Abstract
Proteomic tools offer a new platform for studies of complex biological functions involving large numbers and networks of proteins. Intracellular networks of proteins perform key functions in neurons and glia. The unicellular eukaryote Saccharomyces cerevisiae has been the prototype for eukaryotic proteomic studies, and when combined with genomics, microarrays, genetics, and pharmacology, new insights into the integrated function of the cell emerge. The anatomical complexity of the nervous system both in cell types and in the vast number of synapses introduces novel technical and biological issues regarding the subcellular organization of protein networks. Here we will discuss the technology of proteomics and its applications to the nervous system.
What is proteomics?
The completion of the human genome sequence was a tour de force of technology and international cooperation, but it came as a surprise to many that at first sight we have barely more genes than the fly and worm. Our human complexity must therefore be sought elsewhere, and interest has shifted to studying gene function and the way gene products interact. The large-scale study of proteins encoded by a genome has become known as “proteomics.” Proteomics has recently been expanded to include all manner of protein studies, from yeast two-hybrid protein interaction studies to structure determination by x-ray crystallography, but we suggest that it should be limited to its original interpretation, as part of functional genomics.
In this interpretation, proteomics is justified by two things: the development of high-sensitivity mass spectrometry techniques and the availability of large databases: originally protein databases, then expressed sequence tag (EST) databases, and now complete genome sequences. This combination means that with clever biology and sample preparation, proteins can be identified at the rate of several thousand per day. The problem of making sense of this wealth of information is only beginning to be addressed, but we and others have shown that powerful insights into function can be made.
Proteomics to date is anchored in mass spectrometry and bioinformatics and is about the analysis of many proteins in parallel. Importantly, in almost all cases proteins do not work alone but rather as part of larger complexes; thus proteomics lends itself to the study of the functions performed by these complexes. The proteome is dynamic and varies with time and with cellular location. Thus proteomics is a massive undertaking, and it is misleading to believe that it has an end point in the sense of completing a proteome. To bring clarity, we suggested that proteomics be divided into expression proteomics and interaction (Cell Map) proteomics.
Expression proteomics is the large-scale study of variations in protein expression and is analogous to differential gene expression. So far it is based on the relatively old technique of two-dimensional gel electrophoresis, revitalized by the ability to characterize almost all of the separated protein spots by mass spectrometry. A good gel can separate several thousand proteins, and robots are now commercially available for staining gels, spot excision, and subsequent proteolysis before mass spectrometry. Among the limitations, certain important classes of proteins, such as membrane proteins, do not readily enter gels, and because protein abundance varies over a huge range it is essential to have enrichment strategies if proteins other than “housekeeping” proteins are to be seen. As an alternative to gels, isotope-coded affinity tags (ICATs) and mass spectrometry (Gygi et al., 1999; Ideker et al., 2001) can be used. With this method, a variation in the expression of specific proteins between two samples is detected by differential mass analysis of peptides labeled with stable isotopes. It is important to point out that expression proteomics is not replaced by mRNA microarray methods because there is only a moderate correlation (r = 0.61) between changes in protein abundance and mRNA (Ideker et al., 2001). In addition, microarray methods give no insight into protein modifications such as phosphorylation.
In our opinion, using proteomic approaches to study protein complexes and signaling pathways is likely to provide a better route to understanding how proteins interact to form cellular machines. We termed this Cell Map proteomics, but it is also known as interaction or functional proteomics (Blackstock and Weir, 1999). It is not two-dimensional gel- and image analysis-based and has inherent enrichment of proteins of interest by affinity purification, so many of the problems of expression proteomics are absent. Although yeast two-hybrid approaches for the study of protein interaction have higher throughput, the interaction is not in an authentic cellular context, the incidence of false positives is high, and putative interactions require extensive validation.
Affinity enrichment is widely used in cell map proteomics, and this can be approached through the optimization of various affinity reagents with specificity toward the proteins within a complex of interest (e.g., NMDA receptor complex) (Husi and Grant, 2001a). An alternative generic strategy is the use of engineered affinity tags such as the tandem affinity purification strategy (Rigaut et al., 1999), which shows great promise. This procedure, relying on the ability to express a dual-tagged cDNA in the cell of interest and recover sufficient protein for mass spectrometry, is well established in yeast and mammalian cell lines and could be adapted for studies in the nervous system through various transgenic methods.
Proteins isolated from gels or tagged with stable isotope labels or purified complexes are now rapidly analyzed by mass spectrometry. Proteins can be analyzed at the low femtomole level, which for protein complex purification requires 108-109cells. Protein mass alone is insufficient to identify a protein, so samples are always subjected to proteolysis, usually with trypsin, and the pool of tryptic peptides is analyzed by one of two methods. If the protein is relatively pure, for example a single spot from a two-dimensional gel, then a peptide mass fingerprint generated by matrix-assisted laser desorption mass spectrometry (MALDI), may be sufficient. This approach has the advantage of being rapid but in reality is only successful for pure samples of high abundance and for fully sequenced genomes. It is also not suitable for defining post-translational modifications. Thus, MALDI is often used as a prescreen for the second and more powerful approach of low-flow HPLC tandem mass spectrometry. Here the peptide pool is partially separated by liquid chromatography (LC) and tandem mass spectrometry provides both mass and sequence information on many thousands of peptides, under complete automation. The approach is very powerful and is used for protein complexes, ICAT strategies, and, with additional LC, for the analysis of the whole yeast proteome in a single experiment (Washburn et al., 2001).
As noted, proteomics relies on mass spectrometry and on the availability of large and ever-growing databases. Many excellent software packages are available for searching mass spectrometry data against protein, EST, and genome databases. Most current efforts are going into building integrated sample management and protein identification with intelligent decision-making. Our experience is that at very low levels, manual interaction with database-searching tools is essential. Proposed interactions need validating, and although large data sets are in some ways self-validating, there remains a gap between the rate at which proteins can be identified and the rate at which they can be validated.
Functional proteomics in neuroscience
As with the emergence of any new tool, the old chestnuts in neuroscience will be subject to another round of interrogation. We will briefly discuss some of the more obvious applications and raise issues specific to the nervous system. First, at the level of individual cells in the nervous system, by far the most attention has been historically focused on the electrical properties of neurons and their connections at synapses. And here the majority of this study has focused on ion channels and neurotransmitter receptor subunits, which are well known to be subject to regulatory phosphorylation. Identification of phosphorylation sites has traditionally been performed using radioisotope labeling, which is difficult in tissues. Mass spectrometry, which detects the mass of the phosphate group, is rapidly replacing isotopic methods (Larsen and Roepstorff, 2000) and will guide the generation of new phosphospecific antibodies, which are now a staple of the signal transduction biologist.
Simply identifying the presence of proteins in key compartments within neurons and glia will provide an essential framework for understanding function. The synaptosome fractionation method opened up biochemical approaches to purification of synaptic proteins (Cotman and Matthews, 1971; Gombos et al., 1971; Soifer and Whittaker, 1972). Synaptic compartments [synaptic vesicles and postsynaptic densities (PSDs)] are two notable fractions on the presynaptic and postsynaptic side, respectively. The expression proteomic approach of identification of individual proteins with Edman sequencing and mass spectrometry has so far yielded ∼30 proteins (Walikonis et al., 2000), in contrast to the functional proteomic approach, which suggests that the molecular complexity of the PSD is far higher (Husi et al., 2000). These neuronal fractions and specialized glial structures such as the paranodal axoglial junction are ripe for proteomic analysis.
In the same way that mRNA microarray methods can be used for expression profiling in brain diseases, expression proteomics has similar applications. Microarrays are being widely used in disease profiling (Mirnics et al., 2001) and, in the case of a study of schizophrenia (Mirnics et al., 2000), have led to the identification of several candidate molecules known to be involved with presynaptic function. In contrast, a proteomic study (Edgar et al., 2000) identified a different set of molecules. It is too early to systematically compare the approaches; however, as suggested by experimental systems, the correlation between mRNA and protein levels is modest, and it seems that a more integrative analysis taking into consideration both sets of data with other bioinformatic information will be more productive.
Functional proteomics and molecular networks
The multiprotein complex of intracellular proteins associated with receptors and channels is well known to be involved with signal transduction both at the level of modulating the receptor/channel (Browning et al., 1985; Levitan, 1985) and in functionally driving downstream pathways connecting to plasticity machinery within the neuron (Migaud et al., 1998; Grant and O'Dell, 2001). Mass spectrometry, antibody, and yeast two-hybrid methods are well established tools for characterizing these complexes and are approaches that can be extended to all other nervous system receptors and channels (Husi and Grant, 2001b).
As discussed above, proteomic methods can produce overwhelming quantities of data. These often take the form of lists of proteins such as those found in the NMDA receptor complex (Husi et al., 2000) or lists of interacting proteins found in genome-wide yeast two-hybrid screens (Schwikowski et al., 2000). Bioinformatic annotation of these lists provides a powerful insight into the function of individual molecules as well as new ideas about the molecular basis of cellular functions. For example, approximately one-third of the ∼75 proteins found to be associated with the NMDA receptor were known previously to be involved with the induction of long-term potentiation or long-term depression. This indicates that the general function of these complexes is in the induction of plasticity, but also predicts that associated proteins of unknown physiological function will also participate in plasticity. This message that function is predicted from knowledge of interaction partners also emerged from an analysis of 2709 protein interactions in the yeast S. cerevisiae.
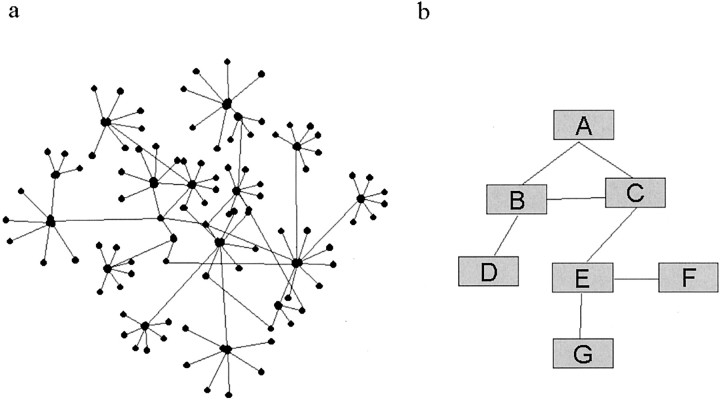
One of the first surprises of this cell-wide study of yeast interactions was that a single large network of 2358 interactions between 1538 proteins was made and the next largest network contained only 19 proteins (Schwikowski et al., 2000; Tucker et al., 2001). Proteins of similar function were close to each other and organized into clusters separated by no more than two other proteins. This allows one to predict the function for a novel protein clustered with those of known function. Figure 1 shows a schematic example of the organization of these networks of individual interacting proteins and the functional group interaction map.
Fig. 1.
Schematic of protein interaction networks in a simple cell. a, A simple hypothetical protein interaction map for 100 proteins. Each protein is shown as ablack dot, and connections between pairs of proteins are indicated by single lines. This is a simple adaptation of a 1200 protein interaction map from Schwikowski et al. (2000).b, Functional group interaction map. Because most interactions are between proteins in the same functional class (Schwikowski et al., 2000), it is possible to summarize the map ina to boxes with a functional definition. In this arbitrary example we have connected various boxes, which on the basis of a published yeast interaction study could be: A, Amino acid metabolism; B, protein degradation;C, cell cycle control; D, signal transduction; E, cell polarity; F, vesicular transport; G, cell structure.
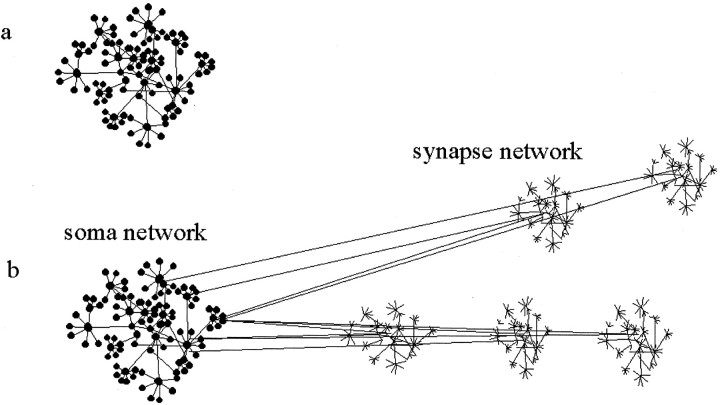
These networks or maps of protein interactions and function have not been produced for neurons to date; however, they will need to take into consideration the dramatic contrast in spatial organization of the two cells, spherical yeast versus stellate neurons. Simply put, the synapse will be the location of specific networks (here referred to as synaptic networks), which are satellites connected to a single network at the soma (soma networks) (Fig. 2). The soma network may share many common features with that described from yeast because that is a generic cell. Moreover, the synaptic networks may have clusters of proteins (functional sets) that are also found within the soma network, such as those involved with vesicular transport, protein synthesis, or signal transduction.
Fig. 2.
A hypothetical neuron interaction map.a, A map of a simple spherical cell as shown in Figure1a. b, A map of interactions superimposed on the subcellular architecture of neurons. The soma network is the same as that for the spherical cell, and five individual synapse protein interaction maps are shown. The synapse networks could under many conditions behave autonomously, and under other conditions interact with the soma network. The spatial organization of neuronal architecture will present novel regulatory features as well as pose methodological challenges.
Molecular network maps of neurons with soma networks connected to synaptic networks will provide a wealth of interesting questions, many of which lend themselves to computational models. A diverse set of proteomic approaches is relevant to the study of protein networks in neurons. The levels of individual proteins, interactions, phosphorylation, and activity (enzyme assays) could be monitored and, where possible, preferably in live cells. The cell biology of protein networks will move away from the study of a single protein and move toward studies of multiple proteins simultaneously. Yeast studies suggest that a highly integrated approach to the study of networks is required where genetics, expression arrays, and proteomics are combined (Ideker et al., 2001). Perhaps the most exciting aspect of studying the function of intracellular molecular networks will be in understanding how they contribute to neuronal networks at the level of circuits and how these intracellular networks regulate behavior.
Footnotes
Correspondence should be addressed to Dr. Seth Grant, Department of Neuroscience, Edinburgh University, 1 George Square, Edinburgh EH8-9JZ, UK. E-mail: seth.grant@ed.ac.uk.
REFERENCES
- 1.Blackstock WP, Weir MP. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–127. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 2.Browning MD, Huganir R, Greengard P. Protein phosphorylation and neuronal function. J Neurochem. 1985;45:11–23. doi: 10.1111/j.1471-4159.1985.tb05468.x. [DOI] [PubMed] [Google Scholar]
- 3.Cotman CW, Matthews DA. Synaptic plasma membranes from rat brain synaptosomes: isolation and partial characterization. Biochim Biophys Acta. 1971;249:380–394. doi: 10.1016/0005-2736(71)90117-9. [DOI] [PubMed] [Google Scholar]
- 4.Edgar PF, Douglas JE, Cooper GJ, Dean B, Kydd R, Faull RL. Comparative proteome analysis of the hippocampus implicates chromosome 6q in schizophrenia. Mol Psychiatry. 2000;5:85–90. doi: 10.1038/sj.mp.4000580. [DOI] [PubMed] [Google Scholar]
- 5.Gombos G, Morgan I, Breckenridge WC, Breckenridge JE, Vincedon G. Isolation and biochemical structure of synaptosomal membrane (in French). C R Seances Soc Biol Fil. 1971;165:499–506. [PubMed] [Google Scholar]
- 6.Grant SG, O'Dell TJ. Multiprotein complex signaling and the plasticity problem. Curr Opin Neurobiol. 2001;11:363–368. doi: 10.1016/s0959-4388(00)00220-8. [DOI] [PubMed] [Google Scholar]
- 7.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 8.Husi H, Grant SG. Isolation of 2000-kDa complexes of N-methyl-d-aspartate receptor and postsynaptic density 95 from mouse brain. J Neurochem. 2001a;77:281–291. doi: 10.1046/j.1471-4159.2001.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- 9.Husi H, Grant SG. Proteomics of the nervous system. Trends Neurosci. 2001b;24:259–266. doi: 10.1016/s0166-2236(00)01792-6. [DOI] [PubMed] [Google Scholar]
- 10.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 11.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 12.Larsen MR, Roepstorff P. Mass spectrometric identification of proteins and characterization of their post-translational modifications in proteome analysis. Fresenius J Anal Chem. 2000;366:677–690. doi: 10.1007/s002160051562. [DOI] [PubMed] [Google Scholar]
- 13.Levitan IB. Phosphorylation of ion channels. J Membr Biol. 1985;87:177–190. doi: 10.1007/BF01871217. [DOI] [PubMed] [Google Scholar]
- 14.Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 15.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 16.Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486. doi: 10.1016/s0166-2236(00)01862-2. [DOI] [PubMed] [Google Scholar]
- 17.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 18.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 19.Soifer D, Whittaker VP. Morphology of subcellular fractions derived from the electric organ of Torpedo. Biochem J. 1972;128:845–846. doi: 10.1042/bj1280845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tucker CL, Gera JF, Uetz P. Towards an understanding of complex protein networks. Trends Cell Biol. 2001;11:102–106. doi: 10.1016/s0962-8924(00)01902-4. [DOI] [PubMed] [Google Scholar]
- 21.Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Washburn MP, Wolters D, Yates JR., III Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]