Abstract
Microtubule-associated serine/threonine kinase 1 (MAST1) is a central driver of cisplatin resistance in human cancers. However, the molecular mechanism regulating MAST1 levels in cisplatin-resistant tumors is unknown. Through a proteomics screen, we identified the heat shock protein 90 B (hsp90B) chaperone as a direct MAST1 binding partner essential for its stabilization. Targeting hsp90B sensitized cancer cells to cisplatin predominantly through MAST1 destabilization. Mechanistically, interaction of hsp90B with MAST1 blocked ubiquitination of MAST1 at lysines 317 and 545 by the E3 ubiquitin ligase CHIP and prevented proteasomal degradation. The hsp90B-MAST1-CHIP signaling axis and its relationship with cisplatin response were clinically validated in cancer patients. Furthermore, combined treatment with a hsp90 inhibitor and the MAST1 inhibitor lestaurtinib further abrogated MAST1 activity and consequently enhanced cisplatin-induced tumor growth arrest in a patient-derived xenograft model. Our study not only uncovers the regulatory mechanism of MAST1 in tumors but also suggests a promising combinatorial therapy to overcome cisplatin resistance in human cancers.
Keywords: Oncology, Therapeutics
Keywords: Head and neck cancer, Protein kinases, Ubiquitin-proteosome system
Introduction
Cisplatin is one of the most compelling chemotherapy agents widely used in the treatment of various forms of human cancers (1). Cisplatin is generally known to eradicate cancer cells by forming DNA cross-links, which prevent DNA synthesis and in turn trigger apoptosis signaling pathways (2). However, responsiveness to cisplatin or platinum-based chemotherapy is often temporary and many cancer patients ultimately relapse with cisplatin-resistant disease (3–5). The mechanisms underlying cisplatin resistance are complex, and include reduction in the intracellular cisplatin level, enhanced damaged DNA repair, and activation of prosurvival signaling pathways often involving activation of the kinase MEK (2, 6). We recently reported that a microtubule-associated serine/threonine kinase 1 (MAST1, also known as SAST170), which is known as a scaffold molecule that links the dystrophin and utrophin complex with microfilaments, is a common critical factor that drives cisplatin resistance in diverse types of human cancer, including head and neck, lung, and ovarian cancers (7, 8). MAST1 confers cisplatin resistance through MEK1 reactivation by replacing its conventional kinase cRaf when cisplatin disrupts the interaction between MEK1 and cRaf (7). Abundance of MAST1 positively correlated with cisplatin resistance in primary tumors obtained from cancer patients who received cisplatin-containing chemotherapy. In addition, we showed that a multikinase inhibitor lestaurtinib inhibits MAST1 kinase activity and overcomes cisplatin resistance in patient-derived xenograft models.
The heat shock protein 90 (hsp90) chaperone machinery is a key regulator of protein homeostasis that maintains the functional and structural integrity of client proteins in human cells (9). Two distinct genes, hsp90AA1/2 and hsp90AB1 encode inducible and constitutively expressed isoforms, hsp90A1/A2 and hsp90B, respectively (10). The chaperoning function of hsp90 is known to be essential during oncogenesis and malignant progression, and is mediated by its interaction with a wide range of proteins including transcription factors, hormone receptors, and protein kinases including AKT, cRaf, and protein kinase D2 (PRKD2), consequently regulating protein maturation and stabilization (11–15). Previous studies indicate that the inhibition of hsp90 with small molecules such as 17-AAG acts synergistically or additively with anticancer agents in the treatment of cancers in mouse models and in humans (16). Inhibitors of hsp90 trigger the effect of anticancer drugs that target client proteins of hsp90 such as human epidermal growth factor receptor 2 (HER2) (17). Hsp90 inhibitors also reverse cisplatin resistance in multiple types of human cancers, including ovarian cancer and bladder cancer, accompanied by inhibition of the AKT and ERK pathways (18, 19).
Subsequent studies have shown that the client proteins of hsp90, which are protein kinases or transcription factors, are often degraded by the multistep, ubiquitin-mediated proteasome pathway. Ubiquitin is primarily activated by an E1 enzyme before being transferred to the E2 ubiquitin–conjugating enzymes. The final transfer of ubiquitin to the substrate requires E3 ubiquitin ligase. The carboxyl terminus of hsc70-interacting protein (CHIP, also known as STUB1) is one of the best-characterized ubiquitin ligases that binds to hsp70 and hsp90 as a cochaperone to control protein stability and function (20, 21). CHIP has been shown to function in the degradation of proteins that are involved in cancer progression. For instance, CHIP efficiently ubiquitinates and downregulates HER2 (22). CHIP also interacts with and mediates ubiquitination and proteasomal degradation of protein arginine methyltransferase 5 (PRMT5) in human prostate cancer cells (23). DNA damage–induced apoptosis suppressor (DDIAS), which is associated with cisplatin resistance in lung cancer, is known to be regulated by CHIP-mediated proteasomal degradation (24). In addition, a reverse correlation between CHIP and cancer progression is observed. For example, downregulation of CHIP expression is observed in colorectal cancer compared with normal tissues and in advanced breast cancer (25, 26).
Although MAST1 is identified as a critical factor that mediates cisplatin resistance by rewiring MEK activation in a cRaf-independent manner, the molecular mechanism by which MAST1 protein is regulated and stabilized to confer cisplatin resistance in cancer cells is entirely unexplored. Deciphering the mechanism of MAST1 regulation in cancer cells will not only contribute to our understanding of the biology of MAST1 regulation but could also offer an effective combinatorial strategy linked to MAST1 to overcome cisplatin resistance. Here, we incorporate multidisciplinary proteomic approaches to identify a unique MAST1 regulatory mechanism that is mediated by the chaperone protein hsp90B and the E3 ubiquitin ligase CHIP, and further demonstrate how this regulatory machinery contributes to cisplatin-resistant tumor progression in human cancers.
Results
Hsp90B interacts with and stabilizes MAST1 in cisplatin-resistant cancer cells.
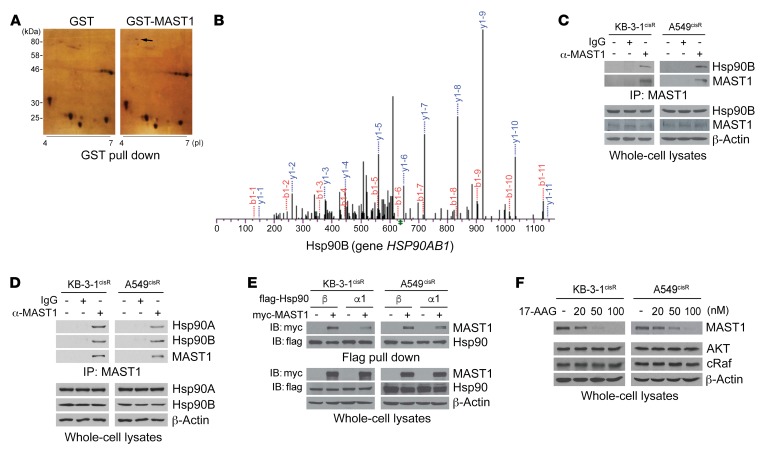
To better understand the mechanism of MAST1 regulation, we investigated the proteins that interact with MAST1 in cells through 2-dimensional (2D) gel electrophoresis–based proteomic profiling. GST or GST-fused MAST1 was pulled down from human 293T cells. The protein captured by GST-MAST1 but not by GST alone was excised from the 2D gel (Figure 1A). Through mass spectrometry–based (MS-based) proteomic analysis, we identified hsp90B as a potential binding partner of MAST1 (Figure 1B). Endogenous interaction between hsp90B and MAST1 was demonstrated by coimmunoprecipitation analysis in cisplatin-resistant sublines of human carcinoma KB-3-1 and lung cancer A549 cells (Figure 1C) (7). We further explored whether the interaction is limited to the hsp90B isoform by comparing the interaction between MAST1 and hsp90B or hsp90A. We found that hsp90A, another isoform of hsp90, also binds to MAST1 in cisplatin-resistant cancer cells (Figure 1D). Although hsp90A binds to MAST1 in cisplatin-resistant cancer cells, the interaction was weaker compared with hsp90B (Figure 1E). In line with the result obtained from proteomics analysis, this indicates that hsp90B is the predominant hsp90 isoform that binds to MAST1. To demonstrate the functional significance of this binding, we examined whether treatment with 17-AAG, a selective inhibitor of hsp90, alters MAST1 protein levels in cells. Interestingly, treatment with 17-AAG diminished MAST1 protein levels in a dose-dependent manner, while the same doses of 17-AAG were insufficient to decrease other known hsp90 client proteins, AKT or cRaf, in cancer cells (Figure 1F). High dose or long duration of 17-AAG treatment eventually decreased protein levels of AKT or cRaf as previously reported (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI125963DS1). However, MAST1 mRNA expression levels remained unchanged by 17-AAG treatment (Supplemental Figure 2). Together, these data suggest that hsp90B interacts with and stabilizes MAST1 protein in cancer cells.
Figure 1. Hsp90B binds to and stabilizes MAST1 in cisplatin-resistant cancer cells.
(A) 2D gel electrophoresis–based proteomic analysis for MAST1-interacting protein identification. 293T cells expressing GST-MAST1 or GST alone were subjected to GST pull down and eluates were separated by 2D gel electrophoresis and visualized by silver staining. Black arrow indicates hsp90B only shown in GST-MAST1 eluates. (B) MS spectra of hsp90B fragment identified by LC-MS/MS. (C) Endogenous interaction between MAST1 and hsp90B was determined by MAST1 coimmunoprecipitation in cisplatin-resistant cancer cells. (D and E) Interaction of hsp90 isoforms with MAST1. Interaction was determined by coimmunoprecipitation. Myc-MAST1 and flag-hsp90B or hsp90A1 were overexpressed in KB-3-1cisR and A549cisR cells in E. (F) Effect of hsp90 inhibition on MAST1 protein level. Cisplatin-resistant cancer cells were treated with increasing concentrations of 17-AAG for 24 hours. MAST1 protein levels were determined by Western blotting. Data are representative of 2 (A and C–E) and 3 (F) independent biological experiments.
Hsp90B confers cisplatin resistance to cancer cells mainly through MAST1 regulation.
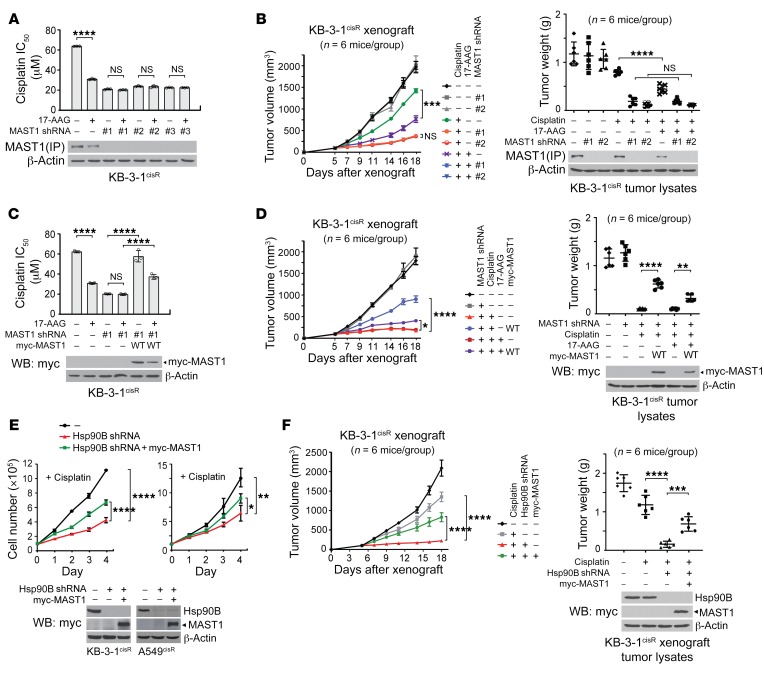
We previously reported that MAST1 plays a pivotal role in driving cisplatin resistance in human cancers (7). Hsp90 inhibitors are considered to be effective in overcoming cisplatin resistance, but their mechanism of action is unclear. To investigate whether hsp90B contributes to cisplatin resistance through MAST1, we examined the effect of targeting hsp90 on cisplatin resistance in the presence and absence of MAST1. Treatment with the hsp90 inhibitor 17-AAG significantly sensitized cisplatin-resistant cancer cells to cisplatin, whereas the effect of 17-AAG on cisplatin resistance was abolished when MAST1 was silenced by stable gene knockdown (Figure 2A and Supplemental Figure 3A). These in vitro observations were confirmed in vivo. 17-AAG treatment sensitized xenograft tumors to cisplatin, and MAST1 protein level was decreased, whereas the effect of 17-AAG was eradicated in mice bearing tumors with MAST1 knockdown (Figure 2B and Supplemental Figure 3B). Moreover, the cisplatin resistance and 17-AAG effect abolished by MAST1 knockdown was restored when MAST1 was rescue-expressed in cells and in xenograft mice, further suggesting that the effect of 17-AAG on cisplatin response is mediated through MAST1 (Figure 2, C and D, and Supplemental Figure 3, C and D). 17-AAG disrupts the function of hsp90 isoforms by binding to the ATP binding pocket (27). Consistent with this, surface plasmon resonance (SPR) revealed that 17-AAG binds to both hsp90A and hsp90B to alter their function (Supplemental Figure 4A). However, knockdown of hsp90B but not hsp90A abolished the role of 17-AAG in attenuating MAST1 protein level, cell viability, and cisplatin resistance (Supplemental Figure 4, B and C). These data suggest that hsp90B but not hsp90A specifically stabilizes MAST1 protein and contributes to cisplatin-resistant cell growth in cancer cells.
Figure 2. Inhibition of hsp90B sensitizes cisplatin-resistant cancer cells to cisplatin through MAST1.
(A) Cisplatin IC50 upon 17-AAG treatment (50 nM, 48 hours) with or without MAST1 knockdown. Cisplatin IC50 values were determined by CellTiter-Glo assay and analyzed by GraphPad Prism 8. (B) Effect of 17-AAG and MAST1 knockdown on tumor growth of cisplatin-treated xenograft mice. Mice were treated with cisplatin (5 mg/kg) and 17-AAG (50 mg/kg) from 5 days after xenograft. Tumor volume (left) and tumor weight (right) for each group and MAST1 expression in tumor lysates are shown. Cisplatin IC50 (C) and cisplatin-resistant tumor growth (D) upon 17-AAG treatment, MAST1 knockdown, and rescue expression of MAST1 WT. Cell viability assay and xenograft assay were performed as in A and B. (E) Cell proliferation of KB-3-1cisR and A549cisR cells with hsp90B knockdown and MAST1 overexpression in the presence of cisplatin. Cells were treated with sublethal doses of cisplatin (5 μg/mL KB-3-1cisR; 2 μg/mL A549cisR) and proliferation was determined by trypan blue exclusion. (F) Effect of hsp90B knockdown and MAST1 overexpression on cisplatin-resistant tumor growth. Mice were treated with cisplatin (5 mg/kg) from 5 days after xenograft. Tumor volume (left) and tumor weight (right) for each group and hsp90B and MAST1 expression in tumor lysates are shown. Data shown are representative of 2 (A–D and F) and 3 (E) independent biological experiments. Data are mean ± SD from 3 technical replicates for A, C, and E; n = 6 for B, D, and F. Error bars represent SEM for tumor volume and SD for tumor weight. Statistical analysis was performed by 2-way ANOVA for B, D, and F (left), and E, and 1-way ANOVA for A and C and B, D, and F (right). *P < 0.05; **P < 0.01; ***P < 0.005; ****P < 0.0001.
We next conducted a study to investigate whether overexpression of MAST1 rescues the cisplatin resistance lost by targeting hsp90B. Ectopic expression of MAST1 significantly restored the cisplatin-induced cell proliferation decrease mediated by hsp90B knockdown in cisplatin-resistant cancer cells (Figure 2E). This was further validated in vivo in xenograft mice. The experimental mice bearing KB-3-1cisR cells with hsp90B knockdown were more sensitive to cisplatin treatment than the control group carrying xenograft tumors with hsp90B. However, overexpression of myc-MAST1 partially but significantly restored the decreased tumor growth and proliferation in vivo (Figure 2F and Supplemental Figure 4D). Both loss- and gain-of-function approaches suggest that hsp90B contributes to cisplatin resistance in cancer cells mainly by regulating MAST1 protein.
Hsp90 impedes ubiquitination of MAST1 at lysine 317/545 and protects MAST1 from proteosomal degradation.
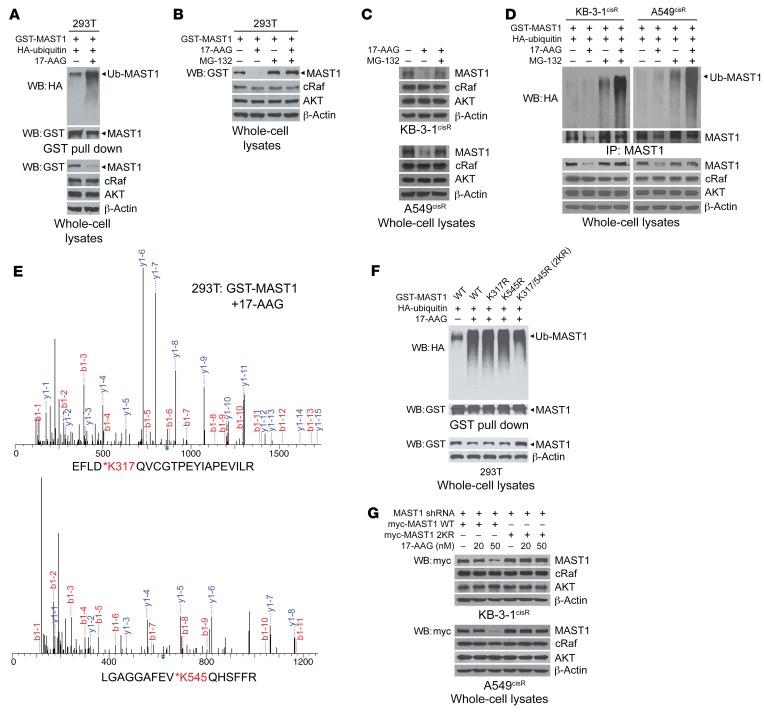
To explore the molecular mechanism by which hsp90B stabilizes MAST1 to confer cisplatin resistance in cancer cells, we first assessed the ubiquitination status of MAST1 in the presence and absence of hsp90. Inhibition of hsp90 resulted in MAST1 ubiquitination and degradation, whereas treatment with proteasome inhibitor MG-132 prevented 17-AAG–mediated MAST1 degradation in 293T cells (Figure 3, A and B). This was further confirmed in cisplatin-resistant cancer cells (Figure 3C). Although longer duration or higher dose of 17-AAG treatment eventually decreased protein levels of AKT or cRaf as shown in Supplemental Figure 1, treatment with 17-AAG induced degradation of MAST1 while the same duration and dose of 17-AAG did not alter the levels of AKT or cRaf in cisplatin-resistant cancer cells. MG-132 treatment increased MAST1 and eventually AKT and cRaf protein levels at a longer time point. This suggests that these proteins are degraded by the proteasome, but that MAST1 is more susceptible to hsp90 or proteasome inhibition than other client proteins, AKT and cRaf, in cells (Supplemental Figure 5). Furthermore, treatment with the proteasome inhibitor MG-132 led to MAST1 ubiquitination, which was further enhanced by hsp90 inhibition (Figure 3D). These results suggest that MAST1 undergoes ubiquitin-mediated proteasomal degradation when hsp90 is functionally abrogated.
Figure 3. Inhibition of hsp90 induces ubiquitination of MAST1 at lysine 317/545 that leads to proteasomal degradation.
(A) Effect of 17-AAG on MAST1 ubiquitination. 293T cells with GST-MAST1 and HA-tagged ubiquitin (Ub) were treated with 17-AAG for 4 hours and subjected to GST pull down. Anti-HA antibody was used to detect ubiquitinated MAST1. (B and C) Effect of 17-AAG on MAST1 proteasomal degradation. Cells were treated with or without MG-132 (10 μM) before addition of 17-AAG (1 μM) in 293T (B) or cisplatin-resistant cancer cells (C), and exogenous or endogenous MAST1 levels were detected, respectively. cRaf and AKT levels are shown for comparison. (D) Effect of 17-AAG on MAST1 ubiquitination and proteasomal degradation in cisplatin-resistant cancer cells. (E) MS spectra of ubiquitinated peptide fragments of MAST1. 293T cells with GST-MAST1 were treated with 1 μM of 17-AAG for 4 hours. Ubiquitination at K317 and K545 in MAST1 was identified using LC/MS-MS. (F) Ubiquitination of MAST1 WT and K317R or/and K545R mutants upon 17-AAG treatment. (G) Degradation of MAST1 WT and K317R/K545R (2KR) upon 17-AAG treatment in KB-3-1cisR and A549cisR cells. MAST1 knockdown cells were transfected with shRNA-resistant MAST1 WT or 2KR and treated with indicated concentrations of 17-AAG for 24 hours. Data shown are representative of 2 (A, B, and F) and 3 (C, D, and G) independent biological experiments.
Through a MS-based proteomic analysis, we identified that MAST1 is specifically ubiquitinated at lysine residues K317 and K545 upon hsp90 inhibition (Figure 3E). To investigate whether these 2 residues are the main ubiquitination sites of MAST1 that mediate proteasomal degradation upon hsp90 inhibition, we generated the lysine-to-arginine mutant forms of MAST1, K317R, K545R, and the double KR mutant K317R/K545R (2KR). Mutation at either single ubiquitination site did not alter the ubiquitination level of MAST1. However, simultaneous mutation at both K317 and K545 resulted in decreased ubiquitination (Figure 3F). Moreover, wild-type (WT) MAST1 was degraded upon 17-AAG treatment in a dose-dependent manner, whereas 2KR MAST1 remained intact (Figure 3G). These data indicate that K317 and K545 are the key ubiquitination sites in MAST1 and that ubiquitination of both residues leads to proteasomal degradation when functional hsp90 is absent.
Hsp90 supports cisplatin-resistant tumor growth through stabilizing MAST1 and preventing its ubiquitination at K317 and K545.
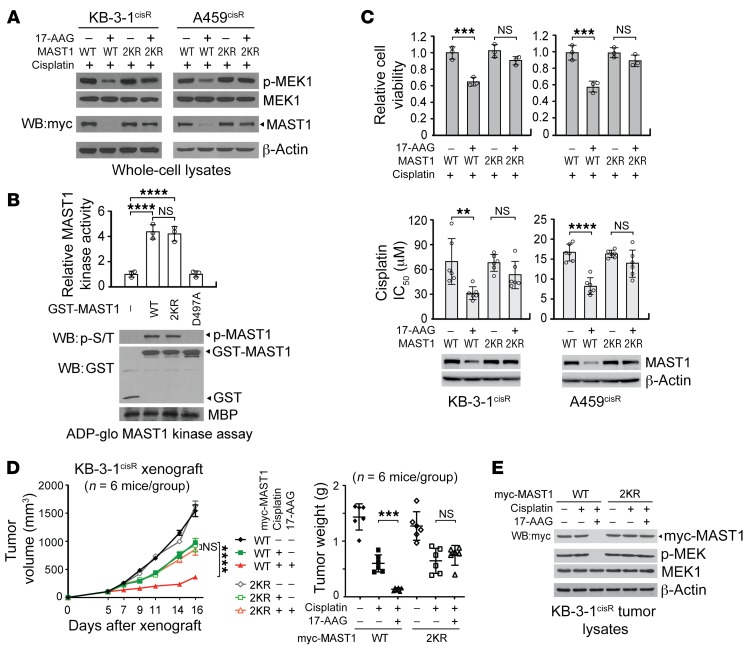
We next investigated whether ubiquitination of MAST1 at K317 and K545 upon hsp90 inhibition is critical for MAST1-mediated cisplatin-resistant cancer cell proliferation and tumor growth. Treatment with 17-AAG decreased the level of MAST1 and attenuated activity of the downstream substrate MEK1, whereas 2KR MAST1 did not respond to 17-AAG in terms of MAST1 level or MEK1 activity (Figure 4A). Mutation of K317 and K545 to arginine in MAST1 did not affect its kinase activity in an in vitro MAST1 kinase assay, suggesting that the decreased MEK1 activity upon MAST1 2KR expression is not a consequence of a change in direct kinase activity (Figure 4B). The effect of MAST1 WT or 2KR was monitored in cells with MAST1 knockdown. These cells showed similar MAST1 activity and cisplatin response compared with cells harboring endogenous MAST1 (Supplemental Figure 6, A and B). While cisplatin-resistant cells expressing MAST1 WT were sensitive to 17-AAG treatment in the presence of cisplatin, expression of MAST1 2KR led the cells to become resistant to 17-AAG treatment, resulting in no significant decrease in cell viability and cisplatin resistance (Figure 4C). This finding was further validated in vivo. While the mice bearing MAST1 WT xenograft tumors responded to 17-AAG treatment, as demonstrated by attenuated tumor growth upon cisplatin exposure and decreased MAST1 levels and MEK1 phosphorylation, the mice bearing MAST1 2KR tumors were resistant to these effects of 17-AAG treatment (Figure 4, D and E, and Supplemental Figure 6C). These data suggest that hsp90 contributes to cisplatin-resistant tumor growth by protecting MAST1 from being ubiquitinated at K315 and K545.
Figure 4. Ubiquitination of MAST1 at K317 and K545 induces MAST1 degradation and cisplatin-mediated cell death upon 17-AAG treatment.
(A) Effect of 17-AAG on MAST1 WT and K317R/K545R (2KR) degradation. Cells were treated with or without 17-AAG (200 nM) and sublethal doses of cisplatin as in Figure 2E. MAST1 expression and MEK1 activation was assessed by immunoblotting. (B) MAST1 in vitro kinase assay of MAST1 WT and 2KR. Kinase activity of GST-MAST1 variants. Kinase dead mutant D497A MAST1 was used as a negative control. (C) Cell viability and cisplatin sensitivity of cisplatin-resistant cells expressing MAST1 WT or 2KR. Cell viability was determined by trypan blue exclusion in cells treated with 200 nM of 17-AAG and sublethal doses of cisplatin for 48 hours. Cisplatin sensitivity is shown as cisplatin IC50, which was determined by CellTiter-Glo assay. (D) Effect of 17-AAG and cisplatin treatment on tumor volume and tumor weight of xenograft mice bearing KB-3-1cisR with MAST1 WT or 2KR overexpression. Mice were treated with cisplatin (5 mg/kg) and 17-AAG (50 mg/kg) from 5 days after xenograft. (E) MAST1 expression and MEK1 activation levels in tumor lysates. WT or 2KR MAST1 was overexpressed in MAST1 knockdown cells for functional assays. Data are mean ± SD from 3 technical replicates for B and C; n = 6 for D. Error bars represent SEM for tumor volume and SD for tumor weight. Data shown are representative of 3 (A and C [top]) and 2 (B–E) independent biological experiments. Data are mean ± SD from 3 technical replicates for B and C; n = 6 for D. Error bars represent SEM for tumor volume and SD for tumor weight. Statistical analysis was performed by 2-way ANOVA for D (left) and 1-way ANOVA for all the rest. **P < 0.01; ***P < 0.005; ****P < 0.0001.
CHIP ubiquitinates MAST1 at K317/K545 and mediates MAST1 degradation.
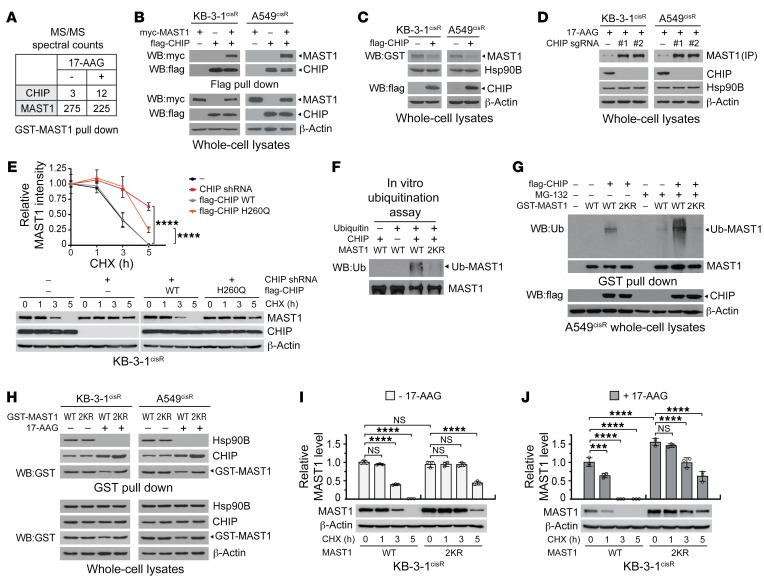
To further explore the molecular mechanism of MAST1 ubiquitination, we sought a potential ubiquitin ligase that binds to MAST1 from the MS-based proteomic analysis shown in Figure 3E. An E3 ubiquitin ligase, CHIP, was identified as a potential binding partner of MAST1 (Figure 5A). Endogenous and exogenous protein interaction between CHIP, MAST1, and hsp90 was confirmed by coimmunoprecipitation in cisplatin-resistant cancer cells (Figure 5B and Supplemental Figure 7A). To examine whether CHIP is involved in MAST1 degradation, we first modified the level of CHIP and monitored any changes in MAST1 protein levels in cisplatin-resistant cancer cells. CHIP overexpression reduced the level of MAST1 and conversely, knockout of CHIP resulted in enhanced MAST1 levels in cisplatin-resistant cancer cells (Figure 5, C and D). However, CHIP modulation did not alter hsp90B levels, suggesting that the effect of CHIP is specific for MAST1. In addition, CHIP modulation consequently altered MEK and ERK phosphorylation levels when the cells were treated with cisplatin (Supplemental Figure 7, B and C). Next, degradation of MAST1 by CHIP was examined by the cycloheximide chase assay. Overexpression of CHIP promoted degradation of MAST1, whereas inhibition of CHIP by genetic knockdown or CHIP H260Q, a mutant lacking ubiquitin ligase activity, lessened the degradation of MAST1 (Figure 5E). These data suggest that CHIP could be a potential negative regulator of MAST1 that induces MAST1 ubiquitination and degradation. Indeed, in vitro and in vivo CHIP ubiquitination assays revealed that CHIP directly mediates the ubiquitination of MAST1 at K317 and K545 in cancer cells (Figure 5, F and G).
Figure 5. CHIP ubiquitinates and degrades MAST1 when unmasked by hsp90B.
(A) MAST1 interacts with CHIP in cells. GST-pull-down samples from 293T were applied to LC-MS/MS. Spectral counts of CHIP and MAST1 in samples treated with or without 17-AAG are shown. (B) Overexpressed CHIP and MAST1 interact in cancer cells. Effect of CHIP overexpression (C) or knockout (D) on MAST1 levels. (E) Comparison of MAST1 protein stability in cells with CHIP modulation was achieved by cycloheximide (CHX) chase assay. Cells with CHIP knockdown were transfected with shRNA-resistant CHIP variants followed by 5 μg/mL CHX treatment for the indicated time. MAST1 amount was determined by density analysis. Representative data are shown. (F) In vitro CHIP ubiquitination assay using purified MAST1 WT or 2KR. (G) Ubiquitination of MAST1 WT and 2KR in cells. GST pull-down samples from A549cisR cells treated with MG-132 (10 μM) were immunoblotted with anti-ubiquitin antibody. (H) Interaction of MAST1 WT or 2KR, hsp90B, and CHIP in the presence or absence of 17-AAG. MAST1 WT and 2KR protein stability in the absence (I) and presence (J) of 17-AAG was determined by cycloheximide chase assay. Data shown are representative of 3 (B–E, I, and J) and 2 (F, G, H) independent biological experiments. Data are mean ± SD from 3 technical replicates. Statistical analysis was performed by 2-way ANOVA for E and 1-way ANOVA for I and J. ***P < 0.005; ****P < 0.0001.
To further investigate the effect of hsp90B on the accessibility of CHIP to MAST1 in cells, we first examined the interaction among CHIP, hsp90B, and MAST1 WT or 2KR in the presence and absence of hsp90B inhibition. Coimmunoprecipitation and SPR showed that hsp90B binds to WT and 2KR MAST1 to a similar degree and that CHIP binding to MAST1 is increased when hsp90B is inhibited by 17-AAG (Figure 5H and Supplemental Figure 7D). We next assessed MAST1 protein stability in the presence and absence of 17-AAG or CHIP by cycloheximide chase assay. Although MAST1 2KR were slightly more stable than MAST1 WT, both were stable when cells contained hsp90B (Figure 5I), whereas the degradation of MAST1 WT was greater than that of MAST1 2KR when cells were treated with 17-AAG (Figure 5J). In addition, MAST1 WT and 2KR were not degraded by CHX when cells lacked CHIP, suggesting that CHIP is the main ubiquitin ligase of MAST1 (Supplemental Figure 7E). These data suggest that the majority of MAST1 is protected from CHIP-mediated degradation by hsp90B and CHIP contributes to MAST1 degradation mainly by ubiquitinating MAST1 at K317 and K545.
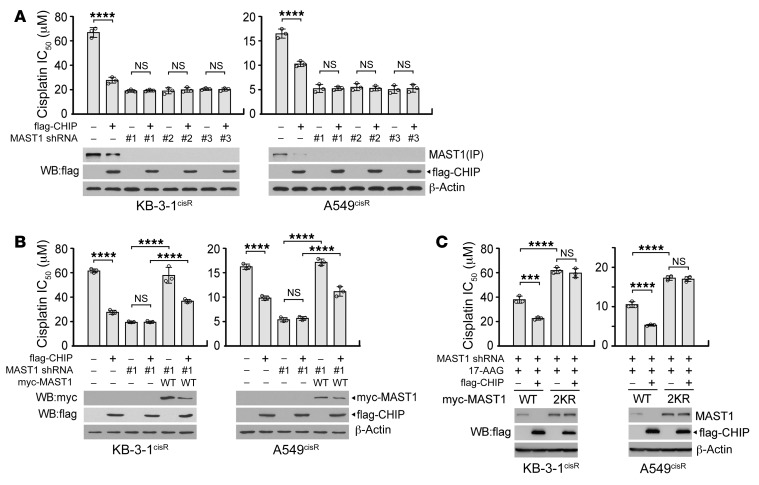
Next, we demonstrated whether CHIP signals through MAST1 and contributes to MAST1-mediated cisplatin resistance in cancer cells. Overexpression of CHIP decreased MAST1 levels and sensitized cisplatin-resistant cancer cells to cisplatin, while MAST1 knockdown abolished and rescue expression of MAST1 WT restored the effect of CHIP on cisplatin resistance (Figure 6, A and B, and Supplemental Figure 8). Moreover, expression of MAST1 2KR prevented MAST1 from being degradated and resulted in increased cisplatin resistance regardless of CHIP expression (Figure 6C). Overall, these data indicate that CHIP contributes to cisplatin resistance predominantly by inducing destabilization of MAST1 through ubiquitination at K317 and K545.
Figure 6. CHIP degrades MAST1, which consequently sensitizes cisplatin-resistant cells to cisplatin.
(A) Effect of CHIP overexpression and MAST1 knockdown on cisplatin sensitivity and MAST1 protein level. (B) Effect of MAST1 WT rescue expression on cisplatin sensitivity and MAST1 protein level in cells with CHIP overexpression and MAST1 knockdown. (C) Effect of CHIP and MAST1 WT or 2KR overexpression on cisplatin sensitivity and MAST1 protein level. KB-3-1cisR and A549cisR cells with flag-CHIP and MAST1 knockdown or WT/2KR overexpression were treated with increasing concentrations of cisplatin in the presence of 17-AAG for 48 hours. Cell viability was determined by CellTiter-Glo assay. Data are mean ± SD from 3 technical replicates and representative of 4 (A) and 2 (B and C) independent biological experiments. Statistical analysis was performed by 1-way ANOVA. ***P < 0.005; ****P < 0.0001.
Expression of hsp90Β positively and CHIP negatively correlates with MAST1 protein level and cisplatin resistance in cancer patients.
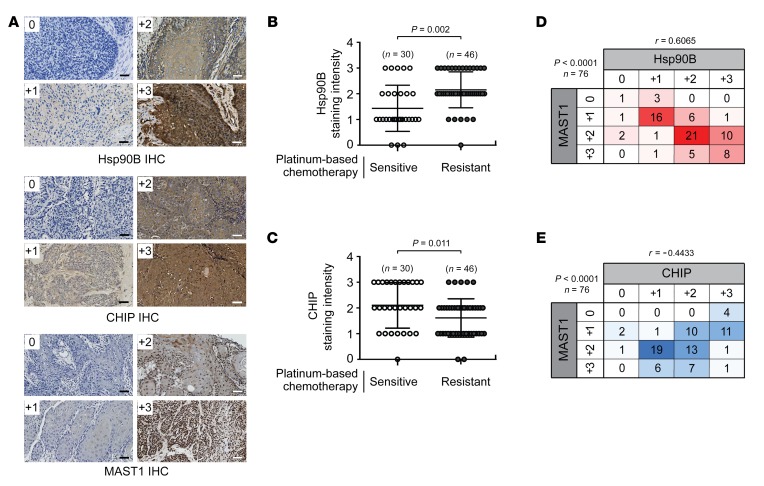
To determine the clinical relevance of our finding, we performed immunohistochemistry (IHC) staining to explore the correlation between hsp90B, CHIP, and MAST1 protein expression levels and cisplatin response in 76 tumor samples collected from head and neck squamous cell carcinoma (HNSCC) patients who received platinum-based (cisplatin or carboplatin) chemotherapy (Figure 7A). hsp90B expression levels were significantly higher in the tumors collected from patients with cisplatin-resistant disease that recurred within 2 years of therapy compared with samples from patients with cisplatin-sensitive tumors who showed no evidence of disease for more than 2 years after platinum-based chemotherapy (Figure 7B). In contrast, CHIP expression levels were higher in tumors collected from the cisplatin-sensitive patient group than in tumors from the cisplatin-resistant patient group (Figure 7C). Furthermore, MAST1 protein expression positively correlated with hsp90B protein levels with an r value of 0.6065, whereas a negative correlation between MAST1 and CHIP protein levels (r = –0.4433) was observed in the same HNSCC tumors from patients who received platinum-based chemotherapy (Figure 7, D and E). Collectively, these data clinically validate our findings and support a functional relationship between hsp90B, CHIP, and MAST1 in cisplatin response of human cancers.
Figure 7. Hsp90B-MAST1-CHIP signaling correlates with cisplatin resistance in human head and neck cancer.
(A) Tumor samples from HNSCC patients who received platinum-based chemotherapy. Representative IHC staining images of hsp90B, CHIP, and MAST1 for 0, +1, +2, and +3 scores are shown. Scale bars = 50 μm. The levels of hsp90B (B) and CHIP (C) were monitored by IHC in tumors from HNSCC patients who were sensitive or resistant to platinum-based (cisplatin or carboplatin) chemotherapy. The correlation matrix heatmaps between MAST1 and hsp90B (D) or CHIP (E) in HNSCC patients who received platinum-based chemotherapy. r represents Pearson’s correlation coefficient. For B and C, n = 30 (open circles; platinum-sensitive group) and n = 46 (closed gray circles; platinum-resistant group); for D and E, n = 76. P values were determined by 2-tailed Student’s t test for B and C, and χ2 test for D and E.
Combinatorial targeting of MAST1 and hsp90 further inhibits MAST1 and sensitizes tumor cells to cisplatin treatment.
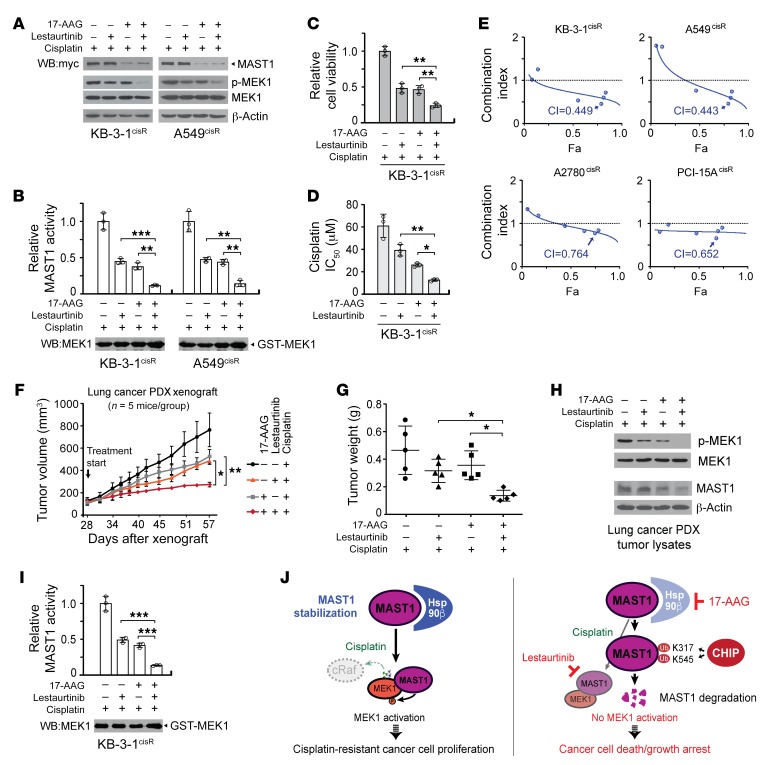
Our studies of the molecular mechanism underlying hsp90B-mediated MAST1 regulation in cisplatin resistance of human cancers suggest an alternative combinatorial therapeutic strategy which could further improve MAST1-based therapy in patients with cisplatin-resistant tumors. We previously identified lestaurtinib as an effective MAST1 inhibitor. Hsp90 inhibitors are currently being actively pursued in clinical trials. Here, we examined whether combined treatment with 17-AAG and lestaurtinib could further improve cisplatin response in vitro and in vivo by attenuating MAST1 activity. Indeed, treatment with 17-AAG and lestaurtinib further attenuated the ability of MAST1 to reactivate MEK1 by decreasing both MAST1 protein level and inhibiting its kinase activity in cisplatin-resistant cancer cells (Figure 8, A and B). The combinatorial targeting of hsp90 and MAST1 by 17-AAG and lestaurtinib synergistically decreased cell viability of diverse cisplatin-resistant cancer cell lines with the combination index (CI) of 0.443–0.764 and further sensitized the cells to cisplatin (Figure 8, C–E, Supplemental Figure 9, A and B, and Supplemental Table 1). Lastly, the combined effect of 17-AAG and lestaurtinib was examined in a patient-derived xenograft (PDX) model of lung cancer. Supporting the results obtained in vitro, administration of 17-AAG in combination with lestaurtinib enhanced the effects of cisplatin in PDX mice by significantly decreasing the tumor growth and tumor proliferation with minimum toxicity (Figure 8, F and G, and Supplemental Figure 10). The significant tumor growth decrease was accompanied by attenuated MAST1 activity in PDX tumors, which was assessed by MEK1 S217/S221 phosphorylation and ADP-Glo MAST1 kinase assay using inactive recombinant MEK1 as a substrate (Figure 8, H and I). These data suggest that combinatorial therapy with 17-AAG and lestaurtinib may further be effective in overcoming cancer cisplatin resistance by targeting both the protein regulatory system and the enzyme activity of MAST1.
Figure 8. Combination of 17-AAG and lestaurtinib further inhibits MAST1 activity and cisplatin-resistant tumor growth.
(A and B) Combination effect of 17-AAG and lestaurtinib on expression and activity of MAST1. KB-3-1cisR and A549cisR cells were treated with 17-AAG (100 nM) and lestaurtinib (100 nM) in the presence of sublethal doses of cisplatin for 24 hours. The kinase activity of MAST1 was assessed by phospho-MEK1 S217/S221 level (A) and ADP-Glo Kinase assay (B). Effect of combinatorial treatment with 17-AAG and lestaurtinib on cell viability (C) and cisplatin sensitivity (D). (E) Combination index (CI) plots are shown for diverse cisplatin-resistant cancer cell lines. (F–H) Effect of cisplatin treatment with the combination of 17-AAG and lestaurtinib on tumor growth of lung cancer PDX mice. Mice were administered 17-AAG, lestaurtinib, and cisplatin from 28 days after xenograft. Tumor volume (F) and tumor weight (G) are shown. (H and I) The kinase activity of MAST1 in PDX tumors. Data shown are representative of 3 (A, C–E, and H) and 2 (B and I) independent biological experiments. Data are mean ± SD from 3 technical replicates for B–D and I. Error bars indicate SEM (F) and SD (G) in 5 mice/group. Statistical analysis was performed by 2-way ANOVA for F and 1-way ANOVA for all the rest. *P < 0.05; **P < 0.01; ***P < 0.005. (J) Proposed model for the stability regulation of MAST1 in human cancers. Left: Hsp90B binds to and stabilizes MAST1, which reactivates MEK1 in the absence of cRaf upon cisplatin treatment and promotes cisplatin-resistant tumor growth. Right: Loss of hsp90B induces ubiquitination of MAST1 at K317 and K545 by CHIP, which triggers its degradation. Combinatorial treatment with 17-AAG and lestaurtinib further diminishes MAST1 kinase activity and attenuates cisplatin-resistant tumor growth.
Discussion
A large proportion of cisplatin-treated cancer patients experience therapeutic failure and tumor recurrence due to the acquisition of cisplatin resistance, which is complex and poorly defined. Understanding key pathway nodes that are crucial for driving resistance can provide a critical step toward circumventing cisplatin resistance in human cancers. Here we delineate the molecular mechanism by which an essential cisplatin resistance driver, MAST1, is stabilized and maintained in cancer cells and mediates cisplatin-resistant tumor growth. MAST1 is stabilized by direct interaction with the chaperone hsp90B in cancer cells, which mediates cisplatin-resistant cancer cell proliferation and tumor growth (Figure 8J, left). Inhibition of hsp90 leads to ubiquitination of MAST1 by the E3 ligase CHIP at K317 and K545, mediating proteasome-mediated degradation. Furthermore, combined pharmacological inhibition of hsp90 and MAST1 further sensitizes cancer cells to cisplatin by attenuating both the protein level and kinase activity of MAST1 (Figure 8J, right). Consistent with our findings, we also provide clinical evidence that MAST1 protein expression levels inversely correlate with CHIP but are positively linked with hsp90B in tumor tissue samples from cancer patients who received cisplatin-containing regimens.
In our study, pharmacological inhibition of hsp90 sensitized cisplatin-resistant cancer cells to cisplatin, resulting in enhanced cell death, whereas removal of MAST1 by genetic knockdown fully diminished the effect of hsp90 inhibitor in terms of cisplatin response. Although hsp90 stabilizes numerous client proteins and prevents their degradation in cancer cells, our findings implicate MAST1 as the predominant client protein that contributes to hsp90-mediated cisplatin resistance in the cancer cells we examined. Previous studies report that targeting hsp90 sensitizes multiple types of cancer cells to cisplatin, including esophageal squamous cell carcinoma, bladder cancer, and non–small cell lung cancer, and that hsp90 inhibitor–induced cisplatin sensitization is accompanied by inactivation of AKT and ERK signaling and degradation of thymidine phosphorylase (19, 28, 29). Overexpression of AKT or mitogen-activated protein kinase kinase 1/2 (MKK1/2) restores cisplatin resistance and the thymidine phosphorylase levels in non–small cell lung cancer cells. Therefore, it is possible that the activity of AKT, not the protein level, is controlled by hsp90 and contributes to cisplatin resistance or that ERK1/2 and its effector thymidine phosphorylase contribute to cisplatin resistance by serving as one of the downstream signaling factors of the MAST1-MEK pathway. In addition, hsp90 inhibition-mediated cisplatin sensitization was accompanied by an increase in the levels of the transcription factor p53 in head and neck cancer cells (30). It is plausible that MAST1 and other cellular protein factors are transcriptionally regulated by p53 and contribute to cisplatin-resistant tumor growth.
In the absence of hsp90B, MAST1 is exposed to the E3 ubiquitin ligase CHIP for ubiquitination at K317 and K545. Ubiquitination-deficient mutation (K to R) at a single site, K317 or K545, did not alter the ubiquitination status of MAST1, which suggests that ubiquitination at both sites is needed for MAST1 to be fully ubiquitinated and degraded. However, 2KR mutants did not completely restore MAST1 protein stability and cisplatin-resistant cancer cell survival, which suggests that additional ubiquitination sites may exist in MAST1 that may contribute to its stability and function. Furthermore, our proteomics analysis and coimmunoprecipitation studies revealed that MAST1 forms a complex with both hsp90B and CHIP. However, CHIP binds to and ubiquitinates MAST1 to a greater extent when hsp90B is inhibited. Hsp90B binds to WT and 2KR MAST1 to a similar degree, suggesting that, while CHIP-MAST1-hsp90B forms a complex, hsp90B binds to MAST1 and interferes with the access of CHIP to MAST1. However, the binding does not occur directly on these 2 ubiquitination sites. MAST1 is degraded even in the presence of hsp90B, although the degree of degradation is greater when cells are treated with 17-AAG. This indicates that not all the MAST1 molecules are occupied by hsp90B and this provides room for CHIP to still bind and ubiquitinate MAST1. In addition, other components may be present in the complex including hsp70 and hsp40, the cochaperones that form an intermediate complex in cells (31, 32). Detailed biochemical studies defining precise stoichiometric interaction and binding motifs on MAST1, hsp90B, and CHIP as well as structural analysis of the complex would be needed to further understand how the interaction between hsp90B and MAST1 prevents MAST1 from being exposed to CHIP for ubiquitination. Microtubule-associated serine/threonine kinase like (MASTL), which shares 23% sequence similarity with MAST1, is reported to be regulated by an E3 ubiquitin ligase HECTD3 in HEK293 cells (33). Whether MASTL is involved in cisplatin resistance in human cancers could be further investigated.
We demonstrated the cross-talk between hsp90B-MAST1-CHIP not only in one type of cancer cell line but in multiple cancer types including cervical, lung, and head and neck cancers. This suggests that hsp90B and CHIP are the common MAST1 regulators in cancer cells and that the hsp90B-MAST1-CHIP signaling axis represent a target for intervention in multiple types of cancer. Moreover, the level of hsp90B showed a stronger correlation with MAST1 than CHIP, which implicates hsp90B as the primary factor that controls MAST1 stability.
Combinatorial treatment with a hsp90 inhibitor and a MAST1 inhibitor further decreased MAST1 activity and sensitized cancer cells to cisplatin treatment. The MAST1 inhibitor lestaurtinib is a potent multikinase inhibitor that has been used in clinical trials and is reported to be generally well tolerated in cancer patients (34). Multiple inhibitors of hsp90 including tanespimycin (17-AAG), retaspimycin (17-AAG hydroquinone), and alvespimycin (17-DMAG) in combination with other anti-cancer agents are undergoing clinical evaluation (35–37). Understanding the molecular basis of the effects of the hsp90 regulatory mechanism on MAST1 will be valuable in applying optimized anti-MAST1 combination therapy to treat cancer patients with tumors that are resistant to cisplatin-based chemotherapy.
Methods
Reagents.
Lentiviral shRNA clones targeting hsp90A, hsp90B, CHIP, and MAST1 were obtained from GE Healthcare Dharmacon. Lentiviral sgRNA clones for CHIP were from Genecopoeia. The sense strand sequences of shRNA were CCAGAATGAAGGAGAACCAGA for hsp90A, GCCTTGGACAAGATTCGCTAT for hsp90B, CCCAAGTTCTGCTGTTGGACT for CHIP and CCACTTCCTCTCCAAACACTT (#1), CCACGGTCTACTTCTATGAAT (#2), and CGTGATGATGAATCACGTCTA (#3) for MAST1. The CHIP sgRNA sequences were GGCCGACTGCCGGCGCGCCC for clone #1 and GCAGCAGCACGAGCAGGCCC for clone #2. Human hsp90B, hsp90A, CHIP, and MAST1 were flag or myc tagged by PCR and cloned into pLHCX and pDEST27 (38). The CHIP H260Q mutant, MAST1 variants including K317R, K545R, K317R/K545R (2KR), D497A, and CHIP or MAST1 shRNA resistant silent mutants were generated using site-directed mutagenesis kit (Agilent Technologies). 17-AAG and lestaurtinib were from LC Laboratories. MG-132 was from EMD Millipore. Cisplatin and myelin basic protein were obtained from Sigma-Aldrich. Recombinant inactive MEK1 was from SignalChem. Cisplatin-resistant cell lines were established from parental cells by continuous exposure to cisplatin, using a modified method (39, 40). Briefly, parental A549 and PCI-15A cells were cultured with a series of increasing concentrations of cisplatin (0.1–4 μM) for a 4-month period. Cells were under cisplatin treatment for 48 hours followed by culturing in complete media to reach 90% confluency before the next round of treatment. Cisplatin was increased every cycle by 0.66 μM and 10%–15% resistant cells were selected out for the next treatment. A2780cisR cells were obtained from Sigma-Aldrich. KB-3-1cisR cells were obtained as described (7, 41). A549cisR, PCI-15AcisR, A2780cisR, and KB-3-1cisR cells are 7.76-, 5.49-, 9.61-, and 7.21-fold more resistant to cisplatin than their parental cells in terms of cisplatin IC50 (7). Cisplatin-resistant cells were maintained in 0.825 μM (A549cisR, PCI-15AcisR), 1.3 μM (A2780cisR), and 1.65 μM (KB-3-1cisR) cisplatin and cisplatin was removed from the media for 3 days prior to conducting experiments. A549 cells were purchased from American Type Culture Collection and PCI-15A cells were obtained as described (42). Lung cancer PDX tumor was obtained as described (43).
Antibodies.
Antibodies against hsp90B (5087), hsp90 (4877/C45G5), myc (2276/9B11), AKT (4685/11E7), cRaf (9422), HA (3724/C29F4), phospho-MEK1 S217/221 (9154/41G9), MEK1 (2352/61B12), phospho-ERK1/2 T202/Y204 (4376/20G11), MEK1 (9107/3A7), and ubiquitin (3936/P4D1) were purchased from Cell Signaling Technology. Antibodies against hsp90A (ab2928), phospho-Ser/Thr (ab17464), Ki-67 (ab92742/EPR3610), hsp90B for IHC staining (ab53497/H90-10), and CHIP for Western blot and IHC staining (ab134064/EPR4447) were obtained from Abcam. Anti-MAST1 antibodies for Western blot (NBP2-17228) and IHC staining (NBP1-81453) were obtained from Novus Biologicals. Antibodies against flag (F7425), B-actin (A1978/AC-15), and glutathione S-transferase (G1160/GST-2) were purchased from Sigma-Aldrich. Anti–myelin basic protein (MBP) antibody (sc-271524/F-6) was from Santa Cruz Biotechnology.
Cell culture.
KB-3-1cisR and 293T cells were cultured in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). A549cisR cells were cultured in RPMI 1640 medium with 10% FBS. Lentivirus and retrovirus production, virus infection for gene knockdown, and protein overexpression in human cells and stable cell selection were performed as previously described (38, 44). 17-AAG treatment conditions that gave a similar drug effect were used throughout the manuscript. The treatment conditions were 1 μM for 4 hours, 100 nM for 24 hours, or 50 nM for 48 hours, depending on experimental readouts.
Two-dimensional electrophoresis.
2D gel electrophoresis was carried out to identify potential MAST1 binding proteins. Briefly, GST and GST-MAST1 WT were overexpressed in 293T cells. Cells were lysed and applied to GST pull down. Eluted proteins were resuspended in rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS, 20 mM DTT, 2% IPG buffer [GE Healthcare], 0.002% bromophenol blue) to a final volume of 120 μL and loaded on 7 cm immobilized pH gradient pH 3–10 NL strips (GE Healthcare). Isoelectric focusing was performed at 300 V for 30 minutes, gradient 1000 V for 1 hour, gradient 5000 V for 90 minutes, and held at 5000 V for 1 hour. Separation in the second dimension was performed by 10% SDS-PAGE. The protein spots were visualized by silver staining. For protein identification, the protein spots of interest were excised from the gel and subjected to mass spectrometry analysis.
Ubiquitination sites mapping.
For determination of the ubiquitination sites of MAST1, GST and GST-MAST1 WT were overexpressed in 293T cells, followed by 1 μM of 17-AAG treatment for 4 hours. Cell lysates were used for GST pull down using glutathione S sepharose 4B beads. The bead bound MAST1 was washed with 20 mM Tris-HCl (pH 8.0). On-bead trypsin digestion was performed and peptides were analyzed by LTQ Orbitrap Hybrid Mass Spectrometer (Thermo Fisher Scientific).
Cell proliferation assays and cisplatin sensitivity analysis.
Cell viability was determined by trypan blue exclusion or CellTiter-Glo assay. Cisplatin-resistant cancer cells were seeded on 6- or 96-well plates and treated with the indicated concentrations of cisplatin with 17-AAG and/or lestaurtinib for the indicated time. Cell viability or proliferation was measured by trypan blue cell counting or CellTiter-Glo Luminescent Viability Assay (Promega).
Drug combination studies.
The combination effect of 17-AAG and lestaurtinib on cisplatin-resistant cell growth was analyzed using CompuSyn 1.0 as described (45, 46). Briefly, the individual dose-effect of each drug was obtained by treating 4 cisplatin-resistant cancer cell lines (cervical cancer KB-3-1cisR, lung cancer A549cisR, ovarian cancer A2780cisR, and head and neck cancer PCI-15AcisR) with 17-AAG or lestaurtinib in the presence of sublethal doses of cisplatin. The median effect dose (Dm) and linear correlation coefficient of the ME-plot (r) were analyzed. Optimal concentration ratios were obtained based on the Dm values and 6 serial dilutions of the optimal ratio were used to measure the cytotoxic effect. Combination index (CI) values were calculated. CI values of less than 1, equal to 1, and greater than 1 indicated synergistic, additive, and antagonistic effects, respectively. The raw data are presented in Supplemental Table 1.
MAST1 kinase activity assay.
The GST-fused MAST1 variants were enriched from cell lysates with glutathione sepharose 4B beads. MAST1 eluted with the addition of reduced glutathione was incubated with inactive MEK1 or MBP in a kinase assay buffer (40 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 200 mM ATP, 0.1 mg/mL BSA) at 30°C for 30 minutes. For detection of MAST1 kinase activity in cisplatin-resistant cancer cells or PDX tumors, endogenous MAST1 was immunoprecipitated using anti-MAST1 antibody and subjected to in vitro kinase assay using inactive MEK1 or MBP as a substrate. The MAST1 kinase activity was determined by ADP-Glo Kinase Assay, phospho-MEK S217/S221 immunoblotting of cell lysates, or autophosphorylation level of MAST1 by phospho-serine/threonine immunoblotting.
Quantitative RT-PCR.
Total RNA from KB-3-1cisR and A549cisR cells was isolated using the RNeasy Kit (Qiagen). Total RNA (1 μg) was converted into cDNA with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was performed on a 7500 Real Time PCR System (Applied Biosystems) using iTaq Universal SYBR Green Supermix (Bio-Rad). Primers used for MAST1 qPCR were forward 5′-TCTCTGGACCGCGC TTTCTA-3′ and reverse 5′-TGAGGCTTTTCCGATTACTGGT-3′. All reactions were performed in triplicate. MAST1 gene level was calculated by comparative delta Ct and normalized using GAPDH gene.
CHIP ubiquitination assays.
In vitro ubiquitination assay was performed using the Human CHIP Ubiquitin Ligase Kit (R&D Systems) according to the manufacturer’s instructions using purified MAST1 WT or K317R/K545R (2KR). Briefly, GST-fused MAST1 WT or 2KR proteins were enriched from 293T cells and used as substrates. In vitro CHIP ubiquitination reaction was initiated by adding purified E1, E2, CHIP, and ubiquitin. The samples were applied to SDS-PAGE and ubiquitinated MAST1 was detected by anti-ubiquitin antibody. For in vivo ubiquitination assay, A549cisR cells were transfected with various constructs as indicated and treated with or without 10 μM MG-132 for 6 hours. Ubiquitination of MAST1 in cancer cells was determined by GST pull down followed by immunoblotting of ubiquitin.
Surface plasmon resonance.
Recombinant hsp90A or hsp90B (1 μM) were coupled to the CM5 sensor chip of a Biacore X100 (GE Healthcare). The indicated amount of 17-AAG was prepared in 0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% vol/vol Surfactant P20 and injected over hsp90A or hsp90B at 30 μL/min for a contact time of 1 minute at 20°C. Single-cycle kinetic analysis was performed for interaction analysis between hsp90 isoforms and 17-AAG. For MAST1 WT-hsp90B or MAST1 2KR-hsp90B binding, recombinant hsp90B at a series of dilutions was injected over a GST-MAST1 WT– or 2KR–coupled (1 μM) sensor chip. After subtracting the average of the buffer blank, the corrected sensorgrams were analyzed by BIAevaluation Software version 2.1 (GE Healthcare).
Xenograft studies.
Animal experiments were performed according to the protocol approved by the Emory University Institutional Animal Care and Use Committee. Nude mice (athymic nu/nu 6-week-old females, Envigo) were subcutaneously injected with 1 × 106 KB-3-1cisR cells with MAST1 or hsp90B knockdown and MAST1 variant overexpression. Cisplatin and 17-AAG were administered 5 mg/kg/3.5 days and 50 mg/kg/5 times a week, respectively, by i.p. injection after xenograft for 11–15 days. For PDX mouse model, tumors from small cell lung carcinoma patients were implanted into the flanks of mice (athymic nu/nu 6-week-old females, Envigo). When the tumor volume reached an average size of 100 mm3, mice were evenly divided into 4 groups and cisplatin (5 mg/kg/3.5 days; i.p. injection), 17-AAG (50 mg/kg 5 times a week; i.p. injection), and lestaurtinib (20 mg/kg 5 times a week; subcutaneous injection) were administered for 29 days. Cisplatin, 17-AAG, and lestaurtinib were dissolved in PBS, 5% Tween 80, and 5% dimethyl sulfoxide containing PBS, and 40% polyethylene glycol 400, 10% povidone, and 2% benzyl alcohol containing PBS, respectively. Tumor volume was measured by caliper measurement of 2 perpendicular diameters and calculated using the formula: 4π / 3 × (width / 2)2 × (length / 2). At the experimental endpoint, tumors were dissected and weighted. Tumor proliferation was determined by Ki-67 IHC staining. For all animal studies, randomization, concealed allocation, and blinding of outcome assessment were used.
Immunohistochemistry staining.
Paraffin-embedded tumor specimens from HNSCC patients were obtained from the Emory Head and Neck Satellite Tissue Bank. The evaluation of human specimens was approved by the IRB of Emory University. Patients’ clinical information was obtained from the pathology files at Emory University Hospital under the guidelines and with approval from the IRB of Emory University. All clinical samples were collected with written informed consent under protocols approved by the Health Insurance Portability and Accountability Act. Tumors from HNSCC patients treated with cisplatin- or carboplatin-containing chemotherapy were used for IHC staining and analysis. IHC staining of hsp90B, CHIP, and MAST1 was performed by incubating the sections with anti-hsp90B antibody (1:500 dilution), anti-CHIP antibody (1:500 dilution), and anti-MAST1 antibody (1:200). Xenograft tumors from mice were stained with anti–ki-67 antibody (1:1000). After primary antibody incubation, the slides were incubated with ImmPRESS reagent, avidin–biotin complex system, and 3,3′-diaminobenzidine. Staining was scored as 0 for no staining, +1 for weak staining, +2 for moderate staining, and +3 for strong staining.
Statistics.
Statistical analysis was performed using Graphpad Prism 8. Data with error bars represent mean ± SD, except for xenograft tumor volume curves, which represent mean ± standard error of the mean (SEM). Statistical analysis of significance was based on 2-tailed Student’s t test for Figure 7, B and C and Supplemental Figures 4A and 7D, χ2 test for Figure 7, D and E, and 1-way or 2-way ANOVA with Bonferroni’s post hoc multiple-comparisons testing for all other remaining data. Statistical analyses are based on a set of assumptions such as homogeneity of variances and normal distribution. P values of 0.05 or less were determined as statistically significant. The variability within each group has been quantified with SD or SEM, and used for statistical comparison.
Study approval.
Animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Emory University, Atlanta, Georgia, USA. Studies using human specimens were reviewed and approved by the IRB of Emory University. Tissue specimens were obtained from the Emory University Head and Neck Satellite Tissue Bank. The study was conducted in compliance with good clinical practice. Written informed consent was obtained from individuals or their guardians prior to the sample collection. Clinical information on the specimens was obtained following the regulation of the Health Insurance Portability and Accountability Act.
Author contributions
CP and JC were responsible for acquisition, analysis, and interpretation of data. JC initiated the study and CP expanded and completed the study. ZGC, TKO, and SL provided critical reagents including cell lines, xenograft models, and RNAi library for the study. KRM collected clinical tumor samples and performed histopathological study. NFS and DMS provided clinical information. CP and JL performed animal experiments. DL and LS performed proteomic analyses. LJ performed cycloheximide chase assay. CP, JC, ACB, JK, YJ and AU performed all other experiments. CP, LJ, and SK designed the study and wrote the paper. Authorship order was determined by the amount of experiments performed and shown.
Supplementary Material
Acknowledgments
We acknowledge Dr. Anthea Hammond for editorial assistance. This study was supported by NIH grants R01 CA207768 and R01 CA175316 (to SK), Department of Defense grant W81XWH-17-1-0186 (to SK), Developmental Funds from the Winship Cancer Institute of Emory University (to SK), Winship IRG-17-181-04 from the American Cancer Society (to LJ), the 2019 Breast Cancer Research Foundation-AACR Career Development Awards for Translational Breast Cancer Research 19-20-26-JIN (to LJ), an Elsa U. Pardee Foundation grant (to LJ), and a UF Health Cancer Center pilot grant (to LJ). SK is a Robbins Scholar, a Georgia Cancer Coalition Scholar, and an American Cancer Society Basic Research Scholar.
Version 1. 08/26/2019
Electronic publication
Version 2. 10/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4110–4123.https://doi.org/10.1172/JCI125963.
Contributor Information
Chaoyun Pan, Email: chaoyun.pan@emory.edu.
Jaemoo Chun, Email: jaemoo.chun@gmail.com.
Dan Li, Email: lidan7126@gmail.com.
Austin C. Boese, Email: austin.c.boese@emory.edu.
Jie Li, Email: lijie19930204@gmail.com.
JiHoon Kang, Email: jihoon.kang@emory.edu.
Yunhan Jiang, Email: yunhan.jiang@ufl.edu.
Lina Song, Email: slna1982@gmail.com.
Nabil F. Saba, Email: nfsaba@emory.edu.
Dong M. Shin, Email: dmshin@emory.edu.
Sagar Lonial, Email: SLONI01@emory.edu.
Lingtao Jin, Email: ljin1@ufl.edu.
Sumin Kang, Email: smkang@emory.edu.
References
- 1.Galanski M. Recent developments in the field of anticancer platinum complexes. Recent Pat Anticancer Drug Discov. 2006;1(2):285–295. doi: 10.2174/157489206777442287. [DOI] [PubMed] [Google Scholar]
- 2.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 3.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giaccone G. Clinical perspectives on platinum resistance. Drugs. 2000;59(Suppl 4):9–17; discussion 37. doi: 10.2165/00003495-200059004-00002. [DOI] [PubMed] [Google Scholar]
- 5.Köberle B, Tomicic MT, Usanova S, Kaina B. Cisplatin resistance: preclinical findings and clinical implications. Biochim Biophys Acta. 2010;1806(2):172–182. doi: 10.1016/j.bbcan.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Galluzzi L, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 7.Jin L, et al. MAST1 drives cisplatin resistance in human cancers by rewiring cRaf-independent MEK activation. Cancer Cell. 2018;34(2):315–330.e7. doi: 10.1016/j.ccell.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, Chamberlain JS. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat Neurosci. 1999;2(7):611–617. doi: 10.1038/10165. [DOI] [PubMed] [Google Scholar]
- 9.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18(6):345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 10.Sreedhar AS, Kalmár E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562(1–3):11–15. doi: 10.1016/s0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 11.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 12.Sato S, Ueki M, Alzheimer’s Disease Neuroimaging Initiative Fast score test with global null estimation regardless of missing genotypes. PLoS One. 2018;13(7):e0199692. doi: 10.1371/journal.pone.0199692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giulino-Roth L, et al. Inhibition of Hsp90 Suppresses PI3K/AKT/mTOR signaling and has antitumor activity in Burkitt lymphoma. Mol Cancer Ther. 2017;16(9):1779–1790. doi: 10.1158/1535-7163.MCT-16-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem. 1995;270(41):24585–24588. doi: 10.1074/jbc.270.41.24585. [DOI] [PubMed] [Google Scholar]
- 15.Azoitei N, et al. HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization. Cancer Res. 2014;74(23):7125–7136. doi: 10.1158/0008-5472.CAN-14-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Xiao L, Wang L, Ruden DM. Hsp90 inhibitors and drug resistance in cancer: the potential benefits of combination therapies of Hsp90 inhibitors and other anti-cancer drugs. Biochem Pharmacol. 2012;83(8):995–1004. doi: 10.1016/j.bcp.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedland JC, et al. Targeted inhibition of Hsp90 by ganetespib is effective across a broad spectrum of breast cancer subtypes. Invest New Drugs. 2014;32(1):14–24. doi: 10.1007/s10637-013-9971-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, et al. Reversing drug resistance of cisplatin by hsp90 inhibitors in human ovarian cancer cells. Int J Clin Exp Med. 2015;8(5):6687–6701. [PMC free article] [PubMed] [Google Scholar]
- 19.Tatokoro M, et al. Potential role of Hsp90 inhibitors in overcoming cisplatin resistance of bladder cancer-initiating cells. Int J Cancer. 2012;131(4):987–996. doi: 10.1002/ijc.26475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballinger CA, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/MCB.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connell P, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 22.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99(20):12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HT, Zeng LF, He QY, Tao WA, Zha ZG, Hu CD. The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim Biophys Acta. 2016;1863(2):335–346. doi: 10.1016/j.bbamcr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Won KJ, et al. Stability of the cancer target DDIAS is regulated by the CHIP/HSP70 pathway in lung cancer cells. Cell Death Dis. 2017;8(1):e2554. doi: 10.1038/cddis.2016.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. CHIP/Stub1 functions as a tumor suppressor and represses NF-κB-mediated signaling in colorectal cancer. Carcinogenesis. 2014;35(5):983–991. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- 26.Jang KW, et al. Ubiquitin ligase CHIP induces TRAF2 proteasomal degradation and NF-κB inactivation to regulate breast cancer cell invasion. J Cell Biochem. 2011;112(12):3612–3620. doi: 10.1002/jcb.23292. [DOI] [PubMed] [Google Scholar]
- 27.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3(3):213–217. doi: 10.1016/S1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 28.Ui T, et al. The HSP90 inhibitor 17-N-allylamino-17-demethoxy geldanamycin (17-AAG) synergizes with cisplatin and induces apoptosis in cisplatin-resistant esophageal squamous cell carcinoma cell lines via the Akt/XIAP pathway. Oncol Rep. 2014;31(2):619–624. doi: 10.3892/or.2013.2899. [DOI] [PubMed] [Google Scholar]
- 29.Weng SH, Tseng SC, Huang YC, Chen HJ, Lin YW. Inhibition of thymidine phosphorylase expression by using an HSP90 inhibitor potentiates the cytotoxic effect of cisplatin in non-small-cell lung cancer cells. Biochem Pharmacol. 2012;84(1):126–136. doi: 10.1016/j.bcp.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Roh JL, Kim EH, Park HB, Park JY. The Hsp90 inhibitor 17-(allylamino)-17-demethoxygeldanamycin increases cisplatin antitumor activity by inducing p53-mediated apoptosis in head and neck cancer. Cell Death Dis. 2013;4:e956. doi: 10.1038/cddis.2013.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickey CA, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J Clin Invest. 2007;117(3):648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–308. doi: 10.1379/1466-1268(2003)008<0303:CALBTC>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Zhou L, Prodromou C, Savic V, Pearl LH. HECTD3 mediates an HSP90-dependent degradation pathway for protein kinase clients. Cell Rep. 2017;19(12):2515–2528. doi: 10.1016/j.celrep.2017.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knapper S, et al. A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood. 2006;108(10):3262–3270. doi: 10.1182/blood-2006-04-015560. [DOI] [PubMed] [Google Scholar]
- 35.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuno A, et al. Clinical evaluation and biomarker profiling of Hsp90 inhibitors. Methods Mol Biol. 2018;1709:423–441. doi: 10.1007/978-1-4939-7477-1_29. [DOI] [PubMed] [Google Scholar]
- 37.Butler LM, Ferraldeschi R, Armstrong HK, Centenera MM, Workman P. Maximizing the therapeutic potential of HSP90 inhibitors. Mol Cancer Res. 2015;13(11):1445–1451. doi: 10.1158/1541-7786.MCR-15-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin L, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27(2):257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Negoro K, et al. Establishment and characterization of a cisplatin-resistant cell line, KB-R, derived from oral carcinoma cell line, KB. Int J Oncol. 2007;30(6):1325–1332. doi: 10.3892/ijo.30.6.1325. [DOI] [PubMed] [Google Scholar]
- 40.Wang HQ, Jin JJ, Wang J. Matrine induces mitochondrial apoptosis in cisplatin-resistant non-small cell lung cancer cells via suppression of β-catenin/survivin signaling. Oncol Rep. 2015;33(5):2561–2566. doi: 10.3892/or.2015.3844. [DOI] [PubMed] [Google Scholar]
- 41.Richert N, Akiyama S, Shen D, Gottesman MM, Pastan I. Multiply drug-resistant human KB carcinoma cells have decreased amounts of a 75-kDa and a 72-kDa glycoprotein. Proc Natl Acad Sci U S A. 1985;82(8):2330–2333. doi: 10.1073/pnas.82.8.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang S, et al. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120(4):1165–1177. doi: 10.1172/JCI40582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owonikoko TK, et al. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J Transl Med. 2016;14(1):111. doi: 10.1186/s12967-016-0861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin L, et al. p90 RSK2 mediates antianoikis signals by both transcription-dependent and -independent mechanisms. Mol Cell Biol. 2013;33(13):2574–2585. doi: 10.1128/MCB.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 46.Zhang N, Fu JN, Chou TC. Synergistic combination of microtubule targeting anticancer fludelone with cytoprotective panaxytriol derived from panax ginseng against MX-1 cells in vitro: experimental design and data analysis using the combination index method. Am J Cancer Res. 2016;6(1):97–104. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.