Abstract
Dermatofibrosarcoma protuberans is a locally aggressive superficial mesenchymal neoplasm. It typically occurs in adulthood, and has been reported to have a slight male predilection. Tumors have a characteristic histopathologic appearance, including: storiform architecture, infiltrative “honeycomb” growth within subcutaneous adipose tissue, and immunoreactivity for CD34. Virtually all molecularly characterized cases to date have been found to harbor a COL1A1-PDGFB fusion product. Following identification of an index patient with a novel COL6A3-PDGFD fusion gene, we undertook a molecular investigation, using a combination of RNA sequencing and fluorescence in situ hybridization (FISH), to assess the prevalence of PDGFD rearrangement in dermatofibrosarcoma protuberans (N = 63). Three additional patients were found to have balanced PDGFD rearrangements. Interestingly, all 4 tumors arose on the breast of females. As a result, we subsequently examined 16 additional cases of primary breast dermatofibrosarcoma protuberans, identifying 2 additional tumors with PDGFD rearrangement. The morphology and immunophenotype of all 6 cases was analogous to those with the canonical COL1A1-PDGFB fusion; none of the cases showed fibrosarcomatous transformation. This study illustrates that the COL6A3-PDGFD fusion product is rare in dermatofibrosarcoma protuberans, and associated with an apparent predilection for breast. An awareness of this variant is important for pathologists, as it will not be detected using conventional reverse transcription polymerase chain reaction or FISH-based diagnostic assays for dermatofibrosarcoma protuberans.
Keywords: breast, COL1A1, COL6A3, dermatofibrosarcoma protuberans, PDGFB, PDGFD
1 |. INTRODUCTION
Dermatofibrosarcoma protuberans is a fibroblastic neoplasm originating in the dermis and/or superficial subcutis.1 It may arise throughout the body, but most commonly involves the trunk and extremities.2 Tumors typically occur in early-to-mid adulthood, although it is known to have a wide range of involvement2,3; it is reported to have a slight male predilection.2,3 The clinical behavior is generally characterized by local aggressiveness.1 Complete surgical resection, with widely negative margins, is generally curative.2
Morphologically, most tumors are composed of spindle cells in a prominent storiform/whorled pattern.2,3 Peripherally, there is often infiltration of subcutaneous adipose tissue yielding a characteristic “honeycomb” pattern. Tumors tend to spare adnexal structures. The cells are monomorphic and generally have only minimal mitotic activity.2 Multiple morphologic variants exist, including, for example, those associated with melanin pigmentation (Bednar tumor),4 giant cells,5,6 prominent myxoid stroma,7 and myoid differentiation.8 A subset of cases may undergo fibrosarcomatous transformation (fibrosarcoma ex-dermatofibrosarcoma protuberans) which is associated with frequent local recurrence (58%), as well as metastasis (15%)9; in addition to areas of conventional dermatofibrosarcoma protuberans, these tumors tend to have a fascicular-herringbone architecture, and increased cellularity and mitotic activity.9 Additionally, a pleomorphic variant of dermatofibrosarcoma protuberans has recently been reported.10 Most tumors exhibit diffuse immunoreactivity for CD34,11 but this may be diminished-to-absent in a proportion of cases with fibrosarcomatous transformation12; myoid differentiation is highlighted by smooth muscle actin.8 Virtually all molecularly characterized cases to date have been reported to have a COL1A1-PDGFB fusion gene. The concomitant chimeric protein causes autocrine activation of the PDGFRB protein-tyrosine kinase, which leads to disinhibited cell proliferation13; consequently, in advanced cases, this provides a rationale for targeted therapy using tyrosine kinase inhibitors, such as imatinib.
Clinical diagnostic testing for PDGFB rearrangement in dermatofibrosarcoma protuberans is generally performed by fluorescence in situ hybridization (FISH), since the wide variability in COL1A1 genomic breakpoints makes routine testing by the reverse transcriptase polymerase chain reaction (RT-PCR) impractical. Recently, targeted next-generation sequencing—particularly in the context of limited sampling, and rare or novel fusion products—has emerged as a reliable and useful molecular diagnostic technique. Herein, we report an index case of dermatofibrosarcoma protuberans with a typical morphology and immunoprofile; however, diagnostic RNA sequencing (RNA-Seq) identified a novel COL6A3-PDGFD fusion gene. We subsequently investigated a series of cases of dermatofibrosarcoma protuberans to characterize the prevalence of this novel fusion variant.
2 |. MATERIALS AND METHODS
2.1 |. Cases
A novel COL6A3-PDGFD fusion transcript was identified by RNA-Seq in the course of routine diagnostic testing of the index tumor (Patient A1); consequently, where feasible, subsequent cases that were morphologically and immunophenotypically compatible with dermatofibrosarcoma protuberans—encountered at Mount Sinai Hospital (RNA-Seq) and Memorial Sloan Kettering Cancer Center (FISH) over approximately a 12-month period—were assessed to determine fusion gene status (Cohort A). As a result of these initial findings, a retrospective archival review was then performed at each of the three participating institutions for cases of dermatofibrosarcoma protuberans specifically arising from the breast and/or upper chest (Cohort B). This study was performed following institutional Research Ethics Board approval.
2.2 |. Immunohistochemistry
Formalin-fixed paraffin-embedded tissue blocks were cut at 4 μm and stained for CD34, desmin, smooth muscle actin, S100, and keratin (AE1/AE3) using standard techniques (Supporting Information Table S1). Appropriate on-slide positive controls were used throughout. Scoring of tumors was based on the percentage of positive cells (0: no staining; 1+: <5%; 2+: 5%−25%; 3+: 26%−50%; 4+: 51%−75%; and 5+: 76%−100%).
2.3 |. RNA sequencing
Total RNA was extracted from formalin-fixed paraffin-embedded tissue scrolls (3–4 per case) using the ExpressArt FFPE Clear RNA Ready kit (Amsbio, Cambridge, MA); it was assessed using the RNA 6000 Nano Bioanalyzer Kit (Agilent, Mississauga, ON) and quantitated using the Qubit RNA HS Assay Kit (ThermoFisher Scientific, Mississauga, ON). An input of 20–100 ng total RNA and the TruSight RNA Fusion Panel were used to prepare the RNA-seq libraries (Illumina, San Diego, CA), following manufacturer’s instructions and as previously described.14 Sequencing of each sample was performed with 76 base-pair paired-end reads on an Illumina MiSeq at 8 samples per flow cell (~3 million reads per sample). The results were then analyzed using the STAR and BOWTIE2 aligners, and Manta and JAFFA fusion callers, respectively.15,16
The expression profile of the COL6A3-PDGFD positive tumors were compared with 34 conventional COL1A1-PDGFB positive tumors available on the same RNA-Seq platform (Table 1); the latter had a morphology and immunophenotype typical of dermatofibrosarcoma protuberans. In addition, an unsupervised hierarchical clustering was performed, which further included a large number of other sarcoma types that has been tested on the same platform (these cases constituted routine clinical cases of sarcoma diagnostically evaluated for the presence of an underlying fusion gene event). The mRNA levels of genes of interest were also examined, including PDGFB, PDGFRA, and PDGFRB, among others.
TABLE 1.
Patient summary
| Patient | Age | Sex | Location | Diagnosis | RNA-Seq | FISH |
|---|---|---|---|---|---|---|
| A1 | 58 | F | Breast, L | DFSP | COL6A3-PDGFD | PDGFD |
| A2 | 29 | F | Breast, L | DFSP | COL6A3-PDGFD | PDGFD |
| A3 | 36 | F | Breast, L | DFSP | COL6A3-PDGFD | PDGFD |
| A4 | 22 | F | Breast, L | DFSP | PDGFD | |
| A5 | 67 | M | Lung* | F-DFSP | COL1A1-PDGFB | |
| A6 | 26 | F | Abdomen | F-DFSP | COL1A1-PDGFB | Negative |
| A7 | 16 | M | Groin | DFSP | COL1A1-PDGFB | Negative |
| A8 | 35 | M | Shoulder | DFSP | PDGFB | |
| A9 | 23 | F | Shoulder | DFSP | PDGFB | |
| A10 | 61 | F | Groin | DFSP | PDGFB | |
| A11 | 49 | M | Neck | F-DFSP | PDGFB | |
| A12 | 50 | F | Back | DFSP | PDGFB | |
| A13 | 45 | F | Groin | DFSP | PDGFB | |
| A14 | 28 | F | Back | DFSP | PDGFB | |
| A15 | 32 | F | Foot | F-DFSP | PDGFB | |
| A16 | 34 | F | Back | F-DFSP | PDGFB | |
| A17 | 28 | F | Thigh | DFSP | PDGFB | |
| A18 | 34 | M | Scalp | DFSP | PDGFB | |
| A19 | 71 | F | Groin | F-DFSP | PDGFB | |
| A20 | 74 | M | Thigh | F-DFSP | PDGFB | |
| A21 | 20 | F | Hand | P-DFSP | PDGFB | |
| A22 | 29 | M | Back | F-DFSP | PDGFB | |
| A23 | 19 | M | Hand | F-DFSP | PDGFB | |
| A24 | 44 | F | Abdomen | DFSP | COL1A1-PDGFB | |
| A25 | 83 | F | Back | DFSP | COL1A1-PDGFB | |
| A26 | 43 | F | Breast | DFSP | COL1A1-PDGFB | |
| A27 | 69 | M | Thigh | DFSP | COL1A1-PDGFB | |
| A28 | 43 | F | Scalp | DFSP | COL1A1-PDGFB | |
| A29 | 56 | M | Chest | F-DFSP | COL1A1-PDGFB | |
| A30 | 36 | F | Back | DFSP | COL1A1-PDGFB | |
| A31 | 25 | M | Thigh | DFSP | COL1A1-PDGFB | |
| A32 | 33 | M | Chest | DFSP | COL1A1-PDGFB | |
| A33 | 50 | M | Back | DFSP | COL1A1-PDGFB | |
| A34 | 47 | F | Thigh | DFSP | COL1A1-PDGFB | |
| A35 | 61 | M | Chest | DFSP | COL1A1-PDGFB | |
| A36 | 79 | M | Chest | F-DFSP | COL1A1-PDGFB | |
| A37 | 39 | M | Chest | DFSP | COL1A1-PDGFB | |
| A38 | 34 | M | Abdomen | DFSP | COL1A1-PDGFB | |
| A39 | 39 | F | Arm | DFSP | COL1A1-PDGFB | |
| A40 | 34 | M | Abdomen | DFSP | COL1A1-PDGFB | |
| A41 | 35 | F | Shoulder | P-DFSP | COL1A1-PDGFB | |
| A42 | 29 | F | Axilla | DFSP | COL1A1-PDGFB | |
| A43 | 17 | F | Breast | DFSP | COL1A1-PDGFB | |
| A44 | 37 | F | Abdomen | DFSP | COL1A1-PDGFB | |
| A45 | 67 | F | Buttock | DFSP | COL1A1-PDGFB | |
| A46 | 49 | M | Thigh | DFSP | COL1A1-PDGFB | |
| A47 | 37 | F | Neck | DFSP | COL1A1-PDGFB | |
| A48 | 41 | F | Groin | DFSP | COL1A1-PDGFB | |
| A49 | 49 | M | Shoulder | F-DFSP | COL1A1-PDGFB | |
| A50 | 61 | M | Abdomen | DFSP | COL1A1-PDGFB | |
| A51 | 53 | M | Sacrum | DFSP | COL1A1-PDGFB | |
| A52 | 53 | M | Buttock | F-DFSP | COL1A1-PDGFB | |
| A53 | 35 | F | Arm | DFSP | COL1A1-PDGFB | |
| A54 | 51 | M | Scalp | DFSP | COL1A1-PDGFB | |
| A55 | 28 | F | Back | DFSP | COL1A1-PDGFB | |
| A56 | 52 | F | Back | DFSP | COL1A1-PDGFB | |
| A57 | 54 | F | Chest | DFSP | COL1A1-PDGFB | |
| A58 | 70 | M | Arm | F-DFSP | COL1A1-PDGFB | |
| A59 | 39 | F | Sacrum | DFSP | COL1A1-PDGFB | |
| A60 | 35 | F | Abdomen | DFSP | PDGFB | |
| A61 | 35 | M | Chest | DFSP | PDGFB | |
| A62 | 26 | F | Hand | DFSP | PDGFB | |
| A63 | 51 | F | Breast | F-DFSP | Negative | |
| B1 | 51 | F | Breast | DFSP | PDGFD | |
| B2 | 38 | F | Breast | DFSP | PDGFD | |
| B3 | 40 | F | Breast | F-DFSP | PDGFB | |
| B4 | 79 | F | Breast | DFSP | PDGFB | |
| B5 | 31 | F | Breast | F-DFSP | PDGFB | |
| B6 | 45 | F | Breast | DFSP | PDGFB | |
| B7 | 48 | F | Chest wall | DFSP | PDGFB | |
| B8 | 38 | F | Breast | DFSP | PDGFB | |
| B9 | 44 | F | Breast | DFSP | PDGFB | |
| B10 | 36 | F | Breast | DFSP | PDGFB | |
| B11 | 40 | F | Breast | DFSP | PDGFB | |
| B12 | 31 | F | Breast | DFSP | PDGFB | |
| B13 | 26 | F | Breast | DFSP | PDGFB | |
| B14 | 34 | F | Breast | DFSP | PDGFB | |
| B15 | 36 | F | Chest wall | DFSP | Negative |
Cohort A: Prospective molecular testing of patients diagnosed with dermatofibrosarcoma protuberans over approximately 1 year. Cohort B: Retrospective molecular testing of patients diagnosed with dermatofibrosarcoma protuberans arising specifically on the breast or upper chest over approximately one decade. Novel variant highlighted in bold.
Abbreviations: DFSP, dermatofibrosarcoma protuberans; F, fibrosarcomatous; P, pigmented; F, female; L, left; M, male; FISH, fluorescence in situ hybridization.
Primary leg with lung metastasis.
2.4 |. Fluorescence in situ hybridization
Fluorescence in situ hybridization for PDGFB and PDGFD was performed as previously described in detail (Supporting Information Table S2).17 Custom bacterial artificial chromosome (BAC) clone probes flanking the target genes were designed using the UCSC genome browser (http://genome.ucsc.edu), and obtained from BAC-PAC sources of Children’s Hospital of Oakland Research Institute (Oakland, CA; http://bacpac.chori.org). DNA from each BAC probe was labeled with fluorochromes by nick translation. Formalin-fixed paraffin-embedded tissue sections were deparaffinized, pretreated, and then hybridized with the denatured probes. Following overnight incubation, the slides were rinsed, stained with 4′,6-diamidino-2-phenylindole (DAPI), mounted, and analyzed using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany).
3 |. RESULTS
3.1 |. Dermatofibrosarcoma protuberans with the novel COL6A3-PDGFD fusion variant has a predilection for breast
Following discovery of a novel COL6A3-PDGFD fusion variant (Patient A1), 62 subsequent cases of dermatofibrosarcoma protuberans were molecularly investigated (N = 63; RNAseq = 43, FISH = 20) over approximately a 1-year interval (Cohort A). The average patient age was 43 years (range, 16–83). There were 37 females and 26 males (ratio, 1.4:1). Common sites of involvement included: back (N = 9), abdomen (N = 7) and groin (N = 5), chest (N = 7) and breast (N = 7), upper arm (N = 7), and thigh (N = 6) (Table 1). Fifteen cases had fibrosarcomatous transformation, and two were pigmented.
Of the 63 tumors examined, suitable RNA and/or DNA were obtained in all cases (100%). Four (6.3%) tumors, including the index patient, contained PDGFD rearrangement; in 3 cases testing was performed by RNA-Seq, which revealed a novel COL6A3-PDGFD fusion variant. Fifty-eight patients (92%) possessed conventional PDGFB rearrangement; in the samples tested by RNA-Seq, all were found to contain the canonical COL1A1-PDGFB fusion product (Table 1). A single tumor lacked any know fusion gene by RNA-Seq.
Having observed that all four cases with PDGFD rearrangement arose on the breast, FISH for both PDGFB and PDGFD was subsequently performed on a second cohort of dermatofibrosarcoma protuberans specifically arising on the breast or upper chest (Cohort B). A total of 15 cases were identified. All were females; and, the average age was 41 years (range, 26–79). Two additional cases were found to harbor PDGFD rearrangement (13.3%); 12 patients possessed conventional PDGFB rearrangement (80%) and one patient was negative for both PDGFB and PDGFD rearrangement. Overall, based on the combined cohorts, a total of six patients were found to have the variant PDGFD fusion partner; each case assessed by RNA-Seq identified COL6A3 as the fusion partner. All PDGFD gene rearrangements detected by FISH had a balanced pattern, with no associated losses or gains/amplifications being noted.
3.2 |. Dermatofibrosarcoma protuberans with the COL6A3-PDGFD fusion variant has typical histologic and immunohistochemical features, but often lacks dermal involvement
All six tumors containing PDGFD rearrangement occurred in females, with an average age of 39 years (range, 22–58)—all arose on the breast. Clinically, none were reported to be protuberant and the average size was 1.5 cm (range, 1.0–2.1 cm). Morphologically, most of the cases were centered in subcutaneous fat. Epidermis was represented in only a single case, and noted to be widely uninvolved; a single case involved dermal collagen (Patient B2); and, a single case infiltrated around breast glandular epithelium (Patient A2). A single case contained a prominent ectatic thin-walled vasculature (Patient B1). In each of the tumors the morphology was similar to those with conventional PDGFB rearrangement. The tumors were comprised of sheets of spindle cells with a prominent storiform pattern, and frequently contained peripheral interdigitation amongst lobules of adipose tissue yielding the so-called “honeycomb” pattern. The cytoplasm was scant and palely eosinophilic to amphophilic. The nuclei were ovoid, and variably elongated and monomorphic; mitotic activity averaged 0.8 per 10 HPFs (range, 0–2 per 10 HPFs). None of the cases showed fibrosarcomatous transformation, pigmentation or myoid nodules.
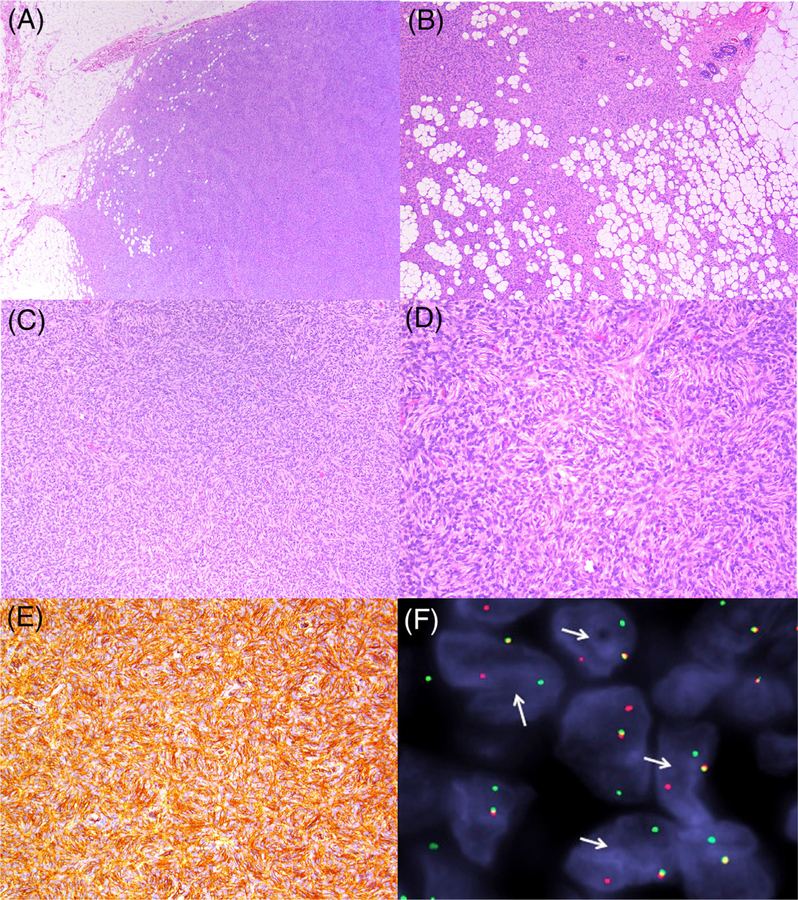
The immunophenotype was likewise similar to conventional dermatofibrosarcoma protuberans. All of the cases diffusely (5+) expressed CD34 (Figure 1); they were negative for desmin, smooth muscle actin, S100, and keratin.
FIGURE 1.
Representative images from dermatofibrosarcoma protuberans with COL6A3-PDGFD fusion gene product: (A) spindle cell neoplasm with relatively well-demarcated peripheral border, and tracking along fibrous septa (patient A4). (B) Tumor infiltrating adipose tissue with a “honeycomb” pattern with involvement, and sparing of glandular epithelium (patient A2). (C) Sheets of spindle cells with a prominent storiform pattern (patient A1). (D) Cells with ample eosinophilic cytoplasm, and bland elongated ovoid nuclei (patient A1). (E) Immunoreactivity for CD34 (patient A2). (F) Fluorescence in situ hybridization showing break-apart signal for PDGFD (arrows; patient A1) [Color figure can be viewed at wileyonlinelibrary.com]
The two cases lacking a fusion gene were likewise morphologically and immunohistochemically typical of dermatofibrosarcoma protuberans.
3.3 |. Dermatofibrosarcoma protuberans with the COL6A3-PDGFD fusion variant clusters together with COL1A1-PDGFB fusion positive tumors, and shows upregulation of PDGFRB expression
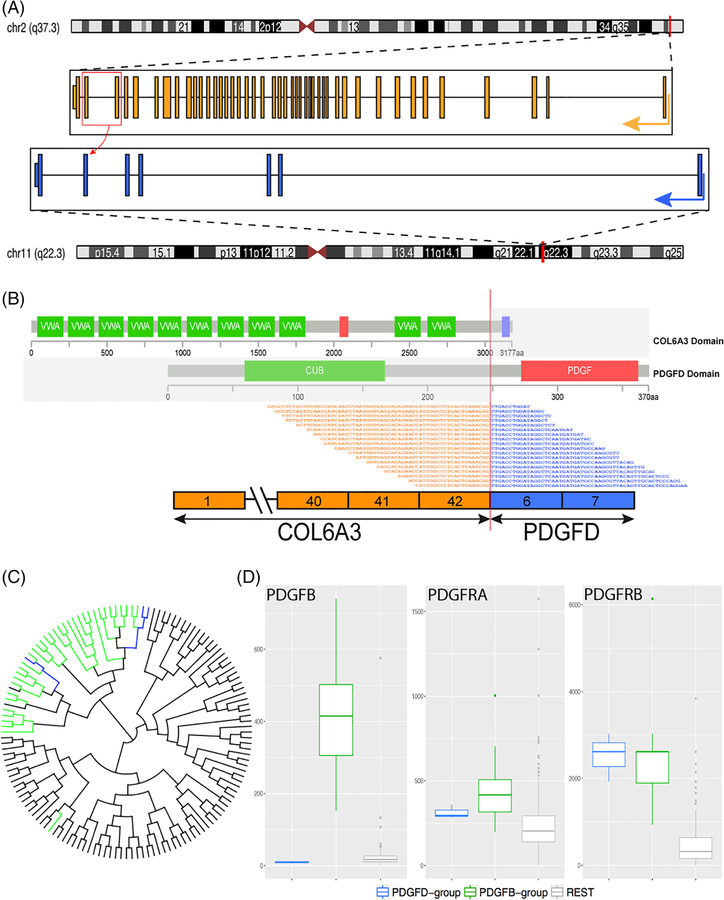
Among the six cases positive for the PDGFD rearrangement, three were tested by RNA-Seq, which showed breakpoints in COL6A3 exon 42 (Patients A1 and A3) and exon 43 (Patient A2) (NCBI Reference Sequence: NM_004369.3) which were fused to PDGFD exon 6 (NM_025208.4) (Figure 2A,B). Rearrangement of PDGFD was independently confirmed by fluorescence in situ hybridization in all six cases (Figure 1). No unbalanced PDGFD rearrangements or amplifications were noted.
FIGURE 2.
COL6A3-PDGFD fusion structure and molecular correlates. (A) Diagrammatic representation of COL6A3 gene on 2q37.3 fused to PDGFD gene on 11q22.3. Red box and curved arrow depict 2 fusion transcript variants in which COL6A3 exons 42 and 43, respectively, are fused with exon 6 of PDGFD. (B) Representative sequences from RNAseq data demonstrating the fusion of COL6A3 exon 42 was fused to exon 6 of PDGFD, with emphasis on the protein domains projected to be retained in the fusion oncoprotein. (C) Unsupervised hierarchical clustering of RNAseq data showing that the 3 COL6A3-PDGFD fusion positive tumors (blue) cluster together with the 34 cases of dermatofibrosarcoma protuberans (green) with the classic COL1A1-PDGFB fusion, and away from other sarcoma types (gray) studied on the same targeted RNAseq platform. (D, left) box plot showing high levels of expression of PDGFB mRNA in the control group (green) compared with the study group (blue) and other sarcomas (gray); (D, center) PDGFRA levels are not increased in any tumor category, while (D, right) PDGFRB mRNA is upregulated in both PDGFD (blue) and PDGFB (green) fusion positive DFSP compared with other sarcoma types (gray) [Color figure can be viewed at wileyonlinelibrary.com]
By unsupervised hierarchical clustering using RNA-Seq data, the three cases with the COL6A3-PDGFD fusion clustered amongst the larger group of dermatofibrosarcoma protuberans with the typical COL1A1-PDGFB fusion, and away from other soft tissue sarcomas available on the same targeted RNA sequencing platform (Figure 2C). Specifically, transcripts were selected above a 20 RPKM threshold expression (calculated by mrfQuantifier), followed by ward.D2 hierarchical cluster analysis and visualized on Dendroscope. The PDGFD gene was not one of the genes represented in our RNA sequencing panel, thus we were not able to evaluate the PDGFD mRNA expression in our cohort. As expected, PDGFB-positive tumors showed overexpression of PDGFB (Figure 2D), while PDGFD-fusion positive tumors did not. However, both fusion-variants showed increased mRNA expression of PDGFRB, while no significant increase of PDGFRA was present (Figure 2D) compared with other sarcoma types.
4 |. DISCUSSION
Dermatofibrosarcoma protuberans is a relatively common superficial sarcoma, which involves diverse anatomic sites and patients across a broad age range. Tumors have a characteristic clinical and histopathologic appearance. Moreover, to date, almost all molecularly characterized cases have been found to possess a COL1A1-PDGFB fusion gene. Herein, we report a novel COL6A3-PDGFD fusion variant in dermatofibrosarcoma protuberans, which appears to have a predilection for breast.
Darier and Ferrand,18 in 1924, are credited with first describing dermatofibrosarcoma protuberans, although earlier reports of the lesion appear to exist.19 Tumors have long been recognized for their distinct clinical presentation and locally aggressive course2,3; the latter initially prompted debate as to whether this merited designation as a true malignancy.3 While the characteristic storiform morphology, adnexal-sparing, and “honeycomb” pattern of subcutaneous infiltration has long-been appreciated,2,3 it was not until the early 1990s that immunoreactivity for CD34 was recognized as distinguishing dermatofibrosarcoma protuberans from related neoplasms.11 Early cytogenetic studies described a supernumerary ring chromosome in a subset of cases20; this subsequently led to the identification of the t(17;22)(q22; q13) event.21–23 Ultimately the COL1A1-PDGFB fusion gene was cloned, which fairly consistently pairs exon 2 of PDGFB with numerous potential exons of COL1A1.13 This fusion product has been identified in 85%−96% of cases.24–30 The incidence appears to increase with the application of more sensitive detection assays; nevertheless, this still implies that a subset of cases may have an alternate fusion gene. This is supported by cytogenetic studies which raise the possibility of other translocation events, including: t(2;17)(q33;q25),31 t (9;22)(q32;q12.2),32 t(5;8)(q13–14;p21),33 and t(X;7)(q21.2;q11.2).34 None of these fusions encompass PDGFD or COL6A3, suggesting these may represent additional potential fusion partners in dermatofibrosarcoma protuberans. Recently an alternative PDGFB fusion partner was identified—COL1A2-PDGFB in a tumor that was otherwise typical of dermatofibrosarcoma protuberans.29
In the routine work-up of a case of dermatofibrosarcoma protuberans using RNA-Seq we discovered a novel COL6A3-PDGFD fusion variant. We proceeded to test a cohort of 63 patients, identifying PDGFD rearrangement in four (6.3%) tumors, while 58 (92%) were confirmed to possess the canonical PDGFB rearrangement (Table 1). Rather surprisingly, all four patients with the variant fusion were women, and each tumor originated from the breast as a non-protuberant, but palpable, subcutaneous mass. As a result, a second cohort of patients—specifically, those with dermatofibrosarcoma protuberans occurring on the breast/chest—was investigated to determine whether this anatomic site may be enriched for the variant PDGFD fusion. A total of 15 additional cases were examined, with two additional cases found to harbor PDGFD rearrangement (13.3%) (Table 1); 12 contained conventional PDGFB rearrangement (80%); and, one patient was negative for both PDGFB and PDGFD rearrangement. All 6 patients with the PDGFD variant fusion were female (100%) and all tumors were located on the breast (100%), predominantly the left (83%). Three of these cases were tested by RNA-Seq, which established COL6A3 as the PDGFD fusion partner. Each of these six cases was morphologically typical of dermatofibrosarcoma protuberans, though dermal involvement could only be documented in a single case. All cases had prominent storiform growth, infiltration of subcutaneous adipose tissue with a “honeycomb” pattern, and minimal nuclear atypia and mitotic activity; all of the cases exhibited diffuse CD34 expression (Figure 1). In addition to facilitating discovery of the variant COL6A3-PDGFD fusion gene, RNA-Seq in this series identified two cases with PDGFB rearrangement that were found to be negative by conventional FISH testing (Patients A6 and A7). This suggests a possible limitation to FISH testing in a minority of cases.
The COL1A1-PDGFB fusion is sufficiently prevalent in dermatofibrosarcoma protuberans that it has led to the suggestion that, in many cases, the absence of this event implies an alternative diagnosis.33 Variant fusions are nevertheless recognized, and with the report of a case with a variant COL1A2-PDGFB fusion it was proposed that constitutive expression of PDGFB is the fundamental mechanism of tumorigenesis in dermatofibrosarcoma protuberans.29 To our knowledge, the COL6A3-PDGFD fusion gene has not previously been characterized, and its relationship to the conventional COL1A1-PDGFB fusion gene is unclear. COL6A3 encodes collagen type VI alpha 3 chain, a microfibrillar component of the extracellular matrix, which is widely present in most connective tissues.35 Fusions involving COL6A3 are known to occur in other neoplasms, notably tenosynovial giant cell tumor, which frequently contains a COL6A3-CSF1 fusion36; in this context, the role of COL6A3 is believed to be that of a strong promoter.37 PDGFD is a member of the platelet-derived growth factor family, a family of disulfide-bonded dimeric isoforms, also known as a mesenchymal growth factor38,39; recently, it has been proposed to have an important role in so-called “epithelial mesenchymal transition.”40 The chimeric proteins arising from COL6A3-PDGFD and COL1A1-PDGFB fusion genes both retain the PDGF domain13,41; and, similar to PDGFB,42 PDGFD binds and activates PDGFR-β38,43; This suggests that, despite a different ligand arising with the variant PDGFD chimera, it may nevertheless result in a similar pattern of autocrine activation via PDGFRB receptor tyrosine kinase signaling as conventional PDGFB. Moreover, by RNA-Seq, tumors with COL6A3-PDGFD fusions show upregulated mRNA expression of PDGFRB.
In summary, we report six novel cases of dermatofibrosarcoma protuberans containing PDGFD rearrangement; RNA-Seq testing in three cases identified COL6A3 as the fusion partner. Whether COL6A3 represents the fusion partner in all PDGFD variants, or whether other genes, such as COL1A1 or COL1A2, are capable of pairing with PDGFD remains to be determined. All cases occurred on the breast of females; nevertheless, in view of the limited size of our initial cohort, this anatomic and sex restriction is perhaps merely coincidental. In each case the morphology and immunophenotype was indistinguishable from those with the conventional COL1A1-PDGFB fusion gene. The only potential difference being relatively infrequent dermal involvement—the tumors appeared largely centered within the subcutis or superficial breast parenchyma. By targeted RNA-Seq the tumors clustered within the same genomic group as dermatofibrosarcoma protuberans containing the conventional COL1A1-PDGFB fusion, suggesting a similar expression profile. With sufficient tissue the diagnosis of dermatofibrosarcoma protuberans is generally straightforward, and predicated on the basis of morphology and immunoreactivity with CD34. In some instances, however, as with the case in limited sampling and/or unusual variants, the diagnosis can be more challenging. Rigorous molecular characterization facilitates both the accurate diagnosis of these neoplasms, as well as an improved understanding of the molecular and biological mechanisms of their tumorigenesis.33 Furthermore, similar to dermatofibrosarcoma protuberans with conventional PDGFB fusions, the variant PDGFD fusion group also showed up-regulation of PDGFRB, which suggests that therapeutic targeting with tyrosine kinase inhibitors, such as imatinib, might be an option in locally advanced or metastatic lesions.44–46
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Ms. Evangeline Agro and Ms. Sharon Crafter for facilitating molecular testing, and to Ms. Grace Murray (Illumina) for providing RNA-Seq assays.
Funding information
Kristin Ann Carr Foundation (CRA), Grant/Award Number: P30-CA008748 (CRA); Cycle for Survival (CRA), Grant/Award Number: P50 CA 140146–01 (CRA)
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- 1.Mentzel T, Pedeutour F, Lazar A, Coindre JM. Dermatofibrosarcoma protuberans In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, eds. WHO Classification of Tumours of Soft Tisue and Bone. Lyon: International Agency for Research on Cancer; 2013:77–79. [Google Scholar]
- 2.Taylor HB, Helwig EB. Dermatofibrosarcoma protuberans. A study of 115 cases. Cancer. 1962;15:717–725. [DOI] [PubMed] [Google Scholar]
- 3.Burkhardt BR, Soule EH, Winkelmann RK, Ivins JC. Dermatofibrosarcoma protuberans. Study of fifty-six cases. Am J Surg. 1966;111(5): 638–644. [DOI] [PubMed] [Google Scholar]
- 4.Bednar B Storiform neurofibromas of the skin, pigmented and nonpigmented. Cancer. 1957;10(2):368–376. [DOI] [PubMed] [Google Scholar]
- 5.Abdul-Karim FW, Evans HL, Silva EG. Giant cell fibroblastoma: a report of three cases. Am J Clin Pathol. 1985;83(2):165–170. [DOI] [PubMed] [Google Scholar]
- 6.Shmookler BM, Enzinger FM, Weiss SW. Giant cell fibroblastoma. A juvenile form of dermatofibrosarcoma protuberans. Cancer. 1989; 64(10):2154–2161. [DOI] [PubMed] [Google Scholar]
- 7.Reimann JD, Fletcher CD. Myxoid dermatofibrosarcoma protuberans: a rare variant analyzed in a series of 23 cases. Am J Surg Pathol. 2007; 31(9):1371–1377. [DOI] [PubMed] [Google Scholar]
- 8.Calonje E, Fletcher CD. Myoid differentiation in dermatofibrosarcoma protuberans and its fibrosarcomatous variant: clinicopathologic analysis of 5 cases. J Cutan Pathol. 1996;23(1):30–36. [DOI] [PubMed] [Google Scholar]
- 9.Mentzel T, Beham A, Katenkamp D, Dei Tos AP, Fletcher CD. Fibrosarcomatous (“high-grade”) dermatofibrosarcoma protuberans: clinicopathologic and immunohistochemical study of a series of 41 cases with emphasis on prognostic significance. Am J Surg Pathol. 1998; 22(5):576–587. [DOI] [PubMed] [Google Scholar]
- 10.Swaby MG, Evans HL, Fletcher CD, et al. Dermatofibrosarcoma protuberans with unusual sarcomatous transformation: a series of 4 cases with molecular confirmation. Am J Dermatopathol. 2011;33(4): 354–360. [DOI] [PubMed] [Google Scholar]
- 11.Aiba S, Tabata N, Ishii H, Ootani H, Tagami H. Dermatofibrosarcoma protuberans is a unique fibrohistiocytic tumour expressing CD34. Br J Dermatol. 1992;127(2):79–84. [DOI] [PubMed] [Google Scholar]
- 12.Goldblum JR. CD34 positivity in fibrosarcomas which arise in dermatofibrosarcoma protuberans. Arch Pathol Lab Med. 1995;119(3): 238–241. [PubMed] [Google Scholar]
- 13.Simon MP, Pedeutour F, Sirvent N, et al. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15(1):95–98. [DOI] [PubMed] [Google Scholar]
- 14.Dickson BC, Sung YS, Rosenblum MK, et al. NUTM1 gene fusions characterize a subset of undifferentiated soft tissue and visceral tumors. Am J Surg Pathol. 2018;42(5):636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Tsai WH, Ding Y, et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-seq data. Nucleic Acids Res. 2016;44(5):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. [DOI] [PubMed] [Google Scholar]
- 17.Kao YC, Sung YS, Zhang L, et al. EWSR1 fusions with CREB family transcription factors define a novel Myxoid Mesenchymal tumor with predilection for intracranial location. Am J Surg Pathol. 2017;41(4): 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darier J, Ferrand M. Dermatofibromes progressifs et récidivants ou fibrosarcomes de la peau. Ann Dermatol Syphilograph. 1924;5: 545–562. [Google Scholar]
- 19.Mopper C, Pinkus H. Dermatofibrosarcoma protuberans. Am J Clin Pathol. 1950;20(2):171–176. [DOI] [PubMed] [Google Scholar]
- 20.Bridge JA, Neff JR, Sandberg AA. Cytogenetic analysis of dermatofibrosarcoma protuberans. Cancer Genet Cytogenet. 1990;49(2): 199–202. [DOI] [PubMed] [Google Scholar]
- 21.Minoletti F, Miozzo M, Pedeutour F, et al. Involvement of chromosomes 17 and 22 in dermatofibrosarcoma protuberans. Genes Chromosomes Cancer. 1995;13(1):62–65. [DOI] [PubMed] [Google Scholar]
- 22.Pedeutour F, Simon MP, Minoletti F, et al. Translocation, t(17;22)(q22; q13), in dermatofibrosarcoma protuberans: a new tumor-associated chromosome rearrangement. Cytogenet Cell Genet. 1996;72(2–3): 171–174. [DOI] [PubMed] [Google Scholar]
- 23.Craver RD, Correa H, Kao YS, Van Brunt T, Golladay ES. Aggressive giant cell fibroblastoma with a balanced 17;22 translocation. Cancer Genet Cytogenet. 1995;80(1):20–22. [DOI] [PubMed] [Google Scholar]
- 24.Sirvent N, Maire G, Pedeutour F. Genetics of dermatofibrosarcoma protuberans family of tumors: from ring chromosomes to tyrosine kinase inhibitor treatment. Genes Chromosomes Cancer. 2003; 37(1):1–19. [DOI] [PubMed] [Google Scholar]
- 25.Patel KU, Szabo SS, Hernandez VS, et al. Dermatofibrosarcoma protuberans COL1A1-PDGFB fusion is identified in virtually all dermatofibrosarcoma protuberans cases when investigated by newly developed multiplex reverse transcription polymerase chain reaction and fluorescence in situ hybridization assays. Hum Pathol. 2008; 39(2):184–193. [DOI] [PubMed] [Google Scholar]
- 26.Segura S, Salgado R, Toll A, et al. Identification of t(17;22)(q22;q13) (COL1A1/PDGFB) in dermatofibrosarcoma protuberans by fluorescence in situ hybridization in paraffin-embedded tissue microarrays. Hum Pathol. 2011;42(2):176–184. [DOI] [PubMed] [Google Scholar]
- 27.Salgado R, Llombart B, Pujol MR, et al. Molecular diagnosis of dermatofibrosarcoma protuberans: a comparison between reverse transcriptase-polymerase chain reaction and fluorescence in situ hybridization methodologies. Genes Chromosomes Cancer. 2011;50(7): 510–517. [DOI] [PubMed] [Google Scholar]
- 28.Ha SY, Lee SE, Kwon MJ, et al. PDGFB rearrangement in dermatofibrosarcoma protuberans: correlation with clinicopathologic characteristics and clinical implications. Hum Pathol. 2013;44(7): 1300–1309. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura I, Kariya Y, Okada E, et al. A novel chromosomal translocation associated with COL1A2-PDGFB gene fusion in Dermatofibrosarcoma Protuberans: PDGF expression as a new diagnostic tool. JAMA Dermatol. 2015;151(12):1330–1337. [DOI] [PubMed] [Google Scholar]
- 30.Karanian M, Perot G, Coindre JM, et al. Fluorescence in situ hybridization analysis is a helpful test for the diagnosis of dermatofibrosarcoma protuberans. Mod Pathol. 2015;28(2):230–237. [DOI] [PubMed] [Google Scholar]
- 31.Sinovic J, Bridge JA. Translocation (2;17) in recurrent dermatofibrosarcoma protuberans. Cancer Genet Cytogenet. 1994;75(2): 156–157. [DOI] [PubMed] [Google Scholar]
- 32.Sonobe H, Furihata M, Iwata J, et al. Dermatofibrosarcoma protuberans harboring t(9;22)(q32;q12.2). Cancer Genet Cytogenet. 1999; 110(1):14–18. [PubMed] [Google Scholar]
- 33.Bianchini L, Maire G, Guillot B, et al. Complex t(5;8) involving the CSPG2 and PTK2B genes in a case of dermatofibrosarcoma protuberans without the COL1A1-PDGFB fusion. Virchows Arch. 2008;452(6): 689–696. [DOI] [PubMed] [Google Scholar]
- 34.Craver RD, Correa H, Kao Y, Van Brunt T. Dermatofibrosarcoma protuberans with 46,XY,t(X;7) abnormality in a child. Cancer Genet Cytogenet. 1995;80(1):75–77. [DOI] [PubMed] [Google Scholar]
- 35.Bonnemann CG. The collagen VI-related myopathies: muscle meets its matrix. Nat Rev Neurol. 2011;7(7):379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.West RB, Rubin BP, Miller MA, et al. A landscape effect in tenosynovial giant-cell tumor from activation of CSF1 expression by a translocation in a minority of tumor cells. Proc Natl Acad Sci U S A. 2006; 103(3):690–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moller E, Mandahl N, Mertens F, Panagopoulos I. Molecular identification of COL6A3-CSF1 fusion transcripts in tenosynovial giant cell tumors. Genes Chromosomes Cancer. 2008;47(1):21–25. [DOI] [PubMed] [Google Scholar]
- 38.Bergsten E, Uutela M, Li X, et al. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nat Cell Biol. 2001;3(5):512–516. [DOI] [PubMed] [Google Scholar]
- 39.Huang W, Fridman Y, Bonfil RD, et al. A novel function for platelet-derived growth factor D: induction of osteoclastic differentiation for intraosseous tumor growth. Oncogene. 2012;31(42): 4527–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Q, Hou X, Xia J, et al. Emerging roles of PDGF-D in EMT progression during tumorigenesis. Cancer Treat Rev. 2013;39(6):640–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greco A, Fusetti L, Villa R, et al. Transforming activity of the chimeric sequence formed by the fusion of collagen gene COL1A1 and the platelet derived growth factor b-chain gene in dermatofibrosarcoma protuberans. Oncogene. 1998;17(10):1313–1319. [DOI] [PubMed] [Google Scholar]
- 42.Simon MP, Navarro M, Roux D, Pouyssegur J. Structural and functional analysis of a chimeric protein COL1A1-PDGFB generated by the translocation t(17;22)(q22;q13.1) in Dermatofibrosarcoma protuberans (DP). Oncogene. 2001;20(23):2965–2975. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Eriksson U. Novel PDGF family members: PDGF-C and PDGF-D. Cytokine Growth Factor Rev. 2003;14(2):91–98. [DOI] [PubMed] [Google Scholar]
- 44.Greco A, Roccato E, Miranda C, Cleris L, Formelli F, Pierotti MA. Growth-inhibitory effect of STI571 on cells transformed by the COL1A1/PDGFB rearrangement. Int J Cancer. 2001;92(3):354–360. [DOI] [PubMed] [Google Scholar]
- 45.Sjoblom T, Shimizu A, O’Brien KP, et al. Growth inhibition of dermatofibrosarcoma protuberans tumors by the platelet-derived growth factor receptor antagonist STI571 through induction of apoptosis. Cancer Res. 2001;61(15):5778–5783. [PubMed] [Google Scholar]
- 46.Maki RG, Awan RA, Dixon RH, Jhanwar S, Antonescu CR. Differential sensitivity to imatinib of 2 patients with metastatic sarcoma arising from dermatofibrosarcoma protuberans. Int J Cancer. 2002;100(6): 623–626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.