Abstract
In Drosophila, the amidated neuropeptide pigment dispersing factor (PDF) is expressed by the ventral subset of lateral pacemaker neurons and is required for circadian locomotor rhythms. Residual rhythmicity in pdf mutants likely reflects the activity of other neurotransmitters. We asked whether other neuropeptides contribute to such auxiliary mechanisms. We used thegal4/UAS system to create mosaics for the neuropeptide amidating enzyme PHM; amidation is a highly specific and widespread modification of secretory peptides in Drosophila. Three different gal4 drivers restricted PHM expression to different numbers of peptidergic neurons. These mosaics displayed aberrant locomotor rhythms to degrees that paralleled the apparent complexity of the spatial patterns. Certain PHM mosaics were less rhythmic than pdf mutants and as severe asper mutants. Additional gal4 elements were added to the weakly rhythmic PHM mosaics. Although addingpdf-gal4 provided only partial improvement, adding the widely expressedtim-gal4 largely restored rhythmicity. These results indicate that, in Drosophila, peptide amidation is required for neuropeptide regulation of behavior. They also support the hypothesis that multiple amidated neuropeptides, acting upstream, downstream, or in parallel to PDF, help organize daily locomotor rhythms.
Keywords: circadian rhythms, Drosophila, neuropeptide, amidation, PHM, gal4, PDF
Relatively little is known about output signals used by circadian pacemaker neurons. In mammals, pacemaker neurons controlling daily locomotion are located in the suprachiasmatic nucleus (SCN), which contains several thousand cells that present several transmitter phenotypes (Silver et al., 1999). The SCN may release diffusible substances to influence behavioral rhythms (Silver et al., 1996), although the identity of such substances remains unknown. In Drosophila, the lateral neurons (LNs) are critical pacemakers regulating daily locomotor rhythms. A ventral subset of LNs (LN-Vs) (Ewer et al., 1992) expresses the neuropeptide pigment dispersing factor (PDF) (Helfrich-Förster, 1995) that is highly related to crustacean pigment dispersing hormone (PDH). Loss of PDF expression produces abnormal locomotor rhythms, and ablation of the PDF neurons produces a very similar phenotype (Renn et al., 1999). These genetic results confirm that LN-Vs are the principal circadian pacemaker neurons in Drosophila (cf. Frisch et al., 1994) and support the hypothesis that pdf encodes the principal circadian transmitter. However, pdf mutant animals are still able to entrain (Renn et al., 1999), and by quantitative measures, their free-running phenotype is less severe than that ofdisco mutants (Dushay et al., 1989; Helfrich-Förster, 1998). In disco mutants, the brain lacks many neurons, including all LNs. Together, these features suggest that additional transmitters are needed to produce normal circadian locomotor rhythms.
To identify such transmitters, we manipulated the biosynthesis of neuropeptides by creating genetic mosaics. In particular, we studied the behavioral consequences of cell-specific deficiencies in C-terminal peptide α-amidation (for review, see Eipper et al., 1993). In vertebrates, >50% of known neuropeptides are amidated; inDrosophila, >90%, including PDF, are amidated (Hewes and Taghert, 2001). α-Amidation is a highly specific modification for secretory peptides, and the two enzymes catalyzing amidation (called PHM and PAL) are found exclusively within luminal vesicular compartments (Eipper et al., 1993). In Drosophila, PHM is encoded by the PHM gene (Kolhekar et al., 1997).PHM mutant animals die as late embryos or young larvae; peptide amidation in both larval and adult stages requires PHM enzyme (Jiang et al., 2000).
The present experimental design had three requirements. The first was to restore sufficient PHM activity to PHM mutant animals to prevent death and so permit the testing of adult behavior. For this, we used the gal4 technique (Brand and Perrimon, 1993). The second was to limit the scope of restored PHM activity compared with its normal patterns and/or levels. For this, we chose gal4lines that featured differing numbers of peptidergic neurons in their expression patterns. The third requirement was to ensure that PHM activity in these mosaic animals was restored within PDF LN-Vs to ask whether neuropeptides other than PDF participate in the regulation of daily locomotion. For this, we analyzed combinations of gal4elements that included pdf-gal4 (Park et al., 2000). The results demonstrate that (1) the normal display of daily locomotor rhythms requires the participation of amidated peptide transmitters, and (2) amidated neuropeptides in addition to PDF are required for this behavioral regulation.
MATERIALS AND METHODS
Genetic strains. The two alleles of PHMused in these studies were described by Jiang et al. (2000). P{lacW}k[07623] is a P-element insertion within the first exon of PHM; here we refer to it asPHM01. PHMP29 contains a ∼1.3 kb deletion generated by mobilizing the k[07623] element; here we refer to it as PHM02. PHM01 is a strong hypomorphic allele, whereasPHM02 appears to be a null. BecausePHM02 also deletes sequences belonging to a neighboring gene (Jiang et al., 2000), we analyzed thetrans-heterozygote combinationPHM01/PHM02. Deficiency stocks were obtained from the Bloomington Stock Center (Indiana University, Bloomington, IN). P{gal4} lines are summarized in Table 1. Thec929-gal4 insertion (at 39C4 of the second chromosome) (Hewes et al., 2000) was recombined onto the chromosome bearing the PHM01 allele. Second chromosome mutations were balanced by In(2LR)O,Cy, y+; third chromosome mutations were balanced by In(3LR)TM3,Sb. Canton-S was used for a wild-type (WT) strain.
Table 1.
Properties of the gal4 lines used in the behavioral experiments
| Name | Position (chromosome)/ nearest gene | Expression pattern | Reference |
|---|---|---|---|
| c929 | 39CD(II)/CG8669 | Secretory cells; many tissues | 1, 2 |
| 36Y | 85C(III)/CG11033 | Secretory cells; many tissues | 1 |
| 386Y | 97D(III)/CG6438 | Secretory cells; many tissues | 3 |
| Appl 3GK | III/5, n.d. | CNS only; low in optic lobes; broadly in central brain | 4 |
| D42 | III/n.d. | ∼200 CNS neurons, including motorneurons | 5 |
| tim(#16) | III/n.d. | Broadly within CNS | 3, 6 |
| tim(#67) | III/n.d. | n.d. | 3 |
| pdf(M) | X/n.d. | Similar to pdf | 7 |
| pdf(N) | X/n.d. | Similar to pdf | 7 |
n.d., Not determined. 1, O'Brien and Taghert, 1998; 2, Hewes et al., 2000; 3, this work; 4, K. White, personal communication; 5, G. Boulianne, personal communication; 6, Kaneko and Hall, 2000; 7, Park et al., 2000.
Molecular biology. Standard molecular methods were used (Maniatis et al., 1982). A UAS-PHM construct was assembled by isolating the EcoRI-KpnI fragment ofPHM cDNA #1 (Kolhekar et al., 1997), blunt-ending theKpnI site, then ligating the insert into the CaSpeR-based vector pP{UAST} at the EcoRI and NotI sites. Several independent germ line transformants were made and recovered by standard procedures (Benveniste and Taghert, 1999). Briefly, DNAs were purified by Qiagen miniprep protocols, according to the manufacturer's recommendations, and injected at a concentration of ∼750 ng/ml into embryos homozygous for the elementP{(Δ2–3)99B} that is a source of transposase activity. Backcrosses of positive transformants to balanced stocks that contained dominant markers were used to identify insertion chromosomes. Southern blot analyses were used to determineP-element copy number. Western blots were used to evaluate basal and induced levels of PHM. In all studies reported here, we used a single-copy, homozygous-viable, second chromosome insertion (C1A) that was recombined onto the chromosome bearing thePHM02 allele.
Single-fly PCR. To confirm the genotype of recombinants, we used the procedure of Gloor et al. (1993). Two pairs of oligonucleotides were used in single or multiplex PCR to determine the presence of PHM mutant alleles, as described by Jiang et al. (2000).
Mapping P-element insertion positions. We used methods described by the Berkeley Drosophila Genome Project (BDGP –http://www.fruitfly.org/about/methods/inverse.pcr.html) to map the position of the P{w+,gal4}386Y insertion. It is found at bp ∼118,100 of GenBank sequence number AE003757. This position is within 320 bp of the 3′ end of amontillado (CG6438), which encodes the Drosophila homolog of the neuropeptide-processing enzyme PC2 (Siekhaus and Fuller, 1999); it is also within 1544 bp of the 5′ end of the neighboring gene CG6425. The position of the P{w+,gal4}36Y insertion was determined by plasmid-rescue methods: a ∼2 kb PstI-flanking fragment was subcloned and sequenced. The insertion site lies at bp ∼144,300 of GenBank sequence number AE003681, within the first intron of CG11033.
Immunocytochemistry. CNS and gut tissues were stained in whole mount using procedures similar to those described in Renn et al. (1999). Rabbit anti-dPHM was used at a 1:750 dilution (Kolhekar et al., 1997); guinea pig anti-PAP (this recognizes an non-PDF epitope on the proPDF precursor) (Renn et al., 1999) was used at 1:1000; rabbit anti-FMRFamide (Taghert and Schneider, 1990) was used at 1:2000; mouse anti-βGAL (Promega, Madison WI) was used at 1:1000. Secondary antibodies (Jackson ImmunoResearch, West Grove, PA) conjugated with Cy3 or Alexa 468 were used at 1:200 or 1:500 dilutions. Tissues were cleared in glycerol, mounted in Vectashield (Vector Laboratories, Burlingame, CA), and examined with an Olympus confocal microscope and Fluoview software or with a Zeiss Axioplan microscope fitted with a Spot CCD camera (Diagnostics Instruments, Sterling Heights, MI). Confocal images were assembled in Adobe Photoshop. Spot camera images were used for quantification of immunosignals. In Adobe Photoshop, histogram values of fluorescence intensity from single cell bodies (less than from equivalent background areas immediately adjacent) were acquired for both anti-PHM and anti-PAP signals.
Behavioral analysis. Locomotor activity rhythms of 1- to 5-d-old adult males were monitored at 25°C as described in Hamblen et al. (1986, 1998). Locomotor performance was measured using analytic software provided by the Brandeis Rhythm Package (http://hawk.bcm.tmc.edu/). We first monitored behavior over 7–8 d in 12 hr light/dark (LD) conditions; free-running activity was then monitored in constant darkness (DD) for a further 9–10 d. The number of activity events was recorded per half-hour bin, and average numbers of activity events per bin, per fly were calculated (cf. Hamblen-Coyle et al., 1989). χ2 periodogram analyses (Sokolove and Bushell, 1978) identified animals displaying behavioral rhythmicity with the following thresholds: power ≥ 10 and width ≥ 2.0 (cf. Ewer et al., 1992; Renn et al., 1999). For more sensitive determinations of free-running periodicities, the activities were subjected to a low-pass digital filter (Dowse and Ringo, 1987), then analyzed by Maximum Entropy Spectral Analysis (MESA) (cf. Dowse and Ringo, 1987; Hamblen-Coyle et al., 1989). Subsequently, a MESA-based signal-to-noise ratio (SNR) for each fly was computed (Dowse and Ringo, 1987) and averaged per genotype. SNR averages were log10-transformed; one-way ANOVAs and post hoc tests (Dunnett's test; all versus control) were performed with InStat (GraphPad, San Diego, CA). All behavioral data studying genotypes that have already been described (e.g.,per01) represent results not previously published.
RESULTS
gal4 patterns
The 36Y-gal4 element is inserted within CG11033, which encodes a protein that contains PHD domain found in a class of transcription factors (Aasland et al., 1995). The36Y-gal4 enhancer pattern has not yet been associated with a specific gene. Aspects of the larval 36Y-gal4pattern were described by O'Brien and Taghert (1998). Using UAS-lacZ expression as a reporter, 36Y-gal4neural expression was the most restricted of any gal4 tested in these experiments, after pdf-gal4. In the adult (n = 20; data not shown), UAS-lacZ was limited to ∼30 neurons scattered in the central brain and several cells within the optic lobes that defined a parallel array. Many of the neurons appear to be peptidergic by cell body position and by double antibody staining (data not shown), including a small number of neurons in the vicinity of the PDF-expressing LN pacemakers.36Y-gal4 expression included strong staining in two peritracheal endocrine cells, called PMa and PMb (O'Brien and Taghert, 1998). PMa is the probable Inka cell homologue (Zitnan et al., 1996).36Y-gal4 expression was also prominent in ring gland, salivary glands, fat body, epidermis, and hindgut (n = 20).
The c929-gal4 element is inserted in thecryptocephal gene (crc, CG8669,dATF-4) (Hewes et al., 2000); however, its expression pattern does not match that of crc, but rather matches that of an adjacent gene (R. S. Hewes and P. H. Taghert, unpublished observations). This pattern (n > 30; data not shown) included ∼100 neurons in the CNS. All (or nearly all)c929-gal4 positive neurons are peptidergic, as judged by double immunostaining with antibodies against diverse neuropeptides and neuropeptide biosynthetic enzymes (R. S. Hewes and P. H. Taghert, unpublished observations). In particular, thec929-gal4 pattern precisely overlapped that of strong PHM immunosignals, with very few exceptions. In adults, thec929-gal4 pattern included a set of neurons similar to that of 36Y-gal4, but typically displayed more cells per area, and the cells were often more densely stained (e.g., the LN area). c929-gal4 was expressed in the peritracheal PMa endocrine cell, but not in cell PMb (cf. O'Brien and Taghert, 1998). The neurilemma of the brain was βGAL-positive in c929 gal4 animals; the pattern also included heterogenous expression in a variety of other tissues, including ring gland cells, epidermis, the gut endocrine system, salivary glands, and fat body (n > 20).
We located the 386Y-gal4 element just 3′ toamontillado (CG6438), which is the Drosophilahomolog of the proprotein processing enzyme PC2 (Siekhaus and Fuller, 1999). As tested with UAS-lacZ, the386Y-gal4 expression pattern was broad and included numerous peptidergic neurons in the CNS (n = 15; data not shown) and secretory cells in the periphery (n = 4). The number of 386Y-positive cells was considerably greater than that of either 36Y- orc929-gal4 patterns. The386Y-gal4 pattern included many neurons in the vicinity of LNs, numerous Kenyon cells, as well as other prominent peptidergic neurons (data not shown). In larval stages, the PDF LN-Vs (usually three or four per hemisphere) co-expressed βGAL (n = 5); in adults, only the large LN-Vs co-expressed βGAL (n = 4; data not shown). Outside the CNS,386Y-gal4 drove heterogeneous expression in numerous tissues; most of this expression was correlated with secretory cell activity (e.g., in gut, in peritracheal cells, and in neurosecretory neurons of the PNS; data not shown).
The pdf(M) and pdf(N)-gal4 elements are independent insertion lines of the same transgene. They produced βGAL patterns that were largely comparable with each other and with the pattern described by Park et al. (2000), namely, restriction to PDF-expressing CNS cells, including strong expression in both large and small LN-Vs, in two to four tritocerebral cells in young imagos and in approximately six abdominal ganglion gut efferents (n = 4 each). The cell bodies of small LN-Vs normally stain weakly for PDF, compared with those of large LN-Vs. However, the strength of reporter gene expression in small LN-Vs appeared very strong in both lines and similar to that of the large LN-Vs. Ectopic lacZ expression was seen in approximately eight neurons of the subesophaegal region in the (M) line and in two small neurons of the protocerebrum in the (N) line (data not shown).
The tim(#16)-gal4 driver produced a very broad and strong expression pattern throughout the adult brain (n = 4) (cf. Kaneko and Hall, 2000). Its expression outside the CNS was not studied (but see Kaneko and Hall, 2000).Appl(3GK)-gal4 produced a strong and diverse pattern throughout the adult brain that included numerous cells in all brain regions (n = 4); the extent of the pattern was comparable with that of 386Y-gal4, but appeared less than that of tim-gal4. c155-gal4(Lin and Goodman, 1994) produced a weak level of expression throughout many brain regions and moderate expression in several large neurons of the subesophageal region (n = 8).
Reverting PHM lethality with thegal4/UAS system
We asked whether any of seven different gal4 drivers could produce sufficient PHM activity in a PHM mutant background to revert the early lethality associated with thePHM phenotype (Jiang et al., 2000). Table2 presents the degree to which these genetic combinations could rescue animals trans-heterozygous for the two alleles, PHM01 andPHM02. Neither of twotim-gal4 lines (#16 or #67) was able to drive UAS-PHM-mediated rescue PHM mutant animals, whereas c929-gal4 and 386Y-gal4provided partial rescue (each ∼67% of expected).36Y-gal4 provided a strong measure of rescue in driving UAS-PHM. Combining 36Y-gal4and c929-gal4 drivers produced a measure of rescue comparable with that of 36Y-gal4 alone. Rescued adult animals of all genotypes that were tested survived the 18 d period of the behavioral assay as well as did the wild-type stock. In the following, we simplify the text by describing the behavior of UAS-PHM-rescued PHM mutant flies with reference only to the gal-4 driver(s) that was used (e.g.,36Y-gal4-rescued animals).
Table 2.
Rescue of PHM lethality produced by differentgal4/UAS-PHM combinations
| Potential rescued PHM mosaic (i.e., Cy+) | 2-159 Cy+ | 2-159 Cy− | % of Cy+ expected | ||||
|---|---|---|---|---|---|---|---|
| y w; | PHM01/UAS-PHM,PHM02; | 36Y-gal4 | /+ | 80 | 138 | 110 | |
| y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | 110 | 381 | 67 | ||
| y w; | PHM01/UAS-PHM,PHM02; | 386Y-gal4 | /+ | 32 | 112 | 67 | |
| y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | 36Y-gal4 | /+ | 28 | 51 | 107 |
| y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | D42-gal4 | /+ | 21 | 74 | 68 |
| y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | Appl(3GK) | /+ | 37 | 59 | 116 |
| y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | tim(2-15916)-gal4 | /+ | 76 | 169 | 93 |
| y w; | PHM01/UAS-PHM,PHM02; | tim(2-15916)-gal4 | /+ | 0 | 85 | 0 | |
| y w; | PHM01/UAS-PHM,PHM02; | tim(2-15967)-gal4 | /+ | 0 | 77 | 0 | |
F2-159: Cy+, The number of rescued PHM mutant animals (adults not displaying the marker for the Balancer chromosome and therefore trans-heterozygous for the two PHMalleles);
Cy−, the number of heterozygous PHMmutant animals (adults displaying the marker for the Balancer chromosome, CyO); % of Cy+ expected, percentage of PHM mutant animals observed and/or expected.
gal4 restricts PHM expression
To visualize the restriction of UAS-PHM by the different gal4 drivers in a PHM mutant background, we stained the brains of rescued adult males (1–10 d old) for PHM immunosignals and compared them with wild-type tissues. Wild-type brains display widespread and heterogeneous PHM staining (n = 15) (Fig. 1). PHM-like immunoreactivity is seen in several large cell bodies, including some in the lateral and dorsal protocerebrum, in subesophageal neuromeres, and among Kenyon cells. It also highlights several organized neuropils, like the lobes of the Mushroom bodies and several sections of the Fan-Shaped body, and appears strongly in dorsal protocerebral neuropil and neuropil surrounding the esophageal foramen.36Y-gal4-rescued animals (n = 8) displayed PHM immunosignals in several identifiable peptidergic neurons [e.g., MP1 and MP2 neurons (O'Brien et al., 1991)] on a generally low background of staining (Figs. 1, 2). With c929-gal4 (n = 6), there was a very similar pattern, but with several additional cell bodies in the dorsal protocerebrum, in the LN cell body region, and in the subesophageal neuromeres. There were also a greater number of stained processes (Fig. 1). The pattern ofc929-gal4/UAS-PHM expression differed slightly from that ofc929-gal4/UAS-lacZ; the latter included numerous cells of the neurilemma, but this feature was not apparent with PHM expression. Finally,386Y-gal4-rescued animals (n = 8) displayed a much broader level of staining throughout the brain, including numerous strongly stained cells in the LN cell body region and throughout the dorsal protocerebrum (Fig. 1). These animals also displayed a greater “background” level of immunostaining, suggesting a general low level of expression in many scattered cells. Figure 2 presents drawings to schematize these generalized cellular patterns.
Fig. 1.
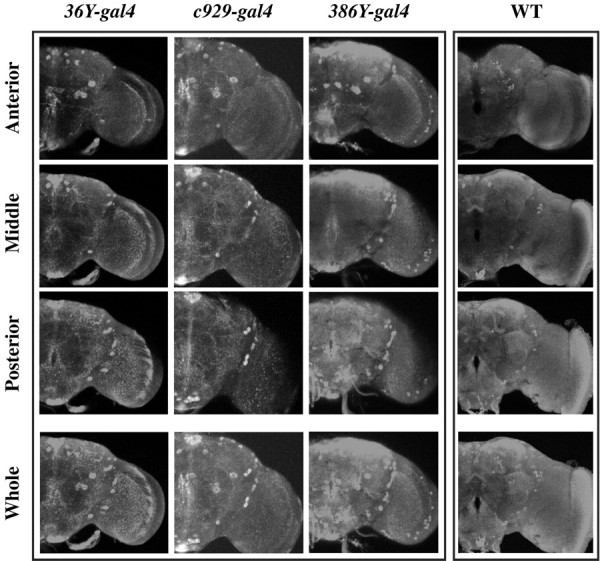
Expression of PHM immunostaining driven by different gal4 drivers in a PHM null background. Each column displays a three-part series of confocal images from a single adult male brain of the genotype indicated. Thebottom panels (Whole) represent overlays of the three-volume sections and depict nearly the entire brain thickness. The three separate scans display anterior,middle, and posterior sections of the brain. For the wild-type (WT) brain,36Y-rescued animals, and c929-rescued animals, section depths were 66, 60, and 60 μm, respectively. For386Y-gal4-rescued animals, section depths were 72, 60, and 60 μm, respectively.
Fig. 2.
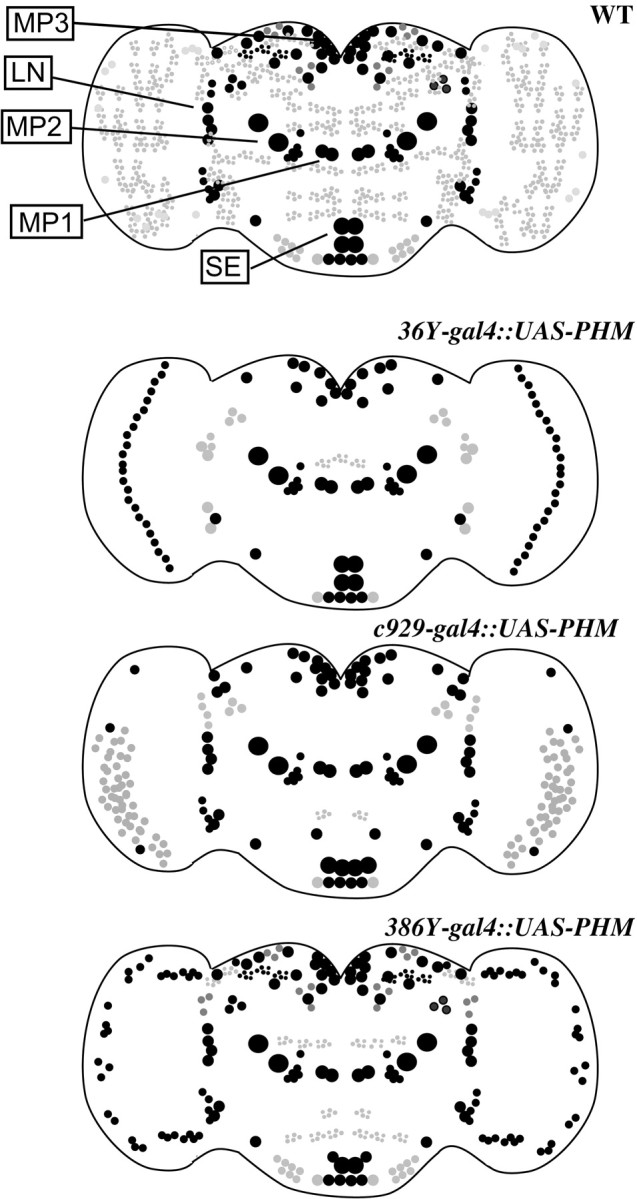
Schematic diagrams of the staining patterns displayed in Figure 1. The genotypes are indicated to theleft of each panel. The sizes and positions of various cell bodies are approximate. Black indicates strong staining; gray indicates weak to moderate staining. Many cell types included in these patterns appeared similar; double antibody staining experiments confirmed that many cellular elements are shared (see text for further details). MP1, MP2,MP3, LN, and SE indicate prominent peptidergic cell groups; most preparations included one or more representatives of each group.
We also asked whether the PDF neurons of the brain of rescued animals were specifically included in any of the above three gal4patterns: 36Y, c929, or 386Y. We performed double antibody labeling on these genotypes for PHM and for identified LN-V neurons by anti-proPDF staining.36Y-gal4 displayed weak PHM expression in one or two large PDF LN-Vs, but not in the small LN-Vs (data not shown).c929-gal4 and 386Y-gal4 displayed expression patterns that were greater than36Y-gal4 and similar to each other; they contained bright PHM immunolabeling in all of the large PDF LN-Vs and the tritocerebral cells, but in not the small LN-Vs (n= 12). Figure 3 shows an example of PHM immunostaining in LN-Vs in a PHM mutant rescued by expression of PHM driven by two gal4 elements,c929-gal4 and 386Y-gal4(n = 4). Even together, these two elements do not generate detectable PHM immunosignals in the small LN-Vs of the adult.
Fig. 3.
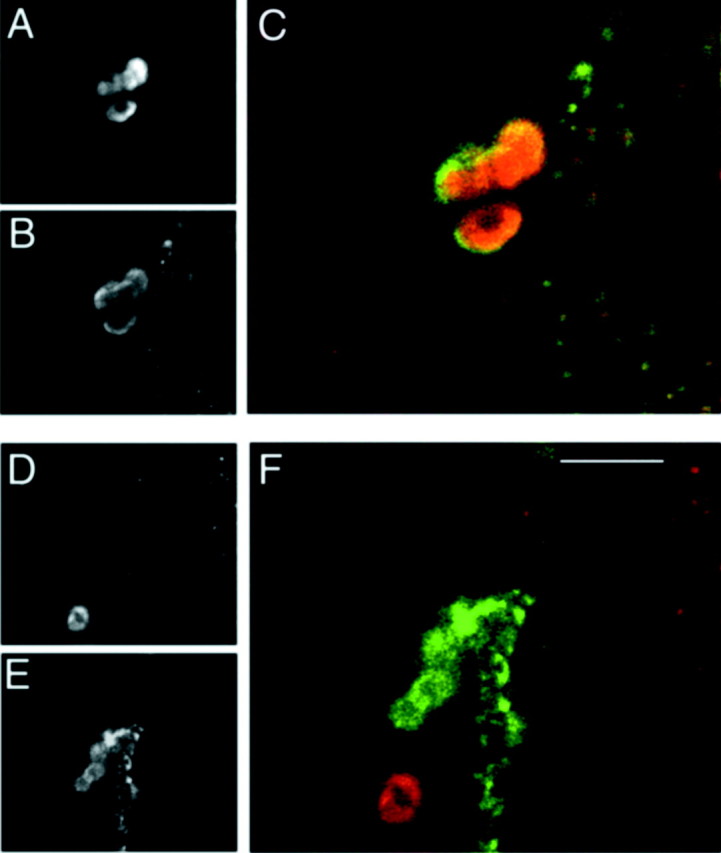
Double-immunostaining to identify PDF neurons included in gal4 expression patterns. Confocal scans of the brain from an adult PHM mutant animal that was rescued by a combination of two gal4 elements (c929-gal4 and386Y-gal4) driving UAS-PHM. The tissue was stained for PHM and proPDF antibodies. Large LN-Vs are stained by both PHM (A andC,red) and proPDF (B andC, green). Small LN-Vs are only stained by proPDF antibodies (E and F,green), but not at all by PHM antibodies (D and F, red). Note another PHM-positive cell body in the vicinity of the small LN-Vs that is not proPDF-positive (F). These images were taken from different focal planes of the same specimen. Scale bar, 20 μm.
PHM expression in PDF neurons
We first studied PHM immunostaining of PDF-expressing neuronal cell bodies in wild-type animals. PHM immunosignals include high level expression in ∼100 brain neurons and a low (perhaps ubiquitous) level of staining in most other cells. We measured the ratio of immunosignal strength (proPDF to PHM) in the three different PDF neurons of the brain, i.e., large LN-Vs, small LN-Vs, and tritocerebral cells in <1-d-old adult male brain hemispheres (n = 10). Although the large LN-Vs and the tritocerebral cells contained detectable PHM immunosignals, the small LN-Vs had little to none (Table3). We next tested the ability of small LN-Vs to accumulate PHM and display PHM immunosignals. We crossedpdf (M) and (N) gal4 lines to UAS-PHMin a wild-type background. In five of five adult brains from each cross, anti-PHM antibody stained the cell bodies (weakly) and processes (moderately) of the small LN-Vs (data not shown).
Table 3.
Quantification of PHM and proPDF immunostaining in PDF neurons
| Cell type | n(cells) | N (specimens) | proPDF (API) | PHM (API) | proPDF/PHM |
|---|---|---|---|---|---|
| Large LN-Vs | 22 | 5 | 54.2 ± 5 | 4.3 ± 0.5 | 13 |
| Small LN-Vs | 17 | 5 | 25.5 ± 2 | 0.1 ± 0.4 | 212 |
| Tritocerebral cells | 4 | 3 | 27 ± 4 | 1.7 ± 1.8 | 15 |
Pixel intensity averages (API) after immunostaining of the three PDF cell types for PHM and for proPDF in brains of young adult males.
The lack of PHM immunosignals in wild-type small LN-Vs prompted us to test for the presence of functional amidating activity in those cells. The anti-PDH (crab PDF) antibody that we use does not discriminate between amidated and nonamidated forms of the peptide; PHMmutant animals are stained normally by this antibody before they die (M. Han and P. H. Taghert, unpublished observations). However, the PT-2 antibody against FMRFamide-like peptides discriminates strongly between amidated and unamidated peptides (Jiang et al., 2000). Therefore, we misexpressed dFMRFa transcripts in small LN-Vs by crossing UAS-dFMRFa flies to each of threepdf-gal4 lines [Bmr(J), M and N] in a wild-type background. We stained resultant larval and adult brains with anti-FMRFa antibody. In five of five specimens of both stages from each cross, both small and large LN-Vs were FMRFamide-immunopositive in both cell bodies and processes (data not shown); large LN-Vs were strongly stained and small LN-Vs were weakly or moderately stained. In summary, the small LN-Vs of WT flies do not contain detectable PHM immunosignals, but they accumulate over-expressed PHM, and they possess endogenous PHM-like amidating activity.
Locomotor behavior under cycling conditions
We compared the behavior of the PHM mosaic flies with that of three genotypes: (1) wild type (WT), (2)per01, and (3)pdf01. We present the analysis in three formats: as tabulated data in Table 4, as average-activity histograms (e.g., Fig.4), and as distributions of SNR values for individuals under constant conditions (Fig.5). The SNR method is described in Materials and Methods. The range of SNR values for WT flies is shown in Figure 5.
Table 4.
Free-running behavior of different genotypes during days 3–9 under constant conditions
| Line | Genotype | N | tau | % R4-a | SNR | Sig.4-b | Events/bin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Standards | 1 | WT | 107 | 23.7 ± 0.1 (91) | 80 | 1.25 ± 0.1 | 4-160 | 17.4 ± 1 | |||||
| 2 | y w; | ; | pdf01 | 45 | 25.4 ± 2 (7) | 16 | 0.50 ± 0.1 | NS | 12 ± 1 | ||||
| 3 | w, | per01 | 29 | 23.0 ± 7.5 (2) | 7 | 0.25 ± 0.2 | 4-160 | 15.3 ± 1 | |||||
| Controls | 4 | y w; | + /UAS-PHM, ; | tim(#16)-gal4 | 14 | 24.1 ± 0.1 (14) | 100 | 0.81 ± 0.2 | 4-160 | 12.4 ± 2 | |||
| 5 | y w; | c929-gal4 | /UAS-PHM | 31 | 24.0 ± 0.1 (23) | 75 | 1.16 ± 0.1 | 4-160 | 20.4 ± 1 | ||||
| 6 | y w; | c929-gal4 | /UAS-PHM,PHM02 | 25 | 23.8 ± 0.1 (15) | 61 | 0.95 ± 0.1 | 4-160 | 22.6 ± 1 | ||||
| 7 | y w; | c929-gal4, | PHM01/y+, CyO | 37 | 23.9 ± 0.1 (27) | 73 | 0.68 ± 0.1 | 4-160 | 16.9 ± 1 | ||||
| 8 | y w; | c929-gal4, | PHM01/UAS-PHM | 28 | 23.6 ± 0.1 (21) | 100 | 1.11 ± 0.1 | 4-160 | 16.1 ± 1 | ||||
| 9 | y w; | y+, | CyO/UAS-PHM,PHM02 | 50 | 23.9 ± 0.3 (41) | 82 | 0.84 ± 0.1 | 4-160 | 16 ± 1 | ||||
| Singlegal4s | 10 | y w; | PHM01/UAS-PHM,PHM02; | 386Y-gal4 | / + | 44 | 24.0 ± 0.4 (37) | 77 | 0.95 ± 0.1 | 4-160 | 8.3 ± 1 | ||
| 11 | y w; | PHM01/UAS-PHM,PHM02; | 36Y-gal4 | / + | 49 | 25.8 ± 0.6 (3) | 6 | 0.29 ± 0.1 | 4-160 | 6.3 ± 1 | |||
| 12 | y w; | c929-gal4, | PHM01/UAS-PHM,PHM02 | 54 | 35.7 (1) | 3 | 0.48 ± 0.1 | — | 19.4 ± 1 | ||||
| Double gal4s | 13 | y w, | pdf-gal4(N); | c929-gal4, | PHM01/UAS-PHM,PHM02 | 60 | 24.5 ± 0.1 (26) | 43 | 0.55 ± 0.1 | NS | 17.5 ± 1 | ||
| 14 | y w, | pdf-gal4(M); | c929-gal4, | PHM01/UAS-PHM,PHM02 | 82 | 23.8 ± 0.2 (18) | 21 | 0.45 ± 0.1 | NS | 11.9 ± 1 | |||
| 15 | y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | 36Y-gal4 | / + | 23 | 26.8 ± 3.3 (5) | 30 | 0.42 ± 0.1 | NS | 11.8 ± 1 | ||
| 16 | y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | 386Y-gal4 | / + | 81 | 23.7 ± 0.1 (66) | 83 | 0.71 ± 0.1 | 4-160 | 13.1 ± 1 | ||
| 17 | y w; | c155-gal4; | c929-gal4, | PHM01/UAS-PHM,PHM02 | 34 | 23.4 ± 0.1 (32) | 72 | 0.48 ± 0.1 | NS | 11.4 ± 1 | |||
| 18 | y w, | c929-gal4, | PHM01/UAS-PHM,PHM02; | D42-gal4 | / + | 38 | 23.7 ± 0.3 (20) | 53 | 0.35 ± 0.1 | NS | 10.5 ± 1 | ||
| 19 | y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | Appl (3GK)-gal4 | / + | 44 | 23.7 ± 0.4 (21) | 48 | 0.42 ± 0.1 | NS | 14.6 ± 1 | ||
| 20 | y w; | c929-gal4, | PHM01/UAS-PHM,PHM02; | tim(#16)-gal4 | / + | 38 | 24.1 ± 0.1 (32) | 84 | 0.76 ± 0.1 | 4-160 | 13.2 ± 1 | ||
| 21 | y w; | PHM01/UAS-PHM,PHM02; | 36Y-gal4 | /tim(#16)-gal4 | 29 | 25.1 ± 0.3 (25) | 84 | 0.89 ± 0.1 | 4-160 | 9.5 ± 1 | |||
tau, Period of the behavioral rhythm in hours. Dunnett'spost hoc analysis, comparing all values to that of line 12 (yw; c929-gal4/UAS-PHM,PHM02) produced the following conclusions:
F4-160: , these stocks different at p < 0.01; NS, these stocks not significantly different. For testing, line 12 was compared separately to each of the four groups (Standards, Controls, Single gal4s, Double gal4s).
Percentage rhythmic by χ-square periodogram analysis: p ≥ 10 and w ≥ 2.
Significance: by ANOVA, SNR averages are significantly different (p < 0.001).
Fig. 4.
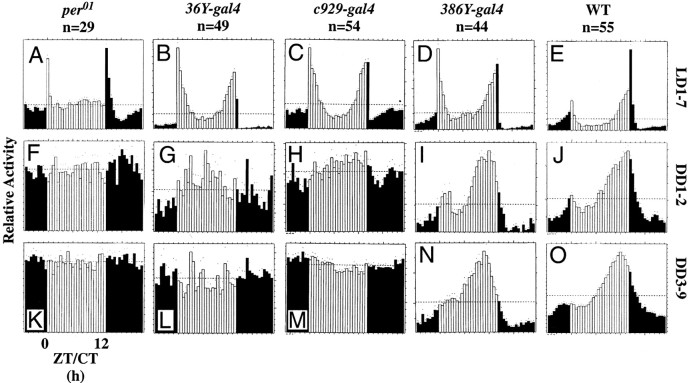
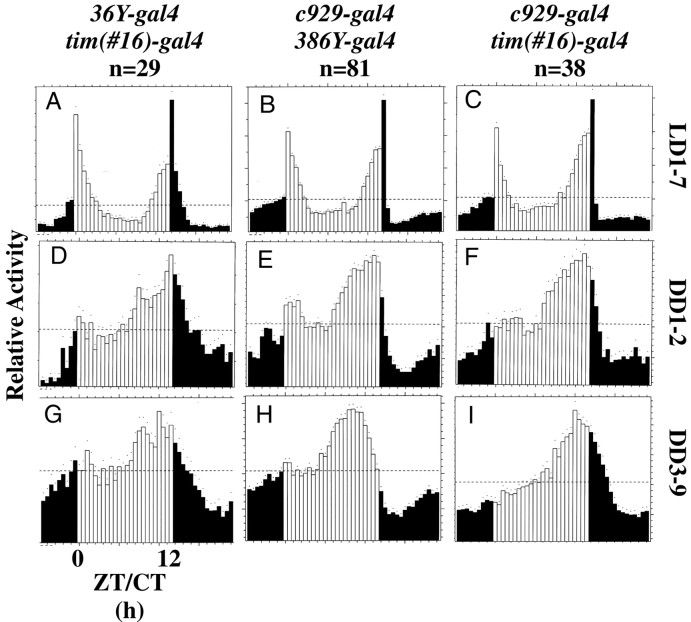
Locomotor activity of normal,per0, and singlegal4:UAS-PHM-rescued PHMmutant flies. Average activity histograms indicating relative levels of locomotion. White and black bars indicate the day and night phases in LD, respectively (Hamblen-Coyle et al., 1989, 1992). n, number of flies tested. For the constant dark (DD) plots (rows 2 and3), white bars designate the subjective day. Dots indicate SEM values for that 30 min time bin with reference to average level of activity per fly. A,F, K,per01; B,G, L,36Y-gal4:UAS-PHM-rescuedPHM mosaics; C, H,M,c929-gal4:UAS-PHM-rescuedPHM mosaics; D, I,N,386Y-gal4:UAS-PHM-rescuedPHM mosaics; E, J,O, Canton-S wild type.A–E, 7 d of LD behavior;F–J, behavior during DD days 1–2;K–O, behavior during DD days 3–9.ZT, Zeitgeber time; CT, circadian time.
Fig. 5.
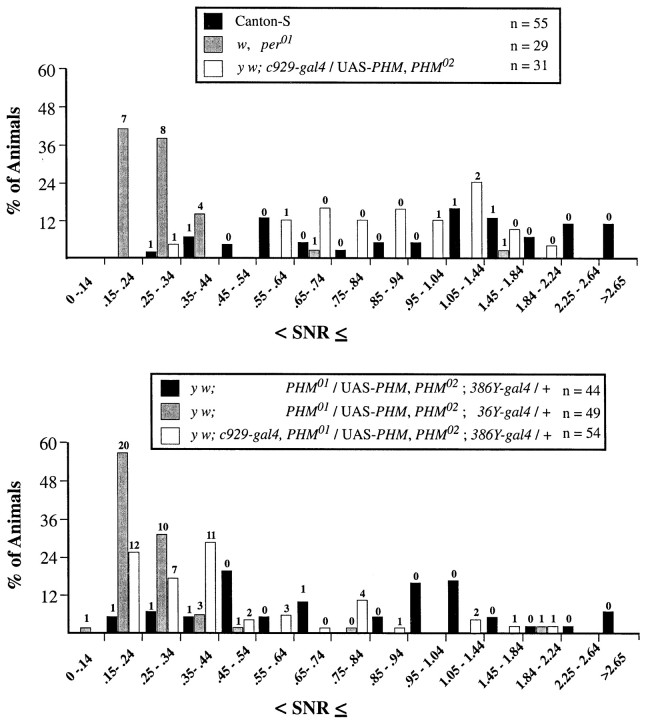
Numerical measures of varying behavioral rhythm strengths in per01, normal, and singlegal4-rescued PHM mutants. Signal-to-noise ratios (SNRs) for the final 7 d (DD days 3–9) of the free running period (see Table 4). The panelsindicate SNR distributions for per01,y w;c929-gal4/UAS-PHM,PHM02, and Canton-S (top); and y w;PHM01/UAS-PHM, PHM02 containing either36Y-gal4,c929-gal4, or386Y-gal4 (bottom). SNR values ≤1.04 were divided into increments of 0.1; between 1.05 and 2.64, SNR values were divided into increments of 0.3; all SNR values >2.65 were grouped together. The ordinate values are the percentage of total flies whose SNR falls within each interval. Thenumbers of flies that were scored arrhythmic by χ-square periodogram analysis are indicated above the histogram bars. For the rhythmic individuals, free-running periods of the different genotypes were calculated by Maximum Entropy Spectral Analyses independently of those in Table 4; they were not significantly different by ANOVA (means: Canton-S, 24.1 ± 0.2 hr;per01, 24.9 ± 1.2 hr; y w; PHM01/UAS-PHM, PHM02;386Y-gal4/+, 23.4 ± 0.3 hr;y w;PHM01/UAS-PHM, PHM02;36Y-gal4/+, 23.9 ± 0.8 hr; y w;c929-gal4/UAS-PHM,PHM02, 23.8 ± 1.1 hr).
Under 12 hr LD cycles, WT males produced two peaks of activity (cf.Hamblen-Coyle et al., 1992). The morning peak was maximal around lights on and anticipated that transition; the evening peak was maximal around lights off and also anticipated that transition (Fig.4E). In contrast, per01flies displayed only transient peaks of activity that were coincident with the light/dark transitions and that showed no anticipatory or sustained nature (Fig. 4A) (cf. Wheeler et al., 1993). Finally, pdf01 flies displayed two peaks of activity. The morning peak lacked anticipation and was very narrow; the evening peak showed anticipation, was sustained, and was phase-advanced relative to that of WT (data not shown) (cf. Renn et al., 1999). The LD behavior of36Y-gal4-rescued flies (Fig.4B) and c929-gal4-rescued flies (Fig. 4C) were similar. Both groups displayed two peaks of activity having normal peak time; locomotor levels accompanying these maxima were sustained, but in neither transgenic line did flies anticipate the morning lights-on transition. In386Y-gal4-rescued animals (Fig.4D), both the morning and evening peaks appeared comparable with those of WT, and the morning peak displayed normal anticipation.
Locomotor behavior under constant conditions
Under conditions of DD, WT animals maintain daily rhythmicity with an activity profile that is temperature-dependent (Majercak et al., 1999). At 25°C, they display a single peak of activity, typically late in the subjective day. Table 4 presents the behavioral analysis of PHM mosaic animals for 7 d under constant conditions (DD days 3–9). Figure 4 presents average group activity plotted separately for DD days 1–2 and 3–9. The majority of WT flies (Table 4, line 1) remained rhythmic over the entire period (Fig. 4J,O); by periodogram analysis, 80% of individuals were rhythmic, and the population displayed an average SNR of 1.25. As previously reported (Wheeler et al., 1993), per01 animals (Table 4, line 3) became largely arrhythmic in the first cycle of DD and remained so (Fig. 4F,K); only 7% ofper01 animals in the current test were rhythmic during DD days 3–9, and their average SNR was 0.25.pdf01 animals (Table 4, line 2) remained rhythmic for DD days 1–2, but only 16% remained rhythmic over DD days 3–9 (data not shown) (cf. Renn et al., 1999); the population had an average SNR of 0.5.
36Y-gal4-rescued flies (Table 4, line 11) resembledper01; they were very weakly rhythmic over DD days 1–2 (Fig. 4G) and DD days 3–9 (Fig.4L). Six percent of these individuals were rhythmic, and the population displayed an average SNR of 0.29.c929-gal4-rescued flies (Table 4, line 12) were also poorly rhythmic during DD days 1–2 (Fig. 4H). During DD days 3–9, c929-gal4-rescued flies were only 3% rhythmic by periodogram (Fig. 4M), but the population displayed a higher SNR than 36Y-gal4-rescued flies (0.49). In this regard they were more comparable withpdf01. During the entire DD period,386Y-gal4 flies displayed a pattern of activity approaching that of WT (Fig. 4I,N, Table 4, line 10).
The distribution of DD days 3–9 SNR averages permits evaluation of the behavior of individuals within a large population. As previously found (Renn et al., 1999), the range of SNR values for WT was 0.3 to >3; in contrast, that for per01 was clustered between 0.2 and 0.4 (Fig. 5A). The range of SNR values forPHM “mosaics” rescued by 36Y-gal4resembled that of per01 (Fig.5B). Values for c929-gal4-rescued flies were also low but included some nearing 0.8.386Y-gal4-rescued flies produced a broad range that largely overlapped that of WT.
The PHM mosaics were constructed by assembling several transgenes. To determine the degree to which the genetic background might influence the behavioral results, we measured locomotor rhythms in six additional control stocks (Table 4, lines 4–9). The latter five contained various subsets of these same transgenes, with or without a balancer chromosome (see Methods, Genetic strains, for definition of balancers). Over-expressing PHM in a wild-type background (Table 4, lines 4 and 5) did not degrade behavioral rhythmicity. By both periodogram and SNR analysis, most control strains appeared less rhythmic than wild-type animals but considerably more rhythmic thanper01 orpdf01 (Fig.6A, Table 4). Of the six control combinations tested, two displayed less rhythmicity than the others. Line 6 (y w;c929-gal4/UAS:PHM,PHM02) produced a nominally lower percentage of rhythmic animals by periodogram analysis, but also displayed a moderately high SNR average (0.95). Line 7 (y w; c929-gal4,PHM[01]/y+,CyO) produced a lower average SNR, but 77% were rhythmic by periodogram.
Fig. 6.
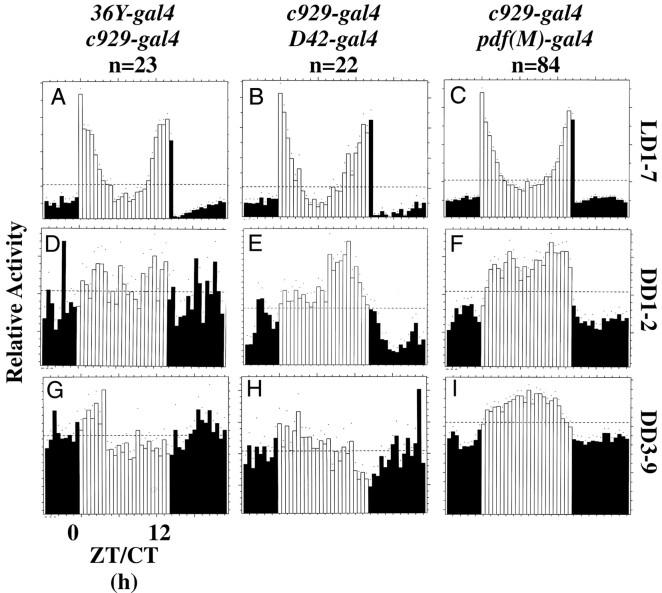
Locomotor activity of doublegal4:UAS-PHM-rescued PHMmutant flies. Average activity histograms for groups of flies, plotted as described in Figure 4. n, number of flies tested. A, D, G,36Y-gal4/c929-gal4:UAS-PHM-rescuedPHM mosaics; B, E,H,c929-gal4/D42-gal4:UAS-PHM-rescuedPHM mosaics; C, F,I,c929-gal4/pdf(M)-gal4:UAS-PHM-rescuedPHM mosaics.
The effect of combining different gal4 elements on LD behavior
We combined gal4 elements with36Y-gal4 or c929-gal4 to try and improve behavior displayed by animals rescued with eithergal4 element alone. D42-gal4 was used because it provides a modest increase in the spatial patterns provided by the36Y- and c929-gal4 elements, specifically by including most motorneurons (G. Boulianne, personal communication). Appl(3GK)-gal4, c155(elav)-gal4 andtim-gal4 were used to effect widespread (i.e., most or all CNS) expression. Finally, pdf-gal4(two distinct lines, called M and N) ensured expression of PHM in all LN-Vs (these neurons secrete the amidated peptide PDF). Combining36Y-gal4, D42-gal4, orpdf(M)-gal4 with c929-gal4 produced LD behavior that was aberrant and similar to that ofc929-gal4 alone (Fig. 6). Combiningpdf(N)-gal4, Appl-gal4, c155-gal4, 386Y-gal4, ortim-gal4 with c929-gal4 produced LD behavior that resembled that of control stocks (Figs. 7,8B,C). Finally, combining tim-gal4 with36Y-gal4 produced LD behavior that resembled that of control stocks (Fig. 8A).
Fig. 7.
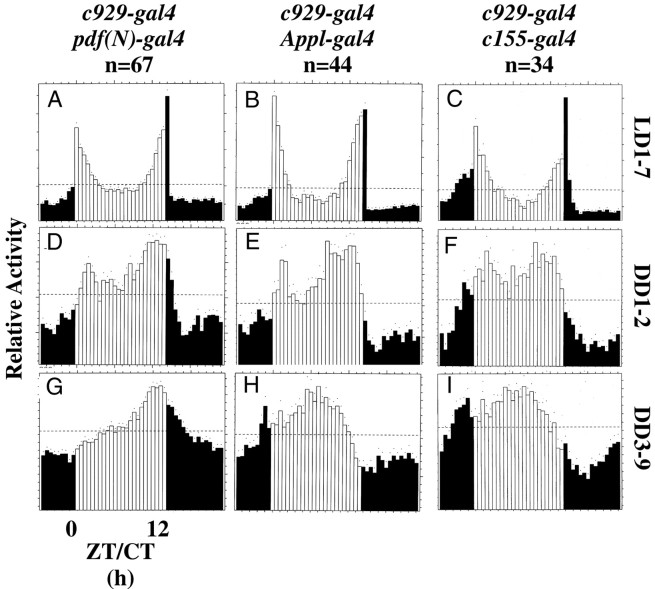
Locomotor activity of doublegal4:UAS-PHM-rescued PHMmutant flies. Average activity histograms for groups of flies, plotted as described in Figure 4. n, number of flies tested.A, D, G,c929-gal4/pdf(N)-gal4:UAS-PHM-rescuedPHM mosaics; B, E,H,c929-gal4/Appl-gal4:UAS-PHM-rescuedPHM mosaics; C, F,I,c929-gal4/c155-gal4:UAS-PHM-rescuedPHM mosaics.
Fig. 8.
Locomotor activity of doublegal4:UAS-PHM-rescued PHMmutant flies. Average activity histograms for groups of flies, plotted as described in Figure 4. n, number of flies tested.A, D, G,36Y-gal4/tim(#16)-gal4:UAS-PHM-rescuedPHM mosaics; B, E,H,c929-gal4/386Y-gal4:UAS-PHM-rescuedPHM mosaics; C, F,I,c929-gal4/tim(#16)-gal4:UAS-PHM-rescuedPHM mosaics.
The effect of combining different gal4 elements on DD behavior
Weak rhythmicity in DD was produced in three combinations ofgal4 elements. Combining 36Y-gal4, D42-gal4, or pdf(M)-gal4 withc929-gal4 did not improve the performance of animals beyond that of c929-gal4 alone (Fig.6G--I, Table 4). CombiningAppl(3GK)-gal4 with c929-gal4increased the percentage of rhythmic individuals (to 48%), but the group retained a low average SNR (0.49) (Fig. 7H, Table 4). Likewise, combining pdf(N)-gal4 withc929-gal4 also increased the percentage of rhythmic flies (to 43%), yet the average SNR remained low (0.55) (Fig. 7G, Table 4). The same was true for combining c155-gal4 withc929-gal4; by periodogram analysis, 72% of these animals were rhythmic, yet they retained a low average SNR (0.48) (Fig.7I, Table 4).
Strong rhythmicity in DD was produced in three combinations ofgal4 elements. Combining tim-gal4 with36Y-gal4 produced 84% rhythmic individuals and a markedly higher average SNR (0.89) than displayed by36Y-gal4 alone (Fig. 8G, Table 4, compare lines 11 and 16). Combining tim-gal4 withc929-gal4 also increased the percentage of rhythmic flies (to 84%) and moderately improved the average SNR (0.76) (Fig.8I, Table 4, compare lines 12 and 20). Combining386Y-gal4 with c929-gal4 produced DD behavior that resembled animals bearing only386Y-gal4 (Fig. 8H, Table 4, compare lines 10 and 16). By MESA, the periods displayed under constant conditions were not significantly different, with the exception of animals containing the 36Y-gal4 element (Table4).
The distribution of SNR values in animals in whichc929-gal4 was combined with either36Y-gal4, D42-gal4, orpdf(M)-gal4 confirmed the hypothesis that none of these combinations significantly improved the rhythmicity ofc929-gal4-rescued animals (Fig.9A). Likewise, addingpdf(N)-gal4, c155-gal4, or Appl-gal4 to c929-gal4 produced only a moderate improvement in the distribution of SNR values (Fig.9B). Both sets of distributions were heavily weighted in the range 0.3–0.5. In contrast, the combination ofc929-gal4 with 386Y-gal4produced a distribution that resembled that of386Y-gal4-rescued flies alone (Fig.9C). Likewise, combining tim-gal4 with either 36Y-gal4 orc929-gal4 produced balanced SNR distributions that more resembled that of WT (Fig. 9C). Table 4 includes the results of statistical analysis comparing each DD days 3–9 SNR data set with that of the c929-gal4-rescued population.
Fig. 9.
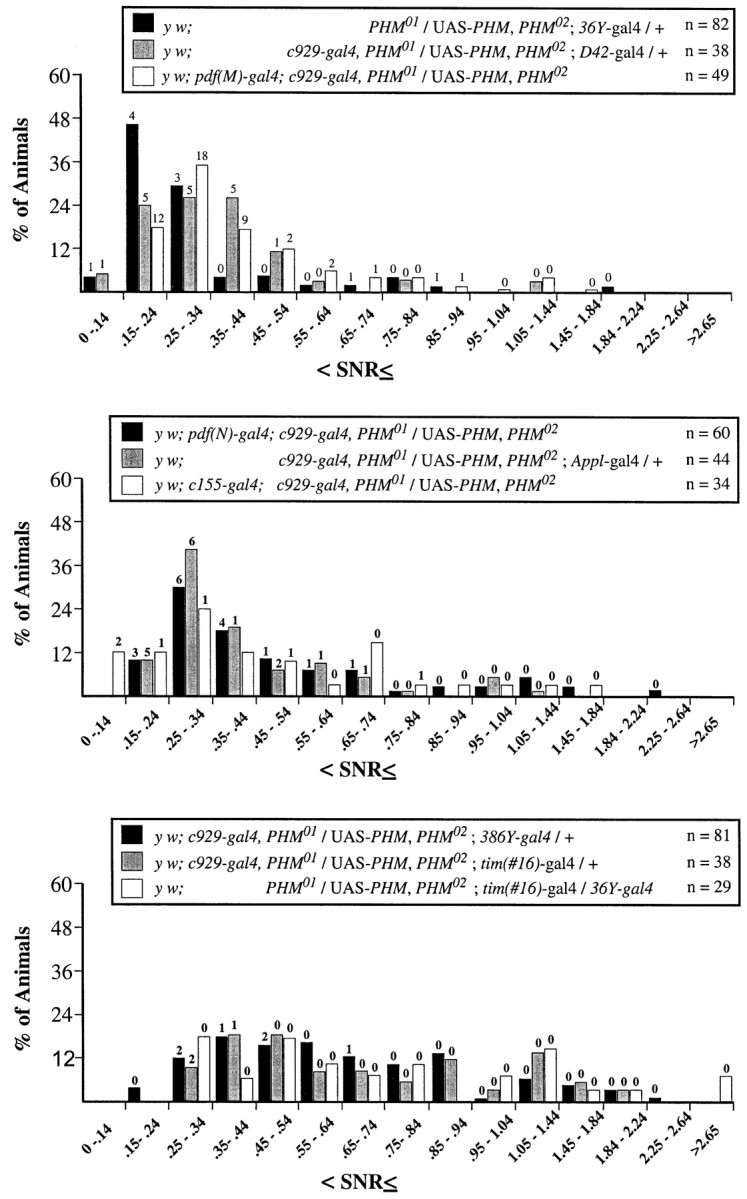
Numerical measures of varying behavioral rhythm strengths in double gal4-rescued PHMmutant flies. SNR values for the final 7 d (DD days 3–9) of the free running period presented as described in Figure 5. Thepanels indicate SNR distributions for y w; PHM01/UAS-PHM, PHM02 containing eitherc929-gal4 and36Y-gal4,c929-gal4 andD42-gal4, orc929-gal4 andpdf(M)-gal4(top); y w;PHM01/UAS-PHM, PHM02 containing eitherc929-gal4 andpdf(N)-gal4,c929-gal4 andAppl-gal4, orc929-gal4 and c155-gal4(middle); and y w;PHM01/UAS-PHM, PHM02 containing eitherc929-gal4 and386Y-gal4,c929-gal4 andtim(#16)-gal4, or36Y-gal4 andtim(#16)-gal4(bottom). The ordinate values are the percentage of total flies whose SNR falls within each interval. Thenumbers of flies that were scored arrhythmic by χ-square periodogram analysis are indicated above the histogram bars. For the rhythmic individuals, free-running periods of the different genotypes were calculated by Maximum Entropy Spectral Analyses, independently of those in Table 4; they were not significantly different by ANOVA (means = y w;c929-gal4,PHM01/UAS-PHM, PHM02;36Y-gal4/ +, 25.4 ± 1.5 hr;y w, pdf-gal4(M); y w;c929-gal4/UAS-PHM,PHM02, 24.4 ± 0.4 hr; y w; c929-gal4,PHM01/ UAS-PHM, PHM02;D42-gal4/ +, 24.2 ± 1.0 hr;y w, pdf-gal4(N);c929-gal4,PHM01/ UAS-PHM, PHM02, 25.2 ± 0.3 hr; y w; c929-gal4,PHM01/ UAS-PHM, PHM02;Appl3GK-gal4/ +, 24.0 ± 0.4 hr;y w; c929-gal4 and c155-gal4, 24.1 ± 0.6 hr; y w;c929-gal4,PHM01/UAS-PHM, PHM02;386Y-gal4/+, 24.1 ± 0.3 hr;y w; c929-gal4,PHM01/ UAS-PHM, PHM02;tim(#16)-gal4/ +, 24.0 ± 0.2 hr;y w;PHM01/UAS-PHM, PHM02;tim(#16)-gal4/36Y-gal4, 24.9 ± 0.4 hr).
Lack of correlation between average activity levels and rhythmicity
We asked whether the average activity levels of different genotypes in DD days 3–9 predicted the strength of behavioral rhythmicity (Table 4). In general, these traits did not appear strongly correlated. Some, but not all, rescued lines displayed normal activity levels. For example, 36Y-gal4-rescued flies displayed the lowest levels of average activity under constant conditions, and this was in accord with their weak rhythmicity (Table4, genotype 11). However, although the level of activity of36Y-gal4-rescued animals was not markedly improved by the addition of tim-gal4, that double transgenic combination (36Y-gal4 andtim(#16)-gal4) produced strong rhythmicity by both periodogram and SNR analysis (Table 4, genotype 20). Likewise,386Y-gal4-rescued flies produced strong measures of rhythmicity in DD days 3–9, yet also displayed low levels of average activity (Table 4, genotype 10).
DISCUSSION
We used a genetic approach to create animal mosaics for PHM, a biosynthetic enzyme that is required for maturation of the majority of neuropeptide transmitters in Drosophila (Jiang et al., 2000). By studying their behavior, we draw two principal conclusions. First, PHM enzyme activity, and hence C-terminal amidation of peptide transmitters, is required for normal circadian locomotion inDrosophila. Second, amidated neuropeptides, in addition to PDF, are required to regulate circuits controlling this behavior.
Neuropeptide amidation is required for daily locomotor rhythms
Our results support the hypothesis that daily locomotor rhythms in flies require signaling by neuropeptides that are C-terminally amidated. We first showed that the gal4/UAS-PHM system predictably controls PHM spatial expression. Next, we found that certain PHM mosaic flies (e.g., 36Y-gal4- andc929-gal4-rescued) were largely arrhythmic under conditions of constant darkness. Our working hypothesis is that such a behavioral disruption is attributable to changes in the normal patterns of peptide amidation. We have not demonstrated the last point directly. It is a premise based on the previous demonstration that manipulation of PHM produces large-scale changes in peptide amidation in both larval and adult stages (Jiang et al., 2000).
A second result supports the conclusion that amidated peptides contribute to daily locomotor rhythms; increasing PHMexpression by combining pdf(N)-gal4 withc929-gal4 improved rhythmic behavior over that displayed by c929-gal4 alone. Although the combination did not completely restore wild-type behavior, the improvement indicates that PHM activity in PDF neurons does contribute to display of rhythmic daily locomotion. We presume that this indicates a requirement for amidation of PDF because the pigment-dispersing activity of PDH on crustacean melanophores is highly dependent (∼300-fold) on the C-terminal amide (Riehm et al., 1985). Whether PDF must be modified as such to display circadian signaling activity is unknown.
Neuropeptides besides PDF are required for daily locomotor rhythms
The strongest evidence for this conclusion comes from the performance of 36Y-gal4- and c929-gal4-rescued flies under constant conditions. Unlikepdf01 flies, these populations displayed little rhythmicity behavior during the first cycle of constant conditions (cf. Renn et al., 1999). Also, the average SNR of36Y-gal4-rescued flies was much less than that ofpdf01 and very comparable with that of per01. Are such deficits attributable to a loss of PHM function or a gain of deleterious function? The pdf neuropeptide gene can produce a gain-of-function phenotype: When pdf was misexpressed by certaingal4 drivers in WT flies, rhythmic locomotor behavior was degraded (Helfrich-Förster et al., 2000). In the case ofPHM, however, two results suggest it is the absence of the enzyme that causes arrhythmicity. First, increasing PHMexpression (by adding tim-gal4 to 36Y-gal4 orc929-gal4) restored near-normal rhythmicity to both arrhythmic lines. Second, misexpression of PHM by driving it with tim-gal4 or with c929-gal4 in a WT did not degrade behavioral rhythmicity. In general, the greater the extent ofPHM gene expression in a PHM mutant background, the more predictable was the degree of behavioral improvement. Interestingly, some combinations restored considerable PHMexpression (e.g., Appl-gal4) but improved behavioral performance only moderately. Together, those experiments suggest that normal PHM expression is required in specific neurons and/or secretory cells for the behavior examined. Thetim-gal4 line produced a broad expression pattern of great complexity. That amount of expression precludes clear definition of places or times by which “additional” PHM restored the functions of circadian regulatory circuits. This point is discussed further in the next section.
Interpretation of gal4 expression patterns
We compared the behavior of stocks that each contained multiple transposons. To improve the scope of the study, we tested five control genotypes that combined subsets of the multiple transposons used in the experimental genotypes. In general, these controls displayed a level of rhythmicity lower than that of WT but greater than those ofPHM mosaics (Table 4). gal4 lines are used primarily to create spatial differences in gene expression (Helfrich-Förster et al., 2000; Waddell et al., 2000). We chose three gal4 lines for this study (36Y,c929, and 386Y), because their expression patterns prominently featured peptidergic neurons of the CNS and secretory cells of peripheral tissues. The three patterns were very similar; the fact that all successfully reverted PHMlethality (Table 2) is probably a reflection of such anatomical similarities. The gal4 patterns also included clear differences in cell number (386Y >c929 > 36Y).
These pattern differences are of interest, because they may reveal specific neurons (or non-neuronal cells) that produce secretory peptides required for circadian behaviors. However, we concluded that the interpretation of where “critical PHM expression” occurs in these experiments is problematic, because there are several ways by which such patterns may defy simple interpretation. For example, two gal4 patterns could appear similar and stable in the adult stage, yet be different because of a transient event during development. In fact, the c929-gal4 pattern is relatively stable in the adult stage but transiently includes the VA neuroendocrine neurons (O'Brien and Taghert, 1998) for only a brief period during adult development (P. H. Taghert, unpublished observations). In such a case, behavioral rescue may reflect temporal, not spatial, differences in gal4-dependent gene expression. A separate problem in the interpretation could arise when two patterns are spatially similar but differ in levels of expression by specific neurons. In that case, the extent of behavioral rescue may reflect quantitative, not spatial, differences in gal4-dependent gene expression.
Given these complexities, we are currently unable to specify in which neurons, beyond the LN-V, PHM activity is required for daily locomotor rhythms. Instead, for subsequent analysis we favor considering candidate amidated neuropeptides directly. From scans of the Drosophila genome, there are at least 23 neuropeptide-encoding genes (Hewes and Taghert, 2001; Vanden Broeck, 2001): This is likely an underestimate because of the difficulty in predicting neuropeptide precursor sequences with accuracy. Of the identified genes, ∼20 encode peptides that are known or are predicted to display C-terminal amidation. Thus, it may be reasonable to systematically address the roles of each of the ∼20 precursors usingDrosophila genetics.
General activity versus the circadian organization of activity in PHM mosaics
Genetic studies of circadian behaviors traditionally strive to establish that a mutant phenotype does not simply degrade the ability to produce movement (Hamblen-Coyle et al., 1989). Here, we analyzed the behavior of animals with large-scale alterations in transmitter profiles throughout the entire nervous system. In one genotype (36Y-gal4/UAS-PHM), rhythmicity under constant conditions was extremely poor (as low as that of per01 animals); activity levels were also lower than in other genotypes tested. Nevertheless, when the locomotor rhythm of 36Y-gal4-rescued animals was restored by addition of tim-gal4, activity levels were not also increased. We propose that the36Y-gal4 transmitter mosaic contains disruptions of distinct neural centers, ones that control the general level of activity and ones that organize rest–activity cycles. A similar point is made considering the results seen withtim-gal4. Addition of tim-gal4 to36Y-gal4-rescued flies produced the greatest restoration of rhythmicity. However, tim-gal4 was by itself unable to revert the lethality of PHM mutants. Therefore, places and times of PHM expression that promote normal vitality do not necessarily equal those promoting circadian behavioral rhythmicity.
The behavior of these PHM mosaics differed from that of clock gene mutants
Flies lacking clock gene function (e.g.,per0) (Wheeler et al., 1993) display light-driven behavior under cycling conditions (Fig.4A), then become arrhythmic during the first cycle of constant conditions (Fig. 4F). ArrhythmicPHM mosaics were different. For example,36Y-gal4-rescued flies, whose rhythmicity under constant conditions was quantitatively as weak as that ofper01 animals, entrained well during LD. Therefore, we conclude that even the most severely arrhythmicPHM mosaic animals we have studied have levels of circadian clock function and output greater than that present in authentic clock mutants.
Evidence for graded levels of rhythmic behavior
The average activity histograms for behavior under constant conditions indicated that different gal4 drivers produced graded levels in circadian locomotor performance. We found evidence for at least three levels. The lowest level was represented by singlegal4 flies (e.g., 36Y); they had the weakest measures of rhythmicity (by periodogram or MESA) and displayed little reproducible variation in the average activity histogram. An intermediate level was seen in certain gal4-combination flies (e.g., c929/D42); these displayed moderate rhythmicity and an average activity peak during early subjective day. The strongest level was seen in other gal4-combination flies (e.g., c929/tim); these were strongly rhythmic, and they displayed a large average activity peak during late subjective day and a rapid decrease in activity during early subjective night. Presumably, these graded levels of performance reflect incremental contributions by different amidated peptides to one or more circuit components. Relating specific peptide systems to separate levels of behavioral performance represents a challenge for future studies.
Footnotes
This work was supported by National Institutes of Health Grants NS-21749 to P.H.T. and GM-33205 to Jeff Hall (Brandeis University, Waltham, MA), National Research Service Award MH11946 to J.H.P., National Science Foundation Grant IBN 97–30003 to P.H.T., and the University of Tennessee New Investigator Supporting Program (J.H.P.). We thank Ning Jiang and Marie Roberts for assistance in creating UAS-PHM transgenic animals and Pam Vanderzalt for assistance with behavioral experiments. We thank Jeff Hall, in whose laboratory preliminary behavioral experiments were performed. We thank Andrea Brand for the pUAST vector and Kalpana White, Maki Kaneko, Jeff Hall, Sarah Smolik, Gabrielle Boulianne, the Bloomington Stock Center, and the Berkeley Drosophila Genome Project forDrosophila stocks. We thank Michael O'Connor for allowing us to use the stock P{w+, UAS-dFMRFa}. We thank Kalpana White and Gabrielle Boulianne for personal communications and Russ Van Gelder, Joel Levine, and Jeff Hall for helpful discussions and for their comments on a draft of this manuscript.
Correspondence should be addressed to Dr. Paul H. Taghert, Box 8108, Department of Anatomy and Neurobiology, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110. E-mail: taghertp@thalamus.wustl.edu.
R. S. Hewes' present address: Department of Zoology, University of Oklahoma, 730 Van Vleet Oval, Norman, OK 73019.
J. H. Park's present address: Department of Biochemistry, Cellular and Molecular Biology, University of Tennessee, Knoxville, TN 37996.
M. A. O'Brien's present address: Promega Corporation, 2800 Woods Hollow Road, Madison WI 53711.
REFERENCES
- 1.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Benveniste RJ, Taghert PH. Cell type-specific regulatory sequences control expression of the Drosophila FMRF-NH2 neuropeptide gene. J Neurobiol. 1999;38:507–520. doi: 10.1002/(sici)1097-4695(199903)38:4<507::aid-neu7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 3.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 4.Dowse HB, Ringo JM. Further evidence that the circadian clock in Drosophila is a population of coupled ultradian period. J Biol Rhythms. 1987;2:65–76. doi: 10.1177/074873048700200106. [DOI] [PubMed] [Google Scholar]
- 5.Dushay MS, Rosbash M, Hall JC. The disconnected visual system mutations in Drosophila melanogaster drastically disrupt circadian rhythms. J Biol Rhythms. 1989;4:1–27. doi: 10.1177/074873048900400101. [DOI] [PubMed] [Google Scholar]
- 6.Eipper BA, Stoffers DA, Mains RE. Biosynthesis of neuropeptides: alpha-amidation. Annu Rev Neurosci. 1993;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 7.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells' influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 9.Gloor GB, Preston CR, Johnson-Schlitz DM, Nassif NA, Phillis RW, Benz WK, Robertson HM, Engels WR. Type I repressors of P-element mobility. Genetics. 1993;135:81–95. doi: 10.1093/genetics/135.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamblen MJ, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zweibel LJ, Konopka RJ, Rosbash M, Hall JC. Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per− mutants. J Neurogenet. 1986;3:249–291. doi: 10.3109/01677068609106855. [DOI] [PubMed] [Google Scholar]
- 11.Hamblen MJ, White NE, Emery PT, Kaiser K, Hall JC. Molecular and behavioral analysis of four period mutants in Drosophila melanogaster encompassing extreme short, novel long, and unorthodox arrhythmic types. Genetics. 1998;149:165–178. doi: 10.1093/genetics/149.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamblen-Coyle M, Konopka RJ, Zwiebel LJ, Colot HV, Dowse HB, Rosbash M, Hall JC. A new mutation at the period locus of Drosophila melanogaster with some novel effects on circadian rhythms. J Neurogenet. 1989;5:229–256. doi: 10.3109/01677068909066210. [DOI] [PubMed] [Google Scholar]
- 13.Hamblen-Coyle MJ, Wheeler DA, Rutila JE, Rosbash M, Hall JC. Behavior of period-altered circadian rhythm mutants of Drosophila in light:dark cycles (Diptera: Drosophilidae). J Insect Behav. 1992;5:417–446. [Google Scholar]
- 14.Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol [A] 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 16.Helfrich-Förster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. J Neurosci. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11:1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hewes RS, Schaefer AM, Taghert PH. The cryptocephal gene (ATF4) encodes multiple basic-leucine zipper proteins controlling molting and metamorphosis in Drosophila. Genetics. 2000;155:1711–1723. doi: 10.1093/genetics/155.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang N, Kolhekar AS, Jacobs PS, Mains RE, Eipper BA, Taghert PH. PHM is required for normal developmental transitions and for biosynthesis of secretory peptides in Drosophila. Dev Biol. 2000;226:118–136. doi: 10.1006/dbio.2000.9832. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 21.Kolhekar AS, Roberts MS, Jiang N, Johnson RC, Mains RE, Eipper BA, Taghert PH. Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J Neurosci. 1997;17:1363–1376. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 23.Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1982. [Google Scholar]
- 25.O'Brien MA, Taghert PH. A peritracheal neuropeptide system in insects: release of myomodulin-like peptides at ecdysis. J Exp Biol. 1998;201:193–209. doi: 10.1242/jeb.201.2.193. [DOI] [PubMed] [Google Scholar]
- 26.O'Brien MA, Schneider LE, Taghert PH. In situ hybridization analysis of the FMRFamide neuropeptide gene in Drosophila. II. Constancy in the cellular pattern of expression during metamorphosis. J Comp Neurol. 1991;304:623–638. doi: 10.1002/cne.903040409. [DOI] [PubMed] [Google Scholar]
- 27.Park JA, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riehm JP, Rao KR, Semmes OJ, Jorenby WH, Hintz MF, Zahnow CA. C-terminal deletion analogs of a crustacean pigment-dispersing hormone. Peptides. 1985;6:1051–1056. doi: 10.1016/0196-9781(85)90427-9. [DOI] [PubMed] [Google Scholar]
- 29.Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons both cause severe abnormalities of circadian behavioral rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 30.Siekhaus DE, Fuller RS. A role for amontillado, the Drosophilahomolog of the neuropeptide precursor processing protease PC2, in triggering hatching behavior. J Neurosci. 1999;19:6942–6954. doi: 10.1523/JNEUROSCI.19-16-06942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 32.Silver R, Sookhoo AI, LeSauter J, Stevens P, Jansen HT, Lehman MN. Multiple regulatory elements result in regional specificity in circadian rhythms of neuropeptide expression in mouse SCN. NeuroReport. 1999;10:3165–3174. doi: 10.1097/00001756-199910190-00008. [DOI] [PubMed] [Google Scholar]
- 33.Sokolove PG, Bushell WN. The χ-square periodogram: its utility for analysis of circadian rhythms. J Theor Biol. 1978;72:131–160. doi: 10.1016/0022-5193(78)90022-x. [DOI] [PubMed] [Google Scholar]
- 34.Taghert PH, Schneider LE. Inter-specific comparison of a Drosophila gene encoding FMRFamide-related neuropeptides. J Neurosci. 1990;10:1929–1942. doi: 10.1523/JNEUROSCI.10-06-01929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. 2001;22:241–254. doi: 10.1016/s0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]
- 36.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler DA, Hamblen-Coyle MJ, Dushay MS, Hall JC. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J Biol Rhythms. 1993;8:67–94. doi: 10.1177/074873049300800106. [DOI] [PubMed] [Google Scholar]
- 38.Zitnan D, Kingan TG, Hermesman JL, Adams ME. Identification of ecdysis-triggering hormone from an epitracheal endocrine system. Science. 1996;271:88–91. doi: 10.1126/science.271.5245.88. [DOI] [PubMed] [Google Scholar]