Abstract
Amyloid-β, the major constituent of senile plaques in Alzheimer's disease, is derived from the amyloid precursor protein (APP) by proteolysis. Kunitz protease inhibitor (KPI) containing forms of APP (APP751/770) interact with a multifunctional endocytic receptor, the low-density lipoprotein receptor-related protein (LRP), which modulates its proteolytic processing affecting production of amyloid-β. We used fluorescence resonance energy transfer (FRET) using labeled LRP and APP in H4 cell line to examine the subcellular localization and the molecular domains involved in the APP-LRP interaction. KPI-containing forms of APP (APP770) demonstrated FRET with LRP that was sensitive to the LRP inhibitor receptor-associated protein (RAP), suggesting an interaction between the extracellular domains of APP770 and LRP. APP695 also interacts with LRP to lesser degree (as measured by extracellular domain probes), and this ectodomain interaction is not altered by RAP. By using C-terminally tagged LRP and APP, we demonstrate a second site of interaction between the C termini of both APP695 and APP770 and the C terminus of LRP, and that the interactions at these regions are not sensitive to RAP. We next examined the possibility that the C-termini APP-LRP interaction was mediated by Fe65, an adaptor protein that interacts with the cytoplasmic tails of LRP and APP. FRET studies confirmed a close proximity between the amino Fe65 phosphotyrosine binding (PTB) domain and LRP cytoplasmic domain and between the carboxyl Fe65 PTB domain and the APP cytoplasmic domain. These findings demonstrate that LRP interaction with APP occurs via both extracellular and intracellular protein interaction domains.
Keywords: amyloid precursor protein, APP, low-density lipoprotein receptor-related protein, LRP, adaptor protein Fe65, fluorescent resonance energy transfer, FRET, protein–protein interaction, secretory pathway, immunocytochemistry, confocal microscopy
The pathological hallmarks of Alzheimer's disease are senile plaques and neurofibrillary tangles. The senile plaques contain amyloid-β, which is generated by proteolysis of the amyloid precursor protein (APP) (for review, seeSelkoe, 1998; De Strooper and Annaert, 2000). APP is a type I integral membrane protein in three major isoforms generated by alternative splicing; APP695 is most abundantly expressed in the neurons, and two other splice variants, APP751 and APP770, contain a Kunitz-type protease inhibitor domain (KPI domain). APP is cleaved by α- or β-secretases, releasing the 100–120 kDa ectodomain of APP (soluble APP) and 10–12 kDa membrane-bound C-terminal fragments. An additional cleavage in the transmembrane domain by γ-secretase results in secretion of 4 kDa amyloid-β peptides.
Three pathways have been described that can lead to the processing of APP into amyloid-β: within the secretory pathway in the endoplasmic reticulum–intermediate compartment pathway (Cook et al., 1997;Hartmann et al., 1997), a trans-Golgi network pathway (Xu et al., 1997), and the endosomal–lysosomal pathway (Haass et al., 1992;Koo and Squazzo, 1994). Recent data suggest that endocytosis of APP holoprotein from the cell surface is an important step in amyloid-β synthesis in some systems (Perez et al., 1999).
The low-density lipoprotein (LDL) receptor-related protein (LRP) is a member of the LDL receptor family and is a ∼600 kDa single transmembrane endocytic receptor. The extracellular domain contains four distinct ligand binding sites. It mediates internalization and degradation of ligands involved in metabolic pathways of lipoproteins and protease–protease-inhibitor complexes (Strickland et al., 1995), including α-2-macroglobulin (Borth 1992), apolipoprotein E (Herz et al., 1988), and KPI domain in tissue factor plasminogen activator (Bu et al., 1992). The binding of ligands to LRP can be blocked by the receptor-associated protein (RAP) (Herz et al., 1991; Williams et al., 1992).
It has been demonstrated previously that LRP can serve as an internalization receptor for the KPI-containing (but not APP695) isoforms of soluble APP (Kounnas et al., 1995). These data led to the hypothesis that the extracellular domains of LRP and APP interact via a ligand–ligand binding domain interaction, and subsequent data supported the idea that this interaction could also mediate APP770-LRP interactions for cell surface APP, leading to APP endocytosis (Knauer et al., 1996; Ulery et al., 2000).
Our current studies examine the interaction of APP and LRP using confocal microscopy-based fluorescence resonance energy transfer (FRET) techniques that provide both exquisite subcellular localization and information about protein–protein interactions in cells. We demonstrate that there are clear interactions at the cell surface and in the secretory pathway between LRP and APP770, and, surprisingly, between LRP and APP695. Studies of interactions of both the ectodomains and the intracellular domains show that there are two distinct sites of interaction between APP and LRP: an RAP-sensitive extracellular interaction between the APP KPI domain and a ligand binding domain of LRP, and an RAP-insensitive intracellular interaction between the C terminus of APP and the C terminus of LRP. The latter may be mediated partly by intracellular adaptor proteins such as Fe65.
MATERIALS AND METHODS
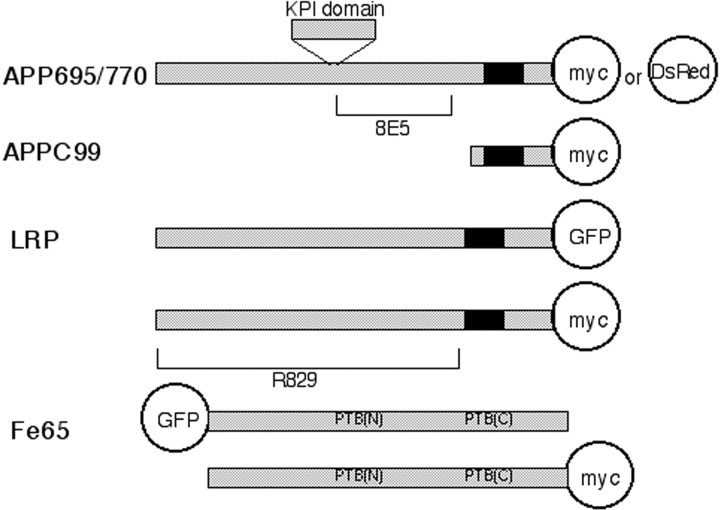
Generation of expression constructs and deletion mutants for human APP, LRP, Fe65, and RAP. Human APP770, APP695 cDNA lacking the termination codon was generated by PCR with a set of primers: 5′-ATAGCTAGCCACCATGCTGCCCGGTTTG-3′ and 5′-TCGTCGACCTGTTCTGCATCTGCTCAAA-3′. The PCR product was digested with the restriction enzymes NheI and SalI and ligated in-frame to the NheI and SalI site of an expression vector: pEGFP-N1 (Clontech, Palo Alto, CA) and pDsRed-N1 (Clontech) (for reagent information, see Fig.1).
Fig. 1.
Reagents used in this study. This scheme shows the constructs used in this study. The membrane-spanning regions are shown in black. Both APP695 and APP770 are tagged at the C terminus with either myc or DsRed. The N-terminus truncated APPC99 is tagged with myc at the C terminus. Full-length LRP is tagged at the C terminus with either myc or EGFP (shown as GFP). The Fe65 construct is tagged either with EGFP at the N terminus or myc at C terminus. For the study of ectodomain interactions, anti-APP antibody (8E5) and anti-LRP antibody (R829), both of which are raised against the extracellular domain of APP or LRP, respectively, are used. For intracytoplasmic interaction, the combination of myc (detected with Cy3) and EGFP or the combination of DsRed and EGFP is used.
To make myc-tagged constructs for APP770, APP770 cDNA lacking the termination codon was generated by PCR and inserted into the HindIII site of pcDNA3.1B (Invitrogen, Carlsbad, CA) in-frame. To make the myc-tagged construct for APP695, human APP695 cDNA lacking the termination codon was generated by PCR with a set of primers: 5′-TTAAAATTTGCTAGCACCGCCATGCTGCCCGG-3′ and 5′-TTTAGCGGCCGCGTTCTGCATCTGCTCAAA-3′. The PCR product was digested withNheI and NotI and ligated into theNheI and NotI site of pcDNA3.1A (Invitrogen).
A deletion mutant containing only the C-terminal 99 amino acids of full-length APP (APPC99) was generated by PCR with a set of primers: 5′GCAAGCTTGCAGAATTCCGACATGACTCAGGA-3′ and 5′-TTCAAGAAACTCGTCTACGTCTTGCGAGCTCCG-3′. The PCR products were digested with HindIII and XhoI and ligated into the HindIII and XhoI site of an expression vector pSecTag2B (Invitrogen).
Authenticity of all PCR-generated constructs was confirmed by DNA sequencing.
Constructs of LRP-EGFP (enhanced green fluorescent protein) was generated from ligating human LRP cDNA digested with restriction enzymes of XhoI and Bsu36I and synthetic oligomers of 5′-TGAGGACGAGATAGGGGACCCCTTGGCAA-3′ and 5′-AGCTTTGCCAAGGGGTCCCCTATCTCGTCC-3′ containing the last part of LRP without a stop codon and HindIII site. These were then inserted into the XhoI and HindIII site of expression vector pEGFP-N1 (Clontech). LRP in pcDNA3.1A (Invitrogen) was also constructed in a similar manner (LRP-myc).
The generation of Fe65-myc, which has myc at its C terminus, has been reported previously (Borg et al., 1996). This construct lacks N-terminal 52 amino acids but maintains the WW and PTB domains. The Fe65-myc construct was digested by SalI and inserted into the SalI site of the pEGFP-C3 vector (Clontech) to generate a GFP-Fe65 construct.
Human RAP cDNA that was originally cloned into pGEX-2T (Amersham Pharmacia Biotech, Piscataway, NJ) vector (Williams et al., 1992) was cut by BamHI and EcoRI, recloned into theBamHI and EcoRI site of the pcDNAINeo vector (Invitrogen), and then further recloned into the BamHI andXhoI sites of the expression vector pcDNA3 (Invitrogen).
Antibodies and proteins. Human RAP was expressed in bacteria as a fusion protein with glutathione S-transferase (GST) as described previously (Williams et al., 1992). Cleavage through thrombin and purification of recombinant RAP was performed as described previously (Williams et al., 1992). The rabbit polyclonal R829 against LRP was raised in the same way as R777 as described previously (Kounnas et al., 1992). Monoclonal antibody 8E5 was raised against 520–668 residues of APP770 and was a gift from Dr. P. Seubert (Elan Pharmaceuticals, South San Francisco, CA). Monoclonal antibody 22C11 raised against 60–100 residues of APP was purchased from Chemicon (Temecula, CA). Mouse monoclonal anti-myc antibody was purchased from Invitrogen. Rabbit polyclonal anti-myc antibody was purchased from Upstate Biotechnology (Lake Placid, NY). Mouse monoclonal antibody 7F1 recognizes RAP (Kounnas et al., 1992).
Cell culture conditions and transient transfection. H4 cells derived from human neuroglioma cells used in this study were obtained from the American Type Culture Collection (Manassas, VA). H4 cells were cultured in OPTI-MEMI with 10% fetal bovine serum, and Chinese hamster ovary (CHO) cells were cultured in Ham's–F-12 with 10% fetal bovine serum. Transient transfection of H4 cells were performed using a liposome-mediated method (FuGene 6; Roche Molecular Biochemicals, Indianapolis, IN). Cells were plated onto four-well chambers 1 d before the transfection. First, a mixture of 1 μg of plasmid DNA and 3 μl of Fugene6 was made in 100 μl of DMEM and left for 15–30 min at room temperature, and then 25 μl of this mixture was added to the medium in each well. The incubation time was from 24 to 48 hr. Double transfection of APP and LRP plasmids was done in the same way. To investigate the possible inhibition by RAP, some cells were also triple-transfected with APP, LRP, and RAP.
Immunohistochemistry. Immunostaining of cells was performed 24–48 hr after transfection. Cells were fixed in 4% paraformaldehyde (PFA) for 10 min, washed in Tris-buffered saline (TBS), pH 7.3, permeabilized by 0.5% Triton X-100 for 20 min, and blocked with 1.5% normal goat serum for 1 hr.
To detect C-terminal interactions, cotransfected cells of APP770-myc (or APP695-myc or APPC99-myc) and LRP-EGFP were used. Cells were immunostained by mouse anti-myc monoclonal antibody (1:1000 dilution; Invitrogen) for 1 hr at room temperature. Cells were then washed three times in TBS and labeled by Cy3-conjugated anti-mouse antibody (10 μg/ml; Jackson ImmunoResearch, West Grove, PA) for 1 hr at room temperature. Immunostained cells were stored in TBS at 4°C until the confocal imaging was performed. To confirm the results, the C-terminal interaction was also studied by APP770-DsRed (or APP695-DsRed) and LRP-EGFP double-transfected cells.
For the N-terminal interactions, cotransfected cells of APP770-myc (or APP695-myc) and LRP-myc were used. These cells were labeled with antibodies against ectodomains of APP and LRP, 8E5 (1:1000) and R829 (1 μg/ml), respectively, and then labeled by Cy3-conjugated anti-mouse antibody and FITC-conjugated anti-rabbit antibodies, respectively.
To confirm the APP-LRP interaction in the secretory pathway, double-transfected cells of APP-myc and LRP-GFP were counterstained with mouse monoclonal anti-GM130 antibody, which stains the Golgi apparatus (1 μg/ml; Becton Dickinson/Transduction Laboratory, San Diego, CA). GM130 was visualized by Cy5-conjugated anti-mouse antibody (10 μg/ml; Jackson ImmunoResearch). Golgi staining was confirmed by confocal microscopy using the 647 line of the krypton–argon laser.
To detect interactions occurring on the cell membrane, cells were incubated with primary antibodies against the extracellular domain (8E5 and R829) in the culture media for 1 hr on ice, then washed with PBS, and fixed in 2% PFA for 10 min without permeabilization. Secondary antibodies conjugated to Cy3 or FITC were applied to visualize the primary antibodies.
RAP treatment was performed in two ways. To study APP-LRP interactions within the secretory pathway, RAP was cotransfected into cells. To observe the transfection efficiency of RAP, RAP was also stained with anti-RAP antibody 7H1 (1:1000) and conjugated with Alexa350 anti-mouse antibody (10 μg/ml; Molecular Probes, Eugene, OR) in the cells triple-transfected cells of APP-DsRed or APP-myc, LRP-EGFP, and RAP. For the cells with APP-myc, rabbit polyclonal anti-myc antibody was used instead. The RAP staining on the triple-transfected cells was confirmed by the 4′,6′-diamidino-2-phenylindole filter of the confocal microscope. To study cell surface APP-LRP interactions, a solution of 500 nm recombinant RAP (Williams et al., 1992) was applied in the culture media 24 hr after transient transfection, and the same assay was performed 24 hr after addition of RAP.
FRET. Immunostaining was observed using a Bio-Rad (Hercules, CA) 1024 confocal microscope mounted on a Nikon (Tokyo, Japan) Eclipse TE300 inverted microscope; the krypton–argon laser (emission, 488 and 568 nm lines) was used to excite the fluorescein or EGFP and Cy3 or DsRed, respectively.
FRET was measured using a method developed for laser-scanning confocal microscopy (Knowles et al., 1999; McLean et al., 2000), which is analogous to the technique used by Rocheville et al. (2000) to study interactions of receptor subunits in cells. The energy transfer was detected as an increase in donor fluorescence (FITC or EGFP) after complete photobleaching of the acceptor molecules (DsRed or Cy3). This technique is also referred to as donor dequenching (Kenworthy and Edidin, 1998; Siegel et al., 2000). The amount of energy transfer was calculated as the percentage of increase in donor fluorescence after acceptor photobleaching; initial scan was obtained at low-laser energy using the 488 line of the krypton–argon laser to record the fluorescein (or EGFP) signal. A second scan was performed with the 568 line, and the area of colocalization was noted. A small part of the cells (∼5 × 5 μm) was then photobleached with intense 568 nm light (laser power 100%) to destroy the acceptor molecules. The cells were then rescanned using 488 nm light. An increase of the fluorescein (or EGFP) within the photobleached area was used as a measure of the amount of FRET present. Exposing single-labeled FITC or EGFP cells to 568 nm light for equivalent times did not alter the amount of fluorescein emission. The ratioFlD2/FlD1(whereFlD2/FlD1indicates the ratio of donor fluorescence after photobleaching to donor fluorescence before photobleaching) was compared with the null hypothesis value of 1.0 by one-group t tests. Comparison of multiple groups was by ANOVA, with Fisher's PLSD post hoctest for significance. FRET can be detected only if the two fluorophores are in close physical proximity: between fluorophores (e.g., EFGP-DsRed) to be less than ∼10 nm. In experiments in which the epitopes are labeled by indirect immunofluorescence, the fluorophores must be <10 nm apart, and the distance between epitopes can be (maximally) 30 nm (e.g., using the R829–8E5 pair) (Chin et al., 2000). This provides a >10-fold increase in resolution compared with double staining with conventional confocal microscopy, which has a resolution of ∼500 nm.
Negative controls for FRET experiments included the following: no FRET was observed between APP-GFP and APP-DsRed when cotransfected; no FRET was observed if the primary or the secondary antibody was omitted; and no FRET was observed when APP-GFP was immunostained with 22C11 against the distal N-terminal end of APP, despite the complete colocalization. Thus, we conclude that the donor dequenching observed after photobleaching of the acceptor reflects FRET.
RESULTS
The H4 human neuroglioma cell line was used in this study, because it has been used for the study of APP processing (Kuentzel et al., 1993) and for investigating the effect of LRP on this process (Ulery et al., 2000; Rebeck et al., 2001). Endogenous APP and LRP, visualized by anti-APP (8E5) and anti-LRP (R829) antibodies, were weakly visible in the cell body of H4 cells using confocal microscopy. Transfection of H4 cells with various LRP and APP constructs revealed similar patterns of immunostaining in the transfected cells when compared with the parental cells. No apparent differences in distribution of the overexpressed proteins compared with endogenous proteins were detected, and, in the current study, the FRET experiments were all performed in H4 cells transfected with plasmids encoding tagged proteins.
Interaction between ectodomains
Previous experiments showed that soluble forms of APP containing the KPI domain is a ligand for LRP (Kounnas et al., 1995; Knauer et al., 1996). We therefore initially investigated FRET between the ectodomains of APP and LRP in intact H4 cells. Cells cotransfected with APP770-myc and LRP were immunostained with monoclonal anti-APP (8E5) and polyclonal anti-LRP (R829) antibodies (both raised against ectodomains) and then labeled by Cy3-conjugated anti-mouse antibody and FITC-conjugated anti-rabbit antibody, respectively. The amount of FRET was calculated as the percentage of increase in donor fluorescence (FITC) after acceptor (Cy3) photobleaching (FlD2/FlD1). The presence of FRET suggests that there is a close apposition between the ectodomains (ratio ofFlD2/FlD1= 1.47 ± 0.06; mean ± SE; n = 14;p < 0.0001) (Fig. 2). Surprisingly, FRET occurs not only on the cell surface but also in intracellular compartments, including the Golgi (confirmed by counterstaining with GM130; data not shown) and endoplasmic reticulum. As a negative control, FRET was assessed using the APP770-myc-transfected cells labeled with monoclonal 8E5 directed against an extracellular APP domain and a polyclonal anti-myc antibody directed against an intracellular domain of APP770-myc; although there was absolute colocalization of the two fluorophores, no FRET was observed because the distance across the membrane is too far to allow FRET. Similarly, exposure of single-labeled FITC stained cells to intense 568 light did result in a change in FITC intensity. Thus, the increase in FITC brightness after acceptor photobleaching indicates a sufficiently close association between the FITC and Cy3 fluorophores to support FRET.
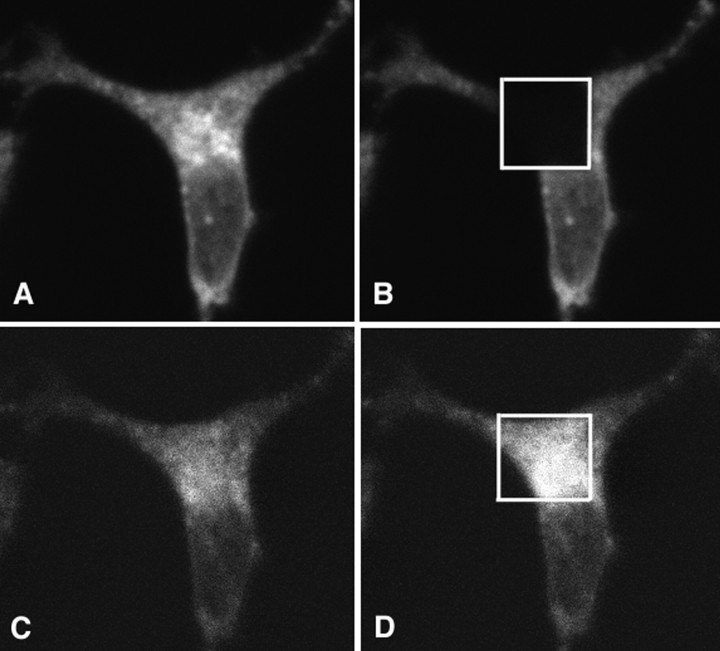
Fig. 2.
Extracellular domains of APP and LRP are closely associated with each other. H4 cells were cotransfected with APP770-myc and LRP-myc, immunostained with anti-APP (8E5) and anti-LRP (R829) antibodies, and visualized with Cy3 and FITC, respectively. Here is shown a typical example of FRET between ectodomains of APP770 and LRP.A, Cy3 (APP770) signal after 568 nm excitation.B, A discrete area of the cell was photobleached using intense 568 nm laser. C, FITC signal (LRP) using 488 nm excitation before photobleaching. D, FITC signal (LRP) using 488 nm excitation after photobleaching of the acceptor fluorophore (Cy3) with intense 568 nm laser light. An increase in donor fluorescence is observed within the discrete area that was photobleached, showing the presence of FRET. In this example, the FITC signal increased by 51%.
To determine whether the APP770-LRP interaction occurred on the cell surface, H4 cells were cotransfected with APP770-myc and LRP. The 8E5 and R829 antibodies against the ectodomains of APP and LRP were then incubated with the transfected cells at 4°C to allow binding but to prevent cellular mediated internalization. Cells were then fixed but not permeabilized, and labeled secondary antibodies were added. APP and LRP were observed on the plasma membrane by confocal microscopy (Fig.3). FRET was also present between APP770 and LRP on the cell membranes, as revealed by increased FITC fluorescence after photobleaching of Cy3. The FRET ratio increase (FlD2/FlD1) of 1.40 ± 0.04 (n = 23; p < 0.0001) was essentially the same as that determined after routine fixation and staining of cells (Fig. 2). This result suggests a close proximity between the ectodomains of APP770 and LRP on the plasma membrane, consistent with an interaction between these two molecules on the cell surface.
Fig. 3.
Extracellular domains of APP and LRP displayed FRET at the cell surface. H4 cells were cotransfected with APP770-myc and LRP-myc, immunostained with anti-APP (8E5) and anti-LRP (R829) antibodies without permeabilization, and visualized with Cy3 and FITC, respectively. APP and LRP on the cell surface were observed.A, Cy3 (APP770) signal using 568 nm excitation.B, Discrete area of the cell was photobleached using intense 568 nm laser. C, FITC signal (LRP) using 488 nm excitation before photobleaching. D, FITC signal (LRP) using 488 nm light after photobleaching the acceptor (Cy3) with intense 568 nm laser light. An increase in donor fluorescence is observed within the discrete area that was photobleached, showing the presence of FRET on the cell membrane. In this example, the FITC signal increased by 38%.
We next asked whether the association of APP770 and LRP occurred via an interaction of APP with the ligand binding region of LRP. This was determined by measuring whether the presence of FRET was sensitive to RAP, because RAP is known to inhibit the binding of all LRP ligands (including soluble APP) to LRP (Kounnas et al., 1995). H4 cells were cotransfected with RAP, APP770-myc, and LRP-GFP. No change in subcellular distribution was observed for APP or LRP. The FRET ratio observed for the APP770-LRP ectodomain interaction was significantly diminished when H4 cells were cotransfected with RAP, APP770-myc, and LRP-GFP (FlD2/FlD1= 1.17 ± 0.03; n = 15; p < 0.005 compared with cells transfected with APP770-myc and LRP-GFP), but surprisingly FRET was not reduced to 1.0 (Fig.4). Thus, the FRET observed between APP770 and LRP is partially, but not completely, inhibited by RAP. Similarly, the APP-LRP interaction at the cell membrane was partially inhibited by exogenous addition of RAP, which was shown as a decrease of FRET ratio (FlD2/FlD1= 1.16 ± 0.01; n = 22; p < 0.001 compared with cells transfected with APP770 and LRP-GFP not treated with RAP). This suggests that, on the plasma membrane and within intracellular compartments such as the Golgi and endoplasmic reticulum, there may be two components of APP770-LRP interactions: one APP770-LRP interaction that is RAP-sensitive, and another APP770-LRP interaction that is RAP-insensitive. The existence of an RAP-insensitive interaction between APP and LRP is interesting, because previous work using soluble forms of APP revealed that RAP was able to block the interaction (Kounnas et al., 1995). The RAP-insensitive interaction could be attributable to the presence of another (RAP-insensitive) site of interaction in the extracellular domains or, more likely, to an intracellular interaction between the C-terminal tails of APP and LRP.
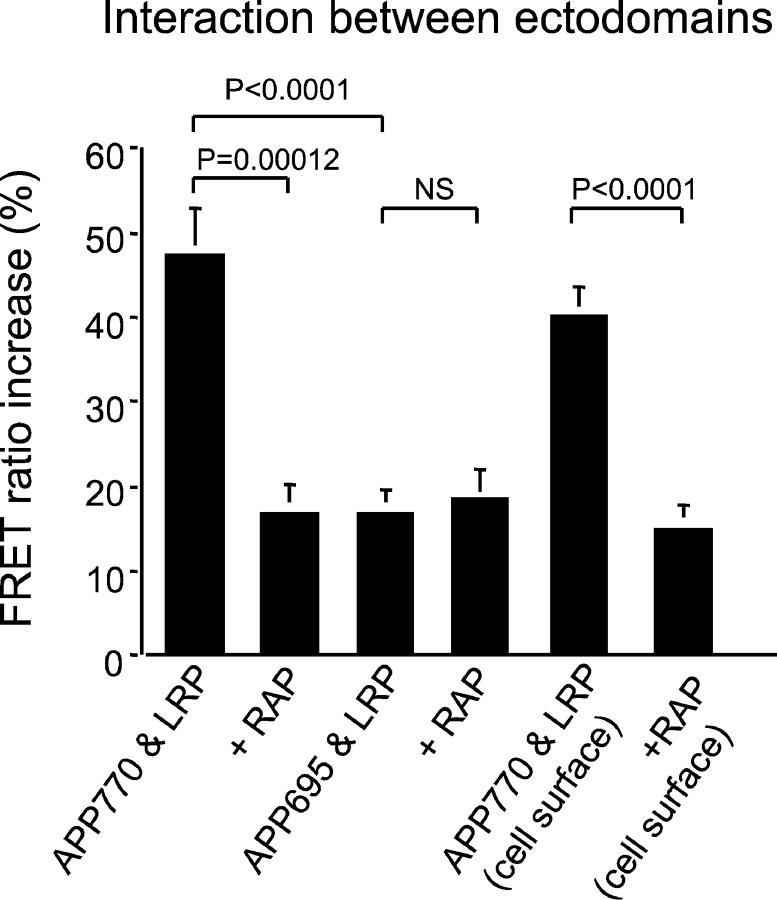
Fig. 4.
The FRET ratio increase of ectodomains is shown as a percentage of increase of donor fluorescence between APP (APP770 and APP695) and LRP. Both APP770 and APP695 demonstrated increases in donor fluorescence significantly above zero (p < 0.0001; one-group t test), but the magnitude of the FRET ratio increase of APP695 was significantly less than that of APP770 (p < 0.0001; ANOVA; Fisher's PLSDpost hoc test). Cotransfection with RAP significantly decreased the FRET ratio between APP770 and LRP (p = 0.00012; ANOVA; Fisher's PLSDpost hoc test) but not APP695 and LRP. The APP695-LRP ectodomain interaction was not affected by RAP. The FRET ratio increase on the cell membrane is almost the same as that of the routinely fixed and permeabilized cells. Cotransfection with RAP also significantly decreased the FRET ratio between APP770 and LRP at the cell surface (p < 0.0001; ANOVA; Fisher's PLSDpost hoc test).
These two possibilities were examined next. To test the former possibility, cells were cotransfected with LRP and APP695-myc, which lacks the KPI domain. We detected FRET between APP695 and LRP (FlD2/FlD1= 1.17 ± 0.03; n = 16; p < 0.0001), which was highly statistically significant, but the magnitude of the FRET was significantly less than that observed for APP770 and LRP (p < 0.0001). Unlike APP770, the APP695-LRP ectodomain interaction was not affected by cotransfection with RAP (FlD2/FlD1= 1.18 ± 0.04; n = 9). Together with the previous experiments, the results are consistent with the hypothesis that there is both an RAP-sensitive and an RAP-insensitive component of the interaction between APP and LRP, and that the RAP-sensitive component is dependent on the presence of the KPI domain.
Interaction between cytoplasmic domains
One possible mechanism that might account for an RAP-insensitive APP-LRP interaction would be an interaction between the intracellular C-terminal domains of LRP and APP. To examine this possibility, cells were transfected with APP and LRP plasmids tagged with either fluorescent markers (EGFP or DsRed) or myc at the C termini. The amount of FRET was again calculated as the percentage of increase in donor fluorescence (EGFP) after acceptor (Cy3 or DsRed) photobleaching. As shown in Figure 5, strong FRET was observed between the C termini of APP770-myc (labeled with Cy3) and LRP-EGFP (FlD2/FlD1= 1.46 ± 0.0.05; mean ± SE; n = 14;p < 0.0001), suggesting the presence of close association between the cytoplasmic domains of these molecules throughout the cell. The same result was obtained with APP770-DsRed and LRP-EGFP (FlD2/FlD1= 1.55 ± 0.05; n = 10; p < 0.0001), excluding the possibility of false positive with the use of secondary antibodies. As a negative control, we also examined FRET between APP770-EGFP and APP770-DsRed and found it to be absent (FlD2/FlD1= 1.01 ± 0.005; n = 10; not significant). Cotransfection with RAP partially inhibited the interaction and significantly decreased the FRET ratio of APP770-myc and LRP-EGFP (FlD2/FlD1 = 1.18 ± 0.04; n = 16; p < 0.0001 compared with no RAP) but again did not reduce the interaction to zero.
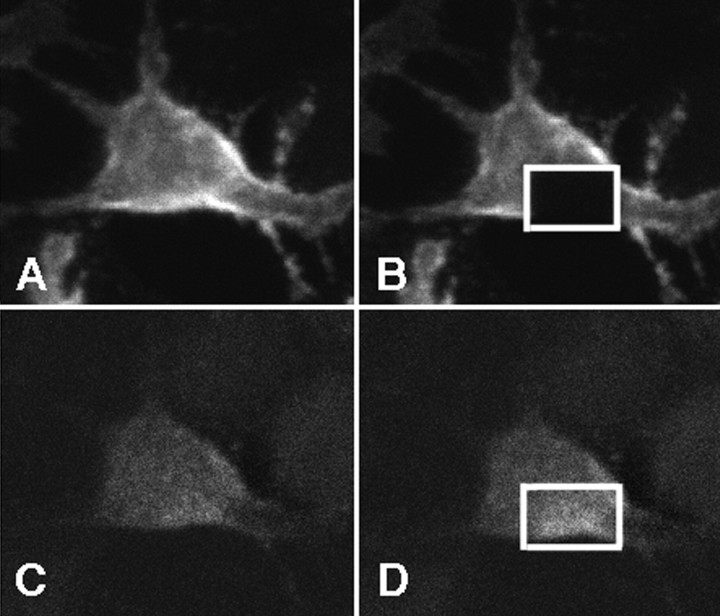
Fig. 5.
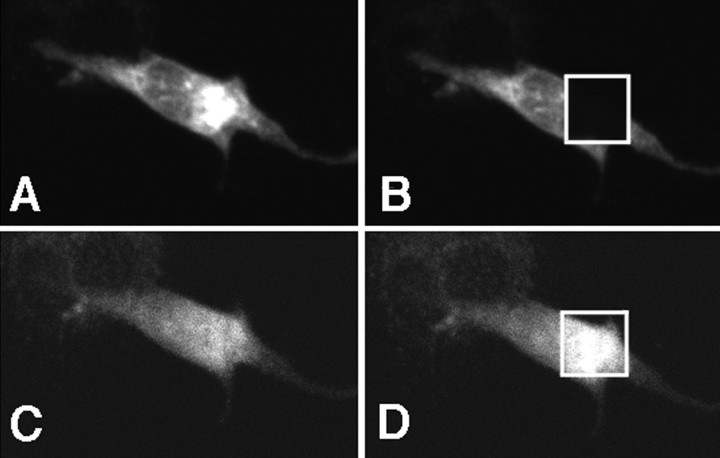
Intracellular domains of APP and LRP are closely associated. H4 cells were transiently transfected with APP770-myc and LRP-EGFP expression constructs. Cells were immunostained by anti-myc antibody conjugated with Cy3. A, Cy3 (APP770) signal using 568 nm excitation. B, Discrete area of the cell was photobleached using intense 568 nm laser. C, EGFP signal (LRP) using 488 nm excitation before photobleaching.D, EGFP signal (LRP) using 488 nm excitation after photobleaching of the acceptor (Cy3) fluorophore with intense 568 nm light. An increase in donor fluorescence is observed within the discrete area that was photobleached, showing the presence of FRET. In this example, the EGFP signal increased by 40%.
We repeated the experiment using APP695 and LRP to test whether the lack of the KPI domain altered the cytoplasmic interaction between APP and LRP. The C terminus of APP695 had an easily detectable and strong FRET with the C terminus of LRP: APP695-myc (labeled with Cy3) and LRP-EGFP (FlD2/FlD1= 1.37 ± 0.06; n = 10; p < 0.0001) (Fig. 6). This ratio was not significantly different from that of APP770. That APP695 interacts with LRP at the C termini was also confirmed using the APP695-DsRed and LRP-EGFP constructs (FlD2/FlD1= 1.53 ± 0.06; n = 14; p < 0.0001). However, unlike the APP770-LRP C-terminal interaction, the APP695-LRP C-terminal interaction was not RAP-sensitive. Cotransfection of RAP did not affect the FRET ratio between APP695 and LRP (for APP695-myc and LRP-EGFP,FlD2/FlD1 = 1.33 ± 0.03, n = 9 , p < 0.0001; for APP695-DsRed and LRP—EGFP,FlD2/FlD1 = 1.56 ± 0.05 , n = 9, p < 0.0001).
Fig. 6.
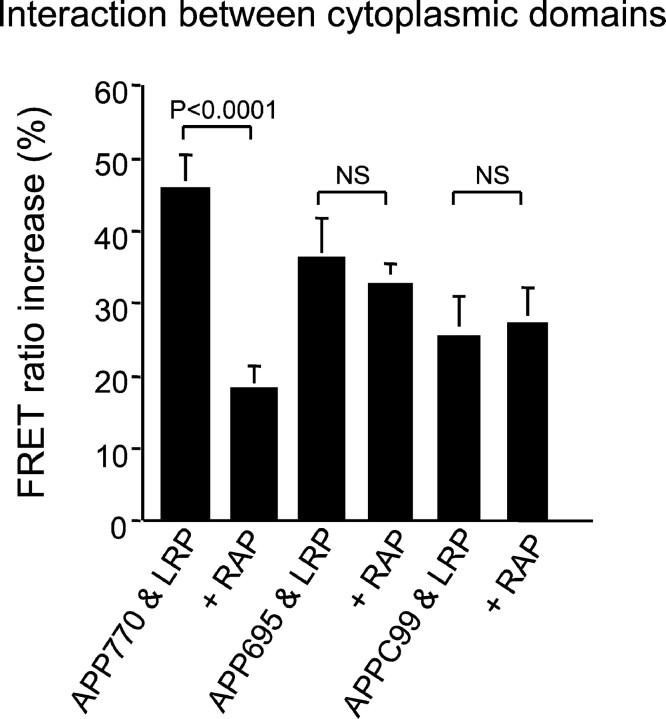
The FRET ratio increase of intracellular domain is shown as a percentage of increase of donor fluorescence between APP and LRP. The FRET ratio increase in donor fluorescence (EGFP) after photobleaching of the acceptor molecule (APP-myc) was measured. The percentage of increase in donor fluorescence was shown in the graph. FRET was present between all of the APP constructs (APP770, APP695, and APPC99) tagged at the C terminal and LRP-EGFP, the ratio being significantly above zero (p < 0.0001; one-group t test). Cotransfection of RAP significantly decreased the FRET ratio increase of APP770 (p = 0.0001; ANOVA; Fisher's PLSDpost hoc test), but there was no significant effect of RAP on APP695 or APPC99-LRP interactions.
These data demonstrate that APP770 interacts with LRP at both an RAP-sensitive and an RAP-insensitive site, whereas APP695 interacts with LRP only at an RAP-insensitive site. As an additional test of the idea that APP-LRP C-terminal interactions occur independently of an interaction in the extracellular domain, we constructed APPC99-myc, which is membrane tethered but does not have an ectodomain. FRET was observed between APPC99-myc (labeled with Cy3) and LRP-EGFP (Fig. 6), although the ratio was smaller than that observed for full-length APP (FlD2/FlD1= 1.26 ± 0.05, for APPC99-myc and LRP-EGFP; n = 17; p < 0.0001). This value did not change after the cotransfection of RAP (FlD2/FlD1= 1.27 ± 0.05; n = 9; p < 0.0001). Thus, the C-terminal domain of APP, in the absence of ectodomain, mediates significant interactions between APP and LRP.
Interactions among APP, LRP, and Fe65
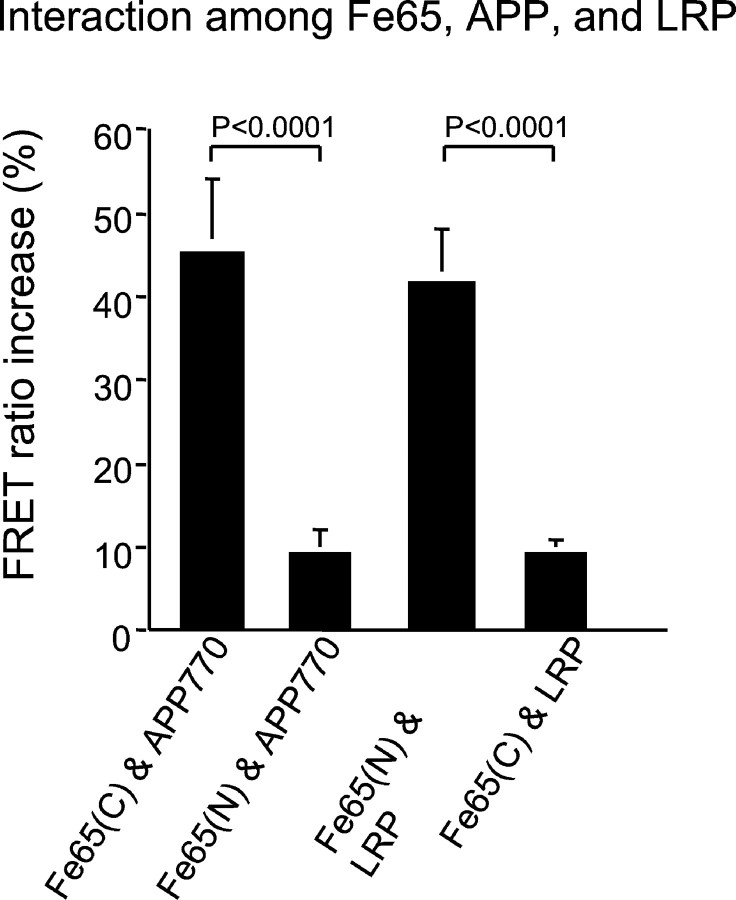
Trommsdorff et al. (1998) used pull-down assays with GST fusion proteins to show that Fe65 associates with the cytoplasmic domain of LRP and suggested a model in which cytosolic adapter proteins such as Fe65 could interact with both APP and LRP, forming a trimeric complex. The model suggests an association of the N-terminal phosphotyrosine binding (PTB) domain of Fe65 with LRP and an association of the C-terminal PTB of Fe65 domain with APP. We used a FRET strategy to test this hypothesis in H4 cells. Two Fe65 fusion proteins were constructed, one containing a myc tag at the C terminus (Fe65-myc) and a second with EGFP coupled to the N terminus (EGFP-Fe65). A close interaction was observed between the C-terminally tagged Fe65-myc and APP770-GFP(FlD2/FlD1= 1.46 ± 0.08; n = 7; p < 0.0001), but a much smaller interaction was observed between the N-terminally tagged EGFP-Fe65 and APP770-DsRed or APP770-myc (FlD2/FlD1= 1.09 ± 0.03; n = 10; p = 0.02) (Fig. 7). In contrast, a close interaction was observed between the N-terminally tagged EGFP-Fe65 and LRP-myc (labeled with Cy3) (FlD2/FlD1= 1.42 ± 0.06; n = 13; p < 0.0001), whereas a much smaller interaction was observed between the C terminus of Fe65-myc and LRP-EGFP (FlD2/FlD1= 1.09 ± 0.02; n = 9; p = 0.001) (Fig. 7). Because the size of theFlD2/FlD1ratio reflects the distance between fluorophores, these data are consistent with a close interaction of the N terminal of Fe65 with LRP and of the C terminal of Fe65 with APP and provide support for a model in which Fe65 interacts with both LRP and APP via cytoplasmic domain interactions.
Fig. 7.
The interaction between Fe65-APP and Fe65-LRP was investigated by FRET. The interaction between Fe65-APP and Fe65-LRP was measured by FRET ratio increase after photobleaching of acceptor molecules. A close interaction was observed between C-terminally tagged Fe65 [shown as Fe65(C)] and APP770, whereas N-terminally tagged Fe65 [shown as Fe65(N)] and APP770 showed a much smaller interaction (p < 0.0001; ANOVA; Fisher's PLSD post hoc test). In contrast, C-terminally tagged Fe65 and LRP showed much less FRET than N-terminally tagged Fe65 and LRP (p < 0.0001; ANOVA; Fisher's PLSD post hoc test).
DISCUSSION
This study examines cellular interactions between different isoforms of APP and LRP. We use in this study an application of digital confocal microscopy combined with FRET (Knowles et al., 1999; McLean et al., 2000; Rocheville et al., 2000). This provides subcellular anatomical resolution with information about protein–protein interactions, allowing the analysis of protein domain structure within the context of cellular systems. Although there are limitations to FRET (for example, the absence of FRET between two epitopes cannot be taken as conclusive evidence against an interaction), FRET is a powerful tool, especially to investigate the morphological relationships of the molecules, and is a complement to biochemical analyses, such as coimmunoprecipitation. FRET has advantages over coimmunoprecipitation in circumstances in which indirect molecular interactions are sought. Even using indirect immunofluorescence (i.e., taking into account the size of the antibody molecules), the greatest distance between two molecules that have detectable FRET is expected to be <30 nm. Direct interactions between fluorophores such as EGFP and DsRed might be expected to be detected only if the fluorophores are less than ∼10 nm apart (Siegel et al., 2000). The FRET ratio is interpreted as a reflection of the distance between fluorophores, so that a larger ratio implies a closer interaction. In this context, the presence of detectable FRET in our study indicates a close interaction between APP and LRP.
The results of the current investigation reveal several novel observations. (1) We demonstrate the presence of a strong interaction between the KPI-containing APP770 and LRP in cells and show that this interaction is partially RAP-sensitive, suggesting that it is mediated by an interaction between an extracellular LRP ligand binding domain and APP770. (2) APP and LRP show a close apposition both at the cell surface and in intracellular organelles, including the Golgi, and the interaction of APP770 and LRP is diminished in these compartments by cotransfection with RAP, suggesting an unexpected interaction of APP and LRP in the secretory pathway. (3) We also surprisingly find an RAP-insensitive interaction between APP695 (which does not contain a KPI domain) and LRP. (4) We show that the C termini of both APP695 and APP770 and LRP can interact, even using the construct encoding truncated APP protein in which the extracellular domains has been deleted, suggesting a second (intracellular) site of protein–protein interaction between APP and LRP. (5) Fe65, a multidomain intracellular adapter protein, shows FRET with both APP and LRP. Together, our data support a model of strong APP-LRP cellular contacts mediated by both direct extracellular and indirect intracellular interactions (Fig.8).
Fig. 8.
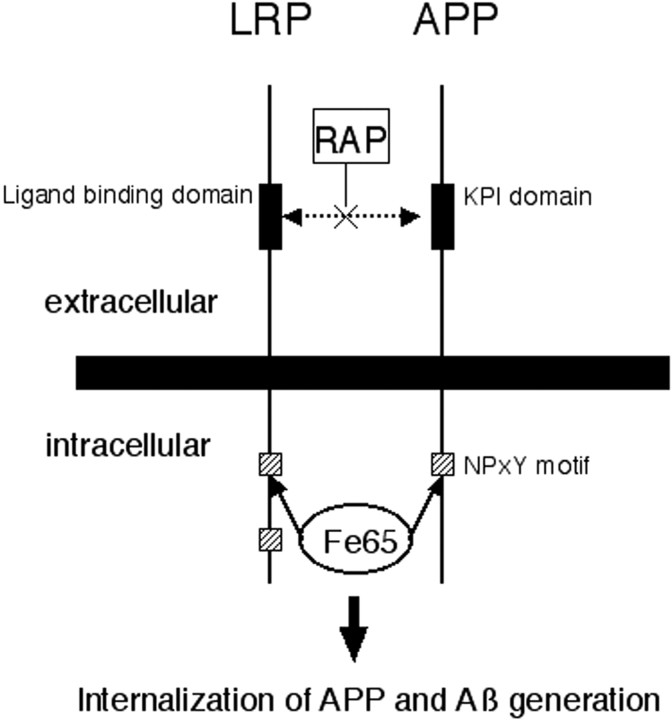
This scheme shows a hypothetical model of interactions of APP, LRP, and the cytoplasmic adaptor protein Fe65. Extracellularly, there may be an interaction between the ligand binding domain of LRP and KPI domain of APP, which is RAP-sensitive. Intracellularly, Fe65 may bind both APP and LRP. We show both interactions occurring in this figure. APP binding to LRP may cause endocytosis of APP and thus modulate amyloid-β (Aβ) synthesis.
Previous studies of APP-LRP interactions from our laboratory have emphasized the role of KPI containing forms of APP because soluble forms of APP containing the KPI domain, but not those lacking the KPI domain, were bound and cleared by LRP (Kounnas et al., 1995). In the present study, we studied the interaction of LRP with full-length, membrane-spanning APP and examined the possibility that LRP interacts with APP770 (a KPI-containing form of APP) or with both APP695 (which does not contain a KPI domain) and APP770. We demonstrate through a series of FRET experiments that there is a close apposition of the extracellular domains of LRP and APP770. This interaction is sensitive to RAP, suggesting that it is a classic ligand–ligand receptor interaction, albeit between two type I transmembrane proteins. FRET between APP and LRP can be detected both at the cell surface and also in intracellular compartments. Interestingly, cotransfection with RAP diminishes the APP770-LRP interaction in the endoplasmic reticulum–Golgi compartments, supporting the possibility that APP-LRP interactions occur in the endoplasmic reticulum–Golgi, suggesting a previously unsuspected interaction in the secretory pathway.
Based on the observation that secreted forms of APP695 do not bind to LRP, we expected there to be little or no interaction of APP695 with LRP (Kounnas et al., 1995). Surprisingly, however, the extracellular domains of APP695 and LRP also show a close approximation. This interaction is not sensitive to RAP, implying that the interaction is mediated by a nonligand binding domain of LRP. In fact, even constructs encoding only the C-terminal portions of APP interact with LRP, showing that APP-LRP protein–protein interactions are mediated by two sites, one located in the ectodomain (between APP770 and LRP) and one between the cytoplasmic termini (APP695 or APP770 and LRP). We interpret these data to suggest that interaction of the intracellular domains of APP and LRP are sufficient to bring the extracellular domains into relatively close proximity, even in instances in which the ligand–ligand binding interaction either is blocked (APP770 in the presence of RAP) or does not occur (APP695). Coimmunoprecipitation experiments demonstrated that both APP751 and APP695 could be pulled down with LRP, although the former appeared to have a fourfold stronger association (Rebeck et al., 2001). In the light of our current data, we hypothesize that, in H4 cells, APP770 interacts with LRP via an ectodomain binding site, as well as via an interaction, possibly indirect, occurring within the cytoplasmic domains of these two molecules. In contrast, APP695 seems to interact with LRP only via the intracellular interaction.
Yeast two-hybrid and isolated protein pull-down experiments (Trommsdorff et al., 1998) suggested a model in which the intracellular portion of APP might interact with LRP via a trimeric complex with Fe65, an intracellular adapter protein. Fe65 contains three distinct interaction domains: one WW and two PTB domains. The WW domain of Fe65 interacts with several proteins, one of which is Mena, the mammalian ortholog of the product of the enabled gene ofDrosophila (Ermekova et al., 1997). The N-terminal PTB domain interacts with LRP (Trommsdorff et al., 1998), and the C-terminal PTB domain interacts with APP (Fiore et al., 1995;Borg et al., 1996; Guenette et al., 1996; Trommsdorff et al., 1998) in isolated systems. Our data showing strong FRET between the N terminus of Fe65 and LRP and the C terminus of Fe65 and APP (but weaker FRET between the N terminus of Fe65 and APP or the C terminus of Fe65 and LRP) support the idea that these interactions are robust in cells. Our data do not rule out the possibility that other adapter proteins might also mediate an APP-LRP cytosolic tail interaction and even that different cell types or tissues may differ in endogenous adapter protein complement. For example, preliminary studies using CHO cells suggest a lesser degree of APP-LRP C-terminal interaction than seen in H4 cells.
Our previous studies of APP-LRP interactions in CHO cells suggested that either genetic deletion of LRP or exogenous RAP treatment could markedly diminish amyloid-β production from APP770-transfected cells but not from APP695-transfected cells (Ulery et al., 2000). Our current studies showed that there is an RAP-sensitive, KPI domain-dependent interaction between APP and ligand binding sites in the LRP ectodomain. Together, we hypothesize that this ectodomain interaction alters the endocytic process to influence amyloid-β generation in the endocytic pathway. Specific analysis of APP-LRP interactions during endocytosis for APP695 and APP770 will be the target of future studies.
Footnotes
This work was supported by National Institutes of Health Grants AG12406 (B.T.H.), AG14473 (G.W.R.), and HL50784 (D.K.S.). A.K. was supported by the Uehara Memorial Foundation.
Correspondence should be addressed to Dr. Bradley T. Hyman, Alzheimer Research Laboratory, Massachusetts General Hospital, 114 16th Street, Room 2009, Charlestown, MA 02129. E-mail:b_hyman@helix.mgh.harvard.edu.
REFERENCES
- 1.Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and Fe65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borth W. Alpha-2-macroglobulin, a multi-functional binding protein with targeting characteristics. FASEB J. 1992;6:3345–3353. doi: 10.1096/fasebj.6.15.1281457. [DOI] [PubMed] [Google Scholar]
- 3.Bu G, Williams S, Strickland DK, Schawartz AL. Low density lipoprotein receptor related protein/a2-macroglobulin receptor is an hepatic receptor for tissue-type plasminogen activator. Proc Natl Acad Sci USA. 1992;89:7427–7431. doi: 10.1073/pnas.89.16.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin JY, Knowles RB, Schneider A, Drewes G, Mandelkow E-M, Hyman BT. Microtubule-affinity regulating kinase (MAPK) is tightly associated with neurofibrillary tangle in Alzheimer brain: a fluorescence resonance energy transfer study. J Neuropathol Exp Neurol. 2000;59:966–971. doi: 10.1093/jnen/59.11.966. [DOI] [PubMed] [Google Scholar]
- 5.Cook DG, Forman MS, Sung JC, Leight S, Kolson DL, Iwatsubo T, Lee VM, Doms RW. Alzheimer's A beta(1–42) is generated in the endoplasmic reticulum/intermediate compartment of NT2N cells. Nat Med. 1997;3:1021–1023. doi: 10.1038/nm0997-1021. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B, Annaert W. Proteolytic processing and cell biological functions of the amyloid precursor protein. J Cell Sci. 2000;113:1857–1870. doi: 10.1242/jcs.113.11.1857. [DOI] [PubMed] [Google Scholar]
- 7.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32874. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 8.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer's amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 9.Guenette SY, Chen J, Jondro Pd, Tanzi RE. Association of a novel human FE65-like protein with the cytoplasmic domain of the beta-amyloid precursor protein. Proc Natl Acad Sci USA. 1996;93:10832–10837. doi: 10.1073/pnas.93.20.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haass C, Koo EH, Mellon A, Hung AY, Selkoe DJ. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann T, Bieger SC, Bruhl B, Tienari PJ, Ida N, Allsop D, Roberts GW, Masters CL, Dotti CG, Unsicker K, Beureitjer L. Distinct sites of intracellular production for Alzheimer's disease Amyloid-β40/42 amyloid peptides. Nat Med. 1997;3:1016–1020. doi: 10.1038/nm0997-1016. [DOI] [PubMed] [Google Scholar]
- 12.Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herz J, Goldstein JL, Strickland DK, Ho YK, Brown MS. 39-kDa protein modulates binding of ligands to low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. J Biol Chem. 1991;266:21232–21238. [PubMed] [Google Scholar]
- 14.Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 A using imaging fluorescence resonance energy transfer. J Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knauer MF, Orland RA, Glabe CG. Cell surface APP751 isoform complexes with protease nexin 2 ligands and is internalized via the low density lipoprotein receptor-related protein (LRP). Brain Res. 1996;740:6–14. doi: 10.1016/s0006-8993(96)00711-1. [DOI] [PubMed] [Google Scholar]
- 16.Knowles RB, Chin J, Ruff CT, Hyman BT. Demonstration by fluorescence resonance energy transfer of a close association between activated MAP kinase and neurofibrillary tangle: implications for MAP kinase activation in Alzheimer's disease. J Neuropathol Exp Neurol. 1999;58:1090–1098. doi: 10.1097/00005072-199910000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Koo EH, Squazzo SL. Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem. 1994;269:21162–21166. [PubMed] [Google Scholar]
- 18.Kounnas MZ, Argraves WS, Strickland DK. The 39-kDa receptor associated protein interacts with two method of the low density lipoprotein receptor family, a2-macroglobulin receptor and glucoprotein 330. J Biol Chem. 1992;267:21162–21166. [PubMed] [Google Scholar]
- 19.Kounnas MZ, Moir RD, Rebeck GW, Bush AI, Argraves WS, Tanzi RE, Hyman BT, Strickland DK. LDL-receptor-related protein, a multifunctional ApoE receptor, binds secreted β-amyloid precursor protein and mediates its degradation. Cell. 1995;82:331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 20.Kuentzel SL, Ali S, Altman RA, Greenberg BD, Raub TJ. The Alzheimer beta-amyloid protein precursor/protease nexin-II is cleaved by secretase in a trans-Golgi secretory compartment in human neuroglioma cells. Biochem J. 1993;295:367–378. doi: 10.1042/bj2950367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean PJ, Kawamata H, Ribich S, Hyman BT. Membrane association and protein conformation of a-synuclein in intact neurons. J Biol Chem. 2000;275:8812–8816. doi: 10.1074/jbc.275.12.8812. [DOI] [PubMed] [Google Scholar]
- 22.Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkow DJ, Chen X, Stokin GB, Koo EH. Mutagenesis identified new signals for b-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Ab42. J Biol Chem. 1999;274:18851–18856. doi: 10.1074/jbc.274.27.18851. [DOI] [PubMed] [Google Scholar]
- 23.Rebeck GW, Moir RD, Mui S, Strickland DK, Tanzi RE, Hyman BT. Association of membrane-bound amyloid precursor protein APP with the apolipoprotein E receptor LRP. Mol Brain Res. 2001;87:238–245. doi: 10.1016/s0169-328x(01)00006-7. [DOI] [PubMed] [Google Scholar]
- 24.Rocheville M, Lange DC, Kumar U, Patel SC, Patel RC, Patel YC. Receptors for dopamine and somatostatin: formation of hetero-oligomers with enhanced functional activity. Science. 2000;288:154–157. doi: 10.1126/science.288.5463.154. [DOI] [PubMed] [Google Scholar]
- 25.Selkoe DJ. The cell biology of the beta-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 26.Siegel RM, Frederiksen JK, Zacharias DA, Chan FK, Johnson M, Lynch D, Tsien RY, Lenardo MJ. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 27.Strickland DK, Kounnas MZ, Argraves SW. LDL receptor-related protein: a multiligand receptor for lipoprotein and proteinase catabolism. FASEB J. 1995;9:890–898. doi: 10.1096/fasebj.9.10.7615159. [DOI] [PubMed] [Google Scholar]
- 28.Trommsdorff M, Borg J-P, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 29.Ulery PG, Beers J, Mikhailenko I, Tanzi RE, Rebeck GW, Hyman BT, Strickland DK. Modulation of beta-amyloid precursor protein processing by the low density lipoprotein receptor-related protein (LRP). Evidence that LRP contributes to the pathogenesis of Alzheimer's disease. J Biol Chem. 2000;275:7410–7415. doi: 10.1074/jbc.275.10.7410. [DOI] [PubMed] [Google Scholar]
- 30.Williams SE, Ashcom JD, Argraves WS, Strickland DK. A novel mechanism for controlling the activity of alpha 2-macroglobulin receptor/low density lipoprotein receptor-related protein. Multiple regulatory sites for 39-kDa receptor associated protein. J Biol Chem. 1992;267:9035–9040. [PubMed] [Google Scholar]
- 31.Xu H, Sweeney D, Wang R, Thinakaran G, Lo AC, Sisodia SS, Greengard P, Gandy S. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc Natl Acad Sci USA. 1997;94:3748–3752. doi: 10.1073/pnas.94.8.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]