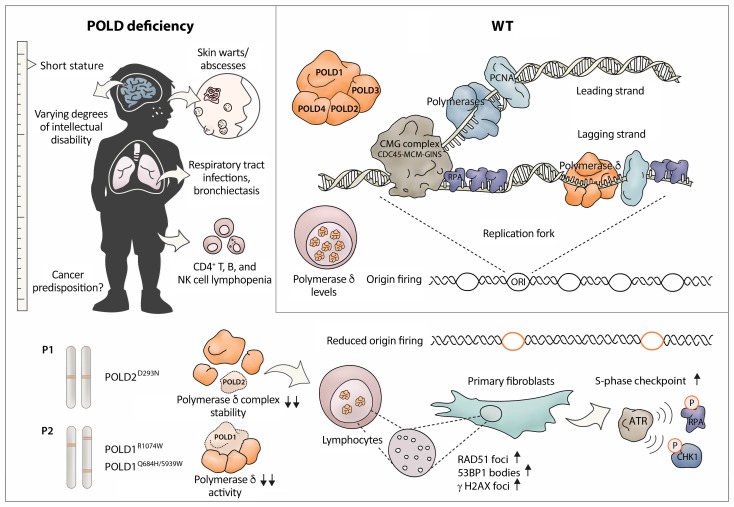
Abstract
Polymerase δ is essential for eukaryotic genome duplication and synthesizes DNA at both the leading and lagging strands. The polymerase δ complex is a heterotetramer comprising the catalytic subunit POLD1 and the accessory subunits POLD2, POLD3, and POLD4. Beyond DNA replication, the polymerase δ complex has emerged as a central element in genome maintenance. The essentiality of polymerase δ has constrained the generation of polymerase δ–knockout cell lines or model organisms and, therefore, the understanding of the complexity of its activity and the function of its accessory subunits. To our knowledge, no germline biallelic mutations affecting this complex have been reported in humans. In patients from 2 independent pedigrees, we have identified what we believe to be a novel syndrome with reduced functionality of the polymerase δ complex caused by germline biallelic mutations in POLD1 or POLD2 as the underlying etiology of a previously unknown autosomal-recessive syndrome that combines replicative stress, neurodevelopmental abnormalities, and immunodeficiency. Patients’ cells showed impaired cell-cycle progression and replication-associated DNA lesions that were reversible upon overexpression of polymerase δ. The mutations affected the stability and interactions within the polymerase δ complex or its intrinsic polymerase activity. We believe our discovery of human polymerase δ deficiency identifies the central role of this complex in the prevention of replication-related DNA lesions, with particular relevance to adaptive immunity.
Keywords: Genetics, Immunology
Keywords: Adaptive immunity, DNA repair, Genetic diseases
Introduction
Eukaryotic genome duplication relies on 3 B-family DNA polymerases: polymerase α, polymerase ε, and polymerase δ (1). Polymerase α is known to have primase activity (2), whereas polymerase ε is thought to synthesize DNA at the leading strand (3) and polymerase δ at the lagging strand (4, 5). More recently, polymerase δ has been shown to also be proficient in leading-strand DNA synthesis (6). Beyond DNA replication, the polymerase δ complex has emerged as a central element in the safeguarding of genome integrity by controlling processes such as break-induced replication (7) and homologous recombination (HR) (8). The catalytic subunit of DNA-dependent polymerase complexes associates with several accessory subunits in multiprotein assemblies. The mammalian polymerase δ complex is a heterotetramer consisting of the catalytic subunit POLD1 and the accessory subunits POLD2, POLD3, and POLD4 (9), which regulate the activity of the complex and have specialized roles in stabilization of the complex, establishment of protein-protein interactions, and regulation of the cell cycle (10, 11). The specific function of the POLD2 subunit has been poorly explored, despite its central structural role in interacting with both POLD1 and POLD3 (12). Interestingly, POLD2 and POLD3 participate in translesion synthesis (TLS) via their interaction with the polymerase ζ complex, a translesion polymerase complex comprising the catalytic subunit REV3L and the accessory subunit REV7 (13, 14).
The catalytic subunit POLD1 and the accessory subunits POLD2 and POLD3 have been shown to be essential in mammalian cellular systems using CRISPR/Cas9 and haploid genetic screens (15, 16). In yeast, POLD1 and POLD2 are also essential (17). This requirement has constrained the generation of polymerase δ–knockout cell lines or model organisms. A recent study in mice demonstrated that haploinsufficiency of POLD3 leads to polymerase δ complex instability and replication stress in vivo (18). In humans, heterozygous germline mutations in the proofreading domain of polymerase δ and polymerase ε have been associated with familial cancer predisposition through a hypermutator phenotype (19, 20). Moreover, heterozygous mutations affecting the polymerase and CysB domains of POLD1 have been found to cause autosomal-dominant mandibular hypoplasia, deafness, and progeroid features (MDP) syndrome (21, 22). Here, we report biallelic mutations in POLD1 and POLD2 leading to reduced functionality of the polymerase δ complex as the underlying cause of an autosomal-recessive syndromic immunodeficiency.
Results
Identification of biallelic mutations affecting the polymerase δ complex.
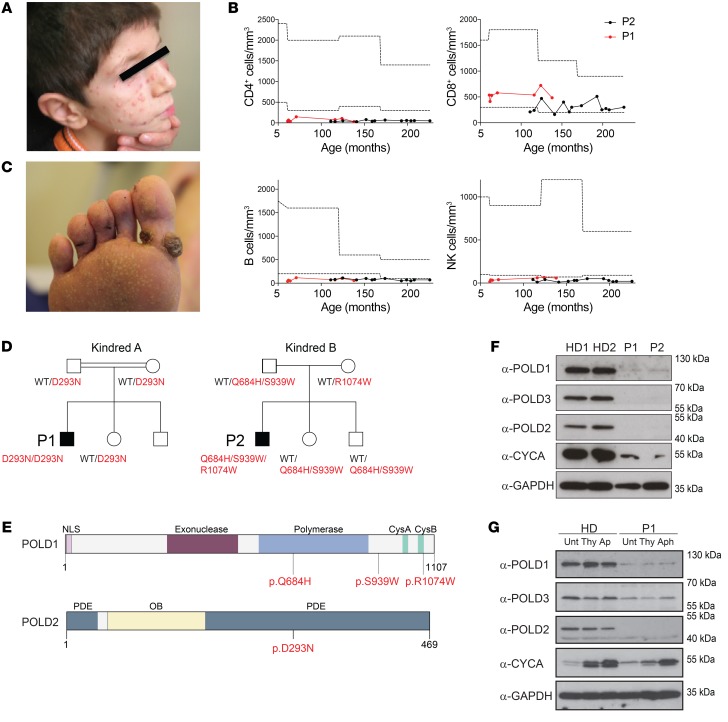
Patient 1 (P1), a 17-year-old male born to consanguineous parents, presented with recurrent upper and lower respiratory tract infections as frequently as 2 to 3 times per month, leading to bronchiectasis since the age of 6 months. He has had chronic molluscum contagiosum–associated lesions on his face since 3 years of age (Figure 1A). At 5 years of age, he started to experience skin abscesses requiring monthly hospitalization. At 4 years of age, the patient had short stature and severe intellectual disability as determined by the Denver test and apparent by speech impairment, attention deficit, and hyperactivity. After starting on regular intravenous Ig (IVIG) replacement, prophylactic antibiotic, and antifungal (trimethoprim-sulfamethoxazole and itraconazole) therapy at 7 years of age, he experienced a reduction in the number of episodes of respiratory infections and skin abscesses. Immunological analyses showed CD4+ T, B, and NK cell lymphopenia (Figure 1B). Patient 2 (P2) is a 24-year-old male born to nonconsanguineous parents. He presented with chronic bronchitis resulting in bronchiectasis and skin warts that were negative for common papilloma virus strains (Figure 1C). The patient had short stature, microcephaly, a low IQ (approximately 70), and hearing impairment. Immunological analyses showed persistent CD4+ T, B, and NK cell lymphopenia (Figure 1B). The patient receives subcutaneous Ig (scIG) treatment, as well as antibiotic and antifungal prophylactic therapy. In-depth immunophenotyping of the main lymphocyte subsets showed a high frequency of CCR7–CD45RA+ TEMRA (effector memory cells reexpressing CD45RA) CD8+ T cells in P1 (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI128903DS1), concomitant with high expression of the CD95 activation marker in CCR7+CD45RA+ naive CD8+ T cells in both P1 and P2 (Supplemental Figure 1B). Despite a reduction in B cell numbers, we found that B cell maturation was largely normal, as judged by the presence of class-switched B cells (CD27+IgD–) (Supplemental Figure 1C) and normal somatic hypermutation (SHM) patterns in the IgA and IgG loci of P1, analyzed by Ig repertoire sequencing (Supplemental Figure 1, D–F). The shared syndromic phenotype in P1 and P2 (Supplemental Table 1), including developmental and immune abnormalities, led us to assume an underlying, undefined syndromic inborn error of immunity (IEI).
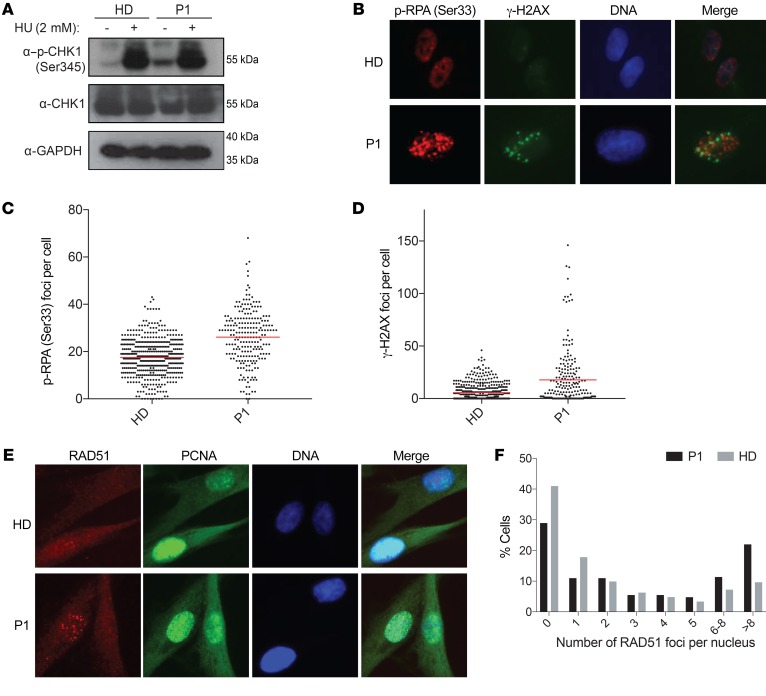
Figure 1. Identification of hypomorphic mutations affecting the polymerase δ complex in patients presenting with syndromic combined immunodeficiency.
(A) Molluscum contagiosum skin infection in P1. (B) Longitudinal peripheral blood CD4+ T cell, CD8+ T cell, B cell, and NK cell counts in P1 and P2. Dotted lines represent the range of reference values. (C) Viral skin warts in P2. (D) Familial segregation of the identified POLD1 and POLD2 mutations in the families of P1 and P2, indicating an autosomal-recessive pattern of inheritance. (E) Domain structure of POLD1 showing the polymerase domain, the exonuclease domain, the nuclear localization signal (NLS) domain, and the cysteine-rich, metal-binding domains CysA and CysB. Domain structure of POLD2 depicting the PDE and oligonucleotide binding (OB) domains. Mutation sites are indicated in red. (F) Protein levels of POLD1, POLD2, POLD3, and CYCA in PBMCs after anti-CD3 and anti-CD28 stimulation for 48 hours. See complete unedited blots in the supplemental material.(G) Protein levels of POLD1, POLD2, and POLD3 in primary fibroblasts. GAPDH was used as a loading control. Cells were untreated (Unt) or synchronized by double-thymidine (Thy) treatment or aphidicolin (Aph) treatment for 24 hours. α, anti.
Genetic analysis of P1 by exome sequencing revealed a homozygous missense variant (p.Asp293Asn; combined annotation–dependent depletion [CADD] score of 28.1) in the POLD2 gene (NM_006230), which displayed perfect segregation under the assumption of autosomal-recessive inheritance (Figure 1D). The POLD2Asp293Asn mutation lies in the phosphodiesterase (PDE) domain (Figure 1E) and is absent from the ExAC and 1,000 Genomes Project public databases. Genetic analysis of P2 by target enrichment sequencing for a panel of IEI genes (23) revealed 3 heterozygous variants in the POLD1 gene (NM_002691) encoding the catalytic subunit of polymerase δ (Figure 1, D and E). Subsequent exome sequencing did not reveal other candidate biallelic variants in P2. The variants p.Gln684His (CADD score of 26), located in the catalytic domain, and p.Ser939Trp (CADD score of 34) in an interdomain region, were inherited in cis on the same allele of maternal origin, whereas the p.Arg1074Trp variant (CADD score of 34) located to the CysB domain on the paternal allele. The POLD1Gln684His and POLD1Arg1074Trp mutations have been reported in the ExAc database, but no homozygotes are present, and the population allele frequency of both variants is below 0.001%. The POLD1Ser939Trp mutation is absent from ExAC and 1,000 Genomes databases. Moreover, we used pLI and pRec scores, computed by ExAC (24), to estimate the likelihood of tolerance to loss of a single or both alleles, respectively, of POLD1 and POLD2. Both genes show low pLI scores of 0.001 and 0.044, respectively, indicating a predicted tolerance to the loss of a single allele, and high pRec scores of 0.998 and 0.950, respectively, indicating a predicted high intolerance for a complete loss of function (24). The tolerance scores for loss of a single allele for POLD1 and POLD2 indicate that both genes are not predicted to present a haploinsufficient phenotype, despite previously reported autosomal-dominant phenotypes (19–22). In familial colorectal cancer, this could be explained by the hypermutator phenotype, in which POLD1 loss of function leads to a functional gain of function (increased mutation rate) (7, 8). Here, we describe 2 patients with biallelic mutations in the POLD1 and the POLD2 subunits of polymerase δ who presented with developmental defects and immunodeficiency.
We assessed the protein levels of the POLD1, POLD2 and POLD3 subunits of polymerase δ in anti-CD3– and anti-CD28–stimulated PBMCs from both P1 and P2. We observed markedly reduced expression of all 3 subunits (Figure 1F), despite mRNA levels of the corresponding subunits that were comparable to those of healthy controls (Supplemental Figure 1, G and H). We did not evaluate POLD4 levels, because of the lack of a satisfactory antibody. Notably, these cells also showed a decrease in the S/G2 cell-cycle phase marker cyclin A (CYCA), indicating a reduced fraction of actively cycling cells. In addition, we analyzed protein levels in primary dermal fibroblasts from P1, which showed reduced levels of POLD1, POLD2, and, to a lesser extent, POLD3. This was independent of CYCA levels, as seen upon enrichment of actively cycling cells (CYCA+) by thymidine or aphidicolin synchronization (Figure 1G). Overall, our results suggest that the identified patient mutations may affect the stability of the polymerase δ complex.
Mutations in POLD1 and POLD2 affect the stability, intrinsic enzymatic activity, and formation of the polymerase δ complex.
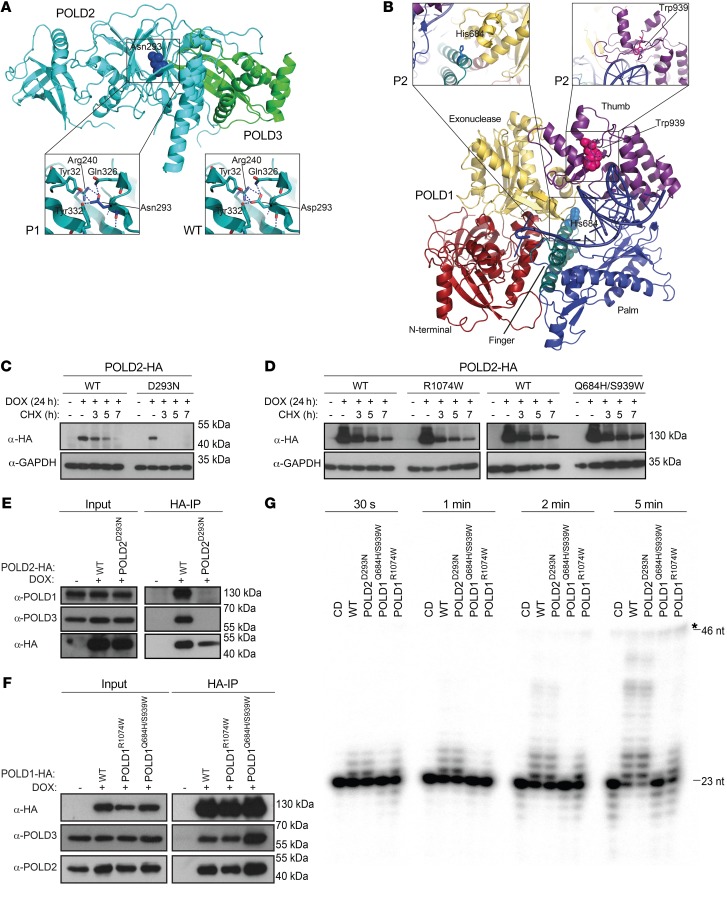
The structure of human POLD2 has been found to be in complex with POLD3 (25), which binds its partner via its N-terminal domain (26). To assess the effect of the POLD2Asp293Asn mutant, we modeled the mutant structure and predicted disruption of electrostatic interactions in the vicinity of Asp293 (Figure 2A). Moreover, we modeled POLD1 mutants using the yeast POLD1 structure (27), which bears a high sequence similarity to human POLD1. Our predictions showed that the POLD1Gln684His/Ser939Trp allele could compromise catalytic domain integrity, given that the Gln684 residue is buried within the catalytic domain and that it resides close to the DNA binding site (Figure 2B), but does not directly affect the catalytic site itself, as the previously described mouse Leu606 mutant showed (ref. 28 and Supplemental Figure 2A). The POLD1Arg1074Trp mutation lies at the C-terminal region of the protein, which is absent from the POLD1 structure model. However, the Arg1074 residue lies in close proximity to a conserved cysteine that mediates interaction with POLD2 in yeast and therefore prompted us to consider interaction defects in the human system (29). In addition, all 4 residues affected by patient mutations show high conservation across eukaryotic evolution (Supplemental Figure 2, B and C). Given the essentiality of the polymerase δ complex, we reasoned that the identified mutants may be hypomorphic in nature through 2 potential mechanisms: reduced intrinsic enzymatic activity and/or reduced complex availability, potentially caused by increased degradation or disrupted assembly of the mutant complex.
Figure 2. The identified mutations affect the stability, interactions, and intrinsic enzymatic activity of the polymerase δ complex.
(A) Overview of the POLD2-POLD3 structure with the indicated position Asp293 (Protein Data Bank [PDB] identifier: 3E0J). The site of the POLD2 mutation surrounding amino acid 293 (Asp293Asn mutant protein) is shown in detail on the left and WT on the right. Polar interactions between residue 293 and nearby residues are indicated with blue dashed lines. Note the predicted loss of 1 interaction with Arg240 upon mutation. (B) The position of the Gln684His and Ser939Trp mutations in the structure of POLD1 (residues 81–984; model on yeast Pol3 PDB ID 3IAY) is shown. The areas surrounding amino acids 684 (left) and 939 (right) in POLD1 are shown in detail. (C and D) Immunoblot analysis of the CHX chase time course of HEK293 cell lines that were DOX-inducible for S-HA–tagged POLD2 WT and POLD2 D293N (C) as well as POLD1 WT, POLD1 R1074W, and POLD1 Q684H/S939W (D). GAPDH was used as a loading control. (E and F) S-HA co-IP from HEK293 cell lines that were DOX-inducible for S-HA–tagged POLD2 and POLD1 and the indicated mutants. (G) Primer extension assay for polymerization activity of recombinant polymerase δ on a radiolabeled, elongated 23-nt primer and 46-nt unlabeled template duplex DNA. The asterisk indicates a background band. CD, catalytically dead.
To assess mutant protein stability, we used an inducible overexpression system of each mutant and WT POLD1 and POLD2 in HEK293 cells and performed cycloheximide (CHX) chase experiments. Our results showed that the stability of POLD2Asp293Asn was reduced compared with WT (Figure 2C), indicating that the reduced levels of polymerase δ observed in P1 cells were due to the identified mutation. The stability of the POLD1 mutants was preserved (Figure 2D). Next, we sought to evaluate whether the identified mutations affected protein-protein interactions within the core complex, such as the reported interactions between POLD2 and POLD3, as well as POLD1 and POLD2. We performed co-IP of streptavidin-hemagglutinin–tagged (S-HA–tagged) versions of WT POLD2 and POLD2Asp293Asn upon inducible overexpression in HEK293 cells. We found a strong reduction in the interaction between POLD2Asp293Asn and POLD1 or POLD3 compared with WT POLD2 (Figure 2E). This reduced interaction was independently observed in a Jurkat T cell line system, in which we stably overexpressed S-HA–tagged POLD2 WT or the POLD2Asp293Asn mutant and performed streptavidin-mediated pulldown using nuclear extracts (Supplemental Figure 2D). Collectively, our data strongly suggest that the interactions within the core polymerase δ complex subunits are probably compromised in P1.
To elucidate the effect of the POLD1 variants on the binding partners POLD2 and POLD3, we performed analogous experiments using the HEK293-inducible overexpression system and co-IP WT POLD1 and the POLD1Arg1074Trp and POLD1Gln684His/Ser939Trp mutants (Figure 2F). In contrast to the POLD2Asp293Asn mutation, all POLD1 mutants showed preserved binding to the POLD2 and POLD3 subunits. Since we did not find aberrations at the level of stability or subunit interactions for any of the POLD1 mutant proteins, we tested whether the intrinsic enzymatic activity of polymerase δ complexes containing the identified mutants was affected. To this end, we expressed and purified WT and mutant heterotetrametric polymerase δ complexes in E. coli (Supplemental Figure 2E). Furthermore, we designed a catalytically dead mutant by introducing the POLD1Asp602Ala and POLD1Asp757Ala double mutation affecting key aspartic acid residues involved in metal ion coordination. Enzymatic in vitro primer extension assays showed reduced polymerase activity in both examined POLD1 mutants, with a more severe reduction for the POLD1Gln684His/Ser939Trp mutant (Figure 2G). POLD2Asp293Asn showed intact activity in vitro. Collectively, our results imply a defective function of the polymerase δ complex in P2 through loss of the polymerase activity of the complex, in contrast to the POLD2Asp293Asn variant in P1, in whom loss of polymerase δ function results from reduced protein stability (Figure 2C).
Polymerase δ–deficient cells show reduced proliferation, cell-cycle delay, and disturbed replication fork dynamics.
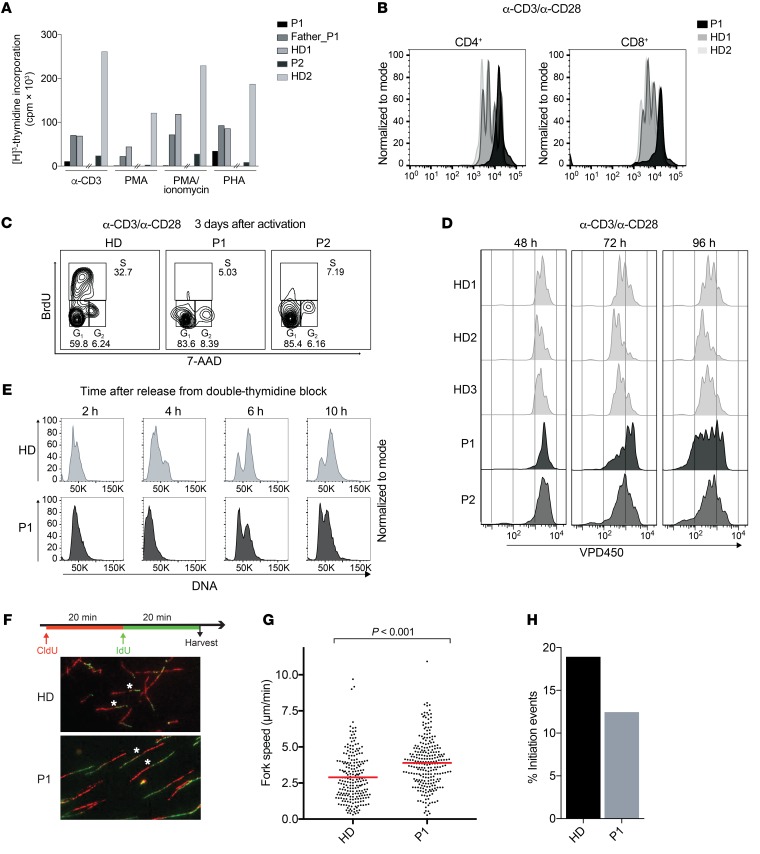
As we observed persistent low lymphocyte counts in the patients (Figure 1B), we explored the connection between polymerase δ mutations and lymphocyte proliferation. In response to proliferative stimuli such as anti-CD3, anti-CD28, phytohemagglutinin (PHA), and PMA-ionomycin, we found that PBMC-derived polymerase δ–deficient T cells from both patients had a reduced proliferative response as measured by [3H]-thymidine uptake or violet proliferation dye 450 (VPD450) (Figure 3, A and B) with intact upregulation of CD25 and CD95 upon T cell receptor (TCR) activation (Supplemental Figure 3A). We hypothesized that these cells may undergo cell-cycle arrest and therefore analyzed cell-cycle dynamics in T cells. Following a 48-hour TCR stimulation, the majority of the patients’ T cells were in G1, whereas approximately 50% of the healthy donor (HD) T cells were actively replicating their DNA as assessed by BrdU incorporation assay (Figure 3C). In P1, given the expansion of TEMRA CD8+ T cells (Supplemental Figure 1A), which are known for their reduced proliferative capacity (30), we aimed to exclude the differentiation stage as a confounding factor for proliferative capacity. To this end, we used in vitro–expanded T cells, which exhibit depleted TEMRA CD8+ T cells (Supplemental Figure 3B). These cells also showed a proliferative delay as compared with healthy control cells (Figure 3D) that was not related to a defect in proximal or distal TCR signaling, as phosphorylation of ERK was intact (Supplemental Figure 3C). We also assessed cell-cycle progression in P1 fibroblasts. P1 and healthy control fibroblasts were synchronized at the G1/S border by double-thymidine block (31), and cell-cycle status was determined upon thymidine release. Analysis of P1 fibroblasts 6 and 10 hours after thymidine release showed slower progression through the S phase when compared with HD fibroblasts (Figure 3E), indicating the presence of a DNA replication–associated defect.
Figure 3. Polymerase δ –deficient cells show reduced proliferation, cell-cycle delay, and disturbed replication fork dynamics.
(A) T cell proliferation as measured by 48-hour thymidine incorporation upon stimulation of PBMCs with anti-CD3, PMA, PMA-ionomycin, and PHA. The hash marks separate independent experiments performed in P1 and P2. (B) T cell proliferation as measured by VPD450 three days after stimulation of PBMCs with anti-CD3 and anti-CD28. Results are representative of 2 independent experiments. (C) Cell-cycle analysis of patient and healthy control T cells as measured by BrdU incorporation. Data are representative of 2 independent experiments. (D) T cell proliferation as measured by VPD450 at the indicated time points after anti-CD3 and anti-CD28 stimulation of in vitro–expanded T cells. (E) Cell-cycle analysis of patient and healthy control fibroblasts upon double-thymidine block and release. (F) Scheme of dual-pulse labeling in DNA fiber analysis and representative images of the fiber analysis experiment performed using patient and HD fibroblasts. Original magnification, ×40. Representative green tracks are marked with an asterisk. Fiber data are representative of 2 independent experiments. Number of fibers counted: 1859 in HDs and 1213 in P1. (G) Replication fork speed in P1 (average of 3.899 μm/min) and HD (average of 2.89 μm/min) fibroblasts as measured by the length of IdU tracks. Statistical significance calculated by Wilcoxon rank sum test. (H) Analysis of initiation events in P1 (12.45%) and HD (18.93%) fibroblasts as measured by the frequency of only IdU-labeled tracks.
We hypothesized that, although intrinsic enzymatic activity did not seem to be impaired in P1, the instability of polymerase δ, as an essential component of the replisome machinery, may lead to disturbance of replication fork dynamics in the cellular context. To assess the patterns of DNA replication in P1 fibroblasts, we used the DNA fiber–labeling technique, which allows direct visualization of single DNA molecules undergoing replication (32). We performed dual-pulse labeling of P1 fibroblasts using 5-chloro-2′-deoxyuridine (CldU) and 5-iodo-2′-deoxyuridine (IdU) (Figure 3F) and observed an increase in the length of polymerized DNA fibers marked by IdU in progressing forks, indicating faster replication fork progression in P1 compared with HD cells (Figure 3G). Increased replication rates have been reported in deficiency of replisome components such as GINS1 (33) and polymerase ε (34) and are regarded as a compensatory mechanism for the concomitant reduction in replication initiation events. We reasoned that instability of polymerase δ may interfere with replisome assembly and therefore lead to reduced replication origin firing. To investigate this, we determined the frequency of de novo fired origins (only IdU-labeled fibers) in P1 fibroblasts and found a reduced number of replication origin initiation events (Figure 3H). Overall, our findings support the hypothesis that reduced levels of polymerase δ compromise cell proliferation and, specifically, S-phase progression, as a result of a rate-limiting amount of an essential component of the replication machinery, leading to disturbed spatiotemporal dynamics of genome duplication.
Polymerase δ mutations lead to replication-associated DNA lesions associated with activation of the S-phase checkpoint.
The disturbed replication dynamics underlying cell-cycle defects in patients’ cells may lead to replication-associated DNA damage and activation of the S-phase checkpoint orchestrated by the Ataxia telangiectasia and Rad3-related protein (ATR) kinase (35). To assess whether polymerase δ deficiency leads to ATR activation, we analyzed the phosphorylation status of the ATR targets p-CHK1 (Ser345) and p-RPA (Ser33). CHK1 phosphorylation on Ser345 could be strongly induced by the replication inhibitor hydroxyurea, but it was already apparent under basal conditions in P1 primary fibroblasts (Figure 4A), indicating persistent replication stress and checkpoint activation in P1 cells at steady state. Furthermore, the number of p-RPA (Ser33) foci was increased from an average of 17 foci per nucleus in HD fibroblasts to an average of 26 foci in P1 fibroblasts in basal conditions (Figure 4, B and C). If replication stress is not processed appropriately, stalled forks can be converted to DNA double-stranded breaks (DSBs). To assess whether replication stress in P1 cells would lead to DSBs, we quantified γ-H2AX foci as a hallmark marker of DNA damage. Strikingly, in P1 fibroblasts, the average number of γ-H2AX foci per nucleus was increased to 18 as compared with 6 in healthy control cells under unchallenged, steady-state conditions (Figure 4, B and D). Polymerase δ has been reported to have a role in various DNA damage repair pathways such as mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), and DSB repair pathways (36). To determine the DNA repair capacity of POLD-deficient cells, we treated P1 cells with different genotoxic compounds. P1 fibroblasts showed no overt sensitivity to an array of genotoxic reagents, which suggests that, despite an unstable POLD complex, its activity was sufficient to ensure proficiency in specific DNA repair pathways (Supplemental Figure 4).
Figure 4. Polymerase δ mutation leads to replication-related DNA lesions associated with activation of the S-phase checkpoint.
(A) Immunoblot analysis of CHK1 phosphorylation at Ser345 in HD and P1 fibroblasts upon treatment with 2 mM hydroxyurea (HU). Total CHK1 and p-CHK1 were run on 2 different gels. GAPDH was used as a sample-processing control. (B) Immunofluorescence staining of p-RPA (Ser33) and γH2AX in HD and P1 fibroblasts. Original magnification, ×40. (C) Quantification of p-RPA (Ser33) foci per nucleus. Average of p-RPA (Ser33) foci: 17.48 (HD) and 26.11 (P1). (D) Quantification of γH2AX foci per nucleus. Average of γH2AX foci: 5.61 (HD) and 17.71 (P1). Number of cells counted in C and D: 481 (HD) and 223 (P1). (E) Immunofluorescence analysis of RAD51 foci and PCNA (S-phase marker) in HD and P1 fibroblasts. Original magnification, ×40. (F) Quantification of RAD51 foci per nucleus. Number of cells counted: 415 (HD) and 273 (P1). Image analysis was performed using CellProfiler, version 2.0.
HR proteins such as RAD51 have well-characterized roles in DNA DSB repair during the S phase but have also recently been implicated more directly in replication processes, revealing a second role as replisome escorting factors for replication fork protection from endonucleases (37) and promotion of replication fork restart. Strikingly, P1 fibroblasts showed increased frequency of RAD51 foci (Figure 4, E and F), suggesting increased activity of RAD51 under conditions of limited availability of polymerase δ.
Polymerase δ deficiency leads to increased 53BP1 nuclear bodies in G1-phase cells.
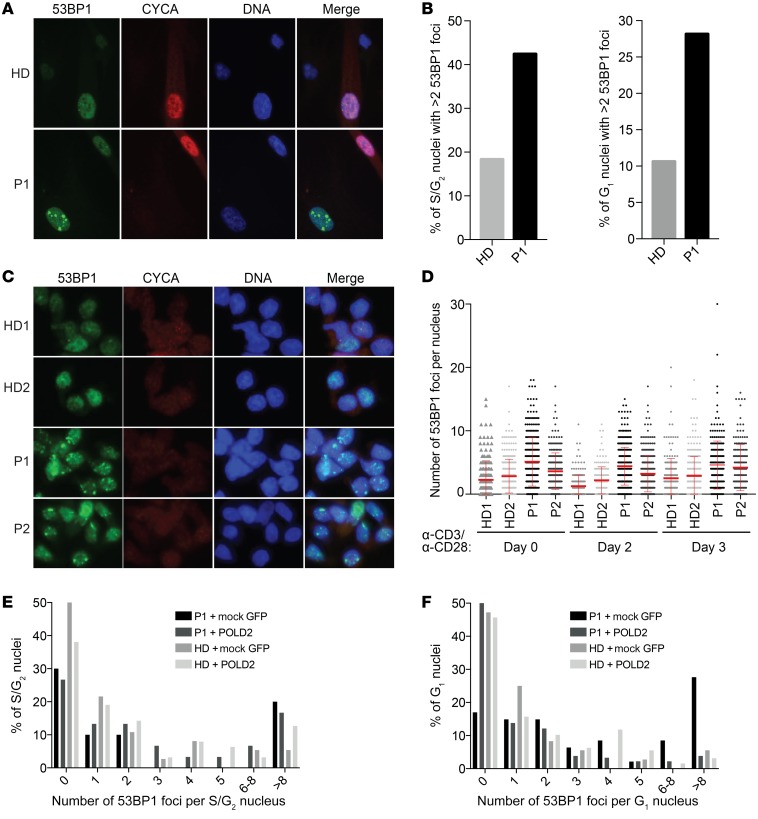
Under-replicated genomic loci have have been shown to be converted to chromatin lesions that are transmitted to daughter cells, resulting in G1 nuclear bodies positive for the DNA damage response (DDR) marker 53BP1, and these lesions are resolved in the next cell cycle (38, 39). Given the presence of replication-associated DNA damage in patients’ cells that partially colocalized with the 53BP1 in S phase (Supplemental Figure 5A), we hypothesized that P1 cells may have under-replicated regions that would persist in G1 cells as nuclear bodies. We co-immunostained cells with antibodies against 53BP1, an established DDR and G1 nuclear body marker, and CYCA. We observed increased numbers of 53BP1 nuclear bodies in G1 (CYCA–) fibroblasts from P1 compared with HD cells (Figure 5A). We also noted that the number of cells with more than 2 foci per nucleus in P1 cells increased in the S and G1 phases by 42.5% and 28.2%, respectively, as compared with 18.3% and 10.6%, respectively for HD cells (Figure 5B). These data suggested that polymerase δ deficiency results in under-replicated regions that are transmitted to the next cell cycle as G1 nuclear bodies and are in line with the identification of polymerase δ as the driver of mitotic DNA synthesis, enabling complete replication of under-replicated regions before cell division (40). We also assessed 53BP1 levels in expanded T cells from both patients and detected increased numbers of foci per cell in both patients compared with healthy controls, with a more pronounced effect in P1 (Figure 5, C and D). To test the direct causality of polymerase δ deficiency on the increased number of G1 nuclear bodies, we stably overexpressed WT POLD2 in P1 fibroblasts. Overexpression of POLD2 in P1 fibroblasts led to stabilization of the polymerase δ subunits POLD1 and POLD3 (Supplemental Figure 5B) and rescued the G1 53BP1 foci (Figure 5, E and F, and Supplemental Figure 5C). Our results suggest that loss of polymerase δ leads to replication stress–associated immunodeficiency and neurodevelopmental abnormalities in this syndrome.
Figure 5. Polymerase δ mutation leads to 53BP1 lesions in G1 that are rescued by transduction of WT POLD2.
(A) Immunofluorescence staining of 53BP1 and CYCA in HD and P1 fibroblasts. Original magnification, ×20. (B) Quantification of 53BP1 foci in S/G2 (CYCA+) and G1 (CYCA–) nuclei. Percentage of cells with more than 2 foci per nucleus, S phase: 18.3% (HD) and 42.5% (P1); G1 phase: 10.6% (HD) and 28.2% (P1). Data are representative of 3 independent experiments. (C) Immunofluorescence staining of 53BP1 foci and CYCA in expanded T cells from HDs, P1, and P2. Original magnification, ×20. (D) Quantification of 53BP1 foci per nucleus from C, unstimulated or 48 hours or 72 hours after anti-CD3 and anti-CD28 stimulation. (E and F) Quantification of 53BP1 foci in (E) S/G2 (CYCA+) and (F) G1 (CYCA–) nuclei of HD and P1 fibroblasts upon transduction with WT POLD2 or GFP control (mock). Number of cells counted: 75 (HD plus mock), 192 (HD plus POLD2), 58 (P1 plus mock), and 213 (P1 plus POLD2). Data are representative of 2 independent experiments. Image analysis was performed using CellProfiler 2.0.
Discussion
Polymerase δ is an essential eukaryotic replicative polymerase, however, the complexity of its activity and the function of its accessory subunits are not well defined. Here, we report on 2 patients with biallelic mutations affecting the POLD1 and POLD2 subunits of polymerase δ, who presented with developmental defects and combined immunodeficiency (CID).
Heterozygous germline mutations in the exonuclease domain of POLD1 have been identified in familial colorectal cancer (19, 41). These exonuclease domain mutations lead to the so-called mutator phenotype found in many human cancers (20). Furthermore, heterozygous mutations in the polymerase domain of POLD1 have been identified in individuals presenting with MDP syndrome (21, 22). POLD1, consequently, has been classified as a tier 1 cancer gene (COSMIC database; https://cancer.sanger.ac.uk/cosmic) with both germline and somatic variants with documented activity in cancer. Functionally validated somatic POLD2 variants are absent from databases such as COSMIC, and, to our knowledge, no mutation has previously been associated with human disease. Loss-of-function intolerance scores for POLD1 and POLD2 indicate that both genes are not predicted to present a haploinsufficient phenotype, despite the autosomal-dominant phenotypes reported in the literature (19–22). In familial colorectal cancer, this could be explained by the hypermutator phenotype, characterized by a genetic loss of function that leads to a functional gain of function, in which mutations in the proofreading domain lead to an increased mutation rate (19, 41). However, in the case of MDP syndrome, the incomplete penetrance of the syndrome remains unexplained.
Since DNA instability syndromes are classically associated with cancer predisposition, it could be expected that the reported patients are likely to develop malignancies. No malignant transformation has been detected in the patients described here, possibly because of largely intact monitorization of genome integrity, which may instead predispose polymerase δ–deficient cells to cell-cycle arrest and senescence. Notably, though, T cell immunodeficiency has a well-established impact on cancer risk due to compromised antitumour immunity and an inability to efficiently clear cancer-associated virus infections (42). Moreover, recent studies on mutational signatures across various cancer entities have identified a distinct mutational signature linked to the related polymerase ε (43). Therefore, despite the cancer-free history of the patients described here, it is quite possible that polymerase δ complex–deficient patients are at heightened risk of cancer development, and we would recommend that patients be monitored closely for any neoplastic changes. A reliable and quantitative assessment of cancer predisposition in human POLD deficiency will require larger cohort sizes and longer clinical follow-up.
In our study, we reveal 2 different mechanisms leading to polymerase δ deficiency. On the one hand, the identified POLD2 mutant leads to severe polymerase δ complex instability, a finding reminiscent of Pold3+/– heterozygous mice (18) and Pole4-null mice and recently reported in POLE1-deficient patients (34). The reduction in polymerase δ availability may restrict replication by interfering with the assembly of replisomes. On the other hand, the identified POLD1 mutants did not affect protein stability but affected the intrinsic polymerase activity of the complex. As shown in Figure 2B and Supplemental Figure 2A, both mutations (p.Q684H and p.S939W) found in cis were located in the vicinity of, but not directly at, the polymerase active site. Our experiments addressed the effect of the combined allele and not the relative contribution of each mutation to the POLD1 catalytic activity. While, presumably, the p.Q684H mutant, residing close to the DNA binding site, may contribute to a larger extent to catalytic inactivity, such mutations may not exhibit the same severe phenotype as a mutation located exactly at the active site, such as the mouse Leu604Gly/Lys mutation. Equivalent to Leu606 in humans, this mutation was described as homozygously lethal in utero in Venkatesan et al. (28) (Supplemental Figure 2A), and, similarly, homozygous human mutations directly affecting the active site of the POLD1 polymerase domain are likely to be nonviable because of the essentiality of DNA replication.
At the cellular level, we observed increased replication fork speed and reduced origin firing in patients’ fibroblasts. A relationship between reduced origin activation and a compensatory higher fork speed has been suggested before in cells of GINS1-deficient patients (33) and polymerase ε–deficient patients (34). Previous reports of patients with deficiency of the replication factor MCM4 did not explore the dynamics of the replication fork (44, 45). Moreover, in our study, the S-phase checkpoint was found to be activated in the patients’ cells to ensure completion of faithful DNA replication with limited polymerase δ complex. Stalled replication forks that fail to restart can be converted to DSBs. In our patients’ cells, DSB repair was activated through HR, apparent by increased RAD51 foci. Besides its well-characterized role in HR, RAD51 has also been recently implicated directly in replication fork protection from endonucleases (37) and in replication fork restart in yeast as an alternative mechanism for polymerase δ–dependent fork restart (46). Therefore, these mechanisms may also be activated in polymerase δ deficiency.
Immunodeficiency associated with DSB repair stems, in many cases, from the necessity of antigen receptor rearrangement (47). However, a separate group of deficiencies is emerging in which immunodeficiency is caused by depletion of essential DNA replication factors. Of note, 2 families with mutations in the replicative polymerase ε have been described as presenting with a CID with decreased lymphocyte proliferation (48, 49). In patients with polymerase δ mutations, we found a reduced lymphocyte expansion capacity, which could be explained by activation of the S-phase checkpoint and an increase in replication-related DNA lesions, such as 53BP1 G1 nuclear bodies, which are known to localize to under-replicated loci. Although rapid lymphocyte expansion processes may make them more vulnerable to replication stress, it is interesting to note that lymphocytes have been previously shown to suppress p53 and ATR during the germinal center reaction in order to tolerate replicative stress (50, 51), but it is unclear whether these mechanisms are active in expanding human peripheral lymphocytes. Moreover, replication timing experiments have shown that the replication program in lymphocytes differs significantly from that in fibroblastic cell lines (52).
Overall, our findings define what we believe to be a novel genetic syndrome caused by polymerase δ deficiency and reveal a previously undefined role, to our knowledge, of polymerase δ in neurodevelopment and lymphocyte biology. Future research will shed light on the differential regulation of checkpoint activation and replication timing in lymphocytes versus other cell types. Replisome components have emerged as key determinants of adaptive immunity that should be systematically analyzed in the context of syndromic immunodeficiency.
Methods
SNP array genotyping.
For P1, homozygosity mapping was performed using the Affymetrix 6.0 SNP array according to the Affymetrix Genome-Wide Human SNP Nsp/Sty 6.0 protocol. Genotype calling was performed using Affymetrix Genotyping Console software. Detection and annotation of the homozygous intervals were performed using HomozygosityMapper (53) and PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) (54).
Exome sequencing and panel sequencing.
For P1, exome sequencing was performed using the TrueSeq Exome Enrichment Kit (Illumina). A multiplexed pool of 12 samples was sequenced on 4 lanes of the Illumina HiSeq2000 sequencing platform in 100-bp paired-end mode. P2 was sequenced using a custom-designed targeted enrichment (Haloplex) approach for a panel of 356 genes, including 248 known IEI genes and IEI candidate genes (design 082014) as described before (23). Reads were demultiplexed and aligned using Burrows-Wheeler Aligner (BWA) software (SourceForge; http://bio-bwa.sourceforge.net/) to the human reference genome assembly GRCh37 (UCSC hg19). Insertion-deletion realignment was performed as well as recalibration based on Genome Analysis Toolkit (GATK) quality scores (55). To call single nucleotide variants (SNVs) and deletion-insertion variants (DIVs), UnifiedGenotyper and GATK Variant quality score recalibration was performed as described previously, with minor modifications (56). Generated lists of SNVs and DIVs were annotated with ANNOVAR (http://annovar.openbioinformatics.org/en/latest/) or SnpEff (SourceForge; http://snpeff.sourceforge.net/). Variants present in 1000 Genomes Project and/or the ExAC database with a minor allele frequency of 0.01 or greater were excluded from further analyses. We used the SIFT (57), PolyPhen-2 (58), and CADD (59) tools for in silico assessment of the effects of the identified mutations.
Cell culture, cell synchronization, and cell line generation.
Human PBMCs were isolated from heparinized blood obtained from the patients and HDs using Ficoll-Hypaque (GE Healthcare). Expanded T cells were obtained upon activation of PBMCs with irradiated feeder cells consisting of a mixture of irradiated allogenic PBMCs and EBV-immortalized B cells. Expanded T cells were grown in RPMI-1640 containing 5% human serum and IL-2 (100 U/mL). Fibroblast cell lines were established from skin biopsies following standard procedures. Cells were cultured at 37°C in a humidified atmosphere with 5% CO2 in glucose-rich DMEM or RPMI-1640 containing 10%–20% inactivated FCS (Life Technologies, Gibco, Thermo Fisher Scientific), 50 units/mL penicillin, 50 μg/mL streptomycin, and 292 μg/mL l-glutamine (all from Gibco, Thermo Fisher Scientific). For cell-cycle synchronization, cells were grown in the presence of 2.5 mM thymidine for 18 hours, released for 8 hours, and treated with 2.5 mM thymidine for an additional 18 hours or treated with 0.45 μM aphidicolin for 24 hours. pTO-SII-HA-GW vectors (60) for N-terminal S-HA–tagged fusion carrying human POLD2, POLD1, and mutant versions thereof were transfected into HEK293 Flp-In-TREx cells (Life Technologies, Thermo Fisher Scientific) together with a Flp recombinase expression plasmid pOG44 (Life Technologies, Thermo Fisher Scientific). Recombinants were selected according to the instructions supplied by the manufacturer, and the resulting cell lines were tested for doxycycline-dependent (DOX-dependent) stable transgene expression.
Flow cytometry antibodies.
The following anti-human monoclonal antibodies were used for flow cytometry–based immunophenotyping: anti–CD3-APC-H7 (clone SK7, 560176, BD Biosciences); anti–CD4-BV605 (clone RPA-T4, 562658, BD Biosciences); anti–CD8-FITC (HIT8a, 555634, BD Biosciences); anti–CD45RA-AF700 (clone HI100, 560673, BD Biosciences); anti–CD19-PerCPCy5.5 (clone HIB19, 45-0199-42, Life Technologies, Thermo Fisher Scientific); anti–IgD-FITC (clone IA6-2, 555778, BD Biosciences); anti–CD95-PECy7 (clone DX2, 561633, BD Biosciences); anti–CD25-PE (clone M-A251, 555432, BD Biosciences); and anti–CCR7-PE-CF594 (clone 150503, 562381, BD Biosciences).
Immunoblot analysis.
Total protein was extracted from cell lines, PBMCs and primary fibroblasts from the affected subjects and healthy controls. Cell lysates were separated by SDS-PAGE and transferred onto PVDF or nitrocellulose membranes. Blots were probed overnight with primary antibodies. Primary antibodies (1:1000 dilution except if indicated otherwise) included anti-POLD1 (1:500; sc-17776, Santa Cruz Biotechnology); anti-POLD2 (HPA026745, MilliporeSigma); anti-POLD3 (A301-244A-M, Bethyl Laboratories); anti-CYCA (sc-751, Santa Cruz Biotechnology); anti-GAPDH (sc-32233, Santa Cruz Biotechnology); anti-p44/42 MAPK (ERK1/2) (4695, Cell Signaling Technology); anti–p-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (4370, Cell Signaling Technology); anti–p-CHK1 (Ser345) (2348, Cell Signaling Technology); anti-CHK1 (sc-56291, Santa Cruz Biotechnology); and anti-HA (H6533, MilliporeSigma). Antibody binding was detected by ECL (Amersham Pharmacia-Biotech).
T cell proliferation assays.
Thymidine incorporation assays were performed as described previously (61). Flow cytometric analysis of cell division of PBMCs was performed using BD Horizon VPD450 according the manufacturer’s instructions.
Cell-cycle analysis.
An assay for BrdU incorporation into patients’ T cells was performed using the FITC BrdU Flow Kit from BD Pharmingen (catalog 559619) following the manufacturer’s instructions.
Immunofluorescence.
Fibroblasts were seeded onto glass coverslips and cultured for at least 24 hours with or without 0.45 μM aphidicolin. After washing with PBS, cells were fixed in either methanol or 4% paraformaldehyde and permeabilized with PBS containing 0.1% Triton X-100. Cells were blocked with PBS containing 4% BSA at room temperature and subsequently incubated with primary antibodies. Cells were incubated with primary anti-p-RPA (Ser33) (1:400; A300-246A-T, Bethyl Laboratories Inc), anti-γH2AX (1:400; 05-636, Millipore), anti-PCNA (1:400; 13110, New England Biolabs), anti-53BP1 (1:400; 612522, BD Biosciences), anti-RAD51 (1:400; PC130, Merck Millipore) and anti-CYCA (1:1000; sc-751, Santa Cruz Biotechnology) antibodies at 4°C overnight. Cells were then washed and incubated with secondary antibodies for 1 hour (Alexa Fluor 488–conjugated goat anti–rabbit IgG, Alexa Fluor 555–conjugated goat anti–mouse IgG and Alexa Fluor 647-conjugated goat anti-rabbit IgG; Thermo Fisher Scientific). GFP-Booster (ATTO448; ChromoTek) was used to increase the GFP signal in transduced fibroblasts. Nuclei were stained with DAPI at room temperature for 15 minutes. Coverslips were mounted in Fluorescent Mounting Medium (Dako). Images were acquired with an Axio Imager M2-1 (Zeiss) or a Deconvolution Microscope (Leica) using a ×40 objective. Quantification of p-RPA, γH2AX, CYCA, 53BP1, and RAD51 foci was performed using CellProfiler image analysis software (https://cellprofiler.org/) (62).
Plasmids.
For stable transduction, cDNA encoding for WT human POLD2 was subcloned into a bicistronic retroviral pMMP vector under the control of a CMV promoter coexpressing POLD2 and the eGFP marker gene via an internal ribosome entry site (IRES) sequence. The Q5 Site-Directed Mutagenesis Kit (New England Biolabs) was used to generate all POLD2 and POLD1 mutants in the respective plasmids. The vectors for bacterial expression (pET303-hpold1 and pCOLA-hPold234) were a gift from Yoshihiro Matsumoto (University of New Mexico, Albuquerque, New Mexico, USA). For inducible expression, human POLD1, POLD2, or POLD3 and mutant versions thereof, as well as GFP control cDNA, were subcloned into pTO-SIIHA-GW vectors for N-terminal S-HA–tagged fusion, using gateway recombination as described previously (63). The Q5 Site-Directed Mutagenesis Kit (New England Biolabs) was used to generate all POLD2 and POLD1 mutants in the respective plasmids.
Recombinant polymerase δ complex purification.
The WT polymerase δ and the identified mutants were purified as follows. The pET303-hPOLD1 plasmid and the pCOLA-234 plasmid encoding POLD2, POLD3, and POLD4 were cotransformed into Rosetta2 cells by electroporation. Protein expression was induced in mid-log–phase cells grown at 16°C by the addition of 1 mM β-d-1-thiogalactopyranoside (IPTG), and the cells were collected 16 hours later. Cells were lysed using a French Press in P200 buffer (40 mM HEPES, pH 7.5, 10% glycerol, 200 mM NaCl, 0.2 mM PMSF, 1 mM benzamidine, 0.1% Triton X-100), and the crude extract was incubated with nickel-NTA beads (QIAGEN), to which polymerase δ was bound via the polyhistidine tag on the p12 subunit. Bound enzyme was eluted with P200 supplemented with 600 mM imidazole, and buffer exchanged back to P200 using Amicon concentrators (MilliporeSigma). WT and mutant amounts were normalized based on Coomassie staining.
Primer extension enzymatic assay.
The reaction buffer contained 40 mM Tris-HCl (pH 7.4), 4 mM MgCl2, 0.1 mg/mL BSA, 5 mM DTT, 25 μM of each dNTP and 10 nM 5′ end–labeled primer-template DNA duplex. Reactions were incubated for the indicated durations at 37°C and stopped by the addition of 95% formamide. Samples were resolved on a 15% polyacrylamide TBE-urea gel, and reaction products were visualized following imaging on a PhosphorImager (Molecular Dynamics). The following oligonucleotides were used as a substrate for the reaction as described by Weedon et al. (21): 5′-CGCGCCGAATTCCCGCTAGCAAT-3′ and 5′-GCGCGGAAGCTTGGCTGCAGAAGATTGCTAGCGGGAATTCGGCGCG-3′.
Fiber assay.
DNA fiber labeling was done as described previously (64). Briefly, patients’ and HD fibroblasts were pulsed labeled with 25 μM CldU for 20 minutes and subsequently pulse labeled with 250 μM IdU for another 20 minutes. After labeling, cells were washed twice with ice-cold PBS, trypsinized, and harvested in cold PBS. Subsequently, cells were lysed with spreading buffer (200 mM Tris-HCl, pH 7.4, 50 mM EDTA, 0.5% SDS) onto microscope slides that were tilted to allow spreading of the DNA fibers. Next, samples were fixed in methanol–acetic acid for 10 minutes and air dried. Immunostaining of CldU and IdU on acid-treated DNA fibers was performed using rat anti-BrdU monoclonal antibody (1:1000; AbD Serotec) and mouse anti-BrdU monoclonal antibody (1:750; BD). Secondary antibodies used were Alexa Fluor 555–conjugated goat anti–rat IgG (1:500; Molecular Probes) and Alexa Fluor 488–conjugated goat anti–mouse IgG (1:500; Molecular Probers). Fibers were imaged using the Axio Imager M2-1 (Zeiss) or a Deconvolution Microscope (Leica) with a ×63 objective. Image analysis was performed with ImageJ software (NIH).
Transduction of patient-derived fibroblasts with WT POLD2.
The pMIG vector containing S-HA–tagged GFP or WT POLD2 was packaged into VSV-G pseudotyped retroviral particles. Patient and healthy control fibroblasts were pretreated with polybrene (0.5 μg/mL; MilliporeSigma) and infected with retrovirus. Eighteen hours after infection, the medium was replaced with fresh culture medium, and the cells were monitored for GFP expression.
Co-IP.
Co-IP was performed using DOX-inducible HEK293-FlpIn cells treated with 1ug/ml doxycycline for 24 hours. After lysis and sonication in buffer I (50 mM HEPES, pH 8.0, 150mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 1mM PMSF) and complete protease inhibitor cocktail (MilliporeSigma), IP was performed with anti-HA beads (A 2095, Milli-poreSigma), followed by 4 washes with buffer I and boiling of beads in SDS-PAGE sample buffer.
For nuclear pulldown assays, stably transduced Jurkat E6.1 cells were subjected to subcellular fractionation essentially as described previously (65). Nuclear extract was used for pulldown with Strep-Tactin agarose beads (2-1201-010, IBA), and bound proteins were eluted with 2.5 mM D-Biotin (A14207/L05109, Alfa Aesar).
Protein modeling and visualization of mutated residues.
The POLD2Asp293Asn mutant was modeled using the POLD2-POLD3 crystal structure (PDB 3E0J, ref. 66) as a template, introducing the mutation with Coot (67). A model of human POLD1 was generated with the Swiss model suite (68), using as a template the yeast POL3 structure (PDB ID 3IAY, chain A) (27), which bears a 50% sequence identity with human POLD1. The resulting model comprises residues 81–984 of the human protein. The POLD1Q684H and POLD1S939W mutants were modeled with Coot. The structure representations were generated with PyMOL (Molecular Graphics System, version 2.0, Schrödinger).
Repertoire sequencing using next-generation sequencing.
Ig heavy chain transcripts (IGH) were amplified from PBMC cDNA in a multiplex PCR and then purified and sequenced using Roche 454 sequencing as previously described (69). Sequences were demultiplexed on the basis of their multiplex identifier sequence, and 10 nucleotides were trimmed from both sides to remove the primer sequence using the ARGalaxy tool (70). Fasta files were uploaded into IMGT/High-V-Quest (71), and subsequently, the IMGT output files were analyzed using ARGalaxy (69). Only productive sequences that were complete, without ambiguous bases, present 2 or more times, with a C subclass that could be defined, were included in the analysis as a single sequence.
Real time quantitative PCR.
RNA was extracted from freshly isolated or frozen PBMCs using the RNeasy Kit (QIAGEN), and reverse transcription was performed using Reverse Transcriptase from Promega using both oligo-DT and random hexamer primers. Quantitative real time PCR for POLD1 and POLD2 mRNA expression was performed using Kappa Sybr Fast qPCR MasterMix ABI Bioprism (Kapa Biosystems) on a 7900HT Fast Real-Time PCR System (Applied Biosciences). The intron-spanning primers POLD1 (5′-TCTCCTCCATCAGTGCCAG-3′ and 5′-AAACGCTGTTTGAAGCGG-3′) and POLD2 (5′-GCTGCAGTTCACACAGCTTC-3′ and 5′-TATGCCCACATTTATGCCAC-3′) were used. Relative RNA expression normalized to GAPDH mRNA abundance was calculated by the relative standard curve/ΔΔCt method (Applied Biosystems).
Study approval.
The present study was approved by the IRB of the Medical University of Vienna. Written consent was obtained from the patients or their parents.
Author contributions
KB conceived, designed, and coordinated the study. CDC, ÖYP, KLW, SB, and KB designed research, analyzed and interpreted data, and wrote the manuscript. CDC, ÖYP, KLW, EG, SW, RCA, ES, and A. Krolo performed research. CDC analyzed genetic data, performed immunophenotyping and T cell proliferation analysis, cell-cycle analysis, immunoblotting, primer extension assays, and immunofluorescence assays and generated the stable overexpression cell lines. ÖYP performed IP experiments, designed and performed immunoblotting, drug sensitivity, fiber assay, and immunofluorescence experiments, and analyzed and interpreted the data. KLW performed cell fractionation and co-IP experiments. A. Krolo analyzed P2 genetics data. KB provided clinical data from P2 and, together with SB, A. Kiykim, EKA, AO, LK, and EFW, interpreted clinical data for both P1 and P2. EG performed protein purification and protein modeling studies. ES identified the POLD2 mutation in P1 and performed T cell signaling analysis. HI performed the Ig mutation analysis. WFP performed the thymidine incorporation assay. MVFB, GSF, JM, and JIL provided scientific input.
Supplementary Material
Acknowledgments
We thank the patients and their families for participation. We thank Yoshihiro Matsumoto (University of New Mexico, Albuquerque, New Mexico, USA) for providing E. coli expression vectors. We thank Ulrich Goldmann for advice on protein modeling, Rui Martins and Melania Zauri for advice on image analysis, and Stavroula Woutsas (all from CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences) for help with genetic analysis of P1. We thank Tatjana Hirschmugl (Ludwig Boltzmann Institute for Lung Vascular Research, Graz, Austria) for graphical artwork displayed in the graphical abstract. This work was funded by the European Research Council (ERC) under the European Union Seventh Framework Program (FP7/2007-2013; ERC grant agreement 310857, to KB), Austrian National Bank (ÖNB Jubilee Fund 16385, to KB), a grant from the Jeffrey Modell Foundation (to KB and MB), and the Austrian Science Fund Lise Meitner Program Fellowship (FWF M1809, to KLW).
Version 1. 08/26/2019
Electronic publication
Version 2. 10/01/2019
Print issue publication
Footnotes
CDC’s present address is: Wellcome Sanger Institute, Hinxton, England.
KLW’s present address is: Instituto Gulbenkian de Ciência, Oeiras, Portugal.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4194–4206.https://doi.org/10.1172/JCI128903.
Contributor Information
Cecilia Domínguez Conde, Email: cc36@sanger.ac.uk.
Özlem Yüce Petronczki, Email: Oezlem.YuecePetronczki@rud.lbg.ac.at.
Safa Baris, Email: safabaris@hotmail.com.
Katharina L. Willmann, Email: kwillmann@igc.gulbenkian.pt.
Enrico Girardi, Email: EGirardi@cemm.oeaw.ac.at.
Elisabeth Salzer, Email: esalzer@cemm.oeaw.ac.at.
Stefan Weitzer, Email: stefan.weitzer@meduniwien.ac.at.
Rico Chandra Ardy, Email: Rico.Ardy@rud.lbg.ac.at.
Ana Krolo, Email: Ana.Krolo@rud.lbg.ac.at.
Hanna Ijspeert, Email: h.ijspeert1@gmail.com.
Ayca Kiykim, Email: dr_gora@yahoo.com.
Elif Karakoc-Aydiner, Email: elifaydiner@gmail.com.
Elisabeth Förster-Waldl, Email: elisabeth.foerster-waldl@meduniwien.ac.at.
Leo Kager, Email: leo.kager@stanna.at.
Giulio Superti-Furga, Email: gsuperti@cemm.oeaw.ac.at.
Javier Martínez, Email: javier.martinez@meduniwien.ac.at.
Joanna I. Loizou, Email: jloizou@cemm.oeaw.ac.at.
Ahmet Ozen, Email: ahmetozen_md@yahoo.com.
Mirjam van der Burg, Email: m.van_der_burg@lumc.nl.
Kaan Boztug, Email: kaan.boztug@rud.lbg.ac.at.
References
- 1.Loeb LA, Monnat RJ. DNA polymerases and human disease. Nat Rev Genet. 2008;9(8):594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 2.Santocanale C, Foiani M, Lucchini G, Plevani P. The isolated 48,000-dalton subunit of yeast DNA primase is sufficient for RNA primer synthesis. J Biol Chem. 1993;268(2):1343–1348. [PubMed] [Google Scholar]
- 3.Pursell ZF, Isoz I, Lundström EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317(5834):127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larrea AA, et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107(41):17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30(2):137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RE, Klassen R, Prakash L, Prakash S. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell. 2015;59(2):163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343(6166):88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Mol Cell Biol. 2008;28(4):1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie B, Mazloum N, Liu L, Rahmeh A, Li H, Lee MY. Reconstitution and characterization of the human DNA polymerase delta four-subunit holoenzyme. Biochemistry. 2002;41(44):13133–13142. doi: 10.1021/bi0262707. [DOI] [PubMed] [Google Scholar]
- 10.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 11.Jain R, Aggarwal AK, Rechkoblit O. Eukaryotic DNA polymerases. Curr Opin Struct Biol. 2018;53:77–87. doi: 10.1016/j.sbi.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Brocas C, Charbonnier JB, Dhérin C, Gangloff S, Maloisel L. Stable interactions between DNA polymerase δ catalytic and structural subunits are essential for efficient DNA repair. DNA Repair (Amst) 2010;9(10):1098–1111. doi: 10.1016/j.dnarep.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111(8):2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RE, Prakash L, Prakash S. Pol31 and Pol32 subunits of yeast DNA polymerase δ are also essential subunits of DNA polymerase ζ. Proc Natl Acad Sci U S A. 2012;109(31):12455–12460. doi: 10.1073/pnas.1206052109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350(6264):1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 18.Murga M, et al. POLD3 is haploinsufficient for DNA replication in mice. Mol Cell. 2016;63(5):877–883. doi: 10.1016/j.molcel.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rayner E, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16(2):71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 21.Weedon MN, et al. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat Genet. 2013;45(8):947–950. doi: 10.1038/ng.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elouej S, et al. Exome sequencing reveals a de novo POLD1 mutation causing phenotypic variability in mandibular hypoplasia, deafness, progeroid features, and lipodystrophy syndrome (MDPL) Metab Clin Exp. 2017;71:213–225. doi: 10.1016/j.metabol.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Erman B, et al. Combined immunodeficiency with CD4 lymphopenia and sclerosing cholangitis caused by a novel loss-of-function mutation affecting IL21R. Haematologica. 2015;100(6):e216–e219. doi: 10.3324/haematol.2014.120980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baranovskiy AG, et al. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle. 2008;7(19):3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem. 2004;279(3):1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 27.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16(9):979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesan RN, et al. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27(21):7669–7682. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giot L, Chanet R, Simon M, Facca C, Faye G. Involvement of the yeast DNA polymerase delta in DNA repair in vivo. Genetics. 1997;146(4):1239–1251. doi: 10.1093/genetics/146.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geginat J, Lanzavecchia A, Sallusto F. Proliferation and differentiation potential of human CD8+ memory T-cell subsets in response to antigen or homeostatic cytokines. Blood. 2003;101(11):4260–4266. doi: 10.1182/blood-2002-11-3577. [DOI] [PubMed] [Google Scholar]
- 31.Chen G, Deng X. Cell synchronization by double thymidine block. Bio Protoc. 2018;8(17):e2994. doi: 10.21769/BioProtoc.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cottineau J, et al. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest. 2017;127(5):1991–2006. doi: 10.1172/JCI90727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellelli R, et al. Polε instability drives replication stress, abnormal development, and tumorigenesis. Mol Cell. 2018;70(4):707–721.e7. doi: 10.1016/j.molcel.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saldivar JC, Cortez D, Cimprich KA. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat Rev Mol Cell Biol. 2017;18(10):622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu JJ, et al. Estrogen promotes the survival and pulmonary metastasis of tuberin-null cells. Proc Natl Acad Sci U S A. 2009;106(8):2635–2640. doi: 10.1073/pnas.0810790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolinjivadi AM, Sannino V, de Antoni A, Técher H, Baldi G, Costanzo V. Moonlighting at replication forks – a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 2017;591(8):1083–1100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- 38.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13(3):243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 39.Harrigan JA, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193(1):97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minocherhomji S, et al. Replication stress activates DNA repair synthesis in mitosis. Nature. 2015;528(7581):286–290. doi: 10.1038/nature16139. [DOI] [PubMed] [Google Scholar]
- 41.Campbell BB, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171(5):1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hauck F, Voss R, Urban C, Seidel MG. Intrinsic and extrinsic causes of malignancies in patients with primary immunodeficiency disorders. J Allergy Clin Immunol. 2018;141(1):59–68.e4. doi: 10.1016/j.jaci.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Martincorena I, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171(5):1029–1041.e21. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gineau L, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122(3):821–832. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hughes CR, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122(3):814–820. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moriel-Carretero M, Aguilera A. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell. 2010;37(5):690–701. doi: 10.1016/j.molcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 47.Slatter MA, Gennery AR. Primary immunodeficiencies associated with DNA-repair disorders. Expert Rev Mol Med. 2010;12:e9. doi: 10.1017/S1462399410001419. [DOI] [PubMed] [Google Scholar]
- 48.Pachlopnik Schmid J, et al. Polymerase ε1 mutation in a human syndrome with facial dysmorphism, immunodeficiency, livedo, and short stature (“FILS syndrome”) J Exp Med. 2012;209(13):2323–2330. doi: 10.1084/jem.20121303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frugoni F, et al. A novel mutation in the POLE2 gene causing combined immunodeficiency. J Allergy Clin Immunol. 2016;137(2):635–638.e1. doi: 10.1016/j.jaci.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 51.Ranuncolo SM, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8(7):705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 52.Letessier A, et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470(7332):120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 53.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37(Web Server issue):W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willmann KL, et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat Commun. 2014;5:5360. doi: 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmetterer KG, et al. Two newly diagnosed HLA class II-deficient patients identified by rapid vector-based complementation analysis reveal discoordinate invariant chain expression levels. Int Arch Allergy Immunol. 2010;152(4):390–400. doi: 10.1159/000288292. [DOI] [PubMed] [Google Scholar]
- 62.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pichlmair A, et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487(7408):486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 64.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giambruno R, et al. Affinity purification strategies for proteomic analysis of transcription factor complexes. J Proteome Res. 2013;12(9):4018–4027. doi: 10.1021/pr4003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baranovskiy AG, et al. X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle. 2008;7(19):3026–3036. doi: 10.4161/cc.7.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.IJspeert H, et al. Evaluation of the antigen-experienced B-cell receptor repertoire in healthy children and adults. Front Immunol. 2016;7:410. doi: 10.3389/fimmu.2016.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.IJspeert H, van Schouwenburg PA, van Zessen D, Pico-Knijnenburg I, Stubbs AP, van der Burg M. Antigen Receptor Galaxy: a user-friendly, Web-based tool for analysis and visualization of T and B cell receptor repertoire data. J Immunol. 2017;198(10):4156–4165. doi: 10.4049/jimmunol.1601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alamyar E, Duroux P, Lefranc MP, Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.