Abstract
The differentiation of precursor cells into neurons has been shown to be influenced by both the Notch signaling pathway and growth factor stimulation. In this study, the regulation of neuronal differentiation by these mechanisms was examined in the embryonic day 10 neuroepithelial precursor (NEP) population. By downregulating Notch1 expression and by the addition of a Delta1 fusion protein (Delta Fc), it was shown that signaling via the Notch pathway inhibited neuron differentiation in the NEP cells, in vitro. The expression of two of the Notch receptor homologs, Notch1 and Notch3, and the ligand Delta1 in these NEP cells was found to be influenced by a number of different growth factors, indicating a potential interaction between growth factors and Notch signaling. Interestingly, none of the growth factors examined promoted neuron differentiation; however, the fibroblast growth factors (FGFs) 1 and 2 potently inhibited differentiation. FGF1 and FGF2 upregulated the expression of Notch and decreased expression of Delta1 in the NEP cells. In addition, the inhibitory response of the cells to the FGFs could be overcome by downregulating Notch1 expression and by disrupting Notch cleavage and signaling by the ablation of the Presenilin1 gene. These results indicate that FGF1 and FGF2 act via the Notch pathway, either directly or indirectly, to inhibit differentiation. Thus, signaling through the Notch receptor may be a common regulator of neuronal differentiation within the developing forebrain.
Keywords: Notch, Delta, growth factors, fibroblast growth factor (FGF), neuroepithelial precursor cells, regulation of differentiation
The complex array of neurons and glia within the nervous system initially arise from the neuroepithelial precursor (NEP) cells found within the neural tube. It now appears that cell interactions are pivotal regulators of neuron differentiation, in addition to allowing cell proliferation and the continual renewal of the precursor population. The Notch receptor, and its ligands Delta and Serrate/Jagged, are membrane-bound proteins that provide an essential mechanism of interaction between adjacent cells. Studies examining the role of Notch signaling during development have found it to be associated with the inhibition of neurogenesis (Austin et al., 1995;Chitnis et al., 1995; Dorsky et al., 1995, Nakamura et al., 2000). However, the exact role of Notch and Delta in mammalian CNS precursor cells remains to be determined.
The Notch receptor is a 300 kDa protein with a single transmembrane domain. The extracellular domain of Notch consists of epidermal growth factor (EGF)-like repeats, which are required for ligand binding, and a second region, the Lin12/Notch repeat region. The intracellular domain also contains highly conserved domains, the RAM23 domain and the cdc10/ankyrin repeat (Fleming 1998), which bind to intracellular proteins that regulate Notch signaling. Delta also contains EGF-like repeats in the extracellular domain; however, receptor binding occurs via the Delta/Serrate/Lag (DSL) domain, at the N terminus of the protein (Fleming 1998).
Analysis of Notch signaling has shown that the intracellular domain is cleaved after ligand binding (Schroeter et al., 1998), a process that appears to be mediated by Presenilin proteins (De Strooper et al., 1999; Song et al., 1999; Steiner et al., 1999). The intracellular domain of Notch is directed to the nucleus (Tamura et al., 1995), where it initiates transcription of certain basic helix-loop-helix (bHLH) genes (Jarriault et al., 1998), resulting in the inhibitory cellular response and the downregulation of Delta expression (Heitzler et al., 1996; Parks et al., 1997). Delta expression is also regulated by various bHLH transcription factors, which are essential for neurogenesis (Ma et al., 1996, 1998; Fode et al., 1998; Casarosa et al., 1999). Although many of the proteins involved in Notch signaling have been discovered, extracellular factors that regulate the pathway remain to be determined.
Various growth factors are also important for the development of the nervous system, many of which inhibit neuronal differentiation. These include fibroblast growth factor 1 (FGF1) and FGF2 (Murphy et al., 1990; Drago et al., 1991), leukemia inhibitory factor (Bonni et al., 1997; Koblar et al., 1998), and bone morphogenetic proteins (Shou et al., 1999). The inhibitory nature of these factors prompted our examination of potential interactions between growth factors and the Notch signaling pathway.
In the present study we have shown that FGF1, FGF2, or Notch signaling in mouse NEP cells inhibited neuron differentiation and that expression of Delta and Notch within these cells was regulated by FGF1 and FGF2. The inhibition of neurogenesis with FGF could be overcome by disrupting Notch signaling in vitro, indicating that inhibition caused by FGF may be regulated by the Notch pathway.
MATERIALS AND METHODS
Reagents. Recombinant FGF2 was obtained from Boehringer Mannheim (Roche, Germany). Recombinant FGF1, brain-derived neurotrophic factor (BDNF), and neurotrophin (NT)-3 were obtained from PeproTech Inc. Recombinant FGF8, transforming growth factor (TGF) β5, and oncostatin M (OSM) were obtained from R&D Systems. Recombinant bone morphogenetic protein (BMP) 2 and BMP4 were obtained from Genetics Institute. CNTF was obtained from Alomone labs (Jerusalem, Israel). Recombinant leukemia inhibitory factor (LIF) was obtained from AMRAD. Each of the growth factors was used at 50 ng/ml unless stated otherwise, except recombinant LIF (specific activity 108 U/mg), which was used at a concentration of 1000 U/ml in culture.
Isolation of embryonic day 10 neuroepithelial precursor cells. Embryonic day 10 (E10) NEP cells were isolated from CBA mice as described previously (Murphy et al., 1990). Cells were dissociated into a single-cell suspension, washed, and resuspended in serum-free media before plating onto tissue culture plates coated with poly-dl-ornithine (Sigma, St. Louis, MO) and laminin (Life Technologies, Gaithersburg, MD). Cells were plated at high cell density, in either 24-well or 96-well plates (Falcon) or Terasaki plates (Bio-Rad, Burlingame, CA) at 200,000, 15,000 and 2,000 cells per well, respectively. The serum-free media was the media hormone mix described previously by Reynolds and Weiss (1992) andGritti et al. (1996). When cultured in the presence of serum, cells were grown in Monomed (Life Technologies) with part A supplement (CSL).
E10 NEP cells were isolated from Presenilin1 null mutant mice (Wong et al., 1997) obtained from Dr. Hui Zheng (Baylor College of Medicine). The genotypes of individual mice were determined by PCR using primers to Presenilin1, 5′ primer, AGCCAAGAACGGCAGCAGCAGCATGACAGGCAGAG, and 3′ primer, CTTCCATGAGCCATTTGCTAAGTGC, and the neo gene CCATTGCTCAGCGGTGCTGTCCAT as described by Qian et al. (1998).
RT-PCR analysis. RNA was purified from tissue and cultured cells using the RNeasy minikit (Qiagen, Hilden, Germany). RNA was reverse transcribed using Superscript II (Life Technologies) with 10 pmol oligo-dT (Promega, Madison, WI) as primer and 1 μg of total RNA. The RNA and oligo-dT were denatured by heating to 70°C before rapidly chilling on ice. The reverse transcription reaction was performed in a volume of 20 μl containing 1× Superscript II buffer, 0.1 mm DTT, and 10 μm each of dATP, dTTP, dCTP, and dGTP for 1 hr at 42°C. For PCR amplification, specific oligonucleotide primer pairs were incubated with 2 μl cDNA and 2.5 U/μl pfu polymerase (Stratagene, La Jolla, CA) in a 100 μl reaction mix that included 1× buffer and 200 μm each of dATP, dTTP, dCTP, and dGTP. Typical cycle conditions were 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C for 35 cycles. Ten microliters of the reaction were then separated on a 1% agarose gel and visualized by ethidium bromide staining.
The primers for RT-PCR analysis were as follows: mouseNotch1 5′ primer, TTACAGCCACCATCACAGCCACACC and 3′ primer, ATGCCCTCGGACCAATCAGA; mouse Notch2 5′ primer, GAGGCGCTCTTCTGCTGTTGAAGA and 3′ primer, ATAGAGTCACTGAGCTCTCGGACAG; mouse Notch3 5′ primer, ACACTGGGAGTTCTCTGT and 3′ primer, GTCTGCTGGCATGGGATA; mouse Delta1, 5′ primer TGTGACGAGCACTACTACGGAGAAG and 3′ primer, AGTAGTTCAGGTCTTGGTTGCAGAA; and mouse β-actin 5′ primer RT-PCR β-actin5′ primer, CTGAAGTACCCATTGAACATGGC and 3′ primer CAGAGCAGTAATCTCCTTCTGCAT.
Northern blot analysis. RNA was isolated from cells cultured in 24-well plates for 24 hr in the presence of various growth factors and serum using the RNeasy minikit (Qiagen). After electrophoresis through a 0.8% agarose–formaldehyde gel, RNA was capillary transferred to Hybond nylon membrane (Amersham, Arlington Heights, IL) overnight using 20× SSC buffer and then fixed to the filter by exposure to UV light using a Stratalinker (Stratagene). The filter was prehybridized for 2 hr at 68°C in a solution containing 4× SSC, 5× Denhardt's solution (0.1% Ficoll, 0.1% polyvinylpyrrolidine, 0.1% BSA), 2.5% SDS, and 100 μg/ml salmon sperm DNA.32P-labeled probes (with a specific activity of at least 106 cpm/ml) were made using random primer DNA synthesis with the NEBlot Kit (New England Biolabs, Beverly, MA) using double-stranded DNA fragments of Notch1 (base pairs 1–889), Notch3 (base pairs 3340–4250), and Delta1 (base pairs 1850–1648). Probes were boiled for 5 min before addition to the hybridization solution and hybridized overnight at 68°C. Unbound probe was removed by washing twice in 2× SSC, 0.1%SDS and then twice in 0.2× SSC, 0.1%SDS for 15 min each wash. Filters were exposed to phosphorimager screens (Molecular Dynamics, Sunnyvale, CA) and scanned by a Molecular Dynamics Phosphorimager. The RNA level of Notch1, Notch3, or Delta1 was quantified using ImageQuant 5.0 (Molecular Dynamics) and by comparison with the total amount of RNA loaded using a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. Filters were stripped of probes for reprobing by boiling in 0.1% SDS.
Treatment with the Delta1 immunoglobulin Fc fusion protein (Delta Fc). The extracellular domain of mouse Delta1, not including the transmembrane domain, was cloned into a derivative of the vector pEF-BOS (Mizushima and Nagata, 1990) containing the Fc region (hinge, CH2, and CH3 domains) of the genomic human IgG1 gene. The resultant plasmid, encoding Delta Fc, was transiently transfected into 293T cells by calcium phosphate transfection. The pEF-BOS vector expressing the Fc region alone was used as a control (control Fc). The transfected cells were lysed in a lysis buffer containing (in mm): 10 HEPES, pH 7.3, 10 NaCl, 1 KH2PO4, 1 CaCl2, 0.5 MgCl2, and 5 NaHCO3, by dounce homogenization on ice. The lysate was centrifuged at 11,000 × g for 10 min to pellet the nuclei and unbroken cells. Production of the Delta Fc protein and the control Fc protein was detected in the supernatant by Western blot analysis with a peroxidase-conjugated polyclonal antibody to human IgG (Dako, Glostrup, Denmark).
E10 NEP cells were cultured in 96-well plates in the media specified for 16 hr before treating with the supernatant obtained from the above cell lysate. The supernatant was added to the cells at a 1:4 dilution in the culture media. The cells were cultured for a further 24 or 48 hr before fixing in 2% paraformaldehyde, staining for microtubule-associated protein 2 (MAP2) and DAPI, and performing cell counts as described below.
Treatment with antisense oligonucleotide. Antisense oligonucleotides were used to downregulate the expression ofNotch1 in the E10 NEP cells. The antisense oligonucleotides were designed against the EGF repeat region of the Notch1gene (Austin et al., 1995) and had an adamentane group attached to the 5′ end via a carbon linker. Nonsense oligonucleotides were used as a control and contained the same ratio of base pairs found in the antisense oligonucleotides but in a random order. The sequence for the antisense oligonucleotide was 5′-GAAGTCATTAACACGCTGCACAC, and for the nonsense oligonucleotide it was 5′-TACATCAGCAGACTACACGAGTC. The oligonucleotides were purified by HPLC and resuspended in a 1% cyclodextrin solution. The adamentane and cyclodextrin enhance the entry of the oligonucleotide into the cell (Epa et al., 2001). Freshly dissected E10 NEP cells were mixed with the oligonucleotides at 5 μm immediately before plating. Cells were plated in 96-well plates and cultured for 48 hr before they were fixed with 2% paraformaldehyde and MAP2-positive cells were counted.
Immunohistochemistry and cell counts. After fixing in 2% paraformaldehyde for 20 min at room temperature, cells were incubated in 100% cold methanol at −20°C for a further 20 min to permeabilize the cells. The cells were washed three times in PBS and blocked by incubating the cells in PBS containing 1% fetal calf serum and 1% normal horse serum for 20 min at room temperature. To stain the MAP2-positive neurons, cells were incubated with a monoclonal anti-MAP2 antibody (Sigma) diluted 1:400 in PBS, 1% FCS for 30 min. After a second wash step, a biotin-conjugated horse anti-mouse Ig secondary antibody (Vector Laboratories, Burlingame, CA) was added to the cells (1:200) for 20 min, followed by a third wash step and incubation with an avidin–horse radish peroxidase complex (Vectorstain Kit, Vector). The MAP2-positive cells were then visualized by the reaction of hydrogen peroxide and 3.3′-diaminobenzidine tetrahydrochloride (DAB) to form a colored product. Cells were washed and stored in PBS containing 0.1% sodium azide. To visualize all cells present in the cultures, the fluorescent DNA binding dye DAPI (Molecular Probes) was added with secondary antibody at a 1:1000 dilution. The cells were then visualized by fluorescence microscopy.
When experiments were performed in 96-well plates, five randomly selected fields of view at 20 magnification were counted. When experiments were performed in Terasaki plates, the entire well was counted. The results were expressed as the number of MAP2-positive neurons produced per 100 DAPI-positive cells counted, with a minimum of 200 cells counted per well. Cell viability was assessed by the nuclear morphology after DAPI staining. In each experiment four wells of each condition were counted, and each experiment was repeated at least three times. All values are expressed as the mean ± SEM.
RESULTS
Delta1 and Notch1, -2, and -3 are expressed in mouse neuroepithelial precursor cells
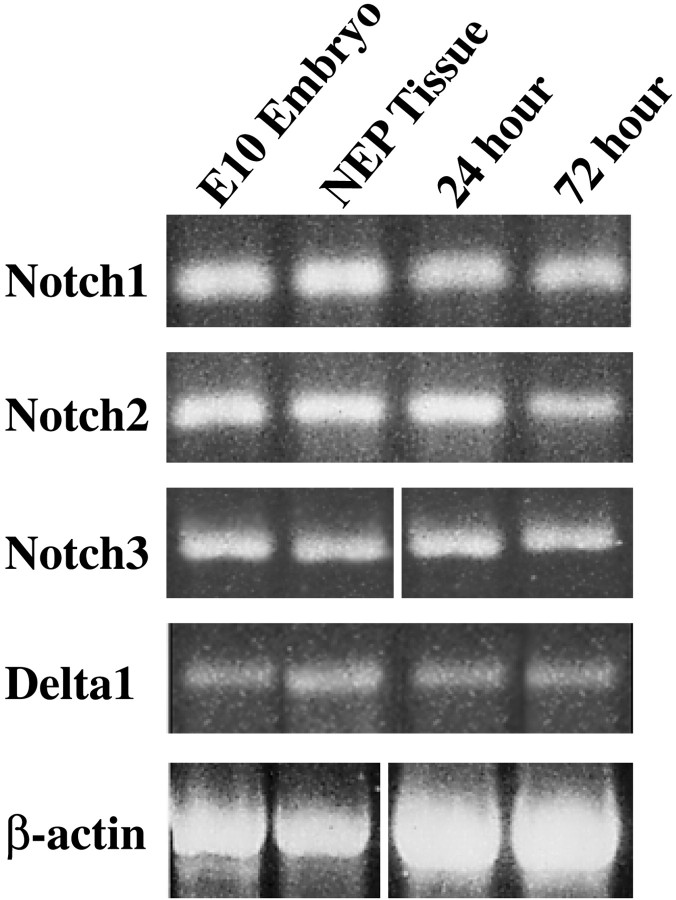
Previous reports indicated that Delta1 expression was first detectable in the mouse CNS at E8 and continued to be expressed in the ventricular zone until approximately E14 (Bettenhausen et al., 1995; Dunwoodie et al., 1997). The various Notch homologs were also reported to be expressed from embryonic day 8 in the mouse nervous system: Notch3 expressed first, followed byNotch2 and Notch1 (Lardelli et al., 1994;Williams et al., 1995; Lindsell et al., 1996; Sestan et al., 1999;Redmond et al., 2000). To determine whether the precursor population from the developing forebrain expressed the various forms of Notch and Delta at the onset of neuronal differentiation, the stage at which they were to be assayed in vitro, RT-PCR analysis was performed on NEP cells isolated from E10 forebrains. mRNA for Notch1,-2, and -3 and Delta1 were all found to be present in the freshly isolated E10 NEP cells and in cells that had been cultured at high cell density in vitro in the presence of serum for 24 and 72 hr (Fig.1).
Fig. 1.
The expression of Notch and Delta in the neuroepithelial precursor cells of the developing mouse nervous system. RT-PCR analysis showing expression of Notch1,Notch2, Notch3, Delta1, and the β-actin control gene in E10 mouse embryos, in E10 NEP cells, and in E10 NEP cells cultured in the presence of serum for 24 and 72 hr.
Delta1 inhibits the differentiation of E10 neuroepithelial precursor cells
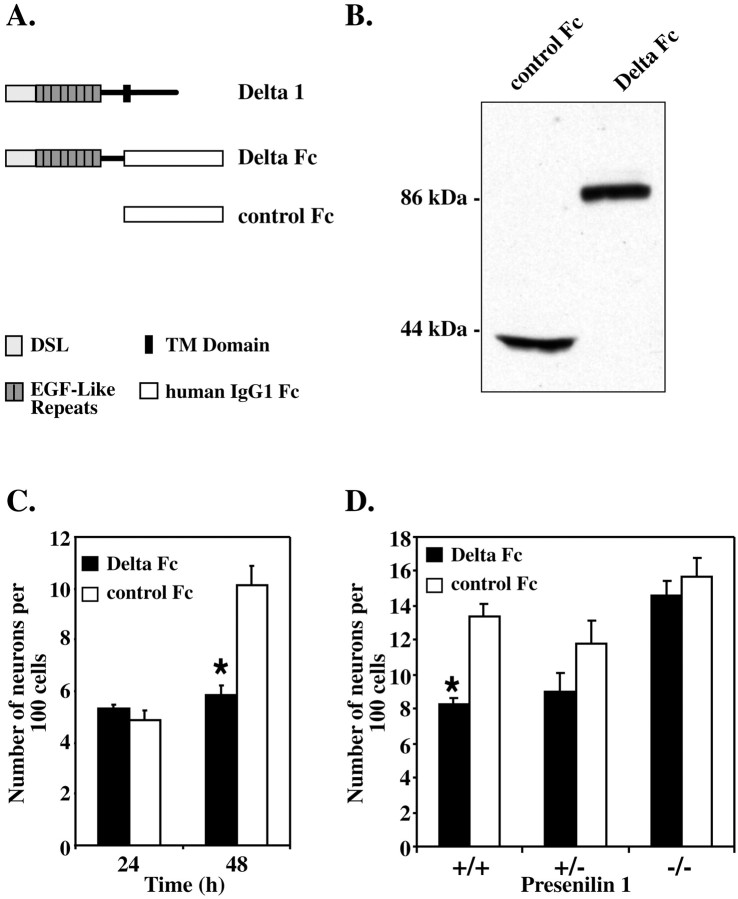
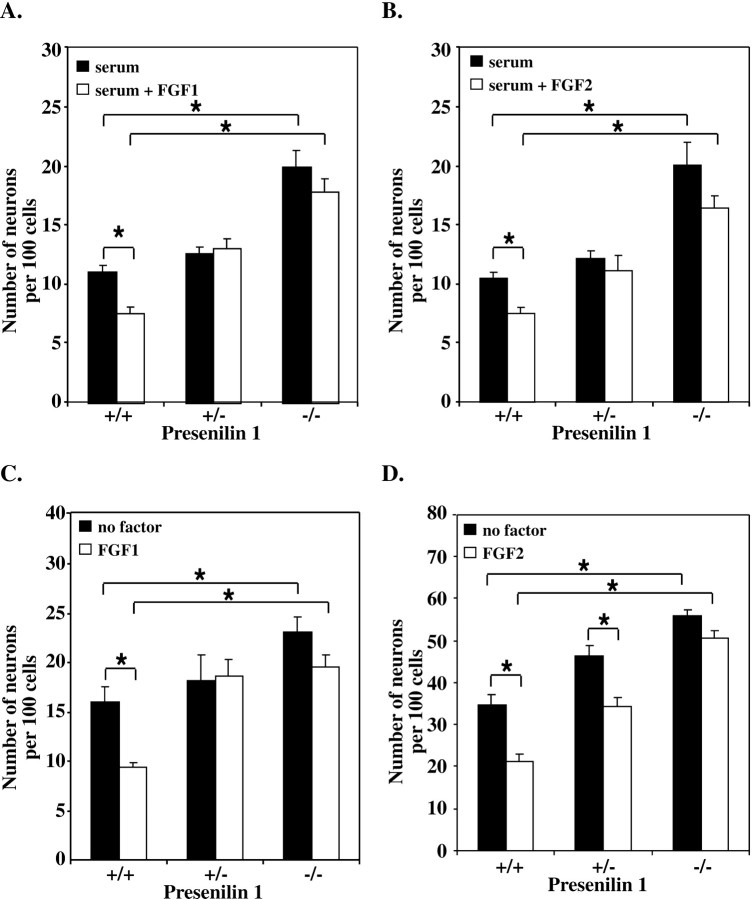
To determine the effect of signaling through Notch on E10 NEP cells in vitro, a fusion protein containing the extracellular domain of Delta1 joined to human IgG1Fc (Delta Fc) was constructed (Fig. 2A). The Delta Fc protein was expressed in transiently transfected 293T cells and detected by Western blot analysis (Fig.2B). The Delta Fc cell lysate, or a lysate from cells transiently transfected with a control IgG1 vector (Fig.2B, control Fc), were added to E10 NEP cells cultured in the absence of serum, and the number of MAP2-positive neurons generated was analyzed. No difference in the number of neurons generated was observed 24 hr after the addition of Delta Fc compared with controls. After 48 hr, however, Delta Fc was able to significantly inhibit neuron differentiation, compared with the controls, to the extent that no new neurons were generated between 24 and 48 hr in the presence of Delta Fc (Fig. 2C). To ensure that the inhibition of differentiation was caused by the Delta Fc protein and not other factors expressed by the 293T cells, the Delta Fc and Fc proteins were purified by size separation chromatography and added to the NEP cells for 48 hr (data not shown). Compared with the neuron number after the addition of Fc cell lysate, the number of neurons produced with the purified Fc protein (21.5 ± 0.2%) and Delta Fc protein (14.5 ± 0.75%) was increased. However, the purified Delta Fc protein inhibited neuron differentiation to the same degree as that observed with the Delta Fc cell lysate. To determine whether Delta Fc was signaling via Notch, Delta Fc was added to cells from Presenilin1 knock-out animals. Presenilin1 is required for the cleavage of the intracellular domain of Notch after ligand binding (Song et al., 1999; Struhl and Greenwald, 1999) and therefore Notch signaling is disrupted in the Presenilin1-deficient cells. The Delta Fc cell lysate was added to NEP cells from wild-type animals (+/+) and animals with a heterozygous (+/−) or homozygous (−/−) deletion inPresenilin1, and the production of neurons after 48 hr in culture was assessed (Fig. 2D). The inhibition of neuronal differentiation observed in the wild-type cells in response to Delta Fc was consistent with that seen in Figure 2C after 48 hr. In contrast, Delta Fc was unable to inhibit the differentiation of NEP from Presenilin1 knock-out animals (Fig. 2D). In addition, although a low level of inhibition was observed by Delta Fc in the Presenilin1 heterozygous cells, this inhibition was not significantly different from the control. These data suggest that Delta Fc signals via Notch to inhibit neuronal differentiation in the neuroepithelial precursor cell population.
Fig. 2.
The extracellular domain of Delta1 inhibits the differentiation of E10 neuroepithelial precursor cells but is unable to inhibit the differentiation of NEP cells with a mutation inPresenilin1. A, Schematic diagram of full-length Delta1, the Delta Fc fusion protein, showing the extracellular domain of Delta1 fused to the human IgG1 Fc, and the control human Fc proteins. The various sequence motifs are defined byshaded boxes and depicted at the bottomof the figure. DSL, Delta/Serrate/Lag domain; TM Domain, transmembrane domain. B, Western blot analysis showing expression of the Delta Fc protein and the control Fc in the cell lysate of transiently transfected 293T cells. The protein was detected using an antibody to human IgG1. C, Assessment of the number of MAP2-positive neurons produced by E10 NEP cells in vitro, after the addition of Delta Fc or control Fc cell lysates. Cells were cultured in the absence of serum in 96-well plates. No difference in neuron production was observed after 24 hr, whereas after 48 hr Delta Fc significantly inhibited neuron differentiation (*p < 0.05; mean ± SEM).D, Delta Fc or control Fc cell lysate was added to wild-type NEP cells (+/+) or NEP cells containing a heterozygous (+/−) or homozygous (−/−) deletion in the Presenilin1 gene. Cells were cultured in the absence of serum in 96-well plates for 48 hr (*p < 0.05; mean ± SEM).
Growth factors can influence the expression of Notch and Delta by E10 NEP cells in vitro
Little is known about the upstream factors that may regulate the Notch–Delta inhibitory pathway. It is possible that various growth factors act to inhibit neurogenesis, at least in part, by regulating the level of Delta or Notch expression. To examine this question more closely, we initially sought to determine which growth factors influenced the neuronal differentiation of the E10 NEP cells. Northern blot analysis then was used to quantify changes in the expression of Notch and Delta induced by these factors.
It was found that the presence of serum in the medium significantly inhibited the differentiation of precursor cells into neurons (Fig.3A) and increased the overall number of cells present in the culture (Fig. 3B). Because of the response observed with serum, we examined the effect of various growth factors on cells cultured in either the presence or absence of serum. Each dose of growth factor that was added to the cells ensured that the cells were maximally stimulated by that growth factor. Members of the fibroblast growth factor family, FGF1, FGF2, and FGF8, were all found to inhibit the neuronal differentiation of the E10 NEP cells and caused a significant increase in cell number in both the presence and absence of FCS (Fig. 3A,B). LIF caused a slight increase, although not significant, in cell number and inhibited neuronal differentiation. However, both of these effects were observed only when serum was present in the cultures, suggesting that certain factors present in FCS are able to alter the response of the cells to LIF. In contrast, OSM, another member of the neurocytokine family, caused a decrease in cell number but had no effect on neuron differentiation. A decrease in cell number occurred when members of the TGFβ superfamily, BMP2, BMP4, TGFβ3, and TGFβ5 were added to the cultures alone. However, with serum present, only TGFβ5 affected cell number. TGFβ5 also caused a slight decrease in the percentage of neurons produced, whereas BMP2, BMP4, and TGFβ3 did not alter neurogenesis. NT-3 decreased the number of cells present in the cultures, whereas BDNF had no effect on either cell number or differentiation (Fig. 3).
Fig. 3.
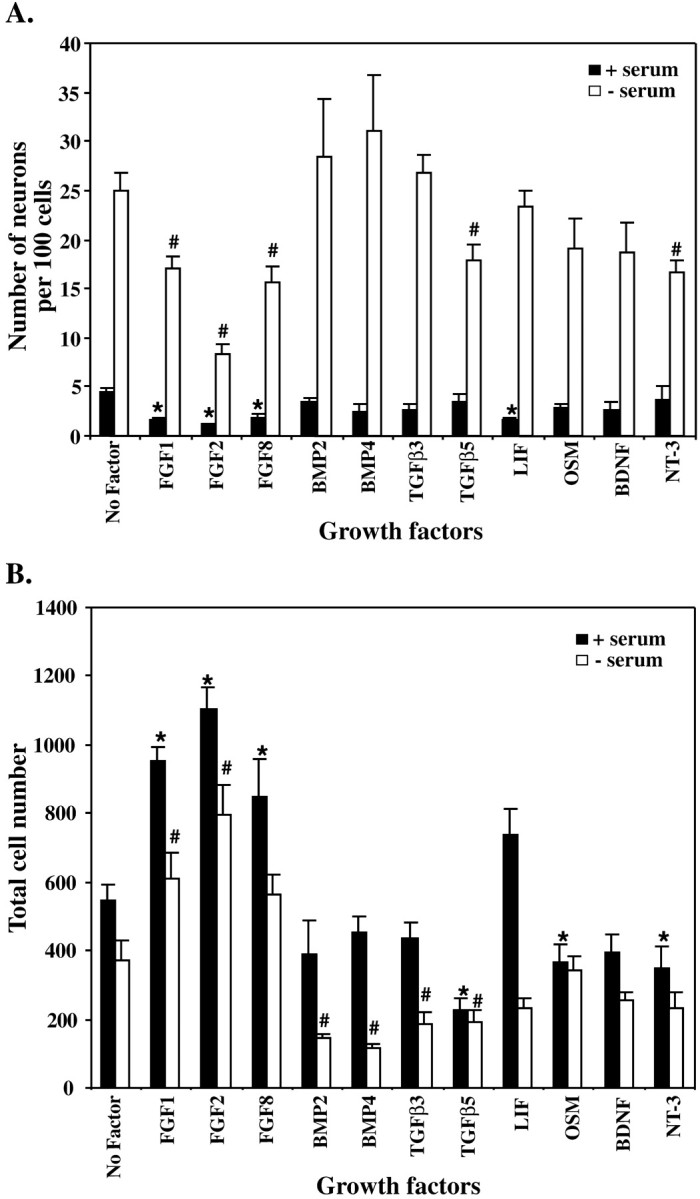
Serum and growth factors affect the neuronal differentiation (A) and overall number of cells (B) generated by E10 NEP cells in vitro. E10 NEP cells were cultured in Terasaki plates for 48 hr in the presence or absence of serum with growth factors.A, The effect on neurogenesis was analyzed by examining the number of MAP2-positive neurons produced per 100 cells.B, The total number of DAPI-positive cells per well. The production of neurons and the number of cells present in cultures without growth factor were compared with those with growth factor added (* denotes significant differences when serum is present; # denotes significant differences when serum is absent; p < 0.05; mean ± SEM). FGF, Fibroblast growth factor;BMP, bone morphogenetic protein; TGFβ, transforming growth factor β; LIF, leukemia inhibitory factor; OSM, oncostatin M; BDNF, brain-derived neurotrophic factor; NT-3, neurotrophin 3.
Overall, none of the growth factors examined caused a significant increase in the percentage of neurons generated in culture, but several, including serum, inhibited the generation of neurons. To determine whether the inhibitory response observed with several of the growth factors was associated with a change in the expression levels ofNotch and Delta, Northern blot analysis was undertaken.
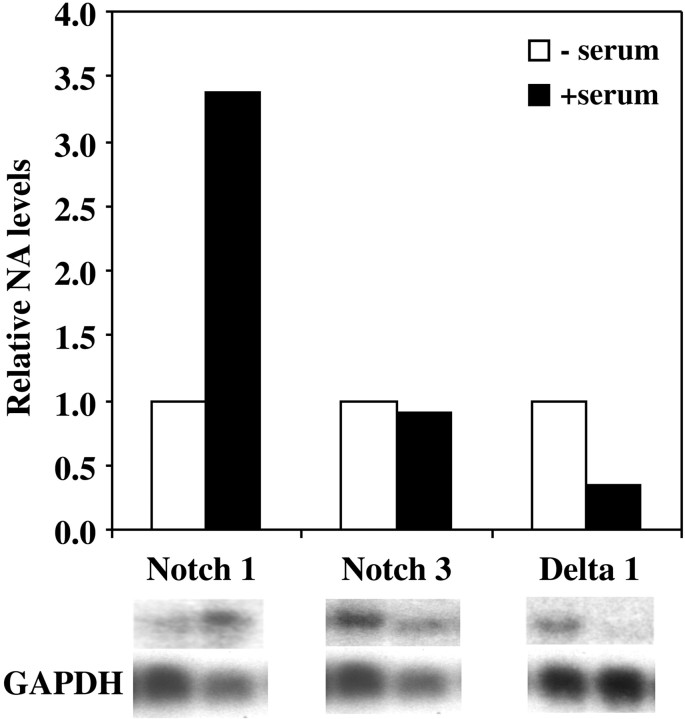
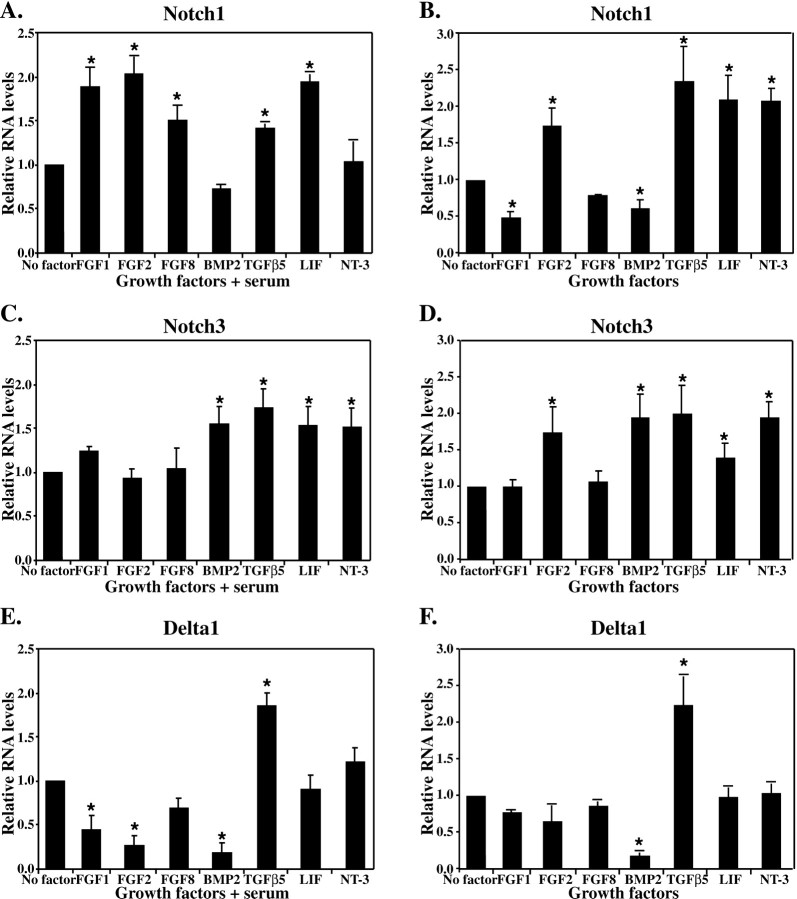
Serum increased the expression of Notch1 and decreased expression of Delta1 in cultured E10 NEP cells but had no effect on Notch3 expression (Fig.4). The increase in Notch1expression observed with serum was further enhanced by the presence of FGF1, FGF2, FGF8, TGFβ5, and LIF (Fig.5A). Interestingly, in the absence of serum, FGF1 significantly decreased Notch1expression (Fig. 5B). BMP2 also decreased Notch1 expression in the absence of serum, whereas it increased the expression ofNotch3 (Fig. 5C,D). Notch3expression was also significantly increased in the absence of serum by TGFβ5, LIF, NT-3, and FGF2 (Fig. 5C,D). In contrast, with serum present FGF2 did not increase Notch3expression (Fig. 5C). Expression of Delta1 was increased only by TGFβ-5, in both the presence and absence of serum (Fig. 5E,F). FGF2 and BMP2 decreased expression of Delta1 in the absence of serum. This decrease was found to be even greater when serum was present and was also observed with the addition of FGF1 and serum. None of the other growth factors examined altered Delta1 expression.
Fig. 4.
Serum alters the expression ofNotch1 and Delta1 but does not affectNotch3 expression in E10 neuroepithelial cells. Cells were cultured in the presence or absence of serum for 24 hr before the RNA was extracted. Northern blot analysis was used to examine gene expression, and the relative level of mRNA expressed was quantified by phosphorimager analysis and comparison with GAPDH expression. Shown is a representative of three independent experiments.
Fig. 5.
Growth factors influence the expression ofNotch1 (A, B),Notch3 (C, D), andDelta1 (E, F). E10 neuroepithelial cells were cultured with growth factors in either the presence (A, C, E) or absence (B, D, F) of serum for 24 hr. Northern blot analysis was used to examine gene expression, and the relative level of mRNA was determined by phosphorimager and comparison with GAPDH expression (*p < 0.05; mean ± SEM).
FGF1 and FGF2 inhibit neurogenesis via the Notch–Delta pathway
To determine whether the upregulation of Notchexpression with FGF1, FGF2, or serum was related to the low neuronal number, an antisense oligonucleotide, designed to downregulate Notch1 expression, was used in the assay. The antisense oligonucleotide, designed against the EGF repeat coding region of the mouseNotch1 gene, has previously been shown to reduce Notch1 expression in neural cells in vitro (Austin et al., 1995). To confirm the specificity and efficacy of the oligonucleotide for Notch1, the antisense oligonucleotide was shown to effectively downregulate Notch1 expression in 293T cells transiently transfected with a Notch1 plasmid, when compared with nonsense controls (Fig.6A). Addition of the antisense oligonucleotide to NEP cells grown in serum alone resulted in a significant increase in the production of MAP2-positive neurons, compared with the nonsense control cultures. FGF1 still exerted some inhibitory effects in the presence of antisense oligonucleotide, in that it did not completely restore the number of neurons generated in the no-factor control. However, when compared with the FGF1 nonsense control, a significant increase in neurons was observed (Fig.6B) with antisense. In contrast, the antisense oligonucleotides were unable to alter neuron number when cells were cultured in 50 ng/ml FGF2 (Fig. 6B). However, when the FGF-2 concentration was lowered to 10 or 5 ng/ml, a significant increase in neuron production was observed with the antisense oligonucleotide compared with nonsense controls (Fig.6C).
Fig. 6.
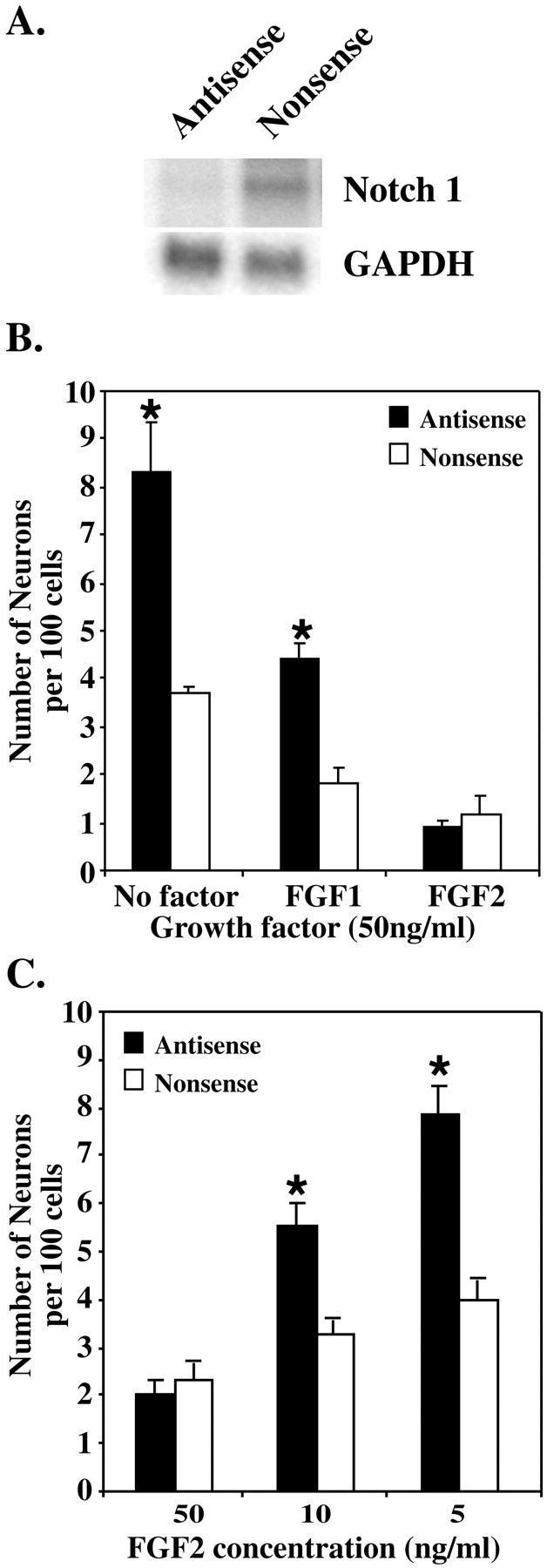
The production of neurons by E10 NEP cells is increased after the downregulation of Notch1 expression by antisense oligonucleotides. A, Northern blot analysis was used to assess the downregulation of Notch1 expression in 293T cells transiently transfected with a Notch1 plasmid. Antisense or nonsense oligonucleotides (5 μm) were added to the cells 24 hr after transfection, and RNA was extracted 48 hr later. B, E10 NEP cells were cultured with 5 μm antisense or nonsense oligonucleotide in the presence of serum alone (No factor) or with serum plus FGF1 or FGF2 (50 ng/ml) for 48 hr. An increase in neuron production was observed after antisense treatment in serum and serum plus FGF1, but not in FGF2. C, E10 NEP cells were cultured with 5 μm antisense or nonsense oligonucleotide in serum plus FGF2 at varying concentrations (50, 10, and 5 ng/ml). An increase in neuron production was observed after antisense treatment at lower concentrations of FGF2 (*p < 0.05; mean ± SEM).
To further establish the role of Notch during FGF signaling, E10 NEP cells from animals with a deletion in the Presenilin1 gene were examined after stimulation with either FGF1 or FGF2. Cells from wild-type animals (+/+) or animals heterozygous (+/−) or homozygous (−/−) for the Presenilin1 deleted allele, were cultured in either the presence or absence of serum and with 50 ng/ml FGF2 or FGF1 for 24 hr (Fig. 7). Both FGF2 and FGF1 significantly inhibited the neuron differentiation of the wild-type (+/+) cells. In contrast, however, FGF2 and FGF1 were unable to inhibit the differentiation of cells from Presenilin1 homozygous knock-out animals, in either the presence (Fig.7A,B) or absence (Fig.7C,D) of serum. In addition, the neuronal differentiation of the Presenilin1 heterozygous cells was not inhibited by FGF1 (Fig. 7A,C) or FGF2 when serum was present (Fig. 7A). However, in the absence of serum, FGF2 continued to inhibit the differentiation of the heterozygous cells (Fig. 7D). Interestingly, compared with the wild-type controls, a significant increase in the production of MAP2-positive neurons was also observed by the Presenilin1 homozygous knock-out cells. An increase in neuron production by the Presenilin1 homozygous knock-out cells also occurred in the presence of FGF1 and FGF2. These results suggest that FGF1 and FGF2 require Notch signaling to inhibit the neuronal differentiation of the E10 NEP cells.
Fig. 7.
FGF2 and FGF1 do not inhibit the neuronal differentiation of NEP cells from Presenilin1 knock-out animals.A, E10 NEP cells from wild-type animals (+/+) or from animals with a heterozygous (+/−) or homozygous (−/−) deletion in Presenilin1 were cultured for 24 hr in the presence of serum and serum plus FGF1 (50 ng/ml). B, Cells were cultured in serum and serum plus FGF2. C, Cells were cultured in serum-free media alone and with FGF1. D, Cells were cultured in serum-free media alone and with FGF2 (*p < 0.05; mean ± SEM).
Delta1 inhibits neuron differentiation in the presence of FGF1, but not in the presence of FGF2
In the absence of serum, there is significant neuronal production in the presence of inhibitory growth factors, especially FGF1 (Fig. 3A). Because these conditions are associated with lowered levels of Notch expression and therefore lowered Notch signaling (Fig. 5B), we examined whether this component of neuronal production could be inhibited if Notch signaling was further activated by adding exogenous Delta Fc to these cultures. Similar to the results obtained in serum-free cultures without any growth factors added (Fig. 2C), the addition of Delta Fc to NEP cells stimulated with FGF1 had no effect on neuron production within the first 24 hr but showed significantly lower numbers of neurons compared with controls at 48 hr (Fig.8A). In contrast, Delta Fc was unable to alter the inhibitory activity of FGF2 at 24 or 48 hr (Fig. 8A). In the presence of FCS, no significant effect of the exogenous Delta on neuron production was observed under any condition (Fig. 8B).
Fig. 8.
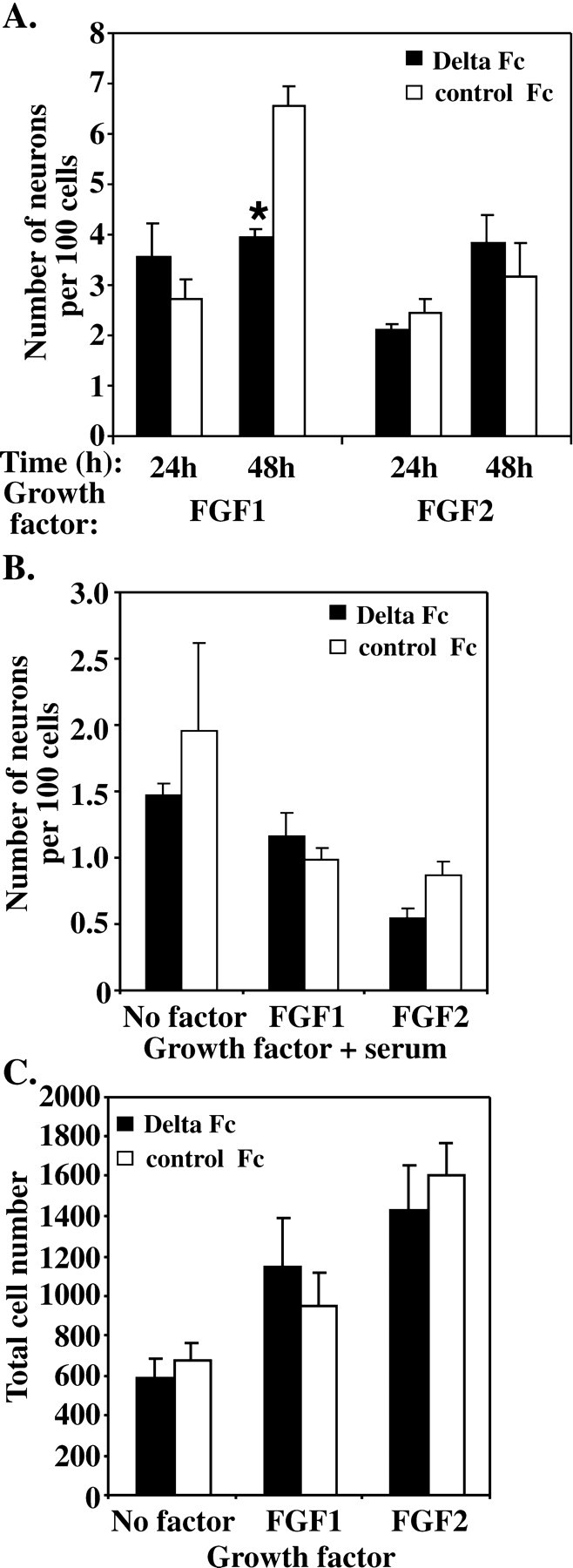
Delta Fc inhibits the differentiation of E10 NEP cells cultured in the presence of FGF1, but not in the presence of FGF2 or serum. A, E10 NEP cells were cultured in serum-free media (No factor) and with either FGF1 (50 ng/ml) or FGF2 (50 ng/ml) for 24 and 48 hr after the addition of the Delta Fc or control Fc cell lysate. No difference in neuron production was observed after 24 hr in culture, but after 48 hr Delta Fc decreased the number of neurons produced in the presence of FGF1, but not FGF2 (*p < 0.05; mean ± SEM) B, E10 NEP cells were cultured in the presence of serum alone (No factor) or serum plus FGF1 or FGF2 (50 ng/ml) for 48 hr after the addition of Delta Fc or control Fc cell lysate. No difference in the number of neurons produced was observed. C, The overall number of cells cultured in the absence of serum (No factor) or with FGF1 or FGF2 was unaltered by the Delta Fc cell lysate. Note the increase in cell number with both FGF1 and FGF2.
To examine whether the inhibitory effect of exogenous Delta on FGF1-stimulated neuronal production was associated with changes to overall growth of the precursor population, the total number of cells present in the culture was examined. No difference in the number of cells was observed between Delta Fc-treated cultures and controls at 48 hr (Fig. 8C), when the cells were cultured either in serum-free media without growth factors or in the presence of FGF1 or FGF2 (50 ng/ml).
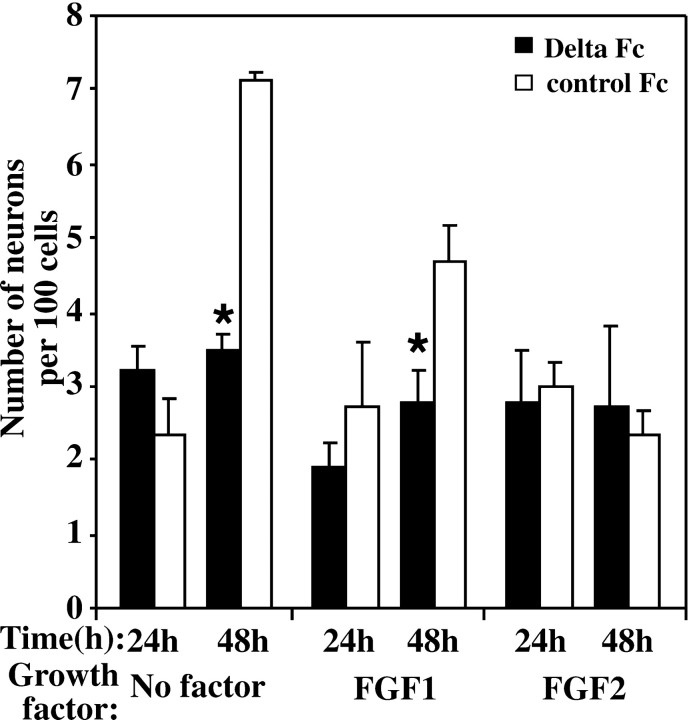
The effect of exogenous Delta was next examined under conditions in which precursor cells were first prevented from differentiating by culturing them in FGF-2 and serum for 24 hr, then allowing neuronal differentiation to occur by removing the growth factor and serum (Fig.9). Significant inhibition of the number of neurons generated was observed 48 hr after serum and growth factor removal compared with controls (Fig. 9). Similarly, if cells were placed in FGF1 after the removal of FGF2 and serum, exogenous Delta was able to significantly decrease the number of neurons generated at 48 hr. If placed back into FGF-2, however, there was no effect of Delta Fc on neuronal production, which remained at very low levels at 48 hr.
Fig. 9.
Delta Fc inhibition of neuron differentiation after removal of serum and FGF2. E10 neuroepithelial cells were cultured for 24 hr in the presence of serum and FGF2 (50 ng/ml). The serum and growth factors were then removed to allow the cells to differentiate, and the Delta Fc or control Fc cell lysate was added. No difference in the number of MAP2-positive neurons produced was observed 24 hr after the addition of Delta Fc compared with controls. Neuron production was decreased 48 hr after replacing the media with serum-free media or FGF1 (50 ng/ml) and adding Delta Fc, but not when the media was replaced with FGF2 and Delta Fc (*p< 0.05; mean ± SEM).
DISCUSSION
The present study shows that at high cell density, which allows for the intimate cell interactions that occur in vivo, the differentiation of neural precursors into neurons is largely regulated by inhibitory signals. Two ligand-activated signaling systems significantly influenced the neuronal differentiation of neuroepithelial precursors: the FGF receptor pathway and the Notch receptor pathway. Furthermore, the evidence suggests that these two systems interact, and the inhibitory action of growth factors, such as FGF1 and 2, on the precursor cells may be mediated via the Notch pathway.
Notch was shown to be directly involved in regulating the step from precursor to neuron, because addition of Delta1 almost totally abrogated the generation of neurons from E10 NEP cells during the 24–48 hr culture period. The action of Notch was confirmed by the increase in neuron production observed when Notch signaling was inhibited, either by downregulating Notch levels with antisense or when examining Presenilin1-deficient NEP cells. Presenilin1 mediates the intracellular cleavage of Notch, and without cleavage Notch signaling is markedly reduced (De Strooper et al., 1999; Song et al., 1999;Struhl and Greenwald 1999). The requirement for Presenilin1 in Notch cleavage remains controversial, given that Presenilin2 may partially compensate for the loss of Presenilin1 (Berechid et al., 1999; Steiner et al., 1999). However, in the developing mouse brain, Presenilin1 is expressed at significantly higher levels than Presenilin2 (Lee et al., 1996), indicating that in NEP cells, Presenilin1 is of primary importance. The inability of Delta Fc to inhibit the differentiation of the Presenilin1 null NEP cells indicates that Notch signaling is disrupted. Furthermore, the increase in neuron production observed by the Presenilin1 null NEP cells suggests that Presenilin1 is required to inhibit neuron differentiation. This is consistent with the premature differentiation of progenitor cells observed by Presenilin1 knock-out mice in vivo (Handler et al., 2000). A role for Notch signaling in the regulation of NEP cell differentiation has been suggested previously, after the examination of animals with a deletion of HES1, a bHLH factor that operates downstream of Notch activation (Jarriault et al., 1995), where an increase in neuron number was observed (Nakamura et al., 2000). Precursor cell differentiation inXenopus and in chick retina is also regulated by Notch (Austin et al., 1995; Chitnis et al., 1995; Dorsky et al., 1997), because downregulation of Notch expression or activity increased neuron number, whereas the overexpression of Delta inhibited neuron differentiation.
During early CNS development, the inhibition of neuron differentiation after Notch signaling potentially allows the precursor population to be maintained. This is consistent with findings that the overexpression of Notch3 during CNS development results in an expansion of the progenitor pool (Lardelli et al., 1996). However, in addition to decreasing neuron number, Notch activation in the developing forebrain in vivohas been shown to promote the differentiation of radial glial cells (Gaiano et al., 2000). Thus, Notch signaling may actively direct the differentiation of precursor cells into alternative cell types. Further experimentation with the NEP cells in vitro may clarify the role of Notch signaling in promoting precursor differentiation.
The identity of the specific Notch receptor activated by the Delta1 Fc protein is uncertain because the NEP cells expressed three of the Notch receptors. The preference of individual ligands for receptors remains unclear, although Jagged1 may preferentially activate Notch2 (Shimizu et al., 1999), whereas Delta1 appears to preferentially activate Notch1 (Weinmaster 1998). The present studies also suggest that Notch1 is of primary importance, because the downregulation of Notch1 with antisense was not compensated by the presence of Notch2 and -3. Furthermore, in situ hybridization analysis has shown that Notch1 is the primary Notch receptor expressed by forebrain NEP cells, whereas here Notch2 and -3 are expressed only weakly (Lardelli et al., 1994; Williams et al., 1995).
The first indication that growth factors may regulate precursor differentiation through regulating Notch activity was the finding that different growth factors altered Notch and Delta expression. With the exception of FGF8, each growth factor that was examined increased the expression of one or two of the Notch homologs, whereasDelta1 expression was decreased in response to FGF1, FGF2, and BMP2 and increased by TGFβ5. Previously, indirect associations between Notch signaling and growth factors have been made by examining the bHLH transcription factors that are involved in Notch signaling. NGF inhibited the DNA binding ability of HES1, thereby allowing the differentiation of PC12 cells (Strom et al., 1997), whereas BMP2, BMP4, and EGF decreased expression of MASH 1 (Ahmad et al., 1998; Shou et al., 1999), which has been implicated in the regulation of Delta expression and is essential for neurogenesis (Chitnis and Kintner, 1996; Casarosa et al., 1999).
The finding that FGF1 and FGF2 stimulated precursor cell proliferation and inhibited neuron differentiation was consistent with previous reports from in vitro culture (Murphy et al., 1990;Kilpatrick and Bartlett, 1993; Ghosh and Greenberg, 1995; Vaccarino et al., 1999) and with the role of FGF2 in vivo as the primary stimulator of precursor cell division in the developing forebrain (Vaccarino et al., 1999). Although an increase in the absolute number of neurons was observed after FGF2 infusion in vivo(Vaccarino et al., 1999), this response was interpreted as being caused by increased number of cell cycles of the precursors, which ultimately generated more neurons. Thus, it appears that precursor proliferation and inhibition of differentiation are inextricably mixed, and this study cannot choose between these as the primary physiological effects of the FGFs. Nevertheless, it appears that Notch signaling is involved in maintaining this state. FGF1 and FGF2 significantly upregulatedNotch1 expression in the NEP cells. Furthermore,Delta1 expression was downregulated by these factors, which is consistent with enhanced Notch signaling (Kunisch et al., 1994;Heitzler et al., 1996). It remains uncertain how Notch signaling is maintained after the downregulation of Delta expression. Other Notch ligands, however, are also expressed at this stage of development (Dunwoodie et al., 1997). In addition, it has been suggested that Delta is cleaved at the cell surface releasing a soluble form of Delta, which may continue to activate Notch (Qi et al., 1999). The link between Notch expression levels and FGF-induced neuronal inhibition was shown by the increased neuronal numbers when Notch signaling was reduced, either by inhibiting the Presenilin1-mediated cleavage of Notch or by downregulating Notch1 expression. These data indicate that the inhibition of differentiation observed in response to FGF1 and FGF2 is mediated, at least in part, via the Notch signaling pathway.
The difference between the biological functions of FGF1 and FGF2 has been difficult to determine. These factors may have distinct roles given that FGF2 is expressed at E9, whereas FGF1 is expressed at E11(Nurcombe et al., 1993). Although the effect of FGF1 and FGF2 on the NEP cells was similar, FGF2 was a more potent inhibitor of neuronal differentiation, and unlike FGF1, Delta Fc was unable to enhance this FGF2 inhibitory response, indicating that FGF2 may stimulate the Notch signaling pathway to a greater extent than FGF1. The differences between FGF1 and FGF2 may be explained by their differential activation of one of the FGF receptors expressed on the NEP cells (Kalyani et al., 1999). A possible mechanism of differential FGF receptor activation involves the binding of heparan sulfate proteoglycans (HSPGs), which regulate ligand-receptor binding and differ in their specificity for the FGFs. At E10 the HSPGs that are produced preferentially bind FGF2, whereas at E12 the binding affinity switches to FGF1 (Nurcombe et al., 1993). Therefore, in the NEP population, FGF2 may be binding with higher affinity, causing the complete activation of the Notch pathway, whereas without the specific HSPGs, FGF1 may not be able to elicit its full effect and therefore Notch signaling is limited.
The inhibitory effect of serum on neuronal differentiation may also be caused by Notch signaling. Serum upregulated Notch1 levels and downregulated Delta1 levels in the NEP cells. In addition, the inhibition observed in response to serum was overcome by downregulating Notch expression and activity (Figs. 6, 7). Interestingly, although serum increased Notch1 expression, Notch3 expression was unaltered. In contrast, BMP2 caused the opposite effect by increasing the expression of Notch3 but not Notch1. Factors such as BMP2 or those present in serum therefore may elicit their effects via specific Notch receptors.
Our data suggest that the primary function of signaling through the Notch receptor is the inhibition of precursor cell differentiation. The growth factors FGF1 and FGF2 appear to act via the Notch pathway to inhibit neuron differentiation. Given the ability of other growth factors to regulate Notch and Delta expression, it is likely that many growth factors regulate neuronal differentiation in a similar manner. Other cellular responses to growth factors, such as proliferation, differentiation, and survival, are probably mediated via separate and distinct pathways. To elucidate this, however, a more detailed analysis of the relationships between the growth factor signaling pathways and Notch signaling is required.
Footnotes
This work was supported by The National Health and Medical Research Council of Australia, The Australasian Spinal Research Trust, and the Co-operative Research Center for Cellular Growth Factors. We thank Dr. Hui Zeng for providing the Presenilin1 knock-out mice and Leanne Stewart and Rachael Borg for assistance with the Presenilin1 knock-out mice. We also thank Dr. Jeff Nye for the gift of the Notch1 expressing plasmid.
Correspondence should be addressed to Dr. Perry F. Bartlett, Development and Neurobiology Group, The Walter and Eliza Hall Institute of Medical Research, Post Office, The Royal Melbourne Hospital, Victoria 3050, Australia. E-mail:bartlett@wehi.edu.au.
REFERENCES
- 1.Ahmad I, Dooley CM, Afiat S. Involvement of Mash1 in EGF-mediated regulation of differentiation in the vertebrate retina. Dev Biol. 1998;194:86–98. doi: 10.1006/dbio.1997.8809. [DOI] [PubMed] [Google Scholar]
- 2.Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- 3.Berechid BE, Thinakaran G, Wong PC, Sisodia SS, Nye JS. Lack of requirement for presenilin1 in Notch1 signaling. Curr Biol. 1999;9:1493–1496. doi: 10.1016/s0960-9822(00)80121-9. [DOI] [PubMed] [Google Scholar]
- 4.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet JL, Gossler A. Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila Delta. Development. 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- 5.Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 6.Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- 7.Chitnis A, Kintner C. Sensitivity of proneural genes to lateral inhibition affects the pattern of primary neurons in Xenopus embryos. Development. 1996;122:2295–2301. doi: 10.1242/dev.122.7.2295. [DOI] [PubMed] [Google Scholar]
- 8.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 10.Dorsky RI, Rapaport DH, Harris WA. Xotch inhibits cell differentiation in the Xenopus retina. Neuron. 1995;14:487–496. doi: 10.1016/0896-6273(95)90305-4. [DOI] [PubMed] [Google Scholar]
- 11.Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signaling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- 12.Drago J, Murphy M, Carroll SM, Harvey RP, Bartlett PF. Fibroblast growth factor-mediated proliferation of central nervous system precursors depends on endogenous production of insulin-like growth factor I. Proc Natl Acad Sci USA. 1991;88:2199–2203. doi: 10.1073/pnas.88.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- 14.Epa WR, Barrett GL, Bartlett PF. Oligonucleotides as inhibitors of protein synthesis. Methods Mol Biol. 2001;169:223–242. doi: 10.1385/1-59259-060-8:223. [DOI] [PubMed] [Google Scholar]
- 15.Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- 16.Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, Guillemot F. The bHLH protein Neurogenin 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron. 1998;20:483–494. doi: 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- 17.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh A, Greenberg ME. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 19.Gritti A, Parati EA, Cova L, Frolichsthal P, Galli R, Wanke E, Faravelli L, Morassutti DJ, Roisen F, Nickel DD, Vescovi AL. Multipotential stem cells from the adult mouse brain proliferate and self-renew in response to basic fibroblast growth factor. J Neurosci. 1996;16:1091–1100. doi: 10.1523/JNEUROSCI.16-03-01091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Handler M, Yang X, Shen J. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development. 2000;127:2593–2606. doi: 10.1242/dev.127.12.2593. [DOI] [PubMed] [Google Scholar]
- 21.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signaling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 22.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signaling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 23.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Israel A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyani AJ, Mujtaba T, Rao MS. Expression of EGF receptor and FGF receptor isoforms during neuroepithelial stem cell differentiation. J Neurobiol. 1999;38:207–224. [PubMed] [Google Scholar]
- 25.Kilpatrick TJ, Bartlett PF. Cloning and growth of multipotential neural precursors: requirements for proliferation and differentiation. Neuron. 1993;10:255–265. doi: 10.1016/0896-6273(93)90316-j. [DOI] [PubMed] [Google Scholar]
- 26.Koblar SA, Turnley AM, Classon BJ, Reid KL, Ware CB, Cheema SS, Murphy M, Bartlett PF. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc Natl Acad Sci USA. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunisch M, Haenlin M, Campos-Ortega JA. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc Natl Acad Sci USA. 1994;91:10139–10143. doi: 10.1073/pnas.91.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 29.Lardelli M, Williams R, Mitsiadis T, Lendahl U. Expression of the Notch 3 intracellular domain in mouse central nervous system progenitor cells is lethal and leads to disturbed neural tube development. Mech Dev. 1996;59:177–190. doi: 10.1016/0925-4773(96)00589-8. [DOI] [PubMed] [Google Scholar]
- 30.Lee MK, Slunt HH, Martin LJ, Thinakaran G, Kim G, Gandy SE, Seeger M, Koo E, Price DL, Sisodia SS. Expression of presenilin 1 and 2 (PS1 and PS2) in human and murine tissues. J Neurosci. 1996;16:7513–7525. doi: 10.1523/JNEUROSCI.16-23-07513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 32.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 33.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy M, Drago J, Bartlett PF. Fibroblast growth factor stimulates the proliferation and differentiation of neural precursor cells in vitro. J Neurosci Res. 1990;25:463–475. doi: 10.1002/jnr.490250404. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nurcombe V, Ford MD, Wildschut JA, Bartlett PF. Developmental regulation of neural response to FGF-1 and FGF-2 by heparan sulfate proteoglycan. Science. 1993;260:103–106. doi: 10.1126/science.7682010. [DOI] [PubMed] [Google Scholar]
- 38.Parks AL, Huppert SS, Muskavitch MA. The dynamics of neurogenic signaling underlying bristle development in Drosophila melanogaster. Mech Dev. 1997;63:61–74. doi: 10.1016/s0925-4773(97)00675-8. [DOI] [PubMed] [Google Scholar]
- 39.Qi H, Rand M, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the Notch ligand Delta by the metalloprotease kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 40.Qian S, Jiang P, Guan XM, Singh G, Trumbauer ME, Yu H, Chen HY, Van de Ploeg LH, Zheng H. Mutant human presenilin 1 protects presenilin 1 null mouse against embryonic lethality and elevates Abeta1–42/43 expression. Neuron. 1998;20:611–617. doi: 10.1016/s0896-6273(00)80999-x. [DOI] [PubMed] [Google Scholar]
- 41.Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 43.Schroeter EH, Kisslinger JA, Kopan R. Notch 1 signaling requires ligand-involved proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 44.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by Notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse Jagged1 physically interacts with Notch2 and other Notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 46.Shou J, Rim PC, Calof AL. BMPs inhibit neurogenesis by a mechanism involving degradation of a transcription factor. Nat Neurosci. 1999;2:339–345. doi: 10.1038/7251. [DOI] [PubMed] [Google Scholar]
- 47.Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner BA. Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci USA. 1999;96:6959–6963. doi: 10.1073/pnas.96.12.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiner H, Duff K, Capell A, Romig H, Grim MG, Lincoln S, Hardy J, Yu X, Picciano M, Fechteler K, Citron M, Kopan R, Pesold B, Keck S, Baader M, Tomita T, Iwatsubo T, Baumeister R, Haass C. A loss of function mutation of presenilin-2 interferes with amyloid beta-peptide production and notch signaling. J Biol Chem. 1999;274:28669–28673. doi: 10.1074/jbc.274.40.28669. [DOI] [PubMed] [Google Scholar]
- 49.Strom A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11:3168–3181. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 52.Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, Hung YT. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat Neurosci. 1999;2:246–253. doi: 10.1038/6350. [DOI] [PubMed] [Google Scholar]
- 53.Weinmaster G. Notch signaling: direct or what? Curr Opin Genet Dev. 1998;8:436–442. doi: 10.1016/s0959-437x(98)80115-9. [DOI] [PubMed] [Google Scholar]
- 54.Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- 55.Wong PC, Zheng H, Chen H, Becher MW, Sirinathsinghji DJ, Trumbauer ME, Chen HY, Price DL, Van der Ploeg LH, Sisodia SS. Presenilin 1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature. 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]