Abstract
BACKGROUND
Adenoid cystic carcinoma (ACC) is a rare malignancy arising in salivary glands and other sites, characterized by high rates of relapse and distant spread. Recurrent/metastatic (R/M) ACCs are generally incurable, due to a lack of active systemic therapies. To improve outcomes, deeper understanding of genetic alterations and vulnerabilities in R/M tumors is needed.
METHODS
An integrated genomic analysis of 1,045 ACCs (177 primary, 868 R/M) was performed to identify alterations associated with advanced and metastatic tumors. Intratumoral genetic heterogeneity, germline mutations, and therapeutic actionability were assessed.
RESULTS
Compared with primary tumors, R/M tumors were enriched for alterations in key Notch (NOTCH1, 26.3% vs. 8.5%; NOTCH2, 4.6% vs. 2.3%; NOTCH3, 5.7% vs. 2.3%; NOTCH4, 3.6% vs. 0.6%) and chromatin-remodeling (KDM6A, 15.2% vs. 3.4%; KMT2C/MLL3, 14.3% vs. 4.0%; ARID1B, 14.1% vs. 4.0%) genes. TERT promoter mutations (13.1% of R/M cases) were mutually exclusive with both NOTCH1 mutations (q = 3.3 × 10–4) and MYB/MYBL1 fusions (q = 5.6 × 10–3), suggesting discrete, alternative mechanisms of tumorigenesis. This network of alterations defined 4 distinct ACC subgroups: MYB+NOTCH1+, MYB+/other, MYBWTNOTCH1+, and MYBWTTERT+. Despite low mutational load, we identified numerous samples with marked intratumoral genetic heterogeneity, including branching evolution across multiregion sequencing.
CONCLUSION
These observations collectively redefine the molecular underpinnings of ACC progression and identify further targets for precision therapies.
FUNDING
Adenoid Cystic Carcinoma Research Foundation, Pershing Square Sohn Cancer Research grant, the PaineWebber Chair, Stand Up 2 Cancer, NIH R01 CA205426, the STARR Cancer Consortium, NCI R35 CA232097, the Frederick Adler Chair, Cycle for Survival, the Jayme Flowers Fund, The Sebastian Nativo Fund, NIH K08 DE024774 and R01 DE027738, and MSKCC through NIH/NCI Cancer Center Support Grant (P30 CA008748).
Keywords: Oncology
Keywords: Head and neck cancer
Introduction
Adenoid cystic carcinoma (ACC) is a rare, often treatment-resistant cancer with a high rate of metastatic spread. Predominantly arising within salivary glands, ACCs may also originate in the trachea, lung, breast, and other sites. ACC tumors are treated with surgery and adjuvant radiation therapy (1), but most cases are characterized by perineural invasion and distant spread that may develop over years to decades. More than 50% of ACCs recur or develop distant metastases over time. Relapsed tumors are generally incurable because no systemic agents to date have been found to be effective. Therefore, overall prognosis has remained poor, with long-term overall survival (OS) rates of 23%–40% (2).
Our knowledge to date of the molecular alterations underlying ACC have been based on several small studies of mainly primary tumors, which have revealed a relatively quiet genome (3, 4), with prevalent alterations including MYB or MYBL1 translocations (28%–59%) (2, 5–7) and a long tail of less frequent mutations (8) that may drive pathogenesis. Recurrent and/or metastatic (R/M) ACC has not been well characterized, and it remains unknown which molecular alterations drive its propensity for metastasis (9). Further progress in the treatment of R/M ACC will require a more comprehensive understanding of the molecular alterations associated with aggressive tumor behavior. Here, we evaluate the comparative genomic landscapes of primary versus R/M ACC in 1045 sequenced cases, a cohort size that approximates the annual incidence of ACC in the United States (approximately 1200 per year). In R/M ACCs, we identify distinct mutational enrichments, patterns of intratumoral genetic heterogeneity, and pathogenic germline alterations, which together reveal previously unrecognized aspects of the “quiet” ACC genome. Taken together, our findings help define a framework governing ACC progression and guide next steps for clinical investigation.
Results
Mutational data were compiled in aggregate on 1045 ACC cases (177 primary tumors in patients with localized disease and 868 cases with R/M disease) (Supplemental Figure 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI128227DS1). Tumors were sequenced with either whole-exome sequencing (WES), whole-genome sequencing (WGS) (n = 193), or targeted next-generation sequencing (NGS) panels (n = 852). A total of 9012 genomic alterations (1259 mutations + 1231 copy number alterations (CNAs) in primary ACC, 5905 mutations + 617 CNAs in R/M ACC) were identified. Head and neck salivary ACCs made up 89.8% of tumors, with the remainder arising in the lung (6.8%) or breast (3.4%) (Supplemental Figure 2). The overall profiles of salivary, lung, and breast ACCs were similar (Supplemental Table 2); similarly, no significant genomic differences were noted based on the location of tumor (primary vs. metastatic site) sampled (Supplemental Table 3). The tumor mutational burden (TMB) for cases profiled with WES/WGS was 0.34 mutations/megabase.
Enriched genetic alterations in R/M ACC.
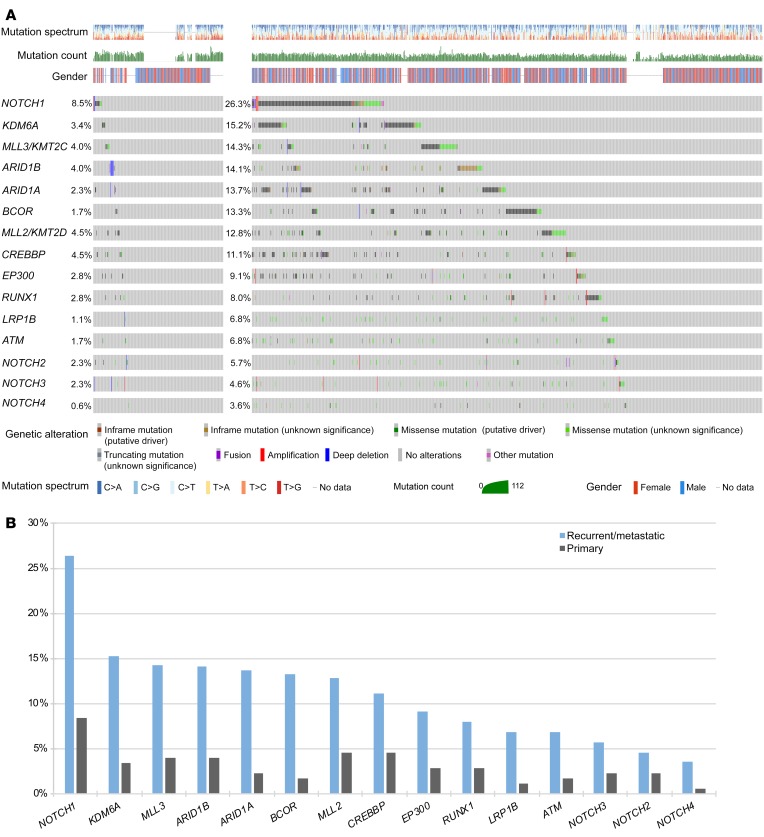
We identified several genes in which alterations were markedly enriched in R/M cases compared with primary ACC cases (Figure 1A and Supplemental Table 4). Mutations in NOTCH1, a critical regulator of cell proliferation and survival, were significantly increased in R/M compared with primary ACC (26.3% vs. 8.5%, OR 3.86, P < 0.0001) (Figure 1B). Very few of the NOTCH1 mutations appeared subclonal based on variant allele frequencies (Supplemental Figure 3). Similar numerical, but nonstatistically significant, enrichments were observed for other members of the Notch family with lower mutational prevalence: NOTCH2, 4.6% vs. 2.3%, OR 2.09, P = 0.17; NOTCH3, 5.7% vs. 2.3%, OR 2.63, P = 0.067; NOTCH4, 3.6% vs. 0.6%, OR 6.59, P = 0.067. In aggregate, 34.1% of R/M cases harbored a mutation in these 4 genes (NOTCH1–4), while 39.6% of R/M cases contained a mutation in the broader Notch pathway (including SPEN and FBXW7).
Figure 1. Comparison of primary vs.
R/M ACC genomic alterations. (A) Oncoprint of primary and R/M ACC. (B) Mutations in cancer genes that are enriched in R/M ACC relative to primary tumors. Statistical comparisons are provided in Supplemental Table 4.
Compared with primary cases, R/M cases were also significantly enriched for alterations in key chromatin-remodeling genes, including KDM6A (15.2% vs. 3.4%, OR 5.12, P = 0.0001), MLL3/ KMT2C (14.3% vs. 4.0%, OR 4.06, P = 0.0005), ARID1B (14.1% vs. 4.0%, OR 4.00, P = 0.0006), ARID1A (13.7% vs. 2.3%, OR 6.87, P = 0.0002), BCOR (13.3% vs. 1.7%, OR 8.92, P = 0.0002), MLL2/KMT2D (12.8% vs. 4.5%, OR 3.10, P = 0.0027), and CREBBP (11.1% vs. 4.5%, OR 2.63, P = 0.011). Other notable R/M enrichment was seen in genes involved in DNA damage repair (ATM, 6.8% vs. 1.7%, OR 4.22, P = 0.016) and tumor suppression (LRP1B, 6.8% vs. 1.1%, OR 6.43, P = 0.010). When correcting for multiple hypothesis testing, all comparisons except for those involving NOTCH2–4 had Benjamini-Hochberg FDR of less than 0.10.
Since most R/M cases were sequenced at higher depth with targeted NGS panels, we assessed the possibility that those mutations enriched in R/M cases might have been mutations with low variant allelic fraction (VAF), below the resolution of WES. None of the mutations that were enriched in R/M cases had VAF of less than 0.05 (a conservative detection threshold in 100× WES; refs. 10–13) in more than 5% of the cases, with the majority between 0% and 2% (Supplemental Table 5). To directly compare the sensitivity of WES (at ~×100) to targeted NGS (at ~×600) for the detection of these enriched mutations, we downsampled the reads from R/M cases sequenced on the ×MSK-IMPACT platform to 100. This minimally altered the resulting VAFs (Supplemental Figure 4), with average change in VAF of 0.011, and only 1 enriched mutation (1/101, or 1%) was not detected at the downsampled depth (Supplemental Table 6). Together, these analyses confirm that the enriched rate of mutations in these genes in R/M cases is unlikely to be an artifact of differences in sequencing depth.
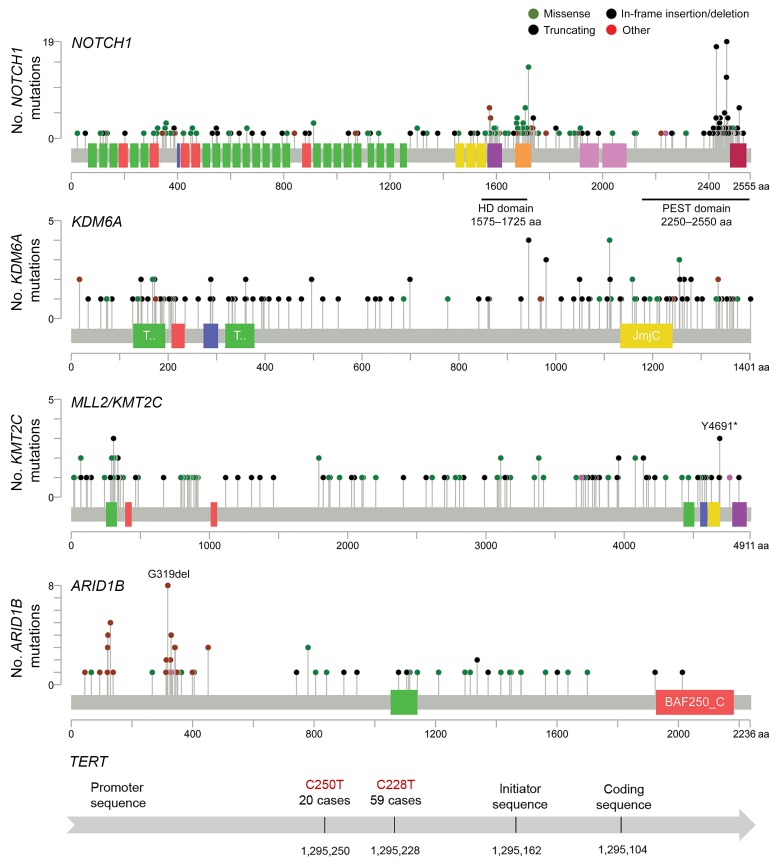
Lollipop plots of key mutated genes are shown in Figure 2. Of the 225 (26.3%) R/M samples with NOTCH1 alterations, 337 distinct NOTCH1 alterations were observed, with 221 (65.6%) considered activating mutations found in hot-spot regions (heterodimerization-negative regulatory region and proline, glutamic acid, serine, threonine-rich [PEST] domain). Overall, 159 unique samples (18.3%) had activating NOTCH1 mutations, indicating the approximate proportion of R/M ACCs anticipated to have an alteration targetable with Notch-directed therapies such as γ secretase inhibitors.
Figure 2. Lollipop plots of mutations in key genes in R/M ACC.
For NOTCH1 in particular, 337 distinct alterations were observed, with 221 (65.6%) found in established hot-spot regions and considered activating mutations. Total number of alterations and incidence by gene are listed in Supplemental Table 4. HD, heterodimerization negative regulatory region; JmjC, Jumonji C; BAF250_C, SWI/SNF chromatin remodeling complex.
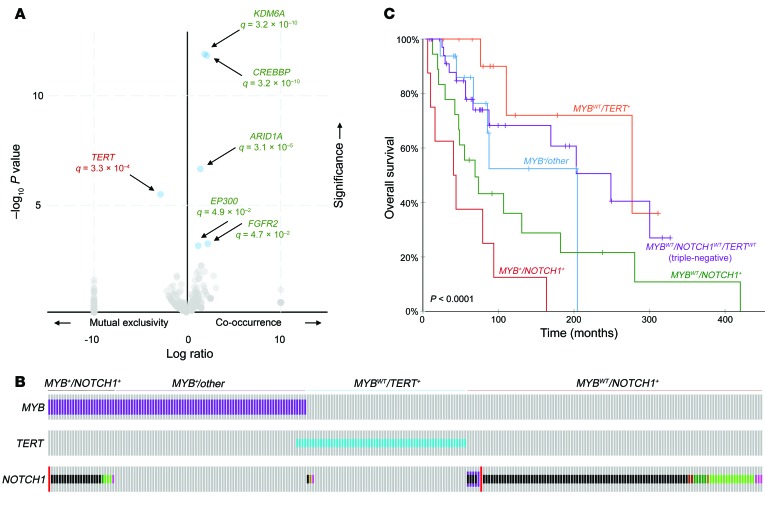
We investigated patterns of mutual exclusivity among mutations in R/M tumors to examine governing patterns of interaction. NOTCH1 mutations were found to be mutually exclusive with TERT promoter mutations well beyond what would be expected by chance (OR 0.62, q = 3.3 × 10–4) (Figure 3A). Conversely, NOTCH1 mutations cooccurred significantly with mutations in chromatin-remodeling genes (KDM6A, OR 1.20, q = 3.2 × 10–10; ARID1A, OR 1.15, q = 3.1 × 10–5; CREBBP, OR 1.35, q = 3.2 × 10–10). Such findings suggest potential cooperative synergy between NOTCH1 and chromatin-remodeling genes to mediate 1R/M phenotype, while cases with altered TERT promoters represent a conspicuously distinct subset.
Figure 3. Distinct molecular ACC subgroups based on genetic alterations.
(A) Volcano plot illustrating genes correlated or anticorrelated with NOTCH1 mutation in R/M ACC. NOTCH1 alterations were found to be highly mutually exclusive with TERT promoter mutations and highly cooccurrent with chromatin-remodeling pathway gene alterations (KDM6A, CREBB, ARID1A, EP300), where q represents the Benjamini-Hochberg FDR statistic. (B) R/M ACC subgroups based on mutually exclusive genetic alterations, including MYB+NOTCH+, MYB+/other, MYBWTTERT+, and MYBWTNOTCH+ divisions. Displayed cases represent 53.2% (305/573) of R/M cases with available data. (C) OS of R/M ACC subgroups. Triple-negative cases are defined as cases lacking alterations in MYB, NOTCH1, or the TERT promoter (MYBWTNOTCH1WTTERTWT). Comparison of the triple-negative subgroup with other subgroups by log-rank test: MYB+NOTCH1+ (P = 5.42 × 10–5), MYBWTNOTCH1+ (P = 0.0269), MYB+/other (P = 0.414), MYBWTTERT+ (P = 0.296).
Of the R/M tumors with available data, 22.2% (129/581) had MYB or MYBL1 rearrangements (identified mainly with NGS encompassing MYB intron 14, or FISH for MYB or MYBL1 rearrangement). The incidence of TERT promoter mutations was significantly elevated in tumors lacking this fusion (OR 3.71, q = 5.6 × 10–3), suggesting that TERT mutations represent an alternate mechanism of ACC pathogenesis that is independent of the hallmark MYB/MYBL1 translocation. It is important to note that many of the cases categorized as negative for MYB rearrangement had only MYB intron 14 profiled and that cases with other rare MYB breakpoints or MYBL1 fusions may not have been identified. However, we observed a similar anticorrelation of TERT promoter mutations and MYB/MYBL1 fusions among the subset of cases profiled with FISH (OR 4.60), supporting the mutual exclusivity of these alterations.
These key alterations, involving MYB, NOTCH1, and TERT, formed distinct molecular subgroups that accounted for more than half (53.2%, or 305/573) of cases with available data: MYB+NOTCH1+, MYB+/other, MYBWTTERT+, and MYBWTNOTCH1+. These proposed molecular subgroups are illustrated in Figure 3B and exhibited significant differences in OS (P < 0.0001) (Figure 3C). In particular, MYBWTNOTCH1+ and MYB+NOTCH1+ status conferred the worst prognosis. Again, we note that most of the MYBWT cases are cases that lack rearrangements involving MYB intron 14, which represents the vast majority, but not all, MYB fusions. The remaining cases, described as triple-negative (MYBWTNOTCH1WTTERTWT), lacked these defining alterations. However, chromatin-remodeling genes were commonly mutated in these cases: 60.1% (160/266) of triple-negative cases harbored at least 1 chromatin-remodeling alteration.
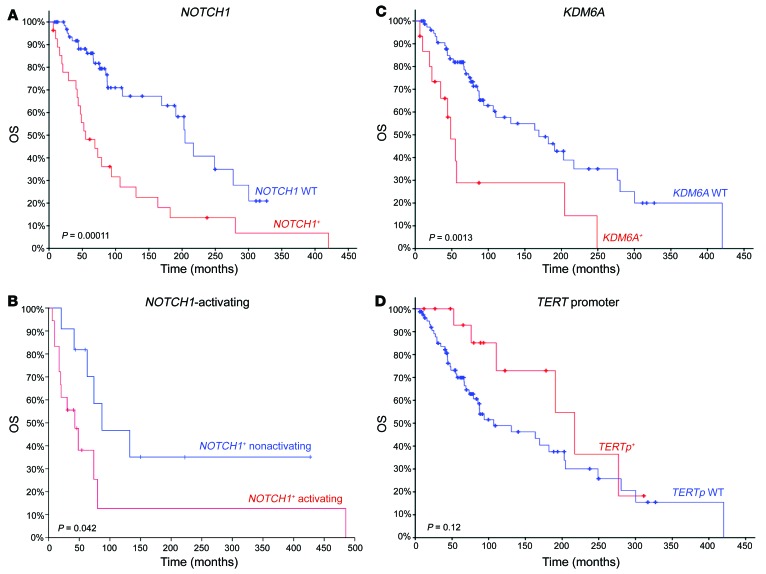
NOTCH1 and KDM6A mutations are associated with poor prognosis.
Clinical data were available for the cohort of 84 R/M ACC cases treated at MSKCC and sequenced using the MSK-IMPACT assay, with median follow-up 8.1 years. NOTCH1-mutant cases exhibited significantly poorer survival outcomes compared with NOTCH1-WT cases (median OS 55.1 vs. 204.5 months, P = 1.10 × 10–4). Among NOTCH1-mutant patients, activating NOTCH1-mutant cases were associated with significantly poorer survival (median OS 31.1 vs. 73.8 months, P = 0.042). KDM6A-mutant cases demonstrated similarly poor OS relative to KDM6A-WT patients (median OS 48.5 vs. 169.3 months, P = 1.32 × 10–3) (Figure 4). Alterations in MYB-NFIB (P = 0.87) or the TERT promoter (P = 0.12) did not appear to have any bearing on survival.
Figure 4. OS comparison for R/M ACC by individual gene.
(A) NOTCH1 mutant vs. NOTCH1 WT, (B) NOTCH1 mutant (activating) vs. NOTCH1 mutant (nonactivating), (C) KDM6A mutant vs. KDM6A WT, and (D) TERT promoter mutant vs. TERT promoter WT.
Actionable alterations beyond NOTCH1.
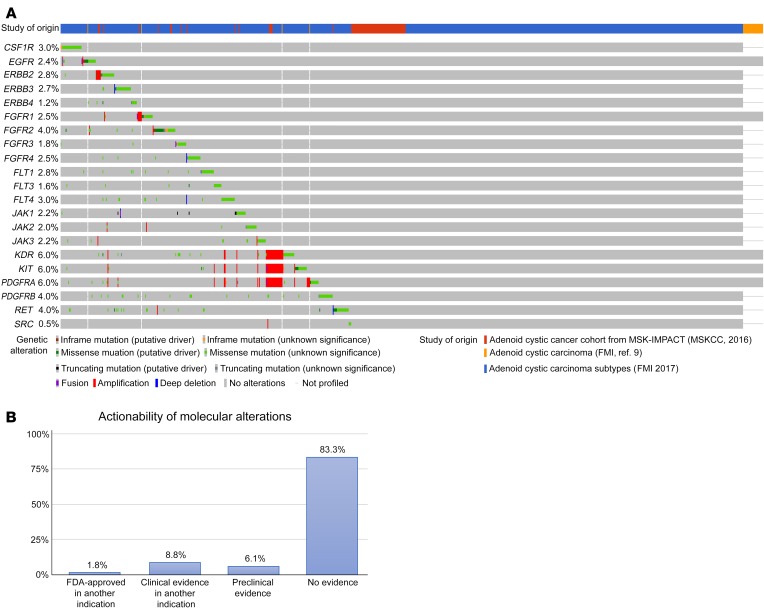
The most common alterations in ACCs are translocations involving MYB or MYBL1, which are not currently targetable. Therefore, at present, the majority of potentially actionable alterations in ACCs are tyrosine kinases, with 346 (40.3%) R/M ACC tumors harboring mutations in genes potentially targetable with available kinase inhibitors (Figure 5A). However, clinical evidence that these targets are clinically actionable remains investigational (14). Only 10.6% of alterations in ACCs in this study had evidence of clinical activity with currently approved or investigational drugs, based on annotation of all mutations in the cohort using OncoKB (Figure 5B).
Figure 5. R/M ACC tumors that harbor gene mutations potentially targetable with available kinase inhibitors.
(A) Oncoprint of R/M ACC genomic alterations stratified by genes potentially targeted by tyrosine kinase inhibitor agents, encompassing 40.3% of R/M ACC cohort. Unpublished data outlined in Supplemental Table 1. (B) Incidence of ACC patients with potentially targetable alterations, derived from OncoKB database. Levels of evidence based upon FDA labeling, NCCN guidelines, expert group recommendations, and biologic response in scientific literature. FMI, Foundation Medicine, Inc.
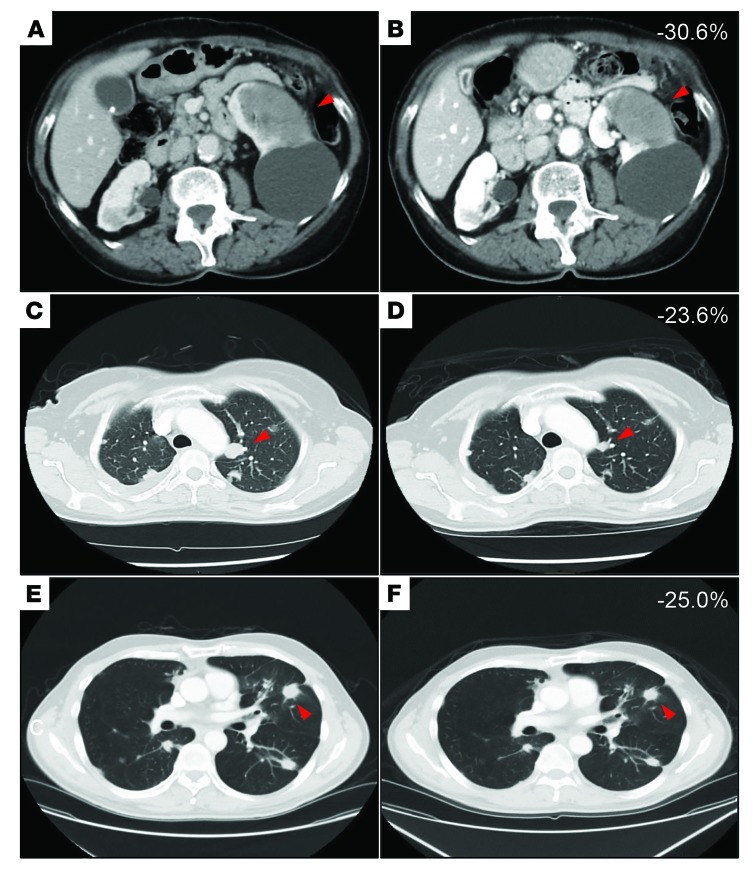
Because of the rarity of this tumor and its diverse profile of molecular alterations, genomic profiling may be invaluable in identifying specific mutations that can lead to clinical benefit among patients being considered for targeted therapeutic approaches. In the MSK-IMPACT cohort (94 patients), 6 of 8 patients with PIK3CA hot-spot mutations were enrolled in phase I basket trials investigating taselisib (a β-sparing PI3K inhibitor) (ClinicalTrials.gov NCT01296555) or alpelisib (an α-specific PI3K inhibitor) (ClinicalTrials.gov NCT01928459). Eligibility required solid tumors with documented PIK3CA mutation and disease progression after at least 1 treatment regimen, with no other available regimens known to provide clinical benefit. In both trials, the primary end point was tolerability and safety. After 2 months on treatment, 5 of 6 (83.3%) enrolled patients had stable disease (SD) and 1 (16.7%) patient had partial response (PR) by RECIST, version 1.1, criteria (15). Five (83.3%) patients displayed tumor-volume reduction (mean 18.8%, range 4.8%–30.6%) while 4 patients (66.7%) had clinical benefit defined as PR or SD lasting more than 6 months (clinical histories and imaging details are provided in Figure 6). Examination of the mutational landscape of all 6 tumors did not reveal clear genetic differences associated with response. These data confirm that certain clinically actionable mutations in ACC identifiable with genomic profiling may result in clinical benefit for patients, although the proportion of ACC patients eligible for such molecularly defined therapies is currently low.
Figure 6. Clinical response of representative metastatic ACC patients with tumors found to have PIK3CA mutations who were enrolled in PI3K basket trials.
Images represent axial CT scans with volume reduction by RECIST criteria. (A) Baseline imaging of a 43-year-old woman with a spleen metastasis. (B) PR after 2 months of treatment with 30.6% volume reduction. (C) Baseline imaging of a 73-year-old woman with metastatic ACC in the left lung. (D) SD after 2 months of treatment with 23.6% volume reduction. (E) Baseline imaging of a 58-year-old man with metastatic ACC in the left lung. (F) SD after 1 month of treatment with 25.0% volume reduction.
Intratumor genetic heterogeneity in ACC.
In 58 ACC samples, we had sufficient WES data available to infer intratumor genetic heterogeneity (ITH), describing clonal and subclonal population (SCP) structure using Fraction and Copy Number Estimate from Tumor/normal Sequencing (FACETS) (16) and PyClone (17). Of these samples, 34.5% had evidence of ITH, with 2 or more clusters of mutationally defined SCP and a mean of 1.5 SCPs per tumor. Furthermore, 6.9% contained 3 or more SCPs (Supplemental Figure 5 and Supplemental Table 7). This distribution of ITH is similar to that observed in hormone receptor–positive breast cancers analyzed with the same methodology (18). These analyses of clonality are performed on bulk sequencing samples and would be expected to lack sensitivity for small SCPs and geographic genetic heterogeneity, thereby likely underestimating the true degree of ITH in ACCs.
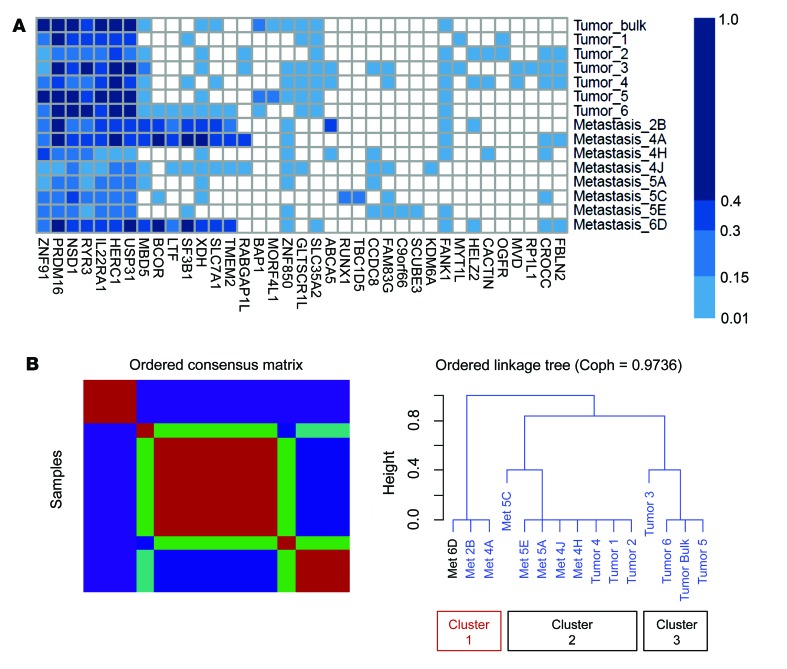
To examine genetic heterogeneity in greater depth, we studied several R/M ACC patients in whom we were able to analyze sequencing data from more than one region and infer mutation clonality using FACETS and PyClone. We performed multiregion WES followed by deeper targeted validation sequencing on a clinically aggressive salivary ACC (primary tumor specimen, 6 subregions of the primary tumor, and 8 distinct lung metastases) (Supplemental Figures 6 and 7). The MYB-NFIB fusion was confirmed to be clonally present via FISH in all regions of the primary and metastatic tumors (Supplemental Figure 8). A total of 16 validated mutations were identified in the primary tumor, consistent with typical somatic mutational load in ACCs (0.53 mutations/megabase). Deep sequencing via orthogonal targeted NGS, with mean target coverage of ×5406 in primary tumor lesions and ×1556 in metastatic lesions confirmed a total of 36 unique mutations across all sites. There was marked genetic heterogeneity and evidence for branching evolution, with only 7 of 36 mutations present in all regions. Of note, 70% (18/26) of the mutations observed in metastatic sites were present in only 1–3 of the 6 subregions in the primary tumor, and 4 mutations were not detected in any part of the primary tumor (Figure 7A). Nonnegative matrix factorization (NMF) of cancer cell fractions (CCFs) inferred by PyClone defined 3 distinct clusters, with primary tumor subregions 1, 2, and 4 most closely related to metastases 5C, 5E, 5A, 4J, and 4H. In contrast, metastases 6D, 2B, and 4A appeared to be significantly divergent (cluster 1) (Figure 7B and Supplemental Figure 9). These findings suggest that certain geographic regions of tumor were more closely related to certain metastatic sites, while other metastatic sites were yet more divergent from the primary tumor. Distinct validated mutations in this cluster would be considered subclonal and included SF3B1, XDH, LTF, and TMEM2, all of which have been implicated in metastasis and poorer prognosis (19–22).
Figure 7. Multiregion sequencing and clonality analysis in a single salivary ACC patient with subsequent lung metastases.
(A) Mutation heatmap demonstrating CCF for each mutation in each site. Primary tumor, 6 subspatial sites, and 8 distant lung metastatic lesions were biopsied and evaluated via high-depth sequencing and orthogonal validation. (B) NMF analysis defines 3 distinct subgroups. Cluster 1 (metastases 6D, 2B, and 4A) diverges significantly from clusters that comprise the primary tumor (bulk) or the tumor subspatial regions. Coph, cophenetic correlation coefficient.
We also analyzed multiregion sequencing data from 2 cases of breast ACC undergoing transformation to high-grade triple-negative breast cancer previously reported by Fusco et al. (23). In both cases, the MYB-NFIB fusion was present and clonal throughout all sampled regions, and in 1 case, a clonal NOTCH1 mutation was present in both regions, indicating the importance of these alterations to tumorigenesis. Concurrently, distinct subclonal mutations unique to high-grade regions were identified, supporting the emergence of subclonal mutations associated with progression to aggressive histology (Supplemental Figure 10).
These findings demonstrate that the “quiet” genome of ACCs, with a low mutational load, belies a more genetically heterogeneous tumor, with subclonal mutations detectable in a subset (34.5%) of tumors sequenced in bulk at standard depth and evidence of branching evolution across spatial regions within the primary tumor and during the development of metastases. In the 1 multiregion sequenced case, the number of mutations discernible in bulk tumor sequencing (even at a very high depth of ×5000) represented less than half of the mutations observed in aggregate across all tumor sites. All 3 studied cases, despite having the canonical MYB-NFIB fusion (presumed to be the early, underpinning driver of tumorigenesis), displayed clonal evolution involving the acquisition and selection of recognized mutations that likely in turn support disease progression. Such findings are consistent with that of spatiotemporal mutational gain by Liu et al. (24) and suggest that the propensity of ACCs to metastasize may be attributable to a surprising degree of tumor genetic heterogeneity, an essential substrate for branching evolution in cancer.
ACCs harbor distinct pathogenic germline mutations.
Germline DNA was analyzed for 90 R/M ACC cases at MSKCC in an anonymized fashion. A total of 38 pathogenic germline mutations were identified, with a mean 0.42 pathogenic variants per sample (Supplemental Table 8). Notably, 33 (36.7%) R/M samples were observed to harbor at least 1 pathogenic germline mutation. The vast majority comprised splicing mutations (94.7%), while frameshift insertions (5.3%) made up the remainder. Thirty-one mutations (34.4%) were found to be potentially clinically actionable, and 30 (33.3%) were high penetrance in nature. Twenty mutations (22.2%) represented germline alterations in DNA-repair pathways; specifically, 4 samples (4.4%) were found to harbor mutations in BRCA1 (c.5152+20T>A, c.5406+8T>C, c.5332+3A>G) or BRCA2 (c.9257–16T>C). Further allele-specific copy number analysis of those cases with BRCA1/2 germline alterations showed no evidence of a second hit, either mutation or loss of the second allele (Supplemental Table 9). Further analysis will be needed to better understand the penetrance of these germline alterations with respect to salivary tissue.
Also of interest, 15 (16.7%) patients harbored MLH1 or MSH6 germline mutations, with implications for microsatellite instability or deficient mismatch repair. However, additional analyses with MSISensor (25) found all cases to be microsatellite stable (Supplemental Table 10), and none exhibited high mutational burden.
Discussion
The innate challenge in ACC management is addressing its high propensity for relapse: no standard treatments exist for R/M disease, and cytotoxic chemotherapy has shown limited efficacy. This treatment resistance is further compounded by ACC’s rarity and its unpredictable, heterogeneous spectrum of disease virulence. Until now, clinical and translational research in ACC has been limited by an incomplete understanding of the molecular alterations associated with aggressive disease: in a tumor with such a quiet genome, what factors drive high rates of relapse and distant metastasis? Here, we present a molecular analysis of the largest cohort to date of R/M ACC and compare it with a sizeable cohort of primary ACC, extending initial observations and identifying relationships that may help guide the next steps of clinical investigation.
We identified a number of mutations in cancer genes that were enriched in R/M ACCs (Figure 1), which belies the standard description of ACC as a tumor with a quiet genome. Compared with primary ACC tumors, in which NOTCH1 mutations are uncommon, we observed evidence of strong selective pressure for NOTCH1 mutations in R/M cases, 26.3% of which harbored this alteration (18.3% had activating mutations). Similarly, multiple genes involved in chromatin modification (KDM, MLL, ARID family of genes) were enriched for mutation in R/M cases (12.8%–15.2%).
NOTCH1 has been studied as both an oncogene and tumor suppressor across numerous tumor types, and dysregulation of its signaling pathway is increasingly recognized as a driver for ACC pathogenesis. Activating NOTCH1 mutations have been previously observed in one cohort as associated with poor prognosis in ACC, including solid histology, liver/bone metastasis, and decreased survival (8). We similarly observed that activating NOTCH1 mutations are associated with poor prognosis (Figure 4) and, furthermore, identifed alterations in other members of the Notch family (NOTCH2, NOTCH3, NOTCH4) (3.6%–5.7%) as well as genes that activate the Notch-signaling pathway, including SPEN (6.7%) and FBXW7 (3.8%). Collectively, such findings solidify the Notch pathway as a central mediator of ACC pathogenesis. Given the augmented presence of Notch alterations in the R/M setting and the association with poor prognosis, these mutations may more specifically facilitate metastatic relapse or mediate disease aggressiveness. The use of γ secretase inhibitors, which prevent cleavage and impede Notch activation, is therefore a promising therapeutic agent. The activity of these agents in Notch-mutant ACCs is currently under active investigation (ClinicalTrials.gov NCT03691207, NCT03422679, NCT01695005).
One possibility suggested by data in this cohort is that of cooperativity between NOTCH1 and chromatin-remodeling pathways, considered gatekeepers to cellular homeostasis and also tumorigenesis (26). While mutations within this family have been previously reported, the consistently higher rate of alterations across multiple key genes (e.g., KDM6A, ARID1A, ARID1B, MLL2, MLL3, BCOR, CREBBP, EP300) among our R/M cases suggests a contributing role to ACC progression. CREBBP and EP300 in particular are known coactivators of MYB, binding to its central transactivation domain to modulate its function (27). Correlative histone aceytyltransferases and demethylases are similarly involved in recruiting NOTCH1 transcriptional regulators during the binding of the Notch transcriptional complex (NTC) with the Notch intracellular domain (NICD) (28–30). We observed a striking cooccurrence between mutations in NOTCH1 and a number of chromatin-remodeling genes (Figure 3A). Cooccurrence between 2 alterations may imply possible biological synergy, whereby only dysfunction in both genes may lead to cancer (31, 32). It is also possible that detection of such enriched cooccurrence would be greater in the R/M phenotype, having undergone selective pressure via prior treatment. Regardless, such findings of chromatin regulator dysfunction partnered with NOTCH1 alterations in ACC suggest that epigenetic mechanisms may promote progression via Notch pathway manipulation. Notably, recent work describes Notch’s dynamic interaction with epigenetic “pioneers” (upstream factors that modulate repressed chromatin states), either as itself a pioneer or as a “settler” (a downstream factor that regulates elements made accessible by other factors) (33).
Conversely, we observed NOTCH1 and TERT promoter mutations to occur in a highly mutually exclusive fashion (q = 3.3 × 10–4) (Figure 3, A and B). Telomerase activity, largely inactive in normal somatic cells, is reconstituted in a wide spectrum of malignancies and enables cancer cells to evade senescence (34). It is interesting that within the context of cancer, NOTCH1 and TERT engage in largely distinct biologic pathways. The enrichment of TERT promoter mutations in R/M ACC cases, independent of NOTCH1 dysregulation, lends compelling support to the existence of a unique TERT-driven subgroup that may benefit from distinct treatment strategies (35).
It is important to note that, although MYB/MYBL1 alterations are prevalent in R/M cases, we did not observe clear associations among MYB status, R/M status, and survival (36). Yet by integrating the above findings, we observed that a network of alterations targeting MYB, NOTCH1, and TERT defines several molecular subgroups that together account for the majority of R/M ACC tumors: MYB+NOTCH1+, MYB+/other, MYBWTTERT+, and MYBWTNOTCH1+ (Figure 3, B and C). Broad therapeutic progress in treating R/M ACC will undoubtedly depend on efforts to target each of these key genes.
In addition, more than 40% of R/M cases contained mutations in tyrosine kinases, potentially targetable with clinically available kinase inhibitors. However, only a fraction of these alterations have clinical evidence supporting their use as biomarkers in cancer. One phase II study of the multikinase inhibitor axitinib demonstrated potential benefit for ACC patients with 4q12 amplification (37). More recently, 2 phase II studies of the multikinase inhibitor lenvatinib in patients with R/M ACC reported an overall response rate of 16%–27% (38, 39), confirming that this approach has the potential to demonstrate clinical benefit in ACC patients. These data suggest that therapeutic approaches that are not directed by genomic data may have limited benefit in tumors such as ACC and that predictive biomarkers are needed to improve response rates.
Another factor driving ACC tumor progression is ITH, which we found was more common than might be expected for a tumor type that generally harbors so few mutations. ITH in cancers results from the expansion of SCPs under selective pressure. More broadly, ITH may be an indication of a tumor’s fitness for evolutionary adaptation and at high levels associates with poorer survival across diverse cancer types (18). These later subclonal events appear to occur independently of early or “trunk” alterations, which may initiate pathogenesis but not propagate it. Despite a quiet genome, evidence from these cases indicates that many primary ACCs nonetheless harbor SCPs that approximate this model of evolution (24) and may drive progression (Supplemental Figure 5). Even in tumors sequenced in bulk at standard depth, there was clear evidence of ITH in approximately one-third of cases, with a distribution very similar to that observed in hormone receptor–positive breast cancer (18).
The multiregion molecular portrait of the primary and metastatic tumor samples from one patient captured over time further illustrated this genomic evolution, where MYB-NFIB fusion was a clonal event, yet subclonal events were associated with metastatic spread. Deep sequencing and NMF clustering analysis depicted 1 cluster comprising exclusively metastatic lesions with distinct mutations (SF3B1, XDH, LTF, TMEM2) that may promote metastasis (Figure 7). SF3B1 mutations are well established in cancer, contributing to alternative splicing and implicated in late metastasis in uveal melanoma (19), while XDH mutations have been shown to promote migration and invasion in hepatocellular carcinoma (20). LTF mutations affect cell motility, while TMEM2 (also known as CEMIP2) mutations appear to confer proinvasive and promigratory potential: both genes are known to be associated with breast cancer metastasis (21, 22). Collectively, such SCPs that drive relapse and metastasis may illustrate why known players such as MYB themselves do not appear to confer poor prognosis (40, 41) or comprehensively explain ACC’s heterogeneous, protracted disease course.
A final, previously unexplored aspect of ACC involves pathogenic germline alterations. We focused our analysis on R/M patients with tumor and matched normal tissue, employing a targeted 76-gene panel that incorporates cancer susceptibility targets. Such selective germline analysis has been suggested for those tumor types with high predisposition, such as prostate cancer (42), and universal germline testing has been proposed for certain malignancies (43). Interestingly, nearly one-third of R/M ACC patients in our study harbored a pathogenic germline variant. The majority of these alterations would not have been detected through standard genetics evaluation and testing based on current clinical guidelines. While anonymization of our data precluded further correlative analyses with clinical outcomes, such knowledge of these additional mutations may further influence preventive or therapeutic decision making. We caution that only a subset (90 patients) of the total cohort underwent germline sequencing and only in an anonymized fashion. Moreover, to date, only one potentially familial case has been reported (44) in what is otherwise considered a noninheritable malignancy, and the penetrance of these alterations in salivary tissues is not known. Although 4 mutations in BRCA1/2 were identified, these appeared monoallelic and therefore have unclear clinical relevance. These preliminary results require further validation in additional data sets before recommending universal germline analysis in ACC patients.
A number of caveats limit the generalizability of our findings. The lack of clinical data for many cases, including stage, histology, type of metastasis, and survival outcomes, precluded more detailed clinical analyses. Because ACC is an orphan disease, sequenced tumors were contributed by a number of institutions and sequenced on different WES, WGS, and targeted NGS platforms. Although our downsampling analysis confirmed that sequencing depth did not seem to affect sensitivity of mutation detection, we note that the presence of these different platforms limited our ability to analyze certain features, such as TMB or germline mutations in all tumors.
In addition, MYB/MLB1 status, a known driver of ACC pathogenesis, though not of prognosis, was not available for all patients. As noted above, MYB rearrangements were primarily assessed on the basis of breakpoints in intron 14 of MYB. While this would be expected to capture the vast majority of MYB fusions involving NFIB or other partners (5), it is now understood that most ACC tumors overexpress MYB, even in the absence of MYB-NFIB fusions, either through other fusion partners, MYBL1 fusions, or other mechanisms (41, 45, 46). Therefore, the cases categorized as MYBWT are most accurately considered as negative for translocations involving MYB intron 14, but some cases may overexpress MYB through other means. MYBL1 in particular was not assessed in NGS panels. Nevertheless, we found that the degree of mutual exclusivity between MYB/MYBL1 and TERT promoter mutations remained unchanged in those samples assessed for translocations more comprehensively with FISH, indicating that profiling of intron 14 is likely to be representative of interactions among the common alterations in ACC. Ultimately, these observations synthesize the largest data set of ACC to date, identifying molecular relationships and pathways to better characterize and target an enigmatic, understudied cancer.
In summary, we identify key genomic drivers in a large-scale R/M ACC analysis, highlighting enrichment in NOTCH signaling, chromatin-remodeling pathways, and TERT promoter mutations among others that underlie ACC oncogenesis. We also demonstrate compelling evidence of genetic evolution and intratumoral heterogeneity despite a quiet genome, which may facilitate the metastatic relapse so commonly observed in ACC. Finally, we identified pathogenic germline mutations in nearly one-third of patients with available data. These alterations may be cooperative or mutually exclusive, underscoring the value of multidimensional, coordinated profiling to advance our understanding of ACC progression. Such insights into ACC’S genomic interplay should help further steer rational therapeutic strategies and ultimately improve patient outcomes.
Methods
We analyzed DNA-sequencing data from unpublished and published cohorts of ACC tumors, in total 10 different data sets (177 primary, 858 R/M cases). Unpublished data sets included those from Sanger/MD Anderson, MSKCC, MSK-IMPACT, and Foundation Medicine (Supplemental Figure 1 and Supplemental Table 1). Published data sets included those of Ho et al. (3), Stephens et al. (4), Martelotto et al. (7), Rettig et al. (9), Mitani et al. (36), and Ross et al (47). For unpublished and published cohorts, each case was verified histologically as ACC (3, 4, 7, 9, 36, 47). Tumor tissue was either snap-frozen or formalin-fixed, paraffin-embedded (FFPE), and cohorts included sequencing data that were collected via WES, WGS, or targeted NGS panels (Supplemental Table 1).
Mutation-analysis pipeline.
In addition to published WES and WGS data, 51 additional samples were subjected to WES. For 16 samples at MSKCC, library preparation and sequencing were performed as previously described via an Illumina HiSeq sequencer using 2 × 125 bp cycles (48). Matching between tumor and normal samples was confirmed via an in-house panel of 118 SNPs together with VerifyBamID (49). Alignment to the hg19 genome build was performed using BWA, version 0.7.10 (50). Insertion/deletion (indel) realignment, recalibration, and duplicate removal were performed using GATK, version 3.2.2 (51). Four independent mutation programs were used to identify single nucleotide variants (SNVs): MuTect; SomaticSniper, version 1.0.4.2; Strelka, version 1; and VarScan, version 2.3.8 (52–55). All potential SNVs and indels underwent orthogonal validation using NimbleGen SeqCapEX target enrichment (Roche), with ×500 and ×250 sequencing depth for tumor and normal DNA, respectively. Allele-specific copy number data were acquired using OncoSNP-SEQ (56). Significant amplifications/deletions and chromosome arm-level alterations were determined using GISTIC 2.0 (57).
For 35 samples at MDACC, tumor and matched normal samples underwent sequencing as previously described via unchained combinatorial probe anchor ligation chemistry with self-assembling DNA nanoball arrays (24). Primary variant calls were made via Cancer Pipeline, version 2.2 (Complete Genomics). Calls were further assessed by the Ingenuity Variant Analysis tool (QIAGEN) and filtered through dbSNP, version 129 (58), the 1000 Genomes Project (59), the NHLBI Exome Sequencing Project (60), and Public Genome Data from Complete Genomics (http://www.completegenomics.com/public-data). Targeted validation sequencing was performed using SureSelect target enrichment (Agilent Technologies) with a probe library designed with 42,087 probes. Enriched target libraries were sequenced on the Illumina HiSeq platform with ×600 median read depth and mapped to the hg19 genome build using BWA. SNV validation was made with GATK (51) and MuTect (52), with VAF of 5% or more in tumor and 5% or less for normal.
Targeted sequencing cohort pipeline.
A total of 94 R/M tumors from patients undergoing treatment at MSKCC were analyzed via Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), a targeted NGS platform that sequences DNA extracted from FFPE tumor samples and matched normal DNA in an environment certified by the Clinical Laboratory Improvement Amendments (CLIA) and approved by the New York State Department of Health. Sequencing both tumor and matched normal DNA in all cases, the assay identifies somatic alterations including SNVs, indels, and structural variants in 410 or 468 genes functionally relevant to cancer or clinically actionable targets and genome-wide copy number. Captured DNA libraries were sequenced to a median exon coverage depth of greater than ×600 using Illumina HiSeq platforms (61).
Additionally, 730 R/M cases were analyzed via Foundation Medicine, a laboratory certified by CLIA, accredited by the College of American Pathologists (CAP), and approved by New York State, as described previously (9, 62). Briefly, 50 ng DNA or more was extracted from FFPE archival tissue containing 20% or greater tumor cell content. Adaptor-ligated DNA underwent hybrid capture for the entire coding region of selected cancer-related genes plus select introns from genes frequently rearranged in cancer (FoundationOne). Mutational prevalence was adjusted for the gene composition of the FoundationOne panel used. Captured libraries were sequenced to a median exon coverage depth of more than 500× using Illumina HiSeq platforms. Resultant sequences were analyzed for short variants (base substitutions, indels), copy number alterations (focal amplifications and homozygous deletions), and select gene rearrangements using the hg19 reference genome (63).
OncoKB (64) was used to categorize missense SNVs as likely oncogenic or of unknown significance. Missense mutations categorized as variants of unknown significance (VUS) in the subset of sequenced samples lacking matched normal DNA sequencing were additionally filtered, including SNVs that were additionally annotated as potentially functional by available databases. Specific parameters for inclusion required one or more of the following annotations: likely oncogenic (OncoKB; ref. 64), medium or high functional impact (MutationAssessor; ref. 65), deleterious/deleterious_low_confidence (Sorting Intolerant from Tolerant [SIFT]; ref. 66), or possibly damaging/probably damaging (PolyPhen2; ref. 67).
Downsampling analysis.
To ensure that R/M mutation data were not enriched simply from deeper sequencing, MSK-IMPACT BAM files for each of the samples were randomly downsampled using samtools to generate 5 independent and distinct BAM files, each with an average coverage of ×100. The downsampled BAM files were then genotyped using pileup (samtools) for mutations detected above the same variant calling thresholds used on the original MSK-IMPACT bam files. The results were tabulated and plotted as a comparison of VAF between downsampled BAM files and the original IMPACT BAM files (Supplemental Figure 4).
Intratumoral genetic heterogeneity.
Using exome and copy number data available from 58 ACCs, tumor purity and allele-specific copy number segmented data were generated using FACETS, version 0.5 (16). This output, together with variant allele frequencies for somatic mutations, was used to estimate cancer cell prevalence for each somatic mutation in a given sample using PyClone, version 0.13.0 (17). All samples harboring 5 or more mutations were included, and intratumor heterogeneity was quantified as the number of genomically distinct clonal or SCPs in each sample, where an SCP represents the outcome of a clonal expansion and is inferred from 2 or more mutations with similar cellular prevalence using a Bayesian clustering approach. A cluster was considered subclonal if the CCF upper CI was greater than 95% (18). Additional qualitative assessment of NOTCH1 mutations across all cases was carried out by plotting a variant allelic frequency (VAF) density histogram for those cases with diploid copy number in the NOTCH1 region.
Multiregion intratumoral heterogeneity in clinically aggressive ACC.
Clonal evolution was further investigated in a single adult salivary ACC patient who developed multifocal distant lung metastases (over 90 distinct lesions) 4 years after definitive therapy (parotidectomy with postoperative proton beam radiation). WES was performed on the primary tumor and 8 distant metastases, with mean target coverage of ×140 and SNVs called as described previously (3). Mutations then underwent orthogonal validation via ultra-deep sequencing in the primary tumor, 6 spatial subregions of the primary tumor, the 8 distant metastatic lesions, and matched normal DNA from peripheral blood, via a custom targeted NGS AmpliSeq library on the Ion Torrent PGM platform (68, 69). An Ion AmpliSeq custom panel was designed with an Ion AmpliSeq Designer (Thermo Fisher) for 165 amplicons targeting the regions of interest. After PicoGreen quantification and quality control by Agilent TapeStation, 20 ng of DNA was used to prepare libraries using the Ion AmpliSeq Library Kit 2.0 and Ion Xpress Barcode Adapters 1–16 Kit according to the manufacturer’s protocols, with 17 cycles of PCR and final extension of 6 minutes. Emulsion PCR was carried out with the Ion OneTouch System and Ion Hi-Q OT2 Kit according to the manufacturer’s protocol. After the emulsion PCR, template-positive Ion Sphere Particles were enriched with the Dynabeads MyOne Streptavidin C1 Beads. Sequencing was then performed with an Ion Torrent Personal Genome Machine (PGM) system using the Ion PGM HiQ Sequencing Kit and Ion 318 Chip. Mean depth of coverage was ×5406.
Sequencing data were aligned using BWA and Picard/GATK to mark duplicates, and HaplotypeCaller, version 2.3.9, was used to annotate variants (51). The ultra-deep sequencing data, together with FACETS, version 0.5 (16), was used to adjust for tumor purity and ploidy, and CCF was determined using PyClone as described above (17). All mutations with VAF greater than 0.01 and CCF less than 2% were included. NMF clustering of CCF was performed in GenePattern (70). Similar analyses of the 2 breast ACCs with high-grade triple-negative transformation were performed on their published mutation calls and copy number findings (23) using PyClone.
To assess the clonality of the MYB-NFIB translocation, FISH was performed using a 3-color probe mix consisting of bacterial artificial chromosomes (BACs) for 5′ MYB (RP11-614H6, RP11-104D9; green), 3′ MYB (RP11-323N12, RP11-1060C14; orange), and 3′ NFIB (RP11-413D24, RP11-589C16; red). Probe labeling, tissue processing, hybridization, posthybridization washing, and fluorescence detection were performed as previously described (3). FISH signals were imaged through the depth of the tissue, and a minimum of 50 discrete nuclei were analyzed within the marked regions.
Germline analysis.
Data from MSK-IMPACT sequenced R/M tumors were anonymized and underwent secondary germline analysis. Germline analysis was performed on normal DNA from peripheral blood and included 76 genes on the MSK-IMPACT panel associated with hereditary cancer predisposition, including all cancer-predisposing genes identified in the American College of Medical Genetics and Genomics (ACMG) guidelines (71). The variant calling procedures were modified for germline mutations (72) and included SNVs, indels, and CNVs (large deletions/duplications). The ACMG scoring scheme (73) was utilized for variant interpretation, where “pathogenic” (based on PVS, PS, PM, and PP scores) variants were reported. All germline mutations of established low-, moderate-, or high-risk (penetrance) were considered clinically actionable for cancer prevention, as defined by Mandelker et al. (42, 74). To evaluate for microsatellite instability, all cases were additionally analyzed with MSISensor (25). MSI scores of less than 3 were considered microsatellite stable, while MSI scores from 3–10 were considered indeterminate, and scores greater than 10 were considered microsatellite unstable. In any cases indeterminate on MSISensor, further orthogonal analysis was performed using MiMSI (75), a multiple instance machine-learning technique to analyze reads from microsatellite sites. BRCA1/BRCA2 second-hit analysis was performed via FACETS (16) by matching tumor and normal-sequencing files in an anonymized fashion by a researcher unconnected with this study.
Therapeutic actionability and clinical levels of evidence.
Each somatic alteration in the 1045 ACC tumors was separately annotated using OncoKB, a precision oncology database that annotates tumor alterations by levels of clinical evidence supporting the use of that mutant allele or other alteration as a predictive biomarker of drug sensitivity to FDA-approved or investigational agents (64).
Statistics.
Clinical, demographic, and pathologic information was collected on all 94 MSKCC R/M ACC patients undergoing treatment at MSKCC. Statistical analysis was performed using SAS 9.3 (SAS Institute). Estimated survival functions were generated via the Kaplan-Meier method and compared with the log-rank test. All statistical tests were 2 sided, and a P value of less than 0.05 was considered statistically significant.
Study approval.
Protocol approval for specimen collection and DNA sequencing was obtained by each respective institution’s IRB. Foundation Medicine received additional IRB approval, including a waiver of informed consent and a HIPAA waiver of authorization, from the Western IRB. The MSKCC IRB provided approval for the analysis of MSKCC patient data as well as the analysis of deidentified data provided from other institutions, including a data transfer agreement with Foundation Medicine. Written, informed consent was obtained from participants prior to inclusion in the study.
Data availability.
The complete genetic data from all cases will be publicly available in cbioportal.org at the time of publication (http://www.cbioportal.org/study/summary?id=acc_2019) (76, 77).
Author contributions
LGTM, NS, and TAC were responsible for study conception. LGTM and NS were responsible for data acquisition and transfer agreements. AO, GJ, AZ, VM, JSRF, BW, NS, TAC, and LGTM carried out bioinformatics analyses; ASH, JT, and LGTM carried out statistical analyses; ASH, ALH, JT, CVM, and LGTM carried out clinical data analysis; and SD and NK carried out pathologic analysis. MGD and LGTM processed biospecimens. ASH, TAC, and LGTM wrote the manuscript, and all authors interpreted data and reviewed the manuscript. JH, MB, MM, JSR, VAM, LC, IG, PT, PH, NA, DBS, PAF, and AKEN interpreted data and reviewed the manuscript. ASH and AO share first author position, with order determined alphabetically.
Supplementary Material
Acknowledgments
We would like to express our deep gratitude to the many patients and families who participated in these studies. We thank Jeffrey Kaufman, Marnie Kaufman, and Nicole Spardy Burr from the Adenoid Cystic Cancer Research Foundation for their scientific leadership and guidance. We remember Barbara Tsipouras and Gabriele Grunewald for their bravery, and we thank Petros Tsipouras and Justin Grunewald for research and patient advocacy. We thank members of the Chan Lab for helpful discussions. We acknowledge the technical support of Magali Cavatore, Kety Huberman,and Agnes Viale in the MSKCC Integrated Genomics Operation core facility, Gouri Nanjangud in the Cytogenetics core facility, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, and the Diagnostic Molecular Pathology Service at MSKCC. We acknowledge funding from the Adenoid Cystic Carcinoma Research Foundation (to NS and TAC), a Pershing Square Sohn Cancer Research grant (to TAC), the PaineWebber Chair (to TAC), Stand Up 2 Cancer (to TAC), NIH R01 CA205426, the STARR Cancer Consortium (to TAC), NCI R35 CA232097 (to TAC), the Frederick Adler Chair (to LGTM), Cycle for Survival (to LGTM), the Jayme Flowers Fund (to LGTM), the Sebastian Nativo Fund (to LGTM), NIH K08 DE024774 and R01 DE027738 (to LGTM), Cedars-Sinai Precision Health (to ASH), the Donna and Jesse Garber Award for Cancer Research (to ASH), and MSKCC through NIH/NCI Cancer Center Support Grant (P30 CA008748).
Version 1. 09/04/2019
Electronic publication
Version 2. 10/01/2019
Print issue publication
Footnotes
Conflict of interest: ALH received funding from Eisai, Bristol-Myers Squibb, Kura Oncology, AstraZeneca, Genentech/Roche, Celldex Therapeutics, Pfizer, Lilly, and Bayer; consulting fees from Bristol-Myers Squibb, Merck, Novartis, AstraZeneca, Regeneron, Sanofi Aventis, Sun Pharma, Eisai, Genentech/Roche, Sanofi Genzyme, and Ayala; and travel reimbursement from Ignyta and Kura Oncology. PH received funding from Stryker, Ethicon, and Medtronic; consulting fees from Loxo/Bayer Oncology; and travel reimbursement from Genentech. DBS received consulting fees from Pfizer, Loxo Oncology, Lilly Oncology, Illumina, Intezyne, and Vividion Therapeutics. JH, MB, MM, JSR, VAM, and LC are employees of Foundation Medicine. JSRF serves on the scientific advisory boards of Volition RX, Paige.AI, Roche Tissue Diagnostics, Ventana, Genentech, InVicro, and Novartis; and received consulting fees from Goldman Sachs (Merchant Banking) and REPARE Therapeutics. TAC is a cofounder of and holds equity in Gritstone Oncology and received research funding from Bristol-Myers Squibb, Eisai, AstraZeneca, and Illumina. LGTM received research funding from Bristol-Myers Squibb, Illumina and AstraZeneca.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4276–4289. https://doi.org/10.1172/JCI128227.
Contributor Information
Angelica Ochoa, Email: ochoaa@mskcc.org.
Gowtham Jayakumaran, Email: jayakumg@mskcc.org.
Ahmet Zehir, Email: zehira@mskcc.org.
Vladimir Makarov, Email: makarovv@mskcc.org.
Martin G. Dalin, Email: martin.dalin@gu.se.
Jie He, Email: Jhe@foundationmedicine.com.
Mark Bailey, Email: markibail@gmail.com.
Meagan Montesion, Email: mmontesion@foundationmedicine.com.
Vincent A. Miller, Email: vmiller@foundationmedicine.com.
Ian Ganly, Email: ganlyi@mskcc.org.
Snjezana Dogan, Email: dogans@mskcc.org.
Nora Katabi, Email: katabin@mskcc.org.
Petros Tsipouras, Email: Petros.tsipouras@gmail.com.
Patrick Ha, Email: patrick.ha@ucsf.edu.
Nishant Agrawal, Email: nagrawal@surgery.bsd.uchicago.edu.
David B. Solit, Email: solitd@mskcc.org.
P. Andrew Futreal, Email: AFutreal@mdanderson.org.
Britta Weigelt, Email: weigeltb@mskcc.org.
Alan L. Ho, Email: hoa@mskcc.org.
Nikolaus Schultz, Email: schultzn@mskcc.org.
References
- 1.Ali S, et al. Long-term local control rates of patients with adenoid cystic carcinoma of the head and neck managed by surgery and postoperative radiation. Laryngoscope. 2017;127(10):2265–2269. doi: 10.1002/lary.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu B, et al. Predictors of outcome in adenoid cystic carcinoma of salivary glands: a clinicopathologic study with correlation between MYB fusion and protein expression. Am J Surg Pathol. 2017;41(10):1422–1432. doi: 10.1097/PAS.0000000000000918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho AS, et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet. 2013;45(7):791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stephens PJ, et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest. 2013;123(7):2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persson M, Andrén Y, Mark J, Horlings HM, Persson F, Stenman G. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc Natl Acad Sci U S A. 2009;106(44):18740–18744. doi: 10.1073/pnas.0909114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitani Y, et al. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin Cancer Res. 2010;16(19):4722–4731. doi: 10.1158/1078-0432.CCR-10-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitani Y, et al. Novel MYBL1 gene rearrangements with recurrent MYBL1-NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin Cancer Res. 2016;22(3):725–733. doi: 10.1158/1078-0432.CCR-15-2867-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrarotto R, et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J Clin Oncol. 2017;35(3):352–360. doi: 10.1200/JCO.2016.67.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross JS, et al. Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next-generation sequencing reveals potential new routes to targeted therapies. Am J Surg Pathol. 2014;38(2):235–238. doi: 10.1097/PAS.0000000000000102. [DOI] [PubMed] [Google Scholar]
- 10.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23(6):703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallman DA, Padron E. Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol Oncol Stem Cell Ther. 2016;9(3):89–95. doi: 10.1016/j.hemonc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann AL, et al. Detailed simulation of cancer exome sequencing data reveals differences and common limitations of variant callers. BMC Bioinformatics. 2017;18(1):8. doi: 10.1186/s12859-016-1417-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu ZK, Shang YK, Chen ZN, Bian H. A three-caller pipeline for variant analysis of cancer whole-exome sequencing data. Mol Med Rep. 2017;15(5):2489–2494. doi: 10.3892/mmr.2017.6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tchekmedyian V, et al. Phase II study of lenvatinib in patients with progressive, recurrent or metastatic adenoid cystic carcinoma. J Clin Oncol. 2019;37(18):1529–1537. doi: 10.1200/JCO.18.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhauer, EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44(16):e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth A, et al. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris LG, et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7(9):10051–10063. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yavuzyigitoglu S, et al. Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123(5):1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Chen GL, et al. Xanthine dehydrogenase downregulation promotes TGFβ signaling and cancer stem cell-related gene expression in hepatocellular carcinoma. Oncogenesis. 2017;6(9):e382. doi: 10.1038/oncsis.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vecchi M, et al. Breast cancer metastases are molecularly distinct from their primary tumors. Oncogene. 2008;27(15):2148–2158. doi: 10.1038/sj.onc.1210858. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, Goodarzi H, Tavazoie SF, Alarcón CR. TMEM2 is a SOX4-regulated gene that mediates metastatic migration and invasion in breast cancer. Cancer Res. 2016;76(17):4994–5005. doi: 10.1158/0008-5472.CAN-15-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusco N, et al. Genetic events in the progression of adenoid cystic carcinoma of the breast to high-grade triple-negative breast cancer. Mod Pathol. 2016;29(11):1292–1305. doi: 10.1038/modpathol.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu B, et al. Spatio-temporal genomic heterogeneity, phylogeny, and metastatic evolution in salivary adenoid cystic carcinoma. J Natl Cancer Inst. 2017;109(10):djx033. doi: 10.1093/jnci/djx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niu B, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2014;30(7):1015–1016. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair SS, Kumar R. Chromatin remodeling in cancer: a gateway to regulate gene transcription. Mol Oncol. 2012;6(6):611–619. doi: 10.1016/j.molonc.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.George OL, Ness SA. Situational awareness: regulation of the myb transcription factor in differentiation, the cell cycle and oncogenesis. Cancers (Basel) 2014;6(4):2049–2071. doi: 10.3390/cancers6042049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oswald F, et al. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21(22):7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yatim A, et al. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48(3):445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulligan P, et al. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol Cell. 2011;42(5):689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 32.Remy E, Rebouissou S, Chaouiya C, Zinovyev A, Radvanyi F, Calzone L. A modeling approach to explain mutually exclusive and co-occurring genetic alterations in bladder tumorigenesis. Cancer Res. 2015;75(19):4042–4052. doi: 10.1158/0008-5472.CAN-15-0602. [DOI] [PubMed] [Google Scholar]
- 33.Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinagre J, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 35.Joseph I, et al. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010;70(22):9494–9504. doi: 10.1158/0008-5472.CAN-10-0233. [DOI] [PubMed] [Google Scholar]
- 36.Rettig EM, et al. Whole-genome sequencing of salivary gland adenoid cystic carcinoma. Cancer Prev Res (Phila) 2016;9(4):265–274. doi: 10.1158/1940-6207.CAPR-15-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho AL, et al. A phase II study of axitinib (AG-013736) in patients with incurable adenoid cystic carcinoma. Ann Oncol. 2016;27(10):1902–1908. doi: 10.1093/annonc/mdw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tchekmedyian V, et al. Phase II study of lenvatinib in patients with progressive, recurrent or metastatic adenoid cystic carcinoma. J Clin Oncol. 2019;37(18):1529–1537. doi: 10.1200/JCO.18.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locati L, et al. Phase II study on lenvatinib (LEN) in recurrent and/or metastatic (R/M) adenoid cystic carcinomas (ACC) of the salivary glands (SG) of the upper aereodigestive tract ( NCT02860936) J Clin Oncol. 2018;36(15):6086. doi: 10.1200/JCO.2018.36.15_suppl.6086. [DOI] [Google Scholar]
- 40.Rettig EM, et al. MYB rearrangement and clinicopathologic characteristics in head and neck adenoid cystic carcinoma. Laryngoscope. 2015;125(9):E292–E299. doi: 10.1002/lary.25356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frerich CA, et al. Transcriptomes define distinct subgroups of salivary gland adenoid cystic carcinoma with different driver mutations and outcomes. Oncotarget. 2018;9(7):7341–7358. doi: 10.18632/oncotarget.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandelker D, et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318(9):825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowery MA, et al. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms. J Natl Cancer Inst. 2018;110(10):1067–1074. doi: 10.1093/jnci/djy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Channir HI, van Overeem Hansen T, Andreasen S, Yde CW, Kiss K, Charabi BW. Genetic characterization of adenoid cystic carcinoma of the minor salivary glands: a potential familial occurrence in first-degree relatives. Head Neck Pathol. 2017;11(4):546–551. doi: 10.1007/s12105-017-0801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drier Y, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48(3):265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brayer KJ, Frerich CA, Kang H, Ness SA. Recurrent fusions in MYB and MYBL1 define a common, transcription factor-driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov. 2016;6(2):176–187. doi: 10.1158/2159-8290.CD-15-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martelotto LG, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J Pathol. 2015;237(2):179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalin MG, et al. Comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin Cancer Res. 2016;22(18):4623–4633. doi: 10.1158/1078-0432.CCR-16-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jun G, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91(5):839–848. doi: 10.1016/j.ajhg.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28(3):311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28(14):1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 55.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yau C. OncoSNP-SEQ: a statistical approach for the identification of somatic copy number alterations from next-generation sequencing of cancer genomes. Bioinformatics. 2013;29(19):2482–2484. doi: 10.1093/bioinformatics/btt416. [DOI] [PubMed] [Google Scholar]
- 57.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ross JS, et al. Comprehensive genomic profiles of metastatic and relapsed salivary gland carcinomas are associated with tumor type and reveal new routes to targeted therapies. Ann Oncol. 2017;28(10):2539–2546. doi: 10.1093/annonc/mdx399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chakravarty D, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 67.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Millat G, Chanavat V, Rousson R. Evaluation of a new NGS method based on a custom AmpliSeq library and Ion Torrent PGM sequencing for the fast detection of genetic variations in cardiomyopathies. Clin Chim Acta. 2014;433:266–271. doi: 10.1016/j.cca.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 69.Nishio SY, Hayashi Y, Watanabe M, Usami S. Clinical application of a custom AmpliSeq library and ion torrent PGM sequencing to comprehensive mutation screening for deafness genes. Genet Test Mol Biomarkers. 2015;19(4):209–217. doi: 10.1089/gtmb.2014.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 71.Kalia SS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 72.Cheng DT, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics. 2017;10(1):33. doi: 10.1186/s12920-017-0271-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hampel H, Bennett RL, Buchanan A, Pearlman R, Wiesner GL, Guideline Development Group. American College of Medical Genetics Genomics Professional Practice Guidelines Committee National Society of Genetic Counselors Practice Guidelines Committee A practice guideline from the American College of Medical Genetics and Genomics and the National Society of Genetic Counselors: referral indications for cancer predisposition assessment. Genet Med. 2015;17(1):70–87. doi: 10.1038/gim.2014.147. [DOI] [PubMed] [Google Scholar]
- 75. Ziegler J. Improved microsatellite instability detection on next-generation sequencing data utilizing deep multiple instance learning. Paper presented at: 30th Anniversary AACR Special Conference Convergence: Artificial Intelligence, Big Data, and Prediction in Cancer; October 14–17, 2018; Newport, Rhode Island, USA. http://www.aacr.org/Meetings/Pages/MeetingDetail.aspx?EventItemID=149 Accessed July 24, 2018.
- 76.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genetic data from all cases will be publicly available in cbioportal.org at the time of publication (http://www.cbioportal.org/study/summary?id=acc_2019) (76, 77).