Abstract
Serine-rich splicing factor 3 (SRSF3) plays a critical role in liver function and its loss promotes chronic liver damage and regeneration. As a consequence, genetic deletion of SRSF3 in hepatocytes caused progressive liver disease and ultimately led to hepatocellular carcinoma. Here we show that SRSF3 is decreased in human liver samples with nonalcoholic fatty liver disease (NAFLD), nonalcoholic steatohepatitis (NASH), or cirrhosis that was associated with alterations in RNA splicing of known SRSF3 target genes. Hepatic SRSF3 expression was similarly decreased and RNA splicing dysregulated in mouse models of NAFLD and NASH. We showed that palmitic acid–induced oxidative stress caused conjugation of the ubiquitin-like NEDD8 protein to SRSF3 and proteasome-mediated degradation. SRSF3 was selectively neddylated at lysine 11 and mutation of this residue (SRSF3-K11R) was sufficient to prevent both SRSF3 degradation and alterations in RNA splicing. Lastly, prevention of SRSF3 degradation in vivo partially protected mice from hepatic steatosis, fibrosis, and inflammation. These results highlight a neddylation-dependent mechanism regulating gene expression in the liver that is disrupted in early metabolic liver disease and may contribute to the progression to NASH, cirrhosis, and ultimately hepatocellular carcinoma.
Keywords: Endocrinology, Hepatology
Keywords: Liver cancer, Obesity, RNA processing
Introduction
Serine-rich splicing factor 3 (SRSF3) is the smallest member of the SR protein family (1, 2) that functions to promote RNA splicing by recruiting components of the spliceosome at constitutive and alternatively spliced exons (3, 4). SRSF3 has also been ascribed a number of cellular functions including controlling cellular proliferation, as it is regulated during G1/S by the E2F transcription factor (5) and controls the G2/M transition of immortal rat fibroblasts (6). These findings have led to the suggestion that SRSF3 is a proto-oncogene and, indeed, it is overexpressed in some tumors (7). Previously, we demonstrated that SRSF3 regulates splicing of the insulin receptor gene (INSR) (8) and plays an important role in hepatocyte differentiation and hepatic glucose and lipid metabolism (9). Furthermore, we have shown that hepatocyte-specific deletion of the SRSF3 gene (SRSF3H-KO) promotes hepatocellular carcinoma (HCC) in aged SRSF3H-KO mice (10, 11), and we and others observed that SRSF3 is reduced in samples of human HCC (10, 11).
The global increase in obesity over the past few decades has caused a corresponding increase in nonalcoholic fatty liver disease (NAFLD) and its more severe form, nonalcoholic steatohepatitis (NASH) (12–16). Both NAFLD and NASH are associated with liver insulin resistance, nonalcoholic cirrhosis, and the development of HCC (17, 18). HCC is currently ranked as the fifth most common cancer worldwide (19, 20) and has a high mortality. It usually arises after years of liver disease and inflammation (19) either due to chronic hepatitis B or C virus (HBV/HCV) infection (21), or alcoholic and nonalcoholic cirrhosis.
The finding that loss of SRSF3 triggers metabolic changes, hepatic fibrosis, and increased liver inflammation (10, 11) that are all features of early liver disease led us to propose that SRSF3 expression might be sensitive to hepatic stress. Indeed, studies have shown that SRSF3 is ubiquitinated under both normal and stressed conditions (22–24). Additionally, it has been reported that SRSF3 is conjugated to the NEDD8 protein, a process termed neddylation, under arsenite-induced stress that is important for stress granule assembly (25). Therefore, we investigated whether SRSF3 is lost in early liver disease. We found that SRSF3 protein expression is lost in early liver disease, including NAFLD, NASH, and cirrhosis, in both humans and mice. We report here that SRSF3 is degraded in the proteasome in response to lipid-induced oxidative stress via neddylation on lysine 11 and that prevention of SRSF3 loss inhibited many of the deleterious changes observed previously. These results identify a neddylation-dependent pathway regulating liver health and may provide novel therapeutic targets that may aid in the treatment of liver diseases.
Results
SRSF3 is reduced in early-stage human liver disease, and cirrhosis.
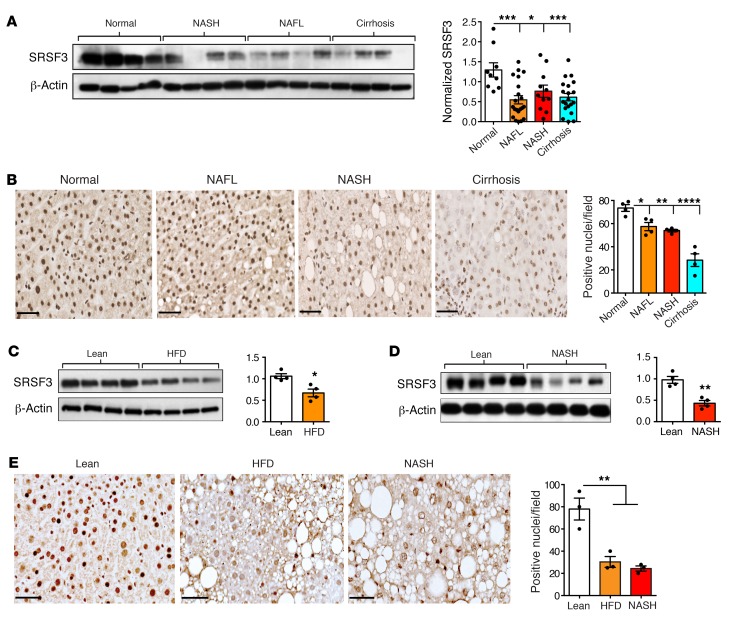
Previously, we reported that hepatocyte-specific deletion of SRSF3 in mice impaired hepatocyte maturation leading to HCC as the mice aged, and that SRSF3 protein expression, but not mRNA expression, was lost in samples of human adenoma and HCC (10, 11). To test whether SRSF3 loss preceded adenoma and/or carcinoma formation, we assessed SRSF3 protein expression in extracts from frozen samples of normal human liver (n = 9), and pathologically confirmed cases of NAFLD (n = 21), NASH (n = 11), and cirrhosis (n = 20). All samples from NAFLD, NASH, and cirrhosis showed reduced levels of SRSF3 protein compared with normal liver by immunoblotting (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI127374DS1). A similar reduction was seen in male and female samples (Supplemental Figure 1B). We confirmed these results by immunohistochemical (Figure 1B) and immunofluorescent (Supplemental Figure 2A) staining of fixed liver sections. The majority of hepatocyte nuclei in normal liver samples stained strongly for SRSF3, and cytoplasmic staining was also observed in some cells, consistent with the reported shuttling of SRSF3 from the nucleus to the cytoplasm during mRNA export (26). In contrast, fewer nuclei in sections from NAFLD and NASH livers stained for SRSF3 and the staining was weaker (Figure 1B). Sections from cirrhotic livers showed very low staining for SRSF3 (Figure 1B).
Figure 1. Loss of SRSF3 expression in NAFLD, NASH, and cirrhosis.
(A) Immunoblotting of SRSF3 in extracts from 4 representative human liver samples from normal individuals and individuals with NASH, NAFL, or cirrhosis. Graph shows quantification of SRSF3 protein level in human liver samples normalized to β-actin (n = 9, 21, 11, and 20 for normal [white], NAFL [orange], NASH [red], and cirrhosis [cyan], respectively). (B) Immunohistochemical staining for SRSF3 on FFPE sections from normal, NAFL, NASH, and cirrhotic livers. Graph shows quantification of SRSF3-positive nuclei per field (brown stain) (n = 4/group). Colors are as in panel A. Scale bars: 50 μm. (C) Immunoblotting of SRSF3 from hepatocytes from obese mice on high-fat diet (HFD) or lean mice on normal chow (Lean). Graph shows quantification of SRSF3 protein levels normalized to β-actin (n = 4/group). Lean mice shown in white, obese mice (HFD) shown in orange. (D) Immunoblotting of SRSF3 from hepatocytes from obese mice on NASH diet (NASH) or lean mice on normal chow (Lean). Graph shows quantification of SRSF3 protein levels normalized to β-actin (n = 4/group). Lean mice shown in white, NASH mice shown in red. (E) Immunohistochemical staining for SRSF3 on FFPE liver sections from lean, obese HFD, or obese NASH diet mice. Graph shows quantification of SRSF3-positive nuclei/field (brown stain) (n = 4/group). Colors are as in panel A. Scale bars: 50 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Tukey’s post hoc testing. All quantified results are presented as mean ± SEM.
SRSF3 is lost in mouse models of NAFLD and NASH.
Having observed the loss of SRSF3 in early-stage liver disease in humans, we wanted to test whether this phenotype could be modeled in mice. We placed mice on a high-fat diet (HFD, 60% fat) for 12 weeks to allow mice to become obese and cause hepatic steatosis. Body weight, liver weight, spleen weight, and fasting glucose levels were all higher in mice on HFD as compared with lean mice (Supplemental Figure 3D). Morphologically, the livers from mice on HFD were larger and paler than livers from lean mice and had a roughened surface (Supplemental Figure 3, A and B). Examination of liver sections from mice on HFD revealed extensive steatosis but with minimal fibrosis restricted to the perivascular regions that was consistent with NAFLD (Supplemental Figure 3B). To examine whether NAFLD caused loss of SRSF3 in hepatocytes, rather than other cells resident in the liver, we purified primary hepatocytes from obese (HFD) and lean (normal chow) mice and immunoblotted cell extracts for SRSF3. We observed loss of SRSF3 in hepatocytes from the HFD mice (Figure 1C) that agreed with the reported loss of SRSF3 in whole liver from HFD mice (27). We also assessed SRSF3 protein expression by immunohistochemistry (Figure 1E) and immunofluorescence (Supplemental Figure 2C) on fixed liver tissue sections from HFD and lean mice. The majority of nuclei in normal samples stained strongly for SRSF3, whereas the nuclei in the HFD samples stained weakly (Figure 1E), as seen with the human NAFLD samples.
A second cohort of mice was maintained on a Western diet for 12 weeks to promote NASH. As in the NAFLD model, body weight, liver weight, spleen weight, and fasting glucose levels were all higher in mice on a NASH diet as compared with lean mice, and morphologically the livers were larger and paler than livers from lean mice (Supplemental Figure 3, A, C, and D). Examination of liver sections from NASH mice revealed less extensive steatosis than the NAFLD livers, consistent with the lower fat content of the diet, but with extensive intralobular fibrosis as detected by Masson’s trichrome and Sirius red staining, consistent with NASH (Supplemental Figure 3C). As observed for the NAFLD mice, SRSF3 was reduced in primary hepatocyte extracts from the NASH mice (Figure 1D). SRSF3 was similarly reduced in whole-liver extracts from NASH mice (Supplemental Figure 2B). This loss of SRSF3 was confirmed by immunohistochemistry and immunofluorescence on fixed liver tissue sections (Figure 1E and Supplemental Figure 2C).
We also investigated the effect of HFD and Western diet on Srsf3 mRNA expression. The Srsf3 gene generates 2 mRNA transcripts through alternative splicing of exon 4; the major transcript (Srsf3-202) skips exon 4 and encodes the full-length SRSF3 protein (Srsf3-FL), and the minor transcript (Srsf3-201) includes exon 4 and contains a premature termination codon (PTC) causing the mRNA to be degraded by nonsense-mediated decay (Srsf3-PTC) (3, 28, 29). RNA from normal, NAFLD, NASH, and cirrhosis liver samples showed higher expression of SRSF3-FL than SRSF3-PTC (Supplemental Figure 4A). RNA from hepatocytes from HFD and NASH mice did not show a difference in Srsf3-FL mRNA levels but showed a significant 3-fold increase in Srsf3-PTC mRNA levels by quantitative PCR (qPCR), although Srsf3-PTC remained a minor isoform (1% to 3%) (Supplemental Figure 4, B and C). The change in the ratio of mRNA isoforms was confirmed by end-point reverse transcription PCR (RT-PCR) using primers spanning exons 3 to 5 (Supplemental Figure 4, B and C). These results also confirmed that the Srsf3-PTC transcript only constitutes a small fraction (<5%) of steady-state mRNA levels in the liver.
Liver disease is associated with altered splicing of the SRSF3 target genes Fn1, Insr, Slk, and Myo1b.
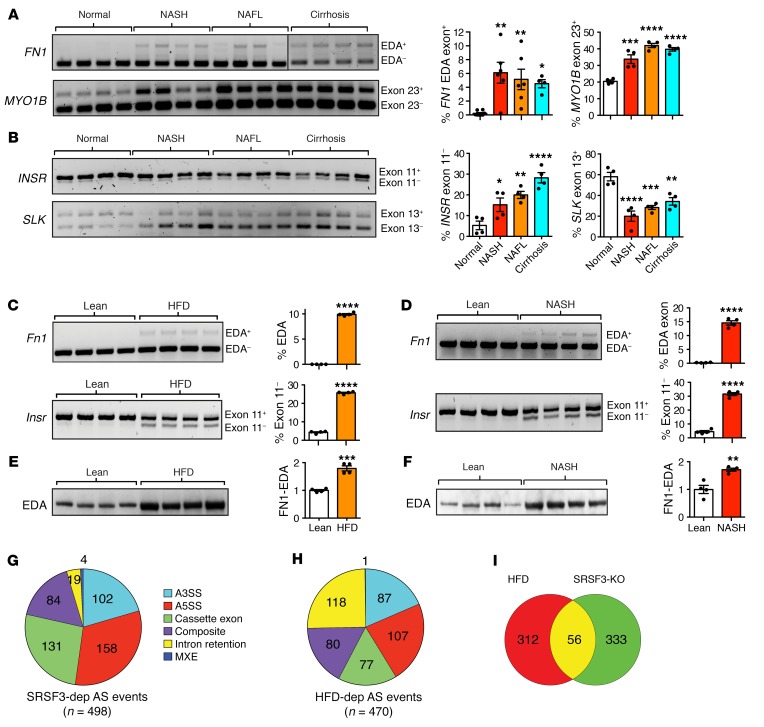
As SRSF3 was reduced in fatty liver disease in both humans and mice, we expected to see alterations in SRSF3 target gene splicing; therefore, we measured splicing of the profibrogenic EDA exon (exon 33) in the fibronectin 1 gene (FN1-EDA), exon 23 in Myo1B, exon 11 in INSR, and exon 13 in the Slk gene, all known SRSF3 targets (10). RNA extracted from livers of frozen samples of human NAFLD, NASH, and cirrhosis showed altered inclusion of EDA-FN1 and Myo1B exons and skipping of INSR and Slk exons (Figure 2, A and B). We also tested splicing of 2 SRSF3 target genes in hepatocytes from the NAFLD and NASH mice. We observed greater inclusion of exon 33 in the Fn1 mRNA (EDA exon) (Figure 2, C and D) and higher expression of the EDA-containing FN1 protein (Figure 2, E and F), which was confirmed by immunofluorescence microscopy (Supplemental Figure 5). Skipping of exon 11 in the Insr mRNA was increased in the RNA from NAFLD (Figure 2C) and NASH (Figure 2D) hepatocytes. These results in both human and mouse liver were consistent with a functional loss of SRSF3.
Figure 2. Alterations in SRSF3 target genes in liver disease.
Analysis of alternative splicing of (A) FN1–EDA exon 33 and MYO1B–exon 23 mRNAs, (B) INSR–exon 11 and SLK–exon 13 mRNAs, in total RNA from normal human liver, NASH, NAFL, and cirrhosis (n = 4/group) by RT-PCR. Colors are as in Figure 1. (C) Analysis of alternative splicing of Fn1 and Insr mRNAs in hepatocytes from lean and obese HFD mice by RT-PCR (n = 4/group). (D) Analysis of alternative splicing of Fn1 and Insr mRNAs in hepatocytes from lean and obese NASH mice by RT-PCR (n = 4/group). Representative gels are shown with the percentage inclusion of EDA and skipping of exon 11. For A–D, graphs show quantification with mean ± SEM (n = 4/group). Statistical significance determined by 1-way ANOVA with Tukey’s post hoc testing (A and B) or Student’s t test (C and D). (E) Immunoblotting of FN1-EDA from hepatocytes from obese mice on high-fat diet (HFD) or lean mice on normal chow (Lean). (F) Immunoblotting of FN1-EDA from hepatocytes from obese mice on NASH diet (NASH) or lean mice on normal chow (Lean). For E and F, graphs show quantification of FN1-EDA protein levels normalized to β-actin. Results are presented as mean ± SEM (n = 4/group). Asterisks indicate statistical significance by Student’s t test. (G) SRSF3-dependent (n = 498) alternative splicing events from hepatocytes with acute loss of SRSF3 by adenoviral expression of CRE or GFP analyzed using JUM. Different colors show different classes of events. A3SS, indicates alternative 3′ splice sites; A5SS, alternative 5′ splice sites; Cassette Exon, cassette exons; Intron Retention, retained introns; MXE, mutually exclusive exons; Composite, complex splicing events that do not fit a single category. (H) HFD-dependent (n = 470) alternative splicing events in hepatocytes from lean mice and obese mice on HFD analyzed using JUM. Colors and categories are as in panel G. (I) Venn diagram showing overlap of genes with altered splicing in obese HFD hepatocytes with SRSF3-dependent events. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To compare the changes in RNA splicing in NAFLD and in response to the loss of SRSF3, we performed RNA sequencing (RNAseq) on total RNA extracted from hepatocytes from livers of mice on HFD or LFD. To assemble a list of SRSF3 splicing target genes in hepatocytes, we extracted RNA from Srsf3-floxed hepatocytes acutely infected with an adenovirus expressing CRE (Adeno-CRE) recombinase or green-fluorescent protein (GFP). Alternative splicing (AS) events were analyzed using JUM (30) and MAJIQ (31, 32). Initially, we identified SRSF3-dependent AS events using RNA from the acute deletion of SRSF3. Inspection of the Srsf3 locus confirmed the deletion of exons 2 and 3 (Supplemental Figure 6A). A total of 3,466 AS events were identified, of which 498 were significantly altered (P < 0.05) after multiple testing correction, in the SRSF3-KO by JUM (Figure 2G and Supplemental Figure 6B). MAJIQ identified 21,458 local splicing variants (LSVs), of which 436 were significantly altered in the KO (P < 0.05) after multiple testing correction. One hundred and sixteen genes were present in both JUM and MAJIQ data sets (Supplemental Figure 6C). When the JUM AS events were broken down by category, cassette exons and alternative 3′ and 5′ splice sites comprised the majority of these events, with a smaller number of composite events, intron retentions, and mutually exclusive exons (Figure 2G). Intron retentions, however, comprised a larger proportion of AS events in the total pool (Supplemental Figure 6B). Analysis of the mouse HFD data using JUM showed a total of 4,712 AS events, of which 470 were significantly altered (P < 0.05) in hepatocytes from obese livers (Supplemental Figure 6D). When broken down by category, cassette exons and alternative 3′ and 5′ splice sites again made up the majority of the significant AS events (Figure 2H). Interestingly, the HFD RNA had a much higher proportion of intron retention events than the SRSF3-KO RNA, which possibly indicated alterations in other splicing factors (28). When the genes subject to AS were compared, 15% of the genes altered by HFD (56 of 368) were also SRSF3 dependent (Figure 2I) that represented a significant enrichment (P < 0.05 by Fisher’s exact test). Thus, the RNAseq results indicated that a significant number of AS events that occurred in livers of obese mice were consistent with loss of SRSF3.
Lipid excess causes oxidative stress and loss of SRSF3.
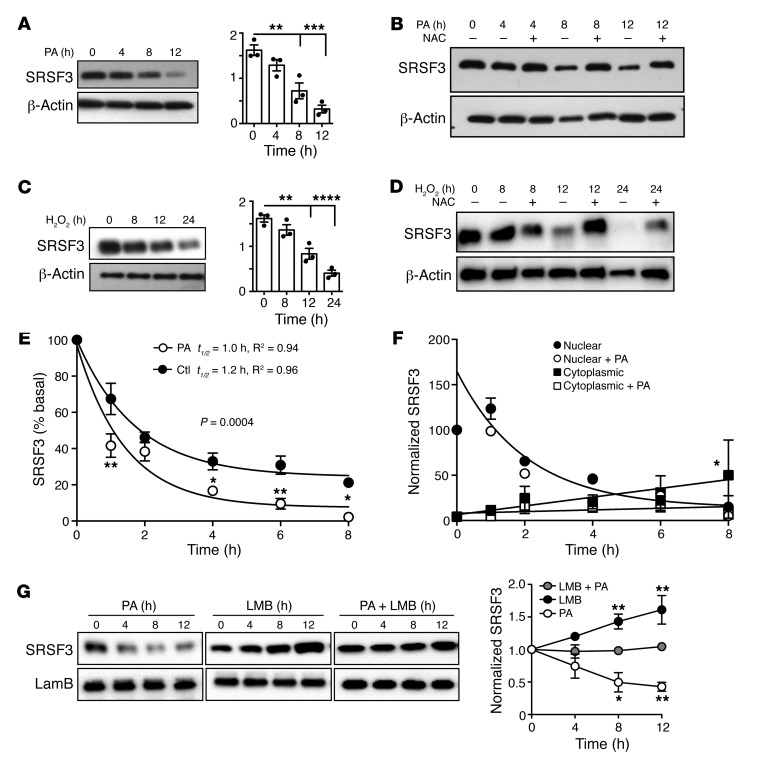
NAFLD and NASH are associated with hepatic steatosis. To test whether lipid overload directly caused loss of SRSF3, we treated human HepG2 cells and mouse primary hepatocytes with palmitic acid (PA) for 4–12 hours. We observed that PA caused a time-dependent loss of SRSF3 in both HepG2 cells (Figure 3A) and primary hepatocytes (Supplemental Figure 7A). As lipid excess causes oxidative stress, we measured the expression of NADPH quinone dehydrogenase 1 (Nqo1), a gene sensitive to oxidative stress, by qRT-PCR and found that PA induced Nqo1 expression (Supplemental Figure 7B). To test whether PA-induced oxidative stress leads to SRSF3 degradation, we pretreated cells with the antioxidant N-acetyl-cysteine (NAC, 10 mM, 15 minutes). Scavenging reactive oxygen species (ROS) with NAC blocked the PA-induced SRSF3 degradation, suggesting that oxidative stress caused SRSF3 degradation (Figure 3B and Supplemental Figure 7C). To test oxidative stress directly, we treated HepG2 cells and primary hepatocytes with H2O2 (500 μM and 500 μM) and found that SRSF3 was degraded in a time-dependent manner (Figure 3C and Supplemental Figure 7D) and the degradation was again blocked by NAC (Figure 3D and Supplemental Figure 7F). As expected, H2O2 induced expression of Nqo1 (Supplemental Figure 7E).
Figure 3. Reduction in SRSF3 protein in response to excess lipid and oxidative stress.
Human HepG2 cells were exposed to (A) 500 μM PA, (B) 500 μM PA in the presence of the antioxidant N-acetyl-cysteine (NAC) to scavenge ROS, (C) 500 μM H2O2, and (D) 500 μM H2O2 in the presence of NAC over time, and extracts were immunoblotted for SRSF3. Graphs in A and C show quantification of SRSF3 protein normalized to β-actin (mean ± SEM, n = 3/group). **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. time 0 by 1-way ANOVA. (E) HepG2 cells were exposed to 500 μM PA or vehicle in the presence of 50 μg/mL cycloheximide for 0–8 hours. Cell extracts were immunoblotted for SRSF3 and results quantified by densitometry. Normalized SRSF3 expression results were fit to a 1-phase exponential decay to calculate the protein half-life (t1/2). The R2 values of the curve fits are given. The 2 curves were significantly different (P = 0.0004). (F) HepG2 cells were exposed to 500 μM PA or vehicle in the presence of 50 μg/mL cycloheximide for 0–8 hours. Cells were separated into nuclear and cytoplasmic fractions and immunoblotted for SRSF3 and normalized to lamin B or β-actin. Nuclear SRSF3 levels again fit a 1-phase exponential decay but there was no significant difference between treatments, so the single-curve fit is shown. Cytoplasmic SRSF3 did not fit an exponential decay, but the data were fit by a linear equation. The 2 treatments had significantly different linear fits. (G) Nuclear and cytoplasmic proteins were isolated from HepG2 cells treated with PA, leptomycin B (LMB), or both (LMB + PA) over time and immunoblotted for SRSF3 and lamin B or β-actin. Graph shows normalized nuclear SRSF3 levels in the 3 groups over time. Cytoplasmic levels of SRSF3 were very low in leptomycin B–treated groups. Results are presented as mean ± SEM (n = 3/group). *P < 0.05, **P < 0.01 vs. cotreatment by 2-way ANOVA.
We also investigated the effect of lipid excess and oxidative stress on Srsf3 mRNA expression. Primary hepatocytes and HepG2 cells exposed to PA or H2O2 for different times did not show a difference in Srsf3-FL mRNA levels but showed significantly increased Srsf3-PTC mRNA levels by qPCR (Supplemental Figure 8, A and B). The change in the ratio of mRNA isoforms was confirmed by end-point RT-PCR (Supplemental Figure 8, C and D). These results also confirmed that oxidative stress induces the Srsf3-PTC transcript (28) but this only constituted a small fraction (<5%) of steady-state mRNA levels even under stressed conditions. We did not observe expression of the truncated SRSF3 protein that had been previously reported (33). To test whether the Srsf3-PTC transcript was indeed regulated by nonsense-mediated decay (NMD) we measured Srsf3 mRNA expression in the presence of the translational inhibitor cycloheximide (CHX, 50 μg/mL), as NMD requires ongoing protein translation. Expression of the major isoform (Srsf3-FL) was much higher than the Srsf3-PTC isoform in both primary hepatocytes and HepG2 cells in the basal state by qPCR (Supplemental Figure 9A). Expression of both isoforms increased over time following CHX treatment, with the minor Srsf3-PTC isoform increasing to approximately the same level as the Srsf3-FL isoform in hepatocytes, but remaining the minor isoform in HepG2 cells (Supplemental Figure 9A). The ratio of isoforms was confirmed by end-point RT-PCR (Supplemental Figure 9B).
SRSF3 protein is rapidly degraded by the proteasome.
We then measured SRSF3 protein stability by treating HepG2 cells with CHX for 1–8 hours. We observed a rapid degradation of SRSF3, with an estimated SRSF3 half-life of 1.2 hours that was accelerated with PA treatment (Figure 3E and Supplemental Figure 10A). We saw a similarly rapid degradation of SRSF3 in mouse primary hepatocytes (Supplemental Figure 10B). To test whether the degradation of SRSF3 was mediated by proteasomes, we treated HepG2 cells and primary hepatocytes with the proteasomal inhibitor MG132 (10 μM) and performed a similar time course over 1–8 hours. There was a strong induction of SRSF3 in cells treated with MG132 alone (Supplemental Figure 10, C and D) and no apparent degradation of SRSF3 over 8 hours in the presence of CHX (Supplemental Figure 10, E and F), indicating that SRSF3 protein was not inherently unstable but was rapidly targeted for proteasomal degradation under normal conditions. SRSF3 shuttles between the nucleus and cytoplasm, so we measured the half-life of SRSF3 in the nucleus or cytoplasm in the presence or absence of PA. The majority of SRSF3 was nuclear (Supplemental Figure 10G). Nuclear SRSF3 levels rapidly dropped following CHX treatment, as seen in the whole cell, but PA did not alter the rate of degradation (Figure 3F and Supplemental Figure 10H). In contrast, cytoplasmic levels increased following CHX treatment but the increase was suppressed by PA treatment (Figure 3F and Supplemental Figure 10I). This suggested that the primary effect of PA occurred in the cytoplasm. So we then tested the effect of blocking nuclear export on SRSF3 levels. HepG2 cells were treated with leptomycin B and PA and nuclear extracts prepared. PA caused the expected decrease in nuclear SRSF3. Furthermore, inhibition of nuclear export with leptomycin B caused an increase in nuclear SRSF3 (Figure 3G). Thus, inhibition of nuclear export prevented the degradation of SRSF3 by PA (Figure 3G), again suggesting that degradation of SRSF3 occurred in the cytoplasm. To test whether lipid-stress reduced multiple splicing factors, we immunoblotted extracts of HepG2 cells that had been treated with PA or H2O2 for increasing times with an antibody that recognizes a conserved epitope in many SR proteins. Only a band at approximately 20 kDa showed a reduction in intensity with PA and H2O2 (Supplemental Figure 11, A and B). This was confirmed to be SRSF3 by re-blotting with an antibody against SRSF3 (Supplemental Figure 11C).
SRSF3 is neddylated in response to PA.
Previous studies have shown that SRSF3 is ubiquitinated under both normal and stress conditions (22–24). To test whether SRSF3 was degraded through the ubiquitination pathway, we treated the primary hepatocytes and HepG2 cells with MG132 to prevent degradation, then tested for ubiquitination of SRSF3. Cell extracts were immunoprecipitated with anti-ubiquitin or anti-SRSF3 antibodies, and then immunoblotted for SRSF3 or ubiquitin, respectively. We were unable to detect ubiquitination of endogenous SRSF3 under any of these conditions (Supplemental Figure 12, A–C). We then overexpressed Flag-tagged SRSF3 (Flag-SRSF3) in HepG2 cells by transfection. Under these conditions, we were able to detect ubiquitinated SRSF3 that was increased in the presence of MG132 (Supplemental Figure 12D). To test whether stress caused by lipid excess could increase ubiquitination of SRSF3, we overexpressed Flag-SRSF3 in HepG2 cells and treated with PA in the presence of MG132. PA-induced stress did not increase ubiquitinated SRSF3 (Supplemental Figure 12E). These results indicated that endogenous SRSF3 was not degraded via the ubiquitin pathway, although ubiquitination could be observed if the protein is overexpressed. We also tested the specificity and efficacy of the antibody used in the immunoprecipitation assay. The immunoblot result showed that more than 90% of the protein was precipitated (Supplemental Figure 12, F–H).
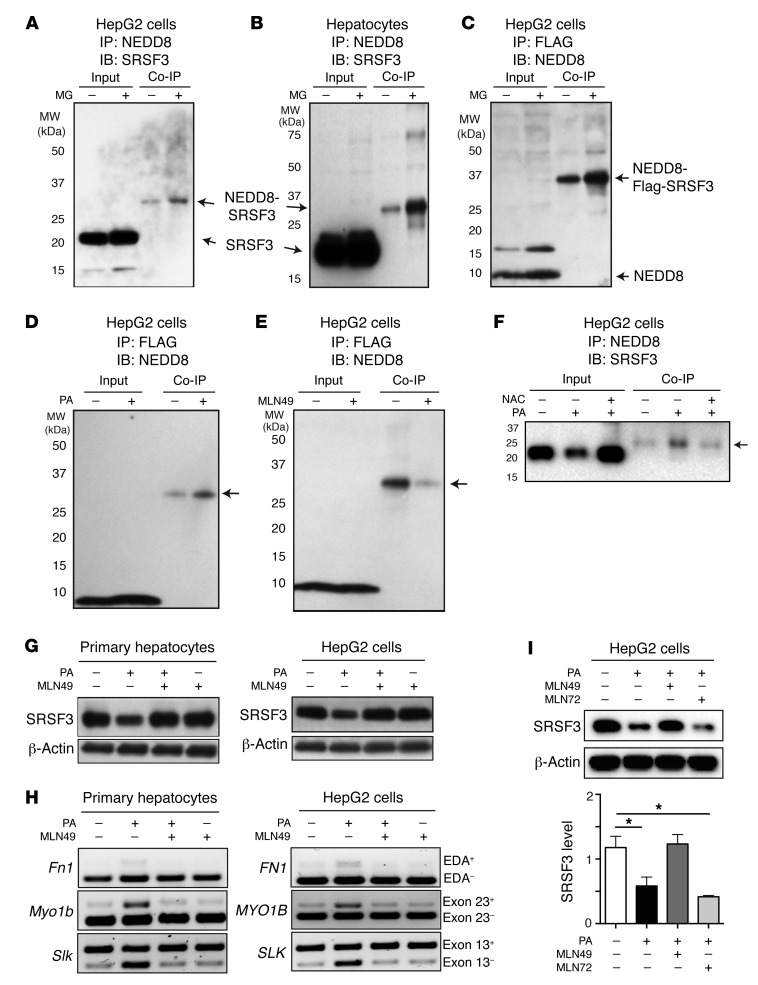
Previously it was reported that arsenite-induced oxidative stress causes neddylation-dependent stress granule formation, and SRSF3 was among the proteins identified as being conjugated to NEDD8 (25). Neddylation is a posttranslational modification similar to ubiquitination and can lead to protein degradation (34). To test whether SRSF3 was neddylated in liver cells, we treated primary hepatocytes and HepG2 cells with MG132, and then tested for conjugation of NEDD8 and SRSF3. Cell extracts were immunoprecipitated with antibodies against NEDD8 and then immunoblotted for SRSF3. An anti-SRSF3 antibody–reactive protein was observed at approximately 30 kDa, corresponding to the expected combined molecular weight of NEDD8 (8 kDa) conjugated to SRSF3 (21 kDa), and levels of this protein increased with proteasomal inhibition by MG132 (Figure 4, A and B). Conversely, we overexpressed Flag-SRSF3 in HepG2 cells, immunoprecipitated with anti-Flag antibodies, and then immunoblotted for NEDD8. A strong NEDD8-reactive protein was observed at approximately 36 kDa, the correct size for NEDD8-conjugated (8 kDa), Flag-tagged SRSF3 (28 kDa) that again increased with proteasomal inhibition, as for the endogenous protein (Figure 4C). We then tested whether lipid-induced stress could increase SRSF3 neddylation. HepG2 cells transfected with Flag-SRSF3 were treated with PA in the presence of MG132. The 36-kDa NEDD8-reactive protein increased with PA treatment, clearly indicating that PA-induced stress promoted SRSF3 neddylation (Figure 4D).
Figure 4. Neddylation of SRSF3 in response to palmitic acid.
HepG2 cells (A) and primary hepatocytes (B) were treated with 10 μM MG132 (MG) for 2 hours. Cell lysates were immunoprecipitated (IP) with anti-NEDD8 antibodies and then immunoblotted (IB) for SRSF3. Input indicates input control, Co-IP indicates the coimmunoprecipitated proteins. The unmodified SRSF3 and NEDD8-SRSF3 conjugate are indicated by arrows. (C) HepG2 cells were transfected with Flag-tagged SRSF3 and then treated with 10 μM MG132; cell lysates were immunoprecipitated with an anti-Flag antibody and then immunoblotted for NEDD8. Monomeric NEDD8 and the NEDD8–Flag-SRSF3 conjugate are indicated by arrows. (D) HepG2 cells were transfected with Flag-tagged SRSF3 and then treated with 10 μM MG132 and 500 μM palmitic acid (PA); cell lysates were immunoprecipitated with an anti-Flag antibody and then immunoblotted for NEDD8. (E) HepG2 cells were transfected with Flag-tagged SRSF3 and then treated with the NEDD8-activating enzyme 1 inhibitor 3 mM MLN4924 (MLN49) for 1 hour and then PA for 12 hours, and cell lysates immunoprecipitated with an anti-Flag antibody and then immunoblotted for NEDD8. The NEDD8–Flag-SRSF3 conjugate is indicated by an arrow in D and E. (F) HepG2 cells were cotreated with the antioxidant N-acetyl-cysteine (NAC) and PA for 12 hours; cell lysates were immunoprecipitated with anti-NEDD8 antibody and then immunoblotted for SRSF3. NEDD8-SRSF3 is indicated by an arrow. (G) Primary hepatocytes or HepG2 cells were treated with 3 mM MLN49 for 1 hour and then PA for 12 hours, and cell lysates were immunoblotted for SRSF3. (H) Primary hepatocytes or HepG2 cells were treated with 3 mM MLN49 for 1 hour and then PA for 12 hours; splicing of the Fn1-EDA exon 33, Myo1b–exon 23, and Slk–exon 13 was analyzed by RT-PCR. (I) HepG2 cells were treated with 3 mM MLN49 and 1 mM MLN7243 (MLN72, ubiquitin-activating enzyme 1 inhibitor) for 1 hour and then PA for 12 hours; cell lysates were immunoblotted for SRSF3. Results are presented as mean ± SEM (n = 3). *P < 0.05 by 1-way ANOVA.
To test whether the neddylation of SRSF3 was necessary for proteasomal degradation, we inhibited the NEDD8-activating enzyme 1 complex (NAE1: APPBP1 and UBA3) by treatment with MLN4924 (35, 36). Inhibition of NAE1 inhibited the conjugation of NEDD8 to exogenous Flag-tagged SRSF3 (Figure 4E). We then tested whether endogenous SRSF3 was neddylated in response to PA-induced oxidative stress. PA caused a decrease in SRSF3 in the whole-cell extract (input) and caused an increase in NEDD8-conjugated SRSF3 (Figure 4F). Scavenging ROS with NAC prevented both the degradation of SRSF3 in the input and also the conjugation to NEDD8 (Figure 4F). Furthermore, we found that MLN4924 blocked the degradation of SRSF3 under PA-induced stress conditions in both primary hepatocytes and HepG2 cells (Figure 4G). To test whether inhibition of neddylation had a functional effect on RNA splicing of SRSF3 target genes, we treated primary hepatocytes and HepG2 cells with MLN4924, and measured splicing of Fn1, Slk, and Myo1b by qRT-PCR. Consistent with the inhibition of SRSF3 degradation, we observed that NAE1 inhibition prevented inclusion of the EDA exon in Fn1, skipping of exon 13 in Slk, and inclusion of exon 23 in Myo1b mRNAs in response to PA in primary hepatocytes and the corresponding FN1, SLK, and MYO1B genes in HepG2 cells (Figure 4, G and H). To exclude a role for ubiquitination in SRSF3 degradation, we treated HepG2 cells with PA in the presence of the NAE1 inhibitor MLN4924 or the specific UAE1 inhibitor MLN7243. The NAE inhibitor, but not the UAE inhibitor, prevented the loss of SRSF3 with PA treatment (Figure 4I), confirming that neddylation, not ubiquitination, was responsible for SRSF3 degradation.
Mutation of lysine 11 prevents SRSF3 protein degradation.
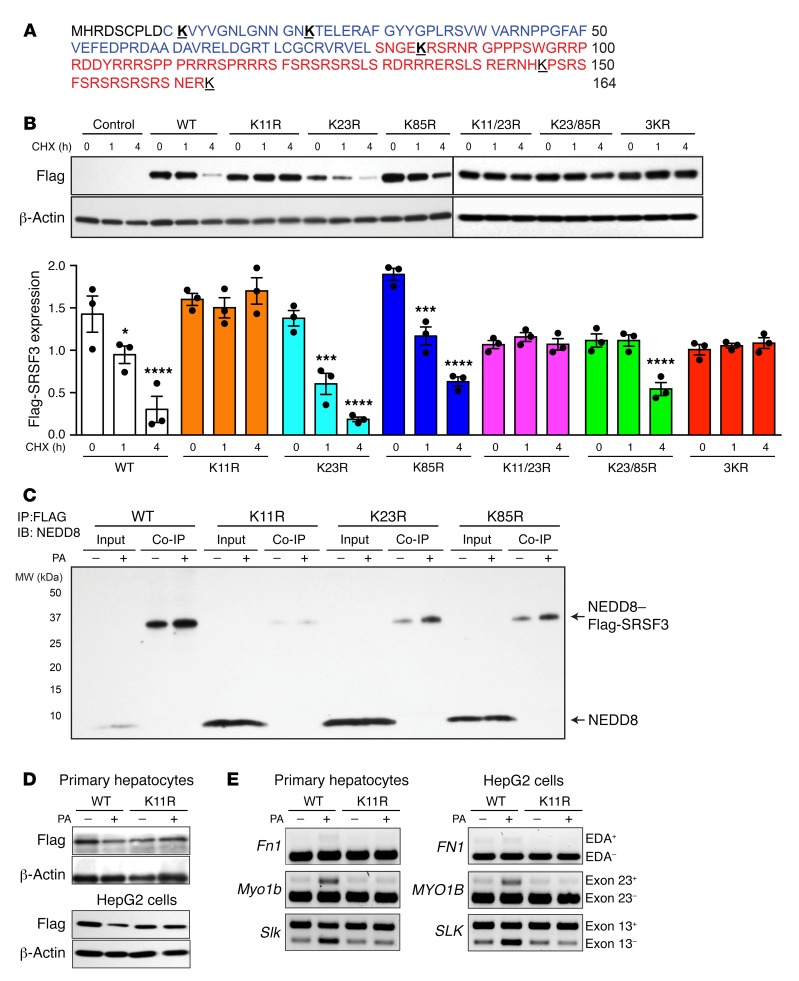
Posttranslational modification by NEDD8 involves conjugation to lysine residues in the target protein. To determine which lysine residues were important for destabilizing SRSF3, we obtained a series of SRSF3 lysine mutants. We measured the half-life of 3 lysine-mutant SRSF3 proteins using CHX (Figure 5, A and B). HepG2 cells were transfected with Flag-tagged wild-type (WT) SRSF3 and lysine-to-arginine mutants in the amino-terminal half of the protein (K11R, K23R, and K85R), as well as double lysine mutants (K11/23R and K23/85R) and a triple lysine mutant (3KR) (25). We observed that WT SRSF3 was rapidly degraded, as seen earlier (Figure 5B), but the K11R mutant was stable and not degraded, as was the double mutant K11/23R and the triple mutant. Mutation of K23 and K85 did not alter SRSF3 stability (Figure 5B). This result suggested that conjugation of lysine 11 was essential for SRSF3 degradation. To demonstrate that lysine 11 was neddylated in response to PA, we transfected WT, K11R, K23R, and K85R mutant SRSF3s into HepG2 cells, and the cells were then treated with PA or vehicle for 12 hours. Cell lysates were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-NEDD8 antibody. We observed that the K11R mutation eliminated basal conjugation of NEDD8 to SRSF3 and prevented the PA-induced neddylation (Figure 5C). The K23R and K85R SRSF3 mutants showed basal and PA-induced neddylation, confirming the stability data (Figure 5C). We then tested the effect of the K11R mutation on the PA-induced degradation of SRSF3 and found that this mutant stabilized SRSF3 and prevented its degradation in primary hepatocytes and HepG2 cells (Figure 5D). We also investigated the ubiquitination of these SRSF3 mutants but could not detect ubiquitinated SRSF3 as we did earlier (Supplemental Figure 13). Consistent with the stabilization of the SRSF3 protein, we observed that expression of SRSF3-K11R prevented inclusion of exon 33 (EDA exon) in Fn1, skipping of exon 13 in Slk1, and inclusion of exon 23 in Myo1b mRNAs in response to PA in primary hepatocytes and the corresponding FN1, SLK, and MYO1B genes in HepG2 cells (Figure 5E). These results indicated that SRSF3 was degraded in response to stress induced by lipid overload via neddylation on lysine 11 and proteasomal degradation.
Figure 5. Mutation of lysine 11 prevented SRSF3 protein degradation.
(A) Amino acid sequence of SRSF3 protein. The RNA recognition motif (RRM) is shown in blue, and the arginine- and serine-rich (RS) domain in red. Lysine residues are bold face and underlined. (B) HepG2 cells were transfected with empty vector (Control), wild-type Flag-SRSF3 (WT), or a series of single lysine-to-arginine mutants (K11R, K23R, K85R), double lysine mutants (K11/23R, K23/85R), or a triple lysine mutant (3KR) as indicated. Cells were treated with cycloheximide (CHX) for 1 and 4 hours, and then lysates were immunoblotted using the anti-Flag antibody. Graphs below show quantification of Flag-SRSF3 levels from transfected HepG2 cells. Results are presented as mean ± SEM (n = 3/group). *P < 0.05, ***P < 0.001, ****P < 0.0001 vs. time 0 by 1-way ANOVA. Colors indicate the different mutants. (C) HepG2 cells were transfected with Flag-SRSF3 or single lysine mutants (K11R, K23R, K85R) as indicated and treated with 500 μM PA for 12 hours. Cell lysates were immunoprecipitated with anti-Flag antibodies and then immunoblotted for NEDD8. Input indicates input control, Co-IP indicates the coimmunoprecipitated complex. Monomeric NEDD8 and the NEDD8–Flag-SRSF3 conjugate are indicated by arrows. (D) Analysis of PA-induced SRSF3 degradation. Primary hepatocytes (top panels) and HepG2 cells (bottom panels) were transfected with Flag-SRSF3-WT or Flag-SRSF3-K11R. Twenty-four hours later, primary hepatocytes or HepG2 cells were treated with 250 μM or 500 μM PA, respectively, for 12 hours. Cells were then lysed and immunoblotted with the anti-Flag antibody. (E) Analysis of PA-induced changes in RNA splicing. Primary hepatocytes (left panels) and HepG2 cells (right panels) were transfected with Flag-SRSF3-WT or Flag-SRSF3-K11R. Twenty-four hours later, cells were treated with 250 μM or 500 μM PA, respectively, for 12 hours. Exon 33 (EDA exon) incorporation in the Fn1 mRNA, Slk–exon 13 skipping, and Myo1b–exon 23 inclusion was analyzed by RT-PCR.
Expression of a neddylation-resistant SRSF3 prevents NASH.
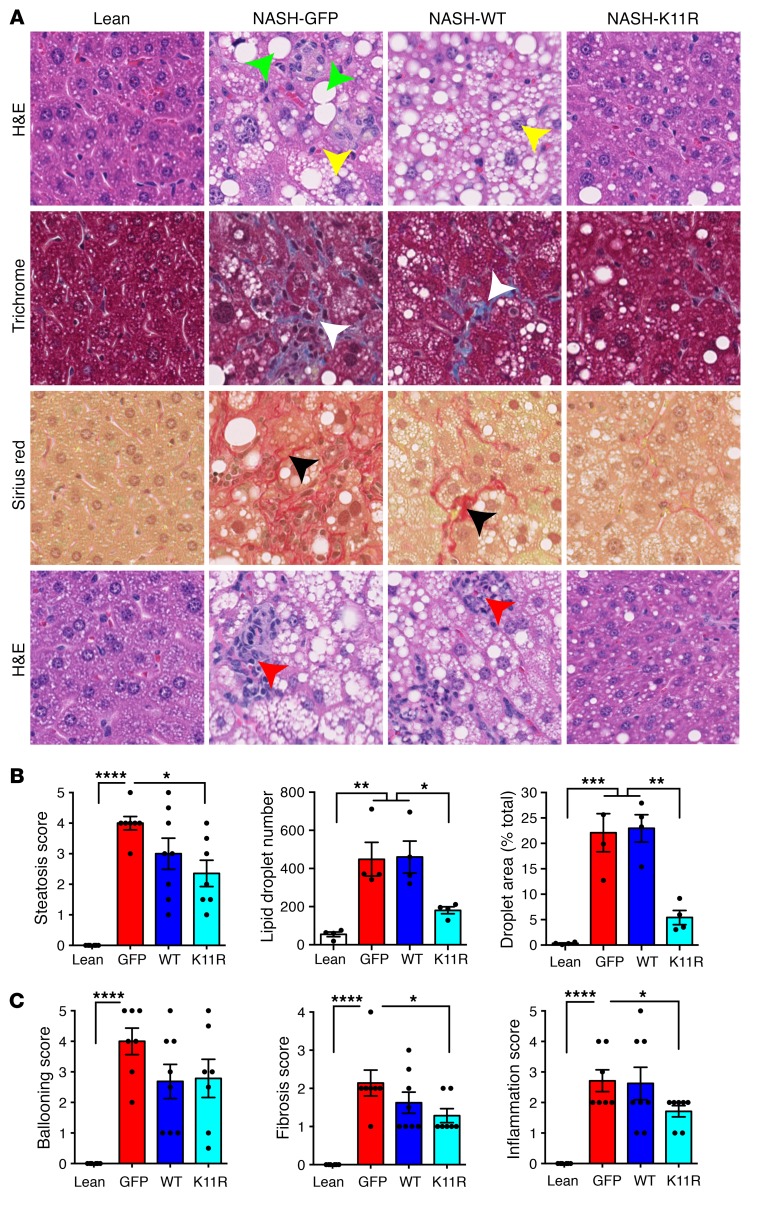
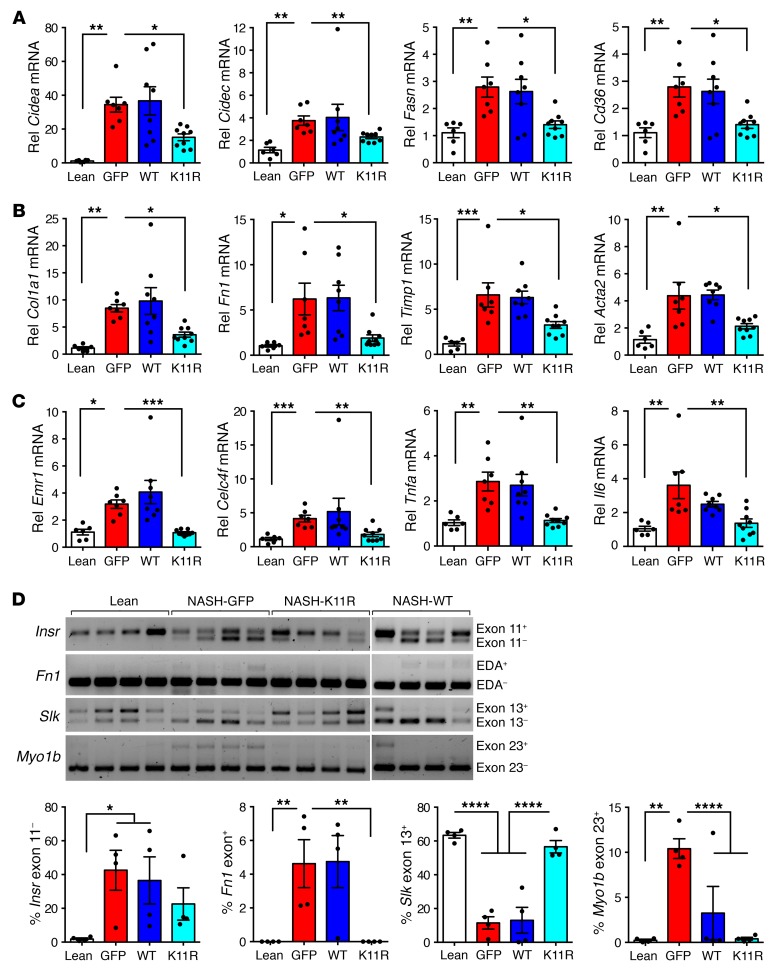
To test whether preventing SRSF3 degradation in vivo would prevent the development of NASH we expressed SRSF3-K11R, WT SRSF3, or GFP as a control, using tail vein injection of AAV serotype 8 (AAV8) in C57BL/6J mice (n = 6–8/group). After infection the mice were placed on the NASH diet for 7 weeks. The body weights of the infected mice increased, livers were pale and larger relative to mice on normal chow as expected, and fasting blood glucose levels were elevated at sacrifice (Supplemental Figure 14, A–C). Hematoxylin and eosin–stained (H&E-stained) sections from livers of GFP-infected mice showed the expected steatosis, whereas the livers from SRSF3-K11R–infected mice showed reduced steatosis (Figure 6, A and B). Mice expressing WT SRSF3 showed an intermediate phenotype. Hepatocyte ballooning, a hallmark of NASH, was increased in the GFP-infected livers (Figure 6, A and C). Surprisingly, expression of WT or SRSF3-K11R did not significantly reduce the number of ballooned hepatocytes, although the trend was to less ballooning (Figure 6C). Parallel sections were stained with Masson’s trichrome and Sirius red for fibrosis. Sections from GFP-infected mice showed extensive intralobular fibrosis but sections from SRSF3-K11R–infected mice showed reduced fibrosis (Figure 6, A and C). Similarly, mice infected with SRSF3-K11R showed a reduction in immune cell infiltration (Figure 6, A and C). The reduction in fibrosis was confirmed by qPCR. GFP-infected mice had elevated Fn1, Col1a1, Timp1, and Acta2 (smooth muscle actin) expression that decreased with SRSF3-K11R expression (Figure 7A). The SRSF3-K11R mice also showed reduced expression of the lipid storage genes Cidea and Cidec, lipid synthesis and transport genes Fasn and Cd36 (Figure 7B), and the inflammatory macrophage and Kupffer cell markers Emr1, Clec4f, Tnfa, and Il6 (Figure 7C). Next, we assessed the effect of mutant lysine 11 on expression of SRSF3 target genes in RNA from isolated hepatocytes. We observed that SRSF3-K11R prevented skipping of exon 11 in the Insr, inclusion of exon 33 (EDA exon) in Fn1, skipping of exon 13 in Slk1, and inclusion of exon 23 in Myo1b (Figure 7D). Thus, overexpression of SRSF3 or a degradation-resistant mutant was sufficient to prevent progressive liver disease in mice on a NASH-inducing diet.
Figure 6. Expression of degradation-resistant SRSF3-K11R reduced liver steatosis, fibrosis, and inflammation in mice on a NASH diet.
(A) Liver sections from lean mice on normal chow (Lean) or mice on a NASH diet for 7 weeks infected with AAV8 expressing GFP (NASH-GFP), wild-type SRSF3 (NASH-WT), or the degradation-resistant K11R-mutant SRSF3 (NASH-K11R). Green arrowheads indicate steatotic hepatocytes and yellow arrowheads indicate ballooned hepatocytes on H&E-stained sections. White and black arrowheads indicate fibrosis on Masson’s trichrome– and Sirius red–stained sections. Red arrowheads indicate inflammatory cell infiltrates on H&E sections. Original magnification ×40. (B) Quantification of steatosis. Histological scoring was performed by a pathologist blinded to the identity of the samples. Lipid droplet number and area were measured using ImageJ (NIH). Lean mice shown in white, GFP-expressing mice in red, WT SRSF3–expressing mice in blue, and SRSF3-K11R–expressing mice in cyan. (C) Histological scoring of hepatocyte ballooning, fibrosis, and inflammation. Results are presented as mean ± SEM (n = 6–8 mice/group). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Tukey’s post hoc testing.
Figure 7. Degradation-resistant SRSF3-K11R attenuated expression of genes involved in lipid metabolism, fibrosis, and inflammation, and impaired SRSF3 target gene splicing in RNA from hepatocytes of mice on a NASH diet.
(A) Expression of lipid storage, synthesis, and transport genes by qPCR. Cell death–inducing DFFA-like effector A (Cidea), cell death–inducing DFFA-like effector C/fat-specific protein 27 (Cidec), fatty acid synthase (Fasn), and fatty acid translocase (Cd36) RNA expression in hepatocytes is shown. (B) Expression of fibrosis genes by qPCR. Collagen 1a (Col1a1), fibronectin (Fn1), tissue inhibitor of metalloprotease (Timp1), and smooth muscle actin (Acta2) RNA expression in hepatocytes is shown. (C) Expression of inflammatory genes by qPCR. Macrophage F4/80 gene (Emr1), Kupffer cell C-type lectin domain family 4 member F (Clec4f), tumor necrosis factor α (Tnfa), and interleukin 6 (Il6) RNA expression in hepatocytes is shown. Lean mice shown in white, GFP-expressing mice in red, SRSF3-WT–expressing mice in blue, and SRSF3-K11R–expressing mice in cyan. (D) Analysis of RNA splicing in infected livers. Primary hepatocytes were generated from lean mice or mice on a NASH diet infected with AAV8-GFP, AAV8-SRSF3-WT, or AAV8-SRSF3-K11R. Splicing of SRSF3 target genes was assessed by RT-PCR as in Figure 2. Representative gels are shown for splicing of the insulin receptor exon 11 (Insr), fibronectin EDA exon 33 (Fn1), STE20-like kinase exon 13 (Slk), and myosin 1b exon 23 (Myo1b) (n = 4/group). Graphs show the percentage inclusion of the spliced exon. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Tukey’s post hoc testing.
Discussion
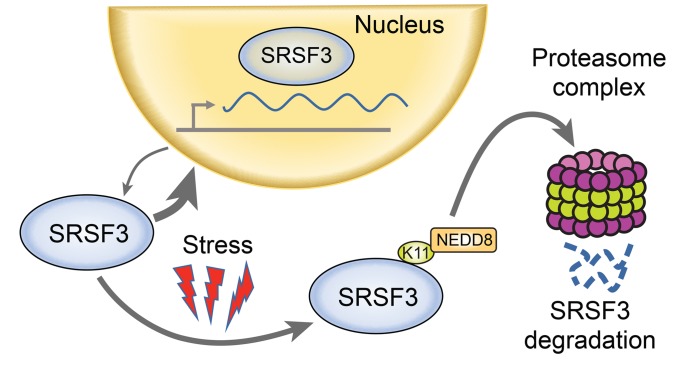
In this study, we report that SRSF3 protein expression was reduced in human NAFLD, NASH, and cirrhotic liver diseases. We also showed that SRSF3 expression was similarly reduced in livers from mouse models of NAFLD and NASH, and we have reported previously that SRSF3 was lost in a CCl4-induced model of liver cirrhosis in mice (10). Additionally, human and mouse livers showed evidence of alterations in alternative RNA splicing of known and potentially novel SRSF3 target genes consistent with a functional loss of SRSF3 in NAFLD and NASH. As the mouse liver faithfully recapitulated the SRSF3 reduction seen in early human liver disease, we used primary mouse hepatocytes and human HepG2 hepatoma cells to investigate the mechanism underlying this loss. We showed that lipid overload led to SRSF3 proteasomal degradation through increased oxidative stress. Mechanistically, this occurred through neddylation of SRSF3 on lysine 11 in the RNA binding domain. Mutation of this single residue, or pharmacological inhibition of neddylation, prevented degradation of SRSF3 and alterations in alternative splicing of downstream target genes. Fractionation experiments showed that the PA-induced SRSF3 degradation appeared to occur mainly in the cytoplasm, as blocking nuclear export prevented degradation. SRSF3 shuttles between the nucleus and cytoplasm associated with target mRNAs, but at any one time most (>90%) SRSF3 is nuclear. NEDD8-dependent degradation of SRSF3 in the cytoplasm explains the low stoichiometry of neddylation at any one time, but the rapid shuttling between the 2 compartments allows total SRSF3 levels to decrease over 8–12 hours (Figure 8). Thus, we have uncovered a neddylation-dependent pathway leading to dysregulation of alternative splicing in fatty liver disease. A genetic knockout of SRSF3 in hepatocytes causes many metabolic changes, including steatosis and fibrosis (9), so it is intriguing to speculate that a functional reduction in SRSF3 may underlie many of the early events in fatty liver disease. Interestingly, deletion of SRSF3 in hepatocytes did not cause hepatocyte ballooning (9), so it is not surprising that expression of the degradation-resistant SRSF3 did not rescue the ballooning phenotype on the NASH diet, and furthermore suggests that hepatocyte ballooning is not triggered by loss of SRSF3. Although these changes are seen in early liver disease, we and others have reported that SRSF3 mRNA and protein are decreased in HCC (10, 11), so these early changes in fatty liver RNA splicing could predispose to the later development of HCC. Our findings are distinct from reported findings in HBV-associated HCC where SRSF3 protein is elevated but mislocalized to the cytoplasm through interaction with the viral Hbx protein (27).
Figure 8. Model of SRSF3 protein degradation.
SRSF3 shuttles between the nucleus and cytoplasm. Cellular stress promotes neddylation of SRSF3 on lysine 11 and targets it for degradation by the proteasome complex.
A number of studies have pointed to a potential role of SRSF3 in other cancers. Unfortunately, many studies rely on SRSF3 mRNA levels as a marker of expression but this is uninformative because SRSF3 autoregulates its own expression, so protein levels do not correlate with RNA (3), and we have shown here that SRSF3 protein degradation is acutely regulated independently of mRNA expression. More importantly if SRSF3 were lost in liver disease, we would expect to observe altered splicing of SRSF3 target genes. Indeed, this is the case, as we observed alterations in splicing of the fibronectin 1 gene FN1, the insulin receptor gene INSR, and 2 genes involved in cytoskeletal rearrangements and epithelial-to-mesenchymal transition — the STE20-like kinase gene SLK (37) and the myosin 1B gene MYO1B (38) — in RNA from NAFLD and NASH subjects. Alternative splicing of exon 11 of the INSR gene gives rise to 2 protein isoforms designated IR-A and IR-B (8, 39–43) and the ratio of these isoforms is altered in many cancers including breast, lung, colon, and liver, to increase expression of IR-A that is a high-affinity receptor for IGF2 (44, 45). At the cellular level, SRSF3 has a role in many aspects of the cell cycle (1, 5, 7, 46), including mitosis (47) and DNA repair (48). In many cell lines, overexpression of SRSF3 will cause tumor acceleration, while knockdown impairs growth. Human papilloma virus protein E2 upregulates SRSF3 expression, which is essential for viral splicing, and hence SRSF3 is overexpressed in many cervical cancers (49). SRSF3 recruits DROSHA to precursor microRNAs to promote microRNA processing (49–51), and SRSF3 regulates expression of both miR-1908 and miR-3131 that have been implicated in oncogenesis (50–54). So there is good evidence that changes in SRSF3 expression or function are linked to carcinogenic progression.
We found that PA-induced stress led to neddylation of SRSF3 and degradation. A previous study investigating the formation of stress granules in U2OS cells reported that the neddylation enzyme UBE2M was essential for granule formation in response to arsenite, and furthermore showed through proteomic profiling that SRSF3 was neddylated on lysine 85 to promote stress granule assembly, as they had previously reported (25, 55). Our results showed that SRSF3 is neddylated on lysine 11 under PA-induced oxidative stress but this modification targeted SRSF3 for degradation. This is distinct from the neddylation on lysine 85 that does not appear to affect protein stability. The neddylation pathway is not as well understood as the analogous ubiquitination pathway, but a few functions have been described (34). Neddylation is increased in HCC and inhibition of the NAE1 activating enzyme with MLN4924 is a promising therapy (56, 57). The first targets identified were the cullin proteins that act as scaffolds for the assembly of ubiquitin E3 ligase complexes. A single conserved lysine in the carboxyl terminus is conjugated with NEDD8 and induces a conformational switch that displaces the inhibitory protein CAND1 and activates the E3 ligase activity of the SCF complex (34). Neddylation mediated by MDM2 inhibits the transcriptional activity of p53 and p73, and neddylation competes with ubiquitination to prevent degradation of PPARγ (58). Conversely, NEDD8 can target proteins for degradation through interaction with the NUB1 protein that recruits NEDD8 to the S5a component of the 19S proteasome (59, 60). Whether SRSF3 is degraded via this NEDD8/NUB1/S5a pathway, and how neddylation responds to oxidative stress remain to be determined. Importantly, when SRSF3 degradation was repressed by overexpression of a degradation-resistant SRSF3-K11R in the liver, steatosis and fibrosis were prevented, which suggested that loss of SRSF3 drove progressive metabolic liver disease.
In summary, our results indicated that neddylation of lysine 11 in SRSF3 is critical for SRSF3 degradation, and thus regulated downstream SRSF3 target genes. The changes in SRSF3 expression and target genes in early liver disease may provide a valuable diagnostic tool, and potentially a new therapeutic target in HCC, as inhibition of SRSF3 degradation prevents progressive liver disease and inhibition of NAE1 by MLN4924 prevents liver cancer cell growth (61).
Methods
Animals and diets.
C57BL/6J mice were purchased from Jackson Laboratories. Mice were housed in a 12-hour light/12-hour dark cycle. Mice had access to food and water ad libitum. Groups of male and female mice were randomly assigned to a HFD (60% fat) to induce NAFLD (D12492, Research Diets Inc.), to a 40% fat plus 0.2% cholesterol diet to induce NASH (D12079B, Research Diets), or normal chow diet. Major lipid components of the NAFLD diet are derived from lard and include 20% palmitic acid, 11% stearic acid, 34% oleic acid, and 29% linoleic acid, resulting in 32% saturated, 36% monounsaturated, and 32% polyunsaturated fat. Major lipid components of the NASH diet are derived from 39% milk fat (23% palmitic acid, 10% stearic, 29% oleic acid, and 2% linoleic acid), 1% corn oil (13% palmitic acid, 3% stearic acid, 31% oleic acid, 52% linoleic acid, and 1% linolenic acid), and 0.2% cholesterol. Body weight and food intake data were recorded weekly.
Isolation of murine primary hepatocytes.
Primary hepatocytes were obtained by 2-step perfusion with liver perfusion medium SC-1 (137 mM NaCl, 5.4 mM KCl, 0.56 mM NaH2PO4•H2O, 0.85 mM Na2HPO4, 10 mM HEPES, 4.2 mM NaHCO3, 0.5 mM EGTA, and 5 mM glucose), followed by digestion medium SC-2 (137 mM NaCl, 5.4 mM KCl, 0.56 mM NaH2PO4•H2O, 0.85 mM Na2HPO4, 10 mM HEPES, 4.2 mM NaHCO3, and 12 mM CaCl2•H2O) containing 0.5 mg/mL collagenase D (Roche). Liver cells were disaggregated by passing through a 100-μm pore nylon mesh Cell Strainer (BD Bioscience) and centrifuged at 100 g for 10 minutes at 4°C. Cells pellets were suspended in 36% Percoll and centrifuged at 60 g for 6 minutes. The resulting cell pellets were washed with SC-2 buffer without collagenase D, the numbers of total viable cells were determined by Trypan blue staining, and cells were plated on collagen-coated plates (Invitrogen). Primary hepatocytes were cultured in William E culture medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C.
Cell culture and transfection.
The HepG2 cell line was obtained from ATCC and maintained in 1× DMEM supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C in an atmosphere of 5% CO2. Transfection of plasmids into HepG2 cells was performed using Lipofectamine 2000 (Invitrogen) and into primary hepatocytes using X-tremeGENE HP DNA transfection regents (Roche) following the manufacturers’ protocols. The series of expression vectors encoding Flag-tagged SRSF3 and the lysine mutants were obtained from Takbum Ohn (Chosun University, Gwangju, Republic of Korea).
Tissue collection and histology.
Human liver samples were obtained from the University of Minnesota Liver Tissue and Cell Distribution Service (UM-LTCDS; Minneapolis, Minnesota, USA). Subject characteristics are provided in Supplemental Table 1. Liver and other tissues were harvested at sacrifice for both histology and RNA extraction. Livers were fixed in formalin for 24 hours followed by washing in 70% ethanol. Paraffin-embedded sections (5 μm) were cut, dewaxed, and stained with H&E. Images were scanned using an Aperio ImageScope and analyzed using the ImageScope software (Leica).
Gene expression.
Total RNA was extracted from the tissues using RNAbee (Tel-Test Inc.) following the manufacturer’s instructions. First-strand cDNA was synthesized using a High-Capacity cDNA Synthesis Kit (Applied Biosystems). Targeted qPCR assays were run in 20-μL triplicate reactions on an MJ Research Chromo4 instrument using iTaq SYBR Green supermix (Bio-Rad). Gene expression levels were calculated after normalization to the housekeeping gene m36B4 using the 2–ΔΔCt method (62) and expressed as relative mRNA levels compared to the control. Primers are listed in Supplemental Table 2.
Immunoblot analysis.
Primary mouse hepatocytes and HepG2 cells were treated with PA (0.25 mM and 0.5 mM, respectively) or H2O2 (0.5 mM), with and without NAC (10 mM) for 0–12 or 0–24 hours (PA and H2O2, respectively), or CHX (50 μg/mL) and MG132 (10 μm) for 0–8 hours as indicated. Cells were pelleted by centrifugation, washed twice with PBS, and lysed with modified RIPA buffer (150 mM NaCl, 50 mM Tris•HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 1 mM sodium orthovanadate, 10 mM NaF, 10 mM glycerophosphate, and 5 mM sodium pyrophosphate) supplemented with protease and phosphatase inhibitors (Roche) for 20–30 minutes on ice. Cell lysates were sonicated and clarified by centrifugation at 12,000 g. In some experiments, cells were fractionated to isolate cytosolic and nuclear protein. In brief, cells were incubated in lysis buffer (10 mM Tris·HCl, pH 7.4, 100 mM NaCl, and 2.5 mM MgCl2) for 15 minutes on ice and then centrifuged at 2,000 g for 10 minutes. Cell lysates were collected as cytosolic protein and the remaining cell pellets were lysed with RIPA buffer, incubated on ice for 20 minutes, and sonicated. The lysate was centrifuged at 15,000 g for 30 minutes to pellet nuclear protein. Equal amounts of cellular protein (10 μg) were separated by SDS-PAGE using 4%−15%/20% Criterion precast gels (Bio-Rad), transferred to PVDF membranes (MilliporeSigma), blocked with 5% nonfat milk for 1 hour at room temperature, and immunoblotted with primary antibodies overnight at 4°C followed by HRP-conjugated secondary antibodies at room temperature for 1 hour, washed 3 times in TBS-Tween 20, and then developed using a chemiluminescent substrate kit (Pierce). Antibodies used for immunoblotting were mouse monoclonal 7B4 anti-SRSF3 (1:1000 dilution, ATCC, CRL-2384), anti-fibronectin mouse monoclonal (1:1000 dilution, ab6328, Abcam), anti–β-actin mouse monoclonal (1:2000 dilution, 3700S, Cell Signaling Technology), anti–lamin B rabbit monoclonal (1:2000 dilution, 13435S, Cell Signaling Technology), HRP-labeled anti-mouse secondary antibody (1:5000 dilution, sc-516102, Santa Cruz Biotechnology), and HRP-labeled anti-rabbit secondary antibody (1:5000 dilution, sc-2357, Santa Cruz Biotechnology).
Immunofluorescence and immunohistochemistry.
Immunofluorescence was performed on formalin-fixed, paraffin-embedded (FFPE) human and mouse liver sections. Tissues were deparaffinized and rehydrated. After incubation with blocking buffer (1% BSA and 5% normal goat serum in PBS) at room temperature for 1 hour, slides were incubated overnight at 4°C with mouse anti-SRSF3 (7B4, 1:100 dilution), rabbit anti-SRSF3 (Abcam, ab73891, 1:100 dilution), or mouse anti-fibronectin (Abcam, ab6328, 1:100 dilution) antibodies in blocking buffer. After washing with PBS, slides were incubated with 1:1000 dilution of Alexa Fluor 488–conjugated anti–rabbit IgG and 1:1000 dilution of Alexa Fluor 555–conjugated anti–mouse IgG (Invitrogen). Nuclei were counterstained with DAPI. Staining was visualized by confocal laser-scanning microscopy (FluoView FV1000, Olympus). Immunohistochemistry was performed on FFPE human and mouse liver sections using the mouse anti-SRSF3 antibody (7B4, 1:500 dilution in 5% normal goat serum overnight at 4°C) and visualized using a Vectastain ABC immunohistology kit (Vector Labs).
Coimmunoprecipitation.
HepG2 and primary hepatocytes were harvested and lysed in RIPA buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) supplemented with protease inhibitors (Roche). Immunoprecipitation was performed by using Protein G beads (Dynabeads, Thermo Fisher Scientific) following the manufacturer’s protocol with slight modification. In brief, 1–2 mg of protein lysate was incubated with RNase A and isotype antibody at 4°C for 30 minutes. Beads and primary antibody were incubated for 10 minutes at room temperature followed by incubation at 4°C overnight. Primary antibody–conjugated beads were incubated with precleared lysates overnight at 4°C. The precipitated protein was eluted in NuPAGE LDS sample buffer followed by heating at 70°C for 10 minutes and immunoblotted using the indicated antibodies (mouse mAb 7B4 anti-SRSF3 antibody, rabbit mAb anti-NEDD8, 2754, Cell Signaling Technology; mouse mAb anti-FLAG, F3165, MilliporeSigma; rabbit mAb anti-FLAG, F7425, MilliporeSigma; and mouse mAb anti-ubiquitin, MAB1510, MilliporeSigma). NAE inhibitor (MLN4924, I-502) was purchased from BostonBiochem, and UAE inhibitor (MLN7243) was provided by Eric Bennett (UCSD).
RNAseq analysis.
Paired-end RNAseq was performed on total RNA isolated from primary hepatocytes purified from livers of obese and lean mice that had been on the HFD or a matched low-fat diet (10% kcal fat, D12450B, Research Diets) for 10 weeks, and on total RNA from primary hepatocytes with acute loss of SRSF3 due to adeno-CRE–mediated excision in Srsf3fl/fl hepatocytes compared with adeno-CRE-GFP–infected hepatocytes. Primary hepatocytes (5X105) were allowed to attach to collagen-coated plates for 6 h then infected with Adeno-Cre or Adeno-Gfp at 10,000 multiplicity of infection. RNA was extracted 24 hours later. All mice were on the C57BL/6J background. Reads were aligned to the mouse (mm10) genome using the STAR-2pass method (63) and AS events identified using JUM (29) and MAJIQ (31, 32). A false-discovery rate of 0.05 was used to identify differential AS events. Mouse genes showing HFD-induced AS events were compared to genes with SRSF3-dependent AS events to identify SRSF3-dependent changes in mouse NAFLD. Significant changes in splicing events by JUM and MAJIQ are provided in Supplemental Table 3. RNAseq data are available through the NCBI’s Gene Expression Omnibus (GEO GSE133398 and GSE133400).
AAV vector production and animal studies.
The pseudotyped AAV8-GFP, AAV8-SRSF3-WT, and AAV8-SRSF3-K11R Flag vectors were produced by the Viral Vector facility core of the Salk Institute for Biological Studies (La Jolla, California, USA). Male C57BL/6J mice were infected by tail vein injection of 1 × 1010 GC AAV8 per mouse. All infected mice were placed on a NASH-inducing diet for 7 weeks and then euthanized for further analysis. We analyzed 4 histological features shown to be independently associated with the diagnosis of NASH in adult liver biopsies: steatosis, hepatocellular ballooning, lobular inflammation, and fibrosis (64). Semiquantitative scores (from 0 to 5) for each feature were assigned to each slide containing 4 cross sections of liver tissue. Steatosis and hepatocellular ballooning were evaluated on slides stained independently with H&E, Masson’s trichrome, or Sirius red. Lobular inflammation was evaluated on H&E-stained sections and fibrosis evaluated using Masson’s trichrome– and Sirius red–stained sections.
Statistics.
Data were analyzed by 1- or 2-way ANOVA followed by Tukey’s multiple-comparisons post hoc test, or 2-tailed Student’s t test as appropriate using Prism (Graph Pad). Normality was assessed by the D’Agostino-Pearson omnibus normality test. Results are expressed as mean ± SEM and considered significant with P less than 0.05.
Study approval.
All animal procedures conformed to the US NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Animal Subjects Committee of UCSD.
Author contributions
As for author order for co-first authors, DK initiated the project and MD joined the project at a later date, but contributed half the data. NJGW and DK developed the hypothesis and designed the studies. DK and MD performed the experiments with the assistance of CS. DK performed the animal studies, the human studies, the tissue staining, the half-life and fractionation experiments, the transfections, and qPCR studies with the assistance of KK, PP, and MK. NW performed the transcriptomic analysis. MD performed the neddylation and ubiquitination experiments. CS, with assistance from KK, PP, and MK, performed animal husbandry and diet administration. GKB assisted with the primary hepatocyte preparation. DB produced and purified the mouse anti-SRSF3 antibody. LJ and RL provided human liver samples. LGE and LJ performed pathological assessment of tissue sections. LGE performed the tail vein viral injections. OO provided the liver RNAseq data from mice on HFD and low-fat diet. DK wrote the first draft of the manuscript and prepared drafts of the figures. NJGW provided financial support from VA and NIH grants, supervised all experiments, edited and proofed the manuscript, and generated the final figures. All authors have approved the final manuscript.
Supplementary Material
Acknowledgments
We would like to acknowledge the assistance of Valeria Estrada, Department of Pathology, UCSD, and the Biorepository and Tissue Technology Shared Resource and the Genomics and Computational Biology Shared Resource at the Moores Cancer Center (supported by NIH CA023100) for tissue processing and sectioning. This work was funded primarily by a VA Merit Review award to NJGW (I01BX000130) and a VA Senior Research Career Scientist Award, and in part by NIH grants to NJGW (HD012303, CA155435, and CA196853). The University of Minnesota Liver Tissue and Cell Distribution Service (UM-LTCDS) was funded by NIH contract HSN276201200017C; the Ontario Tumor Bank, Ontario Institute for Cancer Research (a nonprofit funded by the Government of Ontario, Toronto, Ontario, Canada); the Department of Pathology at the VA San Diego Health System; and the Moores Cancer Center Biorepository at UCSD.
Version 1. 08/08/2019
In-Press Preview
Version 2. 09/16/2019
Electronic publication
Version 3. 10/01/2019
Print issue publication
Funding Statement
This work was funded primarily by a DVA Merit Review award to N.J.G.W. (I01BX000130)
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4477–4491.https://doi.org/10.1172/JCI127374.
Contributor Information
Deepak Kumar, Email: dkumar@ucsd.edu.
Manasi Das, Email: madas@ucsd.edu.
Consuelo Sauceda, Email: cosauceda@ucsd.edu.
Lesley G. Ellies, Email: lellies@ucsd.edu.
Karina Kuo, Email: karina_kuo@yahoo.com.
Purva Parwal, Email: pparwal@ucsd.edu.
Mehak Kaur, Email: mek002@ucsd.edu.
Lily Jih, Email: Lily.Jih@va.gov.
Gautam K. Bandyopadhyay, Email: gbandyopadhyay@ucsd.edu.
Douglas Burton, Email: dwburton@ucsd.edu.
Rohit Loomba, Email: roloomba@ucsd.edu.
Olivia Osborn, Email: oosborn@ucsd.edu.
Nicholas J.G. Webster, Email: nwebster@ucsd.edu.
References
- 1.Corbo C, Orrù S, Salvatore F. SRp20: an overview of its role in human diseases. Biochem Biophys Res Commun. 2013;436(1):1–5. doi: 10.1016/j.bbrc.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 2.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417(1):15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 3.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16(16):5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jumaa H, Nielsen PJ. Regulation of SRp20 exon 4 splicing. Biochim Biophys Acta. 2000;1494(1–2):137–143. doi: 10.1016/s0167-4781(00)00233-5. [DOI] [PubMed] [Google Scholar]
- 5.Kurokawa K, et al. Downregulation of serine/arginine-rich splicing factor 3 induces G1 cell cycle arrest and apoptosis in colon cancer cells. Oncogene. 2014;33(11):1407–1417. doi: 10.1038/onc.2013.86. [DOI] [PubMed] [Google Scholar]
- 6.Jumaa H, Guénet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the Srp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17(6):3116–3124. doi: 10.1128/MCB.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6(7):806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen S, Talukdar I, Webster NJ. SRp20 and CUG-BP1 modulate insulin receptor exon 11 alternative splicing. Mol Cell Biol. 2009;29(3):871–880. doi: 10.1128/MCB.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen S, Jumaa H, Webster NJ. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun. 2013;4:1336. doi: 10.1038/ncomms2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen S, Langiewicz M, Jumaa H, Webster NJ. Deletion of serine/arginine-rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology. 2015;61(1):171–183. doi: 10.1002/hep.27380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elizalde M, et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest. 2014;124(7):2909–2920. doi: 10.1172/JCI74382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison SA, Torgerson S, Hayashi PH. The natural history of nonalcoholic fatty liver disease: a clinical histopathological study. Am J Gastroenterol. 2003;98(9):2042–2047. doi: 10.1111/j.1572-0241.2003.07659.x. [DOI] [PubMed] [Google Scholar]
- 13.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43(2 Suppl 1):S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 14.Ahmed M. Non-alcoholic fatty liver disease in 2015. World J Hepatol. 2015;7(11):1450–1459. doi: 10.4254/wjh.v7.i11.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munteanu MA, Nagy GA, Mircea PA. Current management of NAFLD. Clujul Med. 2016;89(1):19–23. doi: 10.15386/cjmed-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherif ZA, et al. Global epidemiology of nonalcoholic fatty liver disease and perspectives on US minority populations. Dig Dis Sci. 2016;61(5):1214–1225. doi: 10.1007/s10620-016-4143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346(16):1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 18.Torres DM, Harrison SA. Nonalcoholic steatohepatitis and noncirrhotic hepatocellular carcinoma: fertile soil. Semin Liver Dis. 2012;32(1):30–38. doi: 10.1055/s-0032-1306424. [DOI] [PubMed] [Google Scholar]
- 19.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 20.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 21.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44(2):325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10(10):M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udeshi ND, et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol Cell Proteomics. 2012;11(5):148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayabalan AK, et al. NEDDylation promotes stress granule assembly. Nat Commun. 2016;7:12125. doi: 10.1038/ncomms12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Twyffels L, Gueydan C, Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278(18):3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, et al. Alteration of splicing factors’ expression during liver disease progression: impact on hepatocellular carcinoma outcome. Hepatol Int. 2019;13(4):454–467. doi: 10.1007/s12072-019-09950-7. [DOI] [PubMed] [Google Scholar]
- 28.Kano S, et al. Oxidative stress-inducible truncated serine/arginine-rich splicing factor 3 regulates interleukin-8 production in human colon cancer cells. Am J Physiol, Cell Physiol. 2014;306(3):C250–C262. doi: 10.1152/ajpcell.00091.2013. [DOI] [PubMed] [Google Scholar]
- 29.Jiménez M, et al. Splicing events in the control of genome integrity: role of SLU7 and truncated SRSF3 proteins. Nucleic Acids Res. 2019;47(7):3450–3466. doi: 10.1093/nar/gkz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Rio DC. JUM is a computational method for comprehensive annotation-free analysis of alternative pre-mRNA splicing patterns. Proc Natl Acad Sci U S A. 2018;115(35):E8181–E8190. doi: 10.1073/pnas.1806018115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaquero-Garcia J, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5:e11752. doi: 10.7554/eLife.11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norton SS, Vaquero-Garcia J, Lahens NF, Grant GR, Barash Y. Outlier detection for improved differential splicing quantification from RNA-Seq experiments with replicates. Bioinformatics. 2018;34(9):1488–1497. doi: 10.1093/bioinformatics/btx790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kano S, et al. Truncated serine/arginine-rich splicing factor 3 accelerates cell growth through up-regulating c-Jun expression. J Med Invest. 2013;60(3–4):228–235. doi: 10.2152/jmi.60.228. [DOI] [PubMed] [Google Scholar]
- 34.Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9(10):969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin Investig Drugs. 2012;21(10):1563–1573. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Nakatani T, Kamitani T. Inhibition of NEDD8-conjugation pathway by novel molecules: potential approaches to anticancer therapy. Mol Oncol. 2012;6(3):267–275. doi: 10.1016/j.molonc.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway J, Al-Zahrani KN, Pryce BR, Abou-Hamad J, Sabourin LA. Transforming growth factor β-induced epithelial to mesenchymal transition requires the Ste20-like kinase SLK independently of its catalytic activity. Oncotarget. 2017;8(58):98745–98756. doi: 10.18632/oncotarget.21928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohmura G, et al. Aberrant myosin 1b expression promotes cell migration and lymph node metastasis of hNSCC. Mol Cancer Res. 2015;13(4):721–731. doi: 10.1158/1541-7786.MCR-14-0410. [DOI] [PubMed] [Google Scholar]
- 39.Kosaki A, Nelson J, Webster NJ. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J Biol Chem. 1998;273(17):10331–10337. doi: 10.1074/jbc.273.17.10331. [DOI] [PubMed] [Google Scholar]
- 40.Webster NJ, Evans LG, Caples M, Erker L, Chew SL. Assembly of splicing complexes on exon 11 of the human insulin receptor gene does not correlate with splicing efficiency in-vitro. BMC Mol Biol. 2004;5:7. doi: 10.1186/1471-2199-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen S, Talukdar I, Liu Y, Tam J, Reddy S, Webster NJ. Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J Biol Chem. 2010;285(33):25426–25437. doi: 10.1074/jbc.M109.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talukdar I, Sen S, Urbano R, Thompson J, Yates JR, Webster NJ. hnRNP A1 and hnRNP F modulate the alternative splicing of exon 11 of the insulin receptor gene. PLoS ONE. 2011;6(11):e27869. doi: 10.1371/journal.pone.0027869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawrence MC, McKern NM, Ward CW. Insulin receptor structure and its implications for the IGF-1 receptor. Curr Opin Struct Biol. 2007;17(6):699–705. doi: 10.1016/j.sbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr Rev. 2009;30(6):586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- 45.Gallagher EJ, LeRoith D. The proliferating role of insulin and insulin-like growth factors in cancer. Trends Endocrinol Metab. 2010;21(10):610–618. doi: 10.1016/j.tem.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuranaga Y, et al. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Mol Sci. 2018;19(10):E3012. doi: 10.3390/ijms19103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loomis RJ, et al. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33(4):450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X, Zhang P. Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol Cancer. 2015;14:158. doi: 10.1186/s12943-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McFarlane M, Graham SV. Human papillomavirus regulation of SR proteins. Biochem Soc Trans. 2010;38(4):1116–1121. doi: 10.1042/BST0381116. [DOI] [PubMed] [Google Scholar]
- 50.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152(4):844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez N, Cordiner RA, Young RS, Hug N, Macias S, Cáceres JF. Genetic variation and RNA structure regulate microRNA biogenesis. Nat Commun. 2017;8:15114. doi: 10.1038/ncomms15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim K, Nguyen TD, Li S, Nguyen TA. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA. 2018;24(7):892–898. doi: 10.1261/rna.065862.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HR, et al. MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3. Oncotarget. 2017;8(5):8342–8355. doi: 10.18632/oncotarget.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C, et al. An indel polymorphism within pre-miR3131 confers risk for hepatocellular carcinoma. Carcinogenesis. 2017;38(2):168–176. doi: 10.1093/carcin/bgw206. [DOI] [PubMed] [Google Scholar]
- 55.Yoon SP, Kim HH, Kim J, Park RY, Ohn T. Regulation of cellular RNA nano-particle assembly by splicing factor SRp20. J Nanosci Nanotechnol. 2013;13(1):184–187. doi: 10.1166/jnn.2013.6951. [DOI] [PubMed] [Google Scholar]
- 56.Delgado TC, et al. Neddylation, a novel paradigm in liver cancer. Transl Gastroenterol Hepatol. 2018;3:37. doi: 10.21037/tgh.2018.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, et al. Inhibition of neddylation modification by MLN4924 sensitizes hepatocellular carcinoma cells to sorafenib. Oncol Rep. 2019;41(6):3257–3269. doi: 10.3892/or.2019.7098. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Park HS, et al. PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 2016;23(8):1296–1311. doi: 10.1038/cdd.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hipp MS, Raasi S, Groettrup M, Schmidtke G. NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J Biol Chem. 2004;279(16):16503–16510. doi: 10.1074/jbc.M310114200. [DOI] [PubMed] [Google Scholar]
- 60.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276(49):46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 61.Luo Z, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72(13):3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.