-Aldrich | F7425 |
| Mouse monoclonal anti-GFP | Abclonal | AE012 |
| Rabbit polyclonal anti-GFP | ABCAM | ab290 |
| Rabbit polyclonal anti-GFP | Invitrogen | A11122 |
| Rabbit polyclonal anti-GFP | This study | ANTI #76 (Home-Made antibody) |
| Mouse monoclonal anti-CENP-A | Stressgen, Enzo Life Sciences | KAM-CC006 |
| Rabbit polyclonal anti-CENP-A | Upstate | 07–574 |
| Mouse monoclonal anti-CENP-B | Novus Biologicals | H00001059-B01P |
| Rabbit polyclonal anti-CENP-B | abcam | ab25734 |
| Rabbit polyclonal anti-HJURP | Proteintech Group Inc. | 15283–1-AP |
| Rabbit polyclonal anti-GAPDH | ABCAM | ab37168 |
| Rabbit polyclonal anti-GAPDH | Invitrogen | PA1987 |
| Rabbit polyclonal anti-Ubiquitin | Bethyl Laboratories | A300–317A-1 |
| Affinity-purified secondary antibody Alexa Fluor 488 Goat anti-Mouse IgG | Invitrogen | A11001 |
| Affinity-purified secondary antibody Alexa Fluor 594 Goat anti-Mouse IgG | Invitrogen | A11005 |
| Affinity-purified secondary antibody Alexa Fluor 488 Goat anti-Rabbit IgG | Invitrogen | A11008 |
| Affinity-purified secondary antibody Alexa Fluor 594 Goat anti-Rabbit IgG | Invitrogen | A11012 |
| Affinity-purified secondary antibody Alexa Fluor Plus 488 Goat anti-Mouse IgG | Invitrogen | A32723 |
| Affinity-purified secondary antibody Alexa Fluor Plus 555 Goat anti-Rabbit IgG | Invitrogen | A32732 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fetal bovine serum (FBS) | Gibco, Invitrogen | 10082–147 |
| 100 units penicillin/100 μg streptomycin | Gibco, Invitrogen | 15140–148 |
| High-glucose Dulbecco’s modified Eagle’s (DMEM) medium | BioWhittaker | 12–604f |
| Lipofectamine 3000 | Invitrogen | L3000–008 |
| Polyethylenimine (PEI) | Polysciences, Inc. | 23966–2 |
| Complete EDTA-free protease inhibitor cocktail | Roche | 11-873-580-001 |
| Restore Western Blot Stripping Buffer | Thermo Scientific | PI21059 |
| Protein A Sepharose CL-4B | GE Healthcare | 17-0963-03 |
| Anti-Flag M2 affinity gel | Sigma-Aldrich | A2220–1 ML |
| DAPI | Sigma-Aldrich | D9542 |
| Crystal violet solution | SigmaI-Aldrich | HT90132–1L |
| DMEM:DMEM/F12 medium | ATCC | 30–2006 |
| Experimental Models: Cell Lines (sex of cell is indicated.) | ||
| CENP-A−/f RPE-1 cells (female) | (Fachinetti et al., 2017), This study | Dr. Daniele Fachinetti, Dr. Don W. Cleveland |
| CENP-AF/+ RPE-1 cells (female) | (Fachinetti et al., 2017), This study | Dr. Daniele Fachinetti, Dr. Don W. Cleveland |
| HeLa cells (female) | (Niikura and Kitagawa, 2016; Niikura et al., 2015; Niikura et al., 2017) | N/A |
| 293T cells (female) | (Niikura et al., 2016) | N/A |
| Recombinant DNA | ||
| pEGFP-C1 | This study | B161 |
| pEGFP-C1-human CENP-A WT | This study | Dr. Yamini Dalal, B3128 |
| pEGFP-C1-human CENP-A K124A | This study | Dr. Yamini Dalal, B3129 |
| pEGFP-C1-human CENP-A K124Q | This study | Dr. Yamini Dalal, B3130 |
| pcDNA3 | This study | B1512 |
| pcDNA3-Flag-human CENP-A WT | This study | B2854 |
| pcDNA3-Flag-human CENP-A K124R | This study | B2829 |
| pcDNA3-HA-human CENP-A WT | This study | B2852 |
| pcDNA3-HA-human CENP-A K124R | This study | B2836 |
| pBabe-EYFP | This study | B3182 |
| pBabe-EYFP-human CENP-A WT | (Fachinetti et al., 2017), This study | Dr. Daniele Fachinetti, Dr. Don W. Cleveland, B3161 |
| pBabe-EYFP-human CENP-A K124R | (Fachinetti et al., 2017), This study | Dr. Daniele Fachinetti, Dr. Don W. Cleveland, B3164 |
| pQCXIP-EYFP | This study | B3252 |
| pQCXIP-EYFP-human CENP-A WT | This study | B3254 |
| pQCXIP-EYFP-human CENP-A K124R | This study | B3256 |
| pBabe-Flag-human CENP-A WT | This study | B3237 |
| pBabe-Flag-human CENP-A K124R | This study | B3236 |
| pBabe-untagged human CENP-A WT | This study | B3234 |
| pBabe-untagged human CENP-A K124R | This study | B3235 |
| pBabe-Flag-human CENP-A K124R-UbK48R | This study | B3271 |
| pBabe-untagged human CENP-A K124R-UbK48R | This study | B3270 |
| pCGN-HA-Ubiquitin | (Niikura and Kitagawa, 2016; Niikura et al., 2015; Niikura et al., 2017), This study | B2806 |
| pVSV-G | (Fachinetti et al., 2017), This study | Dr. Daniele Fachinetti, Dr. Don W. Cleveland, B3163 |
| pCMV-VSV-G | This study | B1197 |
| pGP | This study | Dr. Kenji Tago (Jichi Medical Univeristy, Japan), B3189 |
| pCI-VSVG | This study | Dr. Kenji Tago (Jichi Medical Univeristy, Japan), B3190 |
| pPAM | This study | B3001 |
| pBabe-puro | (Niikura et al., 2016), This study | Dr. Amruta Ashtekar, Dr. Lawrence S. Kirschner, B3028 |
| pBabe-puro-Cre | (Niikura et al., 2016), This study | Dr. Amruta Ashtekar, Dr. Lawrence S. Kirschner, B3027 |
| psPAX2 | (Niikura et al., 2016), This study | Dr. John Thompson, Dr. Gustavo W. Leone, B3031 |
| pMD2.g | (Niikura et al., 2016), This study | Dr. John Thompson, Dr. Gustavo W. Leone, B3032 |
| Software and Algorithms | ||
| Odyssey CLx Infrared Imaging System | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/ |
| Odyssey Fc Imaging System | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/ |
| FluorChem M system | Proteinsimple | https://www.proteinsimple.com/fluorchem_m.html |
| Image Studio Software | LI-COR Biosciences | https://www.licor.com/bio/products/imaging_systems/ |
| Quantity One 1-D Analysis Software | Bio-Rad | http://www.bio-rad.com/zh-cn/product/quantity-one-1-d-analysis-software?ID=1de9eb3a-1eb5-4edb-82d2-68b91bf360fb |
| ImageJ software | NIH | https://imagej.nih.gov/ij/ |
| Olympus FLUOVIEW FV3000 confocal laser scanning microscope | Olympus | https://www.olympus-global.com/news/2016/nr160405fv3000e.html |
| Acquisition FV31S-SW software | Olympus | https://www.olympus-lifescience.com.cn/en/support/downloads/#!dlOpen=%23detail847250519 |
| Analysis FV31S-DT software | Olympus | https://www.olympus-lifescience.com.cn/en/support/downloads/#!dlOpen=%23detail847250519 |
| cellSens Dimension software Ver. 1. 18 | Olympus | https://www.olympus-lifescience.com.cn/en/software/cellsens/ |
| DM IRE2 motorized fluorescence microscope | Leica | https://www.leica-microsystems.com/applications/life-science/fluorescence/ |
| ARC LAMP power supply HBO100 DC IGN | Ludl Electronic Products, Ltd. | N/A |
| ORCA-ER high-resolution digital CCD camera | Hamamatsu | N/A |
| Openlab version 5.5.2 Scientific Imaging Software | Improvision, PerkinElmer | http://www.perkinelmer.com/lab-products-and-services/resources/cellular-imaging-software-downloads.html |
| Velocity version 6.3 3D Image Analysis Software | Improvision, PerkinElmer | http://www.perkinelmer.com/lab-products-and-services/resources/cellular-imaging-software-downloads.html |
| Molecular Imager Versadoc MP4000 System | Bio-Rad | http://www.bio-rad.com/zh-cn/sku/1708640-molecular-imager-versadoc-mp-4000-system?ID=1708640 |
| Quantity One 1-D Analysis Software | Bio-Rad | http://www.bio-rad.com/zh-cn/product/quantity-one-1-d-analysis-software?ID=1de9eb3a-1eb5-4edb-82d2-68b91bf360fb |
| OpenCFU | https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0054072 | http://opencfu.sourceforge.net/ |
| Other | ||
| Immobilon-FL PVDF Transfer Membrane | EMD Millipore | IPVH00010 |
| NuPAGE™ 4–12% Bis-Tris Protein Gels | Thermo Fisher | NP0335BOX |
| Coomassie brilliant blue R-250 | BBI Life Sciences | CAS 6104-59-2 |
For chemical transfection, cells were transfected with mammalian expression plasmids by using Lipofectamine 3000 (Invitrogen) or linear polyethylenimine (PEI) (Reed et al., 2006). For retrovirus transfection, CENP-A−/F RPE-1 cells were infected with retrovirus produced by transient co-transfection of 293T cells with pBabe-constructs plus helper plasmids, as indicated in the KEY RESOURCES TABLE.
METHOD DETAILS
Immunoblotting
To detect endogenous and exogenous CENP-A proteins in CENP-A−/F RPE-1 cells, immunoblotting was performed as reported previously (Niikura et al., 2016). Cells were suspended in denaturing buffer A1 (20 mM Tris-HCl, pH 7.4; 50 mM NaCl; 0.5% Nonidet P-40; 0.5% deoxycholate; 0.5% SDS; 1 mM EDTA; and complete EDTA-free protease inhibitor cocktail [Roche]) (Wang et al., 2006) or buffer used in the immunoprecipitation assay (see the following section). The cell suspension was sonicated, frozen in liquid nitrogen, and thawed (freeze-thaw process). Before electrophoresis, cell lysates were mixed with SDS sample buffer (Lamb et al., 1995). Immobilon-FL PVDF Transfer Membrane (EMD Millipore) was used for Western blotting, and Restore Western Blot Stripping Buffer (Thermo Scientific) was used to remove primary and secondary antibodies from PVDF membranes to allow chemiluminescent and IRDye Infrared Fluorescent Western blots to be reused. Alternatively, the membrane was incubated with stripping buffer 1 (50 mM Tris-HCl, pH 7.0; 2% SDS; 50 mM DTT) at 70 °C for 30 min, stripping buffer 2 (6.25 mM Tris-HCl, pH 6.7; 2% SDS; 100 mM 2-mercaptoethanol) at room temperature for 10 min plus 50 °C for 30 min, or both of the stripping buffers to remove primary and secondary antibodies. The Odyssey CLx Infrared Imaging System (LI-COR Biosciences), Odyssey Fc Imaging System (LI-COR Biosciences), and/or FluorChem M system (Proteinsimple) were used for detection in the coimmunoblotting analysis. Intensity of band signals was quantified by Image Studio Software (LI-COR Biosciences), Quantity One 1-D Analysis Software (Bio-Rad), and/or ImageJ software (NIH).
CENP-A In Vivo Ubiquitylation Assays
in vivo CENP-A ubiquitylation assays were performed as described previously (Niikura et al., 2016; Niikura et al., 2015; Niikura et al., 2017). To study ubiquitylation of exogenous EYFP-CENP-A, CENP-A−/F REP-1 cells were cotransfected with the indicated expression vector(s) (KEY RESOURCES TABLE) plus pCGN-HA-Ubiquitin (KEY RESOURCES TABLE. and the “Cell Culture and Transfection” section). Cells were collected at 48 h after transfection and lysed in denaturing buffer A1 (20 mM Tris-HCl, pH 7.4; 50 mM NaCl; 0.5% Nonidet P-40; 0.5% deoxycholate; 0.5% SDS; 1 mM EDTA; complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. Proteins were immunoprecipitated overnight at 4°C i n buffer A1; for immunoprecipitation we used Protein A Sepharose CL-4B (GE Healthcare) pre-incubated overnight at 4°C with anti-GFP rabbit pol yclonal antibody (KEY RESOURCES TABLE, ANTI #76) in buffer A1. Immunoprecipitates were washed 4 times with buffer A1, and proteins were eluted with SDS sample buffer (Lamb et al., 1995) and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE).
To study ubiquitylation of exogenous Flag-CENP-A, HeLa cells were cotransfected with the indicated expression vectors (KEY RESOURCES TABLE) plus pCGN-HA-Ubiquitin (KEY RESOURCES TABLE and the “Cell Culture and Transfection” section). Cells were collected at 48 h after transfection and lysed in denaturing buffer A1 (20 mM Tris-HCl, pH 7.4; 50 mM NaCl; 0.5% Nonidet P-40; 0.5% deoxycholate; 0.5% SDS; 1 mM EDTA; complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. Proteins were immunoprecipitated with anti-Flag M2 affinity gel (SIGMA-ALDRICH) overnight at 4°C in buffer A1. Immunoprecipitates w ere washed 4 times with buffer A1, and proteins were eluted with SDS sample buffer (Lamb et al., 1995) and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE).
To study ubiquitylation of exogenous EGFP-CENP-A, HeLa cells were cotransfected with the indicated expression vector(s) (KEY RESOURCES TABLE) plus pCGN-HA-Ubiquitin (KEY RESOURCES TABLE; see Cell culture and transfection). Cells were collected at 48 h after transfection and lysed in denaturing buffer A1 (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% SDS, 1 mM EDTA, complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. Proteins were immunoprecipitated overnight at 4°C in buffer A1; for immunoprecipitation we used Protein A Sepharose CL-4B (GE Healthcare) pre-incubated overnight at 4°C with anti-GFP rabbit polyclonal antibody (KEY R ESOURCES TABLE 1) in buffer A1. Immunoprecipitates were washed 4 times with buffer A1, and proteins were eluted with SDS sample buffer (Lamb et al., 1995) and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE).
Immunoprecipitation Assay
Immunoprecipitation assays were performed as previously described (Niikura et al., 2016; Niikura et al., 2015; Niikura et al., 2017). To study the interaction of exogenous EYFP-CENP-A mutants with endogenous HJURP, cells were cotransfected with the indicated expression vector(s) (KEY RESOURCES TABLE) plus pCGN-HA-ubiquitin (KEY RESOURCES TABLE). Cells were collected at 48 h after transfection, lysed in buffer A1 (20 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.5% Nonidet P-40, 0.5% deoxycholate, 0.5% SDS, 1 mM EDTA, complete EDTA-free protease inhibitor cocktail [Roche]), immunoprecipitated, and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE) as described in CENP-A In Vivo Ubiquitylation Assays of exogenous EYFP-CENP-A. Alternatively, cells were collected at 48 h after transfection and lysed in denaturing buffer B (50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 1.0% Nonidet P-40; 5% skim milk; 1% BSA; 1 mM EDTA; and complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. After lysates underwent centrifugation (10,000 g for 1 min at 4°C), proteins were immunoprecipitated overnight at 4°C i n buffer B; for immunoprecipitation we used Protein A Sepharose CL-4B (GE Healthcare) pre-incubated overnight at 4°C with anti-GFP rabbit pol yclonal antibody (KEY RESOURCES TABLE) in buffer B. Immunoprecipitates were washed 4 times with buffer B, and proteins were eluted with SDS sample buffer (Lamb et al., 1995) and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE).
To study the interaction of exogenous EGFP-CENP-A with endogenous HJURP, HeLa cells were cotransfected with the indicated expression vector(s) (KEY RESOURCES TABLE) plus pCGN-HA-Ubiquitin (KEY RESOURCES TABLE; see Cell culture and transfection). Cells were collected at 48 h after transfection and lysed in denaturing buffer B (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1.0% Nonidet P-40, 5% skim milk, 1% BSA, 1 mM EDTA, and complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. After lysates underwent centrifugation (10,000 g for 1 min at 4°C), proteins were immunoprecipitated overnight at 4°C i n buffer B; for immunoprecipitation we used Protein A Sepharose CL-4B (GE Healthcare) pre-incubated overnight at 4°C with anti-GFP rabbit pol yclonal antibody (KEY RESOURCES TABLE in buffer B. Immunoprecipitates were washed 4 times with buffer B, and proteins were eluted with SDS sample buffer (Lamb et al., 1995) and subjected to Western blot analysis with the indicated antibodies (KEY RESOURCES TABLE).
Immunofluorescence
Indirect immunofluorescent staining was performed as described previously (Niikura et al., 2007; Niikura and Kitagawa, 2016; Niikura et al., 2016; Niikura et al., 2015; Niikura et al., 2017; Niikura et al., 2010; Niikura et al., 2006) with the following minor modifications. Cells were grown for the indicated number of hours or days on coverslip slides after transfection with expression vector(s) (KEY RESOURCES TABLE). Approximately 5.4 × 105 cells per well were seeded on cover glasses (22 mm × 22 mm) put in a 6-well polystyrene plate, and cells were grown for 18 h before transfection.
To detect exogenous CENP-A proteins expressed from pBabe-EYFP-CENP-A (KEY RESOURCES TABLE) in CENP-A−/F RPE-1 cells, cells were cultured and fixed 96 h after transfection with the indicated construct(s) (KEY RESOURCES TABLE). For exogenous CENP-A proteins expressed from pBabe-EYFP-CENP-A, cells transfected with pBabe-Flag-CENP-A or pcDNA3-HA-CENP-A were fixed with 4% paraformaldehyde in PBS at 4°C for 15–30 min and treated with 0.5% Triton X-100 in KB2 (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; and 0.5% BSA) at room temperature for 30 min. Cells were blocked with KB2 and incubated with specific primary antibodies (KEY RESOURCES TABLE) in KB2 at 37°C for 1 h. After cells were washed with KB2, they were incubated with the Alexa Fluor or Alexa Fluor Plus dye-conjugated secondary antibodies (Invitrogen; KEY RESOURCES TABLE) at 37°C for 30 min. Slides were washed three times with blocking buffer and then incubated in KB2 containing 0.5 μg/ml DAPI (Sigma-Aldrich).
To measure the numbers of micronuclei, cells were plated on a 6-well glass slide, fixed with 4% paraformaldehyde, and stained as described above with DAPI and anti-β-tubulin antibody to visualize the cytoskeletal structure. The frequency of the number of interphase micronuclei per cell versus the total number of interphase cells was then measured.
To detect exogenous CENP-A proteins expressed from pEGFP-CENP-A (KEY RESOURCES TABLE) in HeLa cells, cells were cultured and fixed 48 h after transfection with the indicated construct(s) (KEY RESOURCES TABLE). Cells were fixed in methanol at −20°C for 6 min. Cells were treated with 4% goat serum in TBS (25 mM Tris-HCl, pH 7.4; 125 mM NaCl) at room temperature for 10 min and then incubated with specific primary antibodies (KEY RESOURCES TABLE) for 1 h at 37°C. After cells were washed with blocking buffer, they were incubated with the Alexa Fluor dye–conjugated or Alexa Fluor Plus dye–conjugated secondary antibodies (Invitrogen; KEY RESOURCES TABLE). Slides were washed three times with blocking buffer and then incubated in TBS containing 0.5 μg/ml DAPI (SIGMA-ALDRICH).
Image Acquisition, Processing, and Quantitation
We observed cells through an Olympus FLUOVIEW FV3000 confocal laser scanning microscope (Olympus). Image acquisition and processing, including deconvolution and signal quantitation, were performed by Acquisition FV31S-SW (Olympus), Analysis FV31S-DT (Olympus), and cellSens Dimension software Ver. 1.18 (Olympus). Alternatively, we observed cells through a DM IRE2 motorized fluorescence microscope (Leica) equipped with an ARC LAMP power supply HBO100 DC IGN (Ludl Electronic Products, Ltd.), and an ORCA-ER high-resolution digital CCD camera (Hamamatsu). Image acquisition and processing, including deconvolution and signal quantitation, were performed by Openlab version 5.5.2 Scientific Imaging Software (Improvision) and Velocity version 6.3 3D Image Analysis Software (Improvision).
Quantitation of centromere signal intensity in interphase cells was performed manually as described (Niikura and Kitagawa, 2016; Niikura et al., 2016; Niikura et al., 2015; Niikura et al., 2017) (see QUANTIFICATION AND STATISTICAL ANALYSIS), or similar procedures were applied by cellSens Dimension software Ver. 1.18 (Olympus). The integrated signal intensity was calculated by subtracting the fluorescence intensity of the background (measured outside the nucleus). Deconvolved and max-projected images were used for fluorescence intensity measurements.
Colony Outgrowth Assays and Retro-Cre Virus Infection
Colony outgrowth assays were performed as described previously (Fachinetti et al., 2017; Niikura et al., 2010) with the following minor modifications. Cells (2.25 × 105) were plated in a 6-well plate. The next day, 500 to 1000 μl of DMEM medium that contained retro-Cre virus (= filtered through a 0.45 μm membrane filter) was added to an equal volume of DMEM/F12 medium that contained 8 μg/ml polybrene (final 1:1 volume ratio of DMEM:DMEM/F12 medium). After 1 day, 500 or 5000 cells were plated in triplicate on a 6-well plate and cultured in DMEM/F12 medium. For 5000 or 500 cells’ plating, cells were selected with blasticidin S (10 μg/ml) 3–24 days or 3–28 days after virus infection of constructs, respectively. For 5000 or 500 cells’ plating, cells were grown 10 days or 14 days after the plating, respectively. After colony growth, colonies were fixed for 10 min in methanol and stained for 10 min in a crystal violet solution (2.3% crystal violet, 0.1% ammonium oxylate, 20% ethanol; Sigma-Aldrich). The number of colonies was counted with the Molecular Imager Versadoc MP4000 System (Bio-Rad), Quantity One 1-D Analysis Software (Bio-Rad), ImageJ software (NIH), and/or OpenCFU software.
K124R Knock-in Cells (constructed using the CRISPR-Cas9 system)
We generated a CENP-Aflox/K124R[Neo_tk] cell line (in which one allele of CENP-A gene was knocked out by Cre-loxP-mediated excision of exon 3- and 4 and another allele is the K124R mutant) by introducing the K124R mutation into a wild-type CENP-A allele in CENP-Aflox/+ RPE1 cells (Fachinetti et al., 2017) by using aCRISPR-Cas9 mediated gene targeting system. To make a CENP-A targeting plasmid, the oligonucleotides [5’- AGTAACTCGGCCTGCATGTA -3’] encoding gRNA targeting the CENP-A locus were cloned into pLentiCRISPRv2 (Addgene #52961). To generate the left homology arm (LHA) of the donor plasmid, 803 bp DNA fragment (encompassing the CENP-A genomic locus from 98bp upstream of CENP-A exon 3 to 134bp downstream of exon 4) was PCR amplified and cloned into pGEM-T (Promega). A codon, AAG encoding 124th amino acid of CENP-A in exon 4 was replaced with AGG by Quickchange site-directed mutagenesis (Stratagene/Agilent Technologies) to introduce the K124R mutation (LHA-K124R). The 811 bp right homology arm (RHA) (corresponding to 351bp upstream of CENP-A exon 5 to 460bp downstream of the beginning of exon 5) was also PCR amplified. LHA-K124R (or LHA without mutation for making the WT[Neo_tk] allele) and RHA were cloned into the Sa/I/HindIII site and the SpeI/NotI site of the pNeotk-LoxP2 plasmid (gift from Dr. McKinnon), respectively. CENP-Aflox/+ RPE1 cells were co-transfected with the CENP-A targeting plasmid and the donor plasmid using a Neon transfection system (Invitrogen/Fisher Scientific). Transfected cells were selected with 400 μg/ml of G418; then single cells were isolated and individually plated into 96-well plates. Genotypes of neomycin-resistant cells were validated by genomic PCR using specific primers, and the presence of the mutation for K124R mutant was confirmed by sequencing.
Mass Spectrometry
To identify the ubiquitylation site of the ETFP-CENP-A K124R mutant, CENP-A−/F RPE-1 cells were transfected with pQCXIP-EYFP-CENP-A K124R. Cells were collected at 48 h after transfection and lysed in denaturing buffer A1 (20 mM Tris-HCl, pH 7.4; 50 mM NaCl; 0.5% Nonidet P-40; 0.5% deoxycholate; 0.5% SDS; 1 mM EDTA; complete EDTA-free protease inhibitor cocktail [Roche]) by a sonication and freeze-thaw process. Proteins were immunoprecipitated overnight at 4°C in buffer A1; for immunoprecipitation we use d Protein A Sepharose CL-4B (GE Healthcare) pre-incubated overnight at 4°C with anti-GFP mouse monoclonal antibody (KEY RESOURCES TABLE, Abclonal, AE012) in buffer A1. Immunoprecipitates were washed 4 times with buffer A1, and proteins were eluted with SDS sample buffer (Lamb et al., 1995), electrophoresed on an SDS-PAGE gel (KEY RESOURCES TABLE, Thermo Fisher, NP0335BOX), and stained with Coomassie brilliant blue R-250 (KEY RESOURCES TABLE, BBI Life Sciences, CAS 6104-59-2). The gel region of 50–70 kDa was excised and cut out.
MS data acquisition was performed by LC- MS/MS using a nanoLC.2D (Eksigent Technologies) coupled with a TripleTOF 5600+ System (AB SCIEX, Concord, ON). First, samples were chromatographed using a 60 min gradient from 5–80% (mobile phase A: 0.1% (v/v) formic acid, 2% (v/v) acetonitrile; mobile phase B: 0.1% (v/v) formic acid, 98% (v/v) acetonitrile) after direct injection onto a nanoLC Column, 3C18-CL, 75 μm*15 cm (Eksigent Technologies). The gradient was comprised of an increase from 2% to 22% mobile phase B over 40 min, 22% to 35% B in 12 min and climbing to 80% B in 4 min, then holding at 80% B for the last 4 min, all at a constant flow rate of 300 nl/min on an Ekisgent NanoLC system. MS1 spectra were collected in the range 350–1,500 m/z for 250 ms. The 50 most intense precursors with charge state 2–5 were selected for fragmentation, and MS2 spectra were collected in the range 100–2,000 m/z for 100 ms; precursor ions were excluded from reselection for 15 s.
The original MS data were submitted to ProteinPilot Software (version 4.5, AB Sciex) for database searching against UniProt Homo Sapiens database (April 9, 2016, containing 160,566 sequences, http://www.uniprot.org/proteomes/UP000005640) concatenated with reverse decoy database. Trypsin/P was specified as a cleavage enzyme allowing up to 3 missing cleavages, 4 modifications per peptide, and 2–5 charges. Mass error was set to 20 ppm for the first search, 5 ppm for the main search, and 0.02 Da for fragmented ions. False discovery rate (FDR) thresholds for protein, peptide, and modification sites were specified at 1%. The minimum peptide length was set at 7. All the other parameters in ProteinPilot were set to default values. In addition, ProteinProspector (v 5.20.0) was used for search comparisons and protein quantification, as described previously (Fang et al., 2012).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitation and Statistical Analysis of Centromere Signal Intensity in Interphase Cells
Quantitation of centromere signal intensity in interphase cells was performed manually as described (Niikura and Kitagawa, 2016; Niikura et al., 2016; Niikura et al., 2015; Niikura et al., 2017) (see Image Acquisition, Processing, and Quantitation in EXPERIMENTAL MODEL AND SUBJECT DETAILS). As a brief overview, the percentage of remaining signals at the centromeres was quantified by using cellSens Dimension software Ver. 1.18 (Olympus). For this task, use the following formula:
Remaining signals of centromere-kinetochore protein at the centromere-kinetochore
where s is the signal brightness of the selected area, which is confirmed by CENP-B staining; b is the background signal brightness; rsample is the reference CENP-B signals for siRNA(s)-treated cells; and rctrl is the reference CENP-B signals for each WT.
Signals were normalized to each WT, and the mean percentages (±SEM) are shown. ****p < 0.0001 compared to each WT (Student’s t test).
Statistical Analysis of Abnormal Nuclei
More than 200 interphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SEM) are shown based on the summed population: plural + malformed + micro + other abnormal. Other types of abnormal nuclei previously reported (Niikura et al., 2015) (e.g., chromosome bridges), or nuclei of damaged cells or dead cells are presumably due to transfection or other treatments (images not shown). ****p < 0.0001 and ***p < 0.001 when compared with the vector control (Vec: pBabe-EYFP) (Student’s t test). ####p < 0.0001 and ###p < 0.001 when WT and K124R were compared (Student’s t test).
DATA AND CODE AVAILABILITY
The published article includes all datasets/code generated or analyzed during this study.
Highlights.
EYFP tagging induces ubiquitylation at a lysine other than K124 in EYFP-CENP-A K124R.
Flag-tagged and untagged, but not EYFP-tagged, CENP-A K124R mutants are lethal.
Lethality of the K124R mutants is rescued by monoubiquitin fusion.
CENP-A ubiquitylation is essential for centromere localization and viability.
ACKNOWLEDGMENTS
We thank Chao-Jun Li at the at the Model Animal Research Center, Nanjing University for mass spectrometry analysis. We thank Yanmin Dalal, Tatsuo Fukagawa, and current researchers at the Model Animal Research Center, Nanjing University and Greehey Children’s Cancer Research Institute for their helpful discussion, experimental guidance, and reagents. We thank Don W. Cleveland, Daniele Fachinetti, Yanmin Dalal, Minh Bui, Gustavo W. Leone, John Thompson, Lawrence S. Kirschner, Amruta Ashtekar, Ben E. Black, Glennis A. Logsdon, Kenji Tago, and Dawn S. Chandler for their generous gifts of reagents. YN was supported by the 16th Six Big Talent Peaks Fund, Natural Science Foundation of Jiangsu Province, and the Jiangsu Province “Double-First-Class” Construction Fund, and this study was supported by NCI grant R21 CA205659.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
References
- Bui M, Pitman M, Nuccio A, Roque S, Donlin-Asp PG, Nita-Lazar A, Papoian GA, and Dalal Y (2017). Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenetics Chromatin 10, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, and Almouzni-Pettinotti G (2009). HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Logsdon GA, Abdullah A, Selzer EB, Cleveland DW, and Black BE (2017). CENP-A Modifications on Ser68 and Lys124 Are Dispensable for Establishment, Maintenance, and Long-Term Function of Human Centromeres. Dev Cell 40, 104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Kaake RM, Patel VR, Yang Y, Baldi P, and Huang L (2012). Mapping the protein interaction network of the human COP9 signalosome complex using a label-free QTAX strategy. Mol Cell Proteomics 11, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR 3rd, Bassett EA, Wood S, Black BE, and Cleveland DW (2009). Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T, and Earnshaw WC (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30, 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, and Cleveland DW (2007). Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, and Hieter P (1995). Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Niikura Y, Dixit A, Scott R, Perkins G, and Kitagawa K (2007). BUB1 mediation of caspase-independent mitotic death determines cell fate. J Cell Biol 178, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, and Kitagawa K (2016). Immunofluorescence Analysis of Endogenous and Exogenous Centromere-kinetochore Proteins. J Vis Exp, e53732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, and Kitagawa K (2016). CENP-A Ubiquitylation Is Inherited through Dimerization between Cell Divisions. Cell Rep 15, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Ogi H, Abdulle R, Pagala V, and Kitagawa K (2015). CENP-A K124 Ubiquitylation Is Required for CENP-A Deposition at the Centromere. Dev Cell 32, 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Kitagawa R, Ogi H, and Kitagawa K (2017). SGT1-HSP90 complex is required for CENP-A deposition at centromeres. Cell Cycle 16, 1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Ogi H, Kikuchi K, and Kitagawa K (2010). BUB3 that dissociates from BUB1 activates caspase-independent mitotic death (CIMD). Cell Death Differ 17, 1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikura Y, Ohta S, Vandenbeldt KJ, Abdulle R, McEwen BF, and Kitagawa K (2006). 17-AAG, an Hsp90 inhibitor, causes kinetochore defects: a novel mechanism by which 17-AAG inhibits cell proliferation. Oncogene 25, 4133–4146. [DOI] [PubMed] [Google Scholar]
- Reed SE, Staley EM, Mayginnes JP, Pintel DJ, and Tullis GE (2006). Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods 138, 85–98. [DOI] [PubMed] [Google Scholar]
- Srivastava S, and Foltz DR (2018). Posttranslational modifications of CENP-A: marks of distinction. Chromosoma 127, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article includes all datasets/code generated or analyzed during this study.