Abstract
The molluscan Phe-Met-Arg-Phe-amide (FMRFamide)-gated sodium channels (FaNaCs) show both structural and functional similarities to the mammalian acid-sensing ion channels (ASICs). Both channel types are related to the epithelial sodium channels and, although the neuropeptide FMRFamide directly gates the FaNaCs, it also modulates the proton-gating properties of ASICs. It is not yet known whether protons can alter the gating properties of the FaNaCs. We chose to examine this possibility at a site of FaNaC expression in the nervous system of the mollusk Lymnaea stagnalis. We cloned a putative L. stagnalis FaNaC (LsFaNaC) that exhibited a high degree of sequence identity to the Helix aspersa FaNaC (HaFaNaC, 60%), and a weaker homology to the ASICs (ASIC3, 22%). In situ hybridization was used to map the LsFaNaC expression pattern in the brain and to identify the right pedal giant1 (RPeD1) neuron as a site where the properties of the endogenous channel could be studied. In RPeD1 neurons isolated in culture, we demonstrated the presence of an FMRFamide-gated sodium current with features expected for a FaNaC: amiloride sensitivity, sodium selectivity, specificity for FMRFamide and Phe-Leu-Arg-Phe-amide (FLRFamide), and no dependency on G-protein coupling. The sodium current also exhibited rapid desensitization in response to repeated FMRFamide applications. Lowering of the pH of the bathing solution reduced the amplitude of the FMRFamide-gated inward current, while also activating an additional sustained weak inward current that was apparently not mediated by the FaNaC. Acidification also prevented the desensitization of the FMRFamide-induced inward current. The acid sensitivity of LsFaNaC is consistent with the hypothesis that FaNaCs share a common ancestry with the ASICs.
Keywords: Lymnaea, FMRFamide-gated Na+ channel, pH sensitivity, ASIC, epithelial Na+ channel, amiloride, degenerin, cell culture
The neuropeptide Phe-Met-Arg-Phe-amide (FMRFamide) and related peptides have diverse physiological effects, such as regulating heart rate in mollusks (Price et al., 1987), controlling movement inCaenorhabditis elegans (Nelson et al., 1998), and pain modulation in vertebrates (Yang et al., 1985; Waldmann and Lazdunski, 1998). It is believed that in most instances FMRFamide and related neuropeptides work via G-protein-coupled receptors and second messengers (Higgins et al., 1978; Colombaioni et al., 1985; Piomelli et al., 1987; Willoughby et al., 1999a,b). However, in neurons of the snail Helix aspersa it has also been demonstrated that FMRFamide can elicit excitatory effects without activating G-proteins (for review, see Cottrell, 1997). FMRFamide application to these cells directly gated sodium channels with pharmacological properties similar to those of the mammalian epithelial sodium channels (ENaCs): they were highly selective for sodium ions, blocked by amiloride and related drugs, and were insensitive to blockers of other sodium ion channel types. Subsequently, a cDNA encoding an ENaC-related channel was cloned from H. aspersa brain RNA (Lingueglia et al., 1995). Heterologous expression of the channel led to the formation of homotetramers (Coscoy et al., 1998) that responded to both FMRFamide and Phe-Leu-Arg-Phe-amide (FLRFamide) and hence was identified as the H. aspersa FMRFamide-gated sodium channel (HaFaNaC).
Screening of the mammalian CNS for FaNaC homologs identified several new members of the degenerin subgroup of ENaCs that show more sequence identity to FaNaC than do the prototypical ENaCs (García-Añoveros et al., 1997; Waldmann et al., 1997a,b;Chen et al., 1998). These new mammalian channels were gated by decreases in extracellular pH [acid-sensing ion channels (ASICs)], and the expression of some forms in dorsal horn sensory neurons led to the suggestion that they may be important in nociception resulting from acidosis during inflammation (Waldmann and Lazdunski, 1998). Not only were the ASICs somewhat similar in structure to the FaNaCs, but a recent report has demonstrated that many were also modulated by FMRFamide and the structurally related mammalian peptides neuropeptide FF (NPFF) and neuropeptide AF (NPAF) (Askwith et al., 2000). Preapplication of the peptides to dorsal root ganglion sensory neurons or to the heterologously expressed channels increased the duration of the acid-induced inward current by reducing the rate of inactivation, often producing sustained responses.
It has yet to be investigated whether the FaNaCs expressed in molluscan neurons show sensitivity to acid pH, as might be predicted by analogy to ASICs. To study this we set out to identify neurons endogenously expressing an FaNaC in the snail Lymnaea stagnalis. Although FaNaCs have been isolated from two molluscan species [Helix aspersa (HaFaNaC) (Lingueglia et al., 1995) and Helisoma trivolvis (HtFaNaC) (Jeziorski et al., 2000)], the expression of the FaNaC gene in identified neurons has not been demonstrated directly in either of them. Here, we describe the cloning of the L. stagnalis FaNaC (LsFaNaC) and its widespread expression throughout the brain, including in the readily identifiable giant RPeD1 interneuron. RPeD1 cells were subsequently isolated in culture and used to study the peptide and acid sensitivities of the endogenous LsFaNaC.
MATERIALS AND METHODS
Experimental subjects and chemicals. Adult specimens of Lymnaea stagnalis were obtained from Blade Biological (Kent, UK). The animals were kept in large holding tanks containing copper-free water on a 12 hr light/dark cycle and fed lettuce three times a week.
All chemicals were purchased from Sigma (Poole, UK) unless otherwise stated.
Cloning of Lymnaea FMRFamide-gated sodium channel. PCR primers identical to nucleotides 84–105 and 1961–1940 of the HaFaNaC open reading frame (GenBank accession numberX92113) were used to amplify by PCR the entire coding region from theHelix clone (gift of M. Lazdunski). A radiolabeled product was made by replacement of dCTP with α [32P]dCTP in the reaction mix, and this was used as a hybridization probe in the screening of 2 million phages of a Lymnaea stagnalis brain cDNA library (previously described in Vreugdenhil et al., 1988). Three clones were isolated to homogeneity by a further two rounds of screening at high stringency (washes performed with 0.2× SSC, 0.1% SDS at 60°C). DNA from the phages was purified, and inserts were sequenced on both strands using dye-terminator reaction kits following the manufacturer's instructions (Perkin-Elmer, Boston, MA).
In situ hybridization. The protocols for 7 μm tissue section preparation and biotinylated complementary oligonucleotidein situ hybridization were adhered to as previously described (Kellett et al., 1996), using a mixture of 15 different 21–25 mer oligonucleotides as the probe. Negative control hybridizations using a mixture of the complementary (sense) oligonucleotide sequences were performed under identical conditions.
Gene expression detection by RT-PCR. Total cellular RNA was isolated from various Lymnaea tissues using an RNA preparation kit (Qiagen, Bothell, WA) following the manufacturer's instructions. One microgram of each RNA was used in reverse transcription reactions with random hexamers and 10 U of Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). The samples were then used as templates in PCR with primers designed to nucleotides 3001–3025 and 3891–3865 of the LymnaeaFMRFamide-gated sodium channel gene (LsFaNaC; GenBank accession numberAF335548) to amplify an 890 bp product. Samples were separated on a 1% agarose gel, and the identity of bands of the correct size was confirmed by hybridization to a radiolabeled PCR product of the entire 3′ untranslated region (UTR).
Dissection. All dissections were performed in HEPES-buffered saline containing (in mm): 50 NaCl, 1.6 KCl, 2 MgCl2, 3.5 CaCl2, and 10 HEPES, pH 7.9, in distilled water. The CNS, consisting of the circumesophageal ganglionic ring (cerebral, pedal, pleural, parietal, and visceral ganglia) and the buccal ganglia together with a short stretch of esophagus, was isolated from the snail. The preparation was pinned down in a Sylgard-coated dish filled with HEPES-buffered saline with the dorsal surface facing up.
Isolation and culture of RPeD1 neurons. The cell culture procedure was modified after the protocol of Ridgway et al. (1991). Media used included normal saline (NS), antibiotic saline (ABS), defined medium (DM), and conditioned medium (CM). Normal saline used in cell culture experiments contained the same salt concentrations as HEPES-buffered saline described above but was made up in culture grade water (Sigma), whereas ABS also contained gentamycin (150 μg/ml). DM was prepared by mixing 100 ml of special L-15 medium (Life Technologies, Paisley, UK), 80 ml of NS, and 120 ml of culture grade water and by adding glutamine (30 mg), glucose (16.2 mg), and gentamycin (600 μl of 10 mg/ml stock) to the solution. For CM preparation, isolated brains that had been washed extensively in ABS were incubated in DM (two brains per milliliter). After 3 d of incubation, the CM was filter sterilized (Millex-GV, 0.22 μm; Millipore, Bedford, MA). Aliquots of CM (1 ml) were pipetted directly onto culture dishes (Falcon 3001; Becton Dickinson, Rutherford, NJ) coated with poly-l-lysine [15–30 kDa; 1 mg/ml in 15 mm Tris(hydroxymethyl)aminomethane)], and equal amounts of DM were added. The culture dishes were stored at −20°C and thawed 2–3 hr before use.
The isolation of RPeD1 neurons was performed in a laminar flow cabinet after the isolated nervous system was first incubated for 45 min in a protease solution (Sigma type VIII, 1 mg/ml in NS) followed by washing in ABS. Subsequently, the isolated nervous system was pinned out in a dissection dish filled with high-osmolarity DM (30 mmglucose in DM). The RPeD1 neuron was visually identified according to its large size and characteristic position in the right pedal ganglion. Its cell body was exposed by mechanically disrupting the inner connective tissue and then removed, together with a short stretch of its main process, by gentle suction with a Sigmacote-coated, fire-polished micropipette (tip diameter, 150–200 μm) prepared from 1.5 mm glass tubing (GC150T-10; Clark Electromedical Instruments, Reading, UK). After isolation, neurons were transferred onto culture dishes and cultured at 20°C for up to 5 d.
Electrophysiological and pharmacological studies on cultured neurons. For intracellular recordings from isolated RPeD1 neurons, culture dishes containing isolated neurons were placed on the stage of an inverted microscope (Nikon Diaphot) that was equipped with a custom-built, gravity-fed perfusion system. Cells were recorded after 1–3 d in culture, and no changes were seen in the electrical properties of the cells during this period. The culture dishes were perfused with NS at a flow rate of 1–2 ml/min for at least 30 min before the experiment to remove all culture medium. The perfusion was maintained throughout the experiment. RPeD1 cell bodies were impaled with one or two microelectrodes pulled from 1 mm capillaries (GC100F-10; Clark Electromedical Instruments) and filled with saturated potassium sulfate (tip resistance, 20–30 MΩ). The intracellular signals were amplified using an AXOCLAMP2-B amplifier (Axon Instruments), output to a storage oscilloscope (5115 Tektronix), and stored on a DAT recorder (Biologic DTR-1801, Biological Science Instruments, Claix, France). Amplified signals were also digitized using a DigiData 1200 interface (Axon Instruments) and stored on a personal computer. Intracellular recordings were either performed in bridge or twin electrode voltage clamp (TEVC) mode of the AXOCLAMP 2B amplifier controlled by pClamp6 software (Axon Instruments) via the DigiData 1200 interface.
The effects of FMRFamide (0.1 mm), FLRFamide (0.1 mm), and Ser-Asp-Pro-Phe-Leu-Arg-Phe-amide (SDPFLRFamide) (0.1 mm) were tested by focal application from a micropipette (1 sec pulses at 6–12 psi) using a Picospritzer (General Valve, Fairfield, NJ). Amiloride, a selective antagonist for peptide gated Na+ channels, was applied by including it in the perfusion solution at a concentration of 0.1 mm. Na+substitution experiments were performed by replacing 90% of the NaCl in NS with N-methyl-d-glucamine (low Na+ saline). Pipette solutions for the pressure injection of GDP-β-S to test for G-protein-coupled responses contained GDP-β-S (2 mm), KCl (40 mm), and Fast Green (0.1%) in culture grade distilled water (Sigma). The solution was injected into the soma of isolated RPeD1 neurons by applying 500 msec pressure pulses (15 psi) at 1 Hz. The injection was stopped when the soma showed a clear green staining (this usually occurred within 1 min from the start of the injection). The effects of pH on RPeD1 neurons and the FMRFamide-induced inward current in these neurons were studied by either focal application or bath application of saline at pH values of 5.2, 6.0, 6.7, 6.8, and 9.0 in addition to HEPES-buffered saline at pH 7.9. All saline solutions had the same salt concentration as HEPES-buffered saline, pH 7.9 and pH 6.8, but were buffered with 5 mm 2-(4-morpholino)-ethansulfonic acid (MES), pH 5.2, 6.0, and 6.7, or 5 mm Tris, pH 9.0, instead of HEPES.
RESULTS
Cloning of a Lymnaea homolog of the FMRFamide-gated sodium channel
Screening of 2 million phages from a Lymnaea stagnalisCNS cDNA library, using a radiolabeled PCR product encoding the HaFaNaC, identified three positive clones. Sequencing of the clones revealed that all three encoded an identical 4872 bp cDNA that showed an overall 60% identity to the Helix sequence (GenBank accession number X92113). An open reading frame (residues 767–2662) encoding a 71.5 kDa protein (GenBank accession number AF335548) (Fig.1), exhibited 60% identity to theHelix protein and 66% to the Helisoma protein. The open reading frame was flanked by 5′ and 3′ untranslated regions of 766 and 2211 bp, respectively. Comparison of the open reading frame start point with that of the Helix and Helisomaclones showed that LsFaNaC shares a 31 residue N-terminal extension with HtFaNaC that appears to be absent in HaFaNaC. Further screening of the brain cDNA library by PCR revealed no evidence for related genes or alternatively spliced mRNAs that could encode variants of LsFaNaC.
Fig. 1.
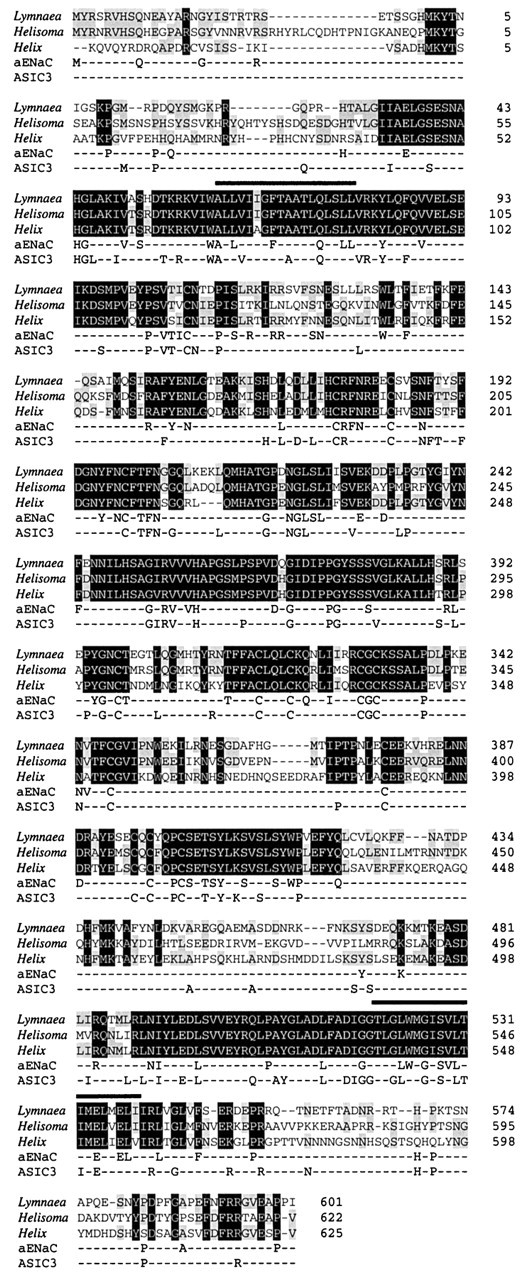
Comparison of FMRFamide-gated sodium channels (FaNaCs) from Lymnaea stagnalis, Helix aspersa, andHelisoma trivolvis (GenBank accession numbers AF335548,X92113, and AF254118, respectively). Alignment of the protein sequences predicted from translation of open reading frames of cDNA sequences. Amino acids conserved in all three FaNaCs are highlighted in black, and those found in only two are highlighted in gray.Numbering on the right represents equivalent amino acid positions within the clones (taking the published start methionine for X92113 as position 1). Overliningshows positions of predicted transmembrane domains. Amino acids of the mammalian epithelial sodium channel α subunit (αENaC, GenBank accession number NM001038) and ASIC3 (GenBank accession numberAF013598) that are conserved in two or more FaNaCs are shown below.
Structural predictions from the primary amino acid sequence suggested a protein with two transmembrane spanning domains (residues 93–111 and 551–571), a cysteine-rich extracellular loop, and intracellular N- and C-terminal domains. The extracellular loop contained seven predicted N-glycosylation signals, five of which were conserved in theHelix protein and two in all members of the superfamily. The loop also contained 15 cysteine residues, 14 of which are conserved in the Helix protein and up to 11 in more distantly related members of the superfamily. The N-terminal domain contained phosphorylation sites for protein kinase C and casein kinase II, as did the C terminus, which also contains a site for protein kinase A. This structure is topologically identical to that predicted for all the members of the amiloride-sensitive epithelial sodium channels. Comparison of the amino acid sequence to other members of the superfamily revealed identities of between 15% [mammalian epithelial sodium channels, αENaC (human): GenBank accession number NM001038] and 22% [mammalian dorsal root acid-sensing ion channel, ASIC 3 (rat): GenBank accession number AF013598] with highest homology in the region covering the predicted second transmembrane domain (up to 46%).
Expression of LsFaNaC in neuronal and other tissues
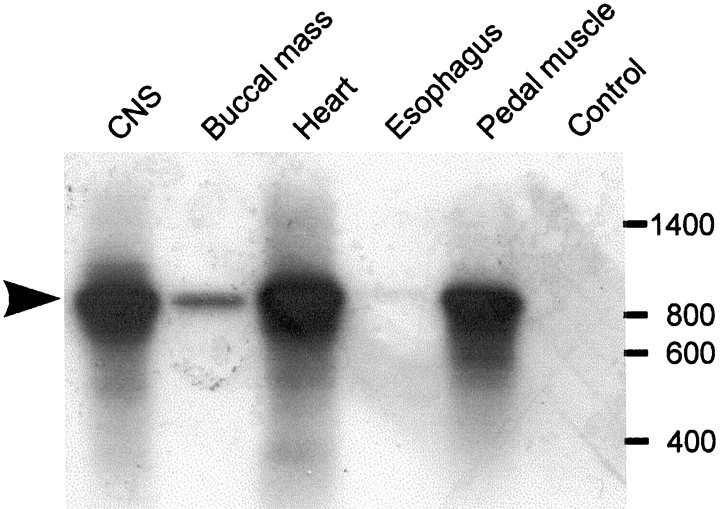
Reverse transcription PCR was used to identify Lymnaeatissues where LsFaNaC is expressed (Fig.2). Reactions performed on total RNA isolated from a number of tissues revealed a widespread distribution of the channel. Especially high levels of expression were detected in the brain, heart, and pedal tissue. Only the brain and heart have been shown previously to definitely contain FMRFamide (Ebberink et al., 1987; Buckett et al., 1990a,b), although there is immunocytochemical evidence for FMRFamergic innervation of the pedal muscle (Schot and Boer, 1982). Low levels of expression were detected in the buccal mass, and no expression was detected in the esophagus.
Fig. 2.
Tissue distribution of LsFaNaC expression inLymnaea. Reverse transcription-PCR reactions that used primers specific for a region within the 3′ UTR of the LsFaNaC cDNA were performed on total RNA derived from five tissues. Products of the correct size (890 bp) were separated by gel electrophoresis, transferred to a nylon filter, and visualized by hybridization with a radiolabeled probe of the identical region of the 3′ UTR. Products were detected in CNS, buccal mass, heart, and pedal muscle, but not in esophagus. The control reaction was performed in an identical manner to the CNS reaction, except reverse transcriptase enzyme was omitted.
In situ analysis of identified neuron expression
In situ analysis of the distribution of the LsFaNaC gene was performed on the serially sectioned CNS (n = 4). More than 400 neurons consistently expressed the gene with staining present in several previously identified giant neurons (Fig.3). Prominent among these was RPeD1, a dopamine-synthesizing member of the respiratory central pattern generator network of Lymnaea (Fig. 3A, arrow) (Syed et al., 1990). A bilaterally symmetrically located but smaller identifiable cell, LPeD1 (Slade et al., 1981), which contains serotonin (Kemenes et al., 1989) was also stained in the opposite left pedal ganglia (data not shown) together with large clusters of medial cells of a wide variety of sizes (Fig. 3A). The expression of LsFaNaC in RPeD1 was consistent with an electrophysiological study in the closely related snail Helisoma, where the homologous cells GDN (giant dopaminergic neuron) showed fast inward currents in response to FMRFamide application (Jeziorski et al., 2000).
Fig. 3.
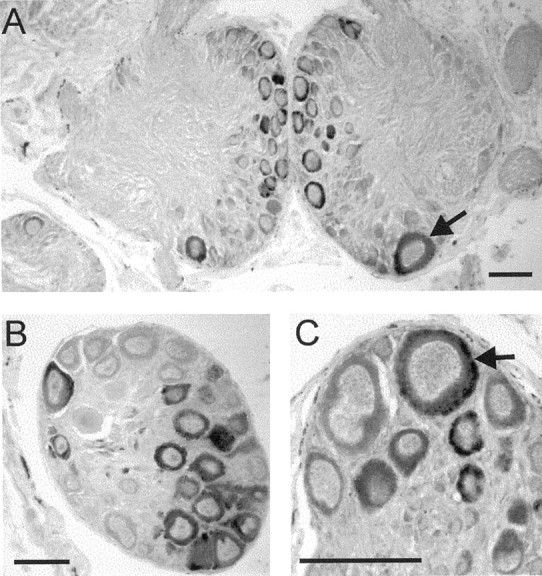
Expression of the LsFaNaC in theLymnaea CNS using in situ hybridization.A, Section through the paired pedal ganglia showing staining of the identified neuron RPeD1 (arrow).B, A cluster of light yellow cells in the right parietal ganglion. C, Expression in a cerebral giant cell (arrow), a modulatory interneuron of the feeding network. Scale bars, 100 μm.
Two serotonin-containing modulatory neurons of the feeding network, the cerebral giant cells (Fig. 3C, arrow) (McCrohan and Benjamin, 1980) were also stained as well as adjacent smaller cells in the anterior lobes of the cerebral ganglia. There were clusters of stained large neurons in the right parietal (Fig. 3B) and visceral ganglion. Some of these correspond to the Light Yellow Cells that are peptidergic neurons likely to be involved in ion and water regulation (Boer and Montagnewajer, 1994). Smaller numbers ofin situ-positive cells occurred in the left parietal, left, and right pleural ganglia with just two pairs of small unidentified cells in the buccal ganglia (data not shown), the main feeding ganglia of the snail (Benjamin, 1983). These results showed that the LsFaNaC is widely distributed in neurons located in a number of different types of behavioral networks.
Electrophysiological characterization of the LsFaNaC
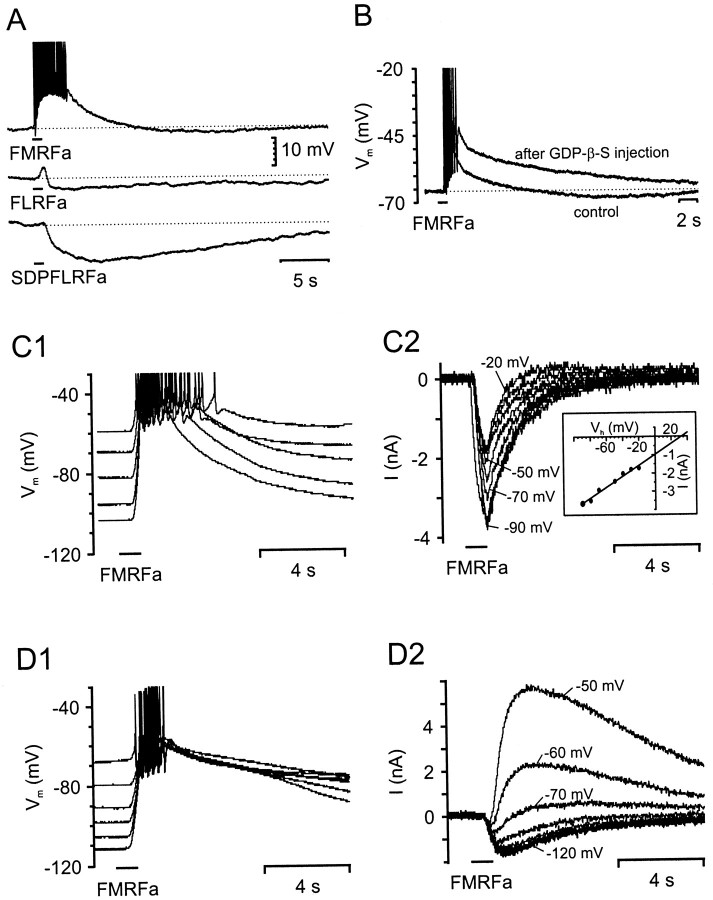
The in situ hybridization study showed that the dopaminergic neuron RPeD1, located in the right pedal ganglion, expresses mRNA for the Lymnaea homolog of the HaFaNaC. This neuron, which is easy to identify because of its large size, was used for studying the LsFaNaC. Isolated RPeD1 neurons in cell culture respond to FMRFamide with an initial fast depolarization followed by a slow hyperpolarization (1 sec pressure pulses; pipette FMRFamide; 0.1 mm;n > 30 cells) (Fig.4A). This was similar to the FMRFamide responses described previously for RPeD1 in the intact nervous system (Skingsley et al., 1993) and in the homologous giant dopaminergic neuron in the pedal ganglia of Helisoma(Jeziorski et al., 2000). The fast depolarizing response was likely to be the response mediated by the FaNaC. Consistent with this was the weaker response obtained with FLRFamide (n = 4 cells) and the absence of a depolarizing response to the N-terminally extended peptide SDPFLRFamide (n = 4 cells) applied at the same concentrations (Fig. 4A). This agonist selectivity was typical of the HaFaNaC (Cottrell, 1997). FLRFamide and SDPFLRFamide both elicited the hyperpolarizing component of the response seen with FMRFamide. This hyperpolarizing response to FMRFamide could be blocked by the injection of the general G-protein blocker GDP-β-S into RPeD1 (Fig. 4B) (n = 6 cells), suggesting the presence of a G-protein-coupled receptor. The depolarizing response to FMRFamide was not blocked, but in fact was consistently enhanced (Fig. 4B), presumably because of the removal of the initial part of the hyperpolarizing current that had a similar onset to the LsFaNaC response.
Fig. 4.
Responses of isolated RPeD1 neurons to focal application of FMRFamide, FLRFamide, and SDPFLRFamide.A, Responses of a single isolated RPeD1 neuron to focal 1 sec applications of FMRFamide, FLRFamide, and SDPFLRFamide (pipette concentration, 0.1 mm each) recorded under current clamp.B, FMRFamide responses (pipette concentration, 0.1 mm) recorded in a RPeD1 neuron before (control) and after the intracellular injection of the G-protein blocker GDP-β-S. Note the increase in the FMRFamide-induced depolarization and the block of the delayed hyperpolarizing response after the injection of GDP-β-S.C1, D1, Voltage dependence of FMRFamide responses (pipette concentration, 0.1 mm) in two different RPeD1 cells recorded under current-clamp conditions. The neuron inC1 showed an exclusively depolarizing response to FMRFamide application, whereas the neuron in D1displayed a biphasic response that consisted of an initial depolarization followed by a delayed hyperpolarization. The hyperpolarizing component readily reversed when the membrane potential was adjusted to values more negative than −70 mV. C2, D2, TEVC recording of series of FMRFamide responses in the same two neurons shown in C1 and D1, respectively. In C2, the holding potential was stepped from −90 to −20 mV in 10 mV increments, whereas holding potentials inD2 were between −120 and −50 mV.
To characterize the LsFaNaC in more detail, FMRFamide responses were recorded in isolated neurons using two-electrode voltage clamp. Brief focal application of FMRFamide caused a fast inward current with a time course that resembled the fast depolarization observed under current clamp. The fast inward current was followed by an outward current at holding potentials more positive than −70 mV that varied in amplitude between individual neurons (Fig. 4, compare D2,C2). The delayed outward current hindered an accurate determination of the reversal potential for the fast inward current in some neurons. However, in RPeD1 neurons with weak or no delayed outward currents, the fast inward current dominated the response at the tested holding potentials, allowing the reversal potential to be determined more accurately (Fig. 4C2). In these cells, the amplitude of the inward current increased linearly at holding potentials between −20 and −90 mV. Responses were not tested at more positive holding potentials because of the strong activation of voltage-gated outward currents. The reversal potential was found to be +40 ± 7 mV (n = 11 cells) by linear extrapolation (Fig. 4C2, insert).
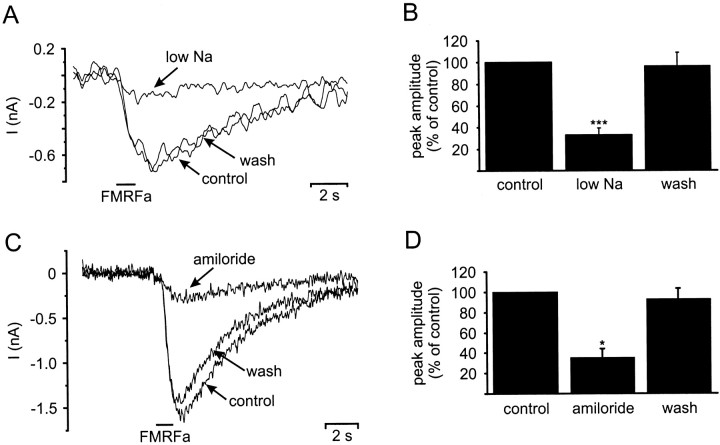
Evidence that Na+ carried the fast inward current was provided by ion substitution experiments under voltage clamp. Replacing 90% of the Na+ ions in normal saline withN-methyl-d-glucamine (low Na+ saline) caused a major reduction in the FMRFamide-induced inward current (Fig.5A). The maximum reduction in the amplitude of the inward current was 90% with a mean reduction of 67 ± 6% (n = 6 experiments performed on four cells) (Fig. 5B). The use of low Na+ saline also caused a shift in the reversal potential of the fast inward current by 58 mV to −18 ± 8 mV (n = 4). This value was identical to the predicted value based on the reversal potential in normal saline and the hypothesis that the fast inward current was predominantly carried by Na+ ions (Lingueglia et al., 1995). Exchanging the low Na+ saline for normal saline reversed the reduction in the peak amplitude of the fast inward current caused by replacement of 90% of the Na+ ions (mean recovery, 96 ± 13%).
Fig. 5.
Sodium dependence and amiloride block of FMRFamide induced inward currents in isolated RPeD1 neurons. A, Replacing 90% of the extracellular Na+concentration with N-methyl-d-glucamine resulted in the reduction of the peak FMRFamide-induced (pipette concentration, 0.1 mm) inward current recorded under TEVC (holding potential, −100 mV) from −0.7 nA (control) to −0.17 nA (low Na). Returning the extracellular Na+ concentration to 50 mm (wash) reversed the effect. B, Summary of six sodium replacement experiments conducted on four cells. Reducing the extracellular Na+ concentration significantly reduced the mean peak amplitude of the FMRFamide-induced inward current to 33 ± 6% of the control value (pairedt test: p < 0.001). The mean peak amplitude returned to 96 ± 12% of the control value, when the extracellular Na+ concentration was raised again to 50 mm. C, Application of amiloride (0.1 mm) decreased the FMRFamide-induced (pipette concentration, 0.1 mm) inward current recorded under TEVC (holding potential, −100 mV) from −1.6 nA (control) to −0.3 nA (amiloride). The block was almost completely reversed after wash-out of amiloride from the bath. D, Summary of three amiloride blocking experiments. Amiloride application blocked a significant proportion of the FMRFamide-induced inward current, reducing the mean peak amplitude to 35 ± 9% of the control value (paired t test;p < 0.05). The amiloride block was reversible, and the mean peak amplitude returned to 92 ± 10% after wash-out of amiloride from the bath. *p ≤ 0.05; ***p ≤ 0.001.
Bath application of amiloride (0.1 mm) to theLymnaea neuron also caused a 65 ± 16% reduction in the fast inward current evoked by FMRFamide pulses in isolated RPeD1 neurons (n = 3 cells) (Fig. 5C,D). The block of the inward current was almost completely reversed (mean recovery, 93 ± 10%) after washout of the amiloride solution (Fig.5D).
Modulation of the FMRFamide response by acidic pH
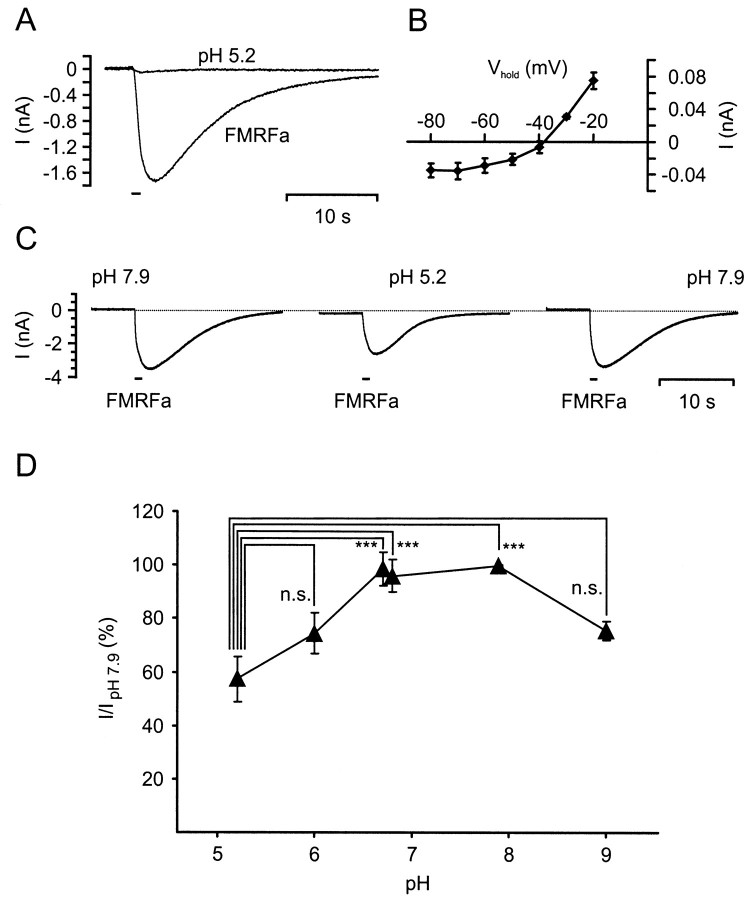
The molecular and electrophysiological characterization of the LsFaNaC clearly demonstrated that it is a member of the Degenerin/ENaC family (DEG/ENaC) family of channels that also includes the H+-gated ASIC channels that are expressed in sensory neurons of the mammalian dorsal root ganglia. The molecular relationship between these channels raised the question of whether protons also gate the LsFaNaC. The application of 1 sec pulses of NS buffered at pH 5.2 caused a very small inward current (mean amplitude, −0.04 ± 0.01 nA; n = 4 cells) in isolated RPeD1 neurons in cell culture, which was considerably weaker than FMRFamide responses in the same neuron (Fig.6A). However, the pH 5.2-induced current was unlikely to be mediated by the LsFaNaC, because its reversal potential (−39 ± 2 mV; n = 4 cells) was significantly more negative than that of the FMRFamide-induced current (+40 mV, see above). Furthermore, the I–Vrelationship for the pH 5.2-induced inward current showed some rectifying behavior at holding potentials more negative than −40 mV (Fig. 6B), which was not seen in the I–Vrelationship for the FMRFamide-induced inward current.
Fig. 6.
Effect of pH 5.2 acidification on FMRFamide-induced inward current in isolated RPeD1 neurons.A, Response of a RPeD1 neuron to focal application of a 1 sec pulse of saline at pH 5.2 recorded under TEVC (trace labeled pH 5.2; holding potential, −100 mV). The pH 5.2 pulse caused a very weak transient inward current. A record of an FMRFamide-induced inward current (1 sec pulse; pipette concentration, 0.1 mm) in the same neuron is shown for comparison. B, MeanI–V plot for the pH 5.2-induced inward current in four RPeD1 cells recorded under TEVC. C, Series of FMRFamide-induced inward currents (pipette concentration, 0.1 mm; holding potential, −100 mV) recorded in a RPeD1 neuron while bathed in saline buffered at pH 7.9 or pH 5.2. D, Summary of the effects of bath pH on the amplitude of FMRFamide-induced inward currents in RPeD1 neurons. Statistical analysis revealed that the reduction by acidification to pH 5.2 was significant compared with values at pH 6.7, 6.8, and 7.9 [ANOVA:F(5,39) = 8.982, p< 0.001; followed by pairwise comparisons using post hoc Tukey honestly significant difference (HSD) tests giving p values ≤ 0.001]. The differences between all other pairs of pH values were not statistically significant (Tukey HSD tests: p values between 0.08 and 0.99).
Despite the lack of evidence for direct H+-gating of the LsFaNaC, altering the pH of the medium had significant modulatory effects on the FMRFamide-activated currents in isolated RPeD1 neurons. This was demonstrated by comparing the responses to 1 sec FMRFamide pulses in saline buffered at pH 7.9 (normal saline) with FMRFamide-induced currents in salines buffered at pH 5.2, 6.0, 6.7, 6.8, and 9.0. Acidification of the medium to pH 5.2 had the most dramatic effect and reversibly reduced the peak amplitude of the FMRFamide-induced inward current to 57 ± 8% (n = 11 cells) of its control value at pH 7.9 (Fig. 6C,D). Saline at pH 6.0 had a weaker blocking effect, reducing the peak amplitude to 74 ± 8% (n = 5 cells), whereas pH 6.7 and pH 6.8 medium had no apparent effect on the FMRFamide-gated current (98 ± 6% and 96 ± 6%; n = 6 cells each) (Fig.6D). A reduction in the peak amplitude of the FMRFamide-gated current to 75 ± 9% (n = 6 cells) of its control value was also observed when the medium was adjusted to an alkaline value of pH 9.0.
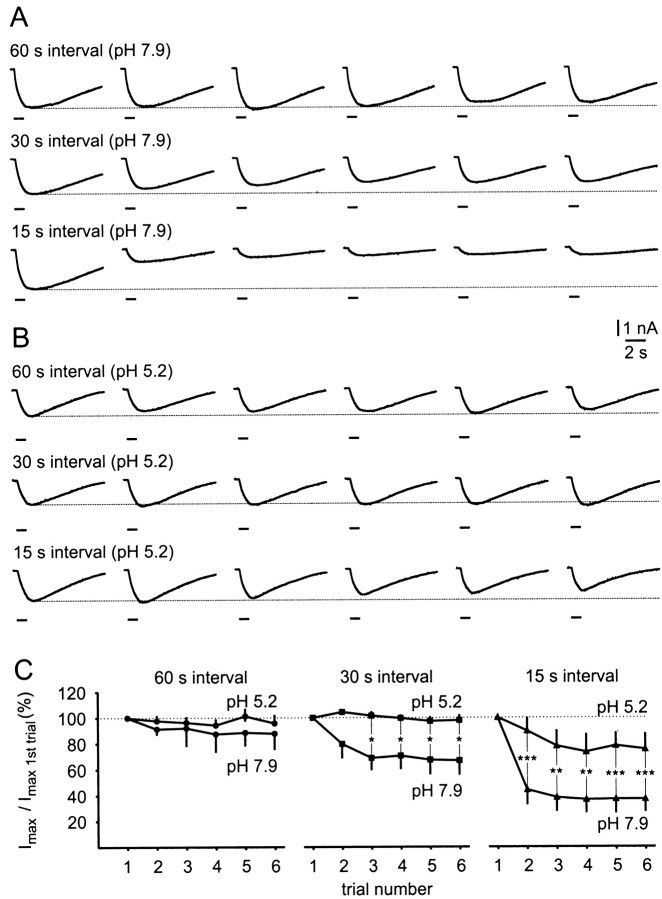
The most dramatic effect of acidification of the medium, however, was a reduction in the desensitizing effects of repeated applications of FMRFamide (Fig. 7). At normal pH (7.9), desensitization was strong at intervals of 15 sec between successive 1 sec pulses of FMRFamide in a six pulse train, less at 30 sec, and not significant at 60 sec intervals (Fig. 7A). Most of the reduction in response occurred between the first and second applications of FMRFamide (Fig. 7C). After acidification of the medium to pH 5.2, no reduction in the amplitude of the response could be detected at 30 sec intervals, and at 15 sec the desensitization was significantly reduced when compared with FMRFamide applications at 15 sec intervals at pH 7.9 (Fig. 7B,C).
Fig. 7.
Desensitization of FMRFamide-induced inward currents in RPeD1 neurons by repeated applications at 60, 30, and 15 sec intervals in bath media at pH 7.9 and pH 5.2. A,B, Sample records of series of inward currents (holding potential, −100 mV) induced in the same RPeD1 neuron by six repeated FMRFamide pulses (1 sec, pipette concentration, 0.1 mm) at intervals of 60, 30, and 15 sec in bath media buffered at pH 7.9 (A) and pH 5.2 (B). Note the substantial desensitization at 15 sec intervals and to a lesser extent at 30 sec intervals in pH 7.9. In contrast, desensitization was weak at 15 sec intervals and absent at 30 sec intervals in pH 5.2.C, Summary of the desensitization data obtained from six individual RPeD1 neurons. Each neuron was tested for desensitization in response to series of six FMRFamide pulses applied at 60, 30, and 15 sec intervals in bath media at pH 7.9 and pH 5.2. The graphs clearly illustrate the difference in desensitization observed in pH 7.9 and pH 5.2. Pairwise comparison between mean peak amplitude values at 15 sec application intervals showed that these values for trials 2–6 were significantly larger in pH 5.2 than in pH 7.9 (t tests;p values 0.001–0.004). Similarly, at 30 sec application intervals, these values were significantly larger in pH 5.2 than in pH 7.9 for trials 3–6 (t test; p values 0.03–0.05). The small differences in desensitization at 60 sec application intervals in pH 5.2 and pH 7.9 were not significant. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
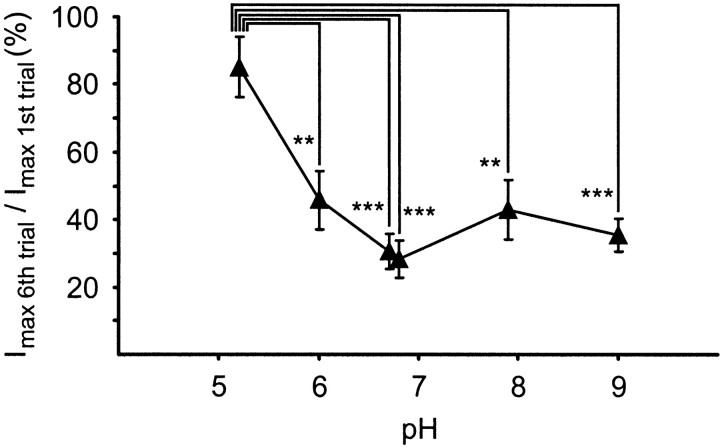
A second set of experiments was performed to study the effect of a wider range of pH changes on the level of desensitization to repeated FMRFamide pulses. The same protocol that produced the maximum desensitization in the previous experiment in HEPES-buffered saline at pH 7.9 (i.e., a train of six FMRFamide pulses at 15 sec intervals) was applied, whereas the bath medium was exchanged for salines buffered at pH 6.8 (HEPES-buffered), pH 5.2, 6.0, and 6.7 (all MES-buffered), and pH 9.0 (Tris-buffered). At pH 7.9, the response to repeated FMRFamide pulses again showed rapid desensitization, and the peak amplitude of the sixth FMRFamide-induced inward current was reduced to 43 ± 9% (n = 6 cells) of the corresponding first response in the series (Fig. 8). The level of desensitization was statistically similar at bath pH values of 9.0, 6.8, 6.7, and 6.0 with responses to the sixth FMRFamide application being reduced to between 28 ± 6% (pH 6.8; n = 6 cells) and 46 ± 9% (pH 6.0; n = 5 cells) of their corresponding control values (Fig. 8). A significant reduction in desensitization was observed only when the bath medium was acidified to pH 5.2. Under these conditions, the amplitude of the last response within a series of six FMRFamide pulses was still 85 ± 9% (n = 6 cells) of the first response (Fig. 8), which is comparable with the result of the previous experiment (Fig.7B).
Fig. 8.
Effects of various bath pH values on desensitization in isolated RPeD1 neurons. Isolated RPeD1 neurons were exposed to series of six FMRFamide pulses (pipette concentration, 0.1 mm, 1 sec) at 15 sec intervals, and the induced inward currents were measured under TEVC (holding potential, −100 mV) in bath media buffered at pH 5.2, 6.0, and 6.7 (MES-buffered), 6.8 and 7.9 (HEPES-buffered), and pH 9.0 (Tris-buffered). The amplitude of the sixth response in each series was expressed as a percentage of the first response, and the mean values for each experiment were plotted against the pH. A statistical analysis revealed that acidification of the bath medium to pH 5.2 significantly reduced desensitization of the FMRFamide-induced inward current compared with all other pH values tested (ANOVA: F(5,29) = 8.684,p < 0.001, followed by post hocTukey HSD tests for pairwise comparison; p ≤ 0.01 for all pairs; **p ≤ 0.01, ***p ≤ 0.001). These tests also showed that none of the differences between the desensitization levels at pH 6.0, 6.7, 6.8, 7.9, and 9.0 were significant (Tukey HSD tests; p values > 0.5).
DISCUSSION
Comparison of the structure of LsFaNaC with other members of the ENaC superfamily of sodium channels revealed many common features. The structure of all the channels was predicted to be that of a protein with two membrane-spanning domains with both the N- and C-terminal domains located within the cell. The most conservation of sequence between all the ENaCs was found in the transmembrane domains, especially the second domain that is known to form the pore of the channel. Mutations within this region lead to altered or lost ion selectivity and amiloride sensitivity (Lingueglia et al., 1995). LsFaNaC shows 93 and 98% sequence identity with HaFaNaC and HtFaNaC, respectively, in these regions and up to 46% identity with the more distantly related ENaCs, indicating that LsFaNaC was indeed a member of this channel superfamily. In both LsFaNaC and HtFaNaC the intracellular N-terminal domain contained a 31 residue extended sequence lacking in HaFaNaC. Whereas Jeziorski et al. (2000) reported that alternative splicing occurs within this region of HtFaNaC, they concluded that the likely site of translational initiation is that predicted by comparison to the sequence of HaFaNaC. Conservation of this N-terminal extension in LsFaNaC, however, would suggest that in bothLymnaea and Helisoma the channel used an alternative start codon. Because this sequence also contains potential phosphorylation sites for several serine–threonine protein kinases, such alternative translational initiation may result in differences in regulation of the channels.
In situ hybridization allowed the identification of many neurons expressing LsFaNaC, including the giant dopaminergic neuron RPeD1. Previous work had shown that this neuron displays a fast depolarizing response to FMRFamide application, as would be predicted for a neuron expressing an FMRFamide-gated Na+ channel (Skingsley et al., 1993). In cell culture, application of FMRFamide induced an inward current in isolated RPeD1 neurons that showed the characteristics of a current that is mediated by a member of the DEG/ENaC superfamily (i.e., reversal potential close to the estimated Na+ reversal potential, high selectivity of the channel for Na+ ions, block by amiloride).
A very similar current was described recently in the giant dopaminergic neuron (GDN) in Helisoma (Jeziorski et al., 2000), which is homologous to the RPeD1 neuron in Lymnaea (Harris and Cottrell, 1995). The same authors also conducted a detailed comparison between the neuronal FMRFamide-induced currents in GDN neurons in the isolated nervous system and FMRFamide-induced currents in oocytes expressing the HtFaNaC. They found only minor differences and concluded that the heterologously expressed channel truly resembles the neuronal channel present in GDN, although they provided no direct evidence for the expression of the HtFaNaC gene in GDN neurons. Considering the strong sequence homology between HtFaNaC and LsFaNaC and the homology between GDN (Helisoma) and RPeD1 (Lymnaea), it would be reasonable to expect very similar results for the heterologous expression of the LsFaNaC.
The molecular relationship between ASICs in mammals and FaNaCs in mollusks prompted us to study the effects of pH changes on FaNaCs. Although acidification of the medium appeared to be unable to directly open the LsFaNaC, it had considerable effects on the characteristics of the FMRFamide-induced current. First, acidification of the medium caused a pH-dependent block of the FMRFamide-gated inward current at pH < 6.0. Some blocking effects were also observed when the medium was adjusted to an alkaline value of pH 9.0. These results are consistent with effects described for the heterologously expressed HaFaNaC (Price and Price, 2000) and more generally with structurally diverse channel types in which changes in extracellular pH can have widely varying outcomes (Hille, 1992; Pasternack et al., 1992;Traynelis, 1998; McLatchie and Bevan, 2001; Shah et al., 2001). Second, acidification to pH 5.2 reduced the desensitization of the response of LsFaNaC to repeated applications of FMRFamide. This is of particular interest because it has been reported recently that application of FMRFamide to the mammalian ASICs substantially reduces their inactivation rate in response to acidification of the medium. This modulation of the ASICs was interpreted as evidence that the peptide-binding site has been at least partially conserved between FaNaC and the ASICs (Askwith et al., 2000). Taking this into account, together with the structural similarity of FaNaCs and ASICs and our demonstration that the rate of desensitization of LsFaNaC is modulated by pH changes, it would suggest that not only is the peptide-binding site likely to be conserved between FaNaCs and ASICs, but also the proton-sensing site. It has been proposed that the weakly conserved region immediately following the last cysteine in the extracellular domain of FaNaC may represent the peptide-binding site, because this region is absent from the other, peptide-insensitive family members (Jeziorski et al., 2000). In light of the recent discovery that ASICs bind FMRFamide and from our results that demonstrate pH modulation of LsFaNaC, it would seem that regions conserved between the FaNaCs and the ASICs may in fact be sites of FMRFamide binding or acid sensing. Such regions should be divergent or nonexistent in those channels, which are not affected by FMRFamide or acid treatment. A comparison of all three FaNaCs with ASIC3 and αENaC reveals that such a region exists, just before TM2 (residues 477–519 in LsFaNaC). In this 43 residue region of near sequence identity within the FaNaCs there is considerably more sequence homology to ASIC3 than there is to αENaC. Mutational analysis of this region and other potential sites should allow the site of ligand binding to be determined. Such a study would elucidate whether acid modulation of the LsFaNaC and acid-gating in ASICs are in fact caused by a conserved, structurally related site or whether the effects are mediated by nonrelated sites.
Footnotes
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council to P.R.B.
S.J.P. and V.A.S. contributed equally to this work.
Correspondence should be addressed to V. A. Straub, Sussex Centre for Neuroscience, University of Sussex, Falmer, Brighton BN1 9QG, UK. E-mail: V.Straub@sussex.ac.uk.
S. J. Perry's present address: Department of Medicine, Duke University Medical Center, Durham, NC 27710.
REFERENCES
- 1.Askwith CC, Cheng C, Ikuma M, Benson C, Price MP, Welsh MJ. Neuropeptide FF and FMRFamide potentiate acid-evoked currents from sensory neurons and proton-gated DEG/ENaC channels. Neuron. 2000;26:133–141. doi: 10.1016/s0896-6273(00)81144-7. [DOI] [PubMed] [Google Scholar]
- 2.Benjamin PR. Gastropod feeding: behavioral and neural analysis of a complex multicomponent system. Sym Soc Exp Biol. 1983;37:159–193. [PubMed] [Google Scholar]
- 3.Boer H, Montagnewajer C. Functional morphology of the neuropeptidergic light yellow cell system in pulmonate snails. Cell Tissue Res. 1994;277:531–538. doi: 10.1007/BF00300226. [DOI] [PubMed] [Google Scholar]
- 4.Buckett KJ, Dockray GJ, Osborne NN, Benjamin PR. Pharmacology of the myogenic heart of the pond snail Lymnaea stagnalis. J Neurophysiol. 1990a;63:1413–1425. doi: 10.1152/jn.1990.63.6.1413. [DOI] [PubMed] [Google Scholar]
- 5.Buckett KJ, Peters M, Dockray GJ, Van Minnen J, Benjamin PR. Regulation of heartbeat in Lymnaea by motoneurons containing FMRFamide-like peptides. J Neurophysiol. 1990b;63:1426–1435. doi: 10.1152/jn.1990.63.6.1426. [DOI] [PubMed] [Google Scholar]
- 6.Chen CC, England S, Akopian AN, Wood JN. A sensory neuron-specific, proton-gated ion channel. Proc Natl Acad Sci USA. 1998;95:10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombaioni L, Paupardin-Tritsch D, Vidal PP, Gerschenfeld HM. The neuropeptide FMRFamide decreases both the Ca2+ conductance and a cyclic 3′,5′-adenosine monophosphate-dependent K+ conductance in identified molluscan neurons. J Neurosci. 1985;5:2533–2538. doi: 10.1523/JNEUROSCI.05-09-02533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coscoy S, Lingueglia E, Lazdunski M, Barbry P. The Phe-Met-Arg-Phe-amide-activated sodium channel is a tetramer. J Biol Chem. 1998;273:8317–8322. doi: 10.1074/jbc.273.14.8317. [DOI] [PubMed] [Google Scholar]
- 9.Cottrell GA. The first peptide-gated ion channel. J Exp Biol. 1997;200:2377–2386. doi: 10.1242/jeb.200.18.2377. [DOI] [PubMed] [Google Scholar]
- 10.Ebberink RHM, Price DA, Vanloenhout H, Doble KE, Riehm JP, Geraerts WPM, Greenberg MJ. The brain of Lymnaea contains a family of FMRFamide-like peptides. Peptides. 1987;8:515–522. doi: 10.1016/0196-9781(87)90018-0. [DOI] [PubMed] [Google Scholar]
- 11.García-Añoveros J, Derfler B, Neville-Golden J, Hyman BT, Corey DP. BNaC1 and BNaC2 constitute a new family of human neuronal sodium channels related to degenerins and epithelial sodium channels. Proc Natl Acad Sci USA. 1997;94:1459–1464. doi: 10.1073/pnas.94.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris SJ, Cottrell GA. Properties of an identified dopamine-containing neurone in culture from the snail Helisoma. Exp Physiol. 1995;80:37–51. doi: 10.1113/expphysiol.1995.sp003833. [DOI] [PubMed] [Google Scholar]
- 13.Higgins WJ, Doble KE, Lesser W, Dunn BM, Price DA. FMRFamide increases the adenylate cyclase activity and cyclic AMP level of molluscan heart. Eur J Pharmacol. 1978;48:425–430. doi: 10.1016/0014-2999(78)90170-x. [DOI] [PubMed] [Google Scholar]
- 14.Hille B. Ionic channels of excitable membranes, Ed 2. Sinauer; Sunderland, MA: 1992. [Google Scholar]
- 15.Jeziorski MC, Green KA, Sommerville J, Cottrell GA. Cloning and expression of a FMRFamide-gated Na+ channel from Helisoma trivolvis and comparison with the native neuronal channel. J Physiol (Lond) 2000;526:13–25. doi: 10.1111/j.1469-7793.2000.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellett E, Perry SJ, Santama N, Worster BM, Benjamin PR, Burke JF. Myomodulin gene of Lymnaea: structure, expression, and analysis of neuropeptides. J Neurosci. 1996;16:4949–4957. doi: 10.1523/JNEUROSCI.16-16-04949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemenes G, Elekes K, Hiripi L, Benjamin PR. A comparison of 4 techniques for mapping the distribution of serotonin and serotonin-containing neurons in fixed and living ganglia of the snail, Lymnaea. J Neurocytol. 1989;18:193–208. doi: 10.1007/BF01206662. [DOI] [PubMed] [Google Scholar]
- 18.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium-channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 19.McCrohan CR, Benjamin PR. Synaptic relationships of the cerebral giant cells with motoneurons in the feeding system of Lymnaea stagnalis. J Exp Biol. 1980;113:351–366. doi: 10.1242/jeb.85.1.169. [DOI] [PubMed] [Google Scholar]
- 20.McLatchie LM, Bevan S. The effects of pH on the interaction between capsaicin and the vanilloid receptor in rat dorsal root ganglia neurons. Br J Pharmacol. 2001;132:899–908. doi: 10.1038/sj.bjp.0703900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson LS, Rosoff ML, Li C. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 1998;281:1686–1690. doi: 10.1126/science.281.5383.1686. [DOI] [PubMed] [Google Scholar]
- 22.Pasternack M, Bountra C, Voipio J, Kaila K. Influence of extracellular and intracellular pH on GABA-gated chloride conductance in grayfish muscle fibres. Neuroscience. 1992;47:921–929. doi: 10.1016/0306-4522(92)90040-9. [DOI] [PubMed] [Google Scholar]
- 23.Piomelli D, Volterra A, Dale N, Siegelbaum SA, Kandel ER, Schwartz JH, Belardetti F. Lipoxygenase metabolites of arachidonic acid as 2nd messengers for presynaptic inhibition of Aplysia sensory cells. Nature. 1987;328:38–43. doi: 10.1038/328038a0. [DOI] [PubMed] [Google Scholar]
- 24.Price DA, Price RB. Structure-activity studies on the Helix FMRFamide-gated sodium channel. Soc Neurosci Abstr. 2000;26:917. [Google Scholar]
- 25.Price DA, Davies NW, Doble KE, Greenberg MJ. The variety and distribution of FMRFamide-related peptides in mollusks. Zool Sci. 1987;4:395–410. [Google Scholar]
- 26.Ridgway R, Syed NI, Lukowiak K, Bulloch AGM. Nerve growth factor (NGF) induces sprouting of specific neurons of the snail, Lymnaea-stagnalis. J Neurobiol. 1991;22:377–390. doi: 10.1002/neu.480220406. [DOI] [PubMed] [Google Scholar]
- 27.Schot LPC, Boer HH. Immuno-cytochemical demonstration of peptidergic cells in the pond snail Lymnaea stagnalis with an antiserum to the molluscan cardioactive tetrapeptide FMRFamide. Cell Tissue Res. 1982;225:347–354. doi: 10.1007/BF00214687. [DOI] [PubMed] [Google Scholar]
- 28.Shah MJ, Meis S, Munsch T, Pape HC. Modulation by extracellular pH of low- and high-voltage-activated calcium currents of rat thalamic relay neurons. J Neurophysiol. 2001;85:1051–1058. doi: 10.1152/jn.2001.85.3.1051. [DOI] [PubMed] [Google Scholar]
- 29.Skingsley DR, Bright K, Santama N, Van Minnen J, Brierley MJ, Burke JF, Benjamin PR. A molecularly defined cardiorespiratory interneuron expressing SDPFLRFamide/GDPFLRFamide in the snail Lymnaea: monosynaptic connections and pharmacology. J Neurophysiol. 1993;69:915–927. doi: 10.1152/jn.1993.69.3.915. [DOI] [PubMed] [Google Scholar]
- 30.Slade CT, Mills J, Winlow W. The neuronal organization of the paired pedal ganglia of Lymnaea stagnalis (L). Comp Biochem Physiol [A] 1981;69:789–803. doi: 10.1016/0300-9629(89)90513-6. [DOI] [PubMed] [Google Scholar]
- 31.Syed NI, Bullock AGM, Lukowiak K. In vitro reconstruction of the respiratory central pattern generator of the mollusk Lymnaea. Science. 1990;250:282–285. doi: 10.1126/science.2218532. [DOI] [PubMed] [Google Scholar]
- 32.Traynelis SF. pH modulation of ligand-gated ion channels. In: Kaila K, Ransom BR, editors. pH and brain function. Wiley-Liss; New York: 1998. pp. 417–446. [Google Scholar]
- 33.Vreugdenhil E, Jackson JF, Bouwmeester T, Smit AB, Van Minnen J, Van Heerikhuizen H, Klootwijk J, Joosse J. Isolation, characterization, and evolutionary aspects of a cDNA clone encoding multiple neuropeptides involved in the stereotyped egg-laying behavior of the fresh water snail Lymnaea stagnalis. J Neurosci. 1988;8:4184–4191. doi: 10.1523/JNEUROSCI.08-11-04184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldmann R, Lazdunski M. H+-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol. 1998;8:418–424. doi: 10.1016/s0959-4388(98)80070-6. [DOI] [PubMed] [Google Scholar]
- 35.Waldmann R, Bassilana F, deWeille J, Champigny G, Heurteaux C, Lazdunski M. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J Biol Chem. 1997a;272:20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 36.Waldmann R, Champigny G, Bassilana F, Heurteaux C, Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997b;386:173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 37.Willoughby D, Yeoman MS, Benjamin PR. Inositol-1,4,5-trisphosphate and inositol-1,3,4,5-tetrakisphosphate are second messenger targets for cardioactive neuropeptides encoded on the FMRFamide gene. J Exp Biol. 1999a;202:2581–2593. doi: 10.1242/jeb.202.19.2581. [DOI] [PubMed] [Google Scholar]
- 38.Willoughby D, Yeoman MS, Benjamin PR. Cyclic AMP is involved in cardioregulation by multiple neuropeptides encoded on the FMRFamide gene. J Exp Biol. 1999b;202:2595–2607. doi: 10.1242/jeb.202.19.2595. [DOI] [PubMed] [Google Scholar]
- 39.Yang HYT, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of 2 brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci USA. 1985;82:7757–7761. doi: 10.1073/pnas.82.22.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]