Abstract
Two distal downstream enhancers controlling astrocyte expression of the human apolipoprotein E (apoE) gene in the brain were identified by analysis of transgenic mice generated with various constructs of the apoE/C-I/C-IV/C-II gene cluster. In wild-type mice, the highest overall levels of apoE mRNA were found in astrocytes in the glomerular layer of olfactory bulbs and in Bergmann glia in the cerebellum. This pattern of expression was reproduced in transgenic mice expressing the entire human apoE gene cluster and also in transgenic mice expressing specific enhancer segments within the cluster. Expression of the human apoE transgene at these sites was specified by two enhancer domains: one enhancer is located 3.3 kb downstream of the apoE gene, and a duplication of this sequence is located 15 kb downstream of the apoE gene. Astrocyte enhancer activity was contained within 620 and 619 bp segments of these domains that show subtle differences in regional expression. In the absence of these distal enhancers, the apoE gene was not expressed in astrocytes. The relatively high levels of apoE expression at specific sites in the olfactory bulb and cerebellum suggest the presence of unique regulatory signals at these locations that may reflect common cellular properties and apoE gene functions. The localization of the two astrocytic enhancers reveals an unexpected complexity in the control of apoE production that is essential to understanding apoE function in the brain.
Keywords: apoE mRNA, astrocyte, olfactory bulb, cerebellum, enhancer, transgenic mouse
Apolipoprotein E (apoE) is a protein (Mr = 35,000) that functions in the CNS. This conclusion is supported by the abundance of apoE mRNA in the brain (Elshourbagy et al., 1985), the presence of immunoreactive apoE in subsets of brain glial cells (Boyles et al., 1985), and the finding of apoE in CSF lipoproteins (Pitas et al., 1987). Several studies suggest that apoE contributes to neural homeostasis. ApoE may help to repair and maintain neuronal integrity after lesion-induced damage (Poirier et al., 1993), acute head injury (Chen et al., 1997), stroke (Slooter et al., 1997), and during aging (Masliah et al., 1995a). ApoE is present in extracellular insoluble amyloid plaques and intracellular neurofibrillary tangles that are characteristic of Alzheimer's disease (Namba et al., 1991).
The presence of either arginine or cysteine at amino acid positions 112 and 158 (Rall et al., 1982) defines three common human apoE variants, E2, E3, and E4 (Zannis and Breslow, 1981;Sing and Davignon, 1985) that affect plaque formation and the progression of neurodegenerative disorders. ApoE4 is an important risk factor for late-onset Alzheimer's disease (Corder et al., 1993; Strittmatter et al., 1993; Roses, 1996). Isoform-specific differences on maintaining neuronal integrity have also been described for apoE. For example, neurite outgrowth by cultured neuronal cells is enhanced by apoE3 and inhibited by apoE4 (Nathan et al., 1994;Sun et al., 1998). Furthermore, apoE3 has a greater neuroprotective effect than apoE4 in aged transgenic mice and after excitotoxin-induced neuronal damage (Buttini et al., 1999). In vivo studies have also supported a role for apoE in learning and memory. ApoE4 is linked to memory deficits associated with human hippocampal disorders (Soininen and Riekkinen, 1996). In addition, synaptic and dendritic alterations correlated with learning deficits are observed in apoE-deficient mice (Masliah et al., 1995b).
Astrocytes are the major site of immunoreactive apoE throughout the CNS (Boyles et al., 1985). ApoE is particularly abundant in Bergmann glia of the cerebellum. Changes in astrocyte protein expression, including an upregulation of apoE synthesis, are initiated by inflammatory and neurodegenerative insults. The production of apoE by hippocampal astrocytes increases dramatically in response to kainic acid excitotoxicity and after surgical lesioning in or near the hippocampus (Page et al., 1998), and apoE accumulates at sites of neuronal degeneration (Ong et al., 1997). Reactive astrocytes have been described that respond to inflammatory mediators, including interleukin-1, tumor necrosis factor, and glucocorticoids (Eddleston and Mucke, 1993;Merrill and Benveniste, 1996; Rutka et al., 1997). However, little is known about the regulatory mechanisms that control the initiation of gene expression in astrocytes in response to inflammatory and degenerative signals. Because the production of apoE by astrocytes appears to be induced in response to multiple physiological signals, we investigated the regulatory elements that specify its expression in these cells.
We used transgenic mouse models generated with constructs of the human apoE gene to identify regulatory domains that direct apoE expression to astrocytes. We found that two enhancers control apoE production in astrocytes and that they have independent differences in the patterns of their activity. The apoE gene promoter cannot direct transcription of the apoE gene in astrocytes in the absence of an astrocyte enhancer. Our results show an unexpected complexity in the regulation of apoE in the brain and permit the use of astrocyte-specific enhancers of apoE expression to help understand the role of apoE in the brain in response to a wide variety of regulatory signals in health and disease.
MATERIALS AND METHODS
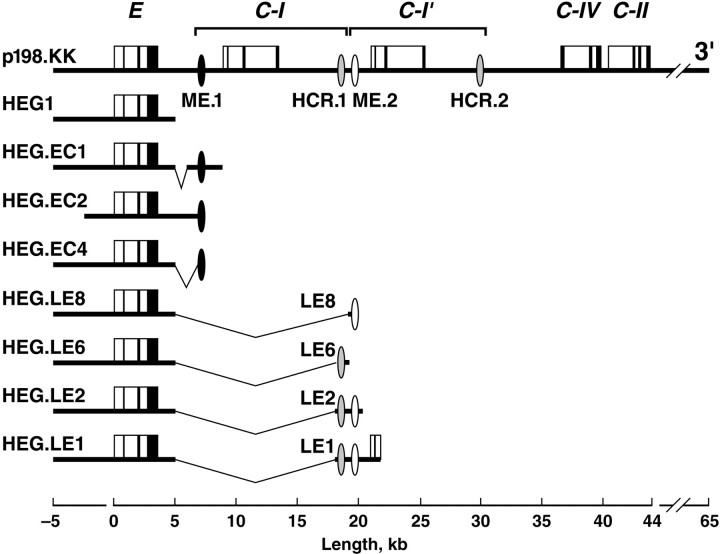
Transgenic mice. Transgenic mice were generated with constructs of the human apoE gene cluster using the inbred FVB/N strain. The apoE3-expressing p198.KK, HEG1, HEG.LE1, HEG.LE2, and HEG.LE6 constructs (Fig. 1) have been described previously (Simonet et al., 1993; Allan et al., 1995a). The LE8 fragment was derived from the LE1 fragment and ligated to the 3′ end of HEG1 to generate HEG.LE8. A single nucleotide substitution was introduced into the codon for amino acid 112 in the fourth exon of the apoE gene in HEG.LE1 to yield an apoE4-expressing construct that was confirmed by nucleotide sequencing and demonstrated to be otherwise identical with the apoE3-expressing HEG.LE1 construct (data not shown). The HEG.EC1 and HEG.EC4 constructs were prepared by ligating 2.6 kb and 620 bp fragments of the apoE/C-I intergenic region to the 3′ end of HEG1; these fragments were generated by PCR extension of oligonucleotides beginning 2.7 and 3.3 kb downstream of the apoE gene. The HEG.EC2 construct was generated bySphI restriction endonuclease digestion of p198.KK.
Fig. 1.
Transgenic constructs. The bold horizontal lines represent human genomic DNA. The apolipoprotein genes are illustrated as solid vertical lines for exons and open boxes for introns, and the scale in kilobases is shown. Transcription is in the 5′ to 3′ orientation for all genes. The 12 kb/11 kb duplication that yielded the apoC-I and apoC-I′ genes is shown as inverted brackets. The locations of hepatocyte-expressing hepatic control regions (HCR.1 and HCR.2) (Allan et al., 1995b, 1997) and astrocyte-expressing multienhancers (ME.1 and ME.2) are indicated by ovals. Theinverted lines illustrate segments downstream of the apoE gene that were deleted in preparing constructs as indicated in Materials and Methods. The HEG.EC1 construct deleted 1.0 kb and the HEG.EC4 construct deleted 1.6 kb from the intergenic region. The LE8 (0.06 kb), LE6 (0.77 kb), and LE2 (1.7 kb) fragments were derived from the LE1 (3.8 kb) fragment and ligated to HEG1. The 3′ end of HEG1 is located 1.7 kb downstream of the apoE gene. The 5′ end of LE1 is located 14.3 kb downstream of the apoE gene. The HEG.EC2 construct consists of an SphI–SphI subfragment of p198.KK.
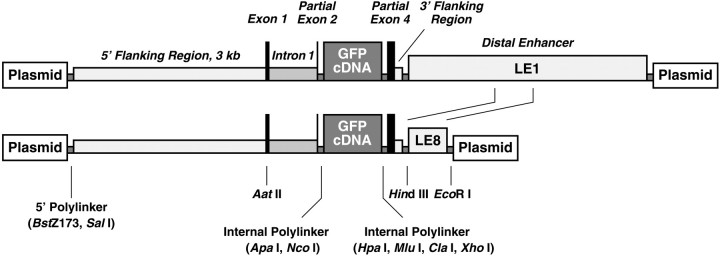
To confirm brain enhancer activity, we modified a transgene expression vector (Fan et al., 1994; Ji et al., 1994) constructed with sequences from the human apoE gene. ApoE gene sequences in the vector include 3.0 kb of 5′ flanking sequence, the first exon, the first intron, and six nucleotides of the second exon ligated to a polylinker that serves as a cDNA insertion site. The cDNA site is followed by the 3′ terminal 124 nucleotides of the human apoE gene fourth exon and 112 nucleotides of 3′ flanking sequence ligated to a polylinker into which the LE1 and LE8 fragments (Fig. 1) were inserted (see Fig. 9). Additional polylinker sites at both ends of the construct permit insertion into and excision from plasmids. An enhanced green fluorescent protein (EGFP) cDNA (Stratagene, San Diego, CA) was inserted into the cDNA site to generate a reporter construct for our studies.
Fig. 9.
Reporter transgene expression vectors. The regulatory components of the vector were derived from the human apoE gene cluster, and details of vector sequences are described in Materials and Methods. The 3.8 kb LE1 and the 0.6 kb LE8 fragments were inserted into the internal flanking polylinker.
To analyze transgene expression, the F1 progeny from two to four different transgenic lines (derived from independent founders) were examined for each construct, including at least two males and two females in each transgenic line. The activity of each enhancer was confirmed by analysis of transgenic mice generated with at least two different subfragments of the p198.KK parent construct. In addition, expression of the heterologous EGFP reporter construct further confirmed the action of the LE1 and LE8 fragments. Thus, multi-enhancer 2 (ME.2) activity was confirmed by analysis of four lines of transgenic mice generated with HEG.LE1, three lines generated with HEG.LE2, and two lines generated with HEG.LE8 (Fig. 1). In addition, three lines from each of two different constructs in which the EGFP vector was ligated to either the LE1 or LE8 fragment were also analyzed (see Fig. 9). Similarly, the activity of ME.1 was confirmed in multiple lines of HEG.EC1, HEG.EC2, and HEG.EC4 transgenic mice (Fig. 1). We show representative data for constructs that reveal the activities of the two different astrocyte enhancers.
In situ hybridization. To prepare tissues, control FVB/N and transgenic mice were perfused with 4% paraformaldehyde. Brains were removed quickly and stored in 4% paraformaldehyde at 4°C for 24–36 hr. Perfused brains were paraffin-embedded, and then 7 μm sections were cut with a rotary microtome, mounted on slides (Superfrost Plus; Fisher Scientific, Pittsburgh, PA), and stored at 4°C until use. Before hybridization, slides were warmed to room temperature and baked at 55°C for 30 min. Paraffin was removed from the slides by two successive 10 min immersions in xylene. Tissue sections were hydrated in a progressive series of 3 min washes in ethanol (100, 90, 70, 50, and 30%), followed by three successive 5 min immersions in 1× PBS (in mm): 137 NaCl, 2.7 KCl, 4.3 Na2HPO4 · 7H2O, 1.4 KH2PO4, pH 7.3. The sections were incubated with 20 μg/ml proteinase K (Boehringer Mannheim, Indianapolis, IN) in 50 mm Tris-HCl, pH 8.0, 5 mm EDTA, and 150 mm NaCl for 15 min at room temperature. Proteolytic activity was stopped by immersion for 10 min in 0.2% glycine in 1× PBS. Tissues were rinsed in 1× PBS for 5 min, fixed in 4% paraformaldehyde in 1× PBS for 5 min, rinsed in 1× PBS for 5 min, and then acetylated with 0.25% acetic anhydride in 0.1 mm triethanolamine buffer at pH 8.0 for 10 min. Tissue sections were given a final rinse in 1× PBS for 5 min and then dehydrated in a progressive series of 3 min washes in ethanol (30, 50, 80, 95, and 100%). RNA probes complementary to nucleotides 492–783 of mouse apoE mRNA and 20–411 of human apoE mRNA were labeled with [α-33P]UTP to a specific activity of at least 2 × 104 Ci/mmol using a RNA transcription kit (Stratagene). Labeled probes were purified through Micro Bio-Spin 30 chromatography columns (Bio-Rad, Hercules, CA).
Tissue sections were incubated for 14–18 hr in a humidified chamber at 42°C with probes diluted to 2.0 × 106 cpm per slide in a buffer containing 50% formamide, 300 mmNaCl, 20 mm Tris, pH 8.0, 5 mm EDTA, 1× Denhardt's solution [0.2% polyvinylpyrrolidone, 0.02% Ficoll, 0.02% bovine serum albumin (BSA)] 10% dextran sulfate, 250 μg/ml sperm DNA, and 0.1 mg/ml tRNA. The sections were washed twice at room temperature in 2× SSC, 1.0 mm EDTA for 10 min, and immersed in 20 μg/ml ribonuclease (RNase) A (Sigma, St. Louis, MO) in 500 mm NaCl and 10 mm Tris, pH 8.0, and 10 U/ml T1 RNase (Boehringer Mannheim) for 1 hr at 37°C. Tissue sections were washed at 55°C in six changes of 0.1× SSC with 1.0 mmEDTA for 4 hr, rinsed twice for 10 min each in 0.5× SSC, and dehydrated. For autoradiography, the slides were exposed at −20°C to β-max film (Amersham, Arlington Heights, IL) for 4 d. For dark-field and bright-field microscopy imaging, the slides were dipped in NTB2 nuclear track emulsion (Eastman Kodak, Rochester, NY), incubated at 4°C for 5 d, and developed with D19 developer (Eastman Kodak). The sections were stained for 2 min with hematoxylin (Fisher Scientific), washed in Scott's Tap Water Substitute (Fisher Scientific) for 3 min and 80% ethanol for 1 min, and then restained with eosin (Fisher Scientific) for 3 min. After dehydration by rinsing in a graded series of ethanol (80, 95, and 100%), the slides were rinsed three times in xylene and overlaid with coverslips.
Cellular localization studies. Immunohistochemistry was done with a tyramide signal amplification (TSA) kit (MA, TSA-fluorescein system, NEL701; TSA-tetramethylrhodamine system, NEL702; NEN, Boston, MA) according to the manufacturer's recommendations with the following changes. Paraffin was removed from the slides by three successive 10 min immersions in 100% xylene. Sections were rehydrated by immersion in 100% ethanol twice for 10 min each, 50% ethanol for 10 min, and H2O twice for 10 min each. Sections were washed three times in PBS–glycine (1× PBS, 0.02 m glycine) containing 1% BSA for 2 min each and were treated with 3% H2O2, 1% BSA in PBS–glycine for 18 min. Slides were rinsed three times in PBS–glycine containing 0.1% BSA for 2 min each and blocked using 3% BSA in PBS–glycine for 60 min at room temperature. Then sections were incubated with a primary antibody for astrocyte-specific glial fibrillary acidic protein (GFAP) or human apoE in 1% BSA in PBS–glycine overnight at 4°C in a humid chamber. Tissues were rinsed three times for 5 min each in PBS–glycine containing 0.1% BSA and blocked with Peroxoblock solution (Zymed, South San Francisco, CA) for 45 sec. After three 5 min washes in PBS–glycine containing 1% BSA, sections were incubated with biotinylated secondary antibodies in 1% BSA in PBS–glycine for 1 hr at room temperature. Slides were washed three times in TNT buffer (0.1m Tris-HCl, 0.15 m NaCl, 0.05% Tween 20) for 5 min each with gentle agitation, incubated in TNB blocking buffer (0.1m Tris-HCl, 0.15 m NaCl, 0.5% blocking reagent supplied in the TSA kit) for 30 min at room temperature, and incubated for 30 min at room temperature in streptavidin–horseradish peroxidase (provided in the TSA kit). Sections were washed three times in TNT buffer for 5 min each with gentle agitation, incubated in fluorescein tyramide or tetramethylrhodamine (supplied in the TSA kit) for 7 min at room temperature, and washed again five times in TNT buffer for 5 min each. Sections were overlaid with coverslips using Vectashield mounting medium for fluorescence (H-1000; Vector Laboratories, Burlingame, CA), and the coverslips were sealed with nail polish. For dual-labeling studies, sequential labelings were performed on each section, including appropriate controls to detect any cross reaction of antibodies. Tissue sections were imaged by fluorescence microscopy. In control experiments in which primary antibody was omitted, no staining of cells could be detected.
RESULTS
Endogenous mouse apoE mRNA expression in the brain
Endogenous apoE mRNA was detected at various levels of expression throughout the brain as shown by autoradiograms after in situ hybridization of both horizontal and sagittal sections from nontransgenic control mice brains (Fig.2A,B). Human apoE mRNA in transgenic mice generated with the complete apoE gene cluster, p198.KK (Fig. 2D,E), had an expression pattern similar to that of endogenous mouse apoE mRNA, although subtle differences in apoE expression were found between transgenic and control brains. The highest level of apoE mRNA in both nontransgenic control (Fig. 2A,B) and p198.KK transgenic (Fig.2D,E) mice was found in the glomerular layer in the olfactory bulb and at the interface of the molecular and granular cell layers in the cerebellum. A strong signal was found at selected ventricular margins, and a less prominent signal that varied in intensity was observed throughout the remaining regions of the brain. These results support the probability that the p198.KK construct contains all of the regulatory sequences needed for complete expression of the apoE gene in transgenic mouse brains. No significant signal was detected with an apoE mRNA sense probe (Fig. 2C) or when the human anti-apoE probe was reacted with a nontransgenic control mouse brain (Fig. 2F), showing that our results with transgene expression are specific for human apoE mRNA.
Fig. 2.
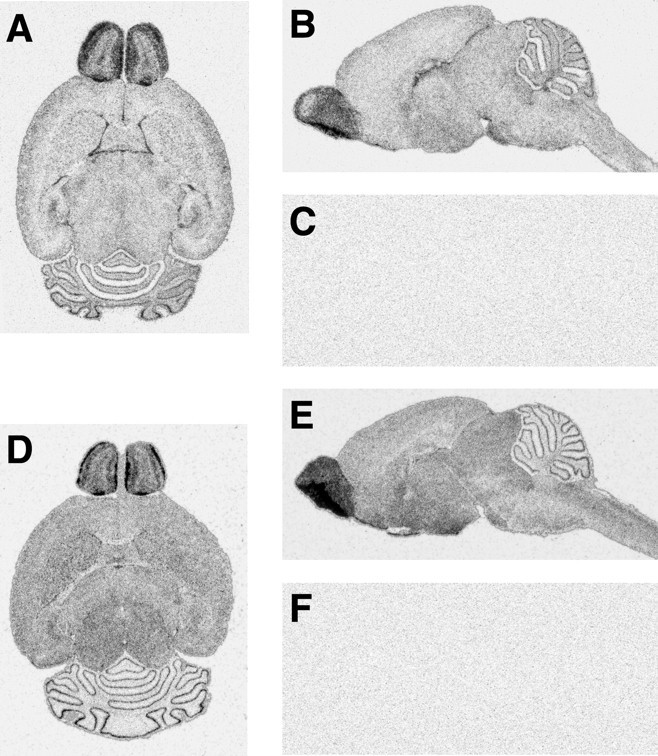
Distribution of expression of endogenous apoE mRNA in nontransgenic control and p198.KK transgenic mouse brains. Horizontal and sagittal sections of nontransgenic control (A, B) and p198.KK transgenic (D, E) mouse brains were radioactively labeled by in situ hybridization and examined by autoradiography. A, B, Distribution pattern of endogenous mouse apoE mRNA (dark grains) as revealed by a mouse antisense probe. C, Hybridization to a sagittal brain section with a sense probe does not produce a signal. D, E,Distribution pattern of p198.KK human transgene apoE mRNA (dark grains) as revealed by a human antisense probe. F, Hybridization to a sagittal nontransgenic control brain section with a human antisense probe does not produce a signal.
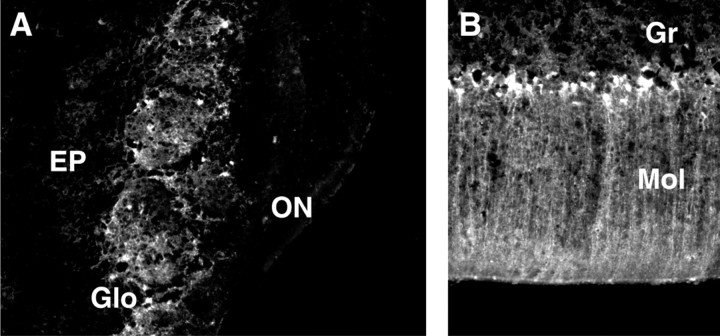
Examination of the olfactory bulbs at higher magnification by dark-field microscopy confirmed the intense expression of apoE mRNA in the glomerular layer and revealed high levels of mouse (Fig.3A) and human (Fig.3B) apoE mRNA in cells bordering both sides of the mitral cell layer. In addition, a strong hybridization signal was detected in scattered cells distributed throughout the olfactory bulb, including the olfactory neuronal layer, external plexiform layer, granular layer, and accessory olfactory bulb. The majority of this signal resembled the distribution of astrocytes in the olfactory bulb, as shown by the reaction of a comparable tissue section with anti-GFAP (Fig.3C). Thus, the glomerular layer is characterized by a prominent GFAP signal that reflects the abundance of astrocytes in this region of the olfactory bulb.
Fig. 3.
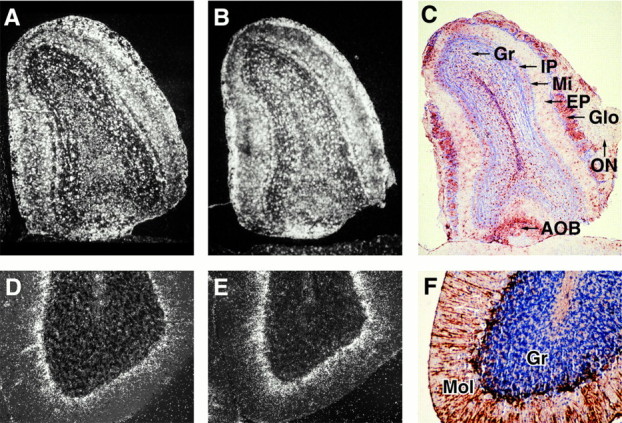
Region-specific expression of endogenous and transgene apoE mRNA in nontransgenic control (A, D) and in p198.KK transgenic (B, E) mouse brains. Tissue sections were labeled by in situ hybridization and examined at 10× or 50× magnification by dark-field microscopy (A, B, D, E). Separate tissue sections were reacted with anti-GFAP, stained with hematoxylin/eosin, and visualized by bright-field microscopy at 50× or 100× magnification (C, F). A, B, D, E, The distribution of endogenous mouse and p198.KK transgenic mouse apoE mRNA is shown by white grains in the olfactory bulb and the cerebellum. C, GFAP-positive cells in the olfactory bulb are indicated by red staining. The granular (Gr), internal plexiform (IP), mitral (Mi), external plexiform (EP), glomerular (Glo), olfactory neuronal (ON) layers, and accessory olfactory bulb (AOB) are indicated. F, GFAP-positive processes radiating into the molecular layer from the interface of the molecular (Mol) and granular (Gr) layers.
In the cerebellum of both nontransgenic control (Fig. 3D) and p198.KK transgenic (Fig. 3E) mice, an intense hybridization signal was detected at the interface of the molecular layer and the granular layer. The signal radiated into the molecular layer, consistent with the morphology of GFAP-positive cellular processes emanating from Bergmann glia cells (Fig.3F). An apoE mRNA hybridization signal was also detected in a few cells scattered throughout the granular region of the cerebellum, compatible with the distribution of protoplasmic astrocytes. No hybridization signal was observed over the nuclei of Purkinje cells at the interface of the molecular layer and the granular layer or over granular neurons (data not shown), indicating that these cells do not normally express detectable levels of apoE. The localization of endogenous mouse and transgenic human apoE mRNA in the mouse cerebellum is similar to the previously reported distribution of immunodetectable apoE in Bergmann glia and protoplasmic astrocytes in the rat cerebellum (Boyles et al., 1985).
An enhancer directs apoE expression in Bergmann glia and olfactory astrocytes
Previous studies have shown astrocytes to be the major site of immunoreactive apoE throughout the brain (Boyles et al., 1985). To determine that astrocytes were the primary sites of apoE mRNA in the brain, we searched for regulatory sequences that control apoE gene expression in this cell type. We investigated the expression of several constructs of the human apoE gene cluster in the brains of transgenic mice by in situ hybridization.
We first examined transgenic mice generated with a 10 kb construct of the intact apoE gene (HEG1) (Fig. 1) having 5 kb of 5′ flanking sequence and 1.7 kb of 3′ flanking sequence. No significant expression of the HEG1 transgene was detected in astrocytes in any region of the brain (Fig. 4A,B). This result agrees with our earlier study in which expression of the HEG1 construct was not detected in the transgenic mouse brain as determined by an RNase protection assay (Simonet et al., 1990). In agreement with these studies, we reconfirmed that the HEG1 construct was expressed in kidney proximal tubule epithelial cells to verify the integrity of the transgene (data not shown).
Fig. 4.
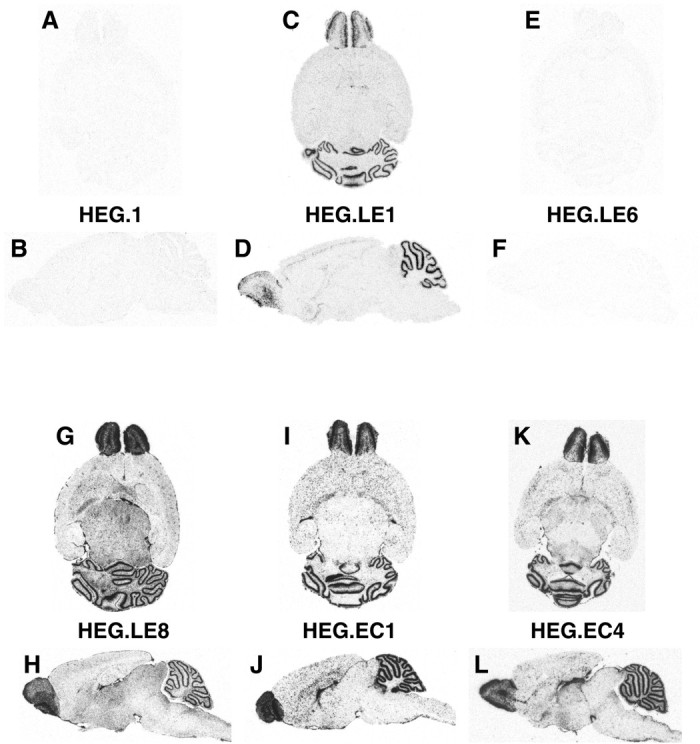
Distribution of expression of human apoE mRNA in transgenic mouse brains. Horizontal (A, C, E, G, I, K) and sagittal (B, D, F, H, J, L) sections from the brains of transgenic mice generated with HEG1, HEG.LE1, HEG.LE6, HEG.LE8, HEG.EC1, and HEG.EC4 constructs were examined by in situ hybridization followed by autoradiography.
We then investigated the activity of an enhancer domain located 15 kb downstream of the apoE gene. Our previous results showed that this domain contains the liver-specific hepatic control region, HCR.1 (Simonet et al., 1993; Dang et al., 1995), with evidence for additional enhancers nearby (Allan et al., 1997). Therefore, transgenic mice generated with the HEG.LE1 construct (Fig. 1) in which the 3.8 kb LE1 fragment was ligated to the 3′ end of HEG1 (Simonet et al., 1993) were analyzed. We found that the HEG.LE1 transgene was expressed at high levels in the olfactory bulbs and the cerebellum, but only low levels of human apoE transgenic mRNA were detected elsewhere in the brain (Fig. 4C,D).
The olfactory bulb expression of the HEG.LE1 construct was prominent in the glomerular layer; additional expression was limited to scattered cells elsewhere in the olfactory bulb (Fig. 5A,B). In the cerebellum, the hybridization signal was relatively intense at the interface of the molecular and granular layers (Fig. 5C). Little or no hybridization signal above background was detected over the large distinctive Purkinje cell bodies (Fig. 5D). The same expression pattern was observed in independent transgenic lines generated with two variants of the HEG.LE1 construct in which one variant expressed the apoE3 isoform (Figs. 4,5), and the other variant expressed the apoE4 isoform (data not shown). This pattern of apoE mRNA expression in astrocytes of the olfactory bulb and cerebellum in HEG.LE1 transgenic mice (Fig. 4C,D) was also observed in multiple lines of transgenic mice generated with the closely related constructs HEG.LE2 (data not shown) and HEG.LE8 (Fig. 4G,H). Thus apoE mRNA expression in these two regions of the brain is consistent between multiple lines of transgenic mice carrying derived subfragments from the same region of the apoE gene cluster.
Fig. 5.
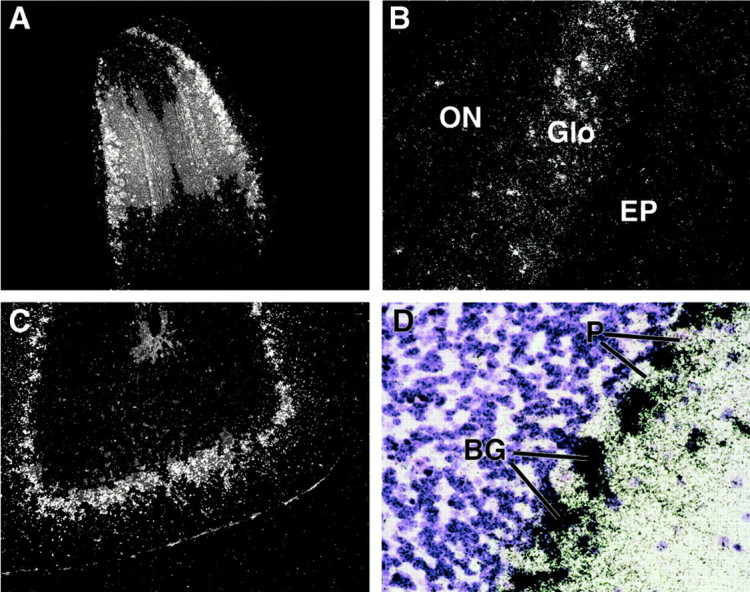
Region-specific expression of human apoE mRNA in transgenic mouse brains after in situ hybridization. Tissue sections from the brains of HEG.LE1 transgenic mice were analyzed by dark-field (A, B, C) or bright-field (D) microscopy. A, B, Olfactory bulb at 10× and 50× magnification. ON, Olfactory neuronal layer; Glo, glomerular layer; EP, external plexiform layer. C, D, Molecular and granular layers of the cerebellum at 50× and 100× magnification. BG, Bergmann glia nuclei; P, Purkinje cell nuclei.
The cellular source of apoE expression in brains of HEG.LE1 mice was confirmed by double immunohistochemistry. Tissue sections were labeled with antibodies to the astrocyte-specific marker GFAP and with antibodies to human apoE. Both human apoE protein and GFAP protein colocalized in astrocytes at the margins of glomeruli in the olfactory bulb (Fig. 6A1,A2,A3) and in Bergmann glia in the cerebellum (Fig.6A4,A5,A6). To ensure the specificity of the antibody for the human apoE protein, control immunohistochemistry for human apoE and GFAP was done on brains of HEG1 mice. No cross reaction of the antibodies to human apoE was detected in the cerebellum of these control transgenic mice (Fig. 6A7), consistent with the in situ hybridization analysis (Fig.4A,B). The only staining observed in the double antibody analysis was for endogenous GFAP (Fig.6A8,A9).
Fig. 6.
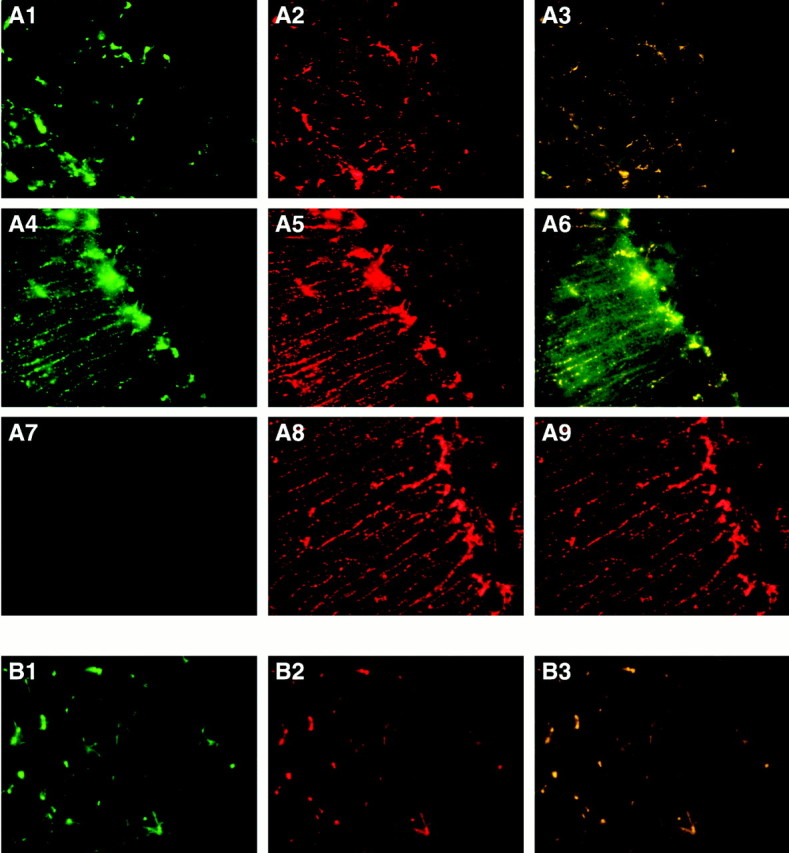
Immunohistochemistry of transgenic mouse brains was detected by fluorescence microscopy. A, Antibody labeling for human apoE protein in HEG.LE1 transgenic mice is shown ingreen (A1, A4, A7), endogenous mouse GFAP is shown in red (A2, A5, A8), and double antibody labeling for both antibodies, which colocalize, is shown inyellow (A3, A6, A9). Sections are shown for double antibody labeling of human apoE and mouse GFAP in astrocytes surrounding glomeruli in the olfactory bulb (A1, A2, A3) and in Bergmann glia of the cerebellum (A4, A5, A6). As a control, sections are shown for dual labeling of human apoE and mouse GFAP in Bergmann glia of the cerebellum (A7, A8, A9) in HEG1 transgenic mice. B, Antibody labeling for human apoE in the thalamus of HEG.LE8 transgenic mice is shown in green(B1), mouse GFAP is shown in red (B2), and double antibody labeling of human apoE and mouse GFAP is shown inyellow (B3).
The HCR.1 liver enhancer sequence has been precisely localized within the 774 bp LE6 fragment of the LE1 domain (Simonet et al., 1993; Dang et al., 1995). Therefore, we examined the brains of HEG.LE6 (Fig. 1) transgenic mice for apoE expression byin situ hybridization to determine if the astrocyte enhancer was located nearby. No transgenic human apoE mRNA was found in HEG.LE6 transgenic mouse brains (Fig. 4E,F); however, the transgene was expressed in the liver as expected (data not shown). Because the astrocyte enhancer activity of HEG.LE1 for apoE expression was not located in the LE6 subfragment, we examined the expression of the apoE gene in other subfragments of the LE1 domain. We found that human apoE mRNA was expressed in brains of transgenic mice generated with HEG.LE2 (data not shown) and with its HEG.LE8 subfragment (Fig.4G,H). There were no detectable differences in the pattern of apoE expression between these two constructs.
All transgenes were expressed at high intensity in astrocytes of olfactory bulb glomeruli and in Bergmann glia of the cerebellum. In HEG.LE8 transgenic mouse brains, apoE mRNA was also detected in astrocytes in white matter of the cerebellum, in multiple layers of the olfactory bulb, and in the thalamus (Fig.7A). However, the 3.8 kb LE1 fragment showed a somewhat restricted pattern of apoE gene expression compared with the expression pattern in mice generated with the 619 bp LE8 subfragment (Fig. 4). These results show that sequences within the LE8 fragment are required for apoE expression in astrocytes throughout the brain and that adjacent sequences may modulate that activity.
Fig. 7.
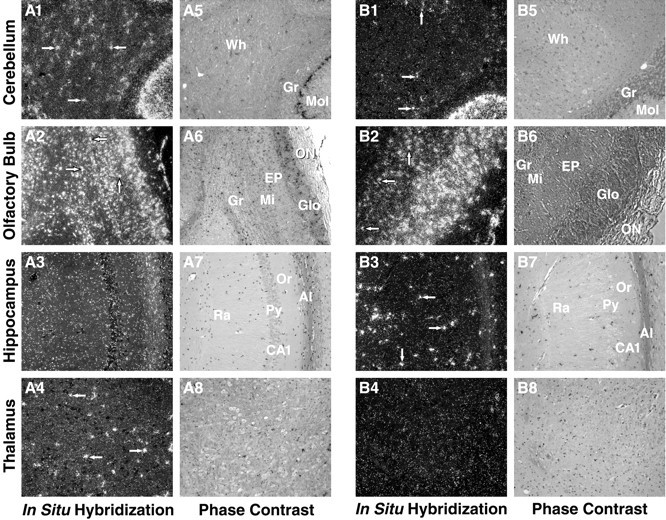
Distribution of expression of endogenous apoE mRNA in ME.2 (in HEG.LE8) (A) and ME.1 (in HEG.EC1 and HEG.EC4) (B) transgenic mouse brains. Horizontal and sagittal sections of transgenic mouse brains were radioactively labeled by in situ hybridization and examined at 50× magnification by dark-field microscopy. The distribution patterns of human transgene apoE mRNA (white grains) are shown as revealed by a human antisense probe. Arrows indicate representative cell bodies showing intense apoE mRNA signal. Signal location and tissue integrity were confirmed by phase-contrast imaging (A5–A8, B5–B8).A5, B5, Cerebellum. Wh, White matter;Gr, granular layer; Mol, molecular layer.A6, B6, Olfactory bulb. Gr, Granular layer;Mi, mitral layer; EP, external plexiform layer;Glo, glomerular layer; ON, olfactory neuronal layer. A7, B7, Hippocampus. Al, Alveus;Or, oriens; CA, cornu ammons; Py,pyramidal; Ra, radiens. A8, B8, Thalamus.
Immunohistochemistry for transgenic apoE and endogenous GFAP in the brains of HEG.LE8 mice confirmed the cellular site of the apoE mRNA signal. Human transgenic apoE protein and endogenous mouse GFAP protein signals colocalized in astrocytes throughout the brain as illustrated by a section from the thalamus (Fig. 6B).
A comparison of the nucleotide sequence of the LE8 fragment with the remainder of the human apoE gene cluster revealed a 620 nucleotide region of homology in the apoE/C-I intergenic region, located 3.3 kb downstream of the apoE gene. We termed this 620 nucleotide region ME.1 and termed the corresponding downstream LE8 sequence ME.2 (Fig.8). The two regions are 95% identical in sequence, suggesting that the astrocyte enhancer activity demonstrated by ME.2 was likely to be preserved in the ME.1 domain. Comparison of the human ME.1 or ME.2 sequence with the mouse apoE/C-I intergenic region (D. Walker and J. Taylor, unpublished observation) revealed a single segment of high homology (Fig.8, ME Mo). The mouse ME is 60% identical in sequence to each of the human ME domains. The highest level of identity between the mouse and human sequences was found in the internal region of the ME domain, suggestive of conserved regulatory sequences in this location. The mouse ME domain is likely to account for all astrocyte enhancer activity in the mouse, because the apoC-I/apoC-I′ gene duplication event did not occur in this species (Raisonnier, 1991).
Fig. 8.

ME domains of the apoE gene cluster in humans and mice. Dark boxes show regions of identity, anddashes show deletions.
We investigated the activity of the upstream ME.1 domain in the duplicated segment of the gene cluster within the apoE/C-I intergenic region. Transgenic mice generated with the HEG.EC1, HEG.EC2, and HEG.EC4 constructs (Fig. 1), all containing the ME.1 domain, were analyzed. They were found to express human apoE mRNA at high levels in astrocytes of the olfactory bulbs and in the cerebellum. ApoE mRNA was found at variable levels in astrocytes elsewhere in the brain, most notably in white matter of the cerebellum, in the external plexiform layer of the olfactory bulb, and in the hippocampus (Fig.4I,J,K,L). The patterns of apoE expression in astrocytes of HEG.EC1 mice (Figs. 4I,J) and HEG.EC2 mice (data not shown) were similar. Subtle but consistent differences were observed between HEG.EC1 (Figs.4I,J) and HEG.EC4 (Figs.4K,L) mice. Immunohistochemistry for transgenic human apoE and endogenous mouse GFAP in the brains of HEG.EC1 mice colocalized in astrocytes (data not shown). These results demonstrate that the ME.1 domain, in addition to the ME.2 domain, controls apoE gene expression in astrocytes throughout the brain.
No significant differences between the ME.1 and ME.2 domains were observed in the intensity of apoE expression in Bergmann glia in the cerebellum and in astrocytes surrounding olfactory glomeruli in transgenic mice generated with minimal length 620 bp (EC4) and 619 bp (LE8) fragments. However, there were notable differences in the intensity of transgene expression observed in other regions of the brain between the ME.1 and ME.2 domains. In white matter of the cerebellum and in thalamic nuclei, apoE mRNA in transgenic mice was increased by the ME.2 domain (in HEG.LE8) (Fig. 4G,H) and decreased by the ME.1 domain [(in HEG.EC1) (Fig.4I,J) and HEG.EC4 (Fig.4K,L)]. Thus, nucleotide differences within the ME.1 and ME.2 domains resulted in altered expression of the apoE gene in subsets of astrocytes.
The astrocyte enhancer activity of the LE1 and LE8 fragments containing the ME.2 domain was confirmed by examining the expression of a heterologous reporter construct in transgenic mice in which green fluorescent protein (GFP) cDNA was placed under the control of apoE promoter sequences and the ME.2 regulatory domain (Fig. 9). For both constructs, intense GFP expression was detected in distinct cells of the glomerular layer in the olfactory bulb having the distribution expected for astrocytes (Fig.10A). Prominent GFP expression was also found in Bergmann glia in the molecular layer of the cerebellum (Fig. 10B). Additional expression of the GFP.LE8 transgene was detected in astrocytes in the white matter of the cerebellum, the external plexiform layer of the olfactory bulb, the hippocampus, and the thalamus (Fig. 11) and was not observed in the GFP.LE1 pattern of expression.
Fig. 10.
Astrocyte enhancer control of reporter gene expression in GFP.LE1 and GFP.LE8 mice. Transgenic expression of the constructs shown in Figure 9 was detected by fluorescence microscopy.A, Olfactory bulb. EP, External plexiform layer;Glo, glomerular layer; ON, olfactory neuronal layer. B, Cerebellum. BG, Bergmann glia;P, Purkinje cells.
Fig. 11.
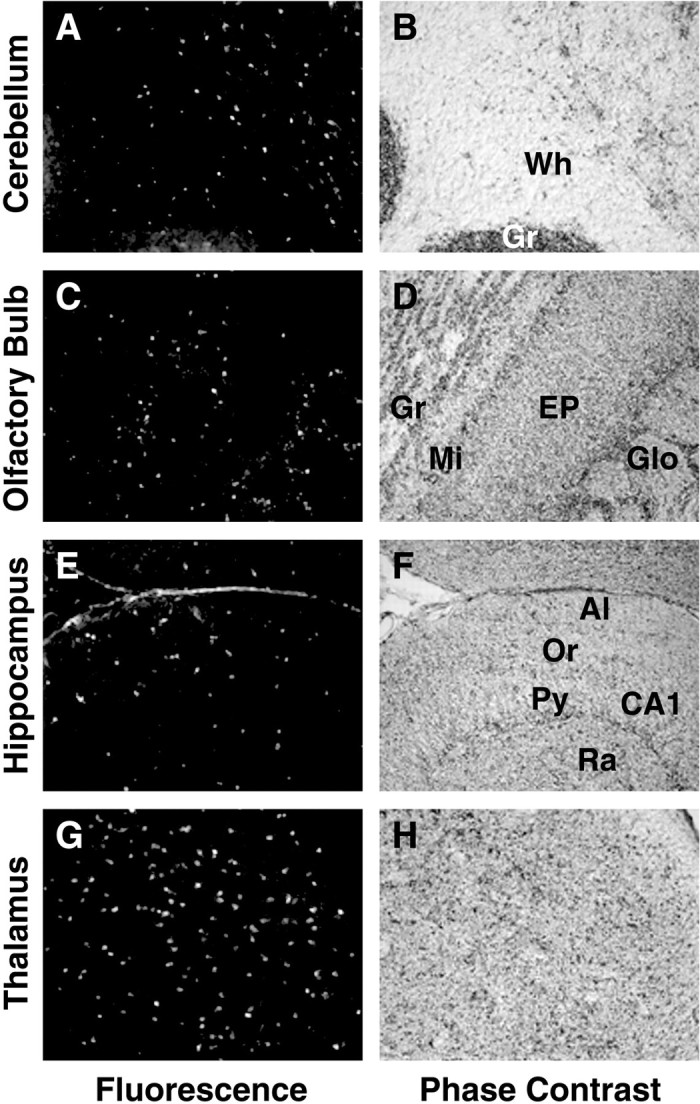
Astrocyte enhancer control of reporter gene expression. Transgenic expression of the GFP.LE8 construct shown in Figure 9 was detected by fluorescence microscopy (A, C, E, G). Signal location and tissue integrity were confirmed by phase imaging (B, D, F, H). A, B, Cerebellum.Wh, White matter; Gr, granular layer. C, D, Olfactory bulb. Gr, Granular layer; Mi, mitral layer; EP, external plexiform layer; Glo, glomerular layer. E, F, Hippocampus. Al, Alveus;Or, oriens; CA, cornu ammons; Py, pyramidal; Ra, radiens. G, H, Thalamus.
DISCUSSION
We report several new findings that are essential for understanding how apoE is regulated in the brain. The highest levels of apoE mRNA in both control and transgenic mice were found in astrocytes of the olfactory bulb and in Bergmann glia of the cerebellum. Analysis of multiple independent lines for each construct minimizes the possibility that differences in apoE mRNA transgene expression between constructs are because of integration artifacts or position effects. In the brain, apoE gene expression under the control of the ME.1 and ME.2 domains was detected only in astrocytes. Our data further suggest that astrocytes are responsible for a majority of apoE expression in the absence of inflammatory signals.
Astrocyte apoE expression was specified by distal regions located 3.3 kb and 15 kb downstream of the apoE gene. These distal sequences are 95% identical in nucleotide sequence, and they probably arose from the duplication event that yielded the two apoC-I genes (Fig. 1). In the absence of ME.1 and ME.2, the apoE promoter failed to direct gene transcription in the brain in vivo. These properties are characteristic of cell-specific enhancers that can be separated from promoters by distances of up to several hundred kilobases (for review, see Dillon and Sabbattini, 2000). The interaction of distal regulatory elements with the transcription complex in a promoter is likely to be mediated by chromatin folding and nucleosome orientation (Workman and Kingston, 1998). Analysis of the ME.1 and ME.2 sequences using the TRANSFAC regulatory sequence database (Heinemeyer et al., 1999) revealed binding motifs for several common transcription factors including CAAT/enhancer-binding protein β (C/EBPβ) and steroid hormone receptors (data not shown). Both factors have been identified in regulatory domains of genes expressed in many genes and cell types, and C/EBPβ has been proposed as a major determinant of combinational regulation for its target genes (Kowenz-Leutz and Leutz, 1999). Identification of the unique combination of ME.1/ME.2 and promoter factors that specify apoE expression in astrocytes remains to be determined.
The finding of exceptionally high levels of apoE mRNA in the olfactory bulb was unexpected. ApoE mRNA was especially prominent in astrocytes in the glomerular layer of both control and transgenic mice. Olfactory glomeruli are spherical modules in which incoming sensory signals from olfactory neuronal axons are processed and consolidated by various interneurons before continuing on through mitral cell dendrites (Mori et al., 1999). Numerous astrocytes spread their processes at the periphery of the glomeruli, partially surrounding small groups of neurites (Valverde and Lopez-Mascaraque, 1991; Treloar et al., 1999). This arrangement would be expected to facilitate delivery of apoE to the glomerular functional unit.
The same distal enhancer domain active in olfactory glomerular astrocytes also directed apoE gene expression at high levels in Bergmann glia of the cerebellum. This finding suggests that the two cell types may have cellular properties in common and that they may be derived from a common progenitor cell type. Both populations of cells may be the mature derivatives of embryonic radial glia (Cameron and Rakic, 1991; Misson et al., 1991). The processes of radial glia serve as scaffolds for the migration of neurons in fetal and neonatal life (Hatten, 1999). Late in rodent embryonic life just before birth, radial glia in developing olfactory protoglomeruli begin a morphological change that ultimately results in mature cells with properties (e.g., GFAP expression) shared by other astrocytes (Treloar et al., 1999). Similarly, radial glia in the developing fetal and neonatal cerebellum undergo a morphological change near the time of birth that results in the subsequent appearance of Bergmann glia (Gasser and Hatten, 1990; Hatten, 1999).
The 3.8 kb LE1 fragment showed a more restricted pattern of apoE gene expression than the subfragments LE2 (1683 bp) and LE8 (619 bp), each of which contains the ME.2 domain. This finding likely reflects the presence of silencing activity in the LE1 fragment, which is not present in the shorter LE2 and LE8 fragments. In support of this possibility, strong silencer sequences in the LE1 fragment have been reported to reduce transgene expression in the liver and other tissues (Simonet et al., 1993). Because no significant difference in apoE mRNA patterns was observed in multiple independent lines expressing the LE2 and LE8 transgenes, silencing activity is most likely located in the 2.7 kb of LE1 that lie outside of the LE2 region.
The HEG.EC4 construct containing the ME.1 domain with no flanking sequences yielded an expression pattern (Fig. 4K,L) that showed a consistent difference from that of ME.2 in the HEG.LE8 construct (Fig. 4G,H). Including flanking sequences with the ME.1 domain (the HEG.EC1 construct) results in a somewhat restricted pattern of apoE expression (Fig.4I,J). These observations suggest that elements in the flanking regions restrict ME.1 activity in a subset of astrocytes. Thus, both ME.1 and ME.2 activities show regional restrictions in their expression pattern because of apparent interactions with regulatory elements in their flanking sequences. Fundamental differences between ME.1 and ME.2 expression patterns also may be because of internal sequence differences between ME.1 and ME.2. It is noteworthy that apoE expression in astrocytes surrounding glomeruli in the olfactory bulb and in Bergmann glia of the cerebellum did not appear to be affected by silencer or modifying activity in the transgenes. This finding supports the possibility of a distinct functional similarity in astrocytes at these two widely separated sites.
The especially high levels of apoE mRNA in astrocytes of olfactory glomeruli may provide a critical production capacity necessary for both development and normal homeostasis in the olfactory bulb. In the 3 month postnatal development period of the mouse, the number and size of glomeruli increase dramatically (Pomeroy et al., 1990). In the adult, many neuronal terminals in each glomerulus undergo degeneration and regeneration as a consequence of the ongoing turnover of the sensory neuroepithelia that occurs throughout life (Graziadei and Monti Graziadei, 1978). The potential role of apoE in facilitating these processes is suggested by the increased amount of apoE that accumulates during the degeneration and regeneration at sites of peripheral nerve injury (Boyles et al., 1989; 1990). The importance of apoE in olfactory function is supported by the correlation of the apoE4 isoform with the profound olfactory deficits associated with Alzheimer's disease (Bacon et al., 1998; Graves et al., 1999). In apoE4-linked Alzheimer's disease, both the ability to identify odors and the threshold of odor detection become impaired (Graves et al., 1999). Furthermore, quantitative assessments of olfaction may be more reliable than global cognitive tests in the assessment of the progression of the disease (Graves et al., 1999). The deleterious effect of apoE4 may be a consequence of its deficiency in supporting neurite remodeling compared with apoE3 (Nathan et al., 1994).
The remarkably high level of apoE mRNA in Bergmann glia of the cerebellum was equally striking. These astrocytic cells are in intimate contact with Purkinje cells, the major GABAergic inhibitory neuron in the cerebellum (Grosche et al., 1999). An individual Bergmann glial cell may form hundreds of organized microdomains that ensheath synapses between axon terminals of parallel fibers and dendritic spines of Purkinje cells. This relationship is intriguing because Purkinje cells are relatively enriched in the apoE receptor 2 (apoER2), a member of the low-density lipoprotein receptor family (Clatworthy et al., 1999), and because apoE is a high-affinity ligand for this receptor (Herz and Willnow, 1995). These findings raise the possibility that glial–neuronal microdomains use rapid apoE ligand–receptor interaction as part of an intercellular signaling pathway or that apoE facilitates active neurite remodeling in this region.
Alternatively, apoE may be involved in the development and homeostasis of the cerebellum by competing with reelin for binding to apoER2. Reelin, a 385 kDa glycoprotein component of the extracellular matrix, is a ligand for apoER2, and reelin binding can be modulated by apoE (D'Arcangelo et al., 1999). Reelin is produced at several sites in the brain, and it plays a key role in determining cellular organization (summarized in Curran and D'Arcangelo, 1998). In the cerebellum, reelin mediates the development of the Purkinje cell layer (Curran and D'Arcangelo, 1998). Postmitotic cells in the external granular layer secrete reelin, and reelin is present in the molecular layer in close association with Purkinje cell dendrites (Miyata et al., 1996). The disruption of the reelin gene resulting in the lack of reelin production in reeler mutant mice (Bar et al., 1995; D'Arcangelo et al., 1995) or the targeted knock-out of apoER2 reelin receptors (Trommsdorff et al., 1999) disrupts neural cell positioning and the development of laminar structures in the brain. These properties suggest that reelin interacts with Purkinje cells to promote assembly of the Purkinje cell layer (D'Arcangelo and Curran, 1998). A similar role for reelin in neuronal organization may exist in other sites in the brain. For example, structural abnormalities have been detected in the olfactory bulbs of reeler mutant mice (Wyss et al., 1980). The production of high levels of apoE by cells derived from radial glia suggests that an interaction between apoE and reelin contributes to neuron progenitor migration and layer development.
Our finding of two astrocyte enhancer domains that direct apoE expression to specific sites of active neurite remodeling in the olfactory bulb and the cerebellum provides a new tool for investigating the functions of apoE in the brain. The potential value of these enhancers was demonstrated by directing expression of a GFP reporter sequence to these sites. These transgene reporter constructs will permit a mapping of the astrocytic domains that express the apoE gene in the olfactory bulb and the cerebellum that should expand our knowledge of apoE action in the brain. The striking pattern of apoE expression in olfactory glomeruli and in the cerebellum suggests a common function for apoE at these sites in regional brain homeostasis that remains to be investigated. Our findings permit the use of astrocyte-specific enhancers of apoE expression to help understand the functions of human apoE isoforms produced by astrocytes and their effects in models of Alzheimer's disease and other neurodegenerative states.
Footnotes
This research was funded in part by National Institutes of Health Grant HL37063. We thank Dale Newland for help with preparing tissue sections, David Walker for advice on in situ hybridization and for help with construct preparation, Shih-Jen Shyh for providing the HEG.LE8 construct, Manuel Buttini for advice on tissue autoradiograms, and David Sanan for help with microscopy and imaging. We also thank Stephen Ordway and Gary Howard for editorial assistance, Steven Gonzalez and John Carroll for help with figures, and Naima Contos for manuscript preparation.
Correspondence should be addressed to Dr. John M. Taylor, The J. David Gladstone Institutes, P.O. Box 419100, San Francisco, CA 94141-9100. E-mail: jtaylor@gladstone.ucsf.edu.
REFERENCES
- 1.Allan CM, Walker D, Segrest JP, Taylor JM. Identification and characterization of a new human gene (APOC4) in the apolipoprotein E, C-I, and C-II gene locus. Genomics. 1995a;28:291–300. doi: 10.1006/geno.1995.1144. [DOI] [PubMed] [Google Scholar]
- 2.Allan CM, Walker D, Taylor JM. Evolutionary duplication of a hepatic control region in the human apolipoprotein E gene locus. Identification of a second region that confers high-level and liver-specific expression of the human apolipoprotein E gene in transgenic mice. J Biol Chem. 1995b;270:26278–26281. doi: 10.1074/jbc.270.44.26278. [DOI] [PubMed] [Google Scholar]
- 3.Allan CM, Taylor S, Taylor JM. Two hepatic enhancers, HCR.1 and HCR.2, coordinate the liver expression of the entire human apolipoprotein E/C-I/C-IV/C-II gene cluster. J Biol Chem. 1997;272:29113–29119. doi: 10.1074/jbc.272.46.29113. [DOI] [PubMed] [Google Scholar]
- 4.Bacon AW, Bondi MW, Salmon DP, Murphy C. Very early changes in olfactory functioning due to Alzheimer's disease and the role of apolipoprotein E in olfaction. Ann NY Acad Sci. 1998;855:723–731. doi: 10.1111/j.1749-6632.1998.tb10651.x. [DOI] [PubMed] [Google Scholar]
- 5.Bar I, Lambert de Rouvroit C, Royaux I, Krizman DB, Dernoncourt C, Ruelle D, Beckers MC, Goffinet AM. A YAC contig containing the reeler locus with preliminary characterization of candidate gene fragments. Genomics. 1995;26:543–549. doi: 10.1016/0888-7543(95)80173-j. [DOI] [PubMed] [Google Scholar]
- 6.Boyles JK, Pitas RE, Wilson E, Mahley RW, Taylor JM. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyles JK, Zoellner CD, Anderson LJ, Kosik LM, Pitas RE, Weisgraber KH, Hui DY, Mahley RW, Gebicke-Haerter PJ, Ignatius MJ, Shooter EM. A role for apolipoprotein E, apolipoprotein A-I, and low-density lipoprotein receptors in cholesterol transport during regeneration and remyelination of the rat sciatic nerve. J Clin Invest. 1989;83:1015–1031. doi: 10.1172/JCI113943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyles JK, Notterpek LM, Anderson LJ. Accumulation of apolipoproteins in the regenerating and remyelinating mammalian peripheral nerve. Identification of apolipoprotein D, apolipoprotein A-IV, apolipoprotein E, and apolipoprotein A-I. J Biol Chem. 1990;265:17805–17815. [PubMed] [Google Scholar]
- 9.Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Mucke L, Mahley RW. Expression of human apolipoprotein E3 or E4 in the brains of Apoe−/− mice: isoform-specific effects on neurodegeneration. J Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cameron RS, Rakic P. Glial cell lineage in the cerebral cortex: a review and synthesis. Glia. 1991;4:124–137. doi: 10.1002/glia.440040204. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Lomnitski L, Michaelson DM, Shohami E. Motor and cognitive deficits in apolipoprotein E-deficient mice after closed head injury. Neuroscience. 1997;80:1255–1262. doi: 10.1016/s0306-4522(97)00007-9. [DOI] [PubMed] [Google Scholar]
- 12.Clatworthy AE, Stockinger W, Christie RH, Schneider WJ, Nimpf J, Hyman BT, Rebeck GW. Expression and alternate splicing of apolipoprotein E receptor 2 in brain. Neuroscience. 1999;90:903–911. doi: 10.1016/s0306-4522(98)00489-8. [DOI] [PubMed] [Google Scholar]
- 13.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late-onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 14.Curran T, D'Arcangelo G. Role of reelin in the control of brain development. Brain Res Rev. 1998;26:285–294. doi: 10.1016/s0165-0173(97)00035-0. [DOI] [PubMed] [Google Scholar]
- 15.Dang Q, Walker D, Taylor S, Allan C, Chin P, Fan J, Taylor J. Structure of the hepatic control region of the human apolipoprotein E/C-I gene locus. J Biol Chem. 1995;270:22577–22585. doi: 10.1074/jbc.270.38.22577. [DOI] [PubMed] [Google Scholar]
- 16.D'Arcangelo G, Curran T. Reeler: new tales on an old mutant mouse. Bioessays. 1998;20:235–244. doi: 10.1002/(SICI)1521-1878(199803)20:3<235::AID-BIES7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 17.D'Arcangelo G, Miao GG, Chen S-C, Soares HD, Morgan JI, Curran T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 18.D'Arcangelo G, Homayouni R, Keshvara L, Rice DS, Sheldon M, Curran T. Reelin is a ligand for lipoprotein receptors. Neuron. 1999;24:471–479. doi: 10.1016/s0896-6273(00)80860-0. [DOI] [PubMed] [Google Scholar]
- 19.Dillon N, Sabbattini P. Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Eddleston M, Mucke L. Molecular profile of reactive astrocytes—implications for their role in neurologic disease. Neuroscience. 1993;54:15–36. doi: 10.1016/0306-4522(93)90380-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elshourbagy NA, Liao WS, Mahley RW, Taylor JM. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc Natl Acad Sci USA. 1985;82:203–207. doi: 10.1073/pnas.82.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan J, Wang J, Bensadoun A, Lauer SJ, Dang Q, Mahley RW, Taylor JM. Overexpression of hepatic lipase in transgenic rabbits leads to a marked reduction of plasma high-density lipoproteins and intermediate-density lipoproteins. Proc Natl Acad Sci USA. 1994;91:8724–8728. doi: 10.1073/pnas.91.18.8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasser UE, Hatten ME. Central nervous system neurons migrate on astroglial fibers from heterotypic brain regions in vitro. Proc Natl Acad Sci USA. 1990;87:4543–4547. doi: 10.1073/pnas.87.12.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graves AB, Bowen JD, Rajaram L, McCormick WC, McCurry SM, Schellenberg GD, Larson EB. Impaired olfaction as a marker for cognitive decline. Interaction with apolipoprotein E ε4 status. Neurology. 1999;53:1480–1487. doi: 10.1212/wnl.53.7.1480. [DOI] [PubMed] [Google Scholar]
- 25.Graziadei PPC, Monti Graziadei GA. The olfactory system: a model for the study of neurogenesis and axon regeneration in mammals. In: Cotman CW, editor. Neuronal plasticity. Raven; New York: 1978. pp. 131–153. [Google Scholar]
- 26.Grosche J, Matyash V, Möller T, Verkhratsky A, Reichenbach A, Kettenmann H. Microdomains for neuron-glia interaction: parallel fiber signaling to Bergmann glial cells. Nat Neurosci. 1999;2:139–143. doi: 10.1038/5692. [DOI] [PubMed] [Google Scholar]
- 27.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 28.Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herz J, Willnow TE. Lipoprotein and receptor interactions in vivo. Curr Opin Lipidol. 1995;6:97–103. doi: 10.1097/00041433-199504000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Ji Z-S, Lauer SJ, Fazio S, Bensadoun A, Taylor JM, Mahley RW. Enhanced binding and uptake of remnant lipoproteins by hepatic lipase-secreting hepatoma cells in culture. J Biol Chem. 1994;269:13429–13436. [PubMed] [Google Scholar]
- 31.Kowenz-Leutz E, Leutz A. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 32.Masliah E, Mallory M, Alford M, Ge N, Mucke L. Abnormal synaptic regeneration in hAPP695 transgenic and APOE knockout mice. In: Iqbal K, Mortimer JA, Winblad B, Wisniewski HM, editors. research advances in Alzheimer's disease and related disorders. Wiley; Chichester, UK: 1995a. pp. 405–414. [Google Scholar]
- 33.Masliah E, Mallory M, Ge N, Alford M, Veinbergs I, Roses AD. Neurodegeneration in the central nervous system of apoE-deficient mice. Exp Neurol. 1995b;136:107–122. doi: 10.1006/exnr.1995.1088. [DOI] [PubMed] [Google Scholar]
- 34.Merrill JE, Benveniste EN. Cytokines in inflammatory brain lesions: helpful and harmful. Trends Neurosci. 1996;19:331–338. doi: 10.1016/0166-2236(96)10047-3. [DOI] [PubMed] [Google Scholar]
- 35.Misson J-P, Takahashi T, Caviness VS., Jr Ontogeny of radial and other astroglial cells in murine cerebral cortex. Glia. 1991;4:138–148. doi: 10.1002/glia.440040205. [DOI] [PubMed] [Google Scholar]
- 36.Miyata T, Nakajima K, Aruga J, Takahashi S, Ikenaka K, Mikoshiba K, Ogawa M. Distribution of a reeler gene-related antigen in the developing cerebellum: an immunohistochemical study with an allogeneic antibody CR-50 on normal and reeler mice. J Comp Neurol. 1996;372:215–228. doi: 10.1002/(SICI)1096-9861(19960819)372:2<215::AID-CNE5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- 38.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer's disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541:163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- 39.Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- 40.Ong W-Y, He Y, Suresh S, Patel SC. Differential expression of apolipoprotein D and apolipoprotein E in the kainic acid-lesioned rat hippocampus. Neuroscience. 1997;79:359–367. doi: 10.1016/s0306-4522(96)00608-2. [DOI] [PubMed] [Google Scholar]
- 41.Page KJ, Hollister RD, Hyman BT. Dissociation of apolipoprotein and apolipoprotein receptor response to lesion in the rat brain: an in situ hybridization study. Neuroscience. 1998;85:1161–1171. doi: 10.1016/s0306-4522(97)00661-1. [DOI] [PubMed] [Google Scholar]
- 42.Pitas RE, Boyles JK, Lee SH, Hui D, Weisgraber KH. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. J Biol Chem. 1987;262:14352–14360. [PubMed] [Google Scholar]
- 43.Poirier J, Baccichet A, Dea D, Gauthier S. Cholesterol synthesis and lipoprotein reuptake during synaptic remodelling in hippocampus in adult rats. Neuroscience. 1993;55:81–90. doi: 10.1016/0306-4522(93)90456-p. [DOI] [PubMed] [Google Scholar]
- 44.Pomeroy SL, LaMantia A-S, Purves D. Postnatal construction of neural circuitry in the mouse olfactory bulb. J Neurosci. 1990;10:1952–1966. doi: 10.1523/JNEUROSCI.10-06-01952.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raisonnier A. Duplication of the apolipoprotein C-I gene occurred about forty million years ago. J Mol Evol. 1991;32:211–219. [PubMed] [Google Scholar]
- 46.Rall SC, Jr, Weisgraber KH, Mahley RW. Human apolipoprotein E. The complete amino acid sequence. J Biol Chem. 1982;257:4171–4178. [PubMed] [Google Scholar]
- 47.Roses AD. Apolipoprotein E alleles as risk factors in Alzheimer's disease. Annu Rev Med. 1996;47:387–400. doi: 10.1146/annurev.med.47.1.387. [DOI] [PubMed] [Google Scholar]
- 48.Rutka JT, Murakami M, Dirks PB, Hubbard SL, Becker LE, Fukuyama K, Jung S, Tsugu A, Matsuzawa K. Role of glial filaments in cells and tumors in glial origin: a review. J Neurosurg. 1997;87:420–430. doi: 10.3171/jns.1997.87.3.0420. [DOI] [PubMed] [Google Scholar]
- 49.Simonet WS, Bucay N, Lauer SJ, Wirak DO, Stevens ME, Weisgraber KH, Pitas RE, Taylor JM. In the absence of a downstream element, the apolipoprotein E gene is expressed at high levels in kidneys of transgenic mice. J Biol Chem. 1990;265:10809–10812. [PubMed] [Google Scholar]
- 50.Simonet WS, Bucay N, Lauer SJ, Taylor JM. A far-downstream hepatocyte-specific control region directs expression of the linked human apolipoprotein E and C-I genes in transgenic mice. J Biol Chem. 1993;268:8221–8229. [PubMed] [Google Scholar]
- 51.Sing CF, Davignon J. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am J Hum Genet. 1985;37:268–285. [PMC free article] [PubMed] [Google Scholar]
- 52.Slooter AJC, Tang M-X, van Duijn CM, Stern Y, Ott A, Bell K, Breteler MMB, Van Broeckhoven C, Tatemichi TK, Tycko B, Hofman A, Mayeux R. Apolipoprotein E ε4 and the risk of dementia with stroke. A population-based investigation. JAMA. 1997;277:818–821. doi: 10.1001/jama.277.10.818. [DOI] [PubMed] [Google Scholar]
- 53.Soininen HS, Riekkinen PJ., Sr Apolipoprotein E, memory and Alzheimer's disease. Trends Neurosci. 1996;19:224–228. doi: 10.1016/0166-2236(96)10027-8. [DOI] [PubMed] [Google Scholar]
- 54.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: High-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Y, Wu S, Bu G, Onifade MK, Patel SN, LaDu MJ, Fagan AM, Holtzman DM. Glial fibrillary acidic protein-apolipoprotein E (apoE) transgenic mice: astrocyte-specific expression and differing biological effects of astrocyte-secreted apoE3 and apoE4 lipoproteins. J Neurosci. 1998;18:3261–3272. doi: 10.1523/JNEUROSCI.18-09-03261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treloar HB, Purcell AL, Greer CA. Glomerular formation in the developing rat olfactory bulb. J Comp Neurol. 1999;413:289–304. [PubMed] [Google Scholar]
- 57.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and apoE receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 58.Valverde F, Lopez-Mascaraque L. Neuroglial arrangements in the olfactory glomeruli of the hedgehog. J Comp Neurol. 1991;307:658–674. doi: 10.1002/cne.903070411. [DOI] [PubMed] [Google Scholar]
- 59.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 60.Wyss JM, Stanfield BB, Cowan WM. Structural abnormalities in the olfactory bulb of the Reeler mouse. Brain Res. 1980;188:566–571. doi: 10.1016/0006-8993(80)90056-6. [DOI] [PubMed] [Google Scholar]
- 61.Zannis VI, Breslow JL. Human very low density lipoprotein apolipoprotein E isoprotein polymorphism is explained by genetic variation and posttranslational modification. Biochemistry. 1981;20:1033–1041. doi: 10.1021/bi00507a059. [DOI] [PubMed] [Google Scholar]