Abstract
Despite progress in intensification of therapy, outcomes for patients with metastatic osteosarcoma (OS) have not improved in thirty years. We developed a system that enabled preclinical screening of compounds against metastatic OS cells in the context of the native lung microenvironment. Using this strategy to screen a library of epigenetically targeted compounds, we identified inhibitors of CDK12 to be most effective, reducing OS cell outgrowth in the lung by more than 90% at submicromolar doses. We found that knockout of CDK12 in an in vivo model of lung metastasis significantly decreased the ability of OS to colonize the lung. CDK12 inhibition led to defects in transcription elongation in a gene length– and expression-dependent manner. These effects were accompanied by defects in RNA processing and altered the expression of genes involved in transcription regulation and the DNA damage response. We further identified OS models that differ in their sensitivity to CDK12 inhibition in the lung and provided evidence that upregulated MYC levels may mediate these differences. Our studies provided a framework for rapid preclinical testing of compounds with antimetastatic activity and highlighted CDK12 as a potential therapeutic target in OS.
Keywords: Genetics, Oncology
Keywords: Cancer, Drug screens, Epigenetics
Introduction
One of the greatest challenges in cancer treatment is the successful targeting and eradication of metastatic disease. Indeed, 90% of cancer-related deaths are caused by metastasis (1), yet paradoxically, many drug discovery efforts are focused on the identification of compounds that can inhibit primary tumor growth and therefore do not target the primary cause of patient mortality. Inherent to this problem is the lack of adequate models that recapitulate the metastatic milieu and that can be exploited to identify potential therapeutic targets as well as to test new inhibitors. While PDX models afford a powerful platform for testing individual compounds or combinations, they may be insufficient to reiterate the complex tumor-microenvironment interactions that are necessary for the establishment of metastases or for the screening of multiple compounds in a high-throughput manner. Hence, there is an urgent clinical need for assays that can more rapidly and reliably test for compounds with antimetastatic activity.
This challenge has never been so pressing as in osteosarcoma (OS), the most common bone tumor of childhood, with a peak incidence in adolescence (2), an age when patients often present with metastatic disease for which treatment options are limited (3). Patients with localized disease have a 65%–70% survival rate (4), while in those with metastatic disease, most commonly to the lung, the chances of survival have not improved beyond 19%–30% (5, 6) despite multiple attempts based on treatment intensification. Indeed, the current standard-of-care treatment regimen used by the Children’s Oncology Group does not stratify for localized and metastatic disease, as the latest large international trial failed to show a significant difference in outcome for therapy intensification in patients with metastatic OS (7).
A major reason for the difficulty in eradicating metastatic OS is the striking genomic complexity of this cancer (8), with a lack of recurrent targetable mutations in either primary or metastatic lesions. Typically, the tumor is rife with structural chromosomal variations with regions of hypermutation (9), including mutations in TP53 or RB1 and associated pathway genes and CDKN2A/B, DLG, ATRX, or members of the PI3K/mTOR pathways (10–13). In fact, the 3 most commonly aberrant pathways are p53, alternative lengthening of telomeres, and PI3K pathways, all of which have been identified through analysis largely of primary tumors, as most cooperative group tumor-banking efforts lack samples from metastatic sites (14). Furthermore, the high degree of genetic instability in OS (8) necessitates an approach that relies not on a single target, but on general processes that are consistently dysregulated and create vulnerabilities in tumor cells.
We recently demonstrated that alterations in the enhancer epigenome are prevalent in metastatic OS cells and endow OS cells with the capacity to colonize and proliferate in the lung (15). Here, we leverage this new knowledge in combination with an assay that permits high-throughput screening in the context of the native lung microenvironment to identify compounds with promising activity against metastatic OS. This effort, conducted in an ex vivo murine model of disseminated OS, revealed CDK12 inhibitors to be effective suppressors of metastatic disease in the lung and provided insights into the mechanism of action of this cyclin-dependent kinase (CDK) in gene transcription.
Results
Ex vivo screening identifies CDK inhibitors as potent suppressors of metastatic OS outgrowth.
With the ultimate goal of targeting the primary cause of mortality in OS patients, we took advantage of a system that enables high-content screening for antimetastatic compounds directly within the context of the lung microenvironment. The approach is based on a lung-explant organ culture model called the pulmonary metastasis assay (PuMA) (16), in which GFP-labeled human metastatic OS cells are seeded into mouse lungs via tail-vein injection. The mouse is euthanized and the lungs insufflated with agarose to preserve the 3D architecture, sectioned, and cultured at an air-fluid interface that maintains the viability of lung cell populations. In this study, we miniaturized the PuMA system to accommodate high-throughput screening of compounds with antimetastatic potential (Figure 1A). Lung explants seeded with GFP-labeled metastatic OS cells were cultured in 96-well format, compounds added to each well, and efficacy monitored via high-throughput confocal imaging using the normalized fluorescent area as a surrogate for metastatic cell growth (Figure 1, A and B). Toxicity to the lung was evaluated through histological methods.
Figure 1. An ex vivo screen identifies compounds that inhibit the growth of metastatic OS.
(A) Outline of the PuMA screen. (B) Left: 96-well plate of lung explants seeded with metastatic OS cells. Center, left: magnified view of 4 individual wells. Fluorescence image of a lung explant seeded with GFP+ OS cells (top center) and corresponding false colored image used for quantification (top right). Bottom center: ×40 magnification of H&E-stained, control-treated lung section after 14 days in PuMA explant culture. Boxed region highlights area of extensive OS cell growth. Bottom right: ×200 magnification of boxed region. (C) Distribution of 112 of the compounds tested, according to class. (D) Fluorescence image of a 96-well plate of lung explants treated for 14 days with 2 doses (high and low) of each compound or vehicle control (DMSO, white boxes). Called hits are boxed in red. Each row (M1–M8) is from a single mouse. (E) Dot plot showing results of all compounds tested at all doses. The dashed line corresponds to 90% reduction of GFP+ area compared with DMSO controls after 14 days of treatment in the PuMA model.
From more than 3000 molecules in the Selleck Chemicals Bioactive Compound Library, based on published potency and our previous findings in neuroblastoma cells (17), we selected 112 compounds with known specificities for a broad set of protein targets that function in transcription regulation and other epigenetic processes (Figure 1C). Each compound was tested at 2 doses for 14 days on lung explants from mice seeded with the metastatic human OS cell-line MG63.3 (18). This cell line was derived from repeated in vivo retrieval of nonmetastatic MG63 cells from the lungs of mice (19). An example of an array of lung-explant tissues exposed to compounds is shown in Figure 1D. Based on a stringent threshold of 90% or greater reduction of GFP positivity after 14 days of treatment, 17 unique hits were identified (Figure 1E and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI127718DS1). These compounds included CDK inhibitors as well as histone deacetylase (HDAC) inhibitors and protein arginine methyltransferase (PRMT) inhibitors.
Among all compounds tested, CDK inhibitors targeting multiple cell cycle– and transcription-related CDKs (1/2/5/9/12) were the most effective, with dinaciclib (20, 21), PHA-767491 (22), and AZD5438 (23), reducing tumor cell growth by more than 99% relative to vehicle-treated explants (Figure 1E and Supplemental Table 1). These data demonstrate the ability of the ex vivo screening approach to identify inhibitors of metastatic OS cell outgrowth in the lung microenvironment.
To identify potential differences in screening ex vivo versus using standard culture, we further performed a parallel screen of all 112 compounds on MG63.3 cells cultured in vitro (Supplemental Figure 1A). All 17 compounds that scored as hits in PuMA scored as hits in vitro, although 27 additional compounds were identified in vitro (Supplemental Figure 1B). Finally, we compared the raw GFP+ area values of the controls, hits, and nonhits in the ex vivo screen and verified the highly significant differences between the hits and controls, with no significant differences between the nonhits and the controls (Supplemental Figure 1C). Thus, direct screening in PuMA can efficiently identify compounds that show efficacy in the most relevant context.
CDK12 inhibitors reduce metastatic cell outgrowth in the lung microenvironment.
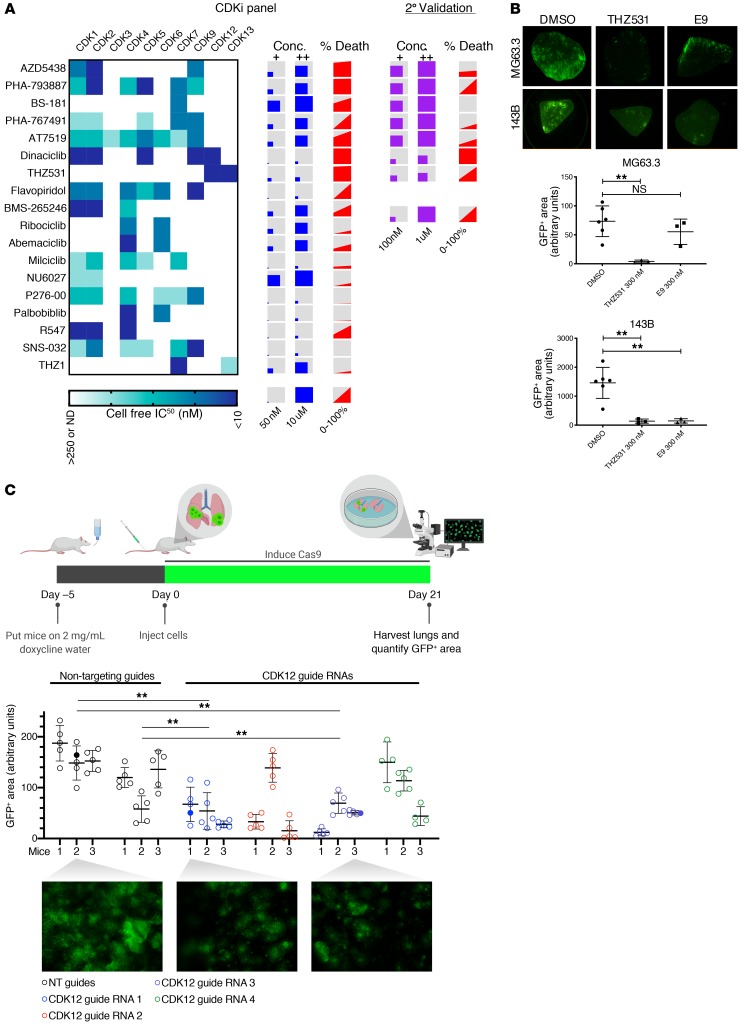
CDK family members are generally expressed at high levels in OS cells. Furthermore, CDK transcript levels are comparable between OS cells cultured in vitro versus in PuMA (Supplemental Figure 2A). To further narrow down the specific CDK vulnerability for metastatic OS, we evaluated 16 different CDK inhibitors from the initial screen in the PuMA system as well as 2 more recently developed compounds with a narrower range of selectivity, THZ1 (24, 25) and THZ531 (26, 27) (Figure 2A). Eight compounds reduced metastatic OS cell outgrowth in the lung by more than 90%; 6 of these, including flavopiridol and dinaciclib, were pan-CDK inhibitors. The remaining 2 were more selective, with BS-181 targeting CDK7 (28) and THZ531 targeting CDKs 12 and 13 (26). Seven of the most active compounds were retested at lower doses than used in the initial screen. In this secondary screen, dinaciclib (targeting CDKs 1,2,5,9,12) and THZ531 were the only 2 compounds that significantly reduced metastatic cell growth at submicromolar doses (Figure 2A). CDK12 is a common target of both compounds, and its expression is elevated in OS patient tumor samples compared with normal (Supplemental Figure 2B). Prior trials with dinaciclib in pediatric tumor models have been disappointing, an effect attributed to its lack of selectivity (29). This prompted us to test E9 (17), a recently developed, highly selective, covalent CDK12 inhibitor that does not target CDK13, but that also noncovalently engages CDK9. When tested on lung explants developed from the metastatic human OS cell line, 143B, these compounds showed activity at submicromolar doses. The MG63.3 model exhibited a similar response to THZ531, although interestingly, was 6-fold less sensitive to E9 than the 143B PuMA model (Figure 2B). Together, these results suggest that agents targeting CDK12 are among the most potent inhibitors of metastatic OS cells we tested, but also that not all OS cell models are equally sensitive to these inhibitors in the context of the lung microenvironment.
Figure 2. CDK inhibitors reduce metastatic cell outgrowth in the lung microenvironment.
(A) Heatmap showing IC50 values for the CDK inhibitors tested, as determined by Selleck chemicals. NN, not determined. Concentrations tested for each compound are represented by the sizes of the squares within the boxes. Percentage death refers to the reduction of the GFP+ area in the compound vs. DMSO control–treated cells at low and high doses. The left side of the gray-red boxes corresponds to the percentage of cell death achieved with the low dose; the right side denotes the percentage of death achieved with the high dose. Right, compound concentrations and corresponding percentages of cell death achieved in a secondary assessment of 7 initial hits. (B) Top: representative GFP images of lung explants seeded with either MG63.3 or 143B cells and treated with the indicated CDK inhibitors for 14 days. Bottom: GFP-based quantification of lung explants from the respective cell line, treated with indicated compound. Data are presented as mean ± SD with at least 3 explants per condition. Ordinary 1-way ANOVA with Tukey’s multiple comparisons test was used to compare across groups. **P < 0.01. Original magnification, ×2. (C) Top: drawing of in vivo CRISPR CDK12 knockout experiment. Middle: quantification of GFP+ area of lungs from each mouse in the experiment. Four to five images were taken per set of lungs and quantified for GFP+ area using ImageJ. Filled dots indicate those in the representative images. Bottom: representative images from indicated lungs. Original magnification, ×2. Data are represented as mean ± SD. Ordinary 1-way ANOVA with Tukey’s multiple comparisons test was used to compare across groups. **P < 0.01.
CDK12 knockout reduces lung metastatic outgrowth in vivo.
To determine whether the inhibition of CDK12 is the mechanism responsible for the observed antimetastatic effect, we performed genetic CDK12 knockout in an in vivo model of pulmonary metastasis. Using GFP-expressing MG63.3 cells engineered to express Cas9 under doxycycline control, we generated 4 unique, inducible CDK12-knockout pools with an additional 2 nontarget guide RNA control pools. In vitro, all 4 knockout pools showed a decrease in CDK12 protein after 7 days of exposure to doxycycline (Supplemental Figure 3A). To determine the effect that loss of CDK12 has on metastatic competence in vivo, we seeded the edited cells via lateral tail-vein injection into the lungs of immunodeficient SCID Beige mice pretreated with doxycycline for 5 days. The mice were then maintained on doxycycline water for the duration of the experiment to ensure Cas9 induction in the xenografted cells. After 21 days, we quantified tumor burden by measuring the GFP-positive area using whole-lung fluorescence microscopy. Multiple guide RNAs targeting CDK12 significantly abrogated metastatic burden when compared with the 2 nontarget controls (Figure 2C). These results indicate that CDK12 is crucial for OS lung metastasis and that its chemical inhibition is likely causing the decreased tumor burden we see in our compound-based experiments.
CDK12 inhibition is broadly effective against multiple human OS cell lines in vitro.
To further delineate the differential sensitivity of CDK12 inhibition in human OS cells, we next tested THZ531 and E9 against a panel of 9 human OS cell lines cultured in vitro, observing dose-dependent selectivity and toxicity in all cell lines (Figure 3A). We observed significant differences in cell viability between THZ531- and E9-treated cells, with the IC50 values for E9-treated cells for the most part (2 to 16 times) significantly lower than those for THZ531-treated cell lines; this was similar to results observed in the PuMA model (Supplemental Figure 4A). To ensure that both E9 and THZ531 efficiently bound to CDK12 in OS cells, we determined the extent of target engagement through a competition assay using a biotinylated derivative of THZ1 (bio-THZ1), a covalent inhibitor of CDK7/12/13 (24) (Figure 3B). Decreased bio-THZ1 binding to CDK12 was seen after both E9 and THZ531 treatment, but at much lower concentrations of E9 compared with THZ531 (50 vs. 200 nM), indicating that E9 engages CDK12 more efficiently than THZ531.
Figure 3. THZ531 and E9 show broad activity against OS cell-line models.
(A) Dose-response curves for OS cells treated with increasing concentrations of THZ531 (left) or E9 (right) for 72 hours. Percentage of cell viability relative to DMSO-treated cells is shown. The data are presented as mean ± SD of triplicate points. (B) Analysis of target engagement in MG63.3 cells following THZ531 or E9 treatment. Cells were treated with THZ531/E9 or DMSO for 6 hours at the indicated concentrations and cell lysates incubated with 1 μM of biotinylated THZ1 (bio-THZ1) overnight, followed by Western blotting to detect CDK12. (C) Colony-formation assays of MG63.3 and 143B cell lines treated with different concentrations of E9 for 12 days. Representative examples are shown (left), with quantification on the right. Results are expressed as mean ± SD. n = 3. **P < 0.01; ****P < 0.0001, 1-way ANOVA with Dunnett’s multiple comparison correction. (D) Western blot analysis of cleaved PARP1 in the indicated cells following treatment with E9 at the indicated doses and times. GAPDH, loading control. (E) Cell-cycle analysis of MG63.3 and 143B cells exposed to 400 nM of E9 for 24 and 48 hours by flow cytometry with propidium iodide (PI) staining. The scale and axes are indicated in the lower left corner. (F) Flow cytometry analysis of γ-H2AX staining in MG63.3 and 143B cells treated with 400 nM E9 for the indicated times.
Although the increased binding of E9 to CDK12 could partly account for the higher cytotoxic effect of this compound in the OS lines tested, this alone could not explain the decreased sensitivity of these cells to THZ531, also a covalent binding inhibitor. To further address the difference in potency between E9 and THZ531, we analyzed the expression of the multidrug resistance family of drug-transporter proteins in our panel of OS cell lines. ABCB1, which was previously shown to mediate THZ531 resistance in neuroblastoma cells (17), was overexpressed in all the OS cell lines tested, in keeping with prior reports of its upregulation in OS cell lines and drug-resistant tissues (Supplemental Figure 4B) (30–34). Addition of the ABCB1 inhibitor tariquidar (35) significantly increased sensitivity to THZ531 in 6 out of 9 cell lines, whereas the sensitivity to E9 did not change substantially upon addition of tariquidar (Supplemental Figure 4, A and C). These data support the role of ABCB1 in the decreased cytotoxic effect of THZ531 in the majority of human OS cell lines tested and, consistent with previous studies (17), indicate that E9 is not a substrate of this efflux protein.
Even with ABCB1 inhibition, the potency of E9 was higher than that of THZ531 for OS cells. E9 treatment caused a significant decrease in colony formation (Figure 3C) and elevated levels of apoptosis, as measured by an increase in cleaved PARP1 expression (Figure 3D). OS cells treated with E9 also underwent G2/M cell-cycle arrest (Figure 3E and Supplemental Figure 4D). Interestingly, we observed a more profound cell-cycle arrest and a delayed increase in apoptosis in MG63.3 cells compared with that in 143B cells. CDK12 depletion affects the expression of DNA damage response (DDR) genes in diverse cancer types, resulting in increased DNA damage (36–38). Similarly, we observed a clear increase in the level of γ-H2AX staining in E9-treated cells, indicating the accumulation of DNA double-strand breaks (DSBs) (Figure 3F and Supplemental Figure 4, E–G). Thus, the potent killing of metastatic OS cells by CDK12 inhibitors may reflect selective toxicity due to their inability to repair DNA lesions, with consequent cell-cycle arrest and apoptosis.
E9 impairs transcription elongation in OS cells.
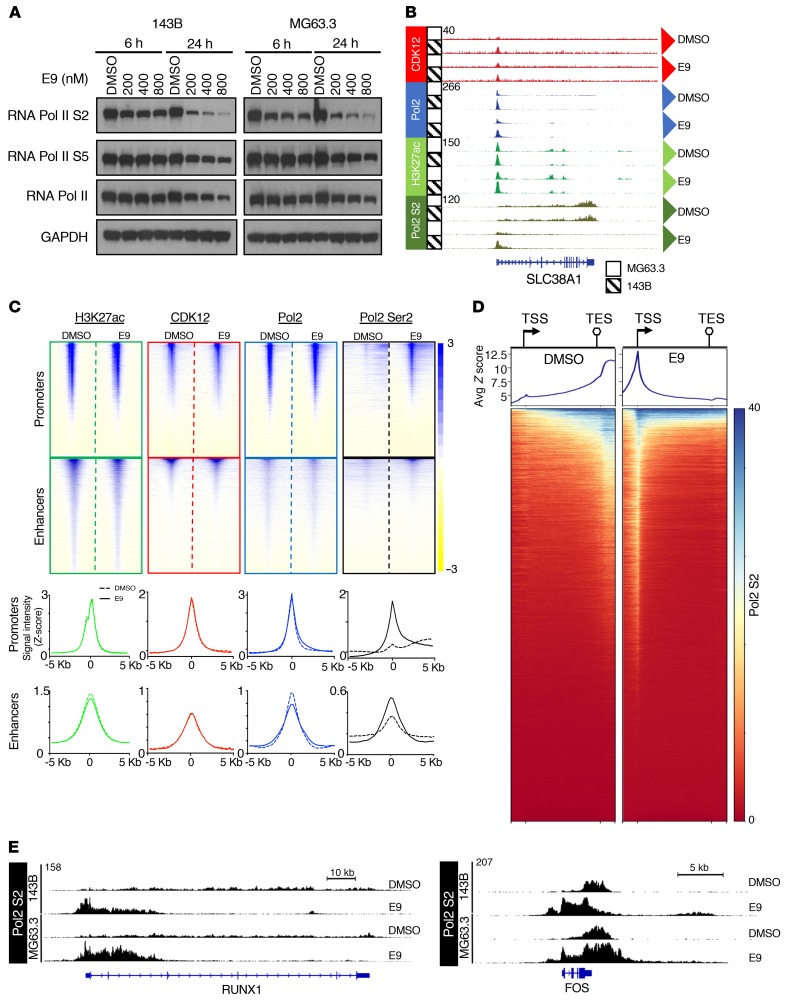
CDK12 phosphorylates serine 2 (S2) within the C-terminal domain (CTD) of RNA polymerase II (Pol II) (39, 40), an essential step in transcription elongation (41). In keeping with this role, E9 led to a dose- and time-dependent decrease in Pol II S2 phosphorylation in 143B and MG63.3 OS cells compared with DMSO-treated cells (Figure 4A). Of note, short-term treatment with E9 (6 hours) had a minimal effect on the transcription initiation-associated serine 5 (S5); however, prolonged treatment (24 hours) resulted in a substantial decrease, likely due to the reduced levels of total RNA Pol II (Figure 4A).
Figure 4. E9 treatment impairs transcription elongation in OS cells.
(A) Western blot analysis of the indicated proteins in 143B and MG63.3 cells treated with DMSO or increasing concentrations of E9 at 6 and 24 hours. GAPDH, loading control. (B) Representative browser views of ChIP-Seq data at the SLC38A1 locus. (C) Windowed heatmaps showing H3K27ac, CDK12, RNA Pol2, and RNA Pol2 Ser2 ChIP-Seq signals ± 5 kb from the TSS in 143B cell line. Below are aggregate plots showing the respective ChIP-Seq signals for the heatmaps depicted above. (D) Metagene analysis of Ser2 ChIP-Seq signal across all genes sorted in descending order based on the average signal per binned region. Windows include 1 kb upstream of the TSS and 1 kb downstream of the TES. (E) Browser views of Ser2 ChIP-Seq signal at RUNX1 and FOS loci in DMSO- and E9-treated OS cells.
Subsequently, to assess the potential effects of CDK12 inhibition on chromatin genome wide, we mapped CDK12, RNA Pol II, and Pol II S2 occupancies using ChIP followed by high-throughput sequencing (ChIP-Seq) analysis in cells treated with either vehicle control (DMSO) or 200 nM of E9 for 6 hours. This dose and time point were chosen to capture the early effects of CDK12 inhibition before the generalized cytotoxicity induced by E9 became apparent. Additionally, because of our prior findings showing enhancer modifications at metastatic OS cells that selectively respond to targeted inhibition (15), we also analyzed the genome-wide chromatin occupancy of H3K27ac, the canonical mark of active enhancers. A representative view of the ChIP-Seq data at an exemplar locus (SLC38A1) is shown in Figure 4B. As expected, in DMSO-treated samples, RNA Pol II bound to nearly all active promoters and, to a lesser extent, enhancers (Figure 4C). Consistent with earlier studies (26), CDK12 signals also coincided with those of RNA Pol II, suggesting that CDK12 primarily binds to actively transcribed genomic regions (Figure 4C and Supplemental Figure 3A).
Upon E9 treatment, we detected only a modest decrease in total Pol II binding compared with that in DMSO-treated cells, with no significant differences in CDK12 or H3K27ac occupancies in either MG63.3 or 143B cells (Figure 4, B and C, and Supplemental Figure 5A). On the other hand, there was a striking increase in Pol II Ser2 binding at promoters and enhancers following E9 treatment (Figure 4, B, C, and E, and Supplemental Figure 5A). To further elucidate the aberrant chromatin binding of phosphorylated Pol II S2 following E9 treatment, we performed a metagene analysis, scaling all genes to the same size and ranking by total Pol II S2 signal (highest to lowest) (Figure 4D). In DMSO-treated cells, Pol II S2 binding was low at the transcription start sites (TSSs) and gradually accumulated to higher levels across gene bodies, peaking at just 3′ of the transcription end site (TES). In contrast, E9 treatment led to a dramatic decrease in Ser2 binding at the 3′ ends of genes, with marked accumulation at the TSS. This lack of Pol II S2 signal across gene bodies and its retention at promoters suggested a block in productive elongation and subsequent early termination. Interestingly, although the vast majority of genes showed a decrease of Ser2 signal in gene bodies, a small number countered this trend with a robust Ser2 signal across the entire gene bodies (see blue at the very top of the heatmaps in Figure 4D and Supplemental Figure 5B). RUNX1 and FOS are 2 exemplary genes that showed these distinct differences in Ser2 occupancy upon E9 treatment (Figure 4E), with RUNX1 losing Ser2 signal in the distal portion of the gene body and FOS gaining Ser2 signal over the entire gene body.
Taken together, these results demonstrate that CDK12 localizes primarily to active promoters and enhancer elements in metastatic OS cells, consistent with findings in Jurkat T cell leukemia cells (26). Moreover, E9 treatment resulted in near-complete elimination of RNA Pol II Ser2 binding from the 3′ ends of most actively transcribed genes, suggesting that E9-induced toxicity in OS cell models originates from an impairment of distal elongation.
CDK12 inhibition affects transcription elongation and RNA processing in OS cells.
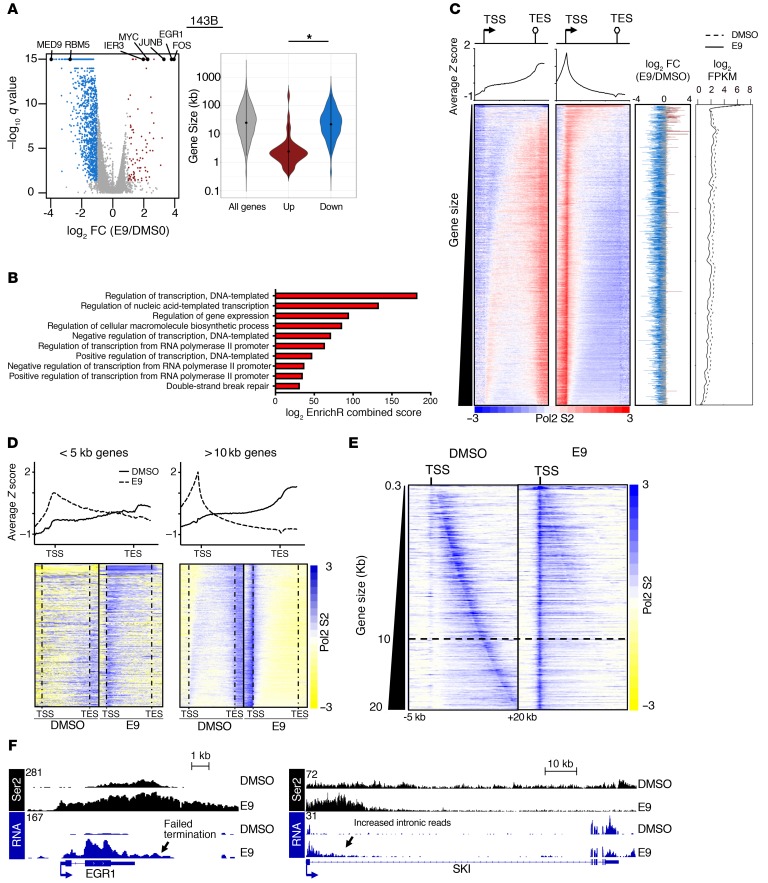
To determine whether the effects observed at the level of chromatin were accompanied by changes in transcript levels, we performed RNA-Seq analysis of E9- and control-treated MG63.3 and 143B OS cells cultured in vitro. In both cell lines, the majority of differentially expressed genes were downregulated upon E9 treatment (Figure 5A and Supplemental Figure 6A). Gene ontology (GO) analyses of downregulated genes revealed significant enrichment of terms related to transcription, biosynthesis, and the DDR (Figure 5B, Supplemental Figure 6B, and Supplemental Table 2).
Figure 5. E9 affects transcript levels in a gene-length– and expression-dependent manner.
(A) Left: volcano plot of RNA-Seq–based expression differences between E9 and DMSO control–treated 143B cells. 1275 Genes showed a decrease of more than 2 fold (q value < 0.05). RNA-Seq for the cell line and condition was performed in triplicate. Right: violin plot of gene sizes for the indicated gene categories in 143B cells. Mann-Whitney U test was used to determine significance. *P < 0.001. (B) GO scores and associated terms from of all genes downregulated more than 2-fold in 143B cells using EnrichR. Terms are ranked based on EnrichR combined scores. (C) Heatmaps of Ser2 ChIP-Seq signal across all expressed genes in DMSO- and E9-treated 143B cells, ranked by gene size. Aggregate plots are shown above. Plotted on the immediate right are corresponding fold changes in transcript levels upon E9 treatment. The line plot on the far right denotes baseline transcript levels and E9-treated transcript levels. All genes are ordered similarly in all plots. (D) Heatmap of Ser2 ChIP-Seq signal in MG63.3 cells in a metagene analysis ± 1 kb of all active genes of the indicated size, ranked by size. Aggregate plots are shown above. (E) Heatmap of Ser2 ChIP-Seq signal in DMSO- and E9-treated 143B cells. Ser2 signals –5 kb and +20 kb of TSSs are shown for all active genes up to 20 kb in length, sorted by increasing gene length. Dark blue signal that runs diagonally from the top to the bottom of the left heatmap corresponds to TESs. (F) Browser view of Ser2 ChIP-Seq and RNA-Seq reads of EGR1 (left) and SKI (right) in E9-and DMSO-treated cells.
Among the upregulated genes, we identified a number of immediate-early genes (IEGs) (65 genes in 143B, 164 in MG63.3 cells), including MYC, FOS, and JUN (Figure 5A). The IEGs were among the highest expressed genes in the transcriptome under normal conditions (Figure 5C and Supplemental Figure 6C). The IEGs were consistently smaller in size (mean length of 3.1 kb) compared with genes downregulated (mean length 25 kb) by E9 treatment (P < 0.001) (Figure 5A and Supplemental Figure 6A). Consistent with their increased expression upon E9 treatment, the TSSs and bodies of short genes (<5 kb) gained high levels of Pol II Ser2 binding, indicative of productive elongation (Figure 5, D and F). Long genes associated with decreased expression due to E9 treatment showed marked loss of Ser2 in gene bodies, indicative of dysfunctional elongation (Figure 5, D and F). Moreover, Ser2 occupancy at short genes in E9-treated cells often extended beyond TESs, suggesting a failure of proper termination (Figure 5, E and F, and Supplemental Figure 6E). Finally, we noted a disproportionately high number of RNA-Seq reads in the first intron of most genes in E9-treated cells, suggesting increased production of short transcripts likely due to early termination (Figure 5F and Supplemental Figure 6F). The concomitant decrease in signal from the first exon suggests that these pre-mRNAs are likely being degraded (Supplemental Figure 6G). Together, these findings demonstrate that CDK12 inhibition with E9 leads to defects in elongation that are dependent on gene length with consequent premature termination, as recently observed in other models (42, 43).
MYC levels correlate with sensitivity to E9 ex vivo.
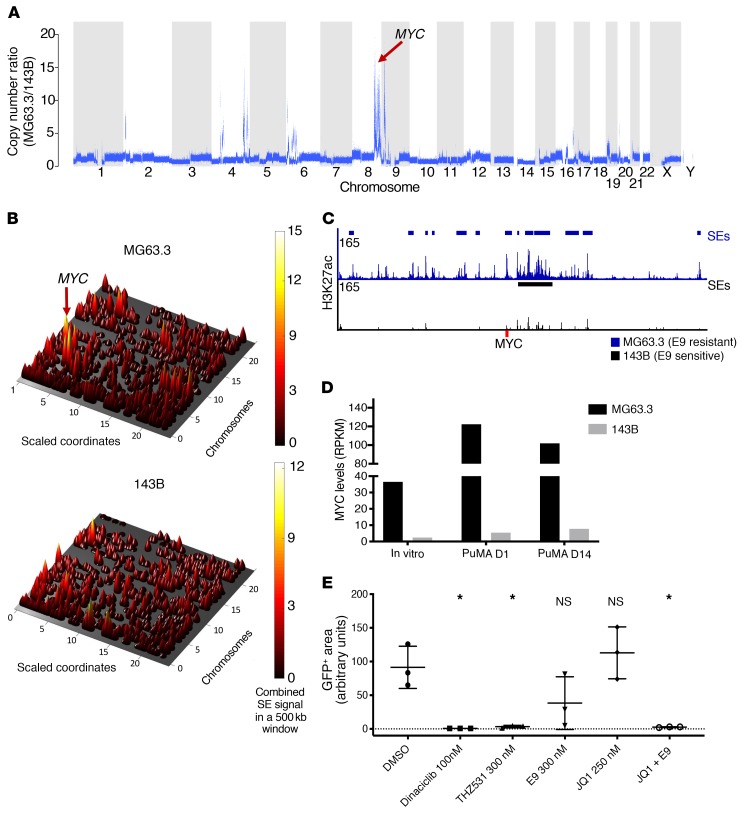
Our studies indicate that OS cell lines cultured in vitro are broadly sensitive to the CDK12 inhibitor E9. However, 2 OS PuMA models (MG63.3 and 143B), while both sensitive to E9 in vitro, differed in their responses in the context of the lung microenvironment, MG63.3 being resistant and 143B sensitive. To delineate the molecular basis of these discrepant phenotypes, we evaluated both the genome and enhancer epigenome in these samples. Whole-genome sequencing revealed that one of the most striking genetic differences between the 2 OS cell lines was amplification of the MYC oncogene: more than 10 times in E9-resistant MG63.3 OS cells compared with the 143B E9-sensitive cells (Figure 6A). ChIP-Seq analysis of enhancer regions using H3K27ac antibodies revealed that the MYC locus had a significantly higher overall enhancer signal in the E9-resistant compared with the E9-sensitive cells, importantly, harboring 12 super-enhancers (SEs) compared with 1 SE in the former (Figure 6, B and C). These combined genetic and epigenetic effects resulted in an 8-fold difference (resistant vs. sensitive) in MYC expression levels in vitro, with a more than 20-fold difference found when the cells were grown in the native lung microenvironment (Figure 6D). To determine whether MYC levels were determinants of ex vivo sensitivity to E9, we combined E9 with the BET inhibitor JQ1, which is known to downregulate MYC-driven transcription programs (44, 45). JQ1 as a single agent had no effect on MG63.3 cells cultured in the PuMA model, consistent with our previous findings of JQ1 treatment on multiple OS lines (15). In contrast, compared with vehicle control–treated MG63.3 cells, those treated with the combination of E9 and JQ1 showed a significant reduction in growth as measured by GFP positivity, comparable to that observed upon treatment with dinaciclib and THZ531 alone (Figure 6E). These results suggest that, in MG63.3 cells, hyperactivation of the MYC-driven oncogenic program due to genetic amplification of MYC coupled with its further epigenetic upregulation likely forms the basis for the increased resistance of these cells to E9 in the PuMA system.
Figure 6. High levels of MYC correlate with insensitivity of OS cells to E9 treatment ex vivo.
(A) CNAs in MG63.3 cells (E9 resistant) relative to 143B cells (E9 sensitive) from whole-genome–sequencing data. (B) Cone plot of H3K27ac SE signals across the epigenomes of MG63.3 and 143B cell lines. (C) Browser views of H3K27ac ChIP-Seq signals at the MYC locus. SE identified in each cell line are indicated. (D) MYC transcript levels in MG63.3 and 143B OS cells cultured in vitro as well as at 1 and 14 days in PuMA model. (E) Relative growth of MG63.3 cells in PuMA lung explants after treatment with the indicated compounds compared with control. Data are presented as mean ± SD of triplicate lung sections. *P < 0.05 versus DMSO control by ordinary 1-way ANOVA with Tukey’s multiple comparison testing.
Discussion
Survival rates for patients with OS have remained stagnant over the past 3 decades, mainly due to the challenge of eradicating metastatic disease and the dearth of targetable mutations in a tumor landscape characterized by extreme genomic instability (8, 46, 47). Here, we provide an improved ex vivo framework for the preclinical screening of potential antimetastatic compounds in the context of the lung microenvironment, an approach that more closely recapitulates metastatic disease than traditional in vitro screening strategies. Through an unbiased screen of compounds that target proteins with broad roles in transcriptional regulation, we identified 2 selective inhibitors of CDK12 — THZ531 and E9 — that potently disrupted metastatic OS cell outgrowth in lung explants. The activity of these agents was not restricted to specific metastatic models, but extended across a broad range of genetically diverse OS cell-line models derived from primary tumors. In addition, when we knocked out CDK12 in vivo using CRISPR/Cas9 to confirm the targets of these inhibitors, we saw a dramatic reduction in the metastatic capacity of the OS cells, confirming that CDK12 appears to be required for successful metastasis.
CDK12 has been shown to regulate the expression of DDR genes and thus facilitate genome stability (36). CDK12 loss-of-function mutations have recently been identified in high-grade serous ovarian carcinoma and metastatic castration-resistant prostate cancer, malignancies with highly unstable genomes (38, 48, 49). In addition, nonmutated CDK12 has been shown to be a vulnerability in EWS/FLI-positive Ewing sarcoma cells, with its inhibition leading to the repression of DDR genes (27). CDK12 does not appear to be mutated in OS (50); therefore, it is very likely that the sensitivity to CDK12 inhibition stems from the remarkable degree of genomic instability that is characteristic of OS. Whether this response is dictated by the high dependence of OS cells on DDR genes for the maintenance of faithful DNA replication needs to be determined.
CDK12-mediated phosphorylation of Ser2 on the CTD of RNA Pol II during transcription facilitates the later stages of elongation (39). Consistent with this model, our analysis of Ser2 phosphorylation as well as chromatin binding, when integrated with analysis of the transcriptome, indicates that the mechanism of E9-induced toxicity in our metastatic OS model is a gene length–dependent impairment of transcription elongation. Moreover, the top-ranked genes affected by E9 treatment encode proteins that function in a number of important biological processes, including transcriptional regulation, oncogenic signaling, and the DDR. OS cells contain multiple genetic and epigenetic alterations that instigate high expression of transcription factors and other key genes that collectively endow fitness and metastatic competence (15). Many of these genes are particularly responsive to CDK12 inhibition by E9, suggesting that the particularly high transcriptional activity of these genes is a general property that may render OS cells selectively vulnerable to CDK12 inhibitors.
Our studies point to defective elongation following CDK12 inhibition with E9, especially affecting longer genes and resulting in aberrant transcriptional termination. Moreover, the increased reads over the first intron suggest that these transcripts could be terminated at alternate polyadenylation sites within the first intron, as recently demonstrated both in embryonic stem cells and cancer cells following CDK12 depletion (42, 43). The reason for the preferential selection for these genes in OS is unclear, but it is highly likely that these longer genes may harbor increased numbers of poly A sites within their proximal introns. This, when coupled with a slowed elongating Pol II, generates the ideal environment for premature termination of Pol II, as has been previously proposed in other models of CDK12 inhibition (42, 43).
It should be stressed that, although the growth inhibitory effects of E9 were comparable across all OS cell lines cultured in vitro, there were pronounced sensitivity differences between OS cells growing in the lung microenvironment. This illustrates the important concept that screening in the relevant microenvironmental context can expose innate sensitivity differences that may not be present under standard culture conditions. Subsequent functional follow-up studies are needed to uncover the basis of these sensitivity differences and to inform the selection of effective drug combinations that could overcome or forestall the emergence of resistance. As demonstrated here, resistance to CDK12 inhibition in the PuMA metastatic disease model can potentially be attributed to genetic and epigenetic mechanisms that together lead to significant (P < 0.001) MYC amplification and overexpression upon OS cell engagement with the lung microenvironment. As we demonstrate, such resistance can be overcome by combining E9 with a bromodomain inhibitor, JQ1. Further validation of this finding would open up the opportunity to regard the genetic or expression status of MYC as a promising biomarker for the prospective identification of patients with metastatic disease who are most likely to respond to CDK12 inhibitors.
Methods
Cell culture
Human OS cell lines (MG63.3-GFP, 143B-GFP, 143B, U2OS, MG63, HOS, SaOS2, SJSA1, G292, and MNNG) were grown in 1× DMEM (Gibco, Thermo Fisher Scientific; 11965-092) supplemented with 10% FBS (Corning, 35-010-CV) and 1% penicillin/streptomycin (Gibco, Thermo Fisher Scientific: 15140-122). All cell lines were routinely tested for mycoplasma. MG63.3-GFP and 143B-GFP were from the lab of Chand Khanna (NIH, Bethesda, Maryland, USA) (19). 143B, U2OS, MG63, HOS, SaOS2, SJSA1, G292, and MNNG were provided by Stuart Orkin (Dana-Farber Cancer Institute).
Mice
Fox Chase SCID Beige mice used in experiments were purchased from Charles River for 2 to 4 weeks prior to experiments and were housed in ultraclean immunocompromised athymic and xerograph core rooms. The number of mice included in each of the described studies was based on extensive past experience in the development and use of murine models of metastases by our group. Each study was designed to minimize unnecessary mouse use, optimize statistical power, and account for known variance in each model system. Within each experiment, mice of the same strain, sex, and age were used for all conditions. Researchers were not blinded to the group assignments of mice.
High-throughput ex vivo PuMA
The method was adapted from the PuMA assay as previously described (16). GFP+ human OS cells (1 × 106) were injected into the tail vein of 10- to 12-week-old female Fox Chase SCID Beige mice (Charles River, strain code 250). Within 20 minutes of injection, mice were euthanized via CO2 inhalation, and the lungs were insufflated with a mixed agarose-media solution through injection via the trachea. The lungs were then allowed to cool on ice for 25 minutes in sterile PBS solution. Once the agarose solidified, transverse serial sections were taken from each lobe of the lung with scissors, yielding between 16 and 20 lung slices per set of lungs. Lung sections were then sliced in half again, horizontally. Sections were arrayed in 96-well plates so that each row of sections was from a single mouse with the rightmost 2 columns being reserved for DMSO control sections. Compounds were arrayed in 2-well pairs going across the row, with the high dose of the compound first and then the low dose second. Single sections were placed on top of B media–soaked Gelfoam (Pfizer, catalog 00300090315085) slices (presoaked overnight at 37°C) that had been placed into the upper compartment of Transwell plates (Corning, CLS3381). The bottom reservoir compartments of the Transwell plates were then filled with fresh media and compound that freely flowed through the permeable membrane and into the Gelfoam. The sections were then cultured for 14 days at 37°C with 5% CO2, with the media being changed every day. After that time, the sections were removed from the Gelfoam and placed into black, clear-bottom plates (VWR 29444-008) for imaging. To quantify metastatic burden in lung sections, fluorescent images were acquired using Operetta High Content Imaging System and Harmony Imaging Software (PerkinElmer) and total GFP+ area for each section was quantified using Acapella Image Analysis software (PerkinElmer). Quantification of efficacy for each compound was measured by dividing the quantified GFP+ area for each section by the average GFP+ area for the 2 respective control sections and then transformed into a percentage.
In vitro screen
GFP+ MG63.3 cells (1 × 103) were plated per well in black clear-bottom plates (Corning, 3603). Compounds were added to the media after the cells adhered overnight. Cells were cultured for 7 days with fresh media and compounds added every day. To assess compound potency, fluorescent images were acquired using Operetta High Content Imaging System and Harmony Imaging Software (PerkinElmer) and total GFP+ area for each well was quantified using Acapella Image Analysis software (PerkinElmer). Quantification of efficacy for each compound was measured by dividing the quantified GFP+ area for each well by the average GFP+ area for DMSO-treated control wells and then transformed into a percentage.
Compounds
Compounds were obtained from the Selleck Chemicals Bioactive Compounds Library (catalogL1700). Of these compounds, approximately 300 were selected based on their declared specificity to target the epigenome. From this list, 112 were chosen based off of previously published high potency and efficacy toward their target. These compounds were diluted to the working concentrations for the screen listed in Supplemental Table 1, which also contains a full list of the compounds used. The Structural Genomics Consortium (University of Toronto, Toronto, Ontario, Canada) also provided their Epigenetics Probes Collection, of which 33 compounds were used, all of which have the concentrations used listed on Supplemental Table 1. Compounds were divided into 3 dosage groups based on potency of enzymatic IC50 toward target listed on the Selleck Chemical website. E9 and THZ531 were obtained from Nathanael Gray (Dana-Farber Cancer Institute). Tariquidar (catalog S8028) was purchased from Selleck Chemicals.
Inducible cell-line generation
MG63.3-GFP cells were transduced to stably express doxycycline-inducible FLAG-tagged Cas9. The pCW-Cas9 vector used was a gift from Eric Lander (Broad Institue, Boston, Massachusetts, USA) and David Sabatini (Whitehead Institute, Boston, Massachusetts, USA) (Addgene plasmid 50661). After puromycin selection (2 μg/mL), the cells were single cell subcloned to generate a clonal population with homogeneously high Cas9 expression upon induction. These cells are referred to as MG63.3-GFP-Cas9i throughout. Single-guide RNAs targeting CDK12 (4 sgRNAs) and nontarget regions (2 sgRNAs) were taken from the genome-wide BRUNELLO library (51). The sgRNAs were cloned into pLV-U6-gRNA-UbC-DsRed-P2A-Bsr, which was a gift from Charles Gersbach (Duke University, Durham, North Carolina, USA) (Addgene plasmid 83919). Lentivirus for each sgRNA was produced using the LentiX Packaging Single Shots (Clontech, 631278) according to the manufacturer’s protocol and was used to transduce MG63.3-GFP-Cas9i cells. After selection with blasticidin (5 μg/mL), cells were split and cultured in the presence or absence of 1 μg/mL doxycycline (Cayman Chemical, 14422). After 7 days, whole-cell lysate was collected from each condition and used for in vitro characterization of CDK12-knockout efficiency via Western blot.
sgRNA sequences
sgRNA sequences were as follows: CDK12-1, forward, KO: CACCGACTGACCGACTGCCTTCTCG; CDK12-1, reverse, KO: AAACCGAGAAGGCAGTCGGTCAGTC; CDK12-2, forward, KO: CACCGATTCACCAGTTCAGTATCTG; CDK12-2, reverse, KO: AAACCAGATACTGAACTGGTGAATC; CDK12-3, forward, KO: CACCGCTAGCAGTCCCATTAAGTCA; CDK12-3, reverse, KO: AAACTGACTTAATGGGACTGCTAGC; CDK12-4, forward, KO: CACCGTGGCCTTCAAACTAGACCGA; CDK12-4, reverse, KO: AAACTCGGTCTAGTTTGAAGGCCAC; nontargeting control-2, forward, KO: CACCGAAAACAGGACGATGTGCGGC; nontargeting control-2, reverse, KO: AAACGCCGCACATCGTCCTGTTTTC; nontargeting control-1, forward, KO: CACCGAAAAAGCTTCCGCCTGATGG; and nontargeting control-1, reverse, KO: AAACCCATCAGGCGGAAGCTTTTTC.
In vivo metastasis experiment
For the in vivo CDK12 knockout experiment, 10- to 15-week-old female SCID Beige mice (Charles River, strain code 250) were used. Mice were placed on water containing 2 mg/mL doxycycline (Cayman Chemical, 14422) for 5 days prior to injection of the cells, and administration was continued for the duration of the experiment. Pretreatment with doxycycline was done in order to prime the metastatic microenvironment for CDK12 knockout in the xenografted cells. Cells (1 × 106) were seeded into the lungs of mice through lateral tail-vein injection, with 3 mice used for each of the 6 total cell lines. The cells were not exposed to doxycycline prior to injection. After 21 days, the mice were euthanized by CO2 inhalation. Their lungs were harvested and imaged by inverted fluorescent microscopy (Leica DM IRB) at × 2.5 magnification. A total of 4 to 5 images were taken per lung to encompass the whole lung area. Each image was quantified for GFP+ area using ImageJ (NIH).
Quantification of metastatic OS cell growth in standard PuMA
Lung sections were imaged on a standard fluorescent microscope system as previously described (15). Lung sections were imaged by inverted fluorescent microscopy (Leica DM IRB) at a magnification of ×2.5. One to 3 images per lung section were taken. Image analysis was performed using ImageJ software to quantify the total GFP+ area per lung section. The metastatic burden was calculated by normalizing the total GFP+ area at the experimental end point to the GFP+ area for each section on the first day of the experiment.
Cell-viability assay and drug combination analysis
For cell-viability assays with single-agent treatment, cells were plated in 96-well plates in triplicates at a seeding density of 2 × 103 cells/well. After 24 hours, the cells were treated with increasing concentrations of the drug, ranging from 10 nM to 10 μM, with DMSO solvent as a control. After 72 hours of incubation, cells were analyzed for cell viability using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, G7570) according to the manufacturer’s protocol. IC50 concentrations were determined using GraphPad Prism 6 nonlinear regression curve fit.
Western blotting
Cells were lysed at 4°C in NP40 buffer (Invitrogen, FNN0021) or RIPA buffer supplemented with protease inhibitors (Roche, 4693159001), phosphatase inhibitors (Roche, P2850), and 1 mM PMSF (Roche, 10837091001). Protein concentrations were measured using the Bio-Rad DC protein assay kit (Bio-Rad, 5000112), and 50 μg of total protein was resolved on precast 4%–12% Bis-Tris gels (Invitrogen, NP0321BOX) and transferred to nitrocellulose membranes (Bio-Rad, 1620113). For CDK12-knockout Western blots, 10 μg of protein was used. The membranes were blocked with 5% dry milk in TBS supplemented with 0.2% Tween-20 (TBS-T) at room temperature for 1 hour and then incubated overnight at 4°C with the following primary antibodies: RNAPII CTD S2 (Bethyl, catalog A300-654A), RNAPII CTD S5 (Bethyl, catalog A300-655A), RNAPII (Santa Cruz Biotechnology Inc., catalog sc-899), cleaved PARP1 (Cell Signaling Technology, catalog 9541), GAPDH (Cell Signaling Technology, catalog 2118S), CDK12 (Cell Signaling Technology, catalog 11973S), cyclophilin B (abcam, catalog ab16045), ABCB1 (Cell Signaling Technology, catalog 12683S), and ABCG2 (Cell Signaling Technology, catalog 4477S). Chemiluminescent detection was performed with the HRP-conjugated secondary antibodies (Santa Cruz Biotechnology Inc., catalog sc-2004; Cell Signaling Technology, catalog 7076S) and developed using Genemate Blue Ultra-Autoradiography film (VWR, 490001-930) or Bio-Rad ChemiDoc Touch Imaging System. Densitometry quantification was performed using Image J. See complete unedited blots in the supplemental material.
Target engagement assay
The biotin-THZ1 (bio-THZ1) pulldown experiment was performed as described previously (24). Briefly, cells were treated with THZ531/E9 or DMSO for 6 hours at the indicated doses. Total cell lysates were prepared as for Western blotting. To pull down CDK12, 1 mg of total protein was incubated with 1 μM bio-THZ1 at 4°C overnight. Subsequently, lysates were incubated with streptavidin agarose (30 μl) for 2 hours at 4°C. Agarose beads were washed 3× with cell lysis buffer and boiled for 10 minutes in 2× gel loading buffer. Proteins were resolved by Western blotting.
Colony-formation assay
Osteosarcoma cells were treated with the indicated concentrations of E9 or DMSO control for 48 hours and then seeded in 6-well plates at clonal density. Cells were incubated for approximately 12 days until the colonies contained a minimum of 50 cells. Colonies were subsequently fixed in methanol, stained with crystal violet (Sigma-Aldrich, C3886) for 30 minutes and air-dried. Clonogenic survival was assessed as percentage of surviving colonies compared with nontreated control.
FACS
For cell-cycle and DNA damage analysis, cells were treated with DMSO or 400 nM of E9. After 24 or 48 hours, cells were trypsinized and fixed in ice-cold 70% ethanol overnight at –20°C. After washing with ice-cold PBS, the cells were incubated in PBS containing 0.5% Tween-20 with γ-H2AX antibody (Cell Signaling Technology, 9718) overnight at 4°C. Cells were subsequently washed in and incubated with Alexa Fluor 488–conjugated secondary antibody (Life Technologies, A11008) for 45 minutes at room temperature and then treated with 0.5 mg/ml RNAse A (Sigma-Aldrich, R6513) in combination with 50 μg/ml propidium iodide (BD Biosciences, 556463). All FACS samples were analyzed on a FACSCalibur flow cytometer (BD Biosciences) using CellQuest software (BD Biosciences). A minimum of 50,000 events was counted per sample and used for further analysis. Data were analyzed using FlowJo software (Tree Star).
Tissue fixing and staining for H&E
Tissue fixing and staining were performed by Case Western Reserve University Histology Services. Lung sections were fixed in 10% formalin overnight before being dehydrated in 80% ethanol. Sections were sliced via microtome and stained for H&E using the following protocol. Slide-mounted sections were placed in Harris Modified Hematoxylin for 15 minutes (Fisher, Sh26-4D). Slides were then moved to tap water and washed 15 times. They were then placed in 1% acid alcohol and washed 8 times. Slides were saturated in lithium carbonate and dipped 5 times. Slides were then washed 15 times in fresh water before a 2-minute wash with 70% ethanol and a 2-minute wash in 95%; they were then placed in eosin for 3 minutes (Fisher, E-511). Slides were then washed in 5 rounds of 100% ethanol for 2 minutes each before being placed in Xylenes for 2 minutes. Slides were then placed in a final round of Xylenes for 2 more minutes before being covered and allowed to dry overnight.
ChIP-Seq
143B-GFP and MG63.3-GFP cells were treated for 6 hours with E9 at 200 nM or DMSO before being crosslinked with formalin. ChIPs were performed using 1 × 107 (H3K27ac) and 4 × 107 (CDK12, RNA Pol2, RNA Pol2 Ser2) crosslinked cells, and sequencing libraries were prepared as previously described (52). The following antibodies were used for ChIP: rabbit anti-H3K27ac (Abcam, 4729), anti-CDK12 (rabbit affinity-purified IgGs directed against a peptide comprising amino acids 201–220 of human CDK12), a gift from Arno Greenleaf (Duke University), RNA Go-ChIP-Grade Purified anti-RNA Polymerase II RPB1 Antibody (BioLegend, 664911), and anti-RNA Pol II subunit B1 (phospho CTD Ser-2) antibody, clone 3E10 (Millipore, catalog 04-1571). ChIP–Seq libraries were sequenced on the 2500 platform at the Case Western Reserve University Genomics Core Facility.
RNA-Seq
Cells were treated with DMSO or 200 nM of E9 for 6 hours. Each line and condition was done in triplicate. RNA extraction was performed with TRIzol (Invitrogen, 15596026) following the manufacturer’s instructions. Total RNA was treated with DNase I (Invitrogen, 18047019). RNA was spiked-in with ERCC RNA Spike-In Mix (Invitrogen, 4456740) and analyzed on an Agilent Bioanalyzer 2100 (G2939BA) for integrity. PolyA+ RNA was isolated using the Illumina TruSeq RNA Sample Preparation Kit (Illumina, RS-122-2001) according to the manufacturer’s protocol. All samples were sequenced on a Novaseq 6000 sequencer at University of Colorado Cancer Center Genomics and Microarray Core (Aurora, Colorado, USA).
Whole-genome sequencing
PCR-free libraries were prepared from MG63.3-GFP and 143B-GFP cells using TruSeq DNA PCR-Free High-Throughput Library Prep Kit (Illumina, 20015963) and following the manufacturer’s instructions. Libraries were pooled and run on the HiSeq 2500 at the Case Western Reserve University Genomics Core to a depth of approximately ×20 coverage.
Data analysis
ChIP-Seq.
The FASTX-Toolkit, version 0.0.13, was used to remove adapter sequences and trim reads with the quality cutoff of 20, excluding any reads shorter than 25 bp. ChIP-Seq reads were aligned to hg19 with Bowtie 2, version 2.0.649, searching for multiple alignments, but reporting the one with the best mapping quality score. PCR duplicates were removed using SAMtools 1.2. Peaks were detected with MACS2 2.1.1.20160309 with an input DNA sample as control, scaling the input data set to match the read depth of the ChIP data set. MACS2 peaks with a q value of greater than 0.001 and overlapping ENCODE blacklisted regions (https://www.encodeproject.org/annotations/ENCSR636HFF/) were filtered out. BigWIGs were generated with DeepTools, version 3.0.1-2-2fa58e8, and normalized to 1× average coverage (RPGC option).
RNA-Seq.
Cutadapt, version 1.14, was used to trim 3′ adapters, discarding trimmed reads shorter than 20 bp. Sickle, version 1.33, was then used to trim reads with the quality cutoff of 20. RNA-Seq reads were aligned to hg19 assembly plus ERCC RNA Spike-In DNA sequences with STAR, version 2.5.3a. Cufflinks, version 2.0.2, was run with genomic bias correction to quantify the levels of transcripts in the hg19 reFLAT annotations file as well as the ERCC transcripts file. Scatter plots of ERCC transcript fragments per kilobase of transcript per million mapped reads (FPKMs) versus their expected concentrations in the spike-in mix were constructed to aid in quality control assessment of the data. The FPKM table was then floored to the background noise level of 0.3. Differential expression analysis between E9-treated and untreated samples was performed by calculating the fold change of mean E9 FPKM over mean DMSO FPKM.
Whole-genome sequencing.
After removing adapter sequences using cutadapt, version 1.8.1, the DNA FASTQ files were aligned to the hg19 reference genome using BWA-mem (version 0.7.17-r1188). The aligned files were then indexed and had PCR duplicates removed using samtools (version 0.1.18). Each BAM file underwent base quality score recalibration using GATK (version 4.0.1.1).
Duplicate gene filtering.
Refseq hg19 genes underwent the following filtering steps prior to use in downstream analyses for each pair of treated and untreated samples to ensure 1-to-1 mapping of genes to TSSs: (a) retrieve RNA-Pol RPKM within 1 kb of each TSS and calculate the minimum of E9 and DMSO RPKMs. In the case of the same gene with multiple TSSs, only retain the gene record with the max RNA-Pol RPKM minimum; (b) in the case of multiple genes with the same TSS, calculate the minimum FPKM between E9 and DSMO for each gene and only retain the gene with the max FPKM minimum.
Metagene analysis.
Duplicate-filtered Refseq hg19 genes were split into positive- and negative-strand genes. Deeptools’ computeMatrix tool was used to generate a normalized, binned matrix of Pol II Ser2 read counts for each gene, covering the region between the TSSs and TESs, along with 1-kb flanking regions on each side. Each gene region was scaled to 5 kb with 50 bp windows used for each bin. The reverse-strand gene matrix was reverse ordered to match the orientation of the positive-strand matrix; the 2 tables were combined and plotted using Deeptools’ plotHeatmap function.
ChIP-Seq heatmap generation.bamToGFF_turbo.py from Bradner Lab (https://github.com/BradnerLab/pipeline) was run on each data set to generate a table of binned, normalized counts covering ± 5 kb of the TSSs. Each region was put into 40 bins with no read extension. Normalized counts were then z scored separately for each data set. The reverse-strand gene matrix was reverse ordered to match the orientation of the positive-strand matrix and both tables were combined, ranked on the middle 4 RNA Pol2 columns in DMSO, and plotted in Java Treeview. Aggregate tracks generated from heatmaps were made by taking the average of all genes within a bin and plotting these across all bins. Plots were generated in PRISM.
Volcano plots.
Cufffdiff, version 2.2.1, was run on E9-treated versus DMSO-treated samples to identify differentially expressed genes with an FDR-adjusted P value (q value) of 0.05 or less. FPKMs were floored to 0.3. log2(E9/DMSO FPKMs) versus –log10(q value) and were plotted for any genes with FPKM of greater than 0.3 in at least 1 sample. Any genes with a q value of less than 0.05 and E9/DMSO FPKMs of greater than 2 were marked in red. Any genes with a q value of less than 0.05 and E9/DMSO FPKMs of less than 0.5 were marked in blue.
Violin plots.
Duplicate filtered gene lists were further filtered to only include expressed genes or genes with FPKM of more than 1 in either DMSO-treated or E9-treated samples. Expressed genes were then narrowed down to only include dysregulated genes (E9/DMSO FPKM > 2 or E9/DMSO FPKM < 0.5). Gene sizes for each of the 3 gene groups (all expressed, upregulated, and downregulated) were plotted as violin plots. Mann-Whitney U test was used to determine significance with P < 0.001.
Gene-size ranked heatmaps.
Duplicate filtered gene lists were further filtered to only include genes with FPKM of greater than 0.3 in DMSO-treated samples and split into positive and negative strand genes. Deeptools’ computeMatrix tool was used to generate a normalized, binned matrix of Pol II Ser2 read counts for each gene, covering the region between the TSSs and TESs, along with 1 kb flanking regions on each side. Each gene region was scaled to 5 kb with 50 bp windows used for each bin. The reverse-strand gene matrix was reverse ordered to match the orientation of the positive-strand matrix and the 2 tables were combined, ranked by gene size in ascending order, and plotted as a heatmap using Java TreeView. Baseline expression (FPKM) as well as log2(E9/DMSO FPKM) were plotted for each gene in the same order as the heatmaps.
GO.
GO enrichment for selected gene sets was performed using EnrichR (53). The EnrichR score is the combined score of the adjusted P value and the z score using Fisher’s exact test.
GSEA.
Gene set enrichment analysis (GSEA) (54) was performed on downregulated genes (>1.5 fold, q < 0.01, from volcano plot) in both the 143B and MG63.3 lines after E9 treatment against the C5 GO gene sets. The top 100 terms for each line are listed in Supplemental Table 2.
Intronic and exonic read comparison.
The first exons and first introns of Refseq genes with DMSO FPKMs greater than 0.3 were retrieved, correcting for strand/transcriptional direction. RNA-Seq RPKMs were calculated in each region, averaged across the 3 RNA-Seq replicates in each group (DMSO and E9) and the averages displayed as boxplots. Mann-Whitney U test was used to compare the E9 and DMSO distributions.
Copy number analysis.
For copy number alteration (CNA) calling, a hg19 interval list using a bin size of 1000 bp was created. GATK was used to call differences in CNAs between MG63.3 and 143B by normalizing the copy number read counts to 143B. All GATK analyses were done in accordance with the GATK best practices.
Statistics
Numerical values in statistically analyzed data are reported as mean ± SD unless otherwise stated. Statistical significance for the ANOVA analysis shown in Figure 2, Figure 6, and Supplemental Figure 1 was determined using ordinary 1-way ANOVA with Tukey’s multiple comparison tests. Statistical significance for the ANOVAs shown in Figure 3 and Supplemental Figure 4 was determined with 1-way ANOVA with Dunnett’s multiple-comparison test correction. Statistical significance for the analysis is shown Figure 5 and Supplemental Figure 6 was determined by Mann-Whitney U test. Statistical analysis was performed with PRISM software (ANOVA) and R (Mann-Whitney U).
Study approval
The animal experiments included in this study were approved by the Case Western Reserve University Institutional Animal Care and Use Committees under protocol 2014-0156.
Data availability
Data sets (RNA-Seq, ChIP-Seq, whole-genome sequencing data) generated and analyzed during this study are included within the published manuscript and were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE132233). RNA-Seq data used in Supplemental Figure 2 were also deposited in that database (GEO GSE74230). The patient data shown in Supplemental Figure 2 are available via The Human Cancer Metastasis Database (HCMDB ID EXP00023, EXP00083; http://hcmdb.i-sanger.com) (55).
Author contributions
This study started with the development of PuMA for high-throughput screening in the Scacheri lab; therefore, all parties agreed that IB would be listed first and PCS would be last. IB, JJM, PCS, MK, REG, and DJA conceived the project. IB, AS, CB, JJM, MK, JL, WDP, ESH, BR, and JMK analyzed data. DJA and YF provided resources. IB, PCS, MK, REG, ZJF, JJM, JMK, and WDP wrote the manuscript.
Supplementary Material
Acknowledgments
The authors acknowledge the Structural Genomics Consortium (SGC) for the epigenetic probes collection used in this study. The authors would like to acknowledge Nathanael Gray for providing the THZ531 and E9 compounds. REG was funded by a Team Path to the Cure grant, and MK was funded by a Rally Foundation grant. The authors would like to thank Richard Sallari for his graphic design support, the QuadW foundation for their financial support, and Dan Weidenthal for his contributions. The authors are also grateful for support from the Case Comprehensive Cancer Center Angie Fowler AYA Cancer Research Fund. Finally, the authors would like to thank Fredrick Schumacher for providing biostatistical insight for the paper. This work was supported by grants RO1 CA193677, RO1 CA204279, RO1 CA160356 (to PCS), RO1 CA197336 (to REG), and F31 CA213965 (to IB) from the National Cancer Institute
Version 1. 09/09/2019
Electronic publication
Version 2. 10/01/2019
Print issue publication
Footnotes
Conflict of interest: REG is a member of the scientific advisory board of the Global Gene Corporation.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(10):4377–4392.https://doi.org/10.1172/JCI127718.
Contributor Information
Ian Bayles, Email: imb10@case.edu.
Malgorzata Krajewska, Email: Malgorzata_Krajewska@dfci.harvard.edu.
W. Dean Pontius, Email: wdp14@case.edu.
Alina Saiakhova, Email: ars51@case.edu.
James J. Morrow, Email: James.J.Morrow@case.edu.
Jim Lu, Email: jimlu@college.harvard.edu.
Zachary J. Faber, Email: zachary.faber@case.edu.
Yuriy Fedorov, Email: yxf204@case.edu.
Ellen S. Hong, Email: esh81@case.edu.
Jaret M. Karnuta, Email: jmk291@case.edu.
Drew J. Adams, Email: dja59@case.edu.
Rani E. George, Email: rani_george@dfci.harvard.edu.
References
- 1.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perkins SM, Shinohara ET, DeWees T, Frangoul H. Outcome for children with metastatic solid tumors over the last four decades. PLoS ONE. 2014;9(7):e100396. doi: 10.1371/journal.pone.0100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyers PA, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23(9):2004–2011. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 5.Meyers PA, et al. Osteogenic sarcoma with clinically detectable metastasis at initial presentation. J Clin Oncol. 1993;11(3):449–453. doi: 10.1200/JCO.1993.11.3.449. [DOI] [PubMed] [Google Scholar]
- 6.Kager L, et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J Clin Oncol. 2003;21(10):2011–2018. doi: 10.1200/JCO.2003.08.132. [DOI] [PubMed] [Google Scholar]
- 7.Marina NM, et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): an open-label, international, randomised controlled trial. Lancet Oncol. 2016;17(10):1396–1408. doi: 10.1016/S1470-2045(16)30214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7(1):104–112. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perry JA, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–E5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriarity BS, et al. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupte A, et al. Systematic screening identifies dual PI3K and mTOR inhibition as a conserved therapeutic vulnerability in osteosarcoma. Clin Cancer Res. 2015;21(14):3216–3229. doi: 10.1158/1078-0432.CCR-14-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheel C, et al. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene. 2001;20(29):3835–3844. doi: 10.1038/sj.onc.1204493. [DOI] [PubMed] [Google Scholar]
- 14.Hingorani P, et al. Current state of pediatric sarcoma biology and opportunities for future discovery: A report from the sarcoma translational research workshop. Cancer Genet. 2016;209(5):182–194. doi: 10.1016/j.cancergen.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrow JJ, et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med. 2018;24(2):176–185. doi: 10.1038/nm.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mendoza A, et al. Modeling metastasis biology and therapy in real time in the mouse lung. J Clin Invest. 2010;120(8):2979–2988. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, et al. Overcoming Resistance to the THZ Series of Covalent Transcriptional CDK Inhibitors. Cell Chem Biol. 2018;25(2):135–142.e5. doi: 10.1016/j.chembiol.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Y, et al. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis. 2009;26(7):599–610. doi: 10.1007/s10585-009-9259-6. [DOI] [PubMed] [Google Scholar]
- 19.Ren L, et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 2015;6(30):29469–29481. doi: 10.18632/oncotarget.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9(8):2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SF, et al. CDK12 inhibition reverses de novo and acquired PARP inhibitor resistance in BRCA wild-type and mutated models of triple-negative breast cancer. Cell Rep. 2016;17(9):2367–2381. doi: 10.1016/j.celrep.2016.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natoni A, et al. Mechanisms of action of a dual Cdc7/Cdk9 kinase inhibitor against quiescent and proliferating CLL cells. Mol Cancer Ther. 2011;10(9):1624–1634. doi: 10.1158/1535-7163.MCT-10-1119. [DOI] [PubMed] [Google Scholar]
- 23.Byth KF, et al. AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther. 2009;8(7):1856–1866. doi: 10.1158/1535-7163.MCT-08-0836. [DOI] [PubMed] [Google Scholar]
- 24.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511(7511):616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159(5):1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat Chem Biol. 2016;12(10):876–884. doi: 10.1038/nchembio.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iniguez AB, et al. EWS/FLI confers tumor cell synthetic lethality to CDK12 inhibition in Ewing sarcoma. Cancer Cell. 2018;33(2):202–216.e6. doi: 10.1016/j.ccell.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali S, et al. The development of a selective cyclin-dependent kinase inhibitor that shows antitumor activity. Cancer Res. 2009;69(15):6208–6215. doi: 10.1158/0008-5472.CAN-09-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorlick R, et al. Initial testing (stage 1) of the cyclin dependent kinase inhibitor SCH 727965 (dinaciclib) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2012;59(7):1266–1274. doi: 10.1002/pbc.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodey B, Taylor CR, Siegel SE, Kaiser HE. Immunocytochemical observation of multidrug resistance (MDR) p170 glycoprotein expression in human osteosarcoma cells. The clinical significance of MDR protein overexpression. Anticancer Res. 1995;15(6B):2461–2468. [PubMed] [Google Scholar]
- 31.Chano T, et al. Differentially expressed genes in multidrug resistant variants of U-2 OS human osteosarcoma cells. Oncol Rep. 2004;11(6):1257–1263. [PubMed] [Google Scholar]
- 32.Suto R, et al. Multidrug resistance mediated by overexpression of P-glycoprotein in human osteosarcoma in vivo. Int J Oncol. 1998;12(2):287–291. [PubMed] [Google Scholar]
- 33.Okada T, et al. Involvement of P-glycoprotein and MRP1 in resistance to cyclic tetrapeptide subfamily of histone deacetylase inhibitors in the drug-resistant osteosarcoma and Ewing’s sarcoma cells. Int J Cancer. 2006;118(1):90–97. doi: 10.1002/ijc.21297. [DOI] [PubMed] [Google Scholar]
- 34.Susa M, et al. Inhibition of ABCB1 (MDR1) expression by an siRNA nanoparticulate delivery system to overcome drug resistance in osteosarcoma. PLoS ONE. 2010;5(5):e10764. doi: 10.1371/journal.pone.0010764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin C, Berridge G, Mistry P, Higgins C, Charlton P, Callaghan R. The molecular interaction of the high affinity reversal agent XR9576 with P-glycoprotein. Br J Pharmacol. 1999;128(2):403–411. doi: 10.1038/sj.bjp.0702807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartkowiak B, Greenleaf AL. Expression, purification, and identification of associated proteins of the full-length hCDK12/CyclinK complex. J Biol Chem. 2015;290(3):1786–1795. doi: 10.1074/jbc.M114.612226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajrami I, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74(1):287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang K, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol Cell Biol. 2015;35(6):928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krajewska M, et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat Commun. 2019;10(1):1757. doi: 10.1038/s41467-019-09703-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dubbury SJ, Boutz PL, Sharp PA. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature. 2018;564(7734):141–145. doi: 10.1038/s41586-018-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muff R, et al. Genomic instability of osteosarcoma cell lines in culture: impact on the prediction of metastasis relevant genes. PLoS ONE. 2015;10(5):e0125611. doi: 10.1371/journal.pone.0125611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovac M, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun. 2015;6:8940. doi: 10.1038/ncomms9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem. 2014;289(13):9247–9253. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu F, et al. SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 sumoylation. Cell Res. 2018;28(6):670–685. doi: 10.1038/s41422-018-0023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Behjati S, et al. Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nat Commun. 2017;8:15936. doi: 10.1038/ncomms15936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanson KR, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9(1):5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt D, et al. Chip-Seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48(3):240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang X, Cairns MJ. Gene set enrichment analysis of RNA-Seq data: integrating differential expression and splicing. BMC Bioinformatics. 2013;14 Suppl 5:S16. doi: 10.1186/1471-2105-14-S5-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng G, Ma Y, Zou Y, Yin A, Li W, Dong D. HCMDB: the human cancer metastasis database. Nucleic Acids Res. 2018;46(D1):D950–D955. doi: 10.1093/nar/gkx1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets (RNA-Seq, ChIP-Seq, whole-genome sequencing data) generated and analyzed during this study are included within the published manuscript and were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE132233). RNA-Seq data used in Supplemental Figure 2 were also deposited in that database (GEO GSE74230). The patient data shown in Supplemental Figure 2 are available via The Human Cancer Metastasis Database (HCMDB ID EXP00023, EXP00083; http://hcmdb.i-sanger.com) (55).