Abstract
We report on the first detection and isolation of B. pseudohinzii (Bordetella pseudohinzii) in laboratory mice in China. Forty‐one B. pseudohinzii strains were isolated from 3094 mice in 33 different laboratory animal facilities in southern China. The isolates were identified through culture and genome sequenceing. Phylogenetic analysis based on the sequences of 16S rRNA and OmpA genes demonstrated that these strains were on the same clade as other B. pseudohinzii strains isolated from mice. Experimental infected mice presented an asymptomatic infection. B. pseudohinzii replicated in both the respiratory tract and the digestive tract. Most importantly B. pseudohinzii shed via feces and infected a group of sentinel mice in a separate cage via cage padding contaminated with B. pseudohinzii‐positive feces, indicating that B. pseudohinzii could transmit efficiently among mice and contaminate environmental facilities. Our study highlights the importance of routine monitoring of the pathogen in laboratory mice and provides vital insights into the transmission of Brodetellae in rodents and human.
Keywords: Bordetella pseudohinzii, isolation, laboratory mice, transmission
1. INTRODUCTION
The genus Bordetella consists of nine species that could infect a variety of hosts, including humans, birds and rodents.1, 2, 3, 4 B. pertussis, the agent of whooping cough in humans, together with B. parapertussis and B. bronchiseptica, are commonly classified as “classical” Bordetella.5 The other six species (Bordetella holmesii, Bordetella trematum, Bordetella avium, Bordetella petrii, Bordetella hinzii and Bordetella pseudohinzii) are classified as “non‐classical” Bordetella.1, 6
Recently, B. pseudohinzii has been identified and isolated from laboratory‐raised mice.7, 8 Whole‐genome analysis has demonstrated that B. pseudohinzii shares high similarity with B. hinzii, which is a causuative agent of respiratory disease in poultry and is associated with several cases of human infection.3, 8, 9, 10 There are 3206 genes present in the genomes of both B. pseudohinzii and B. hinzii, with 570 genes being specific to B. hinzii, and 390 genes being specific to B. pseudohinzii. Therefore, routine diagnostic tests were unable to distinguish between them. It has been reported that B. hinzii has been detected in experimental facilities and is associated with histopathological changes in the lung.4, 11, 12 In light of the similarity between the two species, it has been suggested that the pathogen observed in these studies was actually B. pseudohinzii.4, 8
B. pseudohinzii has been detected and isolated in laboratory mice facilities across the world, including the United States, Japan, Malaysia and Germany.6, 7, 12, 13 This pathogen will cause immunocompetent mice to have a subclinical infection and immunodeficient mice to have rhinitis or pneumonia respiratory symptoms. The current FELASA (Federation for Laboratory Animal Science Associations) recommendations for the health monitoring of mouse colonies in experimental units do not list B. pseudohinzii in their health report form,14 but the The American Association for Laboratory Animal Science (AALAS)/FELASA working group refers to B. hinzii as an “exotic agent” that “should be mentioned when found”.15 Studies using mice as the animal model in China are increasing, but the infection status of B. pseudohinzii in the laboratory animal facilities in China had never been investigated. In the present study, B. pseudohinzii was screened from a total of 3094 mice from 33 facilities and isolated. The characterization of B. pseudohinzii was also studied.
2. MATERIALS AND METHODS
2.1. Sample collection and bacterial isolation
A total of 3094 respiratory secretions were collected from mice from 33 different laboratory animal facilities in Southern China from 2015 to 2018. The specimens were transported on ice to the laboratory within 24 hours of sampling. The samples were inoculated on trypticase soy agar supplemented with 5% sheep blood and cultured under anaerobic conditions at 37°C for 24‐48 hours. The isolates were gram stained, examined by microscope and identified using the ATB Expression microbe identification system (BioMérieux).
2.2. 16S rRNA gene and OmpA gene sequencing and phylogenetic analysis
Seven clinical isolates from different experimental animal facilities were randomly selected for genetic analysis. The genomic DNA was extracted using the bacterial genomic DNA extraction kit (Tiangen Bio‐chemical Technology) according to the manufacturer's protocol. The 16S rRNA gene was amplified using universal primers (forward primer: 5'‐AGAGTTTGATCMTGGCTCAG‐3', reverse primer: 5'‐TACGGYTACCTTGTTACGACTT‐3'). The partial B. pseudohinzii outer membrane protein A (OmpA) gene was amplified using primer pairs as previously described.6 All the primers were synthesized by Sangon Biotech. The PCR conditions were set up in a 50 μL volume containing 5 μL of genomic DNA, 0.4 µmol/L of each primer, 25 μL of Premix ExTaq (Takara, Japan) and 19.4 μL of distilled water as follows: pre‐denaturation at 95°C for 3 minutes; 30 cycles of denaturation at 95°C for 30 seconds; annealing at 58°C for 30 seconds, extension at 72°C for 60 seconds; and finally 72°C for 10 minutes. Then 8 μL of the PCR product was subjected to electrophoresis on a 2% agarose gel. The PCR product was recovered using a DNA purification kit and ligated to the pMDl9‐T vector (Takara, Japan). The positive clones were sequenced. A phylogenetic tree was generated by Mega5 software using the neighbor‐joining method. The robustness of the hypothesis was tested with 1000 nonparametric bootstrap analyses.
2.3. Experimental infection of B. pseudohinzii in mice
The sequences of the OmpA gene in the seven clinical isolates were 100% identical with each other. So one isolate of the seven B. pseudohinzii strains was randomly selected for the experimental infection. The animal infection experiment was approved by the Institutional Animal Care and Use Committee of Guangdong Laboratory Animals Monitoring Institute. Eight‐week‐old SPF female SPF ICR mice were obtained from a commercial supplier (Guangdong Medical Laboratory Animal Center). All the mice were determined to be B. pseudohinzii‐negative by PCR. The mice were randomly divided into 4 groups: inoculated, sentinel, cohabiting, and control. Twenty‐one mice served as the inoculated group. Each mouse in this group was inoculated with 30 μL bacterial suspension at a concentration of 3.0 × 108 CFU/mL. The mice in the control group were inoculated with 30 μL PBS. The animals were observed daily for clinical signs including oral and nasal discharge and action state. To determine the replication of B. pseudohinzii in mice, three animals in the inoculated group were euthanized on each of days 2, 5, 9, 12, 16, 19 and 21 post inoculation (PI). Immediately following euthanasia, a nasal swab, trachea, lung, and cecal contents were collected from each animal. Samples of the corncob padding contaminated with feces from the animals’ caging, and swabs of cage inner walls and outlets were also collected at the same time. All the samples were prepared as homogenates with PBS and the presence of B. pseudohinzii was determined using PCR. When the dirty padding from the inoculated group was identified as positive for B. pseudohinzii by PCR, a single sample of 50 mL of the dirty padding was moved to the sentinel group cage containing 15 mice, and another 15 SPF mice were move into the inoculated group as the cohabiting group. Three mice from each group were euthanized on each of days 7, 14, 21, 28 and 35 post inoculation (PI). The day on which the dirty padding from the inoculated group was identified as B. pseudohinzii positive by PCR was considered as the time of inoculation for the sentinel and cohabiting groups. PI tissue samples were harvested and PCR assays for sequencing the OmpA gene were conducted as above. Two control mice were sacrified on each of days 14, 21, 28 and 35 post inoculation (PI) and tested for B. pseudohinzii by PCR.
3. RESULTS AND DISCUSSION
Bacteriologic cultures of respiratory secretions collected from 3094 mice yielded various kinds of colonies with different morphologies. Hundreds of colonies were isolated from the samples after 24 hours of incubation on trypticase soy agar supplemented with 5% sheep blood. One type of colony displayed a special morphology that was different from that of the common bacteria. These white, round colonies were apparent after 24 hours of incubation under aerobic conditions. They were of medium sized (1‐2 mm), translucent, smooth, convex, and without hemolysis (Figure 1A). Microscopic examination demonstrated that the colonies were gram‐negative, short rod‐shaped bacteria with densely stained poles (Figure 1B). The API 20NE commercial identification kit (bioMérieux) gave the numerical code 0000067, suggesting with a high level of confidence (96.7%) that the bacteria were Bordetella avium. A total of 41 isolates with the same morphology were recovered from tracheal swabs as the dominant organism. All the 41 isolated strains exhibited a consistent profile: code 0000067 on API 20NE.
Figure 1.
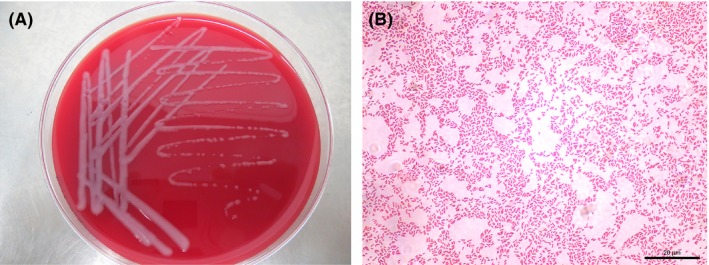
Colony morphology and microscopic examination. A, Colony morphology of one isolated strain cultured on 5% sheep blood tryptone soybean agar medium. B, Gram staining and microscopic examination
To further determine the characteristics of the clinical isolates, seven isolates were randomly selected for genome sequencing and analysis. The sequences of the OmpA genes were 100% identical with that of the previously reported B. pseudohinzii derived from mouse colonies in different countries.4, 7 In addition, the OmpA gene detected in this study had 98% nucleotide sequence homology to B. hinzii F582 and H568 isolated from humans, 97% homology to B. avium, 93% homology to pertussis, and 92% homology to bronchiseptica and parapertussis. A phylogenetic tree based on the sequences characterized all the seven clinical strains as “B. pseudohinzii”. All the isolates formed the same cluster in the OmpA phylogenetic tree (Figure 2A). To confirm the OmpA gene analysis, a phylogenetic analysis based on 16S rRNA sequences was performed and demonstrated that the seven isolates also formed the same group with another B. pseudohinzii strain H4681 and BH370 (Figure 2B). It also indicated that Bordetella hinzii strain 3224 was in the same cluster as B. pseudohinzii. Given the similarity between the two species, it has been suggested that Bordetella hinzii identified in previous reports was actually B. pseudohinzii.4, 8 The phylogenetic analysis performed in the present study also supports that conclusion. The 16S rRNA sequences of seven Bordetella pseudohinzii strains A1, A2, C1, F1, H3, H4 and J1 were deposited in GenBank under accession numbers MK072953 to MK072959, respectively.
Figure 2.
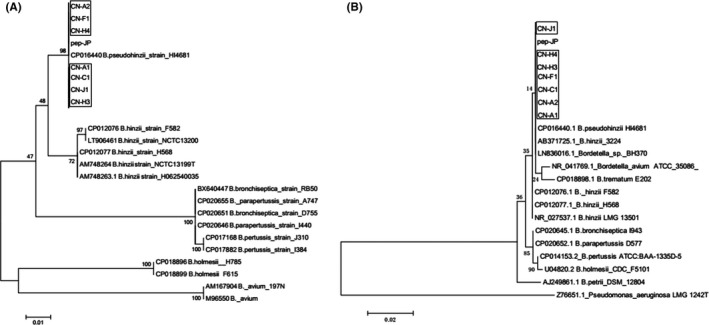
Phylogenetic analysis (neighbor‐joining) based on the outer membrane protein A (OmpA) gene sequence alignment (A) and 16S rRNA gene sequences (B) of seven isolates. Seven Bordetella pseudohinzii strains A1, A2, C1, F1, H3, H4 and J1 were deposited in GenBank under the accession numbers MK072953 to MK072959, respectively
To investigate the replication of B. pseudohinzii in mice, the tissues of the experimentally infected mice were examined for the presence of B. pseudohinzii using PCR. The detection results were summarized in Table 1. Experimental infected mice presented an asymptomatic infection. In the inoculation group, B. pseudohinzii could be detected as early as day 2 PI in the trachea and cecal contents and was consistently present in theses tissues at all the sampling time points throughout the study. The dirty padding was found to be positive on day 12 PI. The bacteria could also be detected in the nasal swabs and lung on days 12 PI, as well as in the trachea and cecal contents. The replication of B. pseudohinzii in trachea and lung of the mice has been demonstrated in previous studies.4, 6 Our novel finding in the present study was that the infected mice shed the bacteria in their feces which contaminated the cage padding. We therefore investigated fecal transmission and replication of B. pseudohinzii in separately caged sentinel mice that received a composite sample of contaminated bedding (bedding used in the inoculation group that tested positive for B. pseudohinzii). After 4 weeks of exposure to the dirty bedding, the sentinel mice were positive to B. pseudohinzii in all the samples tested (Table 1). The B. pseudohinzii‐positive bedding had thus become the new infection source. This result suggested that B. pseudohinzii can transmit through feces and infect other mice. The mice in the cohabiting group living in the same cage as the inoculation group tested positive for Bordetella pseudohinzii in trachea and nasal swabs at 28 days PI, and in all the samples at 35 days PI. All the samples collected from the control mice were negative for B. pseudohinzii throughout the study.
Table 1.
Pathogen detection in mice in three groups
| Days PI | Inoculated group | Sentinel group | Cohabiting group | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples | 2 | 5 | 9 | 12 | 16 | 19 | 21 | 7a | 14 | 21 | 28 | 35 | 7a | 14 | 21 | 28 | 35 |
| Nasal swab | − | − | + | + | + | + | + | − | − | − | + | + | − | − | − | + | + |
| Tracheal | + | + | + | + | + | + | + | − | − | + | + | + | − | − | − | + | + |
| Lung | − | − | − | + | + | + | + | − | − | − | + | + | − | − | − | − | + |
| Cecal contents | + | + | + | + | + | + | + | − | − | − | + | + | − | − | − | − | + |
| Dirty padding | − | − | − | + | + | + | + | − | − | − | + | + | − | − | − | − | + |
| Cage inner wall | − | − | − | + | + | + | + | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ |
| Outlet | − | − | − | − | − | − | − | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ | ∕ |
Dirty padding included feces. +: positive, −: negative, ∕: not determined.
When the dirty padding in the inoculated group was positive for B pseudohinzii by PCR on day 12 PI, a sample of padding was moved to the cage containing the sentinel mice. The day that dirty padding in the inoculated group was determined as positive was considered to be the day of inoculation for the sentinel and cohabiting groups.
Our study is the first to report a natural infection of Bordetella pseudohinzii in captive mouse colonies in China. The infection rate (41/3094) was 1.32%, which is consistent with another study reported in Japan.11 Furthermore, the bacteria could be detected in almost all the mice species studied, including C57BL/6 mice, BALB/c mice, ICR sentinel mice, immunodeficient mice, and transgenic mice. Ten out of 33 facilities had B. pseudohinzii contamination, and one facility tested positive for B. pseudohinzii 5 consecutive times in 3 years, indicating that B. pseudohinzii can be a long‐term contaminant in laboratory animal facilities, which should be addressed as a significant problem.
B. pseudohinzii is a non‐classical Brodetellae species similar to B. avium and B. hinzzi, which makes them hard to distinguish.8 Currently, the identification library of the commercial biochemical identification system contains only two Bordetella species profiles, B. avium and B. bronchiseptica. Thus B. pseudohinzii was identified as B. avium using the commercial kit. Confirmation of the isolates can be reliably performed by nucleotide sequencing. The phylogeny reconstructed using the sequences of 16S RNA and the OmpA gene characterized the isolates as B. pseudohinzii from the Bordetella genus.
Previous studies have demonstrated that B. pseudohinzii infects the respiratory tract of mice and has a potential negative effect on pulmonary research.4 B. pseudohinzii could be detected in the trachea, lung and cecal contents of the experimentally infected mice in the present study. Our results not only confirmed previous studies showing that B. pseudohinzii replicates in the respiratory tract, but also showed that the bacteria could replicate in the digestive tract and shed via feces. Separately caged sentinel mice exposed to the padding contaminated with the feces from the inoculated group tested positive to B. pseudohinzii in all the collected samples after 4 weeks of exposure, indicating that B. pseudohinzii can transmit efficiently among laboratory mice and contaminate environmental facilities. The detection of B. pseudohinzii in one facility 5 consecutive times in 3 years also illustrated that the pathogen can exist for a long time in laboratory animals and facilities. In light of this observation, laboratory animal researchers should pay more attention to B. pseudohinzii and enhance the monitoring of this pathogen to guarantee the quality of their laboratory animals.
To date, B. pseudohinzii has been isolated not only from laboratory mice, but also from wild rats.4, 16 Although there is no evidence that B. pseudohinzii could infect humans, B. hinzii which shares high similarity with B. pseudohinzii has been isolated from humans and is associated with mutiple diseases.9, 10, 17, 18, 19, 20 B. hinzii has been isolated from wild rodents, but the source of transmission remains elusive.16, 21 The isolation of B. pseudohinzii in wild rodents together with our finding that the bacterica can shed through the feces and transmit to other animals raises public health concerns.16 Wild rodents represent an important reservoir of multiple human pathogens.22, 23 Whether B. hinzii can transmit from wild rodents to infect humans requires further investigation in the future. The infection model established in our study may provide important insights into the transmission of Brodetellae in rodents and humans.
CONFLICT OF INTEREST
None.
AUTHOR CONTRIBUTIONS
JW designed and coordinated the overall study. LM, SH and YL carried out the experimental work. FM and LF conducted pathological examinations. MC, JP and YZ analyzed the data. All authors discussed the results and wrote the manuscript.
ACKNOWLEDGEMENTS
This study was supported by a grant from Guangzhou Municipal Science and Technology Project (201904010284). This study was supported by a grant from Guangdong Provincial Science and Technology Project (2018B070714012).
Ma L, Huang S, Luo Y, et al. Isolation and characterization of Bordetella pseudohinzii in mice in China. Animal Model Exp Med. 2019;2:217–221. 10.1002/ame2.12075
Lei Ma and Shuwu Huang contributed equally to this article.
REFERENCES
- 1. Ko KS, Peck KR, Oh WS, Lee NY, Lee JH, Song JH. New species of Bordetella, Bordetella ansorpii sp. nov., isolated from the purulent exudate of an epidermal cyst. J Clin Microbiol. 2005;43:2516‐2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arp LH, Cheville NF. Tracheal lesions in young turkeys infected with Bordetella avium . Am J Vet Res. 1984;45:2196‐2200. [PubMed] [Google Scholar]
- 3. Kattar MM, Chavez JF, Limaye AP, et al. Application of 16S rRNA gene sequencing to identify Bordetella hinzii as the causative agent of fatal septicemia. J Clin Microbiol. 2000;38:789‐794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clark SE, Purcell JE, Sammani S, et al. Bordetella pseudohinzii as a confounding organism in murine models of pulmonary disease. Comp Med. 2016;66:361‐366. [PMC free article] [PubMed] [Google Scholar]
- 5. Linz B, Ivanov YV, Preston A, et al. Acquisition and loss of virulence‐associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genom. 2016;17:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perniss A, Schmidt N, Gurtner C, et al. Bordetella pseudohinzii targets cilia and impairs tracheal cilia‐driven transport in naturally acquired infection in mice. Sci Rep. 2018;8:5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loong SK, Tan KK, Sulaiman S, Wong PF, AbuBakar S. Draft genome of Bordetella pseudohinzii BH370 isolated from trachea and lung tissues of a laboratory mouse. Genom data. 2017;12:69‐70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivanov YV, Linz B, Register KB, et al. Identification and taxonomic characterization of Bordetella pseudohinzii sp. nov. isolated from laboratory‐raised mice. Int J Syst Evol Microbiol. 2016;66:5452‐5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arvand M, Feldhues R, Mieth M, Kraus T, Vandamme P. Chronic cholangitis caused by Bordetella hinzii in a liver transplant recipient. J Clin Microbiol. 2004;42:2335‐2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hristov AC, Auwaerter PG, Romagnoli M, Carroll KC Bordetella hinzii septicemia in association with Epstein‐Barr virus viremia and an Epstein‐Barr virus‐associated diffuse large B‐cell lymphoma. Diagn Microbiol Infect Dis. 2008;61:484‐486. [DOI] [PubMed] [Google Scholar]
- 11. Hayashimoto N, Morita H, Yasuda M, et al. Prevalence of Bordetella hinzii in mice in experimental facilities in Japan. Res Vet Sci. 2012;93:624‐626. [DOI] [PubMed] [Google Scholar]
- 12. Hayashimoto N, Yasuda M, Goto K, Takakura A, Itoh T. Study of a Bordetella hinzii isolate from a laboratory mouse. Comp Med. 2008;58:440‐446. [PMC free article] [PubMed] [Google Scholar]
- 13. Benga L, Benten WP, Engelhardt E, Kohrer K, Gougoula C, Sager M. 16S ribosomal DNA sequence‐based identification of bacteria in laboratory rodents: a practical approach in laboratory animal bacteriology diagnostics. Lab Anim. 2014;48:305‐312. [DOI] [PubMed] [Google Scholar]
- 14. Mähler (Convenor) M, Berard M, Feinstein R, et al. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim. 2014;48:178‐192. [DOI] [PubMed] [Google Scholar]
- 15. Pritchett‐Corning KR, Prins JB, Feinstein R, Goodwin J, Nicklas W, Riley L. AALAS/FELASA working group on health monitoring of rodents for animal transfer. J Am Assoc Lab Anim Sci. 2014;53:633‐640. [PMC free article] [PubMed] [Google Scholar]
- 16. Loong SK, Che‐Mat‐Seri NA, Abdulrazak O, et al. Recovery of Bordetella bronchiseptica sequence type 82 and B. pseudohinzii from urban rats in Terengganu, Malaysia. J Vet Med Sci. 2018;80:77‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fabre A, Dupin C, Benezit F, et al. Opportunistic pulmonary Bordetella hinzii infection after avian exposure. Emerg Infect Dis. 2015;21:2122‐2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fry NK, Duncan J, Edwards MT, et al. A UK clinical isolate of Bordetella hinzii from a patient with myelodysplastic syndrome. J Med Microbiol. 2007;56:1700‐1703. [DOI] [PubMed] [Google Scholar]
- 19. Palacian Ruiz MP, Vasquez Martinez MA, Lopez Calleja AI. Respiratory infection caused by Bordetella hinzii . Arch Bronconeumol. 2013;49:409‐410. [DOI] [PubMed] [Google Scholar]
- 20. Weigand MR, Changayil S, Kulasekarapandian Y, Tondella ML. Complete genome sequences of two Bordetella hinzii strains isolated from humans. Genome Announc. 2015;3:e00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiyipong T, Morand S, Jittapalapong S, Raoult D, Rolain JM Bordetella hinzii in rodents, Southeast Asia. Emerg Infect Dis. 2013;19:502‐503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gajda E, Hildebrand J, Sprong H, Bunkowska‐Gawlik K, Perec‐Matysiak A, Coipan EC. Spotted fever rickettsiae in wild‐living rodents from south‐western Poland. Parasit Vectors. 2017;10:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Obiegala A, Pfeffer M, Pfister K, Karnath C, Silaghi C. Molecular examinations of Babesia microti in rodents and rodent‐attached ticks from urban and sylvatic habitats in Germany. Ticks Tick Borne Dis. 2015;6:445‐449. [DOI] [PubMed] [Google Scholar]


