Abstract
Subcellular mRNA localization, a fundamental mechanism for regulating gene expression, leads to local protein translation that results in the generation of neuronal cell polarity. In this study, we have used P19 embryonic carcinoma cells, which are amenable to transfection, and selection of clonal stable cell lines that are not overexpressing the constructs. We identified the 3′ untranslated region (3′UTR) tau axonal localization signal and examined its effect on tau protein localization in nondifferentiated and neuronally differentiated P19 cells. Using GFP-tagged tau constructs combined with in situ hybridization analysis, we demonstrated colocalization of the targeted tau mRNA and its translated protein in the axon and growth cone. Absence of or mutation in the 3′UTR axonal targeting region of tau mRNA resulted in suppression of tau mRNA localization, and both tau mRNA and tau protein remained in the cell body. Swapping between the 3′UTR tau mRNA axonal localization signal and the 3′UTR MAP2 mRNA dendritic targeting signal proved that the localization of the proteins into the axon or dendrites depends on the specific 3′UTR targeting signals. Moreover, the identification of ribosomal proteins in the axon lends further support to the presence of protein synthetic machinery in the axons, a prerequisite for local translation. It is suggested therefore that the P19 cell system can be used to analyze mutations that affect mRNA transport and local translation and that it has the potential of being used to examine the onset of the neuronal differentiation process.
Keywords: tau protein, tau mRNA, axonal targeting signal, ribosomes, P19 EC cells, neuronal differentiation
Neuronal polarity results from the segregated distribution of molecules and organelles and depends on cytoskeletal organization (Bassell and Singer, 1997; Kiebler et al., 1999). It has been established that MAP2, the high molecular-weight microtubule-associated protein (MAP), is localized in the cell body and dendrites, whereas tau MAP is found mainly in the cell body and axons (Matus et al., 1981; Binder et al., 1985).
The molecular mechanisms responsible for the segregation of MAP proteins into the axons and dendrites are not yet fully understood. Subcellular mRNA localization and local translation within dendrites and axons are posttranscriptional control mechanisms that can explain this segregation and may play a key role in generation and maintenance of neuronal polarity. Recent data, which were obtained using molecular approaches and visualization, have demonstrated the presence of unique mRNA species and local protein synthesis in the dendrites, axons, and their growth cones (Crino and Eberwine, 1996; Kaech et al., 1996;Olink-Coux and Hollenbeck, 1996; Tiedge and Brosius, 1996; Antic and Keene, 1998; Bassell et al., 1998; Ludin and Matus, 1998; Rook et al., 2000). For the majority of the targeted mRNAs, thecis-acting sequences required for their localization are found in the 3′ untranslated region (3′UTR) of the transcript. However, little or no sequence homology was found between these regions. It was suggested therefore that RNA-binding proteins recognize specific secondary structures in the 3′UTR, forming particles that are transported then along the microtubules to specific sites (Bassell and Singer, 1997; Kohrmann et al., 1999; Kiebler and DesGroseillers, 2000).
Our studies have shown that tau mRNA in neuronal primary cultures is localized to the proximal segment of the axon, which depends on 3′UTRcis-acting signals, neuronal binding proteins, and functional microtubules (MTS) (Litman et al., 1993, 1994; Behar et al., 1995). Two of the binding proteins were recently identified by our group as HuD proteins, which belong to the Elav-like RNA-binding protein family that binds to AU-rich regions located within the tau axonal targeting region and determine tau mRNA stability (Good, 1997; Aranda-Abreu et al., 1999; Aronov et al., 1999).
In this work, we used P19 embryonic carcinoma (EC) cells that were generated from mouse embryos after 7 d of gestation (McBurney and Rogers, 1982; McBurney et al., 1988). P19 EC cells are initially multipotent and can differentiate in the presence of retinoic acid (RA) into neuronal cells. We show that these cells are readily transfected, and after the selection of a stable cell lines, which are not overexpressing the constructs, can be used to study the effects of the expressed proteins during neuronal-induced differentiation. Using green fluorescent protein (GFP)-tagged constructs, we identified a fragment containing 240 base pairs from the tau 3′UTR, which were required for axonal targeting. We demonstrate colocalization of the message and its translated protein product in neuronal cell bodies and axons. In P19 lines expressing a construct in which tau axonal targeting region is missing or mutated, the tau message and its translated protein remain in the neuronal cell body. When tau targeting signal is replaced by the MAP2 dendritic localization signal (DTE), the tau message and the translated protein are found in the dendrites (Blichenberg et al., 1999). We have identified ribosomes in the axon of differentiated P19 cells, which lends further support to the presence of protein synthetic machinery in the axons, a prerequisite for local translation.
MATERIALS AND METHODS
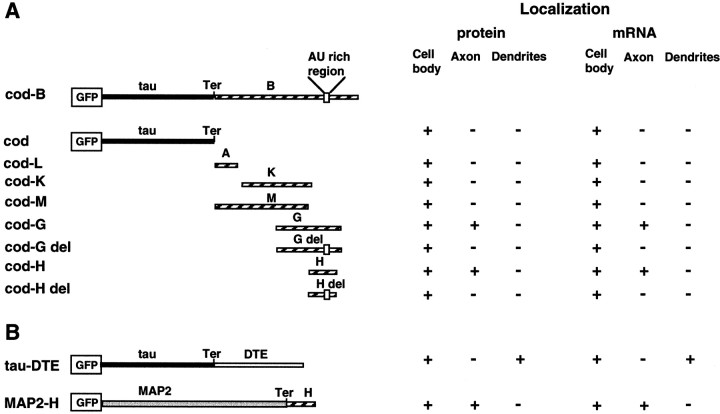
Constructs. All constructs used in this study were cloned in frame into pEGFP-C1 expression vector (Clontech, Palo Alto, CA) and verified by sequence analysis. The schematic representation of the constructs used in this study is shown in Table1. The sizes and locations of the 3′UTR fragments are indicated in the table. The tau–cod construct is the human tau 23 coding region (1134 bp) (Kaech et al., 1996). The tau–cod-H construct includes the 240 bp fragment H from tau 3′UTR (2529–2760) (Sadot et al., 1994; Aronov et al., 1999) cloned downstream of the tau–cod construct. The tau–cod-Hdel was constructed by PCR deletion of 21-AU-rich region from fragment H, as previously described. This region contains the binding site in tau 3′UTR for the HuD stabilization protein (Aranda-Abreu et al., 1999). The tau–cod–MAP2-DTE construct and the reciprocal MAP2–cod–H include a 640 bp fragment of MAP2 3′UTR and a 240 bp fragment of tau 3′UTR, respectively (Blichenberg et al., 1999) (kindly provided by Dr. S. Kindler, Hamburg, Germany). For selection of stable cell lines, we cotransfected with a plasmid containing the puromycin resistance gene, downstream of the pgk promoter (kindly provided by Dr. Peter W. Laird, The Netherlands Cancer Institute, Amsterdam, The Netherlands).
Table 1.
Schematic representation of constructs used in the study
Cell cultures and stable transfections. P19 cells were grown as previously described (Falconer et al., 1992) in MEM containing 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mml-glutamine, in an incubator with 5% CO2. Cotransfection and selection of stable cell lines were aided by FACS sorting, as previously described (Heicklen-Klein et al., 2000). At this stage, the cell lines consisted of ∼90% neuronal cells. Each transfection and cell line selection was repeated at least three times with two different plasmid DNA preparations.
Confocal microscopy analysis of P19 cells. Control P19 cells or stably transfected cell lines were analyzed by Zeiss confocal microscope. For living cells analysis, the cells were washed twice in Ham's F12 medium without phenol-red indicator and containing bovine serum albumin (1 mg/ml) plus 10 mm HEPES, pH 7.5. In the absence of phenol-red, the medium generates substantially less autofluorescence background (Ludin et al., 1996). For high-resolution pictures, the cells were grown on coverslips and transferred to growth chambers to facilitate high-magnification visualization with oil-immersion lenses. GFP-fluorescence signals were observed with GFP-adapted filters. Images were viewed with a Zeiss confocal microscope and analyzed using the LSM confocal imaging software system.
In situ hybridization analysis, RNA probes, and immunohistochemical staining. Differentiated P19 cells were fixed with 4% paraformaldehyde in the presence of 4% sucrose (Litman et al., 1993). A GFP single-stranded RNA probe (452 bp) was synthesized in the sense and antisense orientation, using the appropriate polymerase (T3 or T7 RNA polymerase) in the presence of digoxygenin UTP (RNA transcription kit; Boehringer Mannheim, Mannheim, Germany). In situ hybridization was performed as previously described (Litman et al., 1994). For visualization of the in situhybridization signals and immunostaining with tubulin antibodies, the slides were incubated overnight at 4°C with HRP-conjugated monoclonal anti-Dig (1:500) (Jackson ImmunoResearch, West Grove, PA) together with monoclonal anti-β-tubulin (Biomakor, 1:100). The slides were incubated then for 2 hr at room temperature with a mixture of secondary antibodies including anti-HRP Cy5 (1:100) (Jackson Immuno-Research) and goat anti-mouse Cy3 (1:500) (Jackson ImmunoResearch) for Dig and tubulin antibodies, respectively. For tau immunostaining, tau-1 antibodies were used, followed by goat anti-mouse Cy5 (1:500) (Jackson ImmunoResearch). The coverslips were mounted with mowiol and visualized with an LSM confocal laser scanning imaging system, with 40× objective using a green filter for GFP (excitation, 488 nm; emission, 505–550 nm), a red filter for tubulin (excitation, 545 nm; emission, 560–580 nm), and a far-red filter for mRNA labeled with digoxigenin (excitation, 650 nm; emission, 680 nm). Dendrites were identified by immunochemical staining with polyclonal MAP2 antibodies (kindly provided by Craig C. Garner, University of Alabama, Birmingham, AL), followed by incubation with Cy3-labeled secondary antibodies (Jackson ImmunoResearch). Ribosomes were detected by staining with antibodies to 60S ribosomal subunit (kindly provided by John Hesketh, Department of Biological and Nutritional Sciences, University of Newcastle, Newcastle upon Tyne, UK), followed by incubation with Cy3-labeled secondary antibodies. Control experiments showed that no fluorescent signal is observed in the absence of primary antibodies and that there is no penetrance of signals between the two filters used.
Reverse transcription-(PCR). For quantitative reverse transcription (RT)-PCR, RNA was extracted from undifferentiated or differentiated P19 transfected lines using a Pure kit (Promega, Madison, WI). Extracted RNA (1 μg) was reverse-transcribed in 20 μl with 200 U of Moloney murine leukemia virus reverse transcriptase (Promega) in the presence of random hexamers (0.125 ng/μl), 20 U of RNasin (Promega), and dNTPs at a final concentration of 0.125 mm, for 1 hr at 37°C. PCR was performed on 1 μl of the reverse transcription reaction mixture in a final volume of 50 μl with 2.5 U of Taq polymerase (Promega, according to the manufacturer's instructions) and 20 pmol of the specific primers. The following primers were used to analyze the levels of the RNA isolated from transfected cell lines: for GFP-tau, 5′-ACTACCTGAGCACCCAGTCC-3′ (1089–1108) (Clontech, sequence information) and 5′-TTTGCTGGAATCCTGGTGG-3′ (597–617), to yield a product of 510 bp; and for glyceraldehyde phosphate dehydrogenase (GAPDH), 5′-GCCATCAACGACCCCTTCAT-3′ (118–137) and 5′-TTCACACCCATCACAAACAT-3′ (412–431), to yield a product of 314 bp (Tso et al., 1985). To obtain linear amplification curves, the cDNA mixtures were subjected to 25, 30, and 35 cycles for GFP-tau and 20, 25, and 30 cycles for the GAPDH control, under the following conditions: denaturing at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min. The final extension step was performed at 72°C for 5 min. Then, the samples were separated on a 1% agarose gel and viewed under UV light. Intensities of tau transcripts were normalized to the GAPDH internal control and found to be linearly related to the number of cycles used.
Immunoblot analysis of P19 protein extracts. Proteins were extracted from P19 cells in one volume of lysis buffer consisting of 50 mm Tris, pH 8.5, 1% Triton X-100, 5 mm EDTA, 0.15 m NaCl, and 50 μg/ml phenylmethylsulfonyl fluoride. Cell debris was clarified by centrifugation for 10 min at 16,000 × g at 4°C. Protein samples (25 μg) were resolved by SDS-gel electrophoresis, transferred to nitrocellulose filters, and reacted with tau-1 monoclonal antibody (1:100,000) (Binder et al., 1985) at 4°C for 16 hr. Then, they were visualized with HRP anti-mouse secondary antibodies (Jackson ImmunoResearch) at room temperature for 1 hr and developed using the ECL chemiluminescence procedure.
RESULTS
P19 cells differentiate into neuronal cells showing MAP segregation
The low transfection efficiency of postmitotic neuronal cells (Goslin and Banker, 1997) has hampered studies requiring the expression and translation of exogenous genes. Moreover, it is difficult to follow the targeting of transfected mRNAs and the localization of their translated proteins during the differentiation process in primary cultures. In this study, we overcame this difficulty by using P19 cells transfected with GFP-tagged tau constructs. The schematic representation of the constructs that were used in the current studies for establishing stable cell lines is shown in Table 1. After RA-induced differentiation, the transfected RNA and its translated tagged protein can be subcellularly localized in both living and fixed neuronal cells.
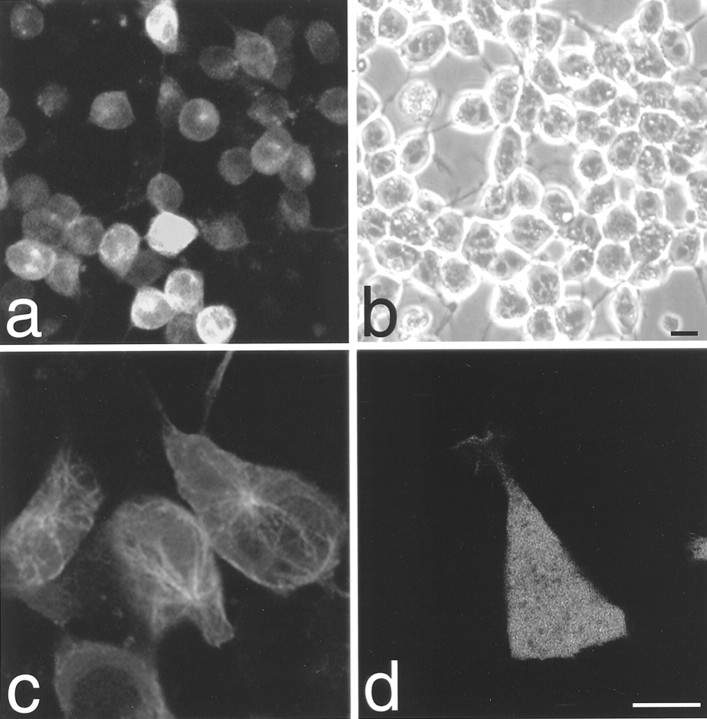
An undifferentiated stable cell line selected after transfection with a GFP-tau–cod-H construct is shown in Figure1a,b. The cells express the fused GFP-tau protein, which is assembled into MTs (Fig. 1c). Before RA-induced neuronal differentiation, a similar pattern of MT staining was observed in all selected cell lines transfected with the different GFP-tau constructs tested (data not shown). These results are similar to previously described findings (Kaech et al., 1996) of a normal distribution of GFP-tau on MTs in non-neuronal HeLa, Chinese hamster ovary, and human hepatoma line PLC, leading us to conclude that the GFP tag does not affect the ability of tau protein to assemble into MTs and can be used to view MTs in living cells with minimal perturbation. In contrast, a control cell line that was selected after transfection with the GFP vector alone shows a diffuse pattern of the GFP protein throughout the cell (Fig.1d).
Fig. 1.
Confocal image analysis of undifferentiated P19 living cell lines. a, Field view of a selected stable cell line transfected with GFP-tau–cod-H construct. b, Phase view of a. Because the photograph shows living cells, not all of the cells in the field are in the same plane; hence, the differences observed in the intensity of GFP fluorescence. Scale bar, 50 μm. c, Distribution of tau protein on MTs in the stable cell line transfected with the GFP-tau–cod-H construct. d, Distribution of P19 cells transiently transfected with the GFP vector alone. Scale bar, 5 μm.
During neuronal differentiation, P19 cells generate an extensive network of processes that can be distinguished as axons or dendrites both morphologically and by the presence of specific cytoskeletal markers, MAP2 in the dendrites and tau in the axons, that are not expressed in the undifferentiated state (Falconer et al., 1992; Tanaka et al., 1992; Heicklen-Klein et al., 2000). The distribution of GFP-tau and MAP2 in a differentiated P19 cell line transfected with GFP-tau–cod-H construct is shown in Figure2. The tau protein is localized in the cell body and axon (Fig. 2a), whereas MAP2 immunostaining is found exclusively in the cell body and dendrites (Fig. 2b). The merged image and the phase view of the same cell are shown in Figure 2, c and d, respectively. The endogenous tau protein localization in differentiated control P19 cells, as stained with monoclonal tau-1 antibodies, is seen in the cell body and axon and is shown in Figure 2e. We therefore conclude that the P19 cell system shows the well known morphological differentiation pattern and segregation of cytoskeletal elements and can be used to study tau mRNA localization. In addition, the transfection experiments indicated that it is possible to obtain a clonal cell line, a useful tool for future biochemical studies.
Fig. 2.

Confocal image analysis of MAP segregation in neuronally differentiated P19 cells. a–d, P19 stable cell expressing GFP-tau–cod-H construct, neuronally differentiated for 9 d. MAP segregation was analyzed by confocal microscopy. Scale bar, 5 μm. a, GFP-tau protein is visible (green) in the soma and axon of a single cell.b, MAP-2 is visible (red) in the soma and dendrite of the same cell. c, Computer-merged analysis of GFP-tau (green) and MAP-2 (red) proteins; GFP-tau and MAP-2 colocalize (orange) in the soma but segregate to the axon and the dendrite, respectively.d, Phase view of the same P19 cell. e, The same cell stained with tau-1 antibodies. Large filled arrowheads denote an axon, large open arrowheads denote dendrites, and small filled arrowheads denote a neuronal cell body.
Tau mRNA and protein localization in neuronal cells is dependent on 3′UTR signals
In undifferentiated P19 cell lines, GFP-tau proteins expressed from the various constructs showed a similar localization and patterns of assembly into MTs. After neuronal differentiation, the localization of the tagged tau protein in living cells differed in the various tested stable cell lines (Fig. 3). In cells transfected with GFP-tau–cod-H, the protein was visible in the cell body and axon (Fig. 3a,b), whereas in cells transfected with GFP-tau–cod (Fig. 3c,d) or GFP-tau–cod Hdel (Fig. 3e,f) the protein remained in the cell body and did not enter the axon. Lines expressing tau coding region linked to fragments A, K, or M, which do not contain the region including the stabilization cis-signal (Table 1), exhibited similar localization to the tau-cod region only, namely, the GFP-tau protein was observed in the cell body only. Similarly, in the cell line expressing the construct GFP-tau–cod-G, the localization that was observed in living neuronal cells was in the cell body and axon, whereas the cell line that was transfected with the construct GFP-tau–cod-Gdel showed expression only in the cell body (data not shown).
Fig. 3.
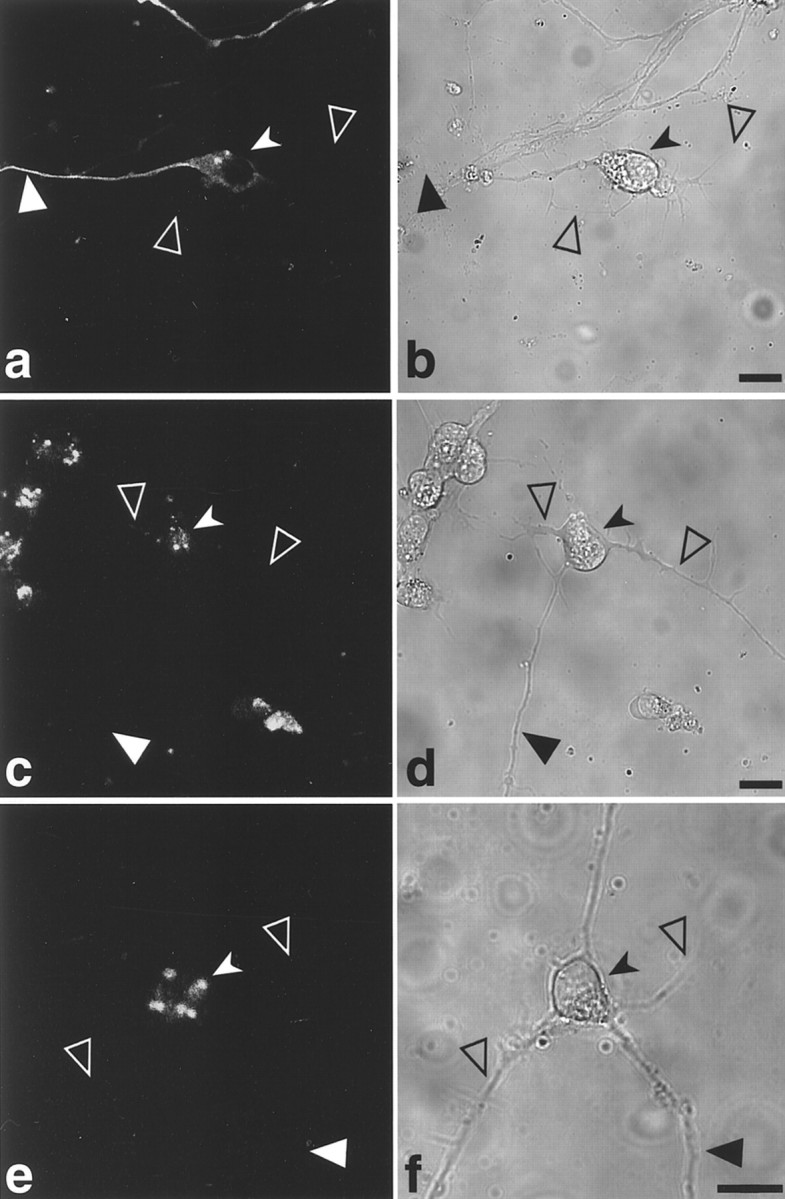
Confocal image analysis of neuronally differentiated P19 living cell lines stably transfected with GFP-tau constructs. Distribution of GFP-tau in cell lines transfected with GFP-tau–cod-H construct (a, b), GFP-tau–cod construct (c, d), and GFP-tau–cod-Hdel (e,f) in differentiated P19 cells. Theright panels show a phase view of the cells. Scale bars, 20 μm. Large filled arrowheads denote an axon,large open arrowheads denote dendrites, and small filled arrowheads denote a neuronal cell body.
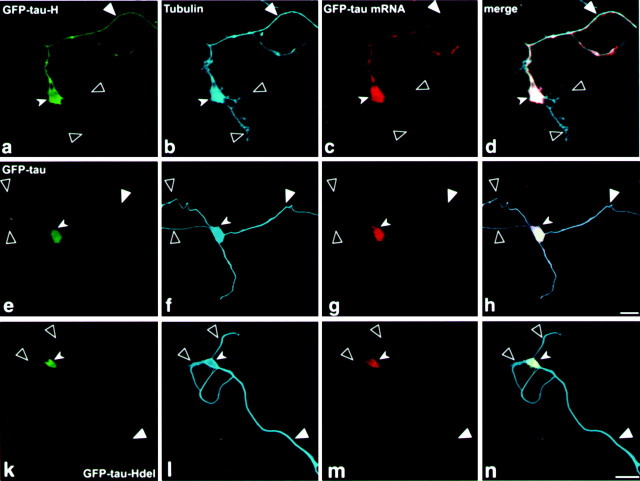
To analyze the distribution of tau message in these cell lines, which will demonstrate a direct connection between the localization of the message and its translated product, in situ hybridization analysis was performed using a GFP probe (Fig.4c,g,k). This probe specifically detects the transfected mRNA and does not detect the endogenous tau mRNA. The results of confocal image analysis (Fig. 4) demonstrate colocalization of the message with its encoded protein (Fig. 4d,h,i). Thus, for example, GFP-tau–cod-H mRNA is present in the cell body and axon, indicating that the mRNA was transported from the cell body into the axon (Fig. 4c). In contrast, the mRNA lacking a 3′UTR axonal signal or with a deletion of the stabilization region (Aranda-Abreu et al., 1999) remains in the cell body (Fig. 4g,k). Deletion of the 21-AU-rich region from fragment H is shown here to be part of the cis-acting signal important for tau mRNA targeting. Control in situ hybridization experiments with GFP-sense probe or GFP-antisense probe on nontransfected cells did not show any signal (data not shown). We conclude that fragment H of 240 bp is necessary and sufficient for the localization of tau mRNA, and the differences in protein distribution result from differences in their mRNA localization, which is dependent on the 3′UTRcis-signals.
Fig. 4.
Confocal microscopy image of P19 cell cultures analyzed by in situ hybridization combined with immunohistochemistry using tubulin antibodies. a–d, Confocal image of a P19 cell line transfected with GFP-tau–cod-H construct. e–h, Confocal image of a P19 cell line transfected with GFP-tau–cod construct. i–l, Confocal image of a P19 cell line transfected with GFP-tau–cod-Hdel construct. a, e, i, GFP-tau fluorescence. b, f,j, P19 cell immunostained with tubulin antibodies.c, g, k, In situ hybridization using a GFP probe labeled with UTP-dig and detected with anti-dig HRP/Cy5. d, h,l, Merged confocal image showing colocalization of GFP-tau fluorescence with tubulin and with GFP-tau mRNA. Scale bars, 10 μm. Large filled arrowheads denote an axon,large open arrowheads denote dendrites, and small filled arrowheads denote a neuronal cell body.
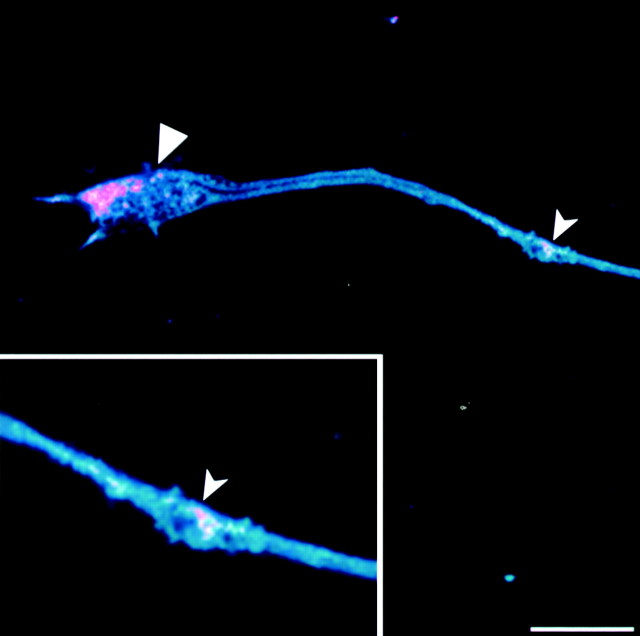
RNA localization in neuronal cells requires a functional MT system (Bassell et al., 1994; Litman et al., 1994). Immunohistochemical staining with tubulin antibodies stains the entire neuronal cell, including the axon and dendrites (Fig.4b,f,j). The merged image from the confocal microscopy analysis, combining the green GFP tau proteins with the cyan MT staining and the red in situ GFP-tau mRNA hybridization signal, shows colocalization that is visualized as a white–cyan color (Fig.4d,h,l). In the GFP-tau–cod-H cell line, the colocalization is seen in the cell body and along the axon, reaching as far as the growth cone (Fig. 4d). Distribution of the message in the axon is observed as a patchy granular form (Fig. 4c), which is seen more clearly in the merged image of the GFP-tau–cod-H cell line (Fig. 4d). This granular distribution, which consists of the moving ribonucleoprotein (RNP) particles, has been described in neurons and oligodendrocytes (Ainger et al., 1993; Knowles et al., 1996; Rook et al., 2000). A higher magnification of a confocal merged image of the axon and a growth cone of the differentiated P19 cell line transfected with GFP-tau–cod-H (Fig. 4d) is shown in Figure5. The granules, as analyzed by in situ hybridization, include tau mRNA, are clearly seen, and are colocalized with the microtubules along the axon (marked bysmall arrowheads). The high density of granules located at the growth cone (marked by large arrowheads) is consistent with the growth activity of a developing axon in early differentiating neurons, which requires MT assembly.
Fig. 5.
Granules observed in the axon and growth cone of a differentiated P19 cell. Merged confocal image at higher magnification of the axon and growth cone analyzed by in situhybridization and tubulin staining of a P19 cell line transfected with GFP-tau–cod-H construct (as shown in Fig. 4). Scale bar, 50 μm.Large arrowhead denotes granules in the growth cone, andsmall arrowheads denote granules along the microtubules.
It has been shown previously that the neuronal granules include protein synthesis machinery (Knowles et al., 1996; Tiedge and Brosius, 1996;Rook et al., 2000). The entire cell is immunostained with tubulin antibodies (Fig. 6Aa). Using antibodies specific for ribosomal proteins, we detected ribosomes in differentiating P19 cells, which are present both in the dendrites and axon (Fig. 6Ac). A higher magnification of an axonal segment shows that the ribosomes reach the growth cone (Fig.6Bc′) and are colocalized in granules with tau mRNA (Fig. 6Bb′). The granules, measured both in the axon and in the growth cone, have a mean particle size of 0.5–0.6 μm, similar to the sizes recorded for myelin basic protein in oligodendrocytes and for β-actin in neurons (Ainger et al., 1993;Knowles et al., 1996; Bassell et al., 1998).
Fig. 6.
Ribosomal proteins are detected in the axon and dendrites of differentiating P19 cells. A, Confocal microscopy image analysis of P19 cell stained with anti-tubulin (a). b, Anti 60s ribosomal proteins. c, In situ hybridization using a tau probe, which detects both the endogenous and transfected tau mRNA. d, Merged confocal image showing colocalization.B, The region boxed in the inset ind is shown in enlarged scale in a′,b′, c′, and d′, corresponding to a, b, c, and d, respectively. Scale bars, 10 μm.Large filled arrowheads denote an axon, large open arrowheads denote dendrites, and small filled arrowheads denote a neuronal cell body.
The tau mRNA and protein levels in transfected cell lines
The localization of mRNA to a particular subcellular compartment, as a mechanism for the control of gene expression, presumably serves the function of minimizing ectopic expression and maximizing local translation at the site at which the protein is functionally required (Carson et al., 1998). Because the tau protein can be regulated at the transcriptional and posttranscriptional levels (Burstein and Greene, 1978; Ginzburg et al., 1982; Sadot et al., 1994), we estimated the levels of GFP-tau protein and RNA in nontransfected and transfected lines by Western blotting and RT-PCR analysis, respectively (Figs. 7,8). The analysis was performed in undifferentiated and differentiated cells on days 8–10, when neuronal differentiation was already at a maximum and ∼90% of the cells had extended processes (Heicklen-Klein et al., 2000).
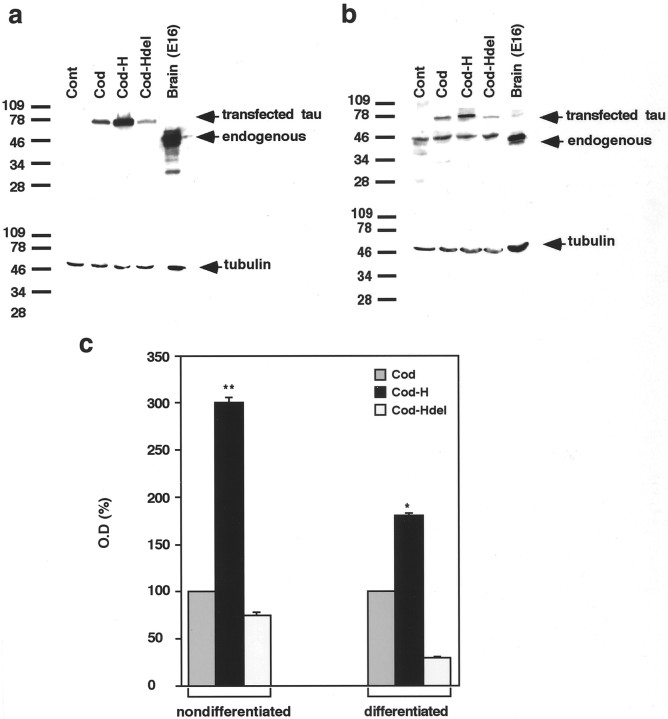
Fig. 7.
Western blot analysis of GFP-tau proteins in stable P19 cell lines. Expression of GFP-tau proteins detected using tau-1 antibody in undifferentiated P19 cells (a) and in P19 cells after differentiation for 8 d (b). The same membrane was used for detection of tubulin as an internal control (a representative blot).c, Quantitative analysis of the results of four experiments, expressed in arbitrary units calibrated to the signal of cells transfected with GFP-tau–cod construct (100%). Values are means ± SEM. Asterisks mark a significant difference in tau protein levels (*p < 0.05, **p < 0.01).
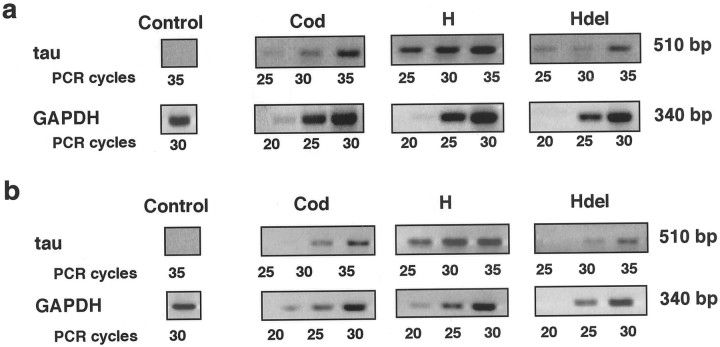
Fig. 8.
RT-PCR analysis of GFP-tau mRNA levels in stable transfected P19 cell lines. Expression of GFP-tau mRNA levels in undifferentiated P19 cells (a) and in P19 cells after differentiation for 8 d (b). A GAPDH signal was used as an internal control. PCR was allowed to proceed for 25, 30, and 35 cycles using GFP-tau primers (yielding a fragment of 510 bp) and for 20, 25, and 30 cycles using GAPDH primers (yielding a fragment of 340 bp). A representative blot from four experiments is shown. The results demonstrate a linear relationship along the cycles. A control experiment, using RNA isolated from nontransfected P19 cells and processed concomitantly, is presented.
The amount of tau protein was assayed using tau-1 antibodies and compared with the amount of tubulin (Fig. 7). Control nontransfected P19 cells, before undergoing neuronal differentiation, do not express endogenous tau, whereas on differentiation endogenous tau protein is detectable and can be distinguished by its size from the tau expressed by the GFP-fused construct. The amount of tau protein in cell lines transfected with the GFP-tau–cod-H construct was significantly (threefold) higher than in cell lines transfected with a construct containing the coding region alone. P19 cells transfected with tau construct containing the deletion of the stabilization signal exhibited a relatively small amount of the protein (35% of GFP-tau–cod). A similar ratio of tau protein was found in the various cell lines after neuronal differentiation, but the amount of protein expressed was reduced. It is important to note that in differentiated P19 cell line transfected with a construct containing fragment H, the levels of GFP-tau protein and endogenous tau protein are similar, whereas in cells transfected with tau–cod or tau–cod-Hdel, the level of the transfected GFP-tau protein is lower than the endogenous protein.
The amounts of RNA in the transfected lines were analyzed by quantitative RT-PCR, using primers specific for GFP and tau, and compared with a standard control of endogenous GAPDH (Fig. 8). The amount of GFP-tau–cod-H mRNA was three times higher than the amount of mRNA derived from cells expressing the coding region only. Cells transfected with a tau construct containing deletion of the stabilization signal had a low level of expression (30% of GFP-tau–cod). These results point to a correlation between the amounts of the mRNAs derived from the constructs and the amounts of the translated proteins, again implying that the presence of fragment H contributes tau mRNA stability and results in higher tau protein levels. From these results, it is possible to suggest that in undifferentiated P19 cells, which are still multipotent, the regulation of tau protein expression might not be identical to that observed in neuronally differentiated cells, which may require additional neuronal-induced proteins. In addition, we showed previously that the activity of cytomegalovirus promoter, which drives the expression of the transfected constructs, is lower in differentiated than in undifferentiated P19 and thus may explain the reduction of tau expression in differentiated P19 cells (Heicklen-Klein et al., 2000).
The 3′UTR axonal and dendritic targeting signals determine dendritic or axonal localization
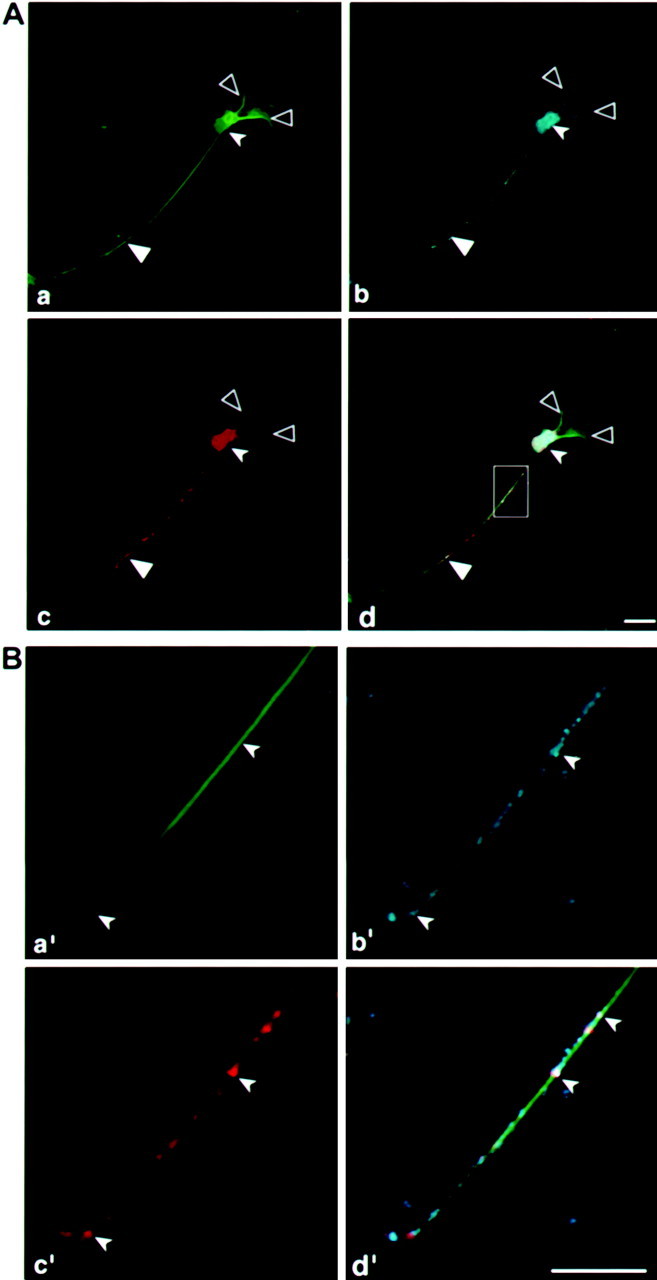
To determine whether the axonal targeting of the tau-H construct is directed by fragment H or influenced by tau–coding sequences as well, we replaced fragment H with a MAP2 localization signal, the DTE fragment that drives MAP2 mRNA into the dendrites (Blichenberg et al., 1999). A cell line expressing GFP-tau–cod-DTE was selected and analyzed after differentiation. The MAP2 localization signal drove the tau message and its encoded protein into the dendrites (Fig.9a,c). The dendritic localization was verified by immunostaining with MAP2 antibodies (Fig. 9b). The complementary experiment, in which a cell line expressing MAP2 coding region is linked to tau axonal targeting fragment H, was selected and is shown in Figure10. As can be seen, the GFP-MAP2 protein is present in the cell body and axon (Fig. 10a). Immunohistochemical analysis with MAP2 specific antibodies (Fig.10b) stains both the axon, in which the transfected protein is localized, and the dendrites in which the endogenous MAP2 is present. The transfected RNA, as tested by in situhybridization with GFP probe, is present only in the cell body and axon (Fig. 10c). These results provided a direct evidence for the colocalization of the targeted message with its translated protein, an axonal or dendritic localization that depends on the specific 3′UTR targeting sequences.
Fig. 9.

MAP2-targeting signal drives tau expression into the dendrites of differentiated P19 cells. Confocal image of a P19 cell line transfected GFP-tau–cod-MAP2-targeting signal. a, Localization of GFP-tau protein in the dendrites. b, P19 cell stained with MAP2 antibodies to visualize the dendrites.c, Localization by in situ hybridization of GFP-tau mRNA with a GFP probe detected with anti-dig HRP/Cy5.d, Merged image of a, b, and c showing the colocalization of GFP-tau protein and mRNA in the dendrites. e, Phase view. Note that the axon seen in e is not stained in a–d because tau is driven to the dendrites. Scale bar, 10 μm. Large filled arrowheads denote an axon, large open arrowheads denote dendrites, and small filled arrowheads denote a neuronal cell body.
Fig. 10.
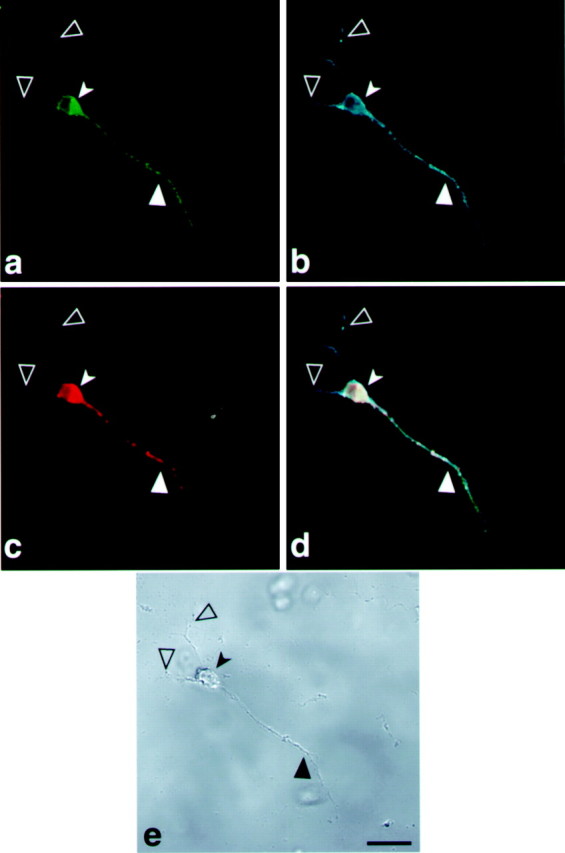
Tau-targeting signal drives MAP2 expression into the axon of differentiated P19 cells. Confocal image of a P19 cell line transfected with a construct containing GFP-MAP2-cod-fragment-H of tau 3′UTR. a, Localization of GFP-MAP2 protein in the axons.b, P19 cell stained with MAP2 antibodies shows the dendrites and axon. c, Localization by in situ hybridization of GFP-MAP2 mRNA with a GFP probe detected with anti-dig HRP/Cy5. d, Merged image ofa, b, and c showing the colocalization of GFP-MAP2 protein and mRNA in the axon.e, Phase view. Scale bar, 10 μm. Large filled arrowheads denote an axon, large open arrowheadsdenote dendrites, and small solid arrowheads denote a neuronal cell body.
DISCUSSION
Posttranscriptional control mechanisms, which play an important role in determining the polarity and activity of neuronal cells, have been investigated in recent years using primary neuronal cultures as a model system (Steward, 1997; Kiebler and DesGroseillers, 2000). Studies have demonstrated the subcellular localization of specific mRNAs in the dendrites and the axon, and in most cases this targeting is dependent on 3′UTR cis-acting signals (Blichenberg et al., 1999). The control of mRNA localization restricts proteins to a particular domain of the polarized cell. The high level of a particular protein at the site of localization facilitates its rapid supply when needed (Wilhelm and Vale, 1993; St Johnston, 1995). Moreover, in neuronal cells, several identified dendritic targeted mRNAs are reportedly upregulated by neuronal activity, and thus, are actively involved in neuronal plasticity (Steward, 1997; Steward et al., 1998; Schuman, 1999; Rook et al., 2000).
A direct connection between the localization of specific mRNAs and their translated proteins has so far been unsuccessful. Recent advances using GFP-tagged proteins have made it possible to study protein movement and localization in living cells (Ludin and Matus, 1998; Kohrmann et al., 1999; Rook et al., 2000). In an attempt to overcome the low efficiency of transfection of postmitotic neuronal cells, we used P19 cells that were stably transfected with GFP-tagged constructs to follow the localization of the expressed tau protein in living neuronal cells. This cell system, which is amenable to transfection, allows the selection of a stable cell line, thereby eliminating the variability attributable to overexpression that is observed often in transient transfection experiments. Moreover, the use of P19 cell lines allows us to study the effect of mRNA localization and protein function during the different stages of neuronal differentiation. P19 EC cells offer a good model system for differentiating neuronal cells that express early and mature neuronal proteins. The segregation of additional neuronal proteins, HuD, GAP-43, VAMP, and SV (the last two being synaptic vesicle proteins), into the axon and dendrites of differentiated P19 cells has also been demonstrated (Finley et al., 1996; Steller et al., 1996; Parnas and Linial, 1997; Mani et al., 2000).
In previous studies using primary cell cultures, we detected tau mRNA localization in the proximal hillock of the axon (Litman et al., 1994). In this study, we were able to demonstrate that fragment H, which includes 240 base pairs, is necessary and sufficient for tau localization in the axon. A summary of the results of tau protein and tau mRNA localization, obtained with the various 3′UTR constructs that were tested, is presented in Table 1. Only fragment H and the longer fragment G showed axonal localization. Other regions of tau 3′UTR, i.e., fragments A, K, and M, were tested similarly but did not indicate targeting (data not shown). The axonal localization of tau protein was abolished in cell lines transfected with constructs that included tau coding region alone or fragment H or G with a deletion in tau mRNA stabilizing signal (Fig. 4) (Aranda-Abreu et al., 1999). We cannot yet separate between the cis-signal(s) responsible for stabilization and the whole 240 bp fragment necessary and sufficient for targeting of tau mRNA, which may suggest that the stabilization step is a prerequisite for localized mRNAs en route to their specific destination within the cell. The fact that fragment G and fragment H showed the same axonal localization suggests that fragment G does not include additional cis-signals that are required for localization. The information obtained by using tagged proteins, together with the results of the in situ hybridization analysis using a specific probe for the transfected construct, provides the first direct evidence for colocalization of the message and its translated protein. When a dendritic localization signal derived from MAP2 3′UTR was attached to tau coding region, both tau mRNA and its translated protein were detected in the dendrite. Conversely, when tau–H fragment was attached to MAP2 coding sequences, both MAP2 mRNA and its translated protein were detected in the axon. These results further demonstrate that the required cis-signals located in the 3′UTR of the targeted message are sufficient and necessary for axonal and dendritic targeting (Figs. 9, 10). Using a P19 cell line and confocal microscope analysis, we could detect tau mRNA along the whole axon as far as the growth cone, a similar localization to that demonstrated for β-actin mRNA in primary neuronal cell cultures (Zhang et al., 1999). The presence of ribosomes in the axon, as necessary components for local protein synthesis is demonstrated in these differentiating cells (Fig. 6). A growing body of evidence is accumulating to challenge the prevailing dogma for the presence of protein synthetic machinery in axons. The presence of ribosomes along mammalian axons was recently shown as focal centers of local translational activity (Koenig and Giuditta, 1999; Koenig et al., 2000). Tau message in the present study is distributed in granular structures that are colocalized along MTs. A preliminary analysis of their protein composition shows that the granules contain the stabilizing protein HuD and a motor protein from the kinesin family (our unpublished observations). Our observation that the localization of tau and MAP2 mRNAs and proteins in the axons and dendrites, respectively, is guided by the axonal and dendritic targeting signals raises the possibility that specifictrans-acting protein factors, including motor proteins, contribute to the directionality of the migration. These proteins may recognize the polarity of the microtubules and bind directly or indirectly to the cis-signals, forming an RNP particle that determines RNA localization. The composition of the RNP proteins remains to be determined (Schnapp, 1999).
The results presented in this study further support the suggested presence of a multistep localization pathway in somatic cells (Wilhelm and Vale, 1993). Our experiments link the presence of the transfected RNA, the first cytoplasmic component in the localization pathway, to its final product, the translated protein in its functional localization. It is tempting to suggest that both the stability and the localization of tau mRNA are coupled to its translation, as observed by its association both with ribosomes and MTs, as previously shown for myelin basic protein (Ueno et al., 1994a,b). In the case of myelin basic protein, the sequence originally identified as the RNA trafficking signal was shown to function by controlling cap-dependent translation (Kwon et al., 1999), while anothercis-sequence from this region is involved in the transport (Munro et al., 1999). In fibroblasts, specific “zip-codes” are involved in localization of β-actin to the leading edge of the cell and its binding to the actin cytoskeleton (Kislauskis et al., 1994). The cis-signals and trans-acting binding proteins may have multiple roles in RNA processing and trafficking, including the steps of nuclear transport, MT-dependent transport in the cytoplasm (Litman et al., 1994; Carson et al., 1997; Kohrmann et al., 1999), and subsequent spatial anchoring and translation (Carson et al., 1998). The protein composition of the granules representing the active transport unit that delivers the mRNAs to their final cellular microdomains and their specific functions remain to be elucidated.
P19 cell lines can be differentiated into different cell lineages that can be used to study the common and the unique components for forming cellular polarity on both the biochemical and the cellular level. Because the stable cell lines produced are homogeneous, no variability in expression or localization is observed (Feng et al., 2000). Our previous results show that tau promoter expression is regulated in P19 during neuronal-induced differentiation, similarly to their in vivo regulation (Heicklen-Klein et al., 2000). The fact that there is a good correlation between the levels of the GFP-tau protein in GFP-tau–cod H cell line and the level of the endogenous tau protein (Fig. 7) suggests that the transfected construct is regulated similarly to the endogenous protein. These results are in agreement with recent information about the expression of GAP-43 gene in P19 cells, which show that failure to express GAP-43 interferes with their neuronal differentiation and stimulates apoptosis (Mani et al., 2000, 2001). Therefore, cell lines can be used to analyze mutations that affect the targeting of specific messages and their ectopic translation, and the function of the proteins can be tested.
Footnotes
This work was supported by grants from the Minerva Foundation (München, Germany); Nella and Leon Benoziyo Center for Neuroscience, Weizmann Institute of Science (Rehovot, Israel); and Grant 1999149 from the United States–Israel Binational Science Foundation (Jerusalem, Israel). I.G. is the incumbent of the Sophie and Richard S. Richard Professorial Chair in Cancer Research.
S.A. and G.A. contributed equally to this project.
Correspondence should be addressed to Irith Ginsburg, Department of Neurobiology, The Weizmann Institute of Science, 76100 Rehovot, Israel. E-mail: irith.ginzburg@weizmann.ac.il.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antic D, Keene JD. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–197. doi: 10.1242/jcs.111.2.183. [DOI] [PubMed] [Google Scholar]
- 3.Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I. Embryonic lethal abnormal vision-like RNA binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J Neurosci. 1999;19:6907–6917. doi: 10.1523/JNEUROSCI.19-16-06907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aronov S, Marx R, Ginzburg I. Identification of 3′ UTR regions implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999;12:131–145. doi: 10.1007/BF02736927. [DOI] [PubMed] [Google Scholar]
- 5.Bassell G, Singer RH. mRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 6.Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 7.Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behar L, Marx R, Sadot E, Barg J, Ginzburg I. Cis-acting signals and trans-acting proteins are involved in tau mRNA targeting into neurites of differentiating neuronal cells. Int J Dev Neurosci. 1995;13:113–127. doi: 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- 9.Binder LI, Frankfurter A, Rebhun LI. The distribution of tau polypeptides in the mammalian central nervous system. J Cell Biol. 1985;101:1371–1378. doi: 10.1083/jcb.101.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci. 1999;19:8818–8829. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burstein DE, Greene LA. Evidence for RNA synthesis-dependent and -independent pathways in stimulation of neurite outgrowth by nerve growth factor. Proc Natl Acad Sci USA. 1978;75:6059–6063. doi: 10.1073/pnas.75.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carson JH, Worboys K, Ainger K, Barbarese E. Translocation of myelin basic protein mRNA in oligodendrocytes requires microtubules and kinesin. Cell Motil Cytoskeleton. 1997;38:318–328. doi: 10.1002/(SICI)1097-0169(1997)38:4<318::AID-CM2>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Carson JH, Kwon S, Barbarese E. RNA trafficking in myelinating cells. Curr Opin Neurobiol. 1998;8:607–612. doi: 10.1016/s0959-4388(98)80088-3. [DOI] [PubMed] [Google Scholar]
- 14.Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 15.Falconer MM, Echeverri CJ, Brown DL. Differential sorting of beta tubulin isotypes into colchicine-stable microtubules during neuronal and muscle differentiation of embryonal carcinoma cells. Cell Motil Cytoskeleton. 1992;21:313–325. doi: 10.1002/cm.970210407. [DOI] [PubMed] [Google Scholar]
- 16.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 17.Finley MFA, Kulkarni N, Huettner JE. Synapse formation and establishment of neuronal polarity by P19 embryonic carcinoma cells and embryonic stem cells. J Neurosci. 1996;16:1056–1065. doi: 10.1523/JNEUROSCI.16-03-01056.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginzburg I, Scherson T, Giveon D, Behar L, Littauer UZ. Modulation of mRNA for microtubule-associated proteins during brain development. Proc Natl Acad Sci USA. 1982;79:4892–4896. doi: 10.1073/pnas.79.16.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Good P. The role of elav-like genes, a conserved family encoding RNA-binding proteins, in growth and development. Sem Cell Dev Biol. 1997;8:557–584. doi: 10.1006/scdb.1997.0183. [DOI] [PubMed] [Google Scholar]
- 20.Goslin K, Banker G. Rat hippocampal neurons in low-density culture. In: Banker G, Goslin K, editors. Culturing nerve cells. MIT; Cambridge, MA: 1997. pp. 251–281. [Google Scholar]
- 21.Heicklen-Klein A, Aronov S, Ginzburg I. Tau promoter activity in neuronally differentiated P19 cells. Brain Res. 2000;874:1–9. doi: 10.1016/s0006-8993(00)02539-7. [DOI] [PubMed] [Google Scholar]
- 22.Kaech S, Ludin B, Matus A. Cytoskeletal plasticity in cells expressing neuronal microtubule-associated proteins. Neuron. 1996;17:1189–1199. doi: 10.1016/s0896-6273(00)80249-4. [DOI] [PubMed] [Google Scholar]
- 23.Kiebler MA, DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 24.Kiebler MA, Hemraj I, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortin J, Dotti CG. The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koenig E, Giuditta A. Protein-synthesizing machinery in the axon compartment. Neuroscience. 1999;89:5–15. doi: 10.1016/s0306-4522(98)00282-6. [DOI] [PubMed] [Google Scholar]
- 28.Koenig E, Martin R, Titmus M, Sotelo-Silveira RJ. Cryptic peripheral ribosomal domains distributed intermittently along myelinated axons. J Neurosci. 2000;20:8390–8400. doi: 10.1523/JNEUROSCI.20-22-08390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-GFP into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon S, Barbarese E, Carson JH. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J Cell Biol. 1999;147:247–256. doi: 10.1083/jcb.147.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litman P, Barg J, Rindzoonski L, Ginzburg I. Subcellular localization of tau mRNA in differentiating neuronal cell culture: implications for neuronal polarity. Neuron. 1993;10:627–638. doi: 10.1016/0896-6273(93)90165-n. [DOI] [PubMed] [Google Scholar]
- 32.Litman P, Barg J, Ginzburg I. Microtubules are involved in the localization of tau mRNA in primary neuronal cell culture. Neuron. 1994;13:1463–1474. doi: 10.1016/0896-6273(94)90432-4. [DOI] [PubMed] [Google Scholar]
- 33.Ludin B, Matus A. GFP illuminates the cytoskeleton. Trends Cell Biol. 1998;8:72–78. [PubMed] [Google Scholar]
- 34.Ludin B, Doll T, Meili R, Kaech S, Matus A. Application of novel vectors for GFP-tagging of proteins to study microtubule-associated proteins. Gene. 1996;95:107–111. doi: 10.1016/0378-1119(95)00899-3. [DOI] [PubMed] [Google Scholar]
- 35.Mani S, Schaefer J, Meiri KF. Targeted disruption of GAP-43 in P19 embryonal carcinoma cells inhibits neuronal differentiation as well as acquisition of the morphological phenotype. Brain Res. 2000;853:384–395. doi: 10.1016/s0006-8993(99)02042-9. [DOI] [PubMed] [Google Scholar]
- 36.Mani S, Shen Y, Schaefer J, Meiri KF. Failure to express GAP-43 during neurogenesis affects cell cycle regulation and differentiation of neuronal precursors and stimulates apoptosis of neurons. Mol Cell Neurosci. 2001;17:54–66. doi: 10.1006/mcne.2000.0931. [DOI] [PubMed] [Google Scholar]
- 37.Matus A, Bernhardt R, Hugh-Jones T. High-molecular weight microtubule-associated proteins are preferentially associated with dendritic microtubules in brain. Proc Natl Acad Sci USA. 1981;78:3010–3014. doi: 10.1073/pnas.78.5.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBurney MW, Rogers BJ. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev Biol. 1982;89:503–508. doi: 10.1016/0012-1606(82)90338-4. [DOI] [PubMed] [Google Scholar]
- 39.McBurney MW, Reuhl KR, Ally AI, Nasipuri S, Bell JC, Craig J. Differentiation and maturation of embryonal carcinoma-derived neurons in cell culture. J Neurosci. 1988;8:1063–1073. doi: 10.1523/JNEUROSCI.08-03-01063.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of heterogenous nuclear ribonucleoprotein A2 response element for RNA trafficking. J Biol Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 41.Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. J Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parnas D, Linial M. Acceleration of neuronal maturation of P19 cells by increasing culture density. Dev Brain Res. 1997;101:115–124. doi: 10.1016/s0165-3806(97)00057-6. [DOI] [PubMed] [Google Scholar]
- 43.Rook MS, Lu M, Kosik KS. CaMKIIα 3′ Untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadot E, Marx R, Barg J, Behar L, Ginzburg I. Complete sequence of 3′-untranslated region of tau from rat central nervous system. J Mol Biol. 1994;241:325–331. doi: 10.1006/jmbi.1994.1508. [DOI] [PubMed] [Google Scholar]
- 45.Schnapp BJ. RNA localization: a glimpse of the machinery. Curr Biol. 1999;9:R725–R727. doi: 10.1016/s0960-9822(99)80468-0. [DOI] [PubMed] [Google Scholar]
- 46.Schuman EM. mRNA trafficking and local protein synthesis at the synapse. Neuron. 1999;23:645–648. doi: 10.1016/s0896-6273(01)80023-4. [DOI] [PubMed] [Google Scholar]
- 47.Steller U, Kohls S, Muller B, Soler R, Muller R, Schlender J, Blohm DH. The RNA binding protein HuD: rat cDNA and analysis of the alternative spliced mRNA in neuronal differentiating cell lines P19 and PC12. Mol Brain Res. 1996;35:285–296. doi: 10.1016/0169-328x(95)00231-g. [DOI] [PubMed] [Google Scholar]
- 48.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 49.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 50.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka Y, Kawahata K, Nakata T, Hirokawa N. Chronological expression of microtubule-associated proteins (MAPs) in EC cell P19 after neuronal induction by retinoic acid. Brain Res. 1992;596:269–278. doi: 10.1016/0006-8993(92)91557-u. [DOI] [PubMed] [Google Scholar]
- 52.Tiedge H, Brosius J. Translational machinery in dendrites of hippocampal neurons in culture. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tso JY, Sun XH, Reece KS, Wu R. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity. Nucleic Acid Res. 1985;13:2485–2502. doi: 10.1093/nar/13.7.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ueno S, Handley VW, Byravan S, Campagnoni AT. Structural features of myelin basic protein mRNAs influence their translational efficiencies. J Neurochem. 1994a;62:1254–1259. doi: 10.1046/j.1471-4159.1994.62041254.x. [DOI] [PubMed] [Google Scholar]
- 55.Ueno S, Kotani Y, Kondoh K, Sano A, Kakimoto Y, Campagnoni AT. The 3′-untranslated region of mouse myelin basic protein gene increases the amount of mRNA in immortalized mouse oligodendrocytes. Biochem Biophys Res Commun. 1994b;204:1352–1357. doi: 10.1006/bbrc.1994.2612. [DOI] [PubMed] [Google Scholar]
- 56.Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of β-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]