Abstract
Metastatic castration-resistant prostate cancer (mCRPC) is a heterogeneous disease with diverse drivers of disease progression and mechanisms of therapeutic resistance. We conducted deep phenotypic characterization of CRPC metastases and patient-derived xenograft (PDX) lines using whole-genome RNA sequencing, gene set enrichment analysis, and immunohistochemistry. Our analyses revealed 5 mCRPC phenotypes based on the expression of well-characterized androgen receptor (AR) or neuroendocrine (NE) genes: AR-high tumors (ARPC), AR-low tumors (ARLPC), amphicrine tumors composed of cells coexpressing AR and NE genes (AMPC), double-negative tumors (i.e., AR–/NE–; DNPC), and tumors with small cell or NE gene expression without AR activity (SCNPC). RE1 silencing transcription factor (REST) activity, which suppresses NE gene expression, was lost in AMPC and SCNPC PDX models. However, knockdown of REST in cell lines revealed that attenuated REST activity drives the AMPC phenotype but is not sufficient for SCNPC conversion. We also identified a subtype of DNPC tumors with squamous differentiation and generated an encompassing 26-gene transcriptional signature that distinguished the 5 mCRPC phenotypes. Together, our data highlight the central role of AR and REST in classifying treatment-resistant mCRPC phenotypes. These molecular classifications could potentially guide future therapeutic studies and clinical trial design.
Keywords: Cell Biology, Oncology
Keywords: Expression profiling, Molecular pathology, Prostate cancer
Introduction
The androgen receptor (AR) regulates cellular programs that promote the survival and proliferation of prostate cancer (PC) cells. Consequently, first-line treatment for metastatic PC centers on inhibiting AR activity through androgen deprivation therapy (ADT), resulting in the suppression of AR target genes and clinical remissions that generally last several years (1). However, ADT is not curative. PC recurs as castration-resistant prostate cancer (CRPC), typically with reactivated AR signaling. Second-generation AR pathway inhibitors (ARIs), such as enzalutamide (ENZ) and abiraterone (ABI), were designed to further repress AR signaling and are primarily used to treat CRPC. Although these agents extend survival, durable complete responses are rare and these therapies also eventually fail (2, 3).
Typically, the vast majority of metastatic CRPC (mCRPC) tumors progress with rising prostate-specific antigen (PSA/KLK3) levels despite standard of care treatment. Moreover, most mCRPC tumors are adenocarcinomas, which have robust AR program activity (4). Though rigorous epidemiological data are lacking, recent studies report that a substantial number of mCRPC tumors progressing on ARIs have lost AR signaling (5). Paralleling increased use of ARIs has been an increase in the proportion of treatment-resistant CRPC metastases that have AR-null phenotypes, i.e. tumors with diffuse small cell or neuroendocrine (NE) characteristics (SCNPC) or the recently described double-negative (DNPC) phenotype that lacks both NE and AR activity (5). A contemporary study evaluating the histology and molecular characteristics of 202 men with mCRPC found that 17% of the evaluable tumors were classified as SCNPC and this phenotype was associated with shortened survival (6). Notably, a subset of tumors exhibited discordance between the pathological assessment of SCNPC and gene expression programs. Other tumors exhibited mixed phenotypes of NE features concurrent with AR activity, raising questions of intratumoral heterogeneity due to mixtures of cell types (6).
In addition to histology, the classification of cancers into subtypes with distinct functional features has been supplemented with transcript profiles. For example, transcript signatures subdivide breast cancers into groups that exhibit distinct outcomes (7). This subclassification, based on gene expression levels, has also been applied to localized PC and shown to associate with treatment outcomes (8, 9). In this study, we sought to characterize the phenotypic diversity of treatment-refractory mCRPC using both histological assessments and gene expression profiling. In addition to SCNPC and DNPC, we identified adenocarcinomas that had high AR activity (ARPC), measurable but low AR activity (ARLPC), and amphicrine tumors comprised of cells exhibiting both AR and NE activity (AMPC). Notably, a subset of DNPC tumors exhibited squamous differentiation. We determined that the relationships of 2 major factors regulating cell differentiation states, AR and RE1 silencing transcription factor (REST), associated with these mCRPC subtypes. We generated a 26-gene transcriptional signature that could be clinically useful for classifying mCRPC phenotypes and prioritizing treatment.
Results
Treatment-refractory mCRPC exhibits diverse phenotypes.
To assess the diversity of phenotypes present following resistance to therapeutics used to treat metastatic PC, we evaluated 98 tumors obtained at rapid autopsy from 55 men between 2003 and 2017. Patient demographics and clinical data are summarized in Supplemental Table 2; supplemental material available online with this article; https://doi.org/10.1172/JCI128212DS1 All patients received ADT. The median duration of treatment was 4.2 years (range, 0.3–15.1 years). Patients also received a variety of other drugs including docetaxel (n = 44; 80%), abiraterone (n = 8; 15%), enzalutamide (n = 4; 7%) or both abiraterone and enzalutamide (n = 17; 31%). Bone, lymph nodes, and liver were the most frequent sites of metastasis.
To provide an initial evaluation of phenotypic diversity, we used histological assessments and immunohistochemical (IHC) analysis of proteins associated with AR-active adenocarcinomas (AR and PSA) and NE differentiation (chromogranin A, CHGA; synaptophysin, SYP). We observed 5 distinct mCRPC phenotypes: adenocarcinomas with near-uniform expression of AR and PSA, and lack of CHGA and SYP expression, classified as ARPC; adenocarcinomas with weak or heterogeneous expression of AR and PSA, and negative for CHGA and SYP, classified as AR-low PC (ARLPC); tumors composed of cells that coexpress AR, PSA, CHGA, and SYP, classified as amphicrine PC (AMPC); tumors with small cell or neuroendocrine histology with CHGA and SYP expression and lack of AR and PSA expression, classified as small cell or neuroendocrine PC (SCNPC); and tumors lacking detectable expression of AR, PSA, CHGA, and SYP, classified as double-negative PC (DNPC; Figure 1A).
Figure 1. Molecular profiling of mCRPC reveals a heterogeneous disease.
(A) IHC of 5 mCRPC sites from patients using antibodies to AR, PSA, CHGA, and SYP. Scale bars: 20 μM. (B) RNA-Seq heatmap of mCRPC specimens acquired through rapid autopsy from 2003–2017 (n = 98). REST-repressed NE genes are listed in the NEURO I panel (top), NE transcription factors are listed in the NEURO II panel (middle), and AR-associated genes are listed in the AR panel (bottom). Results are expressed as log2 fragments per kilobase of transcript per million mapped reads (FPKM) and colored according to scale. (C) Venn diagram showing the number of unique and shared upregulated genes between phenotypes relative to ARPC (up >3-fold; P < 0.05). ARPC (AR-high prostate cancer; AR+/NE–), ARLPC (AR-low prostate cancer; ARlow/NE–), AMPC (amphicrine prostate cancer; AR+/NE+), DNPC (double-negative prostate cancer; AR–/NE–), and SCNPC (small cell or neuroendocrine prostate cancer; AR–/NE+).
Transcriptome profiles associate with mCRPC phenotypes.
Morphologic and IHC analyses are the gold standard for pathologic diagnosis. However, the assessment of transcriptional programs has found utility in subclassifying breast tissue and cancers that exhibit similar histological characteristics (10, 11). We sought to develop a clearer understanding of the mCRPC disease continuum through transcriptome profiling by whole-genome RNA sequencing (RNA-Seq) of 98 mCRPC tumors. Patient metastases were first analyzed and segregated according to expression levels of a gene signature reflecting AR activity (AR panel; Figure 1B). The AR-regulated genes selected for the AR signature are well characterized in the literature and include KLK3, NKX3-1, SLC45A3, and TARP (12–14). Tumors were also analyzed and segregated depending on expression levels of NE-associated genes. We previously demonstrated that NE-associated genes can be separated into REST-repressed genes such as SYP, CHGA, SNAP25, and SRRM4 (NEURO I panel; ref. 15), and transcription factors that regulate NE differentiation, such as SOX2, POU3F2/BRN2, NKX2-1, and LMO3 (NEURO II panel; refs. 15–17). Applying the AR, NEURO I, and NEURO II gene expression sets to the mCRPC tumors clearly defined the 5 mCRPC subtypes (Figure 1B). ARPC tumors expressed AR-regulated genes but also showed heterogeneity with low expression of some NE genes. ARLPCs had attenuated AR expression with concomitant low expression of some AR-regulated genes. AMPCs expressed AR-associated genes and REST-repressed neuronal factors (NEURO I), but lacked expression of the NE-associated transcription factors (NEURO II). DNPC tumors were generally devoid of AR, NEURO I, and NEURO II panel genes. SCNPC tumors lacked AR expression and signaling but expressed both the NEURO I and NEURO II genes. Analysis of 62 corresponding tumor sites through IHC revealed that phenotypic determinations based on AR, PSA, CHGA, and SYP staining mirrored the phenotypic determinations made through RNA-Seq analysis (Supplemental Figure 1). Moreover, our rapid autopsy cohort included 34 patients, each with 2–3 metastatic sites characterized through RNA-Seq. This provided an opportunity to query the intertumoral phenotypic heterogeneity within patients. Of the 34 patients with 2–3 analyzed metastases, 5 patients (14.7%) displayed phenotypic differences between sites. However, this may underestimate the extent of heterogeneity, as 2–3 metastases generally represent a fraction of the total tumor burden. In addition, there can be intratumoral phenotypic heterogeneity that is not readily assessed through bulk RNA sequencing (Supplemental Figure 2).
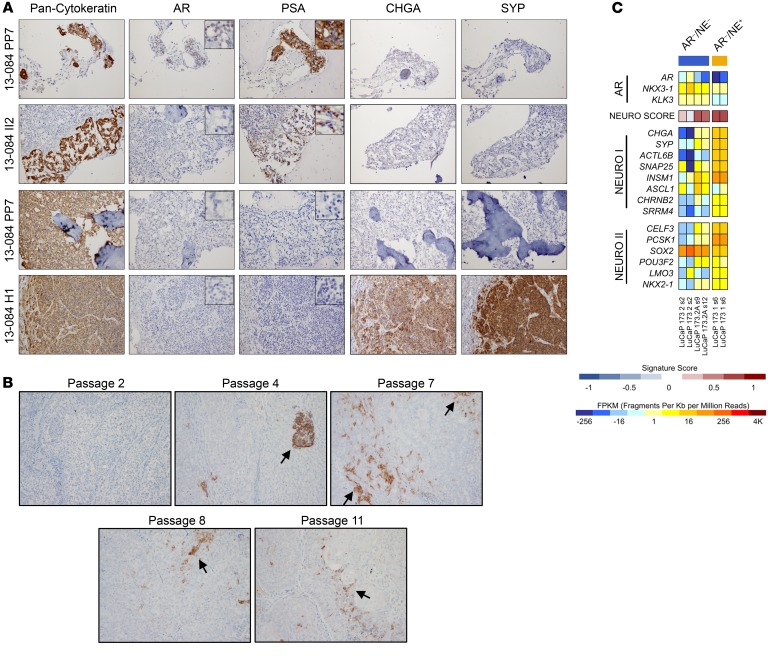
To validate the results of the patient specimen analysis using an orthogonal system, we conducted RNA-Seq and IHC analyses on 18 CRPC LuCaP patient-derived xenograft (PDX) lines. The 5 distinct phenotypes were identified by IHC and accurately segregated according to the AR, NEURO I, and NEURO II gene expression profiles (Supplemental Figure 3, A and B).
To discover novel gene expression profiles for each of the mCRPC phenotypes, we cross-compared the patient metastases RNA-Seq data from the defined phenotypic cohorts (i.e., ARLPC, AMPC, DNPC, and SCNPC) relative to ARPC. This analysis generated a comprehensive list of unique and shared upregulated differentially expressed genes (vs. ARPC, up >3-fold and P < 0.05; Figure 1C and Supplemental Table 3). In addition, this analysis demonstrated that DNPC and SCNPC are markedly different from the other mCRPC phenotypes (806 and 1669 unique upregulated genes respectively; Figure 1C). Notably, the AR-null phenotypes (DNPC and SCNPC) share an additional 590 upregulated genes relative to the AR-expressing phenotypes (Figure 1C). Furthermore, ARLPC, and AMPC share gene expression profiles similar to ARPC (Figure 1C).
Gene Set Enrichment Analysis (GSEA) determined that the 806 upregulated genes unique to DNPC were enriched in Gene Ontology biological process terms for response to external biotic stimulus (P = 4.5 × 10–18), immune system process (P = 3.5 × 10–15), and cornification (P = 5.5 × 10–9). As expected, the 1669 upregulated genes unique to SCNPC were enriched for core neuronal activities such as nervous system process (P = 7.5 × 10–12) and regulation of nervous system process (P = 4.7 × 10–10). Interestingly, the top processes for the 829 upregulated genes in common between SCNPC and DNPC included locomotory behavior (P = 5.0 × 10–07) and cell adhesion (P = 2.1 × 10–07), suggesting changes in metastatic potential common to AR-null phenotypes. The 229 upregulated genes unique to the ARLPC phenotype were enriched for processes in response to external biotic stimulus (P = 3.3 × 10–07) and regulation of inflammatory response (P = 8.4 × 10–06), while the 193 upregulated genes in common between ARLPC, DNPC, and SCNPC phenotypes were significantly enriched in acute inflammatory response (P = 4.0 × 10–11) and defense response (P = 1.3 × 10–06). Finally, GSEA determined that the 111 upregulated genes unique to AMPC were not involved in any significant processes. However, the 250 common upregulated genes between AMPC and SCNPC were involved in ancillary neuronal processes such as neurotransmitter transport (P = 4.4 × 10–10) and synaptic vesicle localization (P = 6.5 × 10–09). Importantly, analysis of the 250 significantly upregulated genes shared between SCNPC and AMPC using the MSigDB C3-Transcription Factor Target database showed that REST was the top transcription factor pathway altered in the gene set (P = 4.2 × 10–35; Supplemental Table 4). Taken together, these data support the use of AR, NEURO I, and NEURO II genes to segregate mCRPC phenotypes and identify biologically relevant pathways that emphasize the heterogeneity of mCRPC.
mCRPC phenotypes represent a disease continuum.
The relationships between different mCRPC phenotypes have not been clearly established though prior studies suggest that SCNPC is often derived from an AR-positive precursor, or share a common progenitor (18, 19). Thus, we investigated these relationships by studying the phenotypic progression of an individual with mCRPC and a complicated treatment history. At diagnosis in 2012, patient 13-084 had a PSA of 159 ng/mL and a prostate biopsy revealed an adenocarcinoma with a Gleason score of 4 + 5 = 9 and IHC demonstrating focal NE differentiation. He was treated with leuprolide and bicalutamide but ensuing CT and bone scans revealed numerous metastases in liver, lung, and bone. Platinum-based chemotherapy was initiated due to the possible presence of SCNPC with cycles of carboplatin/irinotecan (5 cycles) or cisplatin/irinotecan (3 cycles) leading to a PSA decline and stable disease for approximately 8 months. He subsequently received 2 cycles of capecitabine and gemcitabine with eventual PSA progression prior to death (Supplemental Figure 4). At autopsy in 2013, we recovered several metastatic tumors that exhibited different phenotypes, including ARPC, ARLPC, and DNPC sites in bone and SCNPC sites in the liver as determined by IHC (Figure 2A). We developed 2 PDX lines representing SCNPC (LuCaP 173.1) and DNPC (LuCaP 173.2) from acquired patient metastases (5). Previous reports have described the transdifferentiation of an AR-expressing adenocarcinoma PC PDX line to SCNPC (20). We therefore questioned whether the DNPC phenotype is a transition stage. We sought to test the hypothesis that sustained tumor growth through serial passaging of the DNPC LuCaP 173.2 PDX line would lead to SCNPC conversion. Indeed, serial passaging in intact mice of LuCaP 173.2 and staining for SYP expression through IHC showed that first passage PDX tumors were negative for SYP expression, whereas SYP-positive cells were detected by passage 4 and were maintained as a minor cell population through passage 9 (Figure 2B). RNA-Seq of LuCaP 173.2 from passages 2, 9, and 12 indicated that the NEURO I and NEURO II panels of genes were expressed at substantially higher levels in the later passages (Figure 2C). Furthermore, the NEURO scores of LuCaP 173.2 from passages 9 and 12 were similar to SCNPC LuCaP 173.1. These data suggest that mCRPC is a disease continuum, and that although the DNPC phenotype is generally stable, a small proportion of DNPC tumor cells possess an intrinsic plasticity that permits conversion to SCNPC.
Figure 2. Disease progression is a continuum in mCRPC specimens.
(A) IHC of different mCRPC sites from patient 13-084. Site PP7 (bone; ARPC), II2 (bone; ARLPC), PP7 (bone; DNPC), and H1 (liver, SCNPC). Primary antibodies were directed toward pan-cytokeratin, AR, PSA, CHGA, and SYP. Insets for AR and PSA staining are images of the same section using the ×400 objective lens. Original magnification 40×. (B) IHC of LuCaP 173.2 tumor sections from passages 2, 4, 7, 8, and 11 using a SYP antibody. Black arrows point to clusters of cells with SYP positivity. Magnification 100×. (C) RNA-Seq heatmap and NEURO score of LuCaP 173.1 and serial passages from LuCaP 173.2. Results are expressed as log2 FPKM or as enrichment scores and are colored according to scale.
The amphicrine phenotype and relationship with REST expression.
Historically, SCNPC was considered to lack AR activity but recent studies have identified atypical tumors with NE features that express AR and exhibit AR-mediated signaling (6). In addition, classification of the AMPC phenotype based on bulk RNA-Seq or IHC using single markers may be due to tumors comprised of multiple cell types (AR+/NE– and AR–/NE+) or may be due to tumors comprised of individual cells expressing both AR and NE differentiation markers simultaneously. Thus, to establish the existence of AMPC cells in our clinical specimens (i.e., cells with both AR transcriptional output and neuroendocrine features; AR+/NE+), we stained LuCaP PDX tumors and patient metastases with immunofluorescent (IF) antibodies to PSA and SYP. AMPC LuCaP 77CR tumors contained numerous cells coexpressing PSA and SYP (Figure 3A). Though PSA and SYP coexpression normally occurred throughout LuCaP 77CR tumors, we also identified a subset of tumors with focal SYP expression (unpublished observations). Furthermore, we used IF to characterize 6 AMPC metastases from 4 patients. The patient tissues used for IF analysis were adjacent to the specimens used for RNA-Seq. Importantly, IF demonstrated clear PSA and SYP coexpression in patient tumor cells. However, patient 13-042 site M3 and patient 17-033 site J1 showed regions that were mixed of distinct populations of ARPC and SCNPC tumor cells (Supplemental Figure 5).
Figure 3. REST splicing occurs in AMPC and SCNPC phenotypes.
(A) Immunofluorescence of an AMPC LuCaP 77CR tumor using PSA (green) and SYP (red) antibodies. Sections were counterstained with DAPI (blue) and top panels represent LuCaP 77CR PDX sections stained with secondary antibody only. Scale bars: 20 μM. (B) Immunoblot of LuCaP PDX specimens probing for REST, AR, and SYP. ACTB was used as a loading control. Short, 10-second film exposure; long, 5-minute film exposure. (C) PCR of LuCaP PDX specimens using primers specific to REST shows the REST4 insertion sequence appearing in AMPC (LuCaP 77CR) and SCNPC (LuCaP 93, 145.2, and 173.1) but not in DNPC (LuCaP 173.2) or ARPC (LuCaP 86.2 and 73). (D) RNA-Seq heatmap of VCaP cells displaying NE-associated genes (NEURO I and NEURO II) and AR-associated genes. Results are expressed as log2 FPKM and colored according to scale. (E) Immunoblot of C4-2B, VCaP, and LuCaP 93 whole-cell extracts using antibodies against AR, REST, SYP, and ACTB. ACTB was used as a loading control. (F) PCR of C4-2B, VCaP, and NCIH660 cells using primers specific to REST. The upper band represents the REST4 splice variant.
Previously, we interrogated SCNPC patient tumors and PDX lines and determined that SCNPC is associated with loss of REST repressor activity (15). However, the role of REST in other mCRPC phenotypes has not been evaluated. Using representative PDX lines from ARPC (LuCaP 35CR and 96CR), AMPC (LuCaP 77CR), ARLPC (LuCaP 176), DNPC (LuCaP 173.2), and SCNPC (LuCaP 173.1 and 93), we found that full-length REST protein was decreased in both AMPC and SCNPC tumors (Figure 3B). Interestingly, we observed REST species with our REST C-terminus antibody at both approximately 120 KDa and approximately 200 KDa in PDX lysates from ARPC, ARLPC, and DNPC. Full-length REST protein is predicted to be 116 KDa but can be O-glycosylated and readily detected at approximately 200 KDa (21, 22). Additionally, the REST transcript has multiple splice variants that produce truncated proteins (23). Nevertheless, both REST proteins with intact C-terminus repressor domains were diminished in AMPC and SCNPC PDX models. Moreover, AMPC LuCaP 77CR showed robust expression of both AR and SYP protein, whereas ARLPC LuCaP 176 had low AR protein expression (Figure 3B).
Next, we examined alternative splicing of the REST transcript. The RNA splicing factor SRRM4 splices the REST transcript to REST4, resulting in the loss of the C-terminus repressor domain and diminished REST transcriptional repression (15, 24, 25). To examine SRRM4-mediated splicing of REST in mCRPC phenotypes, we conducted PCR using REST primers spanning the SRRM4 splice site and determined that REST4 splicing events occurred exclusively in AMPC and SCNPC LuCaP PDX models (Figure 3C). However, instead of a previously reported 62 bp insertion into the REST transcript (25), band sequencing identified a 50 bp insertion, suggesting a mechanism of REST4 splicing in AMPC and SCNPC phenotypes similar to small cell lung cancer (26). Taken together, these data indicate that the AMPC phenotype arises from the loss of REST transcriptional repression.
To further support the existence of the AMPC phenotype, we interrogated the VCaP PC cell line, which exhibits an amphicrine-like transcript profile. RNA-Seq confirmed that VCaP cells express AR-associated genes and the REST-repressed NEURO I genes but do not express the NEURO II transcription factors that drive the SCNPC phenotype (Figure 3D). Immunoblot analysis determined that VCaP cells express considerable AR and SYP protein and have diminished full-length REST protein expression compared with C4-2B cells (Figure 3E). Immunofluorescence validated that both PSA and SYP are coexpressed in the same VCaP cell (Supplemental Figure 6) and PCR analysis of REST determined that REST transcripts are alternatively spliced to REST4, similar to neuroendocrine NCIH660 cells (Figure 3F). Notably, the growth of VCaP cells is inhibited by ADT or exposure to the AR antagonist enzalutamide (27). In addition, 22Rv1 cells also exhibit features of AMPC as they have attenuated REST expression and appreciable SYP and AR protein expression (28). Taken together, these results confirm the existence of AMPC cells in patient specimens and in CRPC models in vitro and in vivo, and suggest that the AMPC phenotype is still driven by AR activity and is responsive, at least transiently, to AR pathway repression.
REST knockdown in CRPC cells promotes an amphicrine phenotype.
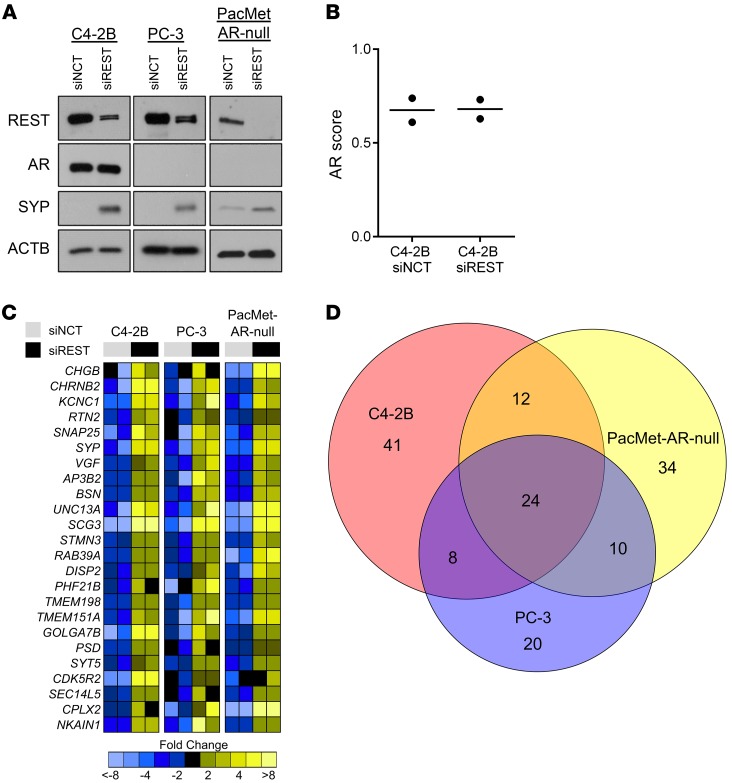
To examine the impact of the loss of REST activity in AR-expressing and AR-null cell lines, we conducted knockdown studies using siRNAs directed to REST (siREST) or a negative control (siNCT) in C4-2B, PC-3, and PacMet AR-null cells. PacMetUT1 cells were modified using CRISPR-Cas9–mediated editing to knockout AR expression and were characterized previously (5, 29). REST knockdown led to increases in the NE-associated protein SYP (Figure 4A). Interestingly, REST depletion in AR-expressing C4-2B cells did not alter AR protein expression or the magnitude of AR transcriptional output (Figure 4, A and B). However, this does not rule out the possibility that complete loss of REST could alter the AR transcriptional output. Transcript profiling by RNA-Seq and subsequent GSEA showed that REST ablation in C4-2B, PC-3, and PacMet AR-null cells led to significant upregulation of known REST-repressed genes (up >3-fold; P < 0.05). Furthermore, the REST pathway was the top altered pathway from the MSigDB C3-Transcription Factor Target database in all siREST transfected cell lines. Surprisingly, the number of genes significantly upregulated with REST knockdown was relatively low across all cell models and only 24 genes were in common between C4-2B, PC-3, and PacMet AR-null cells (Figure 4, C and D; Supplemental Table 5). The REST-repressed genes with increased expression following REST depletion included SYP, SNAP25, CHRNB2 (NEURO I Panel) as well as VGF, SCG3, and CHGB (Figure 4C). However, REST knockdown did not significantly alter the expression of transcription factors and drivers of SCNPC such as NKX2-1, POU3F2, and SOX2 (NEURO II panel) in either AR-expressing or AR-null CRPC cell lines (Supplemental Figure 7). Taken together, we determined that REST loss induces the expression of a limited set of NE-associated genes (NEURO I) and drives PC conversion to the AMPC phenotype with continued evidence of AR activity.
Figure 4. REST knockdown in AR-expressing and AR-null CRPC cell lines.
(A) Immunoblot of REST, AR, SYP, and ACTB using C4-2B, PC-3, and PacMet AR-null cells transfected with either REST siRNA (siREST) or negative control siRNA (siNCT). (B) AR activity scores assessed in C4-2B cells transfected with siNCT (n = 2) or siREST (n = 2) by RNA-Seq. (C) RNA-Seq heatmap of the 24 common upregulated genes (up >3-fold; P < 0.05) between C4-2B, PC-3, and PacMet AR-null cells transfected with siREST or siNCT. Log2 mean-centered ratios of genes are depicted and colored according to scale. (D) Venn diagram describing the interrelationships of all upregulated genes (vs. siNCT; up >3-fold; P < 0.05) identified through RNA-Seq in siREST transfected cell lines.
A subtype of mCRPC exhibits features of squamous cell carcinoma.
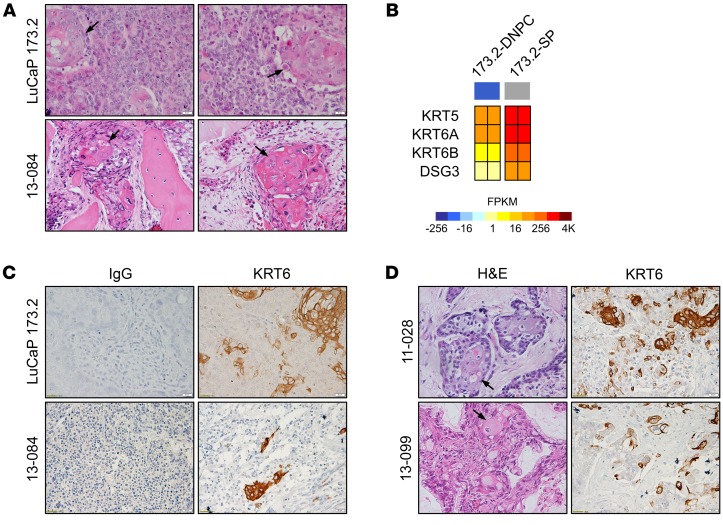
While histologically characterizing the DNPC LuCaP 173.2 PDX model, we observed squamous pearls, which were evidence of focal squamous differentiation (Figure 5A). To determine if the squamous pearls occurred spontaneously during LuCaP 173.2 development or were native to the original malignancy, we evaluated tumors from patient 13-084 and identified squamous pearl structures in adjacent tumor sections of the rib bone metastasis that served as the origin of LuCaP 173.2 (Figure 5A). Squamous pearl cells from LuCaP 173.2 PDX tumors were then isolated using laser capture microdissection and subjected to RNA-Seq and GSEA. Transcriptome analysis determined that 880 genes were upregulated and 29 genes were downregulated in LuCaP 173.2 squamous pearl cells compared with surrounding DNPC tumor cells (FDR < 0.001; Supplemental Table 6). GSEA determined that many of the upregulated genes were enriched in other squamous cancer gene sets, such as RICKMAN_HEAD_AND_NECK (P < 0.0001; ref. 30). Importantly, KRT5, KRT6A, KRT6B, and DSG3 were recently highlighted through ROC curves as biomarkers to differentiate lung adenocarcinoma from lung squamous cell cancer (31). Indeed, our analysis showed that KRT5, KRT6A, KRT6B, and DSG3 were highly expressed in LuCaP 173.2 squamous pearl cells compared with surrounding DNPC tissue (Figure 5B). IHC using a primary antibody specific to KRT6 in LuCaP 173.2 and 13-084 tumor specimens revealed strong KRT6 staining only in tumor cells with squamous pearl morphology (Figure 5C).
Figure 5. DNPC can convert to a squamous phenotype.
(A) H&E staining of mCRPC tissues from LuCaP 173.2 and patient 13-084. Black arrows point to squamous pearl structures. (B) Expression of squamous cell lung cancer associated genes from RNA-Seq of LuCaP 173.2 DNPC cells and squamous pearl (SP) cells isolated by laser capture microdissection. Results are expressed as log2 FPKM and colored according to scale. (C) IHC of specimens from LuCaP 173.2 and patient 13-084 using KRT6 antibody or IgG as a negative control. (D) H&E staining (left panels) and KRT6 IHC (right panels) of DNPC tumor sections from patients 11-028 and 13-099. Scale bars: 20 μM.
Examination of 4 other patients with DNPC metastases identified squamous pearls with positive KRT6 staining in 2 patients: 11-028 and 13-099 (Figure 5D). Interestingly, patient 11-028 had an adenocarcinoma phenotype in the initial prostate biopsy and was subsequently treated with diethylstilbesterol (DES) for 13 months prior to cystoprostatectomy. At the time of cystoprostatectomy, histology and IHC revealed adenocarcinoma with focal basaloid and squamous differentiation in several sections of the prostate as well as a left axillary lymph node that was consistent with squamous carcinoma. The other 2 patients with KRT6-positive metastases, patients 13-084 and 13-099, had primary prostate cancers with no evidence of squamous differentiation, and subsequent hormone therapy led to the appearance of squamous mCRPC. Although DES and hormone therapies have been linked to the development of squamous cancer in the prostate with subsequent squamous metastases (32–36), this report provides evidence for hormone therapy–mediated conversion of ARPC to squamous DNPC at metastatic sites.
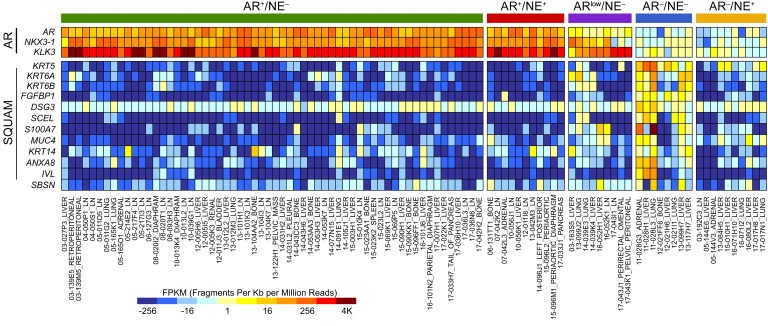
We compared the top significantly (FDR < 0.001) upregulated genes from the LuCaP 173.2 squamous pearl data set with the literature to identify genes with known roles in squamous cell differentiation or other squamous cancers. In addition to KRT5, KRT6A, KRT6B, and DSG3, we also found IVL, SBSN, FGFBP1, SCEL, S100A7, MUC4, KRT14, and ANXA8 to be significantly overexpressed in other squamous cell types (37–44). Importantly, RNA-Seq heatmaps show that these genes are strikingly elevated in subsets of both ARLPC and DNPC patient specimens (Figure 6), suggesting that ARLPC and DNPC phenotypes could be transition states to squamous mCRPC. These results indicate that squamous cell conversion is not a rare occurrence in end-stage disease and should be considered an emerging phenotype following resistance to AR-directed therapy.
Figure 6. Expression of squamous markers is associated with DNPC and ARLPC.
RNA-Seq heatmap of patient specimens (n = 98) highlighting AR-regulated genes and genes associated with squamous pearl cells (SQUAM). Results are expressed as log2 FPKM and colored according to scale.
Transcript signatures define the molecular phenotypes of mCRPC.
The variability in expression of any single marker, both at the biological level and technical level makes tumor classification by immunohistochemistry challenging. Transcript panels for tumor classification have been explored as predictive and prognostic biomarkers for treatment decision-making (45, 46). We leveraged the data generated from the patient metastases and LuCaP PDX models to develop a 26-gene transcriptomic signature for defining treatment-refractory mCRPC phenotypes. Using the aforementioned AR, NEURO I, and NEURO II gene panels, as well as a squamous panel (SQUAM) that includes KRT5, KRT6A, KRT6B, and FGFBP1, we conducted multidimensional scaling (MDS) and cluster analysis of the patient metastases and LuCaP PDX models (Figure 7A and Supplemental Figure 3B). The MDS demonstrated clear distinction between the 5 mCRPC phenotypes (ARPC, ARLPC, AMPC, DNPC, and SCNPC) in both patient specimens and LuCaP PDX models (Figure 7, B and C).
Figure 7. Cluster analysis using AR, NE, and squamous gene expression profiles segregates mCRPC specimens and LuCaP PDX models into the different phenotypes.
(A) RNA sequencing of mCRPC specimens acquired between 2003–2017 (n = 98; modified from Figure 1B). NE genes listed in the NEURO I and NEURO II panels, AR and AR-regulated genes are listed in the AR panel, and squamous associated genes are shown in SQUAM panel. Results are expressed as log2 FPKM and colored according to scale. Multidimensional scaling and cluster analysis of (B) mCRPC specimens (n = 98) and (C) LuCaP PDX models using the 26-gene set depicted in A. The LuCaP analysis was conducted on 18 distinct PDX lines (n = 2 for each line). ARPC (AR+/NE–; green), ARLPC (ARlow/NE–; purple), DNPC (AR–/NE–; blue), AMPC/mixed (AR+/NE+; red), SCNPC (AR–/NE+; yellow).
We further evaluated our 26-gene signature using PolyA RNA-Seq landscapes from 270 CRPC metastases in the Stand Up To Cancer (SU2C) data set (47). Although the SU2C data set contains RNA-Seq from mCRPC tumors earlier in disease progression and from tumors that are responding to treatment, the transcriptional signature segregated the tumors into the 5 mCRPC phenotypes (Supplemental Figure 8, A and B). Interestingly, we detected expression of the squamous-associated genes in 2 DNPC tumors and 2 ARLPC tumors but also observed marked squamous marker expression in 4 ARPC tumors and 1 SCNPC tumor. Thus, removing the squamous genes from the analysis showed a more effective clustering of the tumors into their respective phenotypes (Supplemental Figure 8C). We do not know if the ARPC and SCNPC specimens with squamous aspects represent tumors containing 2 different phenotypes or single phenotypes.
Discussion
The clinical phenotyping of mCRPC has been limited to morphologic and immunohistochemical analyses. Although adenocarcinoma (AR and PSA) and NE (CHGA and SYP) biomarkers have provided some clarity for pathologic classification, the complexity of tumor heterogeneity, and the emergence of new treatment-resistant phenotypes have catalyzed a need for deeper understanding of the mCRPC disease continuum. Moreover, anaplastic tumors or aggressive variants are a clinically defined group of small cell metastatic/CRPC phenotypes with varying degrees of both AR expression (generally AR-null) and NE differentiation (48, 49). The classifications prompted a call for further elucidation of underlying mCRPC biology and more accurate nomenclature that limits confusion between research and medical fields (50). At the transcriptome level, expression signatures for classifying SCNPC have been demonstrated (51), but an encompassing signature that appreciates the spectrum of mCRPC phenotypes has not been identified. In this report, we interrogated end-stage mCRPC patient specimens and treatment-resistant LuCaP PDX models and demonstrated that transcriptome analysis in conjunction with IHC is a powerful method for phenotyping mCRPC in the current era. Our approach led to the characterization of 5 distinct mCRPC phenotypes (AR-high/ARPC, AR-low/ARLPC, amphicrine/AMPC, double-negative/DNPC, and small cell or NE tumors lacking AR expression/SCNPC) and ultimately resulted in a clinically relevant 26-gene transcriptional signature to classify mCRPC biospecimens. Moreover, our data demonstrated that mCRPC is a disease continuum driven by AR, REST, and core SCNPC transcription factor programs; treatment-induced differentiation of DNPC to squamous cell carcinoma is an emerging mCRPC phenotype; AR-low and AR-null phenotypes share common pathways of resistance to AR pathway inhibition that could be exploited for clinical benefit; and loss of REST repressor activity is critical for driving conversion to the AMPC/mixed phenotype but only promotes rather than drives the SCNPC phenotype.
Observations made through our rapid autopsy program support a treatment-induced shift in mCRPC phenotypes with ARI therapies increasing the number of AR-null and AR-low metastases at end-stage disease (5). In addition, our patient, LuCaP PDX, and cell line data suggest that ARPC can transition to ARLPC, AMPC/mixed, DNPC/squamous, or SCNPC to bypass hormone or AR pathway suppression therapies. These results were further verified through our analysis of the SU2C cohort of CRPC metastases (47). Notably, DNPC is a proliferative AR-null intermediate that contains cells with the inherent plasticity and potential to convert to SCNPC or squamous mCRPC (Figure 8 and Supplemental Figure 9). Multiple cell line, murine model, and PDX reports have demonstrated that loss of tumor suppressor proteins, AR-directed therapies, and/or the tumor microenvironment contribute to CRPC cellular plasticity and ARPC to SCNPC transition (16, 20, 52–56). Moreover, Beltran et al. have established that subsets of NE tumors are clonally derived from ARPC (18). Of note, the genomic landscapes of intrapatient CRPC metastases are relatively similar (4). Thus, our analysis indicating that intertumoral phenotypic heterogeneity is not a rare occurrence argues that epigenetic, posttranscriptional, posttranslational, and microenvironment events can contribute to phenotypic diversity in mCRPC. Taken together, our data add further clinical support for the proposed mCRPC disease continuum and demonstrates that treatment-induced selective pressures can change the phenotypic and molecular landscapes of mCRPC.
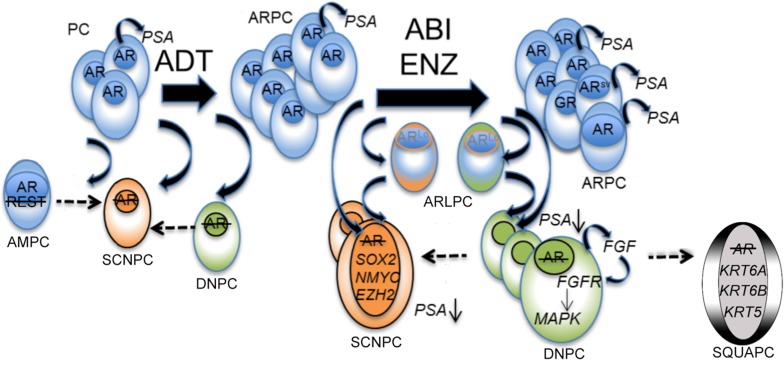
Figure 8. Schematic of the mCRPC disease continuum.
The proposed mechanisms, molecular drivers, and cellular differentiation states following AR pathway inhibition therapy. ADT, androgen deprivation therapy; ABI, abiraterone; ENZ, enzalutamide; PC, hormone-sensitive prostate cancer; ARPC, AR-high prostate cancer; ARLPC, AR-low prostate cancer; SCNPC, small cell or NE prostate cancer; DNPC, double-negative prostate cancer; AMPC, amphicrine prostate cancer; SQUAPC, squamous prostate cancer.
The analyses of DNPC tumors and the LuCaP 173.2 PDX model unexpectedly revealed the appearance of squamous cell pearls within the mass of DNPC tumor cells. RNA-Seq and staining for KRT6 confirmed the molecular nature of the squamous pearls. We hypothesize that only the most differentiated squamous carcinoma cells stain positive for KRT6 and display a cornified GSEA profile and that there exists a proliferating DNPC/squamous intermediary. Support for this hypothesis comes from RNA-Seq and IHC that shows ARLPC metastases and LuCaP 176 significantly enriched for squamous transcriptional profiles but negative for squamous pearl structures and KRT6 staining. The mechanisms of ARLPC/DNPC to squamous transition remain unclear, but future research examining the parallels between prostate and lung cancer lineage plasticity is warranted. For example, prostate and lung epithelial cells can be reprogrammed to small cell NE cancers through induction of the same transcription factor pathways (19). Moreover, lung adenocarcinomas can transition to squamous cell carcinomas through LKB1-loss (57). Our RNA-Seq data show no evidence LKB1/STK11-loss across patient samples but other mechanisms of adenocarcinoma-squamous differentiation are likely. We speculate that there are common molecular pathways driving lung and prostate cancer lineage switching. Taken together, our data demonstrate that treatment-induced ARPC-DNPC-squamous conversion is one potential pathway to bypass AR-suppression strategies.
There are currently no standard treatments for SCNPC, DNPC, and ARLPC phenotypes. However, RNA-Seq and GSEA between mCRPC phenotypes revealed biologically relevant pathways that could be further interrogated for therapeutic benefit. Significantly upregulated genes common to the AR-null and ARLPC phenotypes were enriched for cell adhesion processes, and delving into the pathway revealed CEACAM5 as a top hit. CEACAM5 has been identified as a surface marker of potential utility in directing chimeric antigen T cells in SCNPC (58), and our data reveal that this therapy could also be clinically effective against subsets of ARLPC and DNPC tumors. Moreover, upregulated genes common to ARLPC and AR-null phenotypes are enriched in immune system, inflammatory, and defense responses. Since the phenotype comparisons were relative to ARPC, these data support the notion that the immune cell content of ARLPC and AR-null tumors is strikingly different from that of ARPC tumors. Notably, DNPC-specific immune-related genes included IL8 and CXCR1, which have been reported to promote CRPC metastasis and angiogenesis (59), and genes such as TGFB and RUNX2, which support tumor growth in bone (60). Whether associated with the tumor cells, immune cells, or stromal cells, the presence of enriched immunomodulatory signaling pathways suggests that ARLPC, DNPC, and SCNPC tumors could be more receptive to immunotherapies.
Our group and others have previously demonstrated that loss of REST repressor activity promotes the SCNPC phenotype (15, 24, 61, 62). Here, we confirmed the existence of AMPC cells in vivo and in vitro and demonstrated that AMPC cells express a limited set of neuronal genes that are REST-repressed (NEURO I panel) and maintain AR signaling. Furthermore, transcriptome analysis of mCRPC and LuCaP PDX tumors and siRNA-mediated knockdown of REST in AR-expressing and AR-null cell lines provided compelling evidence that loss of REST repressor activity is critical for conversion to the AMPC/mixed phenotype but does not necessarily drive the SCNPC phenotype. We realize that siRNA treatments transiently relieve REST-mediated transcriptional repression, and that the impact of sustained REST-ablation remains to be determined. However, we hypothesize that epigenetic factors are preventing the expression of core SCNPC transcription factors (NEURO II panel of genes) that would permit SCNPC transdifferentiation in AR-null and AR-expressing cells with REST knockdown. Indeed, the epigenomes of NE and adenocarcinoma tumors are significantly different and EZH2 inhibitors have been shown to reactivate AR expression in some SCNPC models (18, 63). On the other hand, SRRM4-mediated splicing of REST and other neuronal regulators can drive ARPC-SCNPC transition (24). Our results demonstrated that SRRM4-mediated splicing of REST occurs in both AMPC and SCNPC PDX models and in AMPC VCaP cells, suggesting that SRRM4 expression alone is not sufficient to drive SCNPC conversion in all cases. Concordantly, overexpression of SRRM4 converted DU145 cells to a classical SCNPC phenotype but failed to do so in PC-3, 22Rv1, and LNCaP cells (64), implying that multiple hits to the cellular blueprint are required for complete lineage switching. Furthermore, REST directly represses SRRM4 expression and loss of REST activity has been proposed as a feed-forward mechanism for SCNPC conversion (25, 62). Whether loss of REST activity mediates increased SRRM4 expression or increased SRRM4 activity mediates loss of REST requires further examination. Nevertheless, our data clearly show that the homeostatic regulation of the SRRM4-REST axis is required for epithelial differentiation and function.
In summary, our comprehensive analysis of end-stage mCRPC highlights the use of AR and REST transcriptional programs to categorize mCRPC phenotypes in the abiraterone/enzalutamide era. In addition, the data generated in this report could be exploited through biopsy or blood-based biomarkers in future therapeutic studies to define inclusion criteria. This approach could stratify patients according to mCRPC phenotypes and account for the mCRPC disease continuum to implement targeted therapies.
Methods
Tissue acquisition.
Biospecimens were obtained within 8 hours of death from patients who died of metastatic CRPC. Visceral metastases were identified at the gross level, bone biopsies were obtained according to a previously described template (65) from 16–20 different sites, and metastases were identified at a histological level. LuCaP PDX lines were established from specimens acquired at either radical prostatectomy or at autopsy, implanted, and maintained by serial passage in immune compromised male mice (66).
Cell lines.
All cells were maintained at 37°C in humidified Steri-Cult CO2 incubators (Thermo Fisher Scientific). C4-2B (gift from L. Chung, Cedars-Sinai Medical Center, Los Angeles, CA), PC-3 (ATCC), and PacMet AR-null cells (5) were maintained in RPMI-1640 Media (Gibco, Life Technologies) with 10% fetal bovine serum (Atlanta Biologicals). VCaP cells (ATCC) were maintained in DMEM (ATCC) with 10% fetal bovine serum (Atlanta Biologicals).
Transient transfections.
Cells were suspended in Nucleofector Solution V (Lonza) and 5 μL of 50-μM ON-TARGETplus pooled REST siRNA or control siRNA (Dharmacon). Cell suspensions were electroporated using the Nucleofector II device (Lonza) and program T-027 and then replated in complete media. Forty-eight hours after transfection, cells were harvested for RNA or protein for subsequent analysis.
Immunohistochemistry.
The antibodies used in this study are listed in Supplemental Table 1. Five-micron-thick sections of the TMAs were deparaffinized and rehydrated in sequential xylene and graded ethanol. Antigen retrieval was performed in 10 mM citrate buffer (pH 6.0) in a pressure cooker for 30 minutes. Endogenous peroxidase and avidin/biotin were blocked respectively (Vector Laboratories). Sections were then blocked with 5% normal goat-horse-chicken serum, incubated with primary antibody (Supplemental Table 1), incubated with biotinylated secondary antibody (Vector Laboratories), followed by ABC reagent (Vector Laboratories), and stable DAB (Thermo Fisher Scientific). All sections were lightly counterstained with hematoxylin and mounted with Cytoseal XYL (Richard Allan Scientific). Mouse or rabbit IgG was used as negative control. IHC scores were represented as values between 0–200 and were determined as previously described (15).
Immunofluorescence.
Formalin-fixed, paraffin-embedded (FFPE) patient and LuCaP PDX tumors were prepared according to the above methods up to and including antigen retrieval. For cells in culture, VCaP cells were seeded on chamber slides (Nunc Lab-Tek; Thermo Fisher Scientific) in complete media 24 hours prior to fixing. Cells were fixed and permeabilized with ice-cold methanol for 10 minutes and then washed in PBS. Cells and sections were blocked for 1 hour with 5% normal goat-horse-chicken serum, incubated for 1 hour (cells) or overnight (sections) with primary antibodies in blocking buffer, washed, and then incubated with fluorescent secondary antibodies for 45 minutes in the dark. All incubations occurred at room temperature and antibodies used are listed in Supplemental Table 1. Slides were mounted using ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific) and then visualized and imaged at ×20 on an Olympus BX41 Fluorescence Microscope.
Immunoblot analysis.
Protein extracts from LuCaP PDXs and cell lines were obtained using the Nuclear Extract Kit (Active Motif) according to the manufacturer’s protocols. Quantification of total protein was determined using the ProStain Protein Quantification Kit (Active Motif) according to the manufacturer’s protocols. Ten to twenty micrograms of total protein lysate was electrophoresed on 4%–15% Bis-Tris gels (Bio-Rad Laboratories) with 1× Tris/Glycine/SDS Buffer (Bio-Rad Laboratories). The proteins were transferred to PVDF that was blocked with 5% Blotting-Grade Blocker (Bio-Rad Laboratories) in TBS/0.1% Tween-20 and subsequently probed with primary and secondary antibodies (Supplemental Table 1). Proteins were visualized using Clarity Western ECL Substrate (Bio-Rad Laboratories).
RNA isolation.
Total RNA was isolated from 98 tissue samples of CRPC metastases, which had been frozen in OCT (Tissue-Tek) with RNA STAT-60 (Tel-Test). Using an H&E-stained slide for each sample for orientation, 1-mm core punches of tumor were obtained. Alternatively, multiple sections enriched for tumor were cut using a Leica CM3050S cryostat. Total RNA was isolated from flash-frozen LuCaP PDX tissues or cell lines with RNA STAT-60 (Tel-Test) followed by purification with RNeasy Mini Kit (Qiagen) using the manufacturer’s recommended in-solution DNase digestion (Qiagen). The purity and yield of the RNA were determined on a NanoDrop 2000 (Thermo Fisher Scientific). RNA integrity was assessed on a 2100 Bioanalyzer (Agilent Technologies).
PCR and sequencing.
First-strand cDNA synthesis was performed with 1 μg RNA using an Advantage RT-for-PCR Kit (Clontech Laboratories). PCR was performed using either Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen; real-time PCR) or HotStarTaq Plus Master Mix (Qiagen; PCR for sequencing) on a Rotor-Gene Q (Qiagen). PCR primers (Integrated DNA Technologies) specific for REST were adapted from Raj et al. (25). The REST 5′ primer-GAGAACGCCCATATAAATGTGAAC and 3′ primer-CGGGTTACTTCATGTTGATTAGAG were used. The PCR reaction parameters were as follows: 50°C for 2 minutes and 95°C for 2 minutes (one cycle), followed by 30 cycles (standard gels) or 40 cycles (band sequencing) at 95°C for 10 seconds, annealing/extension at T(m) for 30 seconds, and 72°C for 30 seconds; the final extension was 72°C for 7 minutes. REST and REST4 PCR products were visualized after electrophoresis on a 1.2% agarose gel. For sequencing, PCR product bands were dissected out and purified using the QIAquick Gel Extraction Kit (Qiagen). The purified product was then sequenced by Eurofins Genomics using their Standard Read sequencing chemistry.
RNA sequencing.
RNA-Seq libraries were constructed from 1 μg total RNA using the Illumina TruSeq Stranded mRNA LT Sample Prep Kit according to the manufacturer’s protocol. Barcoded libraries were pooled and sequenced on the Illumina HiSeq 2500 generating 50 bp paired end reads. Sequencing reads were mapped to the hg38 human using TopHat v2.1.0 (67). For PDX samples, sequences were also aligned to the mm10 mouse genome and those derived from potential contamination with mouse tissue were removed from the analysis as previously described (68). Gene level abundance was quantified from the filtered human alignments in R using the GenomicAlignments Bioconductor package (69). Differential expression was assessed using transcript abundances as inputs to the edgeR Bioconductor package in R (70). For edgeR analysis, genes filtered for a minimum expression level of at least 1 count per million reads (CPM) in at least 2 samples were used to calculate expression differences using an exact test with a negative binomial distribution, applying a significance level of 0.05 with Benjamin-Hochberg false discovery rate (FDR) adjustment. The RNA-Seq data from this report can be accessed through GEO at GSE126078 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126078).
Pathway analysis.
Gene expression results were ranked by their edgeR statistics and used to conduct Gene Set Enrichment Analysis (GSEA) to determine patterns of pathway activity in different treatment groups. We used the curated pathways from within the MSigDBv6.1. Gene ontology (GO) enrichment of Venn diagram groups was computed using GOrilla with default parameters (71). The target and background list option was used and gene sets with enrichment of P < 0.001 were considered significant.
Multidimensional scaling (MDS) plots.
Specimens were classified according to expression of NEURO I, NEURO II, AR, and SQUAM gene signatures. ARPC and ARLPC phenotypes were differentiated based on AR or KLK3 expression (RNA-Seq) with ALRPC possessing an AR log2 FPKM value less than 4.0 or a KLK3 log2 FPKM value less than 2.0. Phenotypic groups were visualized using classical multidimensional scaling (MDS) calculated with the cmdscale function in R using the expression profiles of the 26 genes from the combined lists of NEURO I, NEURO II, AR, and SQUAM gene signatures. The distance metric was euclidean calculated by dist function on the columns (samples). The RNA-Seq data from the Stand Up To Cancer mCRPC cohort were accessed using dbGaP accession phs000915.v2.p2.
Statistics.
Sample size for each experiment is indicated in the figure legends. Experiments were repeated a minimum of 2 times. Statistical analyses for RNA sequencing, pathway analyses, and MDS were performed as indicated using R software. The enrichment scores were calculated in R using the GSVA package using the 14 genes in the NEURO I and NEURO II gene sets for NEURO scores and a previously published set of AR-regulated genes for AR-activity scores (5, 72). Mean AR-activity scores in transfected C4-2B cells were graphed using GraphPad Prism software.
Study approval.
All rapid autopsy tissues were collected from patients who had signed written informed consent under the aegis of the Prostate Cancer Donor Program at the University of Washington (73). The IRB of the University of Washington approved this study. All patient-derived xenograft experiments were approved by the University of Washington IACUC.
Author contributions
MPL, LGB, PSN, and CM conceived and designed the project. IMC, RC, and AK performed sequencing and bioinformatics analyses. MPL, LGB, LK, BL, YCY, and ACH conducted molecular and cell biology experiments. LDT and RMG provided pathology evaluation. HMN, EC, and CM provided biospecimens and patient-derived xenograft models. CSH, EYY, HHC, EAM, BM, MTS, DWL, and PSN consented patients for the rapid autopsy program and/or provided clinical expertise. MPL, PSN, and CM wrote the manuscript, and all authors reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by a Department of Defense Idea Development Award: Partnering Principal Investigator (W81XWH-17-1-0414;W81XWH-17-1-0415), W81XWH-15-1-0430, PC170431, the Pacific Northwest Prostate Cancer SPORE (P50CA97186), the Department of Defense Prostate Cancer Biorepository Network (W81XWH-14-2-0183), Department of Defense Prostate Cancer Clinical Trials Consortium W81XWH-15-2-0008, National Cancer Institute (NCI) R01 CA230617, NCI P01 CA163227, the Prostate Cancer Foundation, the AACR NextGen Transformative Cancer Research Grant, the Institute for Prostate Cancer Research, and the Richard M. LUCAS Foundation. We would like to thank the patients who generously donated tissue that made this research possible. We would also like to thank Jennifer Conner, Michiyo Dalos, Daniel Sondheim, and the Comparative Medicine Animal Caregivers for assistance with the LuCaP PDX work. Additionally, we would like to thank Paul Lange, Robert Vessella, Funda Vakar-Lopez, Martine Roudier, Xiaotun Zhang, Belinda Nghiem, Jennifer Noteboom, and the rapid autopsy teams in the Urology and Pathology Departments at the University of Washington.
Version 1. 07/30/2019
In-Press Preview
Version 2. 09/16/2019
Electronic publication
Version 3. 10/01/2019
Print issue publication
Footnotes
Conflict of interest: CSH has been compensated through honoraria from Genentech and as a consultant for Astellas, Bayer, Blue Earth Diagnostics, Clovis Oncology, Ferring, Myriad Genetics, Orion Corporation, Janssen, Hinova, and Tolmar; she has research funding from Aragon Pharmaceuticals, Astellas, AstraZeneca, Bayer, Dendreon, Emergent BioSolutions, Genentech, Medivation, Pfizer, and Roche; she has received travel, accommodation, and expenses from Astellas, Bayer, Blue Earth Diagnostics, Clovis Oncology, Ferring, Genentech, Menarini, Myriad Genetics, Orion Pharma GmbH, Hinova, and Pfizer; she has an immediate family member employed by CTI BioPharma with stock and/or other ownership interests in CTI BioPharma. EYY has been compensated as a consultant for Bayer and Merck and has research support from Agensys, Dendreon, Merck, and Seattle Genetics. HHC has research support from Clovis Oncology, Janssen, Sanofi, and Astellas. BM has research support from Janssen, Beigene, and AstraZeneca. MTS has research support from Janssen, AstraZeneca, Zenith Epigenetics, Pfizer, and Hoffmann-La Roche. ACH has research support from eFFECTOR Inc. EC has research support from Sanofi, AstraZeneca, Gilead, Zenith Epigenetics, Genentech, Janssen, Forma Pharmaceuticals, Bayer, and Abbvie. PSN has been compensated as an advisor to Janssen and has provided expert testimony related to drugs sold by Astellas.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4492–4505.https://doi.org/10.1172/JCI128212.
Contributor Information
Mark P. Labrecque, Email: mlabrec@uw.edu.
Ilsa M. Coleman, Email: icoleman@fredhutch.org.
Lisha G. Brown, Email: brown@uw.edu.
Lawrence D. True, Email: ltrue@u.washington.edu.
Lori Kollath, Email: lkollath@uw.edu.
Bryce Lakely, Email: brycel@uw.edu.
Holly M. Nguyen, Email: hmnguyen@u.washington.edu.
Yu C. Yang, Email: ycyang@fredhutch.org.
Rui M. Gil da Costa, Email: rgildaco@fredhutch.org.
Arja Kaipainen, Email: akaipain@fredhutch.org.
Roger Coleman, Email: galasphere2@gmail.com.
Celestia S. Higano, Email: thigano@u.washington.edu.
Evan Y. Yu, Email: evanyu@u.washington.edu.
Heather H. Cheng, Email: hhcheng@uw.edu.
Elahe A. Mostaghel, Email: emostagh@fhcrc.org.
Bruce Montgomery, Email: rbmontgo@u.washington.edu.
Michael T. Schweizer, Email: schweize@uw.edu.
Andrew C. Hsieh, Email: ahsieh@fredhutch.org.
Daniel W. Lin, Email: dlin@u.washington.edu.
Eva Corey, Email: ecorey@u.washington.edu.
Peter S. Nelson, Email: pnelson@fredhutch.org.
Colm Morrissey, Email: cmorriss@u.washington.edu.
References
- 1.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004;25(2):276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 2.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 3.Ryan CJ, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013;368(2):138–148. doi: 10.1056/NEJMoa1209096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar A, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22(4):369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bluemn EG, et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell. 2017;32(4):474–489. doi: 10.1016/j.ccell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal R, et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J Clin Oncol. 2018;36(24):2492–2503. doi: 10.1200/JCO.2017.77.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 8.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113(6):913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irshad S, et al. A molecular signature predictive of indolent prostate cancer. Sci Transl Med. 2013;5(202):202ra122. doi: 10.1126/scitranslmed.3006408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, et al. Correlated metabolomic, genomic, and histologic phenotypes in histologically normal breast tissue. PLoS ONE. 2018;13(4):e0193792. doi: 10.1371/journal.pone.0193792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pareja F, Geyer FC, Marchiò C, Burke KA, Weigelt B, Reis-Filho JS. Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer. 2016;2:16036. doi: 10.1038/npjbcancer.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32(2):399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng WS, Giandomenico V, Pastan I, Essand M. Characterization of the androgen-regulated prostate-specific T cell receptor gamma-chain alternate reading frame protein (TARP) promoter. Endocrinology. 2003;144(8):3433–3440. doi: 10.1210/en.2003-0121. [DOI] [PubMed] [Google Scholar]
- 14.Rickman DS, et al. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res. 2009;69(7):2734–2738. doi: 10.1158/0008-5472.CAN-08-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. SRRM4 expression and the loss of REST activity may promote the emergence of the neuroendocrine phenotype in castration-resistant prostate cancer. Clin Cancer Res. 2015;21(20):4698–4708. doi: 10.1158/1078-0432.CCR-15-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu P, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355(6320):84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bishop JL, et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 2017;7(1):54–71. doi: 10.1158/2159-8290.CD-15-1263. [DOI] [PubMed] [Google Scholar]
- 18.Beltran H, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JW, et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science. 2018;362(6410):91–95. doi: 10.1126/science.aat5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin D, et al. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 2014;74(4):1272–1283. doi: 10.1158/0008-5472.CAN-13-2921-T. [DOI] [PubMed] [Google Scholar]
- 21.Lee N, et al. Interactomic analysis of REST/NRSF and implications of its functional links with the transcription suppressor TRIM28 during neuronal differentiation. Sci Rep. 2016;6:39049. doi: 10.1038/srep39049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Shimojo M, Chai YG, Hersh LB. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Brain Res Mol Brain Res. 2000;80(1):88–98. doi: 10.1016/S0169-328X(00)00129-7. [DOI] [PubMed] [Google Scholar]
- 23.Chen GL, Miller GM. Alternative REST splicing underappreciated. eNeuro. 2018;5(5):ENEURO.0034-18.2018. doi: 10.1523/ENEURO.0034-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol. 2017;71(1):68–78. doi: 10.1016/j.eururo.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 25.Raj B, et al. Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell. 2011;43(5):843–850. doi: 10.1016/j.molcel.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000;60(7):1840–1844. [PubMed] [Google Scholar]
- 27.Tran C, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang YT, et al. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Sci Rep. 2017;7:42795. doi: 10.1038/srep42795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Troyer DA, et al. Characterization of PacMetUT1, a recently isolated human prostate cancer cell line. Prostate. 2008;68(8):883–892. doi: 10.1002/pros.20758. [DOI] [PubMed] [Google Scholar]
- 30.Rickman DS, et al. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–6622. doi: 10.1038/onc.2008.251. [DOI] [PubMed] [Google Scholar]
- 31.Xiao J, et al. Eight potential biomarkers for distinguishing between lung adenocarcinoma and squamous cell carcinoma. Oncotarget. 2017;8(42):71759–71771. doi: 10.18632/oncotarget.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Accetta PA, Gardner WA. Squamous metastases from prostatic adenocarcinoma. Prostate. 1982;3(5):515–521. doi: 10.1002/pros.2990030510. [DOI] [PubMed] [Google Scholar]
- 33.Malik RD, Dakwar G, Hardee ME, Sanfilippo NJ, Rosenkrantz AB, Taneja SS. Squamous cell carcinoma of the prostate. Rev Urol. 2011;13(1):56–60. [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Mixed adenocarcinoma, sarcomatoid carcinoma and adenosquamous carcinoma of the prostate: A case report. Oncol Lett. 2014;8(5):2325–2327. doi: 10.3892/ol.2014.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassler TJ, Orozco R, Bassler IC, Boyle LM, Bormes T. Adenosquamous carcinoma of the prostate: case report with DNA analysis, immunohistochemistry, and literature review. Urology. 1999;53(4):832–834. doi: 10.1016/S0090-4295(98)00418-X. [DOI] [PubMed] [Google Scholar]
- 36.Devaney DM, Dorman A, Leader M. Adenosquamous carcinoma of the prostate: a case report. Hum Pathol. 1991;22(10):1046–1050. doi: 10.1016/0046-8177(91)90014-G. [DOI] [PubMed] [Google Scholar]
- 37.Chao A, et al. Molecular characterization of adenocarcinoma and squamous carcinoma of the uterine cervix using microarray analysis of gene expression. Int J Cancer. 2006;119(1):91–98. doi: 10.1002/ijc.21813. [DOI] [PubMed] [Google Scholar]
- 38.Hamada T, et al. MUC4: a novel prognostic factor of oral squamous cell carcinoma. Int J Cancer. 2012;130(8):1768–1776. doi: 10.1002/ijc.26187. [DOI] [PubMed] [Google Scholar]
- 39.Walts AE, Said JW, Siegel MB, Banks-Schlegel S. Involucrin, a marker of squamous and urothelial differentiation. An immunohistochemical study on its distribution in normal and neoplastic tissues. J Pathol. 1985;145(4):329–340. doi: 10.1002/path.1711450406. [DOI] [PubMed] [Google Scholar]
- 40.Kvedar JC, Manabe M, Phillips SB, Ross BS, Baden HP. Characterization of sciellin, a precursor to the cornified envelope of human keratinocytes. Differentiation. 1992;49(3):195–204. doi: 10.1111/j.1432-0436.1992.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 41.Park GT, Lim SE, Jang SI, Morasso MI. Suprabasin, a novel epidermal differentiation marker and potential cornified envelope precursor. J Biol Chem. 2002;277(47):45195–45202. doi: 10.1074/jbc.M205380200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alam H, Sehgal L, Kundu ST, Dalal SN, Vaidya MM. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell. 2011;22(21):4068–4078. doi: 10.1091/mbc.e10-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, et al. Selective expression of S100A7 in lung squamous cell carcinomas and large cell carcinomas but not in adenocarcinomas and small cell carcinomas. Thorax. 2008;63(4):352–359. doi: 10.1136/thx.2007.087015. [DOI] [PubMed] [Google Scholar]
- 44.Mongiat M, Otto J, Oldershaw R, Ferrer F, Sato JD, Iozzo RV. Fibroblast growth factor-binding protein is a novel partner for perlecan protein core. J Biol Chem. 2001;276(13):10263–10271. doi: 10.1074/jbc.M011493200. [DOI] [PubMed] [Google Scholar]
- 45.Cuzick J, et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol. 2011;12(3):245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sparano JA, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med. 2015;373(21):2005–2014. doi: 10.1056/NEJMoa1510764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abida W, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aparicio AM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res. 2013;19(13):3621–3630. doi: 10.1158/1078-0432.CCR-12-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aparicio AM, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res. 2016;22(6):1520–1530. doi: 10.1158/1078-0432.CCR-15-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Epstein JI, et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol. 2014;38(6):756–767. doi: 10.1097/PAS.0000000000000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai HK, Lehrer J, Alshalalfa M, Erho N, Davicioni E, Lotan TL. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer. 2017;17(1):759. doi: 10.1186/s12885-017-3729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen R, Dorai T, Szaboles M, Katz AE, Olsson CA, Buttyan R. Transdifferentiation of cultured human prostate cancer cells to a neuroendocrine cell phenotype in a hormone-depleted medium. Urol Oncol. 1997;3(2):67–75. doi: 10.1016/S1078-1439(97)00039-2. [DOI] [PubMed] [Google Scholar]
- 53.Deeble PD, Murphy DJ, Parsons SJ, Cox ME. Interleukin-6- and cyclic AMP-mediated signaling potentiates neuroendocrine differentiation of LNCaP prostate tumor cells. Mol Cell Biol. 2001;21(24):8471–8482. doi: 10.1128/MCB.21.24.8471-8482.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labrecque MP, et al. The retinoblastoma protein regulates hypoxia-inducible genetic programs, tumor cell invasiveness and neuroendocrine differentiation in prostate cancer cells. Oncotarget. 2016;7(17):24284–24302. doi: 10.18632/oncotarget.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou M, et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 2017;7(7):736–749. doi: 10.1158/2159-8290.CD-16-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ku SY, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science. 2017;355(6320):78–83. doi: 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han X, et al. Transdifferentiation of lung adenocarcinoma in mice with Lkb1 deficiency to squamous cell carcinoma. Nat Commun. 2014;5:3261. doi: 10.1038/ncomms4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JK, et al. Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc Natl Acad Sci USA. 2018;115(19):E4473–E4482. doi: 10.1073/pnas.1802354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue K, et al. Interleukin 8 expression regulates tumorigenicity and metastases in androgen-independent prostate cancer. Clin Cancer Res. 2000;6(5):2104–2119. [PubMed] [Google Scholar]
- 60.van der Deen M, et al. The cancer-related Runx2 protein enhances cell growth and responses to androgen and TGFbeta in prostate cancer cells. J Cell Biochem. 2010;109(4):828–837. doi: 10.1002/jcb.22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lapuk AV, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227(3):286–297. doi: 10.1002/path.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flores-Morales A, et al. Proteogenomic characterization of patient-derived xenografts highlights the role of REST in neuroendocrine differentiation of castration-resistant prostate cancer. Clin Cancer Res. 2019;25(2):595–608. doi: 10.1158/1078-0432.CCR-18-0729. [DOI] [PubMed] [Google Scholar]
- 63.Kleb B, et al. Differentially methylated genes and androgen receptor re-expression in small cell prostate carcinomas. Epigenetics. 2016;11(3):184–193. doi: 10.1080/15592294.2016.1146851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee AR, Gan Y, Tang Y, Dong X. A novel mechanism of SRRM4 in promoting neuroendocrine prostate cancer development via a pluripotency gene network. EBioMedicine. 2018;35:167–177. doi: 10.1016/j.ebiom.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roudier MP, et al. Phenotypic heterogeneity of end-stage prostate carcinoma metastatic to bone. Hum Pathol. 2003;34(7):646–653. doi: 10.1016/S0046-8177(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 66.Nguyen HM, et al. LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease and serve as models for evaluating cancer therapeutics. Prostate. 2017;77(6):654–671. doi: 10.1002/pros.23313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou J, et al. Phenotypic and transcriptional fidelity of patient-derived colon cancer xenografts in immune-deficient mice. PLoS ONE. 2013;8(11):e79874. doi: 10.1371/journal.pone.0079874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morrissey C, et al. Effects of androgen deprivation therapy and bisphosphonate treatment on bone in patients with metastatic castration-resistant prostate cancer: results from the University of Washington Rapid Autopsy Series. J Bone Miner Res. 2013;28(2):333–340. doi: 10.1002/jbmr.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.