Abstract
Members of the ubiquitous 14-3-3 family of proteins are abundantly expressed in metazoan neurons. The Drosophila 14-3-3ζ gene leonardo is preferentially expressed in adult mushroom bodies, centers of insect learning and memory. Mutants exhibit defects in olfactory learning and memory and physiological neuroplasticity at the neuromuscular junction. Because strong mutations in this gene are lethal, we investigated the nature of the defects that precipitate the learning and memory deficit and the role of the two protein isoforms encoded by leonardo in these processes. We find that the behavioral deficit in the mutants is not caused by aberrant development, LEONARDO protein is acutely required for learning and memory, and both protein isoforms can function equivalently in embryonic development and behavioral neuroplasticity.
Keywords: leonardo, 14-3-3ζ, learning and memory, conditional rescue, inducible transgenes, behavioral neuroplasticity
The 14-3-3s are small acidic molecules found in all eukaryotes and comprise a highly conserved family of proteins classed in two conservation groups (Skoulakis and Davis, 1998; Fu et al., 2000). All 14-3-3 proteins are capable of dimerization mediated by the N-terminal helix (Luo et al., 1995; Wang and Shakes, 1996), with homodimers and heterodimers detected in brain and other tissues (Jones et al., 1995). Each subunit is composed of nine α-helices, forming a negatively charged groove of mostly invariant amino acids (Liu et al., 1995; Xiao et al., 1995). Whether the protein is monomeric or dimeric and dimer composition may regulate their binding selectivity (Fu et al., 2000). Interactions with target proteins are primarily mediated by the motif RSxSpxP (x indicates any amino acid; Sp indicates phosphoserine) (Muslin et al., 1996). Thus, 14-3-3s may act as phosphoserine-binding modules similar to phosphotyrosine-binding Src homology 2 domains (Pawson, 1995). Evidence from Drosophila and other species indicates that 14-3-3s interact physically with Raf and are required for its activation (Skoulakis and Davis, 1998; Fu et al., 2000; Muslin and Xing, 2000). In addition, 14-3-3 proteins associate with PKC, phosphatioylinosinol 3-kinase, and the phosphatase cell division cycle 25 protein, suggesting roles in modulation of activity, specificity, or spatial coordination of many signaling complexes.
Drosophila contains two 14-3-3 genes, one from each conservation group. The leonardo gene encodes two nearly identical protein isoforms, with 88% identity to mammalian ζ, and the D14-3-3ε gene encodes a protein 82% identical to the mammalian ε isoform (Skoulakis and Davis, 1998). Mutations that do not affect the vital functions of the leo gene but compromise preferential expression in adult mushroom bodies and ellipsoid body impair olfactory learning and memory (Skoulakis and Davis, 1998). The mushroom bodies are bilateral clusters of neurons essential for olfactory learning and memory in insects (Davis, 1993; Heisenberg, 1998; Zars, 2000). Their somata lie in the dorsal posterior brain, extending dendrites directly beneath them and fasciculated axons to the brain anterior where they bifurcate, forming the α, α′, β, β′, and γ lobes (Strausfeld, 1976; Crittenden et al., 1998). LEONARDO (LEO) proteins are enriched at the presynaptic side of the embryonic neuromuscular junction. Total lack of LEO leads to embryonic lethality but does not affect synaptogenesis and basic synaptic function (Skoulakis and Davis, 1996; Broadie et al., 1997; Kockel et al., 1997). However, synaptic transmission amplitude and fidelity, facilitation, and potentiation are impaired because of apparent failure in synaptic vesicle mobilization (Broadie et al., 1997). Alternatively, the learning and memory and electrophysiological deficits ofleo mutants may result from subtle, undetectable developmental defects.
To address the question of whether LEO is required for developmental processes or acutely for behavioral neuroplasticity, we attempted conditional rescue of the behavioral phenotype of viable leomutants. To enhance putative developmental defects that may underlie the behavioral phenotype, we rescued animals bearing lethal alleles to adulthood and attempted conditional rescue of their learning and memory deficit. Furthermore, we investigated the role of the two LEO proteins in these processes. The results suggest an acute requirement for LEO in learning and memory supported equally by both isoforms.
MATERIALS AND METHODS
Drosophila culture, strains, and germ line transformation. Drosophila were cultured in standard cornmeal sugar food supplemented with soy flour and CaCl2 at 20–22°C. The lethal leoalleles leoP1375,leoP1188, andleoP2335, as well as the viable allelesleo X1 andleo2.3 have been described previously (Skoulakis and Davis, 1996; Broadie et al., 1997; Kockel et al., 1997;Li et al., 1997). All strains used herein were normalized to theDf(1)yw67c23 (yw) control strain, which was the recipient of P-element-mediated germ line transformation. Transformant lines denoted LI contain P[w+hsleo.15], generated by placing aleonardo cDNA containing exons 1–6,7 under heat shock promoter (hsp) control in the phsCaSpeR vector. Transformant lines denoted LII contain P[w+hsleo.2], a leonardo cDNA containing exons 1–6′,7. Chromosomal localization of the transgenes and their introduction into the mutant backgrounds was achieved with standard genetic crosses. Because theleo gene resides on the second chromosome, lines bearing the construct on the third chromosome were selected for ease of genetic manipulations.
Induction of the transgenes was achieved using programmable cycling incubators (Labline) to deliver daily heat shocks (24–36.5 ± 0.5°C) for 30 min each. For animals raised under continuous cycling conditions (protocol HS A), flies were kept in bottles for 5 d, the parents were removed, and the cultures were maintained under cycling conditions until adults emerged. To obtain rescued homozygotes under noncontinuous transgene induction (protocol HS B), first virgin females and males heterozygous for the P-element-induced mutations and homozygous for the transgene were obtained under conditions of two daily heat shock inductions. They were mated, and the progeny was allowed to develop at 23 ± 2°C. Control cultures were also kept at 23 ± 2°C. Rescue was calculated as the fraction of the expected homozygous or heteroallelic adult flies actually obtained. Unless otherwise indicated, transgene inductions for behavioral, histological, and Western blot analyses were similarly performed in the cycling incubators, but after induction the animals were transferred to the conditions under which they were reared (18°C for the viable transgene bearing strains and controls and 23 ± 2° for lethal homozygotes and heteroallelics raised under protocol HS B) for a 5–6 hr rest period.
Reverse transcription-PCR. Twenty heads, thoraces, and abdomens and 100 μl equivalents of embryos and larvae were homogenized in 100 μl of Trizol, and RNA was prepared as suggested by the manufacturer (Life Technologies, Gaithersburg, MD). For reverse transcription (RT), 20% of the extracted RNA was used per 20 μl of reaction, which contained 2.5 μm each oligo-dT and a random 15-mer and 200 U of Moloney murine leukemia virus H(−) Point Reverse Transcriptase (Promega, Madison, WI). The reaction proceeded as recommended by the supplier (Promega). The PCR reaction contained 10% of each RT, 6.25 nmprimers, and 2.5 U of Taq polymerase (Roche Products, Hertforshire, UK) using 35 cycles at 92°C for 45 sec, 58°C for 1 min, and 72°C for 1 min. Specificity of the reactions was tested with RNased samples before RT, not reverse-transcribed RNA and no nucleic acid inputs. Finally, the identity of the leoI andleoII PCR products was confirmed by restriction analysis. Flies with ablated mushroom bodies were obtained using described methods (DeBelle and Heisenberg, 1994), except that 75 mg/ml hydroxyurea was used to make yeast paste on which the newly hatched larvae fed. The completeness of ablations was verified histologically using the anti-LEO antibody (Skoulakis and Davis, 1996). Batches of adults, which during sampling exhibited >90% ablation, were used for RT-PCR.
Western blot analysis. Three fly heads were homogenized in 30 μl of modified radioimmunoprecipitation assay buffer (0.137m NaCl, 20 mm Tris, pH 8, 10% glycerol, 0.1% SDS, and 0.1% sodium deoxycholate). Samples were boiled for 5 min and centrifuged at 12,000 × g for 10 min and, after addition of Laemli's buffer, 10 μg of protein was loaded per lane for SDS-PAGE and blotting using standard methods. Primary antibodies were used at 1:10,000 and 1:100 dilution for α-LEO and α-syntaxin (antibody 8C3; Developmental Hybridoma Studies Bank, University of Iowa, Iowa City, IA), respectively. The results were visualized with enhanced chemiluminescence (Pierce, Rockford, IL) and quantified using densitometry.
Immunohistochemistry. Frontal paraffin sections (5 μm) of heads were obtained and processed for immunohistochemistry or histology as described previously (Skoulakis and Davis, 1996; Crittenden et al., 1998). The α-LEO, α-DRK, α-FASII, α-DMEF, and α-DAC antibodies and standard hematoxylin–eosin staining were used for structural analysis of mutant brains (Crittenden et al., 1998).
Behavioral analyses. The negatively reinforced olfactory learning assay using aversive odors as conditioned stimuli (CS+ and CS−) and electric shock as the unconditioned stimulus (US), as well as control behavioral assays were performed using established methods (Tully and Quinn, 1985; Skoulakis et al., 1993; Skoulakis and Davis, 1996). Olfactory trap assays were performed essentially as described previously (Ayer and Carslon, 1992), except that each trap was constructed using a 0.5 ml Eppendorf tube with the bottom cut off, inserted bottom to bottom in a similarly cut 1.5 ml Eppendorf tube. The hollow lid of the 1.5 ml tube was filled with 200 μl of 0.8% agarose containing either 0.5 or 0.05% geraniol closed, and the assembly was placed in a 100 × 15 mm Petri dish containing a piece of 3 mm paper moistened with 1 ml of deionized water. Ten male flies were assayed per dish, and their performance was assessed after 48 hr in the dark at 23–24°C. All relevant genotypes were tested in parallel. A performance index (PI) was calculated as the fraction of flies in the trap at the end of the test period. The odorant amounts used were experimentally adjusted to the lowest possible to permit maximal resolution reliably.
Statistical analysis. Untransformed (raw) data were analyzed parametrically with JMP3.1 statistical software package (SAS Institute, Cary, NC) as described previously (Skoulakis et al., 1993;Skoulakis and Davis, 1996). To maintain a constant experiment wise error rate after initial ANOVA, planned multiple comparisons were performed as suggested by Sokal and Rohlf (1981).
RESULTS
Differential expression of leonardo isoforms
The leonardo gene encodes three size classes of transcripts attributable to use of alternative promoters and three polyadenylation sites (Skoulakis and Davis, 1996; Kockel et al., 1997). Alternative splicing of exon 6 or 6′ into the mRNA results in two protein isoforms (LEOI and LEOII) that differ by five amino acids (Fig.1A). Because exons 6 and 6′ are similar in size, alternative inclusion into the mRNA does not contribute to size heterogeneity. To determine the spatial and temporal expression of mRNAs that contain exon 6 (leoI) and 6′ (leoII), we used RT-PCR. Expression of D14-3-3ε (Chang and Rubin, 1997), a message of lower abundance served to monitor the quality of RT-PCR and all PCR sets were performed in duplicate from at least three independent RT reactions.
Fig. 1.
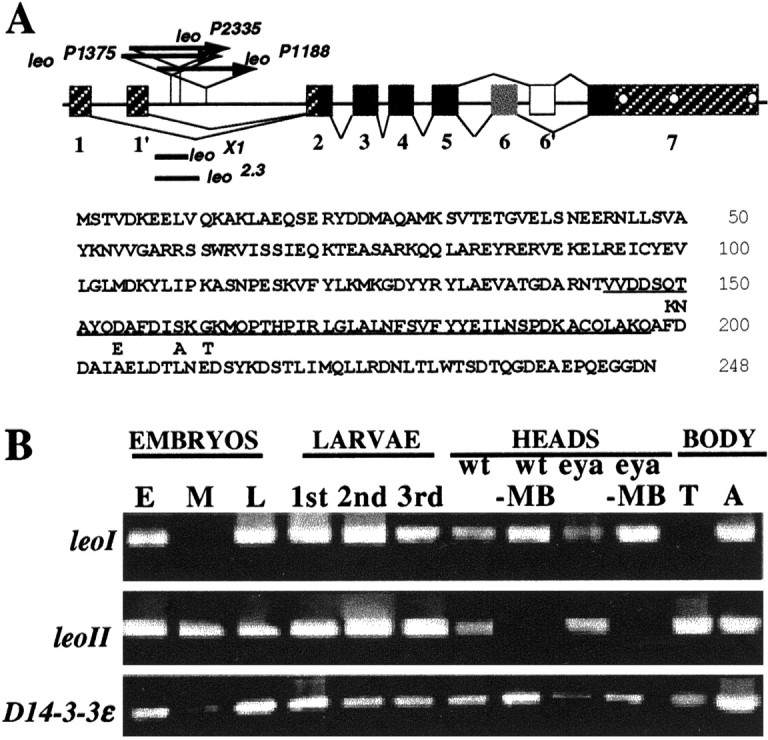
The leonardo gene and isoform-specific temporal and tissue expression. A, Structure of the leo gene. Exons are represented bynumbered boxes, and introns are represented bylines. Untranslated sequences are shown ashatched boxes, and all protein-coding exons are shown with black boxes, except the alternatively spliced exon 6 and 6′ (gray and white boxes, respectively). The polyadenylation sites are indicated by thewhite circles in exon 7. The positions of transposon insertion in the three lethal allelesleoP1375,leoP1188, andleoP2335 are indicated. The sequences deleted in the viable alleles leo X1and leo2.3 are indicated by theblack bars. The sequence of the LEOI (top line) and LEOII (bottom) proteins is shown below the gene structure, with the amino acids encoded by exon 6 (LEOI)underlined and the unique amino acids encoded by the alternative exon 6′ (LEOII) shown under them. B, Differential expression leo mRNAs. RNA was isolated from 0–2 hr (E), 12–14 hr (M), and 18–20 hr (L) embryos, first (1st), second (2nd), and third (3rd) instar larvae, dissected adult heads from control (wt), eyes absent (eya), and hydroxyurea mushroom body-ablated (−MB) control and eyes-absent animals, as well as control thoraces (T) and abdomens (A). The RNA was reverse transcribed, and, after PCR, the products were displayed in a 1.5% agarose gel.
The results of this expression analysis are displayed in Figure1B. Both leoI and leoIItranscripts were present in embryos before activation of the zygotic genome, suggesting that they are deposited in the oocytes maternally. Exclusive presence of leoII transcripts in stage 10–12 embryos indicates preferential splicing of exon 6′ into the mRNA, which may underlie a specific contribution of LEOII to early development. In contrast, both leoI and leoII transcripts were found in late embryos and all larval stages. In adult animals, although both isoforms were present in heads and abdomens, leoI was absent from the thorax.
To determine whether head tissues that require leo function exhibit differential isoform expression (Skoulakis and Davis, 1996;Chang and Rubin, 1997), we subjected flies carrying the eyes-absent mutation and wild-type animals to mushroom body ablation with hydroxyurea (DeBelle and Heisenberg, 1994). Lack of eye tissues did not eliminate one of the isoforms differentially, but leoII was specifically absent from the brains of mushroom body-ablated animals. The results indicate that leoII transcripts are specific to the mushroom bodies, whereas although leoI may be present in these neurons, it is more broadly expressed in the brain. Outside the mushroom bodies, LEO protein is preferentially distributed in the ellipsoid body neurons of the central complex (Skoulakis and Davis, 1996). Because ellipsoid body neurons are not ablated by hydroxyurea (DeBelle and Heisenberg, 1994) and retain LEO immunoreactivity (data not shown), they must contain only leoI transcripts. Interestingly, presence of D14-3-3ε in all tissues and stages tested suggests a broad role in basic cellular functions, and possible colocalization with LEO isoforms may result in heterodimer formation. Together, the differential expression of the twoleonardo mRNAs in embryos and adult tissues suggests functional differences between the two LEO protein isoforms. Therefore, a functional investigation of potential differences between LEOI and LEOII isoforms was necessary before experiments aimed at rescuing the learning–memory deficit of leo mutations.
Transgenic rescue of lethality associated withleo alleles
To investigate potential functional differences of the putative LEO isoforms, we attempted conditional rescue of lethality associated with strong leo alleles (Fig. 1A). These transposon insertions severely compromise all leo expression (Skoulakis and Davis, 1996; Broadie et al., 1997; Li et al., 1997). Because the two isoforms appeared differentially distributed in embryos, these experiments provided an initial measure of possible differences among them and an estimate of the activity and specificity of strains to be used for behavioral analyses. Multiple transgenic lines harboring the leoI (LI) or leoII cDNAs under the hsp70 promoter were used. To assay for inducible expression of the transgenes, all lines were used to rescue the lethality associated with leoP1375. Transcriptional induction of LI and LII transgenes with two 30 min heat shocks daily throughout embryonic larval and pupal stages yieldedleoP1375 homozygotes to varying degrees (Table 1), indicating transgenic rescue of the lethal phenotype. Similar results, albeit lower numbers of rescued animals, were obtained with a single daily induction until adulthood (Philip, 2000). Multiple lines of LI and LII transgenes exhibited rescue under restrictive (18°C) or basal conditions (room temperature), suggesting that transcription in some lines was regulated by genomic elements at the points of insertion (position effects). One transformant line carrying LI and one carrying LII were selected for additional analysis (Table 1, asterisks) based on low basal activity (18°C and room temperature) and high inducibility. However, results obtained with these two were confirmed with additional transformant lines.
Table 1.
Conditional rescue of leoP1375homozygous lethality by isoform-specific transgenes
| Transformant line | % Rescue at 18°C | % Rescue at RT | % Rescue by HS |
|---|---|---|---|
| LI-2.1 | 10.2 ± 6.2 | 100 | 100 |
| LI-2.7 | 21.7 ± 7.5 | 100 | 100 |
| LI-4.1 | 0 | 0 | 28.6 ± 4.8 |
| LI-4.3** | 3.5 ± 1.5 | 5.8 ± 2.4 | 80.4 ± 5.6 |
| LII-1.2 | 0 | 15.2 ± 3.1 | 47.4 ± 3.9 |
| LII-3.0** | 0 | 0 | 21.3 ± 5.2 |
| LII-4.2 | 6.8 ± 2.5 | 23.8 ± 3.6 | 66.7 ± 4.6 |
| LII-8.10 | 16.8 ± 3.7 | 46.7 ± 4.5 | 100 |
| LII-12.0 | 0 | 6.1 ± 3.8 | 12.9 ± 4.1 |
Male and female animals of the genotypesleoP1375/CyO;LEOI (LI) andleoP1375/CyO;LEOII (LII) were mated, and progeny was reared at 18°C, 23 ± 2°C (RT), or under two 30 minute daily inductions (24–36.5 ± 0.5°C heat shocks) throughout development until adult progeny emerged. Percentage of rescue represents the fraction of obtained over expected adultleoP1375 homozygotes.
Using quantitative Western analysis, we estimated the level of LEO protein induced in heads of rescuedleoP1375 homozygotes (Fig.2B, HS protocol A). These homozygotes contained ∼75–80% the amount of LEO present in similarly treated wild-type animals. Interestingly, LEO induced under these conditions perdured at appreciably high levels for 4–6 d (Philip, 2000). Maternal loading ofleoI and leoII presented above, high levels of LEO protein in oocytes (Li et al., 1997), and the stability of induced proteins led to development of a second lethality rescue protocol (HS B). Females raised to adulthood under two daily transgene inductions (HS A protocol) were subsequently mated, and progeny was raised at room temperature (basal conditions for the lines used as defined in Table1). Both protocols resulted in rescue ofleoP1375 homozygotes by LI and LII transgenes (Fig. 2A), albeit with different efficiency, consistent with the lower level of LEO produced under protocol HS B. Neither LI nor LII rescuedleoP1188 orleoP2335 homozygotes to adulthood under either rescue protocol. However,leoP1375/leoP1188andleoP1188/leoP2335, as well asleoP1375/leoP2335(data not shown) heteroallelics were readily obtained with LI under both protocols (Fig. 2A). In contrast, induction of LII did not result in heteroallelics, except when heterozygous with LI. This likely reflected the lower level of expression of the LII-3.0 insertion, as suggested by the lower number of rescued animals recovered with LI/LII heterozygotes compared with those rescued by LI homozygotes (Fig. 2A). In agreement with the later, line LII-8.10 (Table 1) rescued heteroallelics to adulthood (data not shown).
Fig. 2.
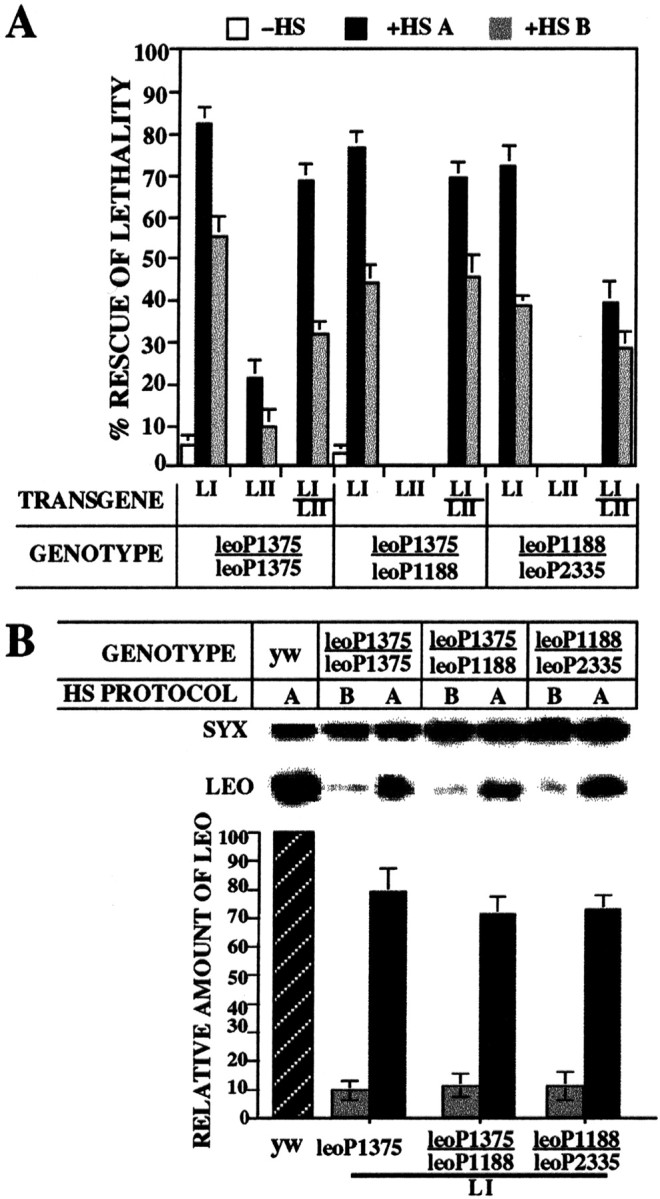
Two rescue protocols yield homozygous or heteroallelic adults that contain contrasting levels of LEO.A, Percentage of rescue of lethal homozygotes and heteroallelics by LEOI (LI), LEOII (LII), and LI/LII transgenes under basal (room temperature; −HS, white bars), two 30 min daily inductions (+HS A, black bars) throughout development, or from induced mothers but reared under basal conditions (+HS B, gray bars). Percentage of rescue represents the fraction obtained over that expected of adult homozygotes or heteroallelics. LII transgenes were not able to rescueleoP1375/leoP1188orleoP1188/leoP2335animals under any conditions (lack of bars). A minimum of 300 animals were scored per cross, and each cross was performed in triplicate. B, A representative quantitative Western blot of head extracts obtained from the indicated animals (GENOTYPE) raised under HS A and HS B conditions (HS PROTOCOL), challenged with the α-LEO (LEO) and α-syntaxin (SYX) antibodies. The amount of syntaxin in the extracts served as a standard to estimate the amount of LEO. Quantification of four independent experiments is in the bar graph below the blot. The level of LEO was normalized over the level of syntaxin in animals raised under HS A (black bars) or HS B (gray bars) protocols and expressed as a fraction of that present in control animals (yw,hatched bars), which was set at 100.
To determine whether the two protocols yield adults with different levels of LEO, we investigated the level of this protein in the heads of rescued homozygotes and heteroallelics (Fig.2B). Significantly, 1- to 2-d-old animals rescued under the two protocols contained drastically different amounts of LEO compared with controls, ∼75–80% under protocol HS A and only 10–15% under protocol HS B. To investigate whether the amount of LEO would increase in response to acute transgene induction,leoP1375 homozygotes were exposed to multiple heat shocks, and the level of protein in their heads was determined (Fig. 3). Although a significant increase in protein was evident after a single 30 min induction, six heat shocks over a period of 40 hr were necessary to accumulate levels of LEO approaching 80% that of control animals. Similar induction profiles were obtained with LI and LII transgenes inleoP1375 (Fig. 3) and heteroallelic animals (Philip, 2000).
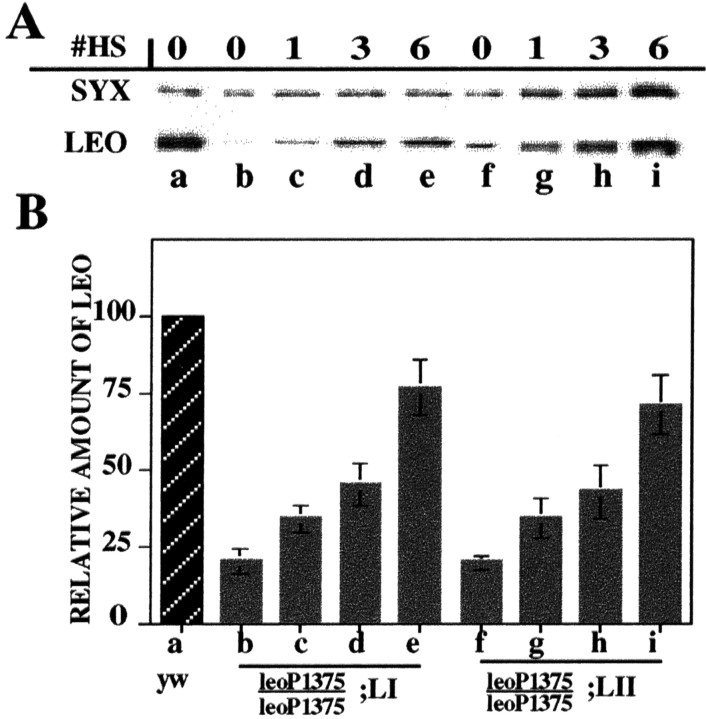
Fig. 3.
Conditional accumulation of LEOI and LEOII proteins in the heads of leoP1375 homozygotes. A, A representative quantitative Western blot of head extracts obtained from animals raised to adulthood under HS B conditions and subjected to zero, one, three, or six inductions (#HS). The accumulation in the heads of such animals was monitored with the α-LEO (LEO) and normalized with the α-syntaxin (SYX) antibody. The mean ± SEM of three independent experiments is shown in B forleoP1375;LI andleoP1375;LII head extracts standardized against the amount in control (yw) animals (hatched bar), which was set at 100. Genotypes and treatments in A indicated with the letters a–iare similarly represented in B.
Collectively, these results indicate that because both LEOI and LEOII can support development to adulthood, under the conditions used the isoforms do not exhibit functional specificity. Furthermore, both LI and LII transgenes are highly inducible and allow manipulation of LEO levels in adult heads over a wide range, and animals thus obtained do not exhibit morphological defects. In addition, these experiments identified highly inducible leo transgenes and methods to obtain animals for behavioral testing as described below.
Inducible rescue of the learning deficit in viableleo mutants
The differential distribution of leo transcripts in adult heads suggested potentially differential roles for LEOI and LEOII in olfactory learning and memory. To investigate whether the behavioral deficit of leonardo viable alleles (Skoulakis and Davis, 1996) could be reversed by conditional induction of the leotransgenes, we introduced both transgenes intoDf(1)yw67c23;leo23, (leo23) andDf(1)yw67c23;leoX1, (leoX1) flies. To ascertain that the transgenes remained inactive during development, all animals including the Df(1)yw67c23 (yw) control strains were raised at 18°C. Because leonardo expression in tissues other than the mushroom bodies and ellipsoid body appears normal in these alleles (Skoulakis and Davis, 1996), quantitative Western blots were not used to monitor LEO levels in the heads of these animals. Transgene induction in animals raised at 18°C was achieved by two 30 min heat shocks delivered 6 hr apart, followed by a 5–6 hr rest period. Accumulation of LEOI and LEOII in the mushroom bodies ofleo23;LI andleo23;LII animals after the rest period was monitored by immunohistochemistry using the anti–LEO antibody. In agreement with previous results (Skoulakis and Davis, 1996), very low levels of LEO protein were present in the mushroom bodies ofleo23;LI andleo23;LII animals. A significant increase of both protein isoforms in the mushroom bodies and ellipsoid body neurons (data not shown) was observed during induction of the respective transgenes, although final accumulation did not equal the amount of LEO in controls (Fig. 4). Similar results were obtained withleoX1;LI andleoX1;LII animals (data not shown). Moreover, lack of LEO during development did not precipitate neuroanatomical aberrations in the brains of mutant animals raised at 18°C (data not shown), determined using multiple antigenic markers (see Materials and Methods).
Fig. 4.
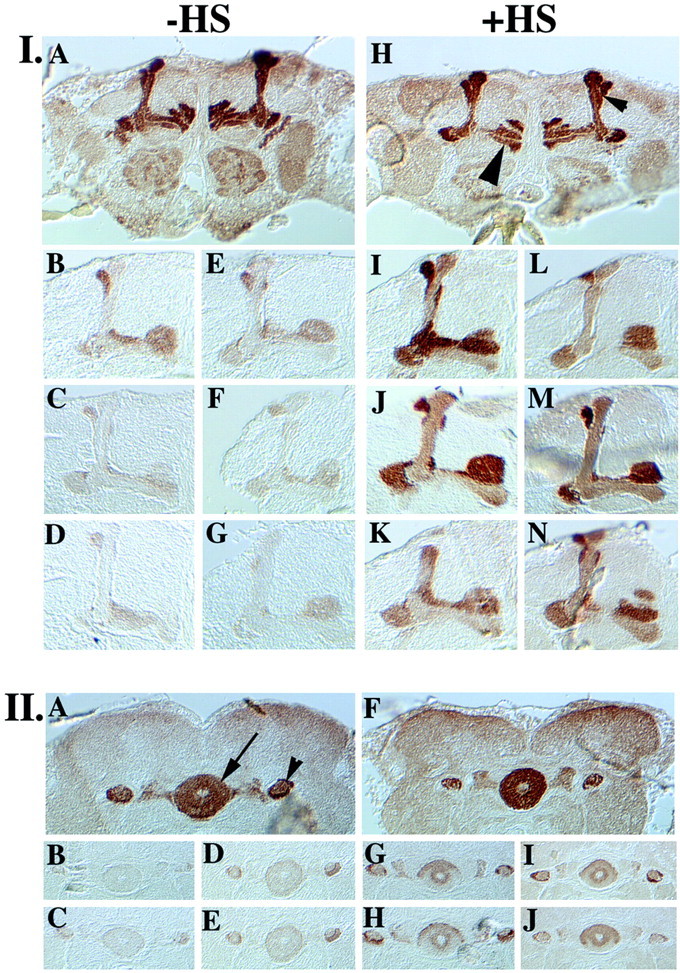
Conditional accumulation of LEO isoforms in the brain of transgenic animals. Paraffin frontal head sections (5 μm) from animals with the genotypes detailed below were challenged with the α-LEO antibody, and the results were visualized by the HRP activity conjugated to the secondary antibody. Control and mutant animals were mounted on the same slide, and slides with all genotypes were processed in parallel. Each genotype was represented by a minimum of nine individuals, and the entire experimental set was repeated three times.I, LEO accumulation in the mushroom body neurons (shown at the level of the lobes). Large arrowhead, The β, β′, γ complex. Small arrowhead, The α, α′complex. Without induction before immunohistochemistry (−HS): A, control animals;B, leo2.3;LI reared at 18°C; C, leoP1375;LI reared under protocol HS B; D,leoP1188/leoP2335;LI reared under protocol HS B; E,leo2.3;LII reared at 18°C;F, leoP1375;LII reared under protocol HS B; G,leoP1188/leoP2335;LI/LII reared under protocol HS B. After two 30 min inductions before immunohistochemistry (+HS): all animals were raised as indicated for the respective genotypes above. Two days after eclosion, the animals were given two 30 min inductions in the cycling incubators delivered 6 hr apart, were allowed a 5–6 hr rest period, and were fixed and processed for immunohistochemistry. H, Control animals; I, leo2.3;LI;J, leoP1375;LI;K,leoP1188/leoP2335;LI;L, leo2.3;LII;M, leoP1375;LII;N,leoP1188/leoP2335;LI/LII.II, LEO accumulation in the ellipsoid body neurons.Arrow, Ellipsoid body. Arrowhead, Mushroom body neuron axonal projections (pedunculi). Animals were raised and treated as indicated above. Without induction:A, control animals; B,leoP1375;LI; C,leoP1188/leoP2335;LI;D,leoP1375;LII; E,leoP1188/leoP2335;LI/LII. After two 30 min inductions: F, control animals;G, leoP1375;LI;H,leoP1188/leoP2335;LI;I,leoP1375;LII; J,leoP1188/leoP2335;LI/LII.
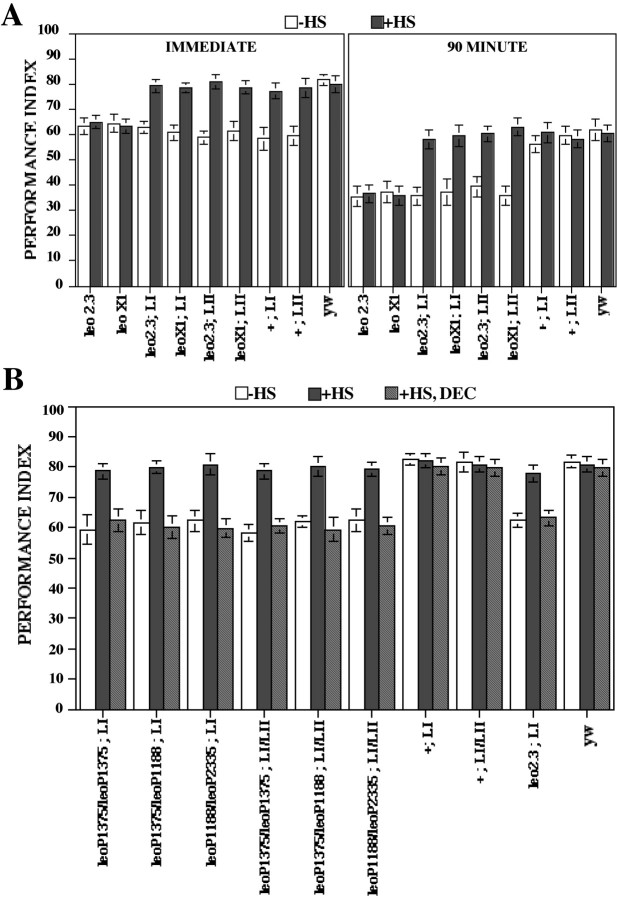
Animals raised at 18°C and ones subjected to the induction and rest period were transferred to 23–24°C 2 hr before behavioral experiments. The growth conditions and temperature shift did not affect the ability of the mutants to perceive the stimuli used for olfactory conditioning compared with similarly treated controls (Table2A). To further investigate their olfactory acuity, the performance of mutants and controls toward an attractive odor, geraniol, was measured using a modified olfactory trap assay (see Materials and Methods). Although an attractive odor is not used in conditioning, this test is a good measure of olfactory acuity. Flies seek and navigate toward the source of an attractive odor, a more complex olfactory task than simple avoidance of an aversive odor. As shown in Table 2A, the performance of experimental animals was not significantly different from controls. Performance of the animals after olfactory conditioning was assessed immediately after training or 90 min later to investigate memory (Fig. 5A). The performance of leo23;LI,leo23;LII,leoX1;LI, andleoX1;LII animals exhibited a significant 30% decrement compared with controls both immediately and 90 min after training, similar to the decrement observed withleo23 andleoX1 animals raised under similar conditions. In contrast, learning and 90 min retention were not significantly different from controls during transgene induction before conditioning. The results suggest that LEOI and LEOII accumulation in the mushroom bodies after transgene induction fully restores the learning and memory deficit of leo23andleoX1 mutants. Interestingly, under the conditions used, both LEOI and LEOII isoforms appear equivalent in rescuing the behavioral deficit of the mutants. Collectively, the results indicate strongly that leonardo gene products are acutely required for mushroom body-dependent olfactory learning and memory.
Table 2.
Task-relevant olfactory and sensorimotor behaviors and olfactory acuity to an attractive odor
| Genotype | Avoidance | Attraction | |||
|---|---|---|---|---|---|
| 90 V | BNZ | OCT | 0.05% GER | 0.5% GER | |
| A | |||||
| leo 2.3 | 78.4 ± 3.7 | 74.7 ± 4.5 | 70.8 ± 4.1 | 44.7 ± 4.3 | 85.6 ± 4.1 |
| leo X1 | 77.6 ± 3.9 | 72.8 ± 4.8 | 70.4 ± 4.4 | 42.4 ± 4.8 | 83.9 ± 4.4 |
| leo2.3;LI | 80.4 ± 4.2 | 74.2 ± 4.3 | 69.9 ± 4.6 | 51.2 ± 5.4 | 88.2 ± 3.5 |
| leoX1;LI | 76.8 ± 2.4 | 76.1 ± 2.3 | 70.6 ± 3.5 | 43.8 ± 5.2 | 86.3 ± 4.5 |
| leo2.3;LII | 77.4 ± 2.8 | 75.7 ± 2.8 | 73.3 ± 3.8 | 42.8 ± 4.7 | 80.4 ± 5.6 |
| leoX1;LII | 79.2 ± 3.4 | 74.1 ± 3.7 | 69.7 ± 4.1 | 44.3 ± 5.0 | 86.7 ± 3.1 |
| +;LI | 76.6 ± 4.2 | 73.4 ± 3.8 | 71.9 ± 4.4 | 45.6 ± 4.7 | 84.4 ± 5.2 |
| +;LII | 79.5 ± 3.2 | 75.9 ± 3.9 | 69.2 ± 3.4 | 41.8 ± 4.4 | 87.6 ± 5.4 |
| yw | 80.1 ± 3.7 | 73.6 ± 3.7 | 68.3 ± 3.9 | 43.7 ± 4.6 | 83.7 ± 4.6 |
| B | |||||
| leoP1375/leoP1375;LI | 77.6 ± 3.8 | 77.9 ± 3.6 | 68.6 ± 3.6 | 41.6 ± 4.6 | 85.6 ± 4.0 |
| leoP1375/leoP1188;LI | 74.3 ± 4.1 | 74.6 ± 3.9 | 67.2 ± 3.9 | 39.4 ± 5.3 | 82.2 ± 4.7 |
| leoP1188/leoP2335;LI | 83.5 ± 3.9 | 77.3 ± 4.2 | 63.4 ± 3.3 | 45.8 ± 4.4 | 87.3 ± 3.7 |
| leoP1375/leoP1375;LI/LII | 73.8 ± 3.6 | 81.3 ± 3.3 | 63.8 ± 4.1 | 43.7 ± 3.8 | 85.8 ± 3.4 |
| leoP1375/leoP1188;LI/LII | 79.5 ± 4.8 | 80.2 ± 3.5 | 66.8 ± 3.8 | 46.7 ± 5.1 | 86.2 ± 4.5 |
| leoP1188/leoP2335;LI/LII | 80.4 ± 3.1 | 73.7 ± 4.1 | 64.6 ± 3.5 | 42.8 ± 4.7 | 83.9 ± 3.4 |
| +;LI | 78.5 ± 4.2 | 78.4 ± 3.8 | 68.2 ± 3.9 | 44.3 ± 4.9 | 86.7 ± 2.9 |
| +;LI/LII | 76.8 ± 3.6 | 76.2 ± 4.1 | 63.7 ± 3.7 | 42.9 ± 4.6 | 84.4 ± 3.2 |
| leo2.3;LI | 78.9 ± 4.7 | 75.3 ± 3.9 | 64.9 ± 3.9 | 43.4 ± 4.2 | 86.3 ± 4.2 |
| yw | 76.3 ± 4.3 | 74.2 ± 4.4 | 65.8 ± 4.2 | 44.6 ± 4.5 | 83.7 ± 4.7 |
A, Mutant and control animals reared at 18°C do not exhibit significant differences in perception of the US–90 V or the CS [benzaldehyde (BNZ) and 3–octanol-(OCT)] used for olfactory conditioning. Additionally, there are no significant differences in perception of a highly diluted attractive odor [geraniol (GER)]. Performance of the animals in these tasks was measured by the performance index calculated as by Skoulakis and Davis (1996) and are shown as PI ± SEM. Thus, rearing the animals at 18°C and the lack of LEO in the mushroom bodies do not precipitate sensory deficits. n > 7 for 90V and BNZ, OCT avoidance, but n > 10 for attraction to GER.
B, The performance (PI ± SEM) of control animals and lethal homozygotes and heteroallelics obtained with protocol HS B in avoidance of the US (90 V) and the CS (BNZ, OCT) is not significantly different, indicating that the nearly total lack of LEO from the mutants does not affect their sensory systems requisite for conditioning.n > 7 for all stocks. In addition, lack of LEO does not alter the attraction of lethal homozygotes and heteroallelics toward diluted geraniol, indicating that lack of LEO does affect odor perception. n > 12.
Fig. 5.
Conditional rescue of the learning and memory deficit of leo mutants. A, Conditional rescue of learning and memory deficits of viable alleles. Learning (IMMEDIATE) and 90 min memory was assessed inleo2.3,leoX1,leo2.3;LI,leo2.3;LII,leoX1;LI,leoX1;LII, and control animals (yw). To investigate potential nonspecific effects of the LI and LII transgenes, flies homozygous for the transgenes but not harboring the mutations in leo were used as additional controls. The mean PI ± SEM is shown for each stock. The performance index was calculated as by Skoulakis and Davis (1996). Learning in mutants reared under conditions that silence the transgenes (18°C; hatched bars) was identical to mutants without the transgenes and significantly different compared with control animals. One-way ANOVA showed significant effects of genotype (F(8, 78) = 16.245;p < 0.001). Planned comparisons showed significant differences among yw and the mutants (p < 0.001) but not among yw and +;LI or +;LII controls or amongleo2.3 andleoX1 compared with uninduced (−HS),leo2.3;LI,leo2.3;LII,leoX1;LI, andleoX1;LII. Similarly, one-way ANOVA indicated significant effects of genotype in 90 min memory (F(8, 74) = 14.925;p < 0.001) and was confirmed for mutants compared with yw (p < 0.001). There were no significant differences among yw and +;LI or +;LII strains. However, learning and 90 min memory of the mutants was not significantly different from controls when animals were trained after transgene induction (black bars), indicating full rescue of the learning and memory deficits. n > 8 for all stocks. B, Reversible conditional rescue of learning deficits of lethal homozygotes and heteroallelics. Learning was assessed in the indicated lethal homozygotes and heteroallelics obtained under protocol HS B without previous transgene induction (−HS, white bars) after transgene induction and recovery (+HS, black bars) or after a lengthy recovery period for the levels of LEO to decay (+HS, DEC, hatched bars). The mean PI ± SEM is shown for each stock. One-way ANOVA indicated significant effects of genotype (F(9, 96) = 14.836; p < 0.001), and subsequent planned comparisons verified significant differences among mutants and control (yw) strains (p < 0.001) but not for yw and +;LI or +;LI/LII flies under uninduced conditions (−HS). n > 8. In contrast, ANOVA did not indicate differences among all strains after transgene induction (+HS), indicating complete rescue of the learning deficit. n > 7. However, decay of the induced LEO protein (+HS, DEC) resulted in significant loss of learning in mutant stocks compared with age-matched controls (p < 0.001) but not among yw and +;LI or +;LI/LII strains. Therefore, the amount of LEO present in the head of mutants at the time of conditioning is essential for learning.
Reversible rescue of learning deficits in lethalleo alleles
Given the behavioral rescue of leo mutants, we wondered whether the learning and memory deficit exhibited by leoviable alleles represents the maximal contribution of LEO-mediated processes in mushroom body-dependent olfactory learning. We used the ability to obtain animals that harbor very low levels of LEO throughout their heads (HS B protocol) to address this question. Preliminary experiments indicated that leoP1375homozygotes rescued under protocol HS A (80% relative level of LEO) (Fig. 2B) and tested 1–2 d after eclosion do not exhibit behavioral deficits (data not shown). Because a substantial number of animals are required for training, allelic combinations with high yields of homozygotes or heteroallelics were selected. The LI and LII transgenic lines were used because they exhibit low basal activity and are highly inducible. Because LII-3.0 does not support rescue to adulthood in sufficient numbers, we used LI/LII heterozygotes.
Western analysis indicated that animals rescued with protocol HS B harbor ∼10–15% of LEO in their heads compared with controls (Fig.2B). To determine whether the remaining protein is differentially localized in the mushroom bodies, we visualized its distribution immunohistochemically. The residual LEO did not accumulate preferentially in the mushroom bodies (Fig. 4I,C, D, F, G) or ellipsoid body (Fig. 4II, B–E) of lethal homozygotes and heteroallelics rescued by LI or LI/LII transgenes under protocol HS B. In fact, the level of LEO in these brain ganglia was nearly undetectable. In contrast, during induction of the transgenes, a significant amount of LEO accumulates in these neurons (Fig.4I, J, K, M,N; II, G–J) but does not attain wild-type levels, in agreement with Western blot results (Fig.3). The nearly complete lack of LEO throughout the adult brain did not result in neuroanatomical anomalies judged by histological and immunohistochemical analyses using multiple markers (see Materials and Methods) to examine the morphology of the mushroom bodies and central brain (Philip, 2000).
To determine whether lack of LEO throughout the animals rescued under protocol HS B precipitated general sensory deficits, we subjected them to behavioral control tests. These leo mutants exhibited normal attraction to geraniol, avoidance of electric shock (US), and avoidance of both aversive odors (benzaldehyde and 3-octanol) used as CS (Table 2B). However, all rescued animals exhibit a 25–30% decrement in olfactory learning (Fig. 5B,open bars). Significantly, the decrease in learning exhibited by the rescued lethal homozygotes and heteroallelics was similar in magnitude with that of leo23;LI animals. Therefore, near lack of LEO throughout the head does not reduce learning further than exhibited byleo23 animals, which lack LEO only in the mushroom bodies. This suggests that theleo23 andleoX1 mutations represent strong mutant alleles with respect to the behavioral phenotype. As withleo23;LI animals, the learning deficit of lethal homozygotes and heteroallelics was fully rescued to control levels by multiple inductions of either LI or LI/LII transgenes. To determine whether restoration of learning ability results from permanent changes attributable to the elevation of LEO, animals were kept at 18°C after induction and trained and tested along similarly treated and aged controls. Restoration of learning during transgene induction decayed back to mutant levels 60–70 hr later compared with age-matched control animals (Fig. 5B, hatched bars). The perdurance of LEO monitored by Western blots (Philip, 2000) necessitated this time for decay, and behavioral training–testing 40–48 hr after induction yielded intermediate learning. Because all flies used in these experiments were less that 8 d old, the actual age of the animals did not affect their performance. Control experiments with transgene induction 1–2 d after eclosion and behavioral training and testing on day 4–5 or induction 4–5 d after eclosion and training–testing on day 7–8 produced identical results (data not shown).
These results indicate that induction of LEO to levels sufficient to restore learning does not precipitate permanent changes but rather that the available amount of protein is acutely essential for this process. Furthermore, elevated LEO expression outside the mushroom bodies and ellipsoid body observed in controls and abrogated in the mutants does not appear essential for learning, the sensory inputs used in these experiments, or for the neuroanatomical integrity of the brain.
DISCUSSION
Genetic analysis of learning and memory in Drosophilahas been highly successful in revealing molecular pathways involved in these processes. Studies have focused on nonvital genes, isolation of viable alleles of essential genes (Davis, 1996; Skoulakis and Davis, 1996; Grotewiel et al., 1997), or transgenic animals carrying in vitro generated mutations (Yin et al., 1994, 1995; Connoly et al., 1996). However, essential genes play cardinal roles in learning and memory (Boynton and Tully, 1992; Skoulakis et al., 1993; Skoulakis and Davis, 1996; Grotewiel et al., 1997; Simon et al., 1998; DeZazzo et al., 2000). Use of hypomorphic viable alleles of essential genes may not reveal their full contribution to learning and memory and may conceal subtle developmental defects. We used a novel method to investigate adult learning and memory effects of an essential gene by obtaining homozygotes for strong lethal alleles by regulated transgene expression. Use of this method to study behavioral roles of essential genes is likely to depend on the nature and vital functions of particular genes. A similar strategy is currently being pursued to investigate behavioral functions of other essential genes (E. M. C. Skoulakis, unpublished results).
Homozygotes for leo loss of function alleles derived from heterozygous mothers die as morphologically normal embryos before hatching because of synaptic transmission defects (Skoulakis and Davis, 1996; Broadie et al., 1997; Kockel et al., 1997; Li et al., 1997). However, embryos that lack maternally deposited leonardoexhibit severe developmental defects (Li et al., 1997). Therefore, the maternally deposited leoI, leoII, andD14-3-3ε mRNAs and their protein products suffice to support development to mature embryos. Because we used mothers heterozygous for leo mutations, activation of the transgenes likely contributes to requirements for leo activity in late embryos, which as shown in Figure 1 contain both leoI andleoII isoforms. However, potential differences in the spatial distribution of the two isoforms, as in the adult head and thorax, could not be resolved with the methods used. Interestingly, both LI and LII transgenes rescue the lethality associated with the weak leoP1375 allele, but only highly expressed transgenes rescued heteroallelic combinations of strong lethal mutations. This suggests that, although both LEOI and LEOII may be required for viability in late embryos, high levels of either protein functionally substitute for the missing isoform in the mutants.
Acute induction of either LI or LII transgenes completely restores learning and memory in leo23 andleoX1 mutant flies. Thus, the behavioral deficit in these animals is unlikely the result of sensory or developmental defects below the threshold of detection but rather are attributable to an acute requirement for LEO in learning–memory. This conclusion is further supported by the reversible rescue of the learning deficit exhibited by lethal homozygotes and heteroallelics. In contrast to leo23 andleoX1 mutants, which lack LEO in mushroom body and ellipsoid body neurons, animals rescued from lethality via protocol HS B contain a negligible amount of LEO throughout their heads. This lack of LEO in lethal homozygotes and heteroallelics should exaggerate putative developmental or sensory deficits present inleo23 andleoX1. However, neither sensory deficits nor anatomical aberrations were detectable in the later, despite the lack of transgene induction in larval or pupal stages. Therefore, either the 10–15% residual LEO suffices for normal larval development and the reorganization of the brain at pupariation or LEO isoforms are not required for these processes. Because transgene induction and LEO accumulation restored the learning deficit of the lethal alleles to control levels but this recovery was eliminated during decay of the protein, LEO is acutely necessary for learning. The multiple transgene inductions necessary to restore learning have been used for rescue of other behavioral mutants (DeZazzo et al., 1999) and may reflect the high level of LEO required for normal neuronal function. Indeed, as inDrosophila (Skoulakis and Davis, 1996), 14-3-3 proteins are highly abundant in vertebrate brains thought to comprise up to 1% of soluble brain protein (Moore and Perez, 1968; Boston et al., 1982; Fu et al., 2000).
Involvement of 14-3-3 proteins in multiple cellular processes may be at least in part the result of multiple isotypes or isoforms present within one cell (Skoulakis and Davis, 1998; Fu et al., 2000). This may be of particular importance in vertebrates in which nine isotypes exhibit primarily overlapping expression patterns, especially in neuronal tissues (Watanabe et al., 1991, 1993; McConnell et al., 1995;Murakami et al., 1996). Similarly, because LEO isoforms and D14-3-3ε appear to be at least partially overlapping, heterodimerization among the three 14-3-3 proteins is possible. In fact, genetic analysis suggested interactions between leonardo andD14-3-3ε gene products critical to embryonic and eye development (Chang and Rubin, 1997).
The distinct expression of leo transcripts in adult ellipsoid body and thorax indicates that LEOI and LEOII may have isoform-specific functions in these tissues and suggest that structural differences between the two isoforms may be reflected in functional specificity. The two LEO isoforms differ by five amino acids in the variable sixth α helix, (Wang and Shakes, 1996; Rittinger et al., 1999). The first two unique amino acids in LEOII (K, N in place of Q, T) are never found at that position among metazoan 14-3-3 isotypes. The third substitution (E in place of D) is present in the vertebrate ζ, β, τ, η, and ς isotypes and the two Caenorhabditis elegans isotypes. Finally, the last two amino acids (A, T in place of S, G) are present in both yeast isotypes but not among animal 14-3-3s (Wang and Shakes, 1996). Thus, the LEOII isoform appears to be a unique ζ isotype. Helix 6 does not appear to be involved in phosphopeptide binding (Rittinger et al., 1999) or dimerization (Liu et al., 1995; Xiao et al., 1995). It is unclear then whether the differences between LEOI and LEOII result in differential dimerization or substrate engagement. The mushroom bodies apparently contain both LEOI and LEOII isoforms and the ellipsoid body contains only LEOI. However, both isoforms rescue equally the olfactory learning and memory deficits of leo mutants; thus, they do not appear to have isoform-specific functions pertinent to these processes. Alternatively, subtle functional differences may have been concealed by elevated transgene expression and the accumulation of a single isoform in the mushroom bodies.
Collectively, the data indicate that monomers and homodimers of either LEO isoform and/or heterodimers with D14-3-3ε are capable of similar physiological roles essential for learning and memory. The results demonstrate that LEO proteins do not contribute to postembryonic developmental processes in the brain. This is expected to enable investigation and identification of signaling molecules engaged by each isoform in the adult mushroom body and ellipsoid body, which in turn may reveal functional differences among them. The role of Raf1 and the Ras/Raf cascade, which requires leonardo function for signaling in developmental processes (Kockel et al., 1997; Li et al., 1997), is of particular interest. Furthermore, these results establish an acute role for 14-3-3 proteins in behavioral neuroplasticity, and, by virtue of the high degree of conservation and similarly elevated neuronal expression, are directly applicable to 14-3-3 function in vertebrates.
Footnotes
This work was supported by National Science Foundation Grant IBN-0080687. We thank Dr. Sumana Datta for valuable discussions and suggestions, Lauren Vrooman for assistance with olfactory assays, and the Developmental Hybridoma Studies Bank (University of Iowa, Iowa City, IA) for antibodies.
Correspondence should be addressed to Efthimios M. C. Skoulakis, Department of Biology, c/o Department of Entomology, Texas A&M University, MS 2475, College Station, TX 77843-2475. E-mail:eskoulakis@bio.tamu.edu.
REFERENCES
- 1.Ayer RK, Carslon J. Olfatory physiology in the Drosophila antenna and maxillary palp:acj6 distinguishes two classes of odorants. J Neurobiol. 1992;8:965–968. doi: 10.1002/neu.480230804. [DOI] [PubMed] [Google Scholar]
- 2.Boston PF, Jackson P, Thomson RJ. Human 14-3-3 protein: Radioimmunoassay, tissue distribution and cerebrospinal fluid levels in patients with neurological disorders. J Neurochem. 1982;38:1475–1482. doi: 10.1111/j.1471-4159.1982.tb07928.x. [DOI] [PubMed] [Google Scholar]
- 3.Boynton S, Tully T. latheo, a new gene involved in associative learning and memoryin Drosophila melanogaster identified from P element mutagenesis. Genetics. 1992;131:655–672. doi: 10.1093/genetics/131.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broadie K, Rushton E, Skoulakis EMC, Davis RL. Leonardo, a Drosophila 14-3-3 protein involved in learning, regulates presynaptic function. Neuron. 1997;19:391–402. doi: 10.1016/s0896-6273(00)80948-4. [DOI] [PubMed] [Google Scholar]
- 5.Chang HC, Rubin GM. 14-3-3ε positively regulates Ras mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 6.Connoly JB, Roberts IJH, Armstrong JD, Kaise K, Forte M, Tully T, O'Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 7.Crittenden JR, Skoulakis EMC, Han K-A, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;5:38–51. [PMC free article] [PubMed] [Google Scholar]
- 8.Davis RL. Mushroom bodies and Drosophila learning. Neuron. 1993;11:1–14. doi: 10.1016/0896-6273(93)90266-t. [DOI] [PubMed] [Google Scholar]
- 9.Davis RL. Physiology and biochemistry of Drosophila learning mutants. Physiol Rev. 1996;76:299–317. doi: 10.1152/physrev.1996.76.2.299. [DOI] [PubMed] [Google Scholar]
- 10.DeBelle SJ, Heisenberg M. Associative odor learning in Drosophila is abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 11.DeZazzo J, Xia S, Christensen J, Velinzon K, Tully T. Developmental expression of an amn(+) transgene rescues the mutant memory defect of amnesiac adults. J Neurosci. 1999;19:8740–8746. doi: 10.1523/JNEUROSCI.19-20-08740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo J, Sandstrom D, de Belle S, Velinzon K, Smith P, Grady L, DelVecchio M, Ramaswami M, Tully T. nalyot, a mutation of the Drosophila Myb-related Adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron. 2000;27:145–158. doi: 10.1016/s0896-6273(00)00016-7. [DOI] [PubMed] [Google Scholar]
- 13.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- 14.Grotewiel MS, Beck CD, Wu K-H, Zhu X-R, Davis RL. Integrin-mediated short-term memory in Drosophila. Nature. 1997;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 15.Heisenberg M. What do the mushroom bodies do for the insect brain? An introduction. Learn Mem. 1998;5:1–10. [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- 17.Kockel L, Vorbraggen G, Jackle H, Mlodzik M, Bohnmann D (1997) Requirement for Drosophila 14-3-3ζ in cell proliferation and Raf-dependent photoreceptor development. Genes Dev 1140–1147. [DOI] [PubMed]
- 18.Li W, Skoulakis EMC, Davis RL, Perrimon N. The Drosophila 14-3-3 protein Leonardo enhances Torso signaling through D-Raf in a Ras1-dependent manner. Development. 1997;124:4163–4171. doi: 10.1242/dev.124.20.4163. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Blenkowska J, Petosa C, Collier RJ, Fu H, Liddington R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature. 1995;376:191–194. doi: 10.1038/376191a0. [DOI] [PubMed] [Google Scholar]
- 20.Luo Z, Zhang X, Rapp U, Avruch J. Identification of the 14-3-3ζ domains important for Self-association and Raf Binding. J Biol Chem. 1995;270:23681–23687. doi: 10.1074/jbc.270.40.23681. [DOI] [PubMed] [Google Scholar]
- 21.McConnell JE, Armstrong JF, Hodges PE, Bard JBL. The mouse 14-3-3ε isoform, a kinase regulator whose expression pattern is modulated in mesenchyme and neuronal differentiation. Dev Biol. 1995;169:218–228. doi: 10.1006/dbio.1995.1139. [DOI] [PubMed] [Google Scholar]
- 22.Moore BW, Perez VJ. Specific acidic proteins of the nervous system. In: Carlson FD, editor. Physiological and biochemical aspects of nervous integration. Prentice-Hall; Englewood Cliffs, NJ: 1968. pp. 343–359. [Google Scholar]
- 23.Murakami K, Situ SY, Eshete F. A gene variation of 14-3-3ζ isoform in rat hippocampus. Gene. 1996;179:245–249. doi: 10.1016/s0378-1119(96)00368-x. [DOI] [PubMed] [Google Scholar]
- 24.Muslin AJ, Xing H. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 2000;12:703–709. doi: 10.1016/s0898-6568(00)00131-5. [DOI] [PubMed] [Google Scholar]
- 25.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 26.Pawson T. Protein modules and signalling networks. Nature. 1995;16:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 27.Philip N. In vivo analysis of 14-3-3 function. MS thesis. Texas A&M University; 2000. [Google Scholar]
- 28.Rittinger K, Budman J, Xu J, Volinia S, Cantley LC, Smerdon SJ, Gamblin SJ, Yaffe MB. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol Cell. 1999;4:153–166. doi: 10.1016/s1097-2765(00)80363-9. [DOI] [PubMed] [Google Scholar]
- 29.Simon AF, Boquet I, Synguelakis M, Preat T. The Drosophila putative kinase Linotte (Derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mech Dev. 1998;76:45–55. doi: 10.1016/s0925-4773(98)00104-x. [DOI] [PubMed] [Google Scholar]
- 30.Skoulakis EMC, Davis RL. Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron. 1996;17:931–944. doi: 10.1016/s0896-6273(00)80224-x. [DOI] [PubMed] [Google Scholar]
- 31.Skoulakis EMC, Davis RL. 14-3-3 proteins in neuronal development and function. Mol Neurobiol. 1998;16:269–284. doi: 10.1007/BF02741386. [DOI] [PubMed] [Google Scholar]
- 32.Skoulakis EMC, Kalderon D, Davis RL. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993;11:197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- 33.Sokal RR, Rohlf FJ. Biometry: the principles and practice of statistics in biological research, Ed 2. Freeman; New York: 1981. [Google Scholar]
- 34.Strausfeld NJ. Atlas of an insect brain, pp 232–242. Springer; New York: 1976. [Google Scholar]
- 35.Tully T, Quinn W. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 36.Wang W, Shakes D. Molecular evolution of the 14-3-3 family. Mol Evol. 1996;43:384–398. doi: 10.1007/BF02339012. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe M, Isobe T, Okuyama T, Ichimura T, Kuwano R, Takahashi Y, Kondo H. Molecular cloning of cDNA to rat 14-3-3η chain polypeptide and the neuronal expression of the mRNA in the central nervous system. Mol Brain Res. 1991;10:151–158. doi: 10.1016/0169-328x(91)90105-7. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe M, Isobe T, Ichimura T, Kuwano R, Takahashi Y, Kondo H. Molecular cloning of rat cDNAs for β and γ subtypes of 14-3-3 protein and developmental changes in expression of their mRNAs in the nervous system. Mol Brain Res. 1993;17:135–146. doi: 10.1016/0169-328x(93)90082-z. [DOI] [PubMed] [Google Scholar]
- 39.Xiao B, Smerdon S, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature. 1995;376:188–191. doi: 10.1038/376188a0. [DOI] [PubMed] [Google Scholar]
- 40.Yin JCP, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 41.Yin JCP, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: Induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 42.Zars T. Behavioral functions of the insect mushroom bodies. Curr Opin Neurobiol. 2000;10:790–795. doi: 10.1016/s0959-4388(00)00147-1. [DOI] [PubMed] [Google Scholar]