Abstract
We are taking a cross-species approach to identify genes that are required for mammalian GABAergic neuron differentiation. On the basis of homeodomain similarity, the vertebrate Pitx genes appear to be orthologs of unc-30, aCaenorhabditiselegans gene necessary for differentiation of the GABAergic phenotype of type D neurons. One of the Pitx genes, Pitx2, is expressed in regions of GABAergic neurogenesis in the mammalian brain. These observations led us to test the functional conservation of the mousePitx2 and worm unc-30 genes using a rescue assay. Pitx2 rescues the GABAergic differentiation defect and partially rescues the axon guidance and behavioral phenotypes of unc-30 mutants, indicating a high degree of functional conservation between these evolutionarily related genes. Previous studies show that UNC-30 directly regulates theunc-25/glutamate decarboxylase gene that encodes the enzyme for GABA synthesis. We find that the promoter regions of the mouse and human genes coding for the 67 kDa glutamate decarboxylase(Gad1) also contain binding sites matching the UNC-30/Pitx2 consensus binding site sequence. We show that these sites specifically bind to Pitx2 protein in vitro and that in transfected neuroblastoma cells, the Pitx2 binding sites contribute to the basal activity of the Gad1 promoter. Furthermore, in cotransfection experiments, we find that Pitx2 strongly activates theGad1 promoter. These results indicate that Pitx2 may regulate Gad1 expression in mammals, suggesting a new role for this key developmental transcription factor as a regulator of GABAergic differentiation during mammalian neural development. Our results suggest that some of the mechanisms regulating GABAergic differentiation are evolutionarily conserved.
Keywords: glutamate decarboxylase, neuron differentiation, Gad67, Gad1, unc-30, Pitx2, GABAergic neuron, mouse development, C. elegans
The differentiation of neurotransmitter phenotypes requires the coordinate regulation of the genes involved in neurotransmitter synthesis and release (Eiden, 1998;Mallet et al., 1998; Goridis and Brunet, 1999; Ernsberger, 2000). Among the phenotypes that develop are GABAergic neurons, the principal inhibitory neurons in the mammalian CNS. These cells play fundamental roles in neuronal function and development. GABAergic differentiation requires several genes, including the glutamate decarboxylases (GADs) for GABA synthesis and the vesicular GABA transporter (VGAT; also known as the vesicular inhibitory amino acid transporter) that packages GABA into synaptic vesicles. In vertebrates, the differentiation of this neurotransmitter phenotype is not well understood.
In contrast, genetic screens in the nematode Caenorhabditis elegans have identified several genes that are required for GABAergic neuron differentiation and function (McIntire et al., 1993a,b). Among them is the homeodomain transcription factor UNC-30, which is necessary and sufficient for specifying the fate of the 19 type D GABAergic motor neurons that control body locomotion (Jin et al., 1994; Hobert et al., 1999). UNC-30 is required for the expression of unc-25/GAD and the vesicular GABA transporter (unc-47/VGAT) in the type D motor neurons (Eastman et al., 1999). UNC-30 regulates these downstream targets through binding sites within the promoters of both genes (Eastman et al., 1999). Moreover, ectopic expression of UNC-30 results in the activation ofunc-25/GAD in non-neural cells (Jin et al., 1994). These studies clearly identified UNC-30 as an immediate upstream regulator ofunc-25/GAD and unc-47/VGAT, revealing its central role in the specification of the GABAergic neurotransmitter phenotype in C. elegans. UNC-30 also regulates other aspects of the terminal differentiation of these neurons, such as axon guidance and synapse formation (Jin et al., 1994). Because of a lack of GABAergic inhibitory input to the body wall muscles, unc-30 mutant worms hypercontract in response to touch, a locomotor phenotype nicknamed “shrinker” (Hodgkin, 1983).
Three mammalian transcription factors (Pitx1–3) contain homeodomains similar to the UNC-30 homeodomain (Semina et al., 1996, 1997; Szeto et al., 1996; Smidt et al., 1997; Gage et al., 1999a). Among them,Pitx2 is expressed in regions of the developing mesencephalon and diencephalon associated with GABAergic neurogenesis (Mucchielli et al., 1996; Kitamura et al., 1997). Within these regions in embryonic day (E) 11.5 to E12.5 mouse embryos, Pitx2 and the Gad1 gene are expressed in similar patterns (Mucchielli et al., 1996; Kitamura et al., 1997; Katarova et al., 2000; Smidt et al., 2000a,b). In the diencephalon, the co-expressing neural progenitors give rise to the zona incerta and the thalamic reticular nucleus, which contain a high percentage of GABAergic neurons (Najlerahim et al., 1990; Benson et al., 1992; Esclapez et al., 1993). Furthermore, the expression of Pitx2 is sustained in these nuclei in newborns, suggesting a long-term role in the differentiation of these neurons (Mucchielli et al., 1996). We also have noted thatPitx2 and Gad expression overlap in basal mesencephalon and superior colliculus (Mucchielli et al., 1996;Katarova et al., 2000). In contrast, the CNS expression ofPitx3 is localized to developing dopaminergic neurons in the mesencephalon (Smidt et al., 1997), and Pitx1 is expressed outside of the developing CNS (Szeto et al., 1996, 1999; Lanctot et al., 1999). The data suggest that Pitx2 is expressed in regions of GABAergic neurogenesis making it a candidate regulator of GABAergic differentiation
The overlapping expression of Gad and Pitx2 and the similarities between the UNC-30 and Pitx2 proteins led us to test the functional equivalence of the two transcription factors in controlling Gad expression and GABAergic differentiation. We report that the expression of mouse Pitx2 in C. elegansactivates unc-25/GAD expression and partially rescues other differentiation defects of the type D GABAergic neurons inunc-30 mutants. The rescue depends on the homeodomain of Pitx2 and on the UNC-30 binding sites in the unc-25/Gad promoter. In the mouse, we find Pitx2 binding sites within theGad1 promoter. Furthermore, our cotransfection experiments show that Pitx2 and UNC-30 can activate the mouse Gad1promoter in mouse and human neuroblastoma cell lines. Taken together, the data show that UNC-30 and Pitx2 are functional homologs. Our results suggest that Pitx2 may be involved in GABAergic neuron differentiation in the mammalian CNS.
MATERIALS AND METHODS
C. elegans genetics, germline transformation, and transgene analysis. Worms were maintained at 22.5°C on NG agar plates as described (Brenner, 1974). unc-30(e596); lin-15(n765ts) mutant animals were used as host in most germline transformation experiments unless otherwise indicated.unc-30(e596) is an early nonsense mutation and behaves as a null mutation of unc-30 (Y. Jin, unpublished data). EachPitx2 expression construct was co-injected at 30–50 ng/μl, along with 50 ng/μl of the Punc-25:: GFP reporter construct (pCZ245) and 50 ng/μl of thelin-15(+)-rescuing plasmid [plin-15(EK)] (Clark et al., 1994), following standard procedures (Mello et al., 1991). pCZ245 contains a 1.4 kbunc-25 promoter that drives the expression of GFP in 4 RME and 19 type D neurons. lin15(n765)ts animals are multivulva at 22.5°C. Transformants were identified as nonmultivulva at 22.5°C. Transformed animals were scored for GFP expression in F1, F2, and subsequent generations using a Zeiss Axioskop with an HQ-FITC filter. Twenty to forty transgenic animals were scored for each independent transgenic line, and at least two independently arising lines were scored for each expression construct.
Construction of the C. elegans expression vectors. The pCZ166 plasmid that expresses mouse Pitx2 in the type D neurons of C. elegans was made by inserting the mousePitx2c cDNA from Mc13–1 (gift of J. Murray, University of Iowa, Iowa City, IA) downstream of a 2.4 kb unc-30 promoter and upstream of the unc-54 3′ untranslated region (UTR) (Fire et al., 1990). For heat-shock inducible expression ofPitx2 in C. elegans, the mouse Pitx2ccDNA was inserted downstream of the promoters Phsp16–41 and Phsp16–2 in pPD49.78 and pPD49.83 respectively, resulting in pCZ202 and pCZ203 (Fire et al., 1990).
Punc-30:: Pitx2-VP16 (pCZ220) and Punc-30:: UNC-30-VP16 (pCZ219) were constructed as follows. The VP16 activation domain was first amplified from pCS2+MTVP16 (Mariani and Harland, 1998) by PCR using the primers 5′ AGCTCATTTCTGAAGAGGAC 3′ and 5′ CTCTACACTAGGTACCCTACTCGTCAA 3′. The resulting 260 bp XhoI and KpnI fragment of VP16 was then fused to Pitx2 at amino acid 320, resulting in Pitx2-VP16 fusion protein that lacks the last four amino acids of Pitx2. UNC-30-VP16 has VP16 fused to UNC-30 at amino acid 340 and lacks the last 19 amino acid residues of UNC-30, which are not required forunc-30 function (Jin et al., 1994). Pitx2-VP16 and UNC-30-VP16 were then used to replace Pitx2 in pCZ166 to generate pCZ220 and pCZ219, respectively.
The truncation mutation in the Pitx2 homeodomain (pCZ214) was made by digesting the Pitx2c cDNA with PstI, followed by klenow fill-in and religation. To generate the T68P mutation in Pitx2 (pCZ215), double-stranded oligonucleotides were generated by annealing two oligos, 5′ AGAGCTGGAAGCCACTTTCCAGAGAAACCGCTACCCA-GACATGTCCCCTCG 3′ and 5′ CGAGGGGACATGTCTGGGTAGCGGTTTCTCTGGAAA GTGGCTTCCAGCTCTTGCA 3′, and were then inserted at the PstI and NruI sites of thePitx2c cDNA.
The Punc-30:: UNC-30-Pitx2(HD) was constructed by PCR amplifying the homeodomain of Pitx2 using the primers 5′ ACTGGAGCTGTCAACAATCTCC 3′ and 5′ CCCGGACTCGAGCTTCCGTAA 3′, and the PCR product was inserted into the unc-30 cDNA digested withXhoI and BsaBI.
Sequence and EMSA analysis of the mouse Gad1promoter. The human Gad1 sequence was obtained from GenBank (accession number AC007405). The sequence of the mouseGad1 promoter region (GenBank accession number AF354680) was determined from a strain 129SvEv genomic clone that was used previously to generate a Gad1 knock-out mouse (Condie et al., 1997). The sequences were compared using the BLAST world wide web server (www.ncbi.nlm.nih.gov/gorf/bl2.html).
A construct for production of a full-length GST fusion protein containing the entire Pitx2c sequence was generated by subcloning anEco47III/HindIII fragment of the cDNA into pGex-KG (Guan and Dixon, 1991). The construct was propagated and the fusion protein was produced in DH5α. Fusion protein induction and purification used standard procedures (Smith and Johnson, 1988). Protein concentration was determined by Bradford assay (Bio-Rad, Hercules, CA) using a BSA standard curve. In the EMSA analysis of the Pitx2 binding sites within the Gad1 promoter, 0.3 pmol of the annealed binding site oligonucleotides were mixed with 0–200-fold molar excess of annealed wild-type competitor oligonucleotides or 250- or 500-fold molar excess of annealed mutant competitor oligonucleotides and incubated on ice for 1 hr in a 20 μl binding reaction. The binding reaction contained 25 mm HEPES, pH 7.9, 84 mm KCl, 5 mm DTT, 10% glycerol, 2.2 μg of poly(dI-dC), and 5 μg of affinity purified full-length GST-Pitx2 fusion protein. The following oligonucleotides were used in these experiments (the sequences are presented for only one of the two oligos used for each binding site): 5′ binding site from Gad1 promoter, GTGCATTCTGGATTACTCATAGGA; 3′ binding site from Gad1 promoter, CCGTGAGCTGGATTTATAATCGCCCTACAAA; hunchback bicoid binding site, CTGCCCATCTAATCCCTTGACGCG; and mutant 5′ binding site from Gad1 promoter, GTGCATTCTGGCGCACTCATAGGA.
Cell culture and transfection experiments. ThePitx2c expression construct (pCI-Pitx2) contained an Eco47III/XhoI fragment of the mousePitx2c cDNA subcloned into the pCI expression vector (Promega, Madison, WI). This expression construct lacks the 38 C-terminal amino acids of the Pitx2 protein. Previous studies have shown that this deletion enhances Pitx2 DNA binding activity with little effect on its transcriptional activity (Amendt et al., 1999). The pCI-unc30 construct contained the full-length unc-30 coding sequence on aStyI/EcoRI fragment of the unc-30 cDNA from pSC24 (Jin et al., 1994). The pCI-unc-30-VP16 expression construct was derived from thePunc-30:: UNC30-VP16 C. elegansexpression vector (see C. elegans methods). The coding sequence was amplified from Punc-30:: UNC30-VP16 by PCR using the following primers: 5′ AGCTAGCCACCATGGATGACAATACGG 3′ and 5′ GCGGCCGCCAATACCATGGTACCCT 3′. The PCR product was subcloned into pGemT (Promega) and moved into pCI as anNheI/NotI fragment.
The Gad1 promoter luciferase reporter plasmid (Gad1-luc) contained a 622 bpAvrII/XbaI fragment from a mouse Gad1genomic clone (GenBank accession number AF354680) cloned into the firefly luciferase reporter vector pGL3Basic (Promega). This fragment contains 506 bp of sequence upstream of the major transcription start site as defined previously by primer extension (Szabo et al., 1996). Mutations in the Pitx2 binding sites in the Gad1 promoter were introduced by site-directed mutagenesis using the Gene Editor Kit (Promega) according to the manufacturer's instructions. Transfections of the wild-type or mutant Gad1-luc vectors contained 500 ng of reporter plasmid. The cotransfections contained 500 ng ofGad1-luc and 400 ng of pCI-unc-30 or pCI-unc-30-VP16 or 200 ng of pCI-Pitx2. Each transfection also contained 10 ng of the internal control vector pRL-SV40 (Promega), encoding the Renilla luciferase under the control of the SV40 promoter. The total amount of DNA in each transfection was made up to 1 μg with pCI plasmid DNA. The mouse Neuro-2A and human SH-SY5Y neuroblastoma cell lines were obtained from the American Type Culture Collection (Manassas, VA). For Neuro-2A cells, the DNA was complexed with 6 μl of lipofectin (Life Technologies, Gaithersburg, MD) in OPTI-MEM (Life Technologies) medium and then incubated with the cells for 5 hr in OPTI-MEM. For SH-SY5Y cells, the DNA mixture was complexed with 4 μl of lipofectamine in OPTI-MEM and incubated with the cells for 5 hr. Then, transfection mixtures were removed, and the cells were returned to incubation medium. In each transfection, 3 × 105 neuroblastoma cells were plated per 35 mm well and transfected 16–20 hr later. Luciferase assays were performed 36–48 hr after transfection using the dual luciferase kit (Promega) and a Turner Designs luminometer. Statistical analysis of the transfection data were performed using Prism Graph Pad software.
RESULTS
Conserved amino acid sequences of the C. elegansUNC-30 and mouse Pitx proteins
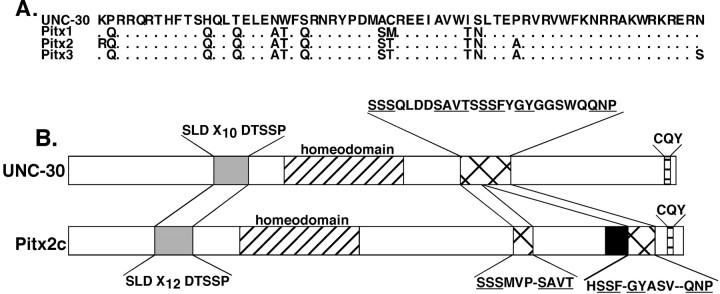
Figure 1A shows an alignment of the UNC-30 homeodomain sequence with the homeodomains of the mouse Pitx proteins. The proteins share an extended homeodomain of 63 amino acids. The amino acid identity to the UNC-30 homeodomain is highest with Pitx1 (84%), followed by Pitx2 and 3 (80% each) (Fig.1A). When we performed a search for sequences in the Pitx proteins that match all of the tripeptide sequences in UNC-30, we found additional regions of similarity between Pitx2 and UNC-30 outside of the homeodomains (Fig. 1B). One such region was found upstream of the homeodomain and consisted of a conserved tripeptide and pentapeptide separated by a similar number of nonconserved residues (shaded boxes in Fig.1B). Another region of similarity was located downstream of the homeodomain. In UNC-30, this region is a contiguous stretch of 26 amino acids, whereas in Pitx2 it is split in two (hatched boxes in Fig. 1B). Near the C terminus of UNC-30 we found a conserved “CQY” tripeptide (Fig.1B). This tripeptide is found near the C termini of all three mammalian Pitx proteins, as well as in the Pitx-related proteins from amphioxis and Drosophila (Lamonerie et al., 1996; Semina et al., 1996, 1997; Szeto et al., 1996; Smidt et al., 1997; Vorbruggen et al., 1997; Yasui et al., 2000). A previously described conserved domain found in the Pitx proteins from vertebrates, amphioxis, and Drosophila as well as other paired-like homeodomain factors from a wide range of species (shown as theblack box in Pitx2 in Fig. 1B) was not found in UNC-30 (Semina et al., 1996; Furukawa et al., 1997; Kitamura et al., 1997; Vorbruggen et al., 1997; Gage et al., 1999a; Yasui et al., 2000). The regions of similarity between the UNC-30 and Pitx2 C terminal regions are of particular interest because this region in Pitx2 has been shown to modulate DNA binding activity and to mediate protein–protein interactions between Pitx2 and the homeodomain factor Pit1 (Amendt et al., 1999). Conservation of these residues in UNC-30 suggests that they may play similar roles in modulating UNC-30 function in the worm.
Fig. 1.
Comparison of Pitx2 and UNC-30 amino acid sequences. A, Alignment of the homeodomains. The four proteins share an extended homeodomain of 63 amino acids. Residues in the Pitx proteins differing from the UNC-30 homeodomain are shown.B, Comparison of the conserved sequences in UNC-30 and Pitx2 that are outside of the homeodomain. The amino acid sequences of the conserved regions are shown above (UNC-30) or below (Pitx2c) the diagrams. In the conserved segment upstream of the homeodomain, X10 and X12 denote the number of nonconserved residues between the conserved tripeptide and pentapeptide. In the case of the conserved 26 amino acid region downstream of the UNC-30 homeodomain (see Results), the conserved residues are underlined.Dashes in the Pitx2c sequences that correspond to the 26 amino acid domain in UNC-30 indicate gaps introduced in generating the alignment to the UNC-30 domain.
Expression of the mouse Pitx2 gene in C. elegans rescues the unc-25/GAD expression and axon guidance defects of unc-30 mutants
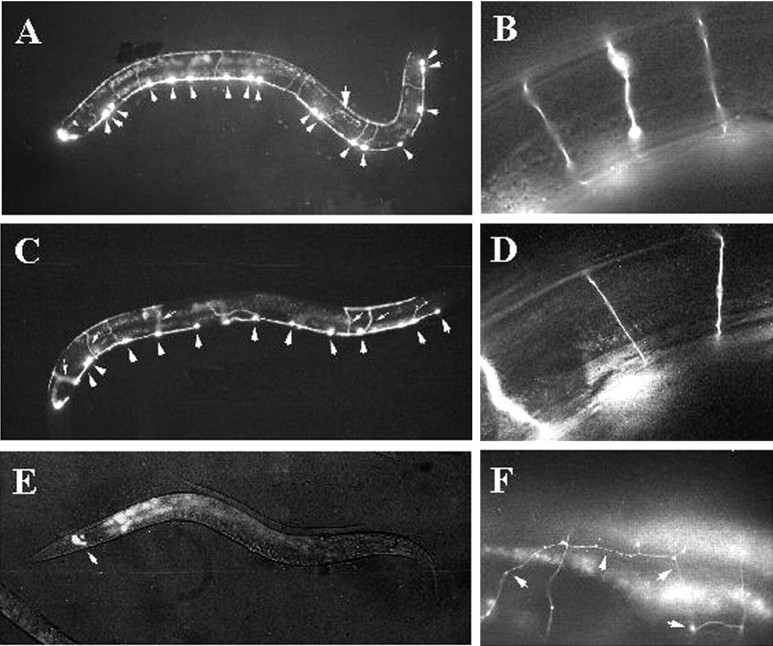
To address the functional significance of the sequence similarity between C. elegans UNC-30 and the mouse Pitx proteins, we examined the effect of expressing mouse Pitx2 in worms. We chose Pitx2 because it is expressed in the developing mouse brain in regions that are rich in GABAergic neurons (Najlerahim et al., 1990; Benson et al., 1992; Esclapez et al., 1993; Mucchielli et al., 1996; Kitamura et al., 1997; Katarova et al., 2000). In unc-30 mutants,unc-25/GAD expression in the 19 type D neurons is completely abolished, and the axons of these D neurons wander and frequently fail to reach their muscle targets (McIntire et al., 1993a; Jin et al., 1994; Wightman et al., 1997). The animals show a shrinker uncoordination phenotype because they lack the reciprocal inhibitory inputs to the body wall muscles. We generated transgenic animals by injecting into unc-30(e596) animals a construct in which the expression of a Pitx2c cDNA is driven by theunc-30 promoter (Punc-30:: Pitx2) (see Materials and Methods). We scored the rescuing activity of mouse Pitx2 using three criteria: (1) the expression of unc-25/GAD, (2) the morphology of the D neuron axon projections, and (3) the movement of unc-30 animals that carry thePunc-30:: Pitx2 transgene. We used aPunc-25:: GFP reporter construct to monitor the activation of unc-25/GAD by Pitx2, and the GFP expression from this reporter also allowed us to examine the morphology of the type D neurons in which unc-25/GAD expression is activated by Pitx2 (Fig. 2). We found that transgenic unc-30 animals carrying Punc-30:: Pitx2 or Punc-30:: UNC-30 transgenes expressed GFP at comparable intensity levels from the Punc-25:: GFP reporter (Fig. 2A–D), indicating that Pitx2 and UNC-30 can activate the unc-25/GAD promoter to a comparable extent. We further quantitated the activation of Punc-25:: GFP by counting the total number of D neurons expressing GFP in the transgenic animals. On average, 13 of 19 type D neurons express GFP in animals carrying Punc-30:: UNC-30, whereas 9 of 19 D neurons are GFP-positive in worms carrying Punc-30:: Pitx2 (Table 1). Not all of the type D neurons express GFP because the transgenes are carried as extrachromosomal arrays and are partitioned in a mosaic fashion. Nonetheless, this result shows that mouse Pitx2 can activate C. elegans unc-25/GAD expression with a comparable ability to UNC-30.
Fig. 2.
Expression of mouse Pitx2 in worms can partially rescue UNC-30 function. A, B,unc-30(e596) worms co-injected with the Punc-30:: UNC-30 and Punc-25:: GFP. A, Activation of Punc-25:: GFP in 17 of 19 type D neurons (arrowheads). Most of the GFP-expressing neurons had normal axonal projections; only one of the 17 GFP-positive axons was misguided (arrow). B, A high-magnification view of the commissural axons showing proper axon guidance in this transgenic unc-30(e596) worm. C, D, unc-30(e596) worms co-injected with Punc-30:: Pitx2 and Punc-25:: GFP. C, Pitx2 activated Punc-25:: GFPexpression in 11 of 19 type D neurons (arrowheads). Thesmall arrows indicate five axons exhibiting the proper trajectories to the dorsal side of the worm.D, A high-magnification view of the normal projection of the commissural axons in this transgenic unc-30 worm.E, unc-30(e596) worms co-injected with Punc-30 vector and the Punc-25:: GFP reporter. Expression of GFP was only detected in the four RME neurons in the head (arrow). F,unc-30(e596) worms carrying a Punc-30:: GFP transgene to show that the commissurals were misguided (arrows).
Table 1.
Rescue of the unc25/GAD expression phenotype inunc-30 mutants by Pitx2 and Pitx2 derivatives
| Transgene | Average number of GFP + neurons | Number of worms scored | Number of transgenic lines |
|---|---|---|---|
| Punc-30∷UNC-30 | 13 | 20 | 2 |
| Punc-30∷Pitx2 | 9 | 40 | 6 |
| Punc-30∷UNC-30∷Pitx2(HD) chimera | 7 | 20 | 4 |
| Punc-30∷Pitx2(T68P) mutant | 6 | 20 | 4 |
| Punc-30∷Pitx2(ΔHD) truncation | 0 | 20 | 6 |
| Punc-30∷Pitx2-VP16 | 11 | 40 | 3 |
| Punc30 vector | 0 | 20 | 4 |
In addition to the activation of unc-25/GAD, Pitx2 expression in worms also partially rescued the axon guidance defects inunc-30 mutants. In wild-type animals, the cell body of each type D neuron is located in the ventral nerve cord, and a nerve process extends anteriorly and bifurcates to form a ventral to dorsal commissural axon projecting to the dorsal nerve cord (Fig.2B) (White et al., 1986). Within the dorsal nerve cord, the nerve process of each D neuron extends both anteriorly and posteriorly and ends when contacting the nerve processes of the neighboring D neurons. In unc-30 mutants, the D neurons show a fully penetrant but variable abnormality in axon morphology. The nerve processes do not follow stereotyped growth paths, and the commissural axons wander and frequently fail to reach the dorsal cord (Fig. 2F). In unc-30 animals expressing Punc30:: UNC-30 transgenes, 73% of the GFP-expressing type D neurons showed normal axon guidance, whereas 27% of the type D neurons in unc-30 animals expressing Pitx2 showed proper axon guidance (Fig. 2A–D,Table 2). Furthermore, the movement of the unc-30 mutant worms carrying the mouse Pitx2 transgene was improved. These transgenic animals did not shrink when touched at the head or tail region, and moved noticeably faster than theunc-30 mutants. The partial rescue of the axon guidance and locomotor defects of unc-30 mutants by Pitx2 indicates that it is able to activate multiple UNC-30 target genes besidesunc-25/GAD, albeit to a lesser degree.
Table 2.
Rescue of the unc-30 axon guidance phenotype
| Transgene | % type D neurons with normal axon trajectories |
|---|---|
| Punc30∷UNC-30 | 73% (n = 55) |
| Punc30∷Pitx2 | 27% (n = 184) |
| Punc-30∷UNC30∷Pitx2(HD) chimera | 61% (n = 142) |
It has been shown previously that ectopic expression of UNC-30 inC. elegans induces ectopic expression of theunc-25/GAD and the unc-47/vesicular GABA transporter genes (Jin et al., 1994; Eastman et al., 1999). To further test whether Pitx2 is a functional UNC-30 homolog, we ectopically expressed Pitx2 under the control of the heat shock promoter. After heat shock, the animals carrying the Phsp16::Pitx2 construct activated Punc-25:: GFP in non-GABAergic neurons and in non-neural cells such as the gut and pharynx (Table3) (data not shown). This result shows that the Pitx2 protein can activate unc-25/GAD expression in neural and non-neural tissues that normally do not expressunc-25/GAD recapitulating the dominant over-expression phenotype seen in worms ectopically expressing unc-30 (Jin et al., 1994).
Table 3.
Pitx2 does not activate the mutant unc-25/GAD reporter, and ectopic Pitx2 drives ectopic unc-25/GAD expression
| Expression construct | Reporter | GFP expression | Number of worms scored | Number of transgenic lines |
|---|---|---|---|---|
| Punc-30∷Pitx2 | Wild-type P unc-25∷GFP | + | 20 | 4 |
| Punc-30∷Pitx2 | Mutant Punc-25∷GFP | − | 20 | 3 |
| Phsp-16-2∷Pitx2 | Wild-type Punc-25∷GFP | Ectopic3-150 | 20 | 12 |
| Phsp-16-2 vector | Wild-type Punc-25∷GFP | − | 20 | 2 |
| Phsp-16-41∷Pitx2 | Wild-type Punc-25∷GFP | Ectopic3-150 | 20 | 3 |
| Phsp-16-41 vector | Wild-type Punc-25∷GFP | − | 20 | 2 |
F3-150: Ectopic expression of Punc-25∷GFP was seen in non-D type neurons, in the gut and pharynx.
Because the bulk of the sequence similarity between Pitx2 and UNC-30 resides in the homeodomain, we asked whether the homeodomain alone accounts for the functional equivalence of Pitx2 and UNC-30. We constructed an UNC-30:: Pitx2(HD) chimeric protein in which we replaced the UNC-30 homeodomain with that of Pitx2 (see Materials and Methods). This chimeric protein [Punc-30:: UNC-30:: Pitx2(HD)] activatedunc-25/GAD expression to the same degree as Pitx2 (Table 1) and rescued the axon guidance defect to the same degree as UNC-30. Sixty-one percent of the type D neurons showed proper morphology in animals expressing the chimeric protein, compared with 73% in animals expressing UNC-30 (Table2). The slight difference in the degree of rescuing activity detected in our transgenic animal studies may reflect protein translation efficiency attributable to the differences of codon usage or the mosaic distribution of extrachromosomal arrays. Taken together, our data support that Pitx2 can functionally substitute for UNC-30, and the homeodomain alone appears to be responsible for the functional equivalence of the two proteins in worms.
The rescue of unc-25/GAD expression requires the UNC-30 binding sites and depends on the Pitx2 homeodomain and its transcriptional activation ability
The UNC-30 protein binds to two bicoid-type homeodomain binding sites located in the unc-25/GAD promoter region (Eastman et al., 1999). When these sites are mutated to CG-rich sequences, UNC-30 binding is eliminated, and unc-25/GAD expression is abolished (Eastman et al., 1999). To determine the specificity of Pitx2 activation of unc-25/GAD, unc-30(e596) worms were co-injected with the Punc-30:: Pitx2 expression construct along with either a wild-type Punc-25:: GFP reporter or a mutant Punc-25:: GFP reporter in which the UNC-30 binding sites were altered to CG-rich sequences. GFP expression was only detected from the wild type unc-25 reporter, supporting that Pitx2 transactivation is dependent on intact UNC-30 binding sites in the unc-25/GAD promoter (Table 3).
Conversely, we tested the dependence of Punc-25:: GFP reporter activation on the Pitx2 homeodomain. We first changed a threonine to proline at amino acid 31 of the Pitx2 homeodomain (Fig. 1A). This mutation is found in patients with Rieger syndrome and results in reduced DNA binding specificity and a loss of transactivation activity in mammalian cells (Semina et al., 1996; Amendt et al., 1998;Hjalt et al., 2001). In our transgenic animal tests, this point mutation reduced, but did not abolish, the rescuing activity of Pitx2. On average 6 of 19 type D neurons express the Punc25:: GFP reporter in animals expressing the Pitx2(T68P) mutant protein, compared with 9 of 19 for the wild-type Pitx2 protein (Table 1). The fact that this mutant Pitx2 had detectable activity in our transgene analysis could be attributable to overexpression of a partially functional protein. Alternatively, it is possible that differences between worm and mammalian cell physiology (for example, lower incubation temperature or differing requirements for cofactors) allow the T68P mutant protein to be functional inC. elegans. We then created a frameshift mutation in the Pitx2 homeodomain such that the protein was truncated at the fifth glutamine residue of the homeodomain (Fig. 1A) (see Materials and Methods). This Punc30:: Pitx2(ΔHD) truncation expression construct did not activate theunc-25:: GFP reporter, demonstrating that Pitx2 activation of the C. elegans unc-25/GAD promoter requires the Pitx2 homeodomain (Table 1).
To further address whether the activation of unc-25/GAD by Pitx2 occurs at the transcription level, we expressed a variant Pitx2 that contains the VP16 activation domain (Materials and Methods). The viral protein VP16 has a strong constitutive transcriptional activation domain that is highly active in a wide range of heterologous proteins in many species, including C. elegans (Sadowski et al., 1988; Sze et al., 1997). We reasoned that a Pitx2-VP16 fusion might increase the transcriptional activity of Pitx2. Indeed, we observed that an average of 11 of 19 type D neurons expressedPunc-25:: GFP in the transgenic animals expressing a Punc-30:: Pitx2-VP16 transgene, compared with 9 of 19 in worms expressing wild type Pitx2 (Table 1). Our data thus support the idea that mouse Pitx2 is likely to activate C. elegans unc-25/GAD gene directly at the transcriptional level.
The promoter regions of the mouse and human Gad1genes contain Pitx2 binding sites
To continue the analysis of Pitx2 and UNC-30 functional conservation, we tested the hypothesis that they could act as regulators of GABAergic neuron gene expression in mammals. In C. elegans, unc-25/GAD is an immediate downstream target of Pitx2 or UNC-30 in the type D GABAergic neurons. These factors act through UNC-30 binding sites in the unc-25/GAD promoter (Table 3) (Eastman et al., 1999). In mammals, two isoforms of the GAD enzyme are co-expressed in GABAergic neurons, each encoded by a separate gene (Gad1 and Gad2) (Erlander et al., 1991; Bu et al., 1992; Esclapez et al., 1993, 1994). We chose to focus on the promoter region of the mouse Gad1 region because it has been well characterized previously (Szabo et al., 1996; Yanagawa et al., 1997). A comparison of the DNA sequences of the mouse and humanGad1 promoter proximal regions revealed conserved putative UNC-30/Pitx2 binding sites (Fig.3A). Three potential binding sites were found. The 5′ site (TAATCC) is a perfect match to the UNC-30/Pitx2 bicoid-type homeodomain consensus (Amendt et al., 1998,1999; Eastman et al., 1999; Hjalt et al., 2001). The 3′ binding site sequences are two adjacent inverted repeats separated by a single nucleotide (GGATTTATAATCG). The two 3′ sites diverge from the consensus at one position in each site (Fig. 3A). The presence of these sites near the mouse and human Gad1 promoters is comparable with the promoter proximal location of the UNC-30 binding sites in the C. elegans unc-25/GAD gene (Eastman et al., 1999).
Fig. 3.
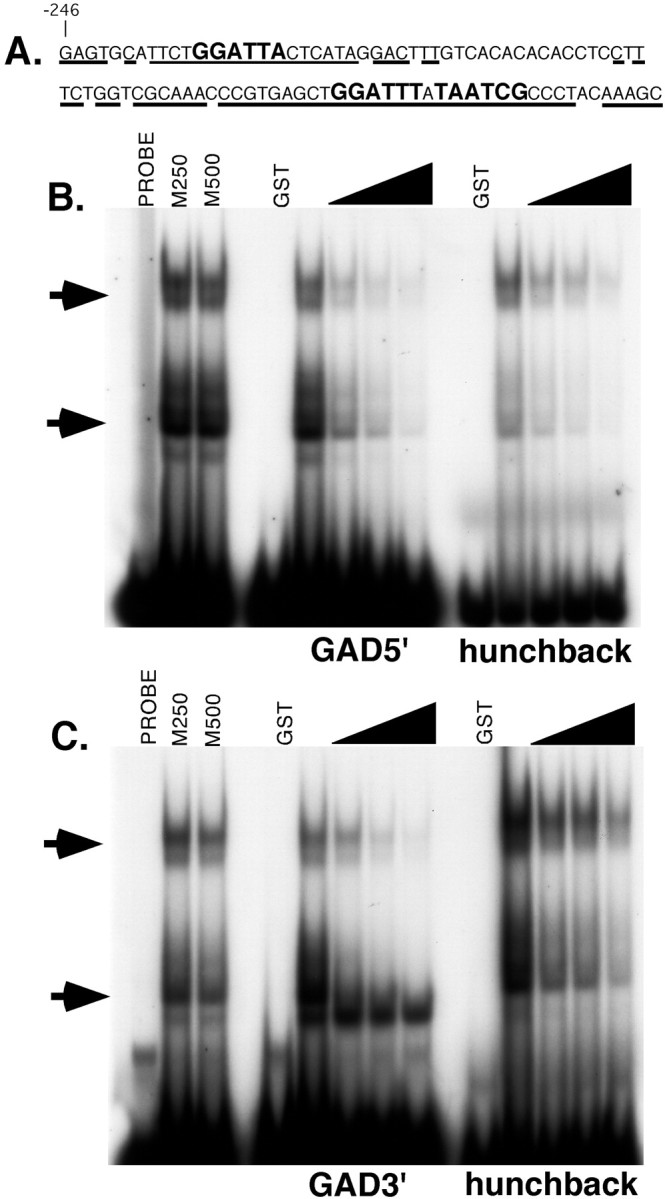
The Pitx2 protein binds to sequences in the mouseGad1 promoter. A, The sequence of the mouse Gad1 promoter region containing the Pitx2 binding sites. The first nucleotide of this sequence is 246 nucleotides upstream of the previously defined major transcriptional start site (Szabo et al., 1996). Underlining indicates the nucleotides that are conserved between the mouse and humanGad1 genes. The Pitx binding sites are shown inlarger letters. B, C, The 5′ site (B) and 3′ site (C) bind to Pitx2 protein specifically. Shifted protein-DNA complexes are indicated by arrows. Reactions containing probe only (Probe), 250-fold or 500-fold excess of mutant competitors (M250, M500), or purified GST protein (GST) are indicated. Increasing amounts (50- to 200-fold molar excess) of unlabeled 5′ site were added as a specific competitor as indicated by the wedges. A bicoid binding site from the Drosophila hunchback promoter was used as a positive control (lanes markedhunchback). The binding of GST-Pitx2 protein to the hunchback promoter site was also competed away by the sites from the mouse Gad1 gene.
Electromobility shift assays (EMSAs) were performed to test the ability of the Pitx2 protein to bind to the putative binding sites in theGad1 promoter. In the first series of EMSAs, oligonucleotides containing the 5′ and 3′ binding sites were incubated with a purified GST-Pitx2 fusion protein. This fusion protein contained the entire Pitx2 protein sequence (Materials and Methods). Each binding site competed with itself for binding to GST-Pitx2, whereas a mutant version of the 5′ site (TGCGCC) failed to compete with either wild-type binding site, demonstrating the sequence specificity of GST-Pitx binding (Fig. 3B,C, lanes M250 and M500). In both cases, affinity purified GST protein alone did not result in a shift (Fig.3B,C). As a positive control, we incubated the GST-Pitx2 protein with a well characterized bicoid binding site from the Drosophila hunchback gene [binding site A3 in Driever and Nusslein-Volhard (1989)]. The site from thehunchback gene also bound to the GST-Pitx2 protein, and this binding was competed away by the 5′ and 3′ binding sites from theGad1 promoter (Fig.3B,C). The multiple shifted protein-DNA complexes detected in the EMSA may be attributable to dimerization mediated by the GST motif in the fusion protein (Tudyka and Skerra, 1997). A similar pattern of shifted probes was seen in the EMSA analysis of the unc-25/GAD promoter using a GST-UNC-30 fusion protein (Eastman et al., 1999). This analysis demonstrated that the conserved binding sites in the mouse Gad1 promoter can specifically bind to the Pitx2 protein.
Pitx2 and UNC-30 activate the mouseGad1 promoter
Transfection analysis was performed to test the contribution of the Pitx/UNC-30 binding sites to Gad1 promoter activity and to test the ability of Pitx2 and UNC-30 to activate expression from this promoter. A 622 bp fragment containing the mouse Gad1promoter as well as flanking upstream and downstream sequences was subcloned, and site-directed mutagenesis was used to modify two of the Pitx2 binding sites in the promoter region (Fig.4A). In both cases, the TAAT motif of the binding sites was replaced with TGCG (Fig.4A). The same mutation introduced into the 5′ Pitx2 binding site (the negative control competitor) eliminated binding to the GST-Pitx2 protein in our EMSA studies of the Gad1promoter (Fig. 3B,C). The wild-type and mutant Gad1 promoter fragments were each subcloned into a luciferase vector (pGL3Basic) to generate the reporter constructs.
Fig. 4.
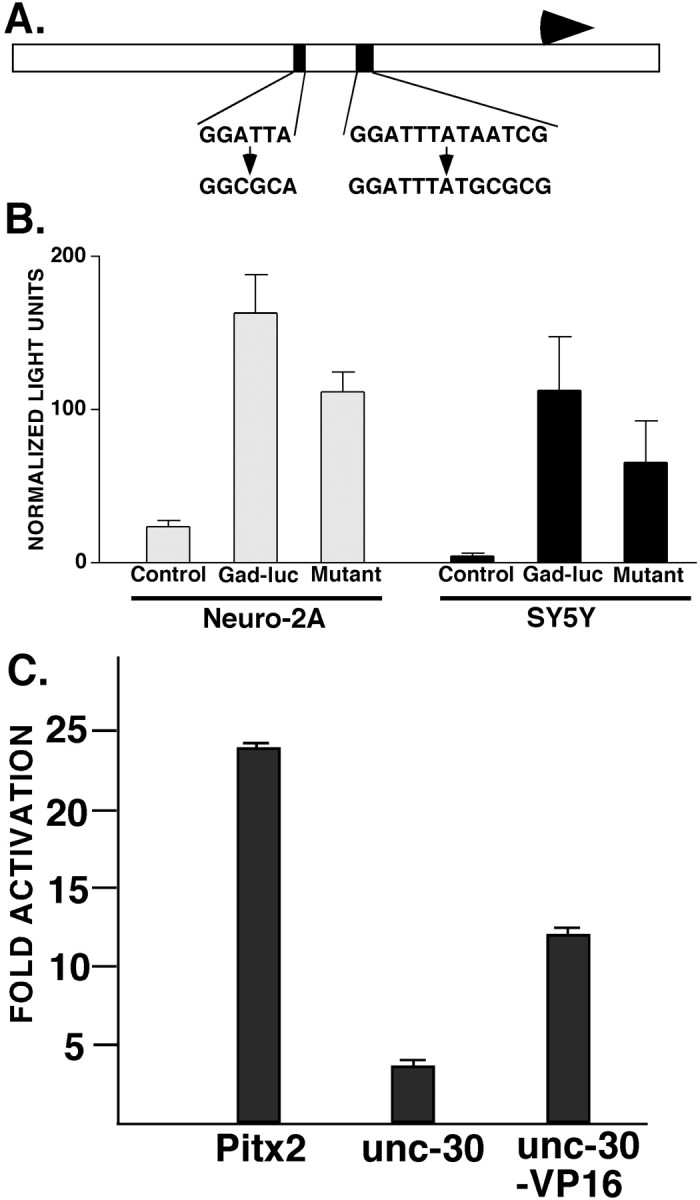
Pitx2 and UNC-30 can activate the mouseGad1 promoter. A, Diagram of the 622 nucleotide region of the Gad1 gene used in the transfection constructs. The Pitx binding sites are indicated (black boxes). The wild-type sequence of each site is shown (top sequence) with the corresponding mutant binding site sequence used in the mutant reporter construct (bottom sequence). The major start site for transcription is indicated by the arrow.B, Expression of the wild-type and mutantGad1-luciferase reporter in Neuro-2A and SH-SY5Y neuroblastoma cell lines. The control values are the activity of the parent pGL3Basic luciferase plasmid alone (values are the mean ± SD). C, Cotransfection of theGad1-luciferase reporter vector with the CMV-Pitx2, CMV-unc-30, or CMV-unc-30-VP16 expression vectors. The fold activation was calculated relative to the activity of theGad1-luciferase reporter plasmid alone (values are the mean ± SD).
The wild-type and mutant Gad1-luciferase reporters were transfected into mouse Neuro-2A and human SH-SY5Y neuroblastoma cells. The reporter was active in both cell lines but to different extents. In Neuro-2A cells, the wild-type Gad1-luciferase plasmid was sevenfold more active than the parent luciferase vector control, whereas in SH-SY5Y cells, this reporter was 25-fold more active than the control plasmid (Fig. 4B). The mutant construct was less active in both cell lines. In the Neuro-2A cells, mutation of the Pitx2 binding sites reduced activity to 68% of wild-type levels, whereas in the SH-SY5Y cells, mutant Gad1-luciferase reporter expression was reduced to ∼58% of wild-type levels (Fig.4B). The reduction in expression of the mutant reporter was significant in both cell lines (p< 0.0001 for Neuro-2A values; p = 0.0006 for SY5Y values; unpaired t test).
We tested the ability of Pitx2 and UNC-30 to activate the mouseGad1 promoter in the Neuro-2A cell line. ThePitx2 and unc-30 cDNAs were placed under the control of the CMV promoter. Cotransfection of the CMV-Pitx2expression vector with the Gad1-luciferase reporter vector resulted in a nearly 24-fold activation of the reporter as compared with the activity of the Gad1 reporter alone (Fig.4C). The CMV-unc-30 vector induced reporter expression approximately fourfold (Fig. 4C). The lower activity of the CMV-unc-30 plasmid compared with the CMV-Pitx2 expression vector may be caused by the inefficient translation of the unc-30 mRNA in mammalian cells. In addition, the UNC-30 factor may lack sequences required for it to act as an efficient transcriptional activator in mammalian cells. Consistent with the latter explanation, we found that a UNC30-VP16 fusion protein (Materials and Methods) was a stronger activator ofGad1-luciferase expression, resulting in an induction of the reporter 13-fold over the control plasmid expression level (Fig.4C). These results show that Pitx2 is a strong activator of the mouse Gad1 promoter. The transfection studies also demonstrate that Pitx2 and UNC-30 can each activate both the mouse andC. elegans Gad genes, further underscoring the functional conservation of the two transcription factors.
DISCUSSION
The C. elegans homeobox gene unc-30 is required for axon guidance and neurotransmitter expression during the terminal differentiation of 19 type D GABAergic neurons (McIntire et al., 1993a; Jin et al., 1994; Wightman et al., 1997). Loss ofunc-30 function leads to a complete loss of GAD expression in these neurons, as revealed by absence of GABA in these cells (McIntire et al., 1993a; Jin et al., 1994). It has been shown previously that the unc-25/GAD promoter is an immediate downstream target of UNC-30, indicating that this factor plays a direct and central role in the regulation of the genes required for the GABAergic phenotype (Eastman et al., 1999). In our genetic rescue experiments, mouse Pitx2 activated unc-25/GAD expression in the type D neurons nearly as well as UNC-30. This Pitx2 rescuing activity was dependent on UNC-30 binding sites in theunc-25/GAD promoter. Moreover, a Pitx2:: VP16 fusion protein showed stronger rescuing activity than Pitx2 alone, further supporting a direct activation of unc-25/GAD by the mouse Pitx2 transcription factor. In addition to rescuing theunc-25/GAD expression defect, Pitx2 was also able to partially specify the normal axon guidance program in the developing type D GABAergic neurons. This result suggests that the mouse Pitx2 homeodomain factor can regulate other unc-30 target genes in addition to unc-25/GAD. Furthermore, the chimeric UNC-30:: Pitx2(HD) showed a similar degree of rescuing activity as Pitx2 and UNC-30, suggesting that the homeodomain largely accounts for the functional equivalence of this protein family. This result was somewhat surprising given that the UNC-30 and Pitx2 homeodomains differ at 12 residues. In other cases, this can be enough divergence to cause differences in homeodomain protein function (Lin and McGinnis, 1992; Chan and Mann, 1993).
Complementing our unc-30 phenotype rescue experiments, analysis of the mouse and human Gad1 promoter regions revealed conserved bicoid-type binding site sequences located proximal to the major transcriptional start site. We demonstrated that a GST-Pitx2 fusion protein bound specifically to these sites in theGad1 gene. Mutation of these binding sites to a sequence that lacks UNC-30/Pitx2 binding activity substantially reducedGad1 promoter activity in mammalian neuroblastoma cells. Furthermore, UNC-30 and Pitx2 activated the Gad1 promoter in transfected neuroblastoma cells. This work suggests that Pitx2 and possibly other bicoid-related homeodomain transcription factors play a role in Gad gene regulation and GABAergic neuron differentiation in mammals. These results also underscore the functional conservation of UNC-30 and Pitx2 in regulating GABAergic differentiation.
We did not test the ability of Pitx1 or Pitx3 to rescue the unc-30 phenotypes or to activate theGad1 reporter plasmid. We anticipate that these paralogs ofPitx2 should behave similarly to Pitx2 in the rescue and gene activation assays. Although the other Pitxfamily members are likely to have similar rescue activities,Pitx2 is the only one that is expressed in appropriate cell types for it to be involved in GABAergic neurogenesis. In the CNS,Pitx3 is expressed in developing dopaminergic neurons in the ventral midbrain, whereas Pitx1 appears not to be expressed in the developing CNS (Szeto et al., 1996, 1999; Smidt et al., 1997;Lanctot et al., 1999).
An important implication of these studies is that Gad1 may be a Pitx2 target gene in the mammalian CNS. Consistent with this, a comparison of Pitx2 and Gad gene expression patterns supports the idea that they are co-expressed in several regions. The expression patterns overlap in the developing zona incerta and reticular nucleus of the ventral thalamus (Mucchielli et al., 1996;Kitamura et al., 1997; Katarova et al., 2000). In the adult, both of these structures are rich in GABAergic neurons (Najlerahim et al., 1990; Benson et al., 1992; Esclapez et al., 1993). Pitx2 andGad1 are also co-expressed in the developing basal mesencephalon and superior colliculus (Mucchielli et al., 1996;Katarova et al., 2000). Outside of the CNS, the two genes are expressed in the dental epithelium during tooth development, and both theGad1 and Pitx2 knock-out mice exhibit defects in palate and tooth development (Asada et al., 1997; Condie et al., 1997;Mucchielli et al., 1997; Gage et al., 1999b; Lin et al., 1999; Lu et al., 1999; Hjalt et al., 2000; St Amand et al., 2000). The comparison of these expression patterns suggests multiple tissues in which Pitx2 may be regulating Gad1 promoter activity. Recently, using an embryonic stem cell-based model of GABAergic differentiation (Westmoreland et al., 2001), we have been able to show that Pitx2 can activate the endogenous Gad1 promoter in neural progenitor cells (J. J. Westmoreland and B. G. Condie, unpublished observations). This observation further supports our hypothesis that Pitx2 is an upstream regulator of GABAergic differentiation in mammals.
Our demonstration that the mouse Pitx2 transcription factor can substitute for the C. elegans UNC-30 protein in drivingunc-25/GAD expression is the first report of a conserved pathway required for the development of a neurotransmitter phenotype. The ability of this factor to rescue unc-25/GAD expression in the worm and to activate the mouse Gad1 gene suggests that it is involved in the differentiation of the GABAergic neurotransmitter phenotype in mammals. It is hoped that this cross-species comparative genetic analysis can be used to highlight additional genes that may be required for development of this neuronal cell type in mammals. In addition, comparative functional studies of mutant Pitx2 proteins in mammalian cells and in the unc-30phenotype rescue assay may lead to new information about the functional domains and residues of this key developmental regulatory factor.
Comparisons of vertebrate and invertebrate embryogenesis have revealed conserved molecular mechanisms controlling the early development of the nervous system. For example, patterning of the mammalian CNS requires the Otx1 and Otx2 homeobox genes, the orthologs of the Drosophila otd (orthodenticle) gene (Simeone, 1998; Acampora et al., 2000). Expression of the humanOtx1 or Otx2 genes in otd mutant flies can partially rescue the otd nervous system phenotypes (Leuzinger et al., 1998; Nagao et al., 1998). Targeted replacement of mouse Otx1 with fly otd also results in a partial rescue of the Otx1 mutant phenotypes (Acampora et al., 1998). The results of the cross-species genetic tests show that the functions of these homeobox factors in early nervous system patterning are conserved. The initial stages of neuronal lineage specification also uses a conserved set of transcription factors; however, cross-species tests of functional conservation for individual genes within this group have not yet been performed (Kageyama et al., 1997;Lee, 1997; Brunet and Ghysen, 1999; Chitnis, 1999; Guillemot, 1999;Hallam et al., 2000). Our results with Pitx2 and UNC-30 indicate that the genetic circuitry regulating the terminal differentiation of neurons may also be highly conserved, with individual genes exhibiting a high level of functional equivalence across species. This suggests that a comparative genetic and developmental approach will be a useful tool to dissect the mechanisms regulating GABAergic neuron differentiation.
In addition to our comparative studies of Pitx2 andunc-30 functions, previous studies of C. elegansneuron differentiation and function have led to insights into the biology of mammalian GABAergic neurons. The mammalian VGAT (also known as the vesicular inhibitory amino acid transporter) was initially cloned using the C. elegans unc-47/VGAT gene as a probe (McIntire et al., 1997; Sagne et al., 1997). The unc-47 gene had been previously identified on the basis of its mutant phenotype as a candidate for the worm VGAT (McIntire et al., 1993a). Additional worm loci may point to conserved regulatory systems involved in mammalian GABAergic development and function. For example, the C. elegans lim-6 LIM-homeobox gene regulates unc-25/GAD expression in the DVB, RIS, and AVL GABAergic neurons (Hobert et al., 1999). This gene is similar to the mammalian Lmx-1a andLmx-1b genes (Hobert et al., 1999). Both of theLmx genes are expressed in the developing CNS and to date have been shown to be involved in formation of the roof plate (Lmx1a) and in mesencephalic dopaminergic neuron differentiation (Lmx1b) (Millonig et al., 2000; Smidt et al., 2000a). It will be of interest to compare the expression patterns of these genes to the distribution of Gad transcripts in the developing CNS. Through this comparative genetic approach we hope to identify additional regulators of GABAergic neuron differentiation in the mouse.
Footnotes
J.M. and Y.J. were supported by National Institutes of Health Grant NS35546 to Y.J. Y.J. is an Alfred P. Sloan research fellow and an Assistant Investigator of Howard Hughes Medical Institute. J.J.W., B.A.M., and B.G.C. were supported by a Medical College of Georgia (MCG) Research Institute grant and an MCG Biomedical Research Support grant to B.G.C. We thank Drs. Jeff Murray, Andy Fire, and Richard Harland for plasmids. We thank Sammy Navarre and Tashon Walker for technical assistance and Dr. Nancy Manley for advice on EMSA and comments on this manuscript.
J.J.W. and J.M. contributed equally to this work.
Correspondence should be addressed to Dr. Brian G. Condie, Institute of Molecular Medicine and Genetics, Medical College of Georgia, 1120 15th Street CB2803, Augusta, GA 30912, E-mail: bcondie@mail.mcg.edu; or Dr. Yishi Jin, Howard Hughes Medical Institute, Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Cruz, CA 95064, E-mail: jin@biology.ucsc.edu.
REFERENCES
- 1.Acampora D, Avantaggiato V, Tuorto F, Barone P, Reichert H, Finkelstein R, Simeone A. Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development. 1998;125:1691–1702. doi: 10.1242/dev.125.9.1691. [DOI] [PubMed] [Google Scholar]
- 2.Acampora D, Gulisano M, Simeone A. Genetic and molecular roles of Otx homeodomain proteins in head development. Gene. 2000;246:23–35. doi: 10.1016/s0378-1119(00)00070-6. [DOI] [PubMed] [Google Scholar]
- 3.Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. Analysis of Pitx2 homeodomain protein activities. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- 4.Amendt BA, Sutherland LB, Russo AF. Multifunctional role of the Pitx2 homeodomain protein C-terminal tail. Mol Cell Biol. 1999;19:7001–7010. doi: 10.1128/mcb.19.10.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asada H, Kawamura Y, Maruyama K, Kume H, Ding RG, Kanbara N, Kuzume H, Sanbo M, Yagi T, Obata K. Cleft palate and decreased brain gamma-aminobutyric acid in mice lacking the 67-kDa isoform of glutamic acid decarboxylase. Proc Natl Acad Sci USA. 1997;94:6496–6499. doi: 10.1073/pnas.94.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson DL, Isackson PJ, Gall CM, Jones EG. Contrasting patterns in the localization of glutamic acid decarboxylase and Ca2+/calmodulin protein kinase gene expression in the rat central nervous system. Neuroscience. 1992;46:825–849. doi: 10.1016/0306-4522(92)90188-8. [DOI] [PubMed] [Google Scholar]
- 7.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. Bioessays. 1999;21:313–318. doi: 10.1002/(SICI)1521-1878(199904)21:4<313::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 9.Bu D-F, Erlander MG, Hitz BC, Tillakaratne NJK, Kaufman DL, Wagner-McPherson CB, Evans GA, Tobin AJ. Two human glutamate decarboxylases, 65-kDa GAD and 67-kDa GAD, are each encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:2115–2119. doi: 10.1073/pnas.89.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan S-K, Mann RS. The segment identity functions of ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev. 1993;7:796–811. doi: 10.1101/gad.7.5.796. [DOI] [PubMed] [Google Scholar]
- 11.Chitnis AB. Control of neurogenesis–lessons from frogs, fish and flies. Curr Opin Neurobiol. 1999;9:18–25. doi: 10.1016/s0959-4388(99)80003-8. [DOI] [PubMed] [Google Scholar]
- 12.Clark SG, Lu X, Horvitz HR. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics. 1994;137:987–997. doi: 10.1093/genetics/137.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condie BG, Bain G, Gottlieb DI, Capecchi MR. Cleft palate in mice with a targeted mutation in the gamma-aminobutyric acid-producing enzyme glutamic acid decarboxylase 67. Proc Natl Acad Sci USA. 1997;94:11451–11455. doi: 10.1073/pnas.94.21.11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophia embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 15.Eastman C, Horvitz HR, Jin Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J Neurosci. 1999;19:6225–6234. doi: 10.1523/JNEUROSCI.19-15-06225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eiden LE. The cholinergic gene locus. J Neurochem. 1998;70:2227–2240. doi: 10.1046/j.1471-4159.1998.70062227.x. [DOI] [PubMed] [Google Scholar]
- 17.Erlander MG, Tillakaratne NJK, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 18.Ernsberger U. Evidence for an evolutionary conserved role of bone morphogenetic protein growth factors and phox2 transcription factors during noradrenergic differentiation of sympathetic neurons. Induction of a putative synexpression group of neurotransmitter-synthesizing enzymes. Eur J Biochem. 2000;267:6976–6981. doi: 10.1046/j.1432-1327.2000.01827.x. [DOI] [PubMed] [Google Scholar]
- 19.Esclapez M, Tillakaratne NJK, Tobin AJ, Houser CR. Comparative localization of mRNAs encoding two forms of glutamic acid decarboxylase with nonradioactive in situ hybridization methods. J Comp Neurol. 1993;331:339–362. doi: 10.1002/cne.903310305. [DOI] [PubMed] [Google Scholar]
- 20.Esclapez M, Tillakaratne NJK, Kaufman DL, Tobin AJ, Houser CR. Comparative localization of two forms of glutamic acid decarboxylase and their mRNAs in rat brain supports the concept of functional differences between the forms. J Neurosci. 1994;14:1834–1855. doi: 10.1523/JNEUROSCI.14-03-01834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa T, Kozak CA, Cepko CL. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA. 1997;94:3088–3093. doi: 10.1073/pnas.94.7.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999a;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- 24.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999b;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 25.Goridis C, Brunet JF. Transcriptional control of neurotransmitter phenotype. Curr Opin Neurobiol. 1999;9:47–53. doi: 10.1016/s0959-4388(99)80006-3. [DOI] [PubMed] [Google Scholar]
- 26.Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 27.Guillemot F. Vertebrate bHLH genes and the determination of neuronal fates. Exp Cell Res. 1999;253:357–364. doi: 10.1006/excr.1999.4717. [DOI] [PubMed] [Google Scholar]
- 28.Hallam S, Singer E, Waring D, Jin Y. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development. 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- 29.Hjalt T, Amendt B, Murray J. PITX2 regulates procollagen lysyl hydroxylase (PLOD) gene expression. Implications for the pathology of Rieger syndrome. J Cell Biol. 2001;152:545–552. doi: 10.1083/jcb.152.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hjalt TA, Semina EV, Amendt BA, Murray JC. The Pitx2 protein in mouse development. Dev Dyn. 2000;218:195–200. doi: 10.1002/(SICI)1097-0177(200005)218:1<195::AID-DVDY17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 31.Hobert O, Tessmar K, Ruvkun G. The Caenorhabditis elegans lim-6 LIM homeobox gene regulates neurite outgrowth and function of particular GABAergic neurons. Development. 1999;126:1547–1562. doi: 10.1242/dev.126.7.1547. [DOI] [PubMed] [Google Scholar]
- 32.Hodgkin J. Male phenotypes and mating efficiency in C. elegans. Genetics. 1983;103:43–64. doi: 10.1093/genetics/103.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372:780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- 34.Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- 35.Katarova Z, Sekerkova G, Prodan S, Mugnaini E, Szabo G. Domain-restricted expression of two glutamic acid decarboxylase genes in midgestation mouse embryos. J Comp Neurol. 2000;424:607–627. doi: 10.1002/1096-9861(20000904)424:4<607::aid-cne4>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 36.Kitamura K, Miura H, Yanazawa M, Miyashita T, Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech Dev. 1997;67:83–96. doi: 10.1016/s0925-4773(97)00110-x. [DOI] [PubMed] [Google Scholar]
- 37.Lamonerie T, Tremblay J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 38.Lanctot C, Gauthier Y, Drouin J. Pituitary homeobox 1 (Ptx1) is differentially expressed during pituitary development. Endocrinology. 1999;140:1416–1422. doi: 10.1210/endo.140.3.6549. [DOI] [PubMed] [Google Scholar]
- 39.Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 40.Leuzinger S, Hirth F, Gerlich D, Acampora D, Simeone A, Gehring WJ, Finkelstein R, Furukubo-Tokunaga K, Reichert H. Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development. 1998;125:1703–1710. doi: 10.1242/dev.125.9.1703. [DOI] [PubMed] [Google Scholar]
- 41.Lin CR, Kioussi C, O'Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 42.Lin L, McGinnis W. Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev. 1992;6:1071–1081. doi: 10.1101/gad.6.6.1071. [DOI] [PubMed] [Google Scholar]
- 43.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- 44.Mallet J, Houhou L, Pajak F, Oda Y, Cervini R, Bejanin S, Berrard S. The cholinergic locus: ChAT and VAChT genes. J Physiol (Paris) 1998;92:145–147. doi: 10.1016/S0928-4257(98)80153-8. [DOI] [PubMed] [Google Scholar]
- 45.Mariani FV, Harland RM. XBF-2 is a transcriptional repressor that converts ectoderm into neural tissue. Development. 1998;125:5019–5031. doi: 10.1242/dev.125.24.5019. [DOI] [PubMed] [Google Scholar]
- 46.McIntire SL, Jorgensen E, Horvitz HR. Genes required for GABA function in Caenorhabditis elegans. Nature. 1993a;364:334–337. doi: 10.1038/364334a0. [DOI] [PubMed] [Google Scholar]
- 47.McIntire SL, Jorgensen E, Kaplan J, Horvitz HR. The GABAergic nervous system of Caenorhabditis elegans. Nature. 1993b;364:337–341. doi: 10.1038/364337a0. [DOI] [PubMed] [Google Scholar]
- 48.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 49.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millonig JH, Millen KJ, Hatten ME. The mouse Dreher gene Lmx1a controls formation of the roof plate in the vertebrate CNS. Nature. 2000;403:764–769. doi: 10.1038/35001573. [DOI] [PubMed] [Google Scholar]
- 51.Mucchielli ML, Martinez S, Pattyn A, Goridis C, Brunet J-F. Otlx2, an Otx-related homeobox gene expressed in the pituitary gland and in a restricted pattern in the forebrain. Mol Cell Neurosci. 1996;8:258–271. doi: 10.1006/mcne.1996.0062. [DOI] [PubMed] [Google Scholar]
- 52.Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C. Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev Biol. 1997;189:275–284. doi: 10.1006/dbio.1997.8672. [DOI] [PubMed] [Google Scholar]
- 53.Nagao T, Leuzinger S, Acampora D, Simeone A, Finkelstein R, Reichert H, Furukubo-Tokunaga K. Developmental rescue of Drosophila cephalic defects by the human Otx genes. Proc Natl Acad Sci USA. 1998;95:3737–3742. doi: 10.1073/pnas.95.7.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Najlerahim A, Harrison PJ, Barton AJL, Heffernan J, Pearson RCA. Distribution of messenger RNAs encoding the enzymes glutaminase, aspartate aminotransferase and glutamic acid decarboxylase in rat brain. Mol Brain Res. 1990;7:317–333. doi: 10.1016/0169-328x(90)90082-o. [DOI] [PubMed] [Google Scholar]
- 55.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 56.Sagne C, El Mestikawy S, Isambert MF, Hamon M, Henry JP, Giros B, Gasnier B. Cloning of a functional vesicular GABA and glycine transporter by screening of genome databases. FEBS Lett. 1997;417:177–183. doi: 10.1016/s0014-5793(97)01279-9. [DOI] [PubMed] [Google Scholar]
- 57.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 58.Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–2116. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- 59.Simeone A. Otx1 and Otx2 in the development and evolution of the mammalian brain. EMBO J. 1998;17:6790–6798. doi: 10.1093/emboj/17.23.6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smidt MP, van Schaick HSA, Lanctot C, Tremblay J, Cox J, van der Kleij AAM, Wolterink G, Drouin J, Burbach JPH. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smidt MP, Asbreuk CH, Cox JJ, Chen H, Johnson RL, Burbach JP. A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat Neurosci. 2000a;3:337–341. doi: 10.1038/73902. [DOI] [PubMed] [Google Scholar]
- 62.Smidt MP, Cox JJ, Schaick HS, Coolen M, Schepers J, Kleij AM, Burbach JP. Analysis of three ptx2 splice variants on transcriptional activity and differential expression pattern in the brain. J Neurochem. 2000b;75:1818–1825. doi: 10.1046/j.1471-4159.2000.0751818.x. [DOI] [PubMed] [Google Scholar]
- 63.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 64.St Amand TR, Zhang Y, Semina EV, Zhao X, Hu Y, Nguyen L, Murray JC, Chen Y. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev Biol. 2000;217:323–332. doi: 10.1006/dbio.1999.9547. [DOI] [PubMed] [Google Scholar]
- 65.Szabo G, Katarova Z, Kortvely E, Greenspan RJ, Urban Z. Structure and the promoter region of the mouse gene encoding the 67-kD form of glutamic acid decarboxylase. DNA Cell Biol. 1996;15:1081–1091. doi: 10.1089/dna.1996.15.1081. [DOI] [PubMed] [Google Scholar]
- 66.Sze JY, Liu Y, Ruvkun G. VP16-activation of the C. elegans neural specification transcription factor UNC-86 suppresses mutations in downstream genes and causes defects in neural migration and axon outgrowth. Development. 1997;124:1159–1168. doi: 10.1242/dev.124.6.1159. [DOI] [PubMed] [Google Scholar]
- 67.Szeto DP, Ryan A, O'Connell S, Rosenfeld M. P-OTX: A PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szeto DP, Rodriguez-Esteban C, Ryan AK, O'Connell SM, Liu F, Kioussi C, Gleiberman AS, Izpisua-Belmonte JC, Rosenfeld MG. Role of the bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tudyka T, Skerra A. Glutathione S-transferase can be used as a C-terminal, enzymatically active dimerization module for a recombinant protease inhibitor, and functionally secreted into the periplasm of Escherichia coli. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vorbruggen G, Constien R, Zilian O, Wimmer EA, Dowe G, Taubert H, Noll M, Jackle H. Embryonic expression and characterization of a Ptx1 homolog in Drosophila. Mech Dev. 1997;68:139–147. doi: 10.1016/s0925-4773(97)00139-1. [DOI] [PubMed] [Google Scholar]
- 71.Westmoreland JJ, Hancock CR, Condie BG. Neuronal development of embryonic stem cells: a model of GABAergic neuron differentiation. Biochem Biophys Res Commun. 2001;284:674–680. doi: 10.1006/bbrc.2001.5031. [DOI] [PubMed] [Google Scholar]
- 72.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode C. elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 73.Wightman B, Baran R, Garriga G. Genes that guide growth cones along the C. elegans ventral nerve cord. Development. 1997;124:2571–2580. doi: 10.1242/dev.124.13.2571. [DOI] [PubMed] [Google Scholar]
- 74.Yanagawa Y, Kobayashi T, Kamei T, Ishii K, Nishijima M, Takaku A, Kobayashi T, Tamura S. Structure and alternative promoters of the mouse glutamic acid decarboxylase 67 gene. Biochemistry. 1997;326:573–578. doi: 10.1042/bj3260573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yasui K, Zhang S, Uemura M, Saiga H. Left-right asymmetric expression of BbPtx, a Ptx-related gene, in a lancelet species and the developmental left-sidedness in deuterostomes. Development. 2000;127:187–195. doi: 10.1242/dev.127.1.187. [DOI] [PubMed] [Google Scholar]