Abstract
Although teleost fish have higher levels of brain aromatase activity than any other vertebrate group, its function remains speculative, and no study has identified its cellular basis. A previous study determined aromatase activity in a vocal fish, the plainfin midshipman (Porichthys notatus), and found highest levels in the telencephalon and lower levels in the sonic hindbrain, which was dimorphic between and within (males) sexes. We have now localized aromatase-containing cells in the midshipman brain both by immunocytochemistry using teleost-specific aromatase antibodies and byin situ hybridization using midshipman-specific aromatase probes. Aromatase-immuno-reactivity and mRNA hybridization signal are consistent with relative levels of aromatase activity in different brain regions: concentrated in the dimorphic sonic motor nucleus, in a band just beneath the periaqueductal gray in the midbrain, in ventricular regions in the hypothalamus, and highest levels in the telencephalon especially in preoptic and ventricular areas. Surprisingly, double-label immunofluorescence does not show aromatase-immunoreactive colocalization in neurons, but instead in radial glia throughout the brain. This is the first study to identify aromatase expression mostly, if not entirely, in glial cells under normal rather than brain injury-dependent conditions. The abundance of aromatase in teleosts may represent an adaptation linked to continual neurogenesis that is known to occur throughout an individual's lifetime among fishes. The localization of aromatase within the intersexually and intrasexually dimorphic vocal-motor circuit further implies a function in the expression of alternative male reproductive phenotypes and, more generally, the development of natural, individual variation of specific brain nuclei.
Keywords: aromatase, estrogen, radial glia, telencephalon, teleost fish, vocal-motor system
The aromatization of circulating androgen to estrogen in the brain is known to be a key mechanism by which testosterone regulates many physiological and behavioral processes, such as brain sexual differentiation, activation of male sexual behavior, and feedback of steroid hormones on gonadotropin secretion (for review, see Balthazart and Ball, 1998). Distribution of aromatase activity in the brain appears to be conserved in vertebrates because highest concentrations are consistently localized in forebrain areas known to control reproduction and sex behavior (Balthazart and Ball, 1998). Teleost fish express the highest levels of brain aromatase activity (100–1000 times greater in the preoptic area–hypothalamus than mammals) (Callard et al., 1990), and high steady-state mRNA levels of brain homogenates corroborate these levels (Gelinas et al., 1998).
Only one study in goldfish attempted to localize aromatase-containing neurons by immunocytochemistry (ICC) using an antiserum to human placental aromatase (Gelinas and Callard, 1997). However, the numbers of aromatase-immunoreactive (IR) cells was not greater than those in rodent and avian studies, which conflicts with known high aromatase activity and aromatase mRNA levels. We have now used species-specific antibodies and mRNA probes to show the anatomical basis for high brain aromatase activity levels in teleosts. Surprisingly most, if not all, aromatase is localized in glial cells.
The plainfin midshipman, Porichthys notatus, exemplifies one of the most broadly studied neuroethological models for brain mechanisms underlying the performance of reproductive tactics among teleosts (for review, see Bass, 1992, 1996; Foran and Bass 1999). Midshipman have two male morphs with divergent spawning and vocal behaviors. Only type I “singing” males acoustically court females, whereas type II “sneaker” males do not court females but instead steal egg fertilizations from singing males. Type I males are divergent from type II males and females (which resemble each other) in a large suite of reproductive-related neuronal traits (Bass, 1992, 1996;Goodson and Bass, 2000). Brain aromatase activity levels resemble other teleosts with the addition of intersexual and intrasexual differences in the hindbrain region, which includes an expansive vocal pacemaker-motoneuron circuitry (Schlinger et al., 1999). One objective of this study was to map the distribution of aromatase-IR cells throughout the brain of midshipman by using an antibody made against a conserved amino acid sequence from known teleost aromatases. Also, we sought to determine whether this antibody would reveal relative numbers of aromatase-IR cells predicted by levels of aromatase activity in this species. To corroborate immunocytochemical localization of aromatase-containing cells, in situ hybridization (ISH) probes based on cloning of a partial aromatase cDNA of midshipman were also used to detect aromatase mRNA throughout the midshipman's brain. The localization of abundant aromatase in glial cells, the only other examples of which are known in mammalian and avian models under brain injury-dependent conditions in situ (Garcia-Segura et al., 1999a,b; Peterson et al., 2001) or in culture (Schlinger et al., 1994), has important implications for neurogenesis and likely neuronal repair, which can continue throughout an individual's lifetime among teleosts (Gelinas and Callard, 1997; Zupanc, 1999).
Parts of this work have been published previously in abstract form (Forlano et al., 2000, 2001).
MATERIALS AND METHODS
Animals. All fish were collected from field sites in Tomales Bay, CA during the 1999 and 2000 summer reproductive season and were held at the University of California Bodega Marine Laboratory in running seawater tanks until shipped to Cornell University. Fish were maintained in artificial seawater tanks until they were killed. All experimental protocols were approved by the Cornell University Institutional Animal Care and Use Committee.
ICC. Two antibodies were generated for this study. The first antibody, made against the peptide CKLQVLESFINESLRFHPVV, was designed a priori, based on a conserved amino acid sequence of known teleost aromatases (see below). A second antibody was made against a peptide sequence deduced from the midshipman aromatase cDNA (see below) and used to confirm the same pattern found with the original antibody.
All fish (nine type I males, 13–17.2 cm; five type II males, 10.2–10.8 cm; eight females, 10.5–13.7 cm) were deeply anesthetized in MS222 (tricaine methanesulfonate; Sigma, St. Louis, MO) and perfused transcardially with teleost Ringer's solution, followed by 4% paraformaldehyde in 0.1 m phosphate buffer (PB). Brains were removed and post-fixed in the same fixative for 1 hr and stored in PB. Before sectioning in the transverse plane at 30 μm on a cryostat, brains were cryoprotected overnight in 30% sucrose in PB and frozen in Cryo-M-bed (Hacker Instruments, Huntington, UK). Sections were collected onto chrom–alum-subbed slides and stored at −80°C until processed as follows: two times for 10 min each in 0.1m PBS, 20 min PBS plus 1.0% bovine serum albumin (BSA) plus 0.3% Triton X-100, overnight (16 hr) in primary antibody solution (anti-aromatase, 400 μl each slide) [ primary antibodies, made by Bethyl Labs Inc. (Montgomery, TX), were diluted 1:5000 (final peptide concentration, 0.2 μg/ml) in PBS plus 0.5% BSA plus 0.3% Triton X-100 plus 0.5% sodium azide], two times for 10 min each in PBS plus BSA, 1 hr in secondary anti-rabbit antibody (Vectastain Rabbit IgG kit; Vector Laboratories, Burlingame, CA) in PBS plus BSA, two times 10 min each in PBS plus BSA, 1 hr avidin–biotin complex (Vectastain Rabbit IgG kit; Vector Laboratories) in PBS plus BSA, two times for 10 min each in PB, 1.5 min diaminobenzidine solution (DAB) (0.05% DAB in 0.1 m PB with 0.3% hydrogen peroxide), two times for 10 min each in PB, dehydrated in a graded series of ethanol, three times for 5 min each in xylene, and coverslipped in Permount (Fisher Scientific, Fair Lawn, NJ). Controls included absence of primary antibody and preabsorption of primary antibody overnight with excess peptide used to generate the antibody. Both controls showed no label in the tissue.
The aromatase antigen was designed based on sequence alignment of known brain [Tilapia (GenBank accession number AF135850),Oreochromis (GenBank accession number AF306786),Danio (GenBank accession number AF120031), andCarassius (GenBank accession number AB009335)] and ovarian [Oryzias (GenBank accession number D82969),Danio (GenBank accession number AF004521),Tilapia (GenBank accession number AF135851), andCarassius (GenBank accession number AB009336)] forms of teleost aromatase located in the GenBank database. After alignment, a sequence was selected that was both highly conserved between brain and ovarian forms of aromatase, as well as displaying a high antigenic index (MacVector Sequence Analysis Software; Oxford Molecular Group). A second antibody (mentioned above) made against the peptide CGENQTENVNYQNLEVLEK was based on the deduced amino acid sequence (residues 39–56) from a partial cDNA of midshipman aromatase (see cloning results). For conjugation to a carrier protein, a Cys residue was introduced at the N terminus of both peptides (shown above).
Also, several other antisera were used with the above protocol to determine the pattern of aromatase immunoreactivity as neuronal or glial: monoclonal mouse anti-human neuronal (Hu) protein antibody (1:1000, 16A11; Molecular Probes, Eugene, OR) as a specific label for neuronal cell bodies (Marusich et al., 1994), monoclonal mouse anti-acetylated tubulin (1:2000, T6793; Sigma) as an axonal marker, rabbit anti-glial fibrillary acidic protein (GFAP) (1:1000, G-9269; Sigma) and monoclonal mouse anti-GFAP (1:1000, MAB360; Chemicon, Temecula, CA) as astrocyte or radial glial markers (mouse anti-GFAP has been shown to work in goldfish; Kalman, 1998), and monoclonal mouse anti-vimentin (VIM) (1:4, 40E-C; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) as a radial glia marker (Alvarez-Buylla et al., 1987). The 40E-C monoclonal antibody developed by A. Alvarez-Buylla was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa (Department of Biological Sciences, Iowa City, IA).
In double-label experiments, both primary antibodies [rabbit anti-aromatase (1:1000) and anti-Hu (1:100) or anti-vimentin (1:1), anti-acetylated tubulin (1:1000), or mouse anti-GFAP (1:100)] and, subsequently, both secondary antibodies were combined and applied as above. Secondary anti-rabbit antibody conjugated to fluorescein (Vector Laboratories) was used to visualize anti-aromatase, and secondary anti-mouse antibody conjugated to Texas Red (Vector Laboratories) was used to visualize anti-Hu, vimentin, GFAP, and acetylated tubulin. Both secondary fluorescence antibodies were used at 1:100 dilution in PBS plus 0.5% BSA. Slides were incubated for 1.5 hr in a 37°C humidified chamber, washed three times for 10 min each in PBS, dipped in ddH2O, and coverslipped with Vectashield (Vector Laboratories). Fluorescently labeled material was visualized on a Nikon (Tokyo, Japan) ES800 compound microscope outfitted with epifluorescence and a double-band pass cube (FITC–Texas Red) to allow for simultaneous visualization of both antibodies. Photographs were taken using Kodak Gold color film (iso 400 for fluorescence, iso 100 for bright field; Eastman Kodak, Rochester, NY) and a 35 mm Nikon mounted camera on the above microscope. Photographs were then scanned at 600 dpi, compiled into plates, and labeled in Adobe Photoshop 4.0 (Adobe Systems, San Jose, CA).
Cloning of midshipman aromatase cDNA. Fish were deeply anesthetized, and brains were dissected into hindbrain, midbrain, and forebrain regions (Schlinger et al., 1999) (Fig.1), rapidly frozen in liquid nitrogen, and stored at −80°C. Because biochemical analyses indicate the highest levels of aromatase activity in the forebrain of these fish (Schlinger et al., 1999), four forebrains of adult females were pooled together for total RNA extraction. Brains were removed from −80°C and immediately ground into a fine powder in liquid nitrogen, transferred into 4 ml of Trizol (Life Technologies, Carlsbad, CA), and mixed by a Tissue Tearer probe for 30 sec, and RNA was purified according to the instructions of the manufacturer. Reverse transcription was as follows: 3.3 μg of total RNA was added to 1 μl of random primers, heated to 70°C for 10 min, chilled on ice 2–3 min, and was reversed transcribed with SuperScript II (Life Technologies) according to the directions of the manufacturer. For reverse transcription (RT)-PCR, two degenerate oligo primers were designed based on conserved regions of goldfish (GenBank accession number AB009335) and Tilapia (GenBank accession numberAF135850) brain aromatase sequence. Forward primer ATGGTNAT(A/C/T)GCNGCNCCNGA(C/T)AC and reverse primer CATNGC(A/G/T)AT (A/G)TG(C/T)TTNCCNAC(A/G)CA were predicted to amplify a 426 bp sequence from the midshipman brain. For amplification of the predicted 426 bp sequence, PCR was performed in a final volume of 100 μl containing 1× reaction buffer, 1.5 mmMgCl2, 200 μm each dNTP, 1 μm each primer, 1 μl Tfl DNA polymerase, and 2 μl of first-strand cDNA reaction. The complete reaction was overlaid with 80 μl of mineral oil and run for 30 cycles under the following conditions: 30 sec at 94°C, 1 min at 50°C, 1 min at 72°C, and 10 min at 72°C. PCR products were run on a 2% agarose gel to confirm predicted size, purified with a Qiagen (Valencia, CA) PCR purification kit, treated with T4 polynucleotide kinase, and blunted with T4 DNA polymerase. This product was then ligated into Bluescript plasmid, which was digested previously with SmaI restriction enzyme and treated with calf intestinal phosphatase. The ligated plasmid was then transformed into competent X-LI-Blue cells and plated onto LB-agarose plates with ampicilin (AMP). Resistant colonies were selected and grown in LB plus AMP (100 μl/ml) overnight, miniprepped, and digested to confirm presence of insert. Two clones with predicted insert were sequenced at the Cornell University Biotechnology Sequencing facility, one of which contained a match to other teleost aromatases.
Fig. 1.
Dorsal view of a midshipman brain indicating areas of transverse sections in Figure 2, posterior to anterior (A–K), which demonstrate anatomical locations of aromatase immunoreactivity. Cb, Cerebellum;M, midbrain; OB, olfactory bulb;OC, occipital nerve; OL, olfactory nerve;PLLN, posterior lateral line nerve; PN, pacemaker neurons; SMN, sonic motor nucleus;SP, spinal cord; T, telencephalon;VM, ventral medullary neurons; V, trigeminal nerve; VIII, eighth nerve; IX, glossopharyngeal nerve; X, vagus nerve.
ISH. The procedure for in situ hybridization detection of aromatase mRNA in midshipman brain was adapted from that published for gonadotropin-releasing hormone mRNA (Grober et al., 1995). Adult midshipman fish (five type I males, 11.5–21 cm; five type II males, 7.5–11.4 cm; five females, 10.9–17.2 cm) were perfused, and brains and gonads (both ovary and testis) were sectioned as above for ICC or dissected fresh and immediately embedded and frozen over dry ice, sectioned at 30 μm, collected onto Superfrost Plus slides (Erie Scientific, Portsmouth, NH), and stored in −80°C. Before hybridization, slides were equilibrated to room temperature for 10 min. Slides with unfixed tissue were immersed in 4% paraformaldehyde for 5 min and rinsed twice in potassium PBS (KPBS), pH 7.2. Both slides that contained both fixed and unfixed tissue were washed two times in KPBS and placed into freshly prepared 0.25% acetic anhydride in 0.1m triethanolamine two times for 5 min each, dehydrated in a series of ethanol washes and chloroform, and air dried.
Hybridization was performed with a mixture of two probes antisense to nucleotides 121–161 (AACCTCTAGGTTTTGGTAATTAACATTT-TCTGTTTGATTC) and 180–220 (GCATTGTGAAGTCAACCACAG-GATGAAACCTCAAAGACTC) of the 426 bp sequence of midshipman aromatase. Oligonucleotides were labeled with a terminal deoxynucleotidyl transferase reaction using [α-33P]d-ATP (specific activity, 1000–3000 Ci/mmol; NEN, Boston, MA). Hybridization solution [4× SSC (1× SSC is 0.15 m sodium chloride and 0.015 m sodium citrate, pH 7.2), 40% deionized formamide, 500 μg/ml denatured calf thymus DNA, 250 μg/ml transfer RNA, 4× Denhardt's solution, 4 mm EDTA, 5 mm sodium phosphate, 10% (w/v) Dextran sulfate, and 1 × 106 cpm total radiolabeled probe (0.5 × 106 cpm each probe)] was placed on each slide (300 μl), coverslipped with parafilm, and incubated overnight (at least 15 hr) in a humidified 37°C chamber. After hybridization, slides were briefly washed twice at 23°C in 1× SCC, washed twice for 30 min at 55°C in 1× SCC in a shaking water bath, and washed once in 1× SCC in 0.1% Triton X-100 at 23°C, briefly rinsed in distilled water and 70% ethanol, and air dried. Slides were then exposed to X-Omat AR film (Eastman Kodak) for 2–3 d at −20°C to confirm signal, subsequently dipped in nuclear emulsion (NTB-2; Eastman Kodak), and exposed for 4 weeks at 4°C. Slides were developed in Kodak D-19 (4 min at 14°C), rinsed in distilled water (10 sec at 14°C), fixed (Kodak GBX fixer; 5 min at 14°C), rinsed in running distilled water (5 min), counterstained in cresyl violet, dehydrated, and coverslipped with Permount. Both unfixed and fixed tissue showed similar signal, although tissue integrity was improved with fixed tissue. Dark-field optics were used to best visualize the overall mRNA signal pattern throughout the brain at 40× magnification, whereas bright-field optics were used to best visualize ISH signal at higher magnification over Nissl-stained tissue. Dark-field photography was performed using Kodak Gold color film (iso 400), after which color prints were scanned at 600 dpi and converted to grayscale, combined into plates, and labeled in Adobe Photoshop 4.0. Bright-field photography for ISH was identical for bright-field ICC (see above).
RESULTS
Distribution of aromatase immunoreactivity
The antibody first used in this study (made against a conserved amino acid sequence of known teleost aromatases) before the midshipman sequence was known labels consistently and specifically within the same brain regions between animals, independent of adult morphotype. The second antibody designed against the midshipman specific sequence gave an identical labeling pattern to the first antibody used. All photomicrographs were taken from tissue labeled with the first antibody. Differences in immunoreactivity between and within sexes will be addressed elsewhere.
Hindbrain
Aromatase-IR cells are concentrated dorsal and dorsolaterally within the sonic motor nucleus (SMN) and the ventral half of the fourth ventricle, whereas fewer aromatase-IR cells are found centrally in the SMN (Figs. 2A,3A–C). Fibers from the majority of these cells project ventrally through the SMN, as well as ventrolaterally along the reticular formation (Figs.2A, 3A,B). Several aromatase-IR cells are also found ventral to the SMN, lateral to and sometimes within the medial longitudinal fasciculus (MLF) (Fig.2A,B). There appears to be a denser population of aromatase-IR cells in the rostral SMN, and some cells extend dorsally along the periventricular region at the level of the vagal motor nucleus (Fig. 2B, Xm). In other areas in the hindbrain, more cells are found just lateral to the MLF, with fibers extending along the reticular formation (Fig.2C,D, RF). Double-label ICC with neural-specific anti-Hu antibody indicates no obvious overlap between aromatase-IR cells and neuronal cell bodies in the SMN (Fig.3C), as well as other regions in the hindbrain.
Fig. 2.
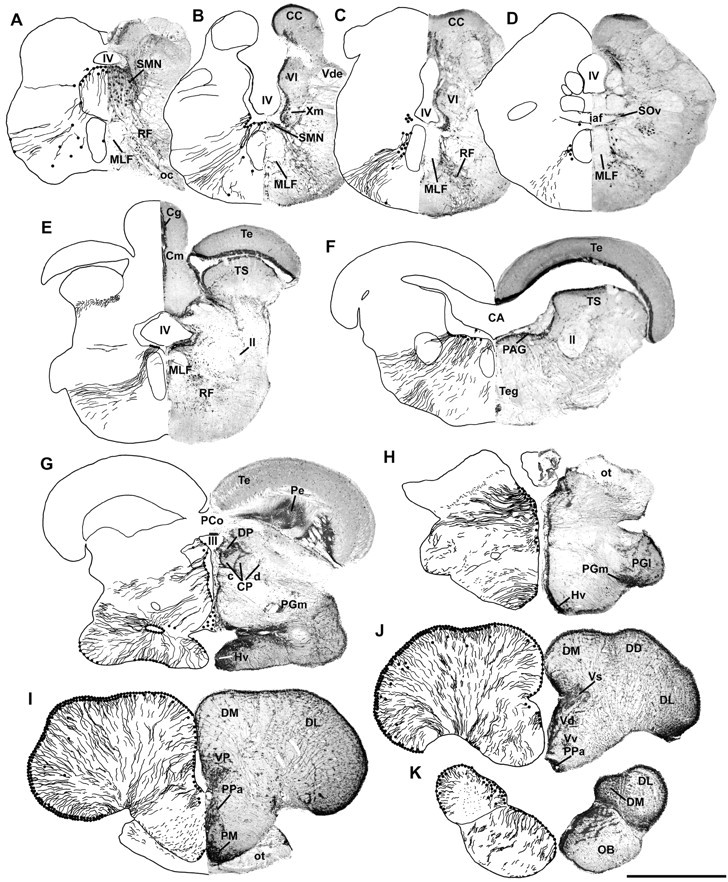
Distribution of aromatase immunoreactivity in the brain of P. notatus. Left half of each section is a camera lucida drawing that shows aromatase-IR cell and fiber distribution; right half is a photomicrograph of ICC processed tissue with Nissl counterstain. A,B, Aromatase-IR cells are concentrated dorsally and dorsolaterally within the sonic motor nucleus (SMN) and around the ventral half of the fourth ventricle (IV). C,D, Aromatase-IR cells remain prominent just lateral to the medial longitudinal fasciculus (MLF) in the rostral hindbrain. E, F, Large numbers of aromatase-IR cells line the periventricular areas of the fourth ventricle (IV) and cerebral aqueduct (CA) in the caudal (E) and rostral (F) midbrain, respectively; label is absent in the midbrain tectum (Te) and torus semicircularis (TS).G, H, Throughout the diencephalon, the third ventricle (III) is lined with aromatase-IR cells, which project ventrolaterally. I,J, Aromatase-IR cells (2–4 layers thick) line the entire periphery of the telencephalic hemispheres, as well as the medial ventricular surface; fiber projections course ventromedially. Aromatase-IR cells are prominent in preoptic areas (PPa, anterior parvocellular; PM, magnocellular).K, Label extends well into the olfactory bulbs (OB) in which numerous IR cells line the dorsomedial edge and fiber projections course ventrolaterally. CC, Cerebellar crest; Cg, granule cell layer of the corpus of the cerebellum; Cm, molecular layer of the cerebellum; CP c/d, compact/diffuse division of the central posterior nucleus; DD, dorsal division of the dorsal telencephalon; DL, dorsolateral telencephalon;DM, dorsomedial telencephalon; DP, dorsal posterior nucleus of the thalamus; Hv, ventral periventricular hypothalamus; iaf, internal arcuate fibers; ll, lateral lemniscus; OC, occipital nerve; ot, optic tract; PAG, periaqueductal gray; PCo, posterior commissure;Pe, periventricular cell layer of the torus semicircularis; PGl, lateral division of nucleus preglomerulosus; PGm, medial division of nucleus preglomerulosus; RF, reticular formation;SOv, ventral division of secondary octaval nucleus;Vd, dorsal nucleus of area ventralis;Vde, descending tract of the trigeminal nerve;Vl, vagal lobe; VP, posterior nucleus of area ventralis of the telencephalon; Vs, supracommissural nucleus of the ventral telencephalon;Vv, ventral nucleus of area ventralis;Xm, vagal motor nucleus. Scale bar, 1 mm.
Fig. 3.
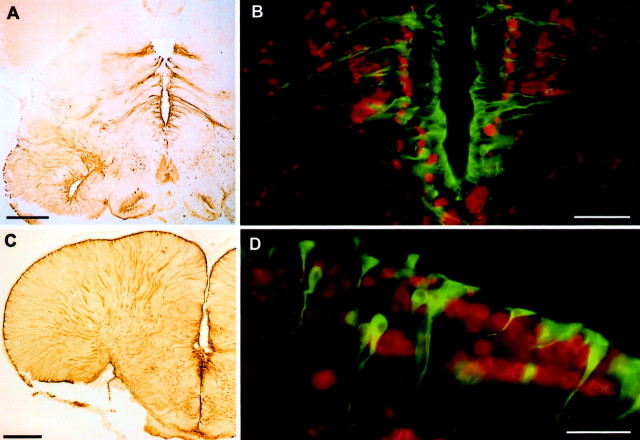
Aromatase immunoreactivity in the hindbrain and midbrain. A, B, Low-magnification photomicrographs of aromatase immunoreactivity visualized by DAB chromogen corresponding to levels near Figure 2, A andB. C, Photomicrograph of the sonic motor nucleus fluorescently double-labeled with anti-aromatase (green) and neuronal specific anti-Hu (red). Note the prominent aromatase-IR fibers throughout the entire sonic motor nucleus. D, Low-magnification photomicrograph of aromatase immunoreactivity visualized by DAB chromogen at level near Figure 2F.E, Photomicrograph of the periventricular region in the midbrain (also near level 2F) fluorescently double-labeled with anti-aromatase (green) and neuronal-specific anti-Hu (red). Scale bars:A, B, D, 500 μm;C, 240 μm; E, 80 μm.
Midbrain and diencephalon
In the caudal midbrain, aromatase-IR cells line the ventral half of the fourth ventricle and fibers course ventrolaterally, just around the MLF (Fig. 2E). Fibers also emerge from the area at the base of the torus semicircularis (Fig. 2E,TS). More rostrally, aromatase-IR cells lie just beneath the periaqueductal gray (PAG), and fibers cover most of the tegmentum (Teg) (Figs. 2F,3D,E). Throughout the midbrain, label is consistently absent in the tectum (Te) and torus semicircularis. In the diencephalon, the third ventricle is completely lined by aromatase-IR cells and fibers mostly project ventrolaterally (Figs. 2G,H,4A,B). The periventricular areas in the hypothalamus also contain labeled cells that project radially, as well as cells that line the ventral edge of the hypothalamus (Fig. 2G,H). Double-label ICC with neural-specific anti-Hu antibody indicates no obvious overlap between aromatase-IR cells and neuronal cell bodies in any regions in the midbrain and diencephalon (Figs. 3E,4B).
Fig. 4.
Aromatase immunoreactivity in the diencephalon and telencephalon. A, Low-magnification photomicrograph of aromatase immunoreactivity visualized by DAB chromogen at level near Figure 2G. Scale bar, 500 μm. B, Photomicrograph of the diencephalon (also near level G) fluorescently double-labeled with anti-aromatase (green) and neuronal-specific anti-Hu (red). Scale bar, 160 μm. C, Low-magnification photomicrograph of aromatase immunoreactivity visualized by DAB chromogen at level near Figure2I. Scale bar, 500 μm. D, Photomicrograph of dorsal telencephalon (also near level 2I) fluorescently double-labeled with anti-aromatase (green) and neuronal-specific anti-Hu (red). Scale bar, 80 μm.
Telencephalon
Compared with other brain regions, the forebrain has by far the greatest numbers of aromatase-IR cells. As seen in the hindbrain and midbrain, aromatase-IR cells are consistently found around ventricular surfaces, and the forebrain is no exception. During development, the telencephalon of teleost fish is formed by an eversion of the ventricular surface (Nieuwenhuys, 1982). Thus, the entire periphery of the telencephalic hemispheres is ventricular surface, and therefore the expression of aromatase-IR cells along this surface is consistent with other brain regions. Labeled cells (two to four thick) line the outer edge of the dorsal and lateral hemispheres, whereas fewer nonventricular cells are found centrally (Figs.2I–K, 4C). Aromatase-IR cells are also located along the median ventricular surface and in the preoptic areas (Fig. 2I, PM). Fibers from most of these cells cover the entire forebrain and appear to converge along the ventrolateral (VL) margin of the telencephalon, as well as in the optic tract (Fig. 2I,J, ot,4C). Aromatase-IR cells and fibers extend well into the olfactory bulb (Fig. 2K, OB). Again, as in other brain regions, there appears to be no overlap between aromatase immunoreactivity and neuronal cell bodies (Fig. 4D), which is additional evidence that the majority of aromatase in these fish is found in glia and not neurons.
Cloning of partial midshipman aromatase cDNA
A predicted 426 bp sequence was amplified from midshipman brain tissue. Figure 5 shows the midshipman partial cDNA sequence with translated amino acid sequence below. Sense and antisense degenerate primers for RT-PCR were designed to the two conserved amino acid regions shown in bold. The amplified region of midshipman brain aromatase contains presumptive functional domains that are conserved among cytochrome P450 enzymes (Hickey et al., 1990; Gelinas et al., 1998), including partial helical region, Ozols peptide region, conserved aromatic region, and partial heme binding region (Fig. 5). The presumed membrane-spanning domain, which is not as conserved as the above functional regions, is likely upstream to the region that was amplified from midshipman brain (Gelinas et al., 1998).
Fig. 5.
Nucleotide and deduced amino acid sequence of aromatase cDNA clone isolated from midshipman brain.Bold amino acids indicate conserved regions of teleost brain aromatase (based on goldfish and Tilapia) to which degenerate primers were made for RT-PCR. Presumed functional domains are underlined and correspond to regions identified in goldfish brain aromatase by Gelinas et al. (1998). I, Helical region (partial); II, Ozols peptide;III, aromatic region; IV, heme binding region (partial).
A comparison of midshipman brain aromatase with other aromatases isolated from other teleost brain and ovarian tissue, as well as zebra finch (ovarian) and human placental aromatase, is shown in Figure6. The antibody first used in this study (designed against a conserved amino acid sequence of known teleost aromatases; see Materials and Methods) corresponds to residues 50–68 and differs from midshipman at positions 50, 52, and 56. A BLAST (National Center for Biotechnical Information) search using the unique midshipman partial cDNA translated protein (without flanking primers) revealed 79% sequence identity to Tilapia mossambica brain aromatase (GenBank accession number AF135850), 76% identity to Oreochromis niloticus brain aromatase (GenBank accession number AF306786), 68% to O. niloticusovarian aromatase (GenBank accession number AF295761), 67% identity toCarassius auratus brain aromatase (GenBank accession numberAB009335), 64% identity to Danio rerio brain aromatase (GenBank accession number AF226619), 64% identity to T. mossambica ovarian aromatase (GenBank accession number AF135851), 57% identity to Poephila guttata (zebra finch) (GenBank accession number S75898), and 57% identity to human placental aromatase (GenBank accession number HUMARM). As expected, highest homologies match with other teleost brain aromatases, although the ovarian form from O. niloticus contains more identities than aromatase isolated from brain in goldfish (C. auratus) and zebrafish (D. rerio). Differences of aromatase isoforms within a single species is exemplified by the high sequence identity of midshipman brain aromatase to Tilapia brain when compared with the relatively low identity to Tilapia ovarian aromatase.
Fig. 6.

Alignment of deduced amino acid sequences of midshipman, P. notatus (Pn), brain aromatase to known teleost brain (br) aromatases [T. mossambica (Tm); O. niloticus (On); C. auratus(Ca); D. rerio (Dr), as well as Tm and On ovarian (ov), zebra finch P. guttata(Pg) (ovarian), and human placental (Hum pl)]. Boxed areas, which includeP. notatus, indicate identical amino acids to midshipman brain aromatase. Bold amino acids indicate conserved regions of teleost brain aromatase (based on goldfish andTilapia) to which degenerate primers were made for RT-PCR. The antibody used in this study before the midshipman sequence was known (see Materials and Methods) corresponds to residues 50–68 and differs from midshipman at positions 50, 52, and 56.
In situ hybridization
Dark-field illumination was the most effective method to view the overall distribution pattern of hybridization signal (Fig.7A–J). The pattern of aromatase mRNA signal was consistent with that found by immunocytochemical localization of the enzyme (compare Figs. 2, 7). Bright-field visualization was also used to identify the location of silver grains in relation to Nissl-stained brain nuclei (Fig.7K–O). Because teleost ovarian tissue is known to produce aromatase, we processed sagittal sections of ovary through the ISH protocol along with brain tissue; we also processed testis. Unlike testis, adult female ovaries at various stages of oocyte development showed robust hybridization with the probes made from brain aromatase (Fig. 7P). This pattern attests to the specificity of the labeling and is entirely consistent with gonadal activity in teleost fish, namely that aromatization occurs in ovarian tissue (Nagahama, 1983).
Fig. 7.

Dark-field (A–J) and bright-field with Nissl counterstain (K–O) visualization of in situ hybridization throughout the brain of P. notatus using probes from partial cDNA cloning of midshipman brain aromatase. Pattern of signal in all brain regions is consistent with that found by ICC (Fig. 2-4).A, Aromatase mRNA signal clearly defines the sonic motor nucleus (SMN) boundary near level Figure2A; heaviest signal is around the dorsal and lateral periphery. B, Aromatase mRNA just below the fourth ventricle (IV) in the rostral medulla near Figure 2E. C, Hybridization signal in the caudal midbrain in between levels at Figure 2, Eand F. D, Aromatase mRNA expression in the diencephalon near Figure 2G. E, Aromatase mRNA expression in the diencephalon–telencephalon transition area near Figure2H. F, The caudal telencephalon between Figure 2, H and I.G, Strong hybridization in the anterior parvocellular division of the preoptic area (PPa), posterior nucleus of area ventralis of the telencephalon (VP), and ventrolateral area (near Fig. 2I).H, Section just caudal to Figure2J at the level of the anterior commissure (ac), which shows very strong punctate signal within thePPa. I, Anterior telencephalon (rostral to Fig.2J) has high aromatase mRNA signal, especially within ventrolateral (VL) and ventral nucleus of area ventralis (Vv), as well as within the optic tract (ot). J, Aromatase mRNA expression in olfactory bulb (OB, near Fig. 2K). Scale bars: A, B, 200 μm;C–J, 400 μm. K, Section through the hindbrain just caudal to the level at Figures 2Band 3B, which indicates high silver grain concentration in the dorsal rostral SMN; signal within the central SMN is much greater than in the MLF. Scale bar, 100 μm. L, Hybridization signal in the midbrain just below the periaqueductal gray (PAG) near the level at Figure 2F. Scale bar, 100 μm. M, ISH signal in the diencephalon at levels between Figure 2, G and H. Notice the clusters of strong signal along III, which expand into the dorsal posterior nucleus of the thalamus (DP). Scale bar, 100 μm. N, ISH signal in the lateral forebrain between levels at Figure 2, Iand J. The ventral and lateral boundaries between VL and DL telencephalon show a strong signal of hybridization. Scale bar, 250 μm. O, Intense ISH signal in Vv and inPPa between levels at Figure 2,I and J. P, ISH signal in a sagittal section through an ovary. Large, discrete clusters of aromatase mRNA hybridization appear between maturing oocytes (O). Scale bar, 500 μm. Hd, Dorsal periventricular hypothalamus; Hv, ventrolateral nucleus of the hypothalamus; tc, tela choroidea;Vm, molecular layer of the valvula; also see Figure2A–K.
Hindbrain
When viewed under dark-field illumination, grains that indicate hybridization to brain aromatase mRNA clearly define the boundary of the SMN (see Fig. 9A). The mRNA signal within the SMN together with that surrounding the SMN and within the reticular formation is concordant with the position of the hindbrain vocal pattern-generating circuit (Fig. 1) (Bass et al., 1994). Similar to ICC labeling (Figs. 2A,B,3A,B), hybridization signal is concentrated in the dorsolateral SMN and is stronger more rostrally where silver grains extend dorsally along the ventricle (Fig.7A,K). Signal within the SMN is also comparatively higher than other surrounding brain areas, such as the MLF (Fig. 7A,K), and dense silver grains from the SMN follow the same ventrolateral pathway into the reticular formation as do aromatase-IR labeled fibers (Figs.2A,B, 3A–C,7A). In the rostral hindbrain, an intense bilateral band forms just outside the periventricular area, below the fourth ventricle (Fig. 7B), as seen in Figure 2E. Hybridization in other areas of the hindbrain correspond to aromatase immunoreactivity in Figure 2, C and D.
Fig. 9.
Colocalization of aromatase and GFAP in the telencephalon of P. notatus by double-label immunofluorescence using anti-aromatase and monoclonal anti-GFAP (MAB360) visualized by secondary fluorescein and Texas Red, respectively. A, Low-magnification photomicrograph that shows aromatase-IR cells (green) along the lateral telencephalic lobe whose projections are double-labeled with anti-GFAP (yellow-orange). Scale bar, 200 μm.B, Single cells with aromatase-IR labeled cell bodies (green) with yellow-orange fiber projections indicating that the same cell is labeled for aromatase and GFAP. Because GFAP immunofluorescence was often more robust in fibers than aromatase, fibers distal to the cell body often appeared morered-orange than yellow when viewed through a double-bandpass filter. Scale bar, 50 μm. C, Abundant colocalization (yellow) of aromatase and GFAP in the anterior parvocellular division of the preoptic area (bottom arrows). Again, fibers from aromatase-IR cells along the midline ventricle are also labeled by MAB360 (top arrow). Scale bar, 200 μm.
Midbrain and diencephalon
Distribution of aromatase mRNA hybridization signal in the midbrain and diencephalon (Fig. 7C,D) also shows the same overall pattern as found by ICC (Figs.2E,G, 3D,4A). Dark-field image of the caudal midbrain identifies a clear signal concentrated between the medial boundary of the torus and the valvula, as well as a strong bilateral signal just beneath the cerebral aqueduct along the periaqueductal gray (Fig.7C,L), first seen in the rostral hindbrain (Fig.7B). The signal is concentrated in a lateral band that outlines the torus and is more diffuse in the ventral tegmentum (Fig.7C). There is a clear absence of hybridization in the optic tectum and torus, as seen by ICC (Figs.2E,F, 3D,7C). Aromatase mRNA is concentrated in the diencephalon along the third ventricle in the thalamus, as well as in the dorsal and ventral periventricular areas of the hypothalamus (Fig.7D,M), similar to that seen in ICC-treated tissue (Fig. 2G, 4A). Aromatase mRNA expression in the diencephalon–telencephalon transition area (near Fig. 2H) shows high signal along the midline ventricle through thalamic and hypothalamic regions and dorsolaterally just beneath the optic tract (Fig. 7E).
Telencephalon
The telencephalon has the most abundant aromatase mRNA signal compared with other brain regions and also shows the same overall pattern as found by ICC. Strongest signal is apparent in preoptic areas along the ventricular midline and in the VL area. Dark-field visualization (Fig. 7F,G) indicates a broad distribution of hybridization throughout the forebrain, with strongest signal in the preoptic areas along the midline ventricle (compare with Fig. 2I) and the VL and dorsolateral (DL) interface (Fig. 7N). Dark-field visualization shows a concentration of silver grains that follow the same pattern as aromatase-IR fibers, which course from the periphery to the VL–DL interface (Fig. 7G–I) that lies near the point of attachment of the tela choroidea (Fig. 7N,tc). Consistent with ICC, hybridization is found along the entire periphery of the telencephalic hemispheres and along ventricular surfaces, as well as in the optic tract (Fig.7G–I). Rostral in the telencephalon, robust signal is found in the anterior parvocellular division of the preoptic area (Fig. 7H,O, PPa), ventral nucleus of area ventralis (Fig.7I,O, Vv), and in the ventral and medial olfactory bulb (Fig. 7J).
Evidence for aromatase expression in glial cells
Because the pattern of aromatase immunoreactivity in P. notatus brain (especially in the telencephalon) appeared reminiscent of radial glia found in other teleosts (Manso et al., 1997;Kalman, 1998) and did not colocalize with neural-specific anti-Hu labeled cells, we used three glial antibodies, rabbit anti-GFAP (G-9269), mouse monoclonal anti-GFAP (MAB360), and mouse monoclonal anti-VIM 40E-C (Developmental Studies Hybridoma Bank) to localize radial glia in the midshipman brain. The pattern of label with all antisera in the telencephalon is strikingly similar to that seen with anti-aromatase (compare Fig. 8 with 2I,J, 4C,D), as well as the staining pattern seen with anti-GFAP and anti-vimentin in other vertebrates, including teleosts (Alvarez-Buylla et al., 1987; Manso et al., 1997; Kalman, 1998). Anti-GFAP (MAB360) labels fibers robustly (Fig.8A–C). Anti-VIM labels fiber projections robustly, but labels few cell bodies (Fig. 8D). In contrast, anti-GFAP (G-9269) labels more cell bodies (Fig. 8E), but fibers distal to the cell bodies are not labeled as heavily. Double-label immunofluorescence with anti-aromatase and mouse anti-GFAP (MAB360) or VIM reveals a colocalization of aromatase immunoreactivity in glial fibers in the forebrain (Fig.9). Direct fiber projections from aromatase-IR cells that line the entire peripheral ventricular zone of the telencephalon are also GFAP and VIM-IR (Fig.9A,B), and some cell bodies that are found medial to the majority of aromatase-IR cells at the ventricular edge of the forebrain are also double-labeled. The anterior parvocellular division of the preoptic area in the forebrain (Fig.9C) and areas of the hypothalamus (inferior lobe and periventricular areas) are also clearly doubled-labeled. In other brain regions, colocalization of aromatase and GFAP and VIM-IR fibers is more limited to lateral and ventrolateral projections. In the midbrain ventricular area and within the SMN of the hindbrain, GFAP and VIM-IR fibers are plentiful, but only few appear colocalized with aromatase-IR fibers. All antisera label glia in areas in which aromatase immunoreactivity never occurs (i.e., optic tectum), and therefore it appears that aromatase is expressed in a subset of radial glia, as well other glial types (found in SMN and along midline ventricles) not labeled by the antisera used in this study.
Fig. 8.
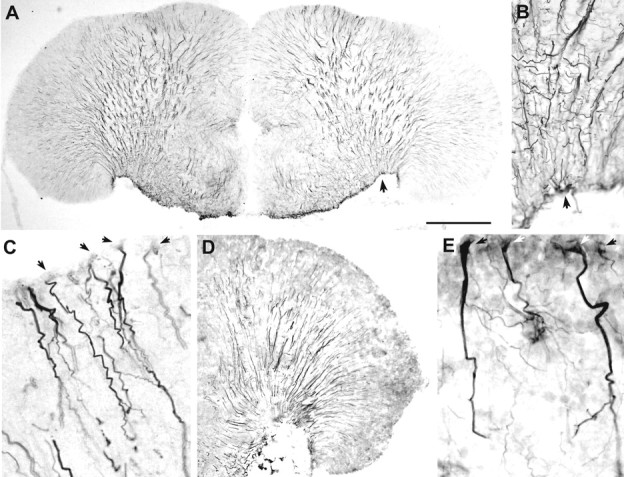
Distribution of radial glia in the telencephalon.A, DAB visualization of radial glia using monoclonal anti-GFAP (MAB360) near the level of Figure 2J. Note the long fibers that extend throughout the lateral telencephalon and converge in the ventrolateral area, virtually identical to the pattern seen with anti-aromatase (Figs.2I,J, 4C).B, Higher magnification of the ventrolateral area fiber pattern in A (arrows indicate same position). C, High magnification of faintly labeled somata (arrows) and darkly labeled fibers in the dorsal telencephalon using MAB360. D, DAB visualization of radial glia in the lateral telencephalon (near the level of Fig.2I) using monoclonal anti-vimentin. Notice the characteristic pattern of labeled fibers similar to anti-GFAP inA. E, Polyclonal anti-GFAP (G-9269) labels glial cell bodies of the same shape and size as anti-aromatase (compare with Fig. 4D). Scale bar:A, 500 μm; B, 100 μm;C, 40 μm; D, 500 μm;E, 35 μm.
DISCUSSION
Although other studies have demonstrated that teleost fishes have the highest levels of aromatase activity compared with any other vertebrate group, this is the first study to corroborate high activity levels with expected relative numbers of aromatase-containing cells throughout the brain of a teleost. This was accomplished, in part, because to our knowledge this is also the first study to use specific antibodies and mRNA probes to localize aromatase-containing cells in the brain. Also, the numbers of aromatase-IR cells and aromatase mRNA in different brain regions correspond well with aromatase activity levels in the brain of this species (i.e., greatest number of aromatase-IR cells in the telencephalon compared with the midbrain and hindbrain). Localization of aromatase immunoreactivity and mRNA in and around the forebrain preoptic nuclei is consistent with studies in other vertebrates.
The use of a teleost-specific aromatase antibody likely accounts for the difference in our results from those of a previous study in goldfish (Gelinas and Callard, 1997). The aromatase enzyme throughout vertebrates shows remarkable variation; our analysis shows only a 57% sequence identity of midshipman brain aromatase to human placental aromatase compared with identities as high as 79% from other teleosts. Thus, as Gelinas and Callard (1997) recognize, it is not surprising that the antibody made against human placental aromatase used in their study might not accurately bind to goldfish brain aromatase. Our antibody did not work for goldfish brain (P. Forlano and A. Bass, unpublished observations), perhaps not surprising given the low sequence identity between midshipman and goldfish brain aromatases and their distant phylogenetic relationship (although the antibody was made to conserved regions of teleost aromatases, it corresponds more to tilapia and midshipman brain, as well as goldfish ovarian aromatase than to goldfish brain aromatase). Also, as shown in goldfish (Gelinas et al., 1998), aromatase expression can vary seasonally, depending on reproductive condition. Perhaps the few goldfish we tested had reduced basal levels of aromatase because of their nonreproductive state. However, our antibody did work on juvenile tilapia (genusOreochromis; Forlano and Bass, unpublished observations). Because of the high sequence identity of midshipman brain aromatase toOreochromis brain aromatase, we tested our specific antibody and ISH probes on Oreochromis brain tissue and found a similar pattern of label as in midshipman with the antibody and ISH probes in the preoptic area (Forlano and Bass, unpublished observations). The sequence identities to different forms of aromatase (ovarian vs brain) might point to differences in expression in glia versus neurons and explain why our antibody labeled tilapia, but not goldfish, brain in a manner similar to that of midshipman.
As we report here, our ISH probes label ovarian aromatase in midshipman. Thus, midshipman aromatase expressed in brain or ovary may be identical forms, or the probes we designed may hybridize to conserved regions in both tissues. If a distinct ovarian form of aromatase exists in midshipman brain, it could be found in neurons in small amounts. Evidence for multiple transcripts of aromatase is seen in goldfish and zebrafish (Callard and Tchoudakova, 1997) and so far are the only examples in vertebrates in which two aromatase transcripts appear to be the result of two different aromatase genes. Neuronal aromatase may exist in midshipman, but the amount we found in glial cells reflects activity levels, whereas neuronal numbers found in goldfish brain does not.
Aromatase localization to glia
An unexpected result of this work is that high levels of aromatase expression are localized in glia and not neurons. Other studies in mammals and birds have shown aromatase expression in glia only after brain injury in adult animals in situ (Garcia-Segura et al., 1999a,b; Peterson et al., 2001) or in culture (Schlinger et al., 1994). This is the first study to provide evidence for brain aromatase expression to be mostly, if not entirely, in glial cells in any vertebrate in unmanipulated animals. Aromatase-IR cells and fibers are glial in appearance, follow the same pattern as radial glia, and are colocalized with vimentin and GFAP-IR fibers in certain brain regions but are never colocalized with Hu-IR neuronal cell bodies or axons labeled by anti-acetylated tubulin. The morphology and staining pattern of radial glia in the adult avian brain using anti-vimentin (the same antiserum used in this study) is remarkably similar to the pattern of aromatase-IR expression in the midshipman brain, namely, somata in the ventricular zone contain fibers that penetrate the forebrain parenchyma for up to several millimeters (Alvarez-Buylla et al., 1987). In general, radial glial cells are characterized by long radial processes that reach the basal surface of the brain and their expression of GFAP and other markers. Radial glia are best known to function in the guidance of migrating neurons but are also known to transform into astrocytes after neurogenesis and migration is completed (for review, see Hartfuss et al., 2001). Moreover, astrocytes are proposed as stem cells in the adult telencephalon (Doetsch et al., 1999), andAlvarez-Buylla et al. (2001) have proposed radial glia to be elongated neuroepithelial cells that give rise to both neurons and glia throughout ontogeny. Recent studies recognize several subtypes of radial glia based on differential antigen expression, and the function of radial glia appears to be dependent on subtype within and between distinct brain regions (Hartfuss et al., 2001). Therefore, it is not surprising that we found colocalization of aromatase immunoreactivity and GFAP and vimentin immunoreactivity in some brain regions and not others, because certain glial subsets may not express the markers we used.
Function of high aromatase levels
Although the functional outcomes of high aromatase levels in the brain of teleosts, and more generally vertebrates, is unknown, we discuss a number of possibilities, including a role in sexual maturation events, as a neurotrophic factor, and as a modulator of steroid-sensitive, neural circuitry.
Although the distribution of estrogen receptors in midshipman brain is unknown, Fine et al. (1990) localized [3H]estradiol-concentrating cells in the closely related toadfish (Opsanus tau, same order and family) to many of the same areas in the forebrain and diencephalon in which we found aromatase expression (area ventralis of the telencephalon, preoptic area, and hypothalamic ventricular regions). Aromatase is known to affect the development of sexually dimorphic brain nucleus of the preoptic area (SDN-POA) and dendritic morphology (Burke et al., 1999) in other vertebrates (for review, seeBeyer, 1999). The SMN, which innervates the sonic swimbladder muscles and is intersexually and intrasexually dimorphic in midshipman (Bass and Marchaterre, 1989; Bass and Baker, 1990; Bass et al., 1996), is enshrouded with aromatase-IR cells and fibers, contains high levels of aromatase mRNA, and probably accounts for most of the aromatase activity found in the hindbrain. Whereas type I male midshipman alone have detectable levels of 11-ketotestosterone (a non-aromatizable androgen in teleost fish), type II males and females have similarly higher testosterone levels than type I males (Brantley et al., 1993). Preliminary results suggest that, like some other vertebrates (Balthazart and Ball, 1998; Gelinas et al., 1998), testosterone can upregulate aromatase expression (Forlano and Bass, 2001); testosterone can also masculinize the sonic motor system (Bass, 1995). We hypothesize that relative levels of aromatase expression in and around this nucleus that correspond to other dimorphic nuclei in the vocal-motor circuit may function to prevent its masculinization by circulating testosterone and therefore may be a key mechanism in both generating and maintaining alternative male phenotypes in this species (Schlinger et al., 1999).
A relationship between relative periods of neurogenesis and aromatase activity in different vertebrate taxa may reflect the role of estrogen as a neurotrophic factor. In mammals, the highest peak of aromatase activity coincides with a critical organizational period of brain development just before birth, followed by a subsequent decrease in activity with age (Lephart, 1996). Songbirds have high aromatase levels in the telencephalon as adults and show dynamic seasonal changes in the song system areas of the brain in which new neurons appear to be recruited at times when song modification occurs (Alvarez-Buylla and Kirn, 1997). Teleost fish have the highest brain aromatase levels of all vertebrates and also show continuous neurogenesis throughout life. The capacity of neurogenesis and plasticity, and regeneration in adult fish resembles properties seen in embryonic mammalian and avian brain (Levine et al., 1994). Possible functions of continual neurogenesis include more efficient replacement of damaged cells in the event of injury and increases in central neuronal elements linked to continual somatic growth (increase in number of peripheral motor and sensory structures). Also, the continual addition of new cells may enable structural changes centrally that may be required for long-term changes in behavior (for review, see Zupanc, 1999). The factors that mediate continual neurogenesis in fish are unknown (Zupanc, 1999). One function of high levels of aromatase in the adult teleost brain may be to provide large quantities of estrogenic compounds to induce continuous cell proliferation (Gelinas et al., 1998). Perhaps the teleost brain, which can grow continuously throughout life, requires the continual production of estrogenic compounds in amounts relative to that seen in upregulation after brain injury in other vertebrate groups.
Estrogen is also known to cause synaptic plasticity and glial activation (Garcia-Segura et al., 1989; Naftolin et al., 1990); recent studies show glia to be steroidogenic and express sex steroid receptors (for review, see Jordan, 1999). In mammals, estrogen is widely documented to act as a growth-stimulating agent, modulate apoptotic cell death, and affect migration of neuroblasts from the subventricular layer and thus their aggregation to form brain nuclei with consequent synapse formation. These trophic effects are mostly seen in distinct brain nuclei that are estrogen-sensitive and become sexually dimorphic in adults (for review, see Beyer, 1999; Wise et al., 2001).Garcia-Segura et al. (1996) first suggested that estrogenic control of insulin-like growth factor may be crucial for the development of sex-specific neuroendocrine circuits and the synaptic plasticity of adult neuroendocrine systems. Coexpression of neurotrophins and estrogen, and their receptors, suggest that both factors work together to differentiate target neurons (Beyer, 1999). Contreras and Wade (1999) demonstrated recently that an aromatase inhibitor administered to 3-d-old zebra finches (a period of active neural development) significantly lowered nerve growth factor (NGF) binding sites in telencephalon brain homogenates, supporting the hypothesis that estrogen influences NGF receptors and therefore brain development. In addition, Dittrich et al. (1999) found 17β-estradiol to prematurely upregulate brain-derived neurotrophic factor (BDNF) in sexually dimorphic song nuclei in juvenile males, and this upregulation was inhibited by an aromatase inhibitor. Thus, regulation of BDNF or other growth factors is one mechanism by which estrogen may influence neural differentiation. In summary, the well established role of glia in neurogenesis, neuroplasticity and repair, as well as recent findings of an intimate interplay between glia, gonadal steroids, and growth factors, would make glial cells prime candidates to execute multiple functions essential for neuronal maturation, plasticity, and repair in all vertebrates.
Midshipman (Brantley et al., 1993; Knapp et al., 1999), as other teleosts (Norris, 1997), have relatively high circulating androgens, so that high aromatase levels in the brain may also function to regulate the amount of steroid(s) that reaches steroid-sensitive neural targets or modify the overall hormonal milieu in circulation. The position of aromatase-IR cells and ISH silver grains are consistently located in ventricular areas throughout the brain, and the location and pattern of projections appears ideal for the conversion of testosterone to estradiol from the cerebral spinal fluid or blood vasculature to bathe the brain in estrogenic compounds. The possible role of glia in teleosts as mediators of brain steroid levels that, in turn, could modulate multiple neuronal functions is consistent with teleosts showing the widest range of plasticity in vertebrate reproductive function at multiple levels of biological organization.
Footnotes
This research was supported by National Institute of Mental Health Predoctoral Training Grant 5 T32 MH15793 (to P.M.F.) and National Science Foundation Grant IBN9987341 (to A.H.B.). We thank Margaret Marchaterre, James Goodson, Matthew Weeg, Wen Wu, Yuko Hara, Ilya Vilinsky, and Cathy Lanning for technical advice, Joseph Sisneros and Margaret Marchaterre for field assistance, Colin Saldanha, Barney Schlinger, Anthony Tramontin, and Neil Segil for helpful suggestions, and U. C. Bodega Marine Laboratory for logistical support.
Correspondence should be addressed to Paul M. Forlano, Department of Neurobiology and Behavior, Seeley G. Mudd Hall, Cornell University, Ithaca, NY 14853. E-mail: pmf4@cornell.edu.
REFERENCES
- 1.Alvarez-Buylla A, Kirn J. Birth, migration, incorporation, and death of vocal control neurons in adult songbirds. J Neurobiol. 1997;33:585–601. [PubMed] [Google Scholar]
- 2.Alvarez-Buylla A, Buskirk DR, Nottebohm F. Monoclonal antibody reveals radial glia in adult avian brain. J Comp Neurol. 1987;264:159–170. doi: 10.1002/cne.902640203. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Buylla A, Garcia-Verdugo JM, Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 4.Balthazart J, Ball G. New insights into the regulation and function of brain estrogen synthase (aromatase). Trends Neurosci. 1998;21:243–248. doi: 10.1016/s0166-2236(97)01221-6. [DOI] [PubMed] [Google Scholar]
- 5.Bass AH. Dimorphic male brains and alternate reproductive tactics in a vocalizing fish. Trends Neurosci. 1992;15:139–145. doi: 10.1016/0166-2236(92)90356-d. [DOI] [PubMed] [Google Scholar]
- 6.Bass AH. Alternate life history strategies and dimorphic males in an acoustic communication system. Fifth Intl Symp Reprod Biol Fish, Fish Symp. 1995;95:258–260. [Google Scholar]
- 7.Bass AH. Shaping brain sexuality. Am Sci. 1996;84:352–363. [Google Scholar]
- 8.Bass AH, Baker R. Sexual dimorphisms in the vocal control system of a teleost fish: morphology of physiologically identified neurons. J Neurobiol. 1990;21:1155–1168. doi: 10.1002/neu.480210802. [DOI] [PubMed] [Google Scholar]
- 9.Bass AH, Marchaterre MA. Sound-generating (sonic) motor system in a teleost fish (Porichthys notatus): Sexual polymorphisms and general synaptology of a sonic motor nucleus. J Comp Neurol. 1989;286:154–169. doi: 10.1002/cne.902860203. [DOI] [PubMed] [Google Scholar]
- 10.Bass AH, Marchaterre MA, Baker R. Vocal-acoustic pathways in a teleost fish. J Neurosci. 1994;14:4025–4039. doi: 10.1523/JNEUROSCI.14-07-04025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass AH, Horvath BJ, Brothers EB. Non-sequential developmental trajectories lead to dimorphic vocal circuitry for males with alternative reproductive tactics. J Neurobiol. 1996;30:493–504. doi: 10.1002/(SICI)1097-4695(199608)30:4<493::AID-NEU5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 12.Beyer C. Estrogen and the developing mammalian brain. Anat Embryol. 1999;199:379–390. doi: 10.1007/s004290050236. [DOI] [PubMed] [Google Scholar]
- 13.Brantley RK, Wingfield J, Bass AH. Sex steroid levels in Porichthys notatus, a fish with alternative reproductive tactics, and a review of the hormonal bases for male dimorphism among teleost fishes. Horm Behav. 1993;27:332–347. doi: 10.1006/hbeh.1993.1025. [DOI] [PubMed] [Google Scholar]
- 14.Burke KA, Kuwajima M, Sengelaub DR. Aromatase inhibition reduces dendritic growth in a sexually dimorphic rat spinal nucleus. J Neurobiol. 1999;38:301–312. [PubMed] [Google Scholar]
- 15.Callard GV, Tchoudakova A. Evolutionary and functional significance of two CYP19 genes differentially expressed in brain and ovary of goldfish. J Steroid Biochem Mol Biol. 1997;61:387–392. doi: 10.1016/s0960-0760(97)80037-4. [DOI] [PubMed] [Google Scholar]
- 16.Callard GV, Schlinger BA, Pasmanik M. Nonmammalian vertebrate models in studies of brain-steroid interactions. J Exp Zool Suppl. 1990;4:6–16. doi: 10.1002/jez.1402560404. [DOI] [PubMed] [Google Scholar]
- 17.Contreras ML, Wade J. Interactions between nerve growth factor binding and estradiol in early development of the zebra finch telencephalon. J Neurobiol. 1999;40:149–157. doi: 10.1002/(sici)1097-4695(199908)40:2<149::aid-neu2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Dittrich F, Feng Y, Metzdorf R, Gahr M. Estrogen-inducible, sex-specific expression of brain-derived neurotrophic factor mRNA in a forebrain song control nucleus of the juvenile zebra finch. Proc Natl Acad Sci USA. 1999;96:8241–8246. doi: 10.1073/pnas.96.14.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 20.Fine ML, Keefer DA, Russel-Mergenthal H. Autoradiographic localization of estrogen-concentrating cells in the brain and pituitary of the oyster toadfish. Brain Res. 1990;536:207–219. doi: 10.1016/0006-8993(90)90027-9. [DOI] [PubMed] [Google Scholar]
- 21.Foran CM, Bass AH. Preoptic GnRH and AVT: Axes for sexual plasticity in teleost fish. Gen Comp Endocrinol. 1999;116:141–152. doi: 10.1006/gcen.1999.7357. [DOI] [PubMed] [Google Scholar]
- 22.Forlano PM, Bass AH. Sex steroid modulation of brain aromatase mRNA expression in a vocal fish. Soc Neurosci Abstr. 2001;27:1081. [Google Scholar]
- 23.Forlano PM, Myers DA, Bass AH. Aromatase distribution in the brain of a vocalizing fish with three adult morphs. Soc Neurosci Abstr. 2000;26:1522. [Google Scholar]
- 24.Forlano PM, Deitcher DL, Bass AH. Distribution of aromatase mRNA in the brain and gonads of a polymorphic vocal teleost. Horm Behav. 2001;39:331. [Google Scholar]
- 25.Garcia-Segura LM, Olmos G, Robbins RJ, Hernandez P, Meyer JH, Naftolin F. Estradiol induces rapid remodeling of plasma membranes in developing rat cerebrocortical neurons in culture. Brain Res. 1989;498:339–343. doi: 10.1016/0006-8993(89)91113-x. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Segura LM, Duenas M, Fernandez-Galaz MC, Chowen JA, Argente J, Naftolin F, Torres-Aleman I. Interaction of the signaling pathways of insulin-like growth factor I and sex steroids in the neuroendocrine hypothalamus. Horm Res. 1996;46:160–164. doi: 10.1159/000185016. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Segura LM, Wozniak A, Azicoita I, Rodriques JR, Hutchinson RE, Hutchinson JB. Aromatase expression by astrocytes after brain injury: implications for local estrogen formation in brain repair. Neuroscience. 1999a;89:567–578. doi: 10.1016/s0306-4522(98)00340-6. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Segura LM, Naftolin F, Hutchinson JB, Azcoitia I, Chowen JA. Role of astroglia in estrogen regulation of synaptic plasticity and brain repair. J Neurobiol. 1999b;40:574–584. [PubMed] [Google Scholar]
- 29.Gelinas D, Callard GV. Immunolocalization of aromatase- and androgen receptor-positive neurons in the goldfish brain. Gen Comp Endocrinol. 1997;106:155–168. doi: 10.1006/gcen.1997.6891. [DOI] [PubMed] [Google Scholar]
- 30.Gelinas D, Pitoc GA, Callard GV. Isolation of a goldfish brain cytochrome P450 aromatase cDNA: mRNA expression during the seasonal cycle and after steroid treatment. Mol Cell Endocrinol. 1998;138:81–93. doi: 10.1016/s0303-7207(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 31.Goodson JL, Bass AH. Forebrain peptides modulate sexually polymorphic vocal circuitry. Nature. 2000;403:769–772. doi: 10.1038/35001581. [DOI] [PubMed] [Google Scholar]
- 32.Grober MS, Myers TR, Marchaterre MA, Bass AH, Myers DA. Structure, localization, and molecular phylogeny of a GnRH cDNA from a paracanthopterygian fish, the plainfin midshipman (Porichthys notatus). Gen Comp Endocrinol. 1995;99:85–99. doi: 10.1006/gcen.1995.1088. [DOI] [PubMed] [Google Scholar]
- 33.Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- 34.Hickey GT, Krasnow JS, Beattie WG, Richards JS. Aromatase cytochrome P450 in rat ovarian granulosa cells before and after luteinization: adenosine 3,5-monophosphate dependent and independent regulation. Cloning and sequencing of rat aromatase cDNA and 5′ genomic DNA. Mol Endocrinol. 1990;4:3–12. doi: 10.1210/mend-4-1-3. [DOI] [PubMed] [Google Scholar]
- 35.Jordan CL. Glia as mediators of steroid hormone action on the nervous system: an overview. J Neurobiol. 1999;40:434–445. doi: 10.1002/(sici)1097-4695(19990915)40:4<434::aid-neu2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 36.Kalman M. Astroglial architecture of the carp (Cyprinus carpio) brain as revealed by immunohistochemical staining against glial fibrillary acidic protein (GFAP). Anat Embryol. 1998;198:409–433. doi: 10.1007/s004290050193. [DOI] [PubMed] [Google Scholar]
- 37.Knapp R, Wingfield JC, Bass AH. Steroid hormones and parental care in the plainfin midshipman fish (Porichthys notatus). Horm Behav. 1999;35:81–89. doi: 10.1006/hbeh.1998.1499. [DOI] [PubMed] [Google Scholar]
- 38.Lephart ED. A review of brain aromatase cytochrome P450. Brain Res Rev. 1996;22:1–26. [PubMed] [Google Scholar]
- 39.Levine EM, Hitchcock PF, Glasgow E, Schechter N. Restricted-expression of a new paired-class homeobox gene in normal and regenerating adult goldfish retina. J Comp Neurol. 1994;348:596–606. doi: 10.1002/cne.903480409. [DOI] [PubMed] [Google Scholar]
- 40.Manso MJ, Becerra M, Becerra M, Anadon R. Expression of a low-molecular-weight (10 kDa) calcium binding protein in glial cells of the brain of the trout (Teleostei). Anat Embryol. 1997;196:403–416. doi: 10.1007/s004290050108. [DOI] [PubMed] [Google Scholar]
- 41.Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 42.Naftolin F, Garcia-Segura LM, Keefe D, Leranth C, MacLusky NJ, Brawer JR. Estrogen effects on the synaptology and neural membranes of the rat hypothalamic arcuate nucleus. Biol Reprod. 1990;42:21–28. doi: 10.1095/biolreprod42.1.21. [DOI] [PubMed] [Google Scholar]
- 43.Nagahama Y. The functional morphology of teleost gonads. In: Hoar WS, Randall DJ, Donaldson EM, editors. Fish physiology. Academic; New York: 1983. pp. 223–275. [Google Scholar]
- 44.Nieuwenhuys R. An overview of the organization of the brain of actinopterygian fishes. Am Zool. 1982;22:287–310. [Google Scholar]
- 45.Norris DO. Vertebrate endocrinology. Academic; San Diego: 1997. [Google Scholar]
- 46.Peterson RS, Saldanha CJ, Schlinger BA. Rapid upregulation of aromatase mRNA and protein following neural injury in the zebra finch (Taeniopygia guttata). J Neuroendocrinol. 2001;13:317–323. doi: 10.1046/j.1365-2826.2001.00647.x. [DOI] [PubMed] [Google Scholar]
- 47.Schlinger BA, Amur-Umarjee S, Shen P, Campagnoni AT, Arnold AP. Neuronal and non-neuronal aromatase in primary cultures of developing zebra finch telencephalon. J Neurosci. 1994;14:7541–7552. doi: 10.1523/JNEUROSCI.14-12-07541.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schlinger BA, Greco C, Bass AH. Aromatase activity in hindbrain vocal control region of a teleost fish: divergence among males with alternative reproductive tactics. Proc R Soc Lond B Biol Sci. 1999;266:131–136. [Google Scholar]
- 49.Wise PM, Dubal DB, Wilson ME, Rau SW, Liu Y. Estrogens: trophic and protective factors in the adult brain. Front Neuroendocrinol. 2001;22:33–66. doi: 10.1006/frne.2000.0207. [DOI] [PubMed] [Google Scholar]
- 50.Zupanc GKH. Neurogenesis, cell death and regeneration in the adult gymnotiform brain. J Exp Biol. 1999;202:1435–1446. doi: 10.1242/jeb.202.10.1435. [DOI] [PubMed] [Google Scholar]