Abstract
Both positive and negative feedback loops of transcriptional regulation have been proposed to be important for the generation of circadian rhythms. To test the sufficiency of the proposed mechanisms, two differential equation-based models were constructed to describe theNeurospora crassa and Drosophila melanogastercircadian oscillators. In the model of the Neurosporaoscillator, FRQ suppresses frq transcription by binding to a complex of the transcriptional activators WC-1 and WC-2, thus yielding negative feedback. FRQ also activates synthesis of WC-1, which in turn activates frq transcription, yielding positive feedback. In the model of the Drosophila oscillator, PER and TIM are represented by a “lumped” variable, “PER.” PER suppresses its own transcription by binding to the transcriptional regulator dCLOCK, thus yielding negative feedback. PER also binds to dCLOCK to de-repressdclock, and dCLOCK in turn activates pertranscription, yielding positive feedback. Both models displayed circadian oscillations that were robust to parameter variations and to noise and that entrained to simulated light/dark cycles. Circadian oscillations were only obtained if time delays were included to represent processes not modeled in detail (e.g., transcription and translation). In both models, oscillations were preserved when positive feedback was removed.
Keywords: circadian, Drosophila, Neurospora, PER, CLOCK, FRQ, positive feedback, simulation
Circadian rhythms reflect oscillating expression of genes, one or a few of which act as clock components, or core genes. The mechanisms by which core genes generate oscillations have been the subject of extensive experimental investigation. Negative feedback loops, involving indirect mechanisms of transcriptional repression, are now known for a few organisms—notably Drosophila melanogaster andNeurospora crassa. In Drosophila the transcriptional activators dCLOCK and CYCLE form a heterodimer that activates per and tim transcription (Darlington et al., 1998; Rutila et al., 1998; Lee et al., 1999; Bae et al., 2000). This heterodimer appears to be bound by PER and TIM to mask its DNA binding activity (Lee et al., 1999;Bae et al., 2000) and thereby repress per andtim transcription. In Neurospora, thewhite-collar proteins WC-1 and/or WC-2 activatefrq transcription (Crosthwaite et al., 1997). The WC proteins may be bound by FRQ, and this may be the mechanism whereby FRQ represses frq transcription (Dunlap, 1999).
Recent results indicate that positive feedback also characterizes these circadian oscillators. In Drosophila, dCLOCK protein represses dclock transcription. Repression is mediated by dCLOCK–CYCLE heterodimers (Bae et al., 2000). PER and TIM activate dclock (Bae et al., 1998). A positive feedback loop can be envisioned as follows. Ifdclock expression is slightly activated, then the resulting activation of per and tim transcription by dCLOCK–CYCLE results in binding of dCLOCK–CYCLE by PER and TIM. Thus, repression of dclock is relieved, and the level of dCLOCK increases further. In Neurospora, increases in FRQ levels are followed by increases in WC-1 levels (Lee et al., 2000). Therefore, a positive feedback loop can be envisioned as follows. If wc-1 expression is initially activated, activation of frq transcription by WC-1 will increase the level of FRQ, which will in turn further increase the level of WC-1. A major question is the functional role of positive feedback in these systems. Intuitively, it appears that positive feedback might “cancel out” negative feedback. However, time delays between transcription and appearance of functional gene product (Garceau et al., 1997; Luo et al., 1998; Glossop et al., 1999) might minimize the conflict. Clearly, the interacting feedback loops with multiple time delays make it difficult to understand intuitively the mechanism of oscillation of even a simple configuration of core genes. Mathematical modeling is emerging as a powerful tool to supplement experimental work and provide insights into the operation of such gene networks (Leloup and Goldbeter, 1998, 2000;Leloup et al., 1999; Ruoff et al., 1999;Scheper et al., 1999; Gonze et al., 2000;Smolen et al., 2000a, 2000b).
The goal of the present investigation was to construct qualitative, differential equation-based models incorporating the above positive and negative feedback loops. Two models were constructed (Fig.1) that represent, forNeurospora and Drosophila, positive and negative feedback of core genes on their own expression. Both models succeeded in simulating circadian oscillations that were robust to parameter variation and to stochastic noise in biochemical reaction terms. The oscillations would entrain to simulated light pulses or light/dark cycles. Positive feedback was not essential for simulation of circadian oscillations, although it may be important for driving “output” circadian genes regulated by core gene products. However, time delays between transcription and appearance of functional protein were required to be of considerable length (∼7 hr).
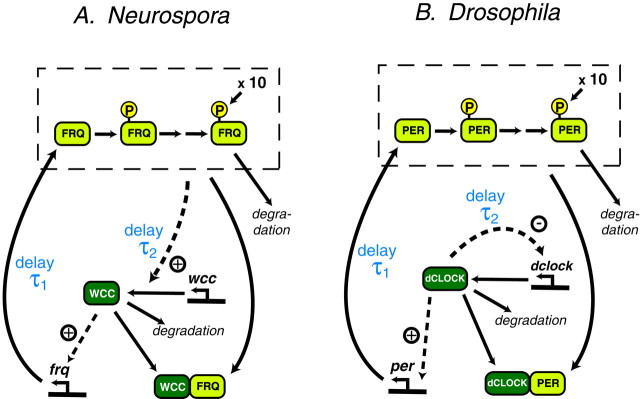
Fig. 1.
Models for the circadian oscillators in Neurospora and Drosophila. A, Neurospora. The FRQ gene product is multiply phosphorylated before degradation. WCC represents a complex of WC-1 and WC-2. All forms of FRQ are assumed equally competent at repressingfrq transcription (dashed box) via binding to WCC. Degradation of WCC is included. The time delay τ1lies between changes in the level of WCC and resultant changes in the level of FRQ. τ2 lies between changes in the level of FRQ and resultant changes in the level of WCC. B,Drosophila. “PER” represents a combination of PER and TIM levels. PER is multiply phosphorylated before degradation. All forms of PER are assumed equally competent at repressing the transcription of per (dashed box) via binding to dCLOCK. Degradation of dCLOCK is included. The time delay τ1 lies between changes in the level of dCLOCK and resultant changes in the level of PER. τ2 lies between a change in the level of free dCLOCK and the subsequent change in the level of total dCLOCK because of regulation of clocktranscription.
MATERIALS AND METHODS
In the models of the Neurospora andDrosophila oscillators presented below, we do not consider separate concentration variables for the nuclear and cytoplasmic compartments. Rather, for simplicity, we model only the concentrations of macromolecular species averaged over a whole cell. Therefore, all concentrations are referenced to the total cell volume. Absolutein vivo concentrations are not well known for circadian proteins. We assumed standard nanomolar units.
A model of the Neurospora circadian oscillator. The model of the circadian oscillator inNeurospora that incorporates interactions between FRQ and the WC proteins is schematized in Figure 1A. The WC-1 and WC-2 proteins are found together in a complex in vivo(Talora et al., 1999; Denault et al., 2001). Recent evidence indicates that FRQ interacts with this complex and may thereby block its activation of frqexpression (Denault et al., 2001). We use WCC (white-collar complex) as an abbreviation for the WC-1/WC-2 heterodimer, and we assume WCC activates frq expression. Our model contains a negative feedback loop in which FRQ repressesfrq transcription by binding and sequestering WCC, and a positive feedback loop in which activation of FRQ synthesis by WCC increases the level of FRQ, which in turn further increases the level of WCC. Experimentally, increases in FRQ are followed by increases in WC-1 (Lee et al., 2000) and thus plausibly in WCC.
The model incorporates a cascade of sequential phosphorylation events for FRQ protein, with only the most highly phosphorylated form being degraded (Fig. 1A). The number of FRQ phosphorylations that occur before degradation appears to be rather high (Garceau et al., 1997) but is not known definitely. Ten phosphorylations were assumed, as indicated by the notation “×10” in Figure1A. F0–10 denotes FRQ monomers with 0, 1, …, 10 phosphorylations, respectively. Phosphorylations of FRQ were assumed to be sequential and irreversible. In the absence of detailed characterization of the enzymes involved, it was assumed for simplicity that the phosphorylations were all performed by a single kinase. Phosphorylation rates are given by Michaelis–Menten expressions in which all species of FRQ that are not fully phosphorylated can act to saturate the kinase. Michaelis–Menten terms represent degradation of fully phosphorylated FRQ and of WCC. All forms of FRQ are assumed able to form a complex with WCC. We use F-WCC to denote an FRQ–WCC complex.
The first differential equation represents synthesis and removal of unphosphorylated FRQ, F0:
| Equation 1 |
In this equation, the first term represents synthesis of F0. The rate of synthesis of F0 is proportional to a function, RsF, of the concentration of free WCC (Eq. 2). The second term in Equation 1 represents removal of F0 via phosphorylation, with a maximal velocity vph and a Michaelis constant Kph. In the denominator, the quantity TOTUNPHOS is a sum over the concentrations of all forms of FRQ that have <10 phosphorylations, whether bound to WCC or not. The use of this quantity reflects the assumption that a single kinase catalyzes all phosphorylations of FRQ. This kinase can therefore bind, and be saturated by, all species of FRQ that are not fully phosphorylated. The third term in Equation 1, −R , represents removal of F0 by association with WCC to form a complex F–WCC. The association process is assumed second-order, with rate constant kf. Thus, R = kf[F0] [WCC]. Dissociation of F–WCC is neglected for simplicity.
In the first term of Equation 1, the function RsF takes the following form:
| Equation 2 |
This form makes the rate of FRQ synthesis, vsF ∗ RsF, a saturable and delayed function of the level of free WCC. A justification for Equation 2 is as follows. The rate of transcription of frqis assumed proportional to the probability that WCC is bound to an upstream regulatory sequence. This probability is assumed to be a saturable function of [WCC], yielding:
| Equation 3 |
Because our model does not consider separate cytoplasmic and nuclear compartments, the concentrations in Equation 3, as in all other equations, are averaged over the total cell volume. To go to Equation2, it is assumed that the rate of FRQ synthesis is proportional to the rate of frq transcription, but with a time delay τ1, taken as 7 hr. Much of τ1 is needed to account for the ∼4 hr delay between the time courses offrq mRNA and FRQ protein during circadian oscillations (Garceau et al., 1997; Luo et al., 1998). τ1 would also include time needed for WC-1 and WC-2 to move to the nucleus and regulate frq.
The use of time delays such as τ1 to represent slow biochemical processes not modeled in detail is an effective way of encapsulating slow processes whose details are too complex or uncertain to model (Smolen et al., 1999). We used distributed, rather than discrete, time delays. With a distributed delay, the derivative of a variable depends on the averaged values of one or more variables over a specified range of previous time (MacDonald, 1989). For example, if the range of previous time is denoted by (t1, t2), a distributed delay for a variable X(t) whose rate of change depends on a function of itself is given by:
| Equation 4 |
Distributed delays are general enough to encapsulate transcription, translation, or transport. For example, if movement of mRNA from a transcription site to translation sites relies on active transport with a range of transport times for individual mRNAs, a distributed delay is a proper modeling framework.
In equations defining delayed functions, angled brackets indicate that the quantity within is averaged over a time window centered at a mean delay τ (τ1 in Eq. 2). For convenience, the mean delay may be termed “the delay.” The width of the time window is denoted δ. In Equation 4, δ = (t2 − t1). δ is stated as a fraction of τ. In Equation 2, the standard value of δ was taken to be 100% of τ1. Such a large value is consistent with experimental results. Garceau et al. (1997) and Luo et al. (1998) found that frq mRNA exhibits a rather narrow oscillation, with a width at half-maximum on the order of 6–8 hr, whereas the rising phase alone of oscillation in FRQ protein appears somewhat longer than this, lasting ∼8–10 hr. These findings suggest a large variability in the times during which individualfrq mRNAs are translated. This corresponds to a large δ.
The differential equation for the rate of change of the level of free WCC, the transcriptional activator complex, is as follows:
| Equation 5 |
In this equation, the first two terms represent synthesis and degradation of WCC. In the denominator of the second term, WCCtot denotes the total concentration of WCC, both free and bound to dCLOCK. The third term, −∑ R , represents removal of free WCC by association with any of the 11 species of FRQ (unphosphorylated, or with 1–10 phosphorylations). The sum runs over 11 association rates, one for each species of FRQ. Each association process is assumed second-order, with the same rate constant kf in all cases. For example, R = kf[F10] [WCC].
FRQ appears to activate the synthesis of one of the WC proteins, WC-1, post-transcriptionally (Lee et al., 2000). The expression of the second white-collar gene, wc-2, does not display circadian oscillations (Crosthwaite et al., 1997). Because WC-2 is in excess over WC-1 (Denault et al., 2001), activation of WC-1 synthesis should correspond to increasing the level of WCC. Therefore, to reflect the activation of WC-1 synthesis by FRQ, the rate of synthesis of WCC in Equation5 is proportional to a delayed function RsW of the total concentration of FRQ not complexed to WCC, [Ftot]. [Ftot] = [F0] + [F1] + ⋯ + [F10]. Analogously to Equation 2, a saturable function was used:
| Equation 6 |
The delay τ2 appears to be large (∼7–8 hr;Lee et al., 2000). We used a value of 7 hr. The window width δ was taken as 30% of the mean delay.
Phosphorylated species of FRQ can be formed and removed by phosphorylation reactions, except that removal of F10 is by degradation. Also, these species can be removed by association with WCC to form FRQ–WC complexes. Each differential equation for these species therefore has three terms:
| Equation 7 |
| Equation 8 |
Within complexes of FRQ and WCC, FRQ can also be phosphorylated. The variables C are used to denote F-WCC complexes with 0, 1, …, 10 FRQ phosphorylations. In Equations 1, 7, and 8, the variable TOTUNPHOS is equal to [C ] + [C ] + … + [C ] + [F0] + [F1] + … + [F9]. There is no evidence to suggest the kinetics of FRQ phosphorylation depend on whether FRQ is complexed with WCC or not. Therefore, for simplicity, the same velocities and Michaelis constants are assumed to govern phosphorylation of FRQ in both cases. For the variables [C ] through [C ], differential equations analogous to Equation 7can therefore be written. One additional term is added to account for degradation of these complexes if the WCC component is degraded. This term has the same form as that in Equation 5 for the degradation of free WCC:
| Equation 9 |
| Equation 10 |
The differential equation for the rate of change of [C ] needs another term, to account for degradation of the complex upon degradation of fully phosphorylated FRQ. This term is analogous to that in Equation 8 for the degradation of fully phosphorylated FRQ:
| Equation 11 |
For most simulations with the Neurospora model (Eqs.1, 2, and 5-11), a standard set of parameter values was used, with exceptions noted in the text or figure legends. The standard set is:
Experimental data to estimate parameter values is lacking, except in the case of the delays τ1 and τ2, which were chosen as discussed above. Therefore, to obtain standard values for the other parameters, it was necessary to rely on trial-and-error variation. Values were found that allowed simulation of stable circadian oscillations robust to small parameter changes and simulation of entrainment to light pulses. Parameter values for the model of the Drosophila oscillator were obtained in the same manner.
A model of the Drosophila circadian oscillator.The PER-TIM heterodimer inhibits per and timexpression by binding to a heterodimer of the transcriptional activators dCLOCK and CYCLE and hindering binding of dCLOCK–CYCLE to the per and tim promoter regions (Lee et al., 1999; Bae et al., 2000). Because CYCLE is present in excess (Bae et al., 2000) it is likely that the concentration of dCLOCK–CYCLE is proportional to that of dCLOCK. Thus, the interaction of dCLOCK with CYCLE was not modeled explicitly. Rather, it was assumed that activation of per andtim is governed by the level of dCLOCK not complexed with PER–TIM. dclock transcription is activated by PER and TIM acting in concert. The activation is actually a de-repression. The PER–TIM heterodimer binds to and sequesters dCLOCK, which otherwise would repress its own gene (Glossop et al., 1999;Bae et al., 2000). Our model assumes this mechanism.
TIM alone does not appear to regulate transcription. Also, the time courses of PER and TIM proteins are similar in shape and largely overlap (Lee et al., 1998; Bae et al., 2000). Because of these considerations, the dynamics of PER and TIM may be represented by a single “lumped” variable, “PER,” which can be thought of as an “average” of PER and TIM levels. Combining PER and TIM into a single variable is also economical. If PER and TIM were modeled separately the number of differential equations would increase greatly. Therefore, in our model, only the variable “PER” was used. Binding of dCLOCK with PER forms an inactive complex, denoted by P–C.
The model of the circadian oscillator in Drosophila, which includes binding of PER to dCLOCK, is schematized in Figure1B. It embodies the negative feedback loop in which PER binds dCLOCK and therefore deactivates per transcription. In addition, it embodies the positive feedback loop that operates as follows. Activation of per transcription by dCLOCK results in sequestration of dCLOCK by binding to PER. Thus, repression ofdclock by dCLOCK is relieved, and the level of dCLOCK increases further.
The model assumes sequential phosphorylation events for PER, with only the most highly phosphorylated form being degraded (Fig.1B). The number of PER phosphorylations that occur is considerable (Edery et al., 1994; Zeng et al., 1996). Ten was assumed as was done above for FRQ. Michaelis–Menten terms represent degradation of PER and dCLOCK. All forms of PER, irrespective of phosphorylation, are assumed able to bind dCLOCK. The resulting model has 23 dependent variables—the same as in our model of the Neurospora oscillator. The equations are almost identical to that model.
The first differential equation represents synthesis and removal of unphosphorylated PER, P0. The rate of synthesis of P0 is given as a function, RsP, of the concentration of free dCLOCK:
| Equation 12 |
In this equation, the first two terms represent synthesis and phosphorylation of P0. In the first term, the function RsP takes the form:
| Equation 13 |
This form makes the rate of PER synthesis a saturable, delayed function of free dCLOCK. The mean delay τ1 was taken as 8 hr. Much of τ1 can be accounted for by the ∼4 hr delay between the time courses of per mRNA and PER protein (Zerr et al., 1990; Vosshall et al., 1994; Glossop et al., 1999). An additional component of τ1 is evident as an experimental offset between the time course of the per transcription rate and the level of per mRNA. Surprisingly, this delay is on the order of 2 hr (So and Rosbash, 1997). Similar delays exist between the time courses of tim transcription,tim mRNA, and TIM protein (So and Rosbash, 1997). Movement of dCLOCK into the nucleus and binding to DNA would also add to τ1. In the denominator of the second term in Equation 12, TOTUNPHOS is a sum of the concentrations of all forms of PER with <10 phosphorylations, whether bound to dCLOCK or not. The third term in Equation 12, −R , represents removal of P0 by association with dCLOCK to form a complex P–C. The association process is assumed second-order, thus R = kf[P0] [dCLOCK]. Dissociation of P–C is neglected for simplicity.
The differential equation governing [dCLOCK] is:
| Equation 14 |
In this equation the first two terms represent synthesis and degradation. The third term, −∑ R , represents removal of free dCLOCK by association with the species of PER with 0–10 phosphorylations. For example, R = kf[P10] [dCLOCK]. In the first term, the function RsC was chosen to reflect the repression of dCLOCK synthesis:
| Equation 15 |
In Equation 15, τ2 represents the delay between changes in free dCLOCK and subsequent changes in the rate of appearance of new dCLOCK protein. Thus, τ2 encompasses dCLOCK transcription and translation. A value of 5 hr was used for τ2. A caveat is that τ2 has not been experimentally determined. Equation 15 embodies the key difference between the Drosophila and Neurospora models. In Equation 15, dCLOCK synthesis is repressed by dCLOCK protein. The analogous Equation 6 in the Neurospora model embodies activation of WCC synthesis by FRQ protein.
Phosphorylated species of PER can be formed and removed by phosphorylation reactions, except that for P10, removal is by degradation. Also, these species can be removed by association with dCLOCK to form P–C complexes. Each differential equation for these species therefore has three terms:
| Equation 16 |
| Equation 17 |
Within complexes of PER and dCLOCK, PER can also be phosphorylated. The variables C are used for P–C complexes with 0, 1, …, 10 PER phosphorylations. Differential equations analogous to Equations 9-11 in theNeurospora model govern the concentrations of these complexes:
| Equation 18 |
| Equation 19 |
| Equation 20 |
For most simulations with the Drosophila model (Eqs.12-20), a standard set of parameter values was used. The standard set is:
As in the model of the Neurospora oscillator, the standard value of the window width δ was taken to be 100% of the mean delay for τ1 and 30% of the mean delay for τ2.
A list of all the parameters in the models of Figure 1, together with summaries of their biochemical significance, is presented in Table1.
Table 1.
Parameters of the models
| Parameter | Biochemical significance |
|---|---|
| τ1 | Time delay between a change in the concentration of transcriptional activator (WCC or dCLOCK) and the resulting change in the synthesis rate of transcriptional repressor (FRQ or PER). |
| τ2 | Time delay between a change in the concentration of FRQ (in Neurospora) or dCLOCK (inDrosophila) and the resulting change in the synthesis rate of transcriptional activator (WCC or dCLOCK). |
| vsF, vsP,vsW, vsC | Maximal velocities of synthesis of FRQ, PER, WCC, and dCLOCK proteins, respectively. |
| vph | Maximal velocity of phosphorylation of each site on FRQ or PER. |
| vdF, vdP,vdW, vdC | Maximal velocities of degradation of FRQ, PER, WCC, and dCLOCK proteins, respectively. |
| K1,K2 | Michaelis constants.K1 describes the regulation of FRQ synthesis (inNeurospora) or PER synthesis (in Drosophila) by the transcriptional activators WCC or dCLOCK, respectively.K2 describes the regulation of WCC synthesis (inNeurospora) or dCLOCK synthesis (in Drosophila) by FRQ or dCLOCK, respectively. |
| Kph | Michaelis constant governing the phosphorylation of each site on FRQ or PER. |
| KdF, KdP,KdW, KdC | Michaelis constants governing the degradation of FRQ, PER, WCC, and dCLOCK, respectively. |
| kf | Forward rate constant governing formation of the FRQ-WCC complex (in Neurospora) or the PER-dCLOCK complex (in Drosophila). |
For numerical integration of the equations, the forward-Euler method was used with storage of averages for use in calculations of delayed quantities. Integration time steps were reduced until no significant difference was seen after further reduction. Final step sizes were 4–5 × 10−5 hr. All models were programmed in FORTRAN 77 and simulated on a Compaq XP1000 workstation. Programs are available from the authors upon request.
RESULTS
Models incorporating feedback loops and time delays can represent the Drosophila and Neurospora circadian oscillators
Simulation of oscillations in Drosophila
The Drosophila model (Fig. 1B) readily simulated large-amplitude circadian oscillations in the level of total PER and total dCLOCK (Fig. 2, top panel). Here and subsequently, “total” protein concentration is the sum over all phosphorylation states and over free and bound states (e.g., for PER, the sum of the model variables [P0] through [P10] and [C ] through [C ]). The period of the simulated oscillations is 23.6 hr. Effects of light were not simulated in Figure 2, so these oscillations correspond to a rhythm in constant darkness. The oscillatory pattern in Figure 2 is stable over time and to modest changes in parameters as discussed further below.
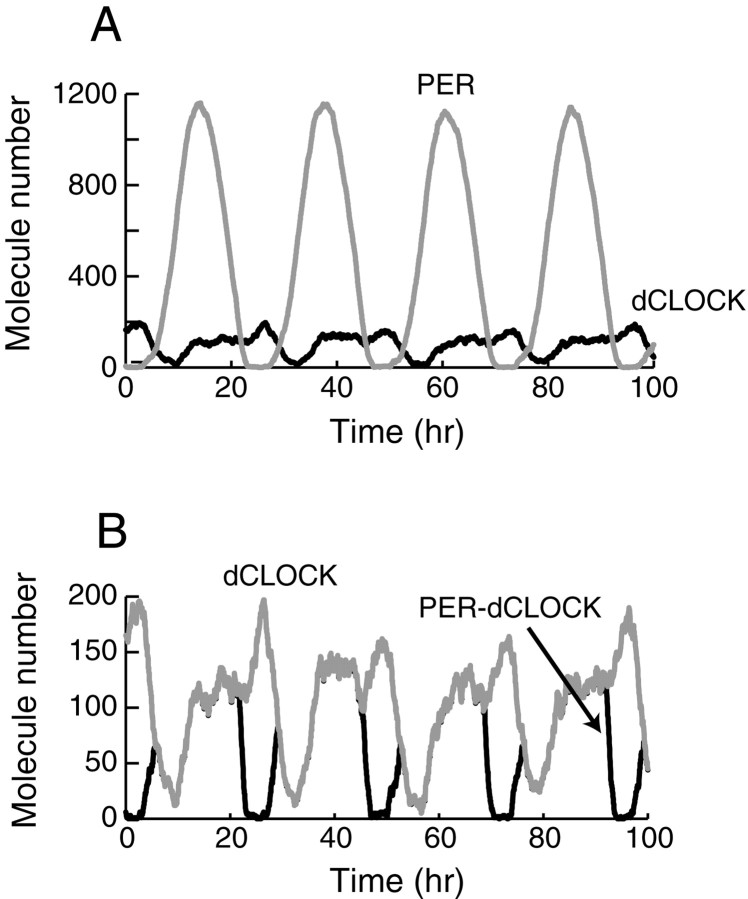
Fig. 2.
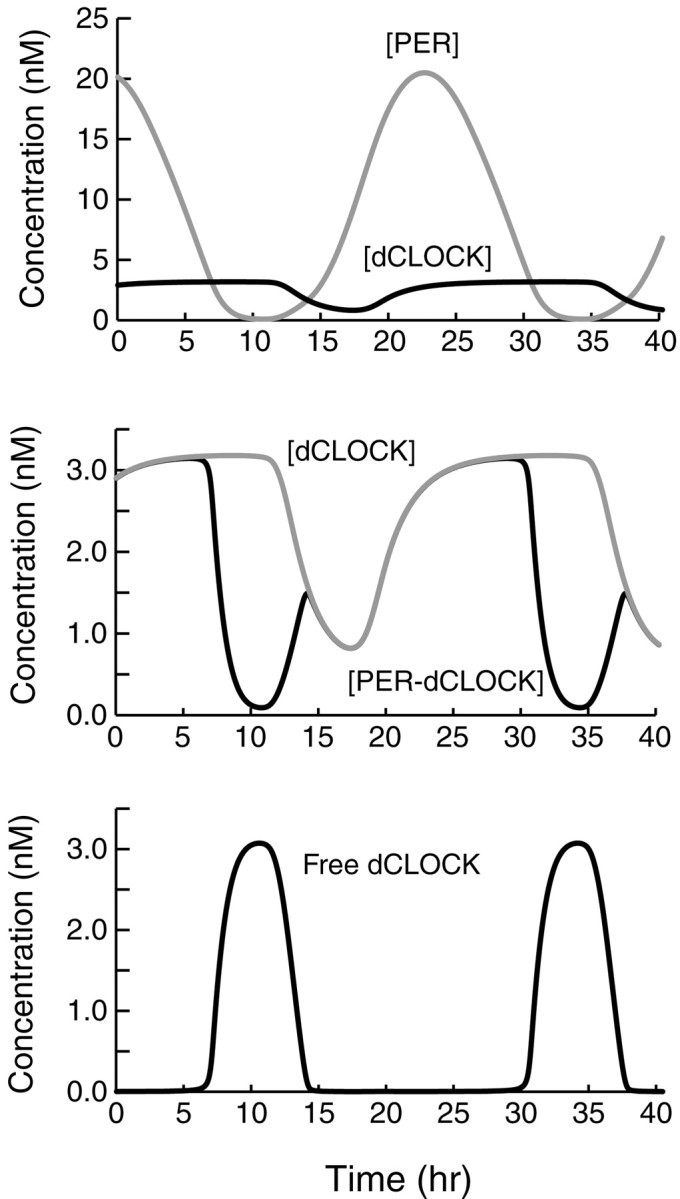
Simulation of circadian oscillations inDrosophila. Subsequent to an initial transient (data not shown), the model converged to oscillations that are independent of initial conditions. The standard set of parameter values (Materials and Methods) was used. Top panel, Time courses are displayed for the level of total PER (top, gray time course) and for the level of total dCLOCK (bottom, blacktime course). Middle panel, Expanded view displaying in more detail the dynamics of dCLOCK (top, gray time course) and of the level of the PER-dCLOCK complex (blacktime course—often overlying the dCLOCK time course). Bottom panel, Time course of the level of free dCLOCK not complexed with PER.
The oscillations of PER concentration in Figure 2 are quite symmetric in appearance, with a gradual rise and fall. The time course of the concentration of total dCLOCK and that of the complex of PER and dCLOCK (the sum of [C ] through [C ]), are displayed in the middle panel of Figure 2. The oscillations of Figure 2 qualitatively agree with experiment in that the average level of dCLOCK is considerably less than that of PER (Bae et al., 2000).
The bottom panel of Fig. 2 illustrates the time course of free dCLOCK (not bound to PER), which is only at significant levels during the intervals when the level of total PER is below total dCLOCK. When [PER] rises at the beginning of an oscillation, the formation of PER–dCLOCK complex rapidly eliminates free dCLOCK. The loss of free dCLOCK acts to terminate per transcription. Phosphorylation and degradation of PER continue, and after several hours a decline in [PER] leads to dissociation of the PER–dCLOCK complex and the regeneration of free dCLOCK. Free dCLOCK can then activateper transcription and [PER] can again rise. It is evident from comparing the top and bottom panels of Fig. 2 that a considerable delay, on the order of the parameter τ1, separates changes in the level of free dCLOCK (such as the increase beginning at t = 6 hr) from the resulting changes in the rate of synthesis of PER protein (the increase beginning at t = 11 hr).
Oscillations of significant amplitude in the concentration of total dCLOCK are seen (Fig. 2, top panel). These oscillations reflect the indirect regulation of dCLOCK synthesis by PER, through sequestration of dCLOCK and de-repression ofdclock transcription. In the middle panel of Figure 2, a circadian cycle of the level of PER–dCLOCK complex displays two humps. The first hump, peaking at t = 13 hr, is caused by the binding of newly synthesized PER to dCLOCK. [PER–dCLOCK] then declines because dCLOCK, both free and in complex, is being degraded. At t = 17 hr dCLOCK synthesis begins, and dCLOCK can bind with PER, so [PER-dCLOCK] rises again, to the second maximum at t ≅ 28 hr. It is not yet known experimentally whether such dynamics characterize the level of a PER–TIM–dCLOCK complex.
Experimentally, oscillations in PER during 24 hr light/dark cycles tend to peak at approximately zeitgeber time (ZT) 20 (Edery et al., 1994; Lee et al., 1998), where ZT0 is lights on and ZT12 is lights off. The dCLOCK time course is clearly later than that of PER throughout the day, peaking at approximately ZT0 (Bae et al., 2000). In the oscillations of Figure 2, a delay on the order of the parameter τ2, ∼7 hr, separates the start of each increase in PER from the start of the resulting increase in dCLOCK. Thus, both in experiment and simulation, there is a significant lag between the PER and dCLOCK time courses.
One shortcoming of the simulated oscillations in Figure 2 is that as the level of PER rises at the beginning of an oscillation, PER–dCLOCK complex begins to form immediately, rather than after an interval of a few hours (Saez and Young, 1996). In the model, newly formed PER can always complex readily with dCLOCK. To more adequately represent the kinetics of PER–dCLOCK (or PER–TIM–dCLOCK) complex formation, a more detailed model would be necessary and would have to include kinetic expressions for PER transport into the nucleus.
The delay parameter τ1 is critical. Decreasing τ1 decreased the oscillation period. For example, a τ1 of 3 hr corresponds to a period of 12.8 hr with other parameters as in Figure 2. The period is more sensitive to τ1 than to any other parameter, and eliminating τ1 abolished oscillations. τ1 appeared essential, because oscillations could not be restored by varying other parameters. In contrast, if τ2 was eliminated, oscillations were preserved. The period is relatively insensitive to τ2. Decreasing τ2 acts to decrease the lag between increases or decreases in [PER] and the resulting increases or decreases in [dCLOCK].
Simulation of oscillations in Neurospora
The Neurospora model (Fig. 1A) readily simulated large-amplitude circadian oscillations in the levels of total FRQ and total WCC (Fig. 3, top panel). The period of these oscillations is 24.0 hr. Effects of light were not simulated in Figure 3, so these oscillations correspond to a free-running rhythm in constant darkness. The oscillatory pattern in Figure 3 is stable over time, and the oscillations are stable to modest changes in parameters (see Fig. 6). Decreasing τ1 decreased the oscillation period. Elimination of τ1 abolished oscillations. Again, τ1 appeared essential, because oscillations could not be restored by varying other parameters. For the standard parameter values, elimination of τ2 also abolished oscillations, but these could be restored by increasing vsFfrom 8.0 to 16.0 nm/hr.
Fig. 3.
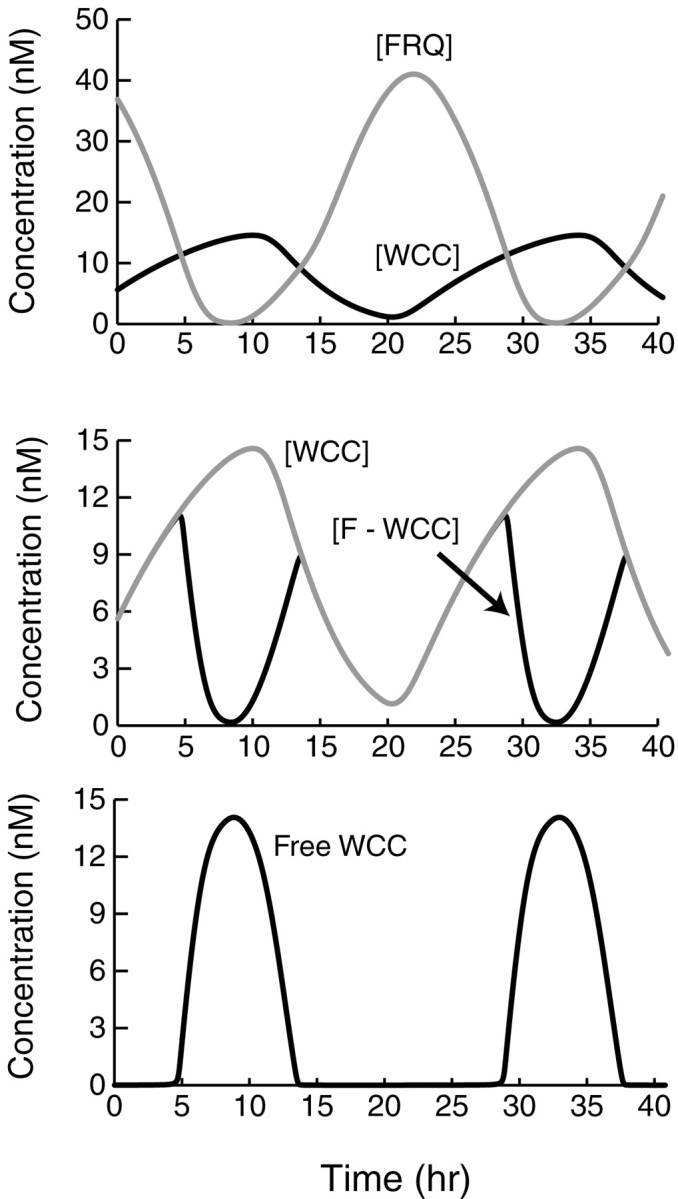
Simulation of circadian oscillations inNeurospora. Subsequent to an initial transient, the model converged to oscillations that are independent of initial conditions. The standard set of parameter values (Materials and Methods) was used.Top panel, Time courses are displayed for the level of total FRQ (top, gray time course) and for the level of total WCC (black time course). Middle panel, Expanded view displaying in more detail the dynamics of WCC (top, gray time course) and of the level of the FRQ–WCC complex (black time course—often overlying the WCC time course). Bottom panel, Time course of the level of free WCC not complexed with FRQ.
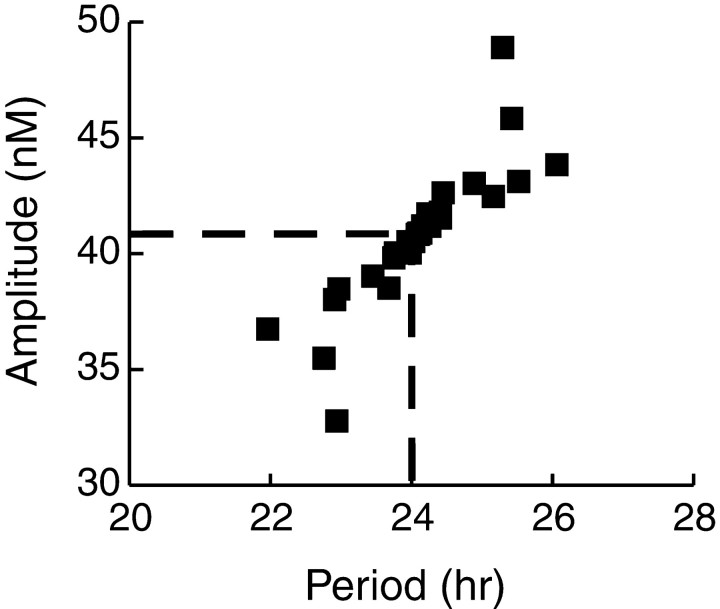
Fig. 6.
Robustness of the Neurospora model to parameter variation. A scatter plot displays the periods and amplitudes of simulated circadian oscillations in [FRQ]. To generate these oscillations, each individual parameter in the set of standard parameter values was increased or decreased by 15%. There are 13 parameters, therefore 27 data points including the control with all parameter values standard. The location of the control data point is marked by the intersection of the horizontal andvertical dashed lines. Four additional simulations are also included. In these simulations, the widths of the time delays τ1 and τ2 were increased or decreased by 15% from their standard values.
The oscillations in Figure 3 display a gradual rise and fall in the concentration of total FRQ. The middle panel of Figure 3 displays in more detail the time courses of total WCC and of the complex of FRQ and WCC (the sum of [C ] through [C ]). As [FRQ] rises, WCC is rapidly bound so that soon essentially all WCC is complexed with FRQ. Over an oscillation, the average concentration of WCC is ∼40% that of FRQ. Experiments have not yet established the relative levels of FRQ and WCC. In Fig. 3 (top), the oscillations of FRQ and WCC are approximately antiphase. A large value of τ2 is required for this relationship. Experimental oscillations of FRQ and WC-1 are also approximately antiphase (Lee et al., 2000).
Figure 3, bottom panel, illustrates the time course of the concentration of free WCC. Free WCC is only at significant levels during the intervals when [FRQ] is below [WCC]. When [FRQ] rises at the beginning of an oscillation, the formation of FRQ–WCC complex rapidly eliminates free WCC. The loss of free WCC acts to terminatefrq transcription. Phosphorylation and degradation of FRQ continue, and after ∼15 hr a decline in FRQ leads to dissociation of the FRQ–WCC complex and the regeneration of free WCC. Free WCC can then activate frq transcription, and [FRQ] can again rise. It is evident from comparing the top and bottom panels of Figure 3 that a considerable delay, on the order of the parameter τ1, separates changes in the level of free WCC (such as the increase beginning at t = 28 hr) from the resulting changes in the rate of synthesis of FRQ protein (the increase beginning at t = 33 hr).
Comparison of simulated and experimental oscillations
Figure 4A compares experimental time courses of Neurospora FRQ and WC-1 levels (Lee et al., 2000) with time courses of FRQ and WCC simulated by our model (Fig. 3). Absolute in vivoconcentrations of these proteins are not known. Thus, the simulated time courses were scaled vertically to match the magnitude of the experimental time courses, and specific units were not used for amounts of proteins in Figure 4. The experimental WC-1 time course is used because no experimental time course for WCC, the complex between WC-1 and WC-2, is available. The overall shape of the simulated FRQ time course, including the relative timing of the peak versus the minimum, appears qualitatively consistent with experiment when the random variability between experimental oscillations is considered (Garceau et al., 1997; Lee et al., 2000). The shape of the simulated WCC time course is quite similar to that of the WC-1 time course. Experimentally, because WC-2 is in excess (Denault et al., 2001), WC-1 levels should determine the levels of WCC, so that one might expect the shapes of WCC and WC-1 time courses to be similar. Our model is qualitative and does not include separate cytoplasmic and nuclear compartments. The dynamics of FRQ and of the WC proteins are expected to differ between compartments, for example, WC-2 and WCC are almost entirely nuclear proteins (Denault et al., 2001). A more detailed model would be required to capture these effects. For this reason, and because of the considerable random variation between experimental FRQ oscillations, more detailed fitting of simulated time courses to experimental data via extensive parameter variation was not performed.
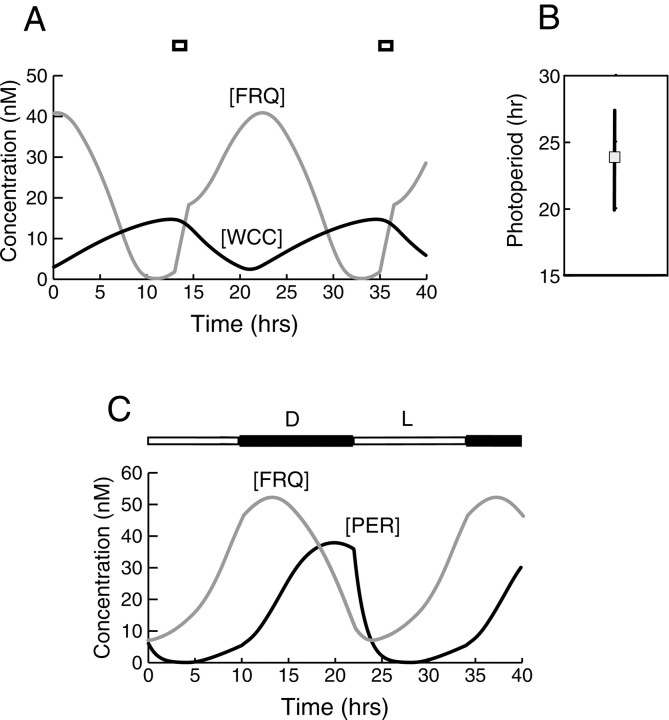
Fig. 4.
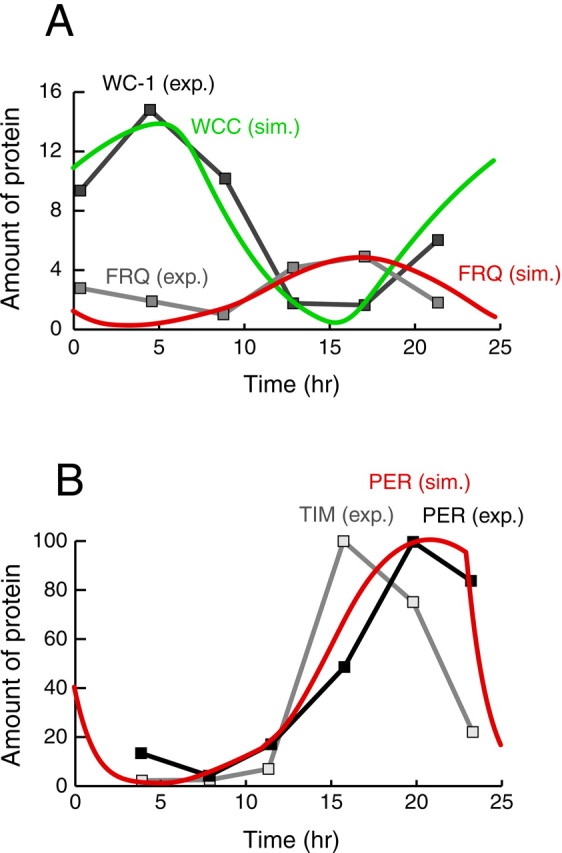
Comparison of simulated and experimental time courses of circadian gene product levels. A,Neurospora. The simulated time course of FRQ (red) is compared with experimental time course (gray with squares). The simulated time course of WCC (green) is compared with the experimental WC-1 time course (black withsquares). Data is from Lee et al. (2000), their Figure 1B. B, Drosophila. The simulated time course of PER (red) is compared with experimental time courses of PER and TIM (black and gray). Data is from Lee et al. (1998), their Figure 2B.
Figure 4B compares experimental time courses ofDrosophila PER and TIM levels (Lee et al., 1998) with the simulated time course of PER. Both experimental time courses are shown because, as discussed in Materials and Methods, “PER” in our model represents an average of PER and TIM. Experimental time courses are during a 12 hr light/dark cycle. Therefore, as discussed in the following section, the simulated time course is also during a 12 hr light/dark cycle (parameters as in Fig.5C). The locations of the peak and minimum, and the shape of the rising phase, are similar in the experimental and simulated time courses. The decline of the simulated time course is somewhat steeper than experiment, however.
Fig. 5.
Entrainment of circadian oscillations simulated by the Neurospora model (A, B) and theDrosophila model (C). A, Entrainment of the oscillations of Figure 3 by simulated light pulses. The interstimulus interval was 20 hr. Short square bars mark the effect of each light pulse, which was assumed to induce an increase in FRQ synthesis. Each 90 min increase initiates an upstroke in total FRQ. The time course of total WCC is also shown. B, Limits of entrainment for the interstimulus interval of the simulated light pulses of A. The free-running oscillation period (24.0 hr) is marked. C, Entrainment of the oscillations of theDrosophila model (Fig. 2) and the Neurosporamodel (Fig. 3) by a 12 hr light/dark cycle. The light and dark phases are indicated by the overbar. Each light phase was assumed to induce a first-order degradation of phosphorylated PER. The degradation rate constant was 0.9 hr−1. Each light phase also decreased the degradation velocity for dCLOCK, vdC, to 1.5 nm/hr. ForNeurospora, each light phase induced a constant synthesis of unphosphorylated FRQ (3.0 nm/hr).
The Neurospora and Drosophila models can simulate entrainment by light
Models of circadian rhythms must be able to simulate responses of the rhythm to light pulses or light/dark cycles (Leloup and Goldbeter, 1998, 2000; Scheper et al., 1999;Gonze et al., 2000). In Neurospora, an increase in frq transcription is observed after a light pulse. Increased frq mRNA levels persist for somewhat longer than 60 min (Crosthwaite et al., 1995). Thus, each light pulse was modeled as an increase in the first term on the right-hand side of Equation 1, which denotes the rate of synthesis of unphosphorylated FRQ (the variable F0). A delay of a few hours, not explicitly included in simulations, would separate light pulses from increases in FRQ synthesis, because of processing and transport of frq mRNA (Garceau et al., 1997).
In one set of simulations, light pulses were modeled by increasing the velocity of synthesis (vsF) of unphosphorylated FRQ, F0, to a constant rate of 10.0 nm/hr for 1.5 hr. Parameter values were otherwise as in Figure 3. Figure 5A demonstrates entrainment of oscillations by these simulated light pulses, with an interstimulus interval (ISI) of 22 hr. The brief increases in FRQ synthesis are each seen to initiate upstrokes of the level of total FRQ from a very low value. ISIs of longer than the free-running period (24 hr) can also entrain the oscillations of Figure 3. With the stimulus parameters of Figure5A, the limits of entrainment for the ISI are 20–27 hr, as illustrated in Figure 5B.
In Drosophila, light enhances the degradation of phosphorylated TIM (Myers et al., 1996; Zeng et al., 1996). Because the Drosophila model does not have a separate variable for the level of TIM, the degradation of phosphorylated PER was enhanced to simulate the effect of light. The effect of such enhancement is to free dCLOCK from the PER–dCLOCK complex, allowing dCLOCK to regulate transcription. In vivo, the degradation of TIM may analogously release free dCLOCK. The effect of light on subsequent transcription of core circadian genes might therefore be similar in simulations with our model and in vivo. To simulate light exposure, first-order degradation rate constants (−kdeg[Pi]) were assumed for the phosphorylated forms of PER (the variables [P1], …, [P10] and [C ], …, [C ]) during the light phase of light/dark cycles or during light pulses. When PER complexed with dCLOCK was degraded, it was assumed that free dCLOCK is released.
Figure 5C demonstrates entrainment of the simulatedDrosophila and Neurospora circadian oscillations to a light/dark cycle. The cycle used was 12 hr. During the 12 hr of “light,” a first-order degradation rate constant of 0.9 hr−1 was assumed for all phosphorylated forms of PER. Because the enhanced PER degradation acted to advance the phase of the succeeding peak in the concentration of free dCLOCK, and then the phase of the next peak in [PER], a compensating parameter change was necessary to maintain an oscillation period of 24 hr. It was therefore assumed that dCLOCK degradation was reduced during the light phase (vdC was decreased to 1.5 nm/hr). This change enhanced and prolonged the succeeding peak of free dCLOCK and the next peak in [PER]. It appears plausible that during the light phase, enhanced PER degradation could use elements of the pathway mediating dCLOCK degradation, competitively reducing vdC. For Neurospora, each light phase was modeled as addition of a constant synthesis rate of FRQ to Equation1 (3.0 nm/hr). In Figure 5C, discontinuities in the slope of the time course of total PER are evident that coincide with the onset of “light” and the imposition of the degradation process. The switch from dark to light occurs on the falling portion of the PER time course, as it does experimentally (Lee et al., 1998; Bae et al., 2000). With the stimulus parameters of Figure 5C, the shape of the time course of total PER during a simulated oscillation is significantly different from the time course in constant darkness (Fig. 2). With the light/dark cycle, the falling portion of the time course is considerably steeper than the rising portion. Experimentally, during entrainment to light/dark cycles, the falling portions of the PER and TIM time courses are indeed steeper than the rising phases (Marrus et al., 1996; Lee et al., 1998). Comparing the FRQ and PER time courses in Figure 5C, it is seen that the light phase coincides with the rising portion of the FRQ time course but with the falling portion of the PER time course. This agrees with observation (Dunlap, 1999). Entrainment of the oscillations of the Drosophila model by light pulses could also be simulated (data not shown). Entrainment to pulses with an ISI considerably shorter than 24 hr was readily obtained. However, entrainment to pulses with an ISI longer than ∼25 hr was not obtained.
Simulated circadian oscillations are robust to parameter variation
Biochemical parameters are expected to vary somewhat from cell to cell and from one member of a species to another. Nevertheless, individual Neurospora orDrosophila are generally observed, in constant darkness, to sustain circadian rhythms with similar period. Because circadian rhythmicity is well preserved from one individual to the next, it is important that in a model of circadian rhythmicity, small parameter variations such as might be expected among individuals should not cause large changes in the period or amplitude of simulated circadian oscillations.
For simulations to test robustness of oscillations to small parameter variations, the standard parameter value sets (Materials and Methods section) were used as the starting point. With theNeurospora and Drosophila models, each individual parameter was increased and decreased by 15% of its standard value. For each model, there are 13 parameters, including the delays τ1 and τ2. Therefore, 27 simulations were performed including the control with standard parameter values. Figure6 plots the period and amplitude of these simulations for the Neurospora model. The amplitude was measured as the peak-to-minimum difference in the oscillation of the concentration of total FRQ. In four additional simulations, also plotted, the widths of the distributed delays τ1–2 were also increased and decreased by 25% from their standard values.
Oscillations were preserved in all 31 simulations, and their appearance never varied dramatically from the control oscillations with no parameter change (Fig. 3). The period of the oscillations, as well as their amplitude, never varied by >25% from the control values of 24.0 hr and 40.9 nm. The largest amplitude increase occurred for a 15% increase in vsF, which increased the amplitude to 48.9 nm. Decreasing τ1 by 15% produced the largest period decrease (to 21.9 hr). Increasing τ1 by 15% produced the largest period increase (to 26.1 hr) and a significant amplitude increase (to 43.8 nm).
With the Drosophila model, changing any individual parameter by 15% also produced only modest changes (not >25%) in oscillation amplitude and period. The oscillation period was most sensitive to τ1 and vph. Overall, the lack of dramatic variability in the above simulations suggests that the models presented here are sufficiently robust to parameter variation that they can be regarded as reasonable representations of biochemical mechanisms responsible for circadian oscillations inNeurospora and Drosophila.
Parameter variation can simulate effects of interference with FRQ or PER phosphorylation
Some of the changes observed after parameter variation can simulate experimental protocols that vary the kinetics of FRQ or PER phosphorylation and thereby alter circadian periods. The period of the simulated Neurospora oscillations could be increased to 30.6 hr by decreasing vph from 38 to 16 nm/hr. Recent experiments have demonstrated that slowing the phosphorylation of FRQ with kinase inhibitors or mutating one of the FRQ phosphorylation sites can lengthen the circadian period to ≥30 hr (Liu et al., 2000). Therefore, the simulations gave the expected qualitative result.
With the Drosophila model, a substantial increase in vph can simulate the effect of thedoubletime-S mutation, which greatly shortens the oscillation period. The doubletime-S mutation may increase the activity of a kinase that phosphorylates PER and thereby stimulate premature degradation of PER (Kloss et al., 1998; Price et al., 1998). To simulate this mutation, vph was increased from 23 to 50 nm/hr. The simulated period decreased to 18.6 hr—a value similar to the reported doubletime-S period of 18 hr (Price et al., 1998). Another mutation,perS, also shortens the oscillation period to ∼19 hr, and this shortened period is associated with an increased instability of PER (Marrus et al., 1996). This instability may be attributable to accelerated phosphorylation of PER (Marrus et al., 1996). Thus, the simulation of thedoubletime-S mutation may also represent theperS mutation.
Robustness of circadian oscillations to stochastic fluctuations is parameter-dependent
Recently, the importance of testing models of circadian rhythmicity for robustness to stochastic noise has been emphasized (Barkai and Leibler, 2000; Smolen et al., 2000a, 2000b). This noise is attributable to random variations in the numbers of molecules caused by the random timing of individual biochemical reaction events. We performed qualitative simulations with the Drosophilamodel to estimate the average numbers of core gene product molecules needed to sustain oscillations. For smaller average molecule numbers, random fluctuations are relatively larger and overwhelm the periodic oscillation pattern. Experiments may demonstrate that circadian oscillations are sustained with smaller molecule numbers than those needed to sustain oscillations in any particular model. In that case, the model will need to be altered.
To simulate stochastic fluctuations in molecule numbers, we proceeded as follows. The standard parameter value set (see Materials and Methods) was used as the starting point. However, enzyme reaction velocities and Michaelis constants in Equations 12-20 were rescaled so that the units of the concentration variables were no longer nanomolar, but rather absolute numbers of molecules. To accomplish this scaling, the parameters vsP, vsC, vph, vdP, vdC, K1, K2, Kph, KdP, and KdC were all multiplied by a common factor. The rate constant kf governing association of PER and dCLOCK is always multiplied by two concentrations. Therefore, proper scaling of kf required dividing by the common factor. The value of the factor, 60, was determined by trial and error to yield oscillations not too degraded by stochastic noise. The peak levels of total PER during oscillations were close to 1000 molecules.
Second, the simulation time step was empirically adjusted to be sufficiently small (5 × 10−6 hr) so that the probability of each biochemical reaction in Equations 12-20 was never larger than 2%. Therefore, in any time step the chance that the copy number of any given molecular species will change by 1 is never larger than the product of 2% and the number of reactions that create or destroy that species. By using such a small time step, the probability of more than one reaction event occurring in any time step can be considered negligible. These reaction probabilities were computed by multiplying the time step with the terms in Equations 12-20 that give the rates of the specific reactions. There are 47 independent reaction terms, which are not enumerated separately here.
Third, at each time step a separate random number was generated for each independent reaction. Each random number was chosen from a uniform distribution over (0, 1). If the random number for a reaction was less than the probability of that reaction, then the reaction was assumed to occur, and the copy numbers of the molecular species involved were changed by 1 or −1. Otherwise, the copy numbers were not changed. If the time step is small so that the probability of more than one reaction occurring per time step is negligible, then the above scheme is an explicit simulation of the master equation governing the evolution of all the molecule numbers, and is accurate. To verify accuracy, simulations were repeated with the time step halved, and no significant differences in dynamics were seen.
Figure 7A illustrates that the model of the Drosophilia oscillator can simulate circadian oscillations of total PER and dCLOCK levels when stochastic noise is included as described above.
Fig. 7.
Robustness of the Drosophila model to stochastic noise. A, Simulation of circadian oscillations. Time courses are displayed for the numbers of molecules of all forms of PER (top, gray time course) and of dCLOCK (bottom, black time course). Parameter values in the standard set were scaled to convert units from a nanomolar concentration to number of molecules. Randomness in the timing of single-molecule synthesis and degradation events was incorporated as discussed in the text. The time step was kept small (5 × 10−6 hr) so that during no time step did the probability of any given reaction occurring exceed 2%. B, Expanded view displaying oscillations of the numbers of molecules of dCLOCK (gray time course) and of the PER-dCLOCK complex (black time course).
The oscillations are robust in that the amplitude and period do not show a large amount of random variation with this choice of parameters. The amplitude is seen to fluctuate, but when the simulation was continued for 50 oscillations, the SD of the amplitude was modest, 9% of the mean of 1014 molecules. The mean value of the oscillation period was 23.5 hr, almost identical to that without fluctuations. The SD in the period was small, 5% of the mean over 50 oscillations. On a short time scale (a few hours) fluctuations in the total number of PER molecules are quite small (apparent in Fig.7A). Figure 7B illustrates in more detail the dynamics of total dCLOCK and of PER–dCLOCK complexes (C ). On the time scale of a few hours, somewhat larger fluctuations are evident for these species. Nevertheless, the circadian rhythm in dCLOCK is robust.
If the simulation of Figure 7 was repeated with a lower scale factor for converting concentrations to molecule numbers, the average copy numbers of molecular species were lower, and circadian oscillations were degraded more appreciably by noise. If the scale factor was reduced from 60 to 20, average copy numbers were reduced by ∼70%. Oscillations in the level of PER were still preserved, and circadian variation in the level of dCLOCK was evident. However, the SD of the PER oscillation amplitude was larger, 20% of the mean over 50 oscillations. Circadian oscillations in PER were preserved for scale factors as low as 3 (peaks were at 50–70 molecules), but oscillations in dCLOCK were overwhelmed by fluctuations. If the scale factor was reduced to 1, then circadian oscillations in PER were also degraded by fluctuations. These results serve to indicate magnitudes of average molecule numbers compatible with simulation of oscillations by the model of Figure 1B. Experiments will need to estimate the average numbers per nucleus of PER and dCLOCK molecules to determine more rigorously whether proposed models of circadian rhythmicity inDrosophila are sufficiently robust to stochastic fluctuations. Analogous simulations (data not shown) were performed with the Neurospora model, and qualitatively similar behavior was observed.
Oscillations of circadian period can be simulated when positive feedback is eliminated
Because regulation of the rate of synthesis of the transcriptional activators dCLOCK and WCC is central to the positive-feedback loops in both models, simulations were performed with the concentrations of total dCLOCK and WCC fixed, eliminating positive feedback. Other parameter values were standard, except vph was increased to 30.0 nm/hr in the Drosophila model. With CLKtot fixed at 1.5 nm and WCCtot fixed at 3 nm, large-amplitude circadian oscillations in the levels of total PER and total FRQ were seen. The maxima of the oscillations were 17.2 nm for PER and 25.0 nm for FRQ, and the minima were near zero. The periods were 23.7 hr for PER and 23.9 hr for FRQ.
The robustness of these oscillations to changes in parameter values was examined as for oscillations with positive feedback (i.e., as in Fig. 6). As before, changes of >25% in amplitude or period were never observed when any parameter was increased or decreased by 15%. This result was also found when the fixed levels of dCLOCK or WCC were increased or decreased by 15%. These simulations indicate that, as suggested by previous models (Leloup and Goldbeter, 1998; Gonze et al., 2000), robust circadian oscillations could be sustained by only the negative feedback loops in which PER and FRQ repress their own transcription and then are slowly phosphorylated and degraded, relieving repression. For both models without positive feedback, entrainment to light pulses with an ISI significantly different from 24 hr could also be simulated. Because positive feedback does not appear essential for simulating circadian oscillations or for simulating entrainment to light, the question arises whether any function can be attributed to the positive feedback loops (see Discussion).
DISCUSSION
Circadian oscillations in Neurospora andDrosophila can be simulated by models incorporating positive and negative feedback and time delays
Two similar models were developed, which incorporate several features common to the circadian rhythm generators inNeurospora and Drosophila. These features include: (1) time delays to represent the intervals between changes in the concentrations of proteins that regulate transcription and changes in the rates of appearance of gene products, and (2) positive and negative feedback loops. In both Neurospora andDrosophila, a gene product, FRQ or PER (with TIM), represses transcription of its own gene, creating a negative feedback loop. But, positive feedback loops are also present in the Drosophilaand Neurospora circadian rhythm generators (Crosthwaite et al., 1997; Dunlap, 1999;Glossop et al., 1999; Bae et al., 2000;Lee et al., 2000).
The models for Neurospora and Drosophila (Fig.1A,B) include both the positive and negative feedback loops. Both models incorporate complex formation between transcriptional activators and repressors, and the feedback loops are “interlocked” in that complex formation is critical for both the positive and negative feedback. In Neurospora, the activator is WCC, a variable that represents a complex of WC-1 and WC-2, and inDrosophila, the activator is dCLOCK. The repressor is FRQ inNeurospora, and in Drosophila it is “PER,” a variable that represents an average or combination of PER and TIM levels. In both models, the repressor (FRQ or PER) represses its own transcription by complexing with the activator (WCC or dCLOCK). This repression constitutes a negative feedback loop. In both models, the repressor (FRQ or PER) also stimulates further synthesis of the activator (WCC or dCLOCK). The activator in turn stimulates further synthesis of the repressor, and a positive feedback loop is thereby formed. The only significant difference between theNeurospora and Drosophila models is the effect of activator–repressor complex formation. In the Neurosporamodel, complex formation blocks the activation of frqtranscription by WCC and the activation of WCC synthesis by FRQ. In theDrosophila model, complex formation represses pertranscription and de-represses dclock transcription.
Both models simulate circadian oscillations of core gene product concentrations (Figs. 2, 3). The oscillations can be entrained to simulated light pulses (Fig. 5). The oscillations are robust to variations in parameter values (Fig. 6). Robustness of oscillations to stochastic noise in biochemical reactions was demonstrated for some parameter values (Fig. 7), although further experimental work is needed to assess how accurately these parameter values describe oscillationsin vivo. These successful simulations of circadian oscillations in Neurospora and Drosophila suggest that despite the simplifications inherent in their construction, the models presented here capture the salient features of the processes underlying circadian rhythmicity.
Simulations suggest that time delays, but not positive feedback, are essential to generate circadian oscillations
The simulations in Figures 2 and 3 suggest that at least one long time delay between a change in the concentration of a transcription factor and a consequent change in the concentration of a regulated gene product is essential for circadian oscillations inDrosophila and Neurospora. The parameter τ1 played this role in simulations. τ1denotes the delay between a change in the concentration of transcriptional activator (dCLOCK or WC) and the resulting change in the synthesis rate of transcriptional repressor (PER or FRQ) and has a value of 7–8 hr.
Qualitatively, one function of τ1 may be to keep the positive and negative feedback loops from “cancelling out” each other. Without a delay, the effect of complex formation between activator and repressor and consequent sequestration of activator would be canceled out by increased synthesis of activator. For example, inDrosophila sequestration of dCLOCK by PER and TIM is vital for repression of per and tim transcription. But sequestration of dCLOCK also derepresses dclock, so more dCLOCK is synthesized. Without time delays, sequestration of dCLOCK and increased dCLOCK synthesis tend to abrogate each other's effect. The delay τ1 offsets these events so they do not directly compete against each other. However, when positive feedback was eliminated by fixing the concentration of WCC or dCLOCK, we found that τ1 was still needed for oscillations. Therefore, one can also think of τ1 as playing an essential role within the negative-feedback loop. In our models, unlike previous models that relied on negative feedback (Leloup et al., 1998;Gonze et al., 2000), the phosphorylation state of FRQ or PER does not affect its ability to repress transcription of its own gene. Therefore, our models lack an implicit time delay that was present in the earlier models, in which only the most highly phosphorylated forms of FRQ or PER could repress transcription. Removal of the implicit delay from the negative-feedback loop tends to abolish oscillations. The explicit delay τ1 is needed to compensate.
The second long time delay in the Neurospora andDrosophila models, τ2, is also important, because it acts to offset the time courses of FRQ and WCC, and of PER and dCLOCK. Experimentally, oscillations in WC-1 lag oscillations in FRQ by ∼10 hr, and are almost antiphase (Lee et al., 2000). In simulations, a long τ2 is necessary to give a lag of this order (Fig. 3) (τ2 = 7 hr).
A model of a generic (not organism-specific) circadian oscillator, relying on a single time delay for autorepression of a core gene by its product, has recently been presented (Lema et al., 2000). In our models, τ1 accounts for most of the delay in the autorepression of per or frq and is thus analogous to the single delay in Lema et al. (2000).
It has previously been suggested that both positive and negative feedback are necessary for sustaining circadian oscillations (Crosthwaite et al., 1997; Hastings, 2000). In fact, Hastings (2000) suggested that an oscillator based on a single negative-feedback loop would progressively dampen over time and that addition of positive feedback would increase stability of oscillations to stochastic fluctuations. With both the Neurospora and Drosophila models, circadian oscillations could be simulated when the total concentration of transcriptional activator (WCC or dCLOCK) is held fixed. Under these conditions only the negative feedback loop in which FRQ or PER represses its own transcription is operative. Negative feedback, coupled with slow phosphorylation of FRQ or PER before degradation, suffices to drive oscillations that are robust to modest variations in parameter values. Earlier models based on negative feedback alone were also able to sustain oscillations indefinitely (Leloup and Goldbeter, 1998; Goldbeter et al., 2000). Therefore, positive feedback loops do not appear essential for circadian oscillations per se. Whether positive feedback increases robustness to stochastic fluctuations, as suggested byHastings (2000), remains to be evaluated.
An alternative possibility is that positive feedback is required to regulate “output,” or clock-controlled, genes (CCGs). CCGs are not part of the core feedback loops, but they are responsible for circadian variation in organism behaviors such as locomotion. InDrosophila, the positive feedback loop appears essential to drive circadian oscillations in the level of total dCLOCK, and inNeurospora, the positive feedback loop appears essential for oscillations in the level of total WC-1. Positive feedback may therefore be essential to drive variations in the expression of CCGs regulated by dCLOCK or WC-1. At least one Drosophila CCG, for pigment-dispersing factor, is known to be regulated by dCLOCK (Park et al., 2000). Whether WC-1 regulates the expression of CCGs in Neurospora is not yet known.
Future directions for experiment and modeling
Further experiments are needed to elucidate the mechanisms responsible for the long time delays that appear essential for operation of the circadian oscillators in Drosophila and inNeurospora—the parameters τ1 and τ2 in our models. In particular, τ1 must be on the order of 6–8 hr for our models to simulate circadian oscillations. In Neurospora, there is evidence from a transgenic system that τ1 can be as short as 3 hr (Merrow et al., 1997). It is, however, evident that during normal circadian oscillations τ1 is longer. During normal oscillations, one component of τ1 (the delay between frq mRNA and FRQ protein) is ∼4 hr (Garceau et al., 1997). It is not evident why, during normal oscillations, τ1 should differ from its value in the transgenic system of Merrow et al. (1997). ForDrosophila, the value of 8 hr assumed for τ1appears reasonable, insofar as one component of (the lag between the time courses of per mRNA and PER protein) is ∼5–6 hr (Saez and Young, 1996; Glossop et al., 1999).
Questions have been raised whether frq inNeurospora, and per and tim inDrosophila, are in fact core circadian genes whose loss of function leads to a loss of circadian rhythmicity. InNeurospora mutants devoid of functional frq, rhythmic conidiation with circadian period can be observed (Lakin-Thomas and Brody, 2000) although these rhythms often display a wide range of periods and poor temperature compensation (Iwasaki and Dunlap, 2000). In transgenicDrosophila with constitutive per ortim expression, behavioral circadian rhythmicity is sometimes seen (Lakin-Thomas, 2000). It has been pointed out that previous screens for Drosophila andNeurospora circadian mutants are biased towards genes whose only important function is to sustain circadian rhythmicity, so that core genes also involved in other biochemical pathways might not have been identified (Lakin-Thomas, 2000). We cannot exclude the possibility that the present models, as well as earlier models based on oscillations of FRQ, PER, and TIM gene products, lack some unidentified essential core genes and feedback loops. However, because alterations in the kinetics of FRQ or PER phosphorylation (Price et al., 1998; Liu et al., 2000) or of PER–TIM interaction (Gekakis et al., 1995) affect circadian period, it is likely that these genes are components of core oscillators. To explain rhythmicity in the absence of these proteins, there may be redundancy in oscillators. For example, there may be a second uncharacterized circadian oscillator in Drosophilaand Neurospora that can sustain behavioral rhythmicity by itself, at least in some individuals. In the presence of functional FRQ, PER, and TIM, these latent oscillators might be overridden or phase locked by the primary oscillators based on FRQ, PER, and TIM.
Previous models of circadian rhythm generation in Drosophilaand Neurospora have included both protein phosphorylation and negative feedback, but have generally not relied on positive feedback (Leloup and Goldbeter, 1998; Leloup et al., 1999; Ruoff et al., 1999; Gonze et al., 2000) (but see Tyson et al., 1999 for a qualitative Drosophila model with a different proposed positive-feedback loop). Nor have they relied on time delays (but seeScheper et al., 1999 and Lema et al., 2000 for generic models based on delays). We believe that a somewhat complex model is needed to capture the dynamics of negative and positive feedback at the level of transcriptional regulation, as well as the time delays that characterize these processes. Perhaps the qualitative models presented here for the circadian oscillators based on transcriptional regulation of frq and ofper/tim begin to address this need.
Positive or negative feedback involving interactions between transcriptional activators and repressors may also underlie circadian rhythms in other organisms. For example, in the cyanobacteriumSynechococcus, the transcriptional activator KaiA appears to enhance the expression of the transcriptional repressors KaiC and/or KaiB (Iwasaki and Dunlap, 2000). In mammals, there also appear to be interacting positive and negative feedback loops, which are based on interactions between the transcriptional activator CLOCK with isoforms of PER and/or cryptochrome proteins (Kume et al., 1999; Shearman et al., 2000). Therefore, models similar to ours might be useful for describing circadian rhythm generation in Synechococcus, mammals, or other organisms.
Footnotes
This work was supported by National Institutes of Health Grants R01 RR11626 and P01 NS38310. We thank P. Hardin for his comments on this manuscript.
Correspondence should be addressed to John H. Byrne, Department of Neurobiology and Anatomy, W. M. Keck Center for the Neurobiology of Learning and Memory, The University of Texas–Houston Medical School, P. O. Box 20708, Houston, TX 77225. E-mail:John.H.Byrne@uth.tmc.edu.
REFERENCES
- 1.Bae K, Lee C, Sidote D, Chuang K, Edery I. Circadian regulation of a Drosophila homolog of the mammalian clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae K, Lee C, Hardin PE, Edery I. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J Neurosci. 2000;20:1746–1753. doi: 10.1523/JNEUROSCI.20-05-01746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkai N, Leibler S. Circadian clocks limited by noise. Nature. 2000;403:267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- 4.Crosthwaite SK, Dunlap JC, Loros JJ. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 5.Crosthwaite SK, Dunlap JC, Loros JJ. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 6.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors PER and TIM. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 7.Denault DL, Loros JJ, Dunlap JC. WC-2 mediates WC-1–FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 2001;20:109–117. doi: 10.1093/emboj/20.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 9.Edery I, Zwiebel L, Dembinska M, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garceau NY, Liu Y, Loros J, Dunlap JC. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 11.Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 12.Glossop N, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 13.Gonze D, Leloup JC, Goldbeter A. Theoretical models for circadian rhythms in Neurospora and Drosophila. C R Acad Sci III. 2000;323:57–67. doi: 10.1016/s0764-4469(00)00111-6. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin BC. Advances in enzyme regulation, vol 3. Oxford; Pergamon: 1965. Oscillatory behavior in enzymatic control processes. pp. 425–438. [DOI] [PubMed] [Google Scholar]
- 15.Hastings MH. Circadian clockwork: two loops are better than one. Nat Rev Neurosci. 2000;1:143–146. doi: 10.1038/35039080. [DOI] [PubMed] [Google Scholar]
- 16.Iwasaki H, Dunlap JC. Microbial circadian oscillatory systems in Neurospora and Synechoccus: models for cellular clocks. Curr Opin Microbiol. 2000;3:189–196. doi: 10.1016/s1369-5274(00)00074-6. [DOI] [PubMed] [Google Scholar]
- 17.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene doubletime encodes a protein closely related to human casein kinase Iɛ. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 18.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 19.Lakin-Thomas PL. Circadian rhythms, new functions for old clock genes? Trends Genet. 2000;16:135–142. doi: 10.1016/s0168-9525(99)01945-9. [DOI] [PubMed] [Google Scholar]
- 20.Lakin-Thomas PL, Brody S. Circadian rhythms in Neurospora crassa: lipid deficiencies restore robust rhythmicity to null frequency and white-collar mutants. Proc Natl Acad Sci USA. 2000;97:256–261. doi: 10.1073/pnas.97.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 22.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19:5316–5323. doi: 10.1128/mcb.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K, Loros JJ, Dunlap JC. Interconnected feedback loops in the Neurospora circadian system. Science. 2000;289:107–110. doi: 10.1126/science.289.5476.107. [DOI] [PubMed] [Google Scholar]
- 24.Leloup JC, Goldbeter A. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J Biol Rhythms. 1998;13:70–87. doi: 10.1177/074873098128999934. [DOI] [PubMed] [Google Scholar]
- 25.Leloup JC, Goldbeter A. Modeling the molecular regulatory mechanism of circadian rhythms in Drosophila. Bioessays. 2000;22:84–93. doi: 10.1002/(SICI)1521-1878(200001)22:1<84::AID-BIES13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 26.Leloup JC, Gonze D, Goldbeter A. Limit cycle models for circadian rhythms based on transcriptional regulation in Drosophila and Neurospora. J Biol Rhythms. 1999;14:433–448. doi: 10.1177/074873099129000948. [DOI] [PubMed] [Google Scholar]
- 27.Lema MA, Golombek DA, Echave J. Delay model of the circadian pacemaker. J Theor Biol. 2000;204:565–573. doi: 10.1006/jtbi.2000.2038. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Loros J, Dunlap JC. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci USA. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo C, Loros JJ, Dunlap JC. Nuclear localization is required for function of the essential clock protein FRQ. EMBO J. 1998;17:1228–1235. doi: 10.1093/emboj/17.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald N. Biological delay systems: linear stability theory. Cambridge UP; Cambridge: 1989. [Google Scholar]
- 31.Marrus SB, Zeng H, Rosbash M. Effect of constant light and circadian entrainment of perS flies: evidence for light-mediated delay of the negative feedback loop in Drosophila. EMBO J. 1996;15:6877–6886. [PMC free article] [PubMed] [Google Scholar]
- 32.Merrow MW, Garceau N, Dunlap JC. Dissection of a circadian oscillation into discrete domains. Proc Natl Acad Sci USA. 1997;94:3877–3882. doi: 10.1073/pnas.94.8.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 34.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. doubletime is a novel Drosophila clock gene that regulates PERIOD accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 36.Ruoff P, Vinsjevik M, Mohsenzadeh S, Rensing L. The Goodwin model: simulating the effect of cycloheximide and heat shock on the sporulation rhythm of Neurospora crassa. J Theor Biol. 1999;196:483–494. doi: 10.1006/jtbi.1998.0846. [DOI] [PubMed] [Google Scholar]
- 37.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 38.Saez L, Young MW. Regulation of nuclear entry of the Drosophila clock proteins PERIOD and TIMELESS. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 39.Scheper T, Klinkenberg D, Pennartz C, van Pelt J. A mathematical model for the intracellular circadian rhythm generator. J Neurosci. 1999;19:40–47. doi: 10.1523/JNEUROSCI.19-01-00040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst G, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 41.Smolen P, Baxter DA, Byrne JH. Effects of macromolecular transport and stochastic fluctuations on the dynamics of gene networks. Am J Physiol. 1999;277:C777–C790. doi: 10.1152/ajpcell.1999.277.4.C777. [DOI] [PubMed] [Google Scholar]
- 42.Smolen P, Baxter DA, Byrne JH. Modeling transcriptional control in gene networks—methods, recent results, and future directions. Bull Math Biol. 2000a;62:247–292. doi: 10.1006/bulm.1999.0155. [DOI] [PubMed] [Google Scholar]
- 43.Smolen P, Baxter DA, Byrne JH. Mathematical modeling of gene networks. Neuron. 2000b;26:567–580. doi: 10.1016/s0896-6273(00)81194-0. [DOI] [PubMed] [Google Scholar]
- 44.So WV, Rosbash M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 1997;16:7146–7155. doi: 10.1093/emboj/16.23.7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talora C, Franchi L, Linden H, Ballario P, Macino C. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 1999;18:4961–4968. doi: 10.1093/emboj/18.18.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyson JJ, Hong CI, Thron CD, Novak B. A simple model of circadian rhythms based on dimerization and proteolysis of PER and TIM. Biophys J. 1999;77:2411–2417. doi: 10.1016/S0006-3495(99)77078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vosshall LB, Price JL, Sehgal A, Saez L, Young MW. Block in nuclear location of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 48.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 49.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]