Abstract
Context
Adipose tissue inflammation and dysregulated energy homeostasis are key mechanisms linking obesity and cancer. Distinct adipose tissue depots strongly differ in their metabolic profiles; however, comprehensive studies of depot-specific perturbations among patients with cancer are lacking.
Objective
We compared transcriptome profiles of visceral adipose tissue (VAT) and subcutaneous adipose tissue (SAT) from patients with colorectal cancer and assessed the associations of different anthropometric measures with depot-specific gene expression.
Design
Whole transcriptomes of VAT and SAT were measured in 233 patients from the ColoCare Study, and visceral and subcutaneous fat area were quantified via CT.
Results
VAT compared with SAT showed elevated gene expression of cytokines, cell adhesion molecules, and key regulators of metabolic homeostasis. Increased fat area was associated with downregulated lipid and small molecule metabolism and upregulated inflammatory pathways in both compartments. Comparing these patterns between depots proved specific and more pronounced gene expression alterations in SAT and identified unique associations of integrins and lipid metabolism–related enzymes. VAT gene expression patterns that were associated with visceral fat area poorly overlapped with patterns associated with self-reported body mass index (BMI). However, subcutaneous fat area and BMI showed similar associations with SAT gene expression.
Conclusions
This large-scale human study demonstrates pronounced disparities between distinct adipose tissue depots and reveals that BMI poorly correlates with fat mass–associated changes in VAT. Taken together, these results provide crucial evidence for the necessity to differentiate between distinct adipose tissue depots for a correct characterization of gene expression profiles that may affect metabolic health of patients with colorectal cancer.
Obesity is a major public health challenge, as it is strongly associated with metabolic disorders and multiple types of cancer, including colorectal cancer (CRC) (1, 2). Although the underlying mechanisms are incompletely understood, obesity-associated adipose tissue inflammation and dysregulated energy homeostasis present well-described molecular links between obesity and cancer (3, 4). Rodent and human studies provide intriguing evidence of enhanced macrophage infiltration and inflammatory expression patterns in obese adipose tissue (5–7). These cellular and molecular alterations presumably result from the inability of adipose tissue to healthily expand in response to excessive caloric supply. The increased expression of antiangiogenic molecules in adipose tissue of obese individuals strengthens these observations (8). As a result, obese adipose tissue manifests a local and systemic, chronic inflammatory response, accompanied by oxidative stress, that fuels metabolic disorders (9–11), cancer development, and potentially cancer progression (12, 13). Additionally, obesity is associated with perturbations of hormone levels and the expression of hormone receptors that systemically orchestrate energy homeostasis (14–16) and are considered major drivers of carcinogenesis (17, 18).
Whereas the evidence linking obesity and its metabolic mediators with CRC risk is strong, there are only limited and contradictive data on the associations with recurrence and mortality after CRC diagnosis (19–21). The issues of body composition, including excessive body fat, are important for cancer survivors, and inconsistent findings of previous studies may be the result of inadequate assessment of anthropometric data (22, 23). Studies have primarily focused on the body mass index (BMI, kg/m2), an anthropometric measurement that allows categorization of individuals as underweight (<18.5 kg/m2), normal weight (18.5 to 24.9 kg/m2), overweight (25 to 29.9 kg/m2), or obese (>30 kg/m2) (24). These categories are widely used to stratify individuals at high risk for diseases associated with metabolic complications, even though the BMI poorly reflects the crucial aspect of body fat distribution (25). In fact, the discrimination between distinct adipose tissue compartments allows for a more precise risk prediction, as an excess of visceral adipose tissue (VAT) is more strongly associated with unfavorable health conditions compared with the accumulation of subcutaneous adipose tissue (SAT) (26, 27). These findings are attributed to distinct cellular structures as well as higher metabolic and inflammatory activity of VAT compared with SAT (28, 29).
Previous studies have identified specific factors that are differentially expressed in distinct adipose tissues or that are associated with body fatness, contributing to the current understanding of the triad of obesity, adipose tissue inflammation and dysregulated energy homeostasis, and metabolic disorders (30, 31). However, these studies have mainly focused on rodent models (32), human SAT (33), only men or women (34), or on subjects with obesity (35), whereas only a few small-scale studies investigating human VAT on a genome-wide level are available (8, 36–40). Moreover, there is a lack of evidence disentangling the disparities between anthropometric measurements in reflecting obesity-associated alterations within adipose tissue compartments, in particular on the background of CRC.
In this study, we present transcriptome data of mesenteric VAT and abdominal SAT from 233 patients with CRC in combination with direct quantification of VAT and SAT mass. To our knowledge, this is the largest study that comprehensively compared genome-wide gene expression patterns in human VAT and SAT of patients with CRC and investigated depot-specific gene expression alterations in association with distinct anthropometric measurements, including BMI, the most commonly used proxy for obesity. These data provide crucial evidence for depot-specific profiles that may affect metabolic health and prognosis of patients with CRC.
Materials and Methods
Study population
This study population is comprised of patients with CRC enrolled in the ColoCare Study (ClinicalTrials.gov no. NCT02328677). The ColoCare Consortium is a multicenter, international prospective cohort for interdisciplinary studies of CRC prognosis and outcomes recruiting newly diagnosed patients with CRC (stage I to IV; International Classification of Diseases, ICD-10, C18 to C20) undergoing surgical resection, with sites at the Fred Hutchinson Cancer Research Center (Seattle, WA), H. Lee Moffitt Cancer Center and Research Institute (Tampa, FL), National Center for Tumor Diseases (Heidelberg, Germany), and Huntsman Cancer Institute (Salt Lake City, UT) (41–43).
This study involved n = 233 patients with CRC recruited between October 2010 and March 2015 at the Division of Preventive Oncology, National Center for Tumor Diseases in Heidelberg, Germany, and was approved by the Institutional Review Board of the Medical Faculty at the University of Heidelberg. Written informed consent was obtained by all participants. Demographic and clinical characteristics on sex, age, tumor site, tumor stage, and receipt of neoadjuvant therapy were obtained from patients’ medical records and pathological reports. Data on individual height, weight 1 year before surgery, and weight at baseline for BMI calculation were collected using questionnaires. Baseline data were verified by reviewing surgical anesthesia records. In case of discrepancy, the data from anesthesia protocols were taken, as these were recorded by the attending anesthetist. Anesthesia records were further viewed to abstract information on administration of glucocorticoids during surgery.
Adipose tissue quantification
For diagnostic and staging purposes, contrast-enhanced CT scans were recorded at the Department of Radiology, University Hospital Heidelberg before or after surgery. Quantification of adipose tissue compartments was performed as previously described (44, 45). Briefly, an axial image slice was chosen between vertebral body L3 and L4 for area-based adipose tissue quantification using a semiautomatic volume tool (Multimodality Workplace, Syngo volume tool, Siemens Healthcare, Erlangen, Germany). Total fat area and visceral fat area (VFA) were specified by manually tracing the abdomen circumference and the abdominal wall. Measurement thresholds of each region of interest were set to the attenuation limits of −190 to −30 Hounsfield units to selectively measure adipose tissue. Subcutaneous fat area (SFA) was calculated by subtracting VFA from the total fat area. Interrater reliability of CT-based adipose tissue quantification was demonstrated previously (46).
Adipose tissue collection
Mesenteric VAT and abdominal SAT samples were collected during primary tumor resection and processed as previously described (47). Briefly, SAT was resected during surgery at the abdominal section, and VAT was removed from the resected colorectal segment. Fresh samples were snap frozen in liquid nitrogen within 45 minutes and stored at −80°C. For further processing, samples were split into aliquots on a cooled cryostat (Leica Microsystems, Wetzlar, Germany; VAT samples) or manually cut within the gas phase of liquid nitrogen (SAT samples) to avoid sample thawing. Representative VAT sections were reviewed by a pathologist, and samples demonstrating lymph nodes were excluded from further processing. Owing to low RNA quality, few samples were repeatedly processed (two times, 14 VAT, 18 SAT samples; three times, 7 VAT, 3 SAT samples; four times, 1 SAT sample).
RNA isolation and gene expression profiling
Whole RNA was isolated from fresh-frozen VAT and SAT aliquots using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. In summary, ceramic beads were added to the samples, and tissues were homogenized using the Precellys 24 tissue lyser (Peqlab, Erlangen, Germany). Genomic DNA was sequestered using DNA spin columns, and flow-through was further processed. Proteins were digested by proteinase K and removed with chloroform extraction. The remaining DNA was eliminated by DNase I. Whole RNA was subsequently isolated on RNA spin columns. RNA concentrations were measured using the Epoch microplate dpectrophotometer (BioTek, Winooski, VT), and RNA quality was assessed by determining the RNA integrity number using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). We did not reuse previously frozen RNA samples. Transcriptomic profiling was performed on the HumanHT-12 Expression BeadChip according to the manufacturer’s protocol (Illumina, San Diego, CA) at the DKFZ Genomics and Proteomics Core Facility (Heidelberg, Germany). Analysis was conducted within 1 (84 VAT, 52 SAT samples), 2 (60 VAT, 48 SAT samples), 3 (46 VAT, 25 SAT samples), or 4 (19 VAT samples) years after sample collection. We did not observe any differences in SAT gene expression by storage time (time from sample collection to analysis; analyzed by Kendall correlation). In VAT, nine transcripts showed significantly decreased median expression signals over time. None of these transcripts showed significant associations in our analyses. Raw gene expression data will be publically available from the ArrayExpress repository at the time of publication (accession no. E-MTAB-8013).
Differential gene expression of five genes that showed significant associations in our analyses (SAA1, IGF1, IL6, CCL2, and CXCL12) was validated by quantitative real-time polymerase chain reaction in a randomly selected subset of paired VAT and SAT samples (n = 15 patients for SAA1, IL6, CCL2, CXCL12; n = 20 patients for IGF1; patients were selected via R function sample). Complementary DNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. The quantitative real-time polymerase chain reaction was run on LightCycler® 480 instrument II (Roche Diagnostics, Basel, Switzerland) using the Power SYBR® Green PCR master mix kit according to the manufacturer’s protocol (Applied Biosystems, Carlsbad, CA). The crossing point values of genes of interest and the reference gene (RPL27) were calculated by LightCycler® 480 software 1.5.0 (Roche Diagnostics). The samples were measured in triplicates, and single deviating replicates (SD >0.5) were excluded. Relative gene expression was quantified by comparing the crossing point values of the gene of interest and the reference gene. (48).
Statistical analysis
Data processing and statistical analyses were performed using the statistical software R 3.3.2 (49). Spearman correlations were calculated to evaluate associations between continuous variables. The Wilcoxon rank sum test was performed to compare anthropometric measurements by sex and tumor site (binary variable), and the Kendall correlation was performed to compare anthropometrics by stage (ordinal variable).
Raw gene expression data were processed prior to statistical analyses. Gene expression was missing for 189 data points (1.2 × 10−5 %) of 135 random samples (86 VAT, 49 SAT samples) and 44 transcripts. The number of missing values per sample ranged from one (in 99 samples) to five (in 3 samples). Transcripts with most frequent missing gene expression values were LOC100133005 (transcript 7400097, 43 missing values) and MIR1282 (transcript 2490543, 26 missing values). Missing expression values were imputed using the nearest neighbor averaging method (package impute, function impute.knn). Data were further transformed using the variance stabilizing transformation method (package lumi, function lumiT), normalized using the robust spline normalization method (package lumi, function rsn), and adjusted for batch effects using the function ComBat (package sva) (50, 51). For gene expression interpretation, transcripts yielding values <8 were considered indistinguishable from background noise and unquantifiable.
Transcriptome data from VAT and SAT were compared by combining the Wilcoxon signed rank test and the Wilcoxon rank sum test as samples were partly paired (package robustrank, function mw.mw.2.perm) (52). Relative fold changes were presented as differences in median gene expression. Obesity-dependent adipose tissue gene expression was assessed using multivariate linear regression analyses with individual gene expression as the dependent variable and the variable of interest as the explanatory variable. The following anthropometric measures were tested individually as explanatory variables: VFA or SFA, and BMI. Multivariate analyses were adjusted for age (years), sex (women/men), tumor site (colon/rectum), tumor stage (I to II/III to IV; patients with carcinoma in situ or no malignancy were excluded), and neoadjuvant treatment (yes/no). Transcriptome analyses were adjusted for multiple testing using the Benjamini and Hochberg correction (53).
Enrichment of gene sets was analyzed by gene set enrichment analysis (GSEA) using the HTSanalyzeR package (54, 55). Genes were ranked by fold changes calculated by comparing median gene expression (VAT vs SAT) or multivariate regression analyses (gene expression vs anthropometric measurement). To identify differentially regulated pathways, we used predefined gene sets from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The number of permutations was set to 1000. P values were adjusted for multiple testing using the Benjamini and Hochberg correction.
In exploratory survival analyses, Cox proportional hazard models were used to investigate the associations of selected genes with overall survival after 24 months of follow-up (package survival). The models were adjusted for age, sex, VFA, tumor stage, and neoadjuvant treatment. Analyses were conducted for the total study population and separately for patients with stage I, II, or III tumors and for patients with stage IV tumors. P values were adjusted for multiple testing using the Holm correction (56).
All statistical tests were two-sided, and P values or adjusted P values <0.05 were considered significant.
In total, 48,107 statistical tests were performed to compare expression levels between VAT and SAT controlling the false discovery rate at 5%. With a total sample size of 233 patients, we could achieve an average power of >90% assuming a fold change of at least 1.5 between the two tissue samples within 5% of all tested probes. If we assume a fold change of at least two within 0.25% of all tested probes, the average power increases to >99%. The average power is the expected proportion of rejected false null hypothesis among the set of false null hypotheses. Statistical power was determined using the FDRsampsize package (57).
Tissue sampling validation
To evaluate for potential confounding of anti-inflammatory effects of glucocorticoid administration during surgery on adipose tissue, we examined whether patients who received a high dose of glucocorticoid (dexamethasone, 4 mg for prevention of postoperative nausea and vomiting) presented with a distinct inflammatory gene expression profile compared with patients who did not receive glucocorticoid intraoperatively. Evaluation of genome-wide expression and specific glucocorticoid receptor target expression (CCL2, IL1B, IL1A, IL6, and PTGS2) (58–60) by Wilcoxon rank sum test demonstrated no significant differences in VAT or SAT transcriptomics between glucocorticoid groups (61).
Results
Cohort characteristics
Key demographics, clinical characteristics, and data availability are presented in Table 1 and in an online repository (61). The study included 233 patients with CRC with 151 (65%) men and a median age of 63 years (range, 22 to 98 years; interquartile range, 56 to 72 years). Sex and age were significantly, and tumor stage and neoadjuvant therapy were modestly, associated with anthropometric indices (61) and were thus included as adjustment variables in the regression models described in the following sections. We did not observe an association between tumor site and anthropometric indices (61), but we included the variable as a confounding factor, as the tumor location determined the location of adipose tissue sampling.
Table 1.
Demographic and Clinical Characteristics of the Study Cohort
| Total (n = 233) | Individuals With VAT Data (n = 209) | Individuals With SAT Data (n = 125) | Individuals With VAT, SAT, and CT Data (n = 72) | |
|---|---|---|---|---|
| Sex, n (%) | ||||
| Women | 82 (35) | 78 (32) | 36 (29) | 22 (31) |
| Men | 151 (65) | 131 (68) | 89 (71) | 50 (69) |
| Age, y | 63 (56–72) | 64 (55–72) | 63 (55–70) | 64 (54–70) |
| BMI, kg/m2 | 26.1 (23.7–29.2) | 26.1 (23.7–29.2) | 26.6 (24.3–30.3) | 26.4 (24.2–28.8) |
| VFA, cm2 | 167 (116–236) | 165 (110–232) | 175 (117–251) | 182 (116–253) |
| SFA, cm2 | 206 (159–255) | 206 (158–256) | 209 (162–255) | 203 (161–255) |
| CT scan availability, n (%) | 153 (66) | 137 (66) | 91 (73) | 72 (100) |
| Weight change within 1 y before surgery (%) | −2.4 (−6.7 to 0) | −2.4 (−6.7 to 0) | −1.1 (−6.7 to 0) | −3.0 (−6.7 to 0) |
| Weight change, data availability, n (%) | 209 (90) | 189 (90) | 112 (90) | 65 (90) |
| Tumor site, n (%) | ||||
| Colon | 108 (46) | 100 (48) | 54 (43) | 33 (46) |
| Rectum | 125 (54) | 109 (52) | 71 (57) | 39 (54) |
| Tumor stage, n (%) | ||||
| I | 38 (16) | 32 (15) | 17 (14) | 8 (11) |
| II | 77 (33) | 70 (33) | 37 (30) | 23 (32) |
| III | 65 (28) | 59 (28) | 37 (30) | 24 (33) |
| IV | 45 (19) | 41 (20) | 27 (22) | 17 (24) |
| Carcinoma in situ or no malignancy | 8 (3) | 7 (3) | 7 (6) | 0 |
| Neoadjuvant therapy, n (%) | ||||
| Yes | 74 (32) | 64 (31) | 44 (35) | 25 (35) |
| No | 159 (68) | 145 (69) | 81 (65) | 47 (65) |
Data presented as median (interquartile range) for continuous variables and frequency (percentage) for categorical variables.
Transcriptomic differences between VAT and SAT
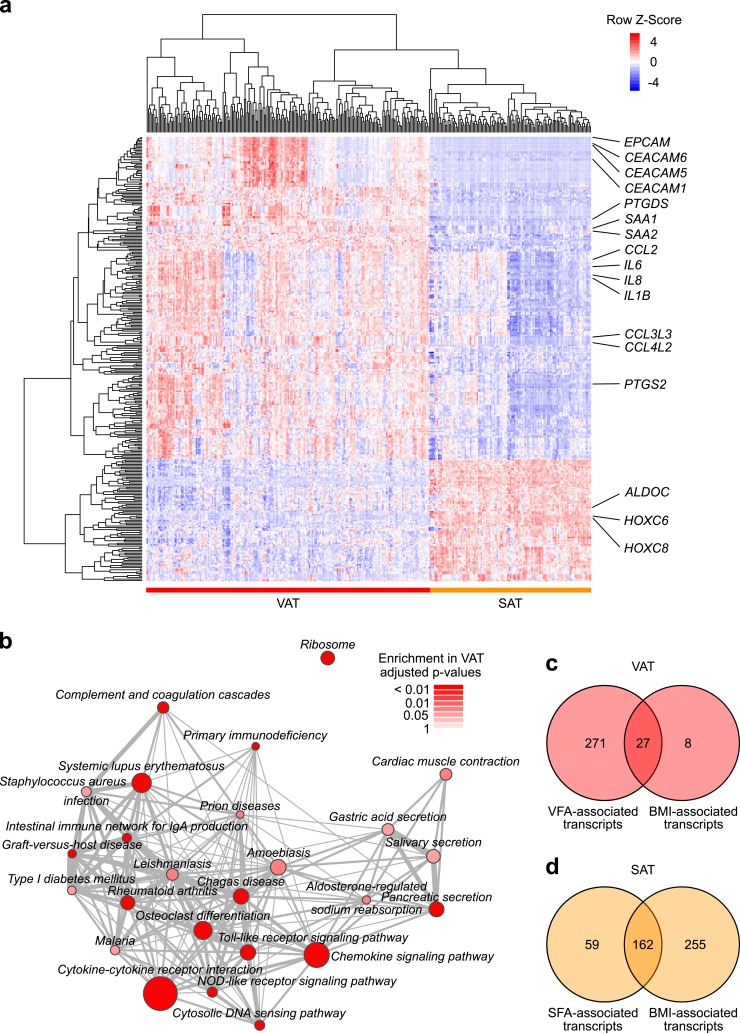
Whole-transcriptome profiles of VAT and SAT samples were available for n = 209 and n = 125 patients, respectively. We compared genome-wide gene expression of both adipose tissue compartments and identified 14,922 differentially expressed transcripts (61). Prominent cytokines and chemokines (e.g., IL6, IL8, CCL2, CXCL12, IL32), as well as genes involved in cell adhesion (e.g., EPCAM), the synthesis of prostaglandins (e.g., PTGDS), and key regulators of metabolic homeostasis (e.g., SAA1, IGF1), were significantly upregulated in VAT compared with SAT (Fig. 1a). In contrast, genes encoding metabolizing enzymes (e.g., ALDOC, ALDH2) and homeobox family genes (e.g. HOXC6) were more highly expressed in SAT (Fig. 1a).
Figure 1.
Transcriptomic differences between VAT and SAT. (a) Heat map of differential gene expression between VAT and SAT. Genes with an adjusted P value <0.05 and relative fold change >0.5 and < -0.5 are displayed. (b) GSEA results of gene sets (KEGG gene set collection) significantly differentially enriched between VAT and SAT. Nodes are sized in relationship to gene set size. Edges denote overlap between gene sets. Analyses were adjusted for multiple testing. (c) VAT transcripts significantly associated with VFA and BMI. (d) SAT transcripts significantly associated with SFA and BMI.
These observations were corroborated by GSEA, as inflammatory pathways were highly enriched in VAT (e.g., Cytokine–cytokine receptor interaction, enrichment score of 0.65, adjusted P < 0.001; Chemokine signaling pathway, enrichment score of 0.65, adjusted P < 0.001) (Fig. 1b). In contrast, metabolic pathways were enriched in SAT, although these pathways did not reach statistical significance after adjustment for multiple testing (e.g., Lysine degradation, enrichment score of −0.7, adjusted P = 0.09; Insulin signaling pathway, enrichment score of −0.47, adjusted P = 0.22).
We performed exploratory analyses to investigate the association between the top 10 genes that were significantly upregulated in VAT and overall survival of patients with CRC. We did not identify any significant associations in the total study population (n = 103 alive vs 27 deceased) or in stage-stratified subgroups (stage I/II/III, n = 87 alive vs 13 deceased; stage IV, n = 16 alive vs 14 deceased) (data not shown). The strongest (not significant) association with overall survival was observed for PTGDS in patients with stage I/II/III tumors (hazard ratio, 5.84, 95% CI, 1.52-22.53) (data not shown).
VFA-associated transcriptomic alterations in VAT
In multivariate regression analysis of n = 134 patients, we investigated the association between measurements of VFA and genome-wide gene expression in VAT. We identified 298 transcripts that significantly correlated with VFA (61). Genes encoding for adipokines (LEP, SAA1), the angiogenesis inhibitor TNMD, the steroid metabolizing enzyme HSD11B1, and NQO1, a gene involved in oxidative stress response, were upregulated with increasing VFA, whereas key enzymes of lipid metabolism (e.g., PPAP2B, AGPAT9) were downregulated. We did not observe an increase in gene expression of prominent mediators of adipose tissue inflammation, such as IL6, IL1B, or CCL2.
In GSEA, we observed similar metabolic expression patterns, as Metabolic pathways, Glycerophospholipid metabolism, and pathways of small molecule metabolism (e.g., Valine, leucine and isoleucine degradation) were downregulated with increasing VFA. In contrast, inflammatory pathways were enriched with increasing VFA (e.g., Toll-like receptor signaling pathway and Chemokine signaling pathway) (61).
SFA-associated transcriptomic alterations in SAT
Following the same approach, we analyzed the association between SFA and gene expression in SAT. In multivariate regression analysis of n = 87 patients, 221 transcripts significantly correlated with SFA (61). Similar to visceral fat, LEP, TNMD, NQO1, and HSD11B1 were strongly upregulated with increasing SFA. Furthermore, genes encoding for proteins of cell migration processes (e.g., ITGB5, ITGB2) and the chemokine CXCL16 were upregulated with increasing SFA, whereas genes encoding for enzymes of the lipid metabolism and the glucose metabolism were downregulated (e.g., AGPAT9, ADH1B). However, similar to VAT, prominent markers of adipose tissue inflammation did not show differential expression in SAT.
In line with our observations in VAT, inflammatory pathways were enriched among upregulated genes (e.g., Toll-like receptor signaling pathway and Cytokine–cytokine receptor interaction) (61). Accordingly, pathways of lipid metabolism and small molecule metabolism were most strongly enriched among downregulated genes in GSEA (e.g., Glycerophospholipid metabolism and Valine, leucine, and isoleucine degradation). Moreover, pathways of cellular movement and immune cell signaling were enriched among genes uniquely upregulated in SAT but not in VAT [e.g., Cell adhesion molecules (CAMs), Leukocyte transendothelial migration, and Natural killer cell–mediated cytotoxicity].
Depot-specific differences in gene expression alterations
To compare the findings described above between VAT and SAT in the most stringent subset of patients, we analyzed n = 72 patients, where paired SAT and VAT gene expression data as well as information on VFA and SFA were available. This patient subset did not differ in demographic or clinical characteristics from the overall study population (Table 1). In these analyses, 57 VAT transcripts were uniquely associated with VFA with no evidence for specific gene set patterns (61). In contrast, twice as many transcripts (115 transcripts) showed an association with SFA in SAT (61). These included genes encoding for the chemokine CXCL16, the integrins ITGB2 and ITGB5, LEP, and enzymes of the lipid metabolism (e.g., LPIN). Two transcripts were differentially expressed in both compartments (C6 and MSTO1).
BMI-associated gene expression alterations in VAT and SAT
BMI is the most commonly used measure of obesity. We thus evaluated the association of BMI with fat area measurements and with depot-specific gene expression patterns in comparison with the respective fat areas. Self-reported BMI correlated with VFA (ρ = 0.65, P < 0.001) and SFA (ρ = 0.71, P < 0.001). However, we identified high variabilities among patients with visceral obesity, as patients with highest VFA (quartile 4, >236 cm2) had BMI values ranging from normal weight to obese (22.0 to 37.8 kg/m2).
In multivariate regression analysis of n = 134 patients, we identified 35 transcripts in VAT that correlated with self-reported BMI. Among these, 27 transcripts overlapped with significant transcripts from VFA analyses above (9%) (Fig. 1c), but showed weaker correlations with BMI than with VFA. In GSEA, we observed similar gene set enrichment patterns in association with BMI compared with VFA (61).
Multivariate regression analysis of SAT gene expression was based on n = 87 patients. We detected 417 transcripts that correlated with self-reported BMI, of which 162 transcripts overlapped with SFA analyses above (73%) (Fig. 1d). These included genes encoding for LEP, metabolizing enzymes (e.g., AGPAT9), integrins (e.g., ITGB2) and the chemokine CXCL16. Additionally, we observed that SAT gene expression of two macrophage markers, CD68 and CD163, was upregulated with increasing BMI. In GSEA, we identified similar enrichment patterns in association with BMI compared with SFA (61).
Discussion
This study provides new insights into the complex interplay between body fat distribution and metabolic gene expression patterns in human adipose depots that may affect metabolic health and disease outcome among patients with CRC. Our data corroborate previous studies of enhanced inflammatory gene expression patterns in VAT, distinguish between depot-specific transcriptome profiles in association with fat area indices, and provide evidence that BMI poorly correlates with gene expression in VAT (Table 2).
Table 2.
Summary of Transcriptomic Differences Between VAT and SAT
| VAT | SAT | |
|---|---|---|
| Basal gene expression profile | Cytokine–cytokine receptor interaction↑ e.g., IL6, IL8, CCL3L3 | Lysine degradation↑ae.g., ALDH2, ALDH3A2, ALDH9A1 |
| Chemokine signaling pathway↑ e.g., IL8, CCL3L3, CCL4L2 | Pyruvate metabolism↑ae.g., ACSS2, ALDH2, ALDH3A2 | |
| Arachidonic acid metabolism↑ae.g., PTGDS, PTGS2, ALOX5 | Insulin signaling↑ae.g., PTPRF, LIPE, FASN | |
| Cell adhesion molecules (CAMs)↑ae.g., EPCAM,bCEACAM6,bCEACAM1b | Fatty acid degradation↑ae.g., ALDH2, ALDH3A2, ADH1B | |
| Adipokines that systemically regulate metabolic homeostasis↑cSAA1, IGF1 | ||
| Gene expression profile in association with VFA and SFA, respectively | Valine, leucine, and isoleucine degradation↓ MUT, BCKDHB, ALDH6A1 | Valine, leucine and isoleucine degradation↓ HADH |
| Glycerophospholipid metabolism↓ AGPAT9 | ||
| Glycerophospholipid metabolism↓ae.g., PPAP2B, GPD1L, AGPAT9 | Fatty acid degradation↓ HADH, ADH1B, ECI1 | |
| Glycolysis/gluconeogenesis↓ ADH1B, ADH1A | ||
| Toll-like receptor signaling pathway↑ IRKA1, MAP2K4 | Toll-like receptor signaling pathway↑ SPP1, MAP2K1 | |
| Cytokine–cytokine receptor interaction↑ e.g., LEP, PLEKHO2, INHBB | Cytokine–cytokine receptor interaction↑ LEP, CXCL16 | |
| Chemokine signaling pathway↑ CXCL16, MAP2K1 | ||
| Leukocyte transendothelial migration↑ ITGB2 | ||
| Natural killer cell–mediated cytotoxicity↑ ITGB2, HCST, MAP2K1 | ||
| Adipokines that systemically regulate metabolic homeostasis↑cLEP, SAA1 | Adipokines that systemically regulate metabolic homeostasis↑cLEP | |
| BMI as an anthropometric proxy | Poor correlation with VFA among viscerally obese | Strong correlation with SFA across all individuals |
| Poor representation of VFA-associated gene expression profile | Good representation of SFA-associated gene expression profile |
Selected KEGG pathways from GSEA and top three corresponding genes (adjusted P value <0.05, ranked by relative fold changes) are presented. Arrows represent the direction of regulation.
P value in GSEA >0.05.
Not included in KEGG pathway, but with evident biological function.
No KEGG pathway.
VAT compared with SAT displayed pronounced gene expression patterns of key mediators of inflammatory signaling and metabolic homeostasis, confirming a recent CRC study, where we have reported similar findings on the metabolite level (47). Increased gene expression of key enzymes of prostaglandin synthesis aligns with our previous findings of elevated levels of free arachidonic acid in VAT, a transmitter of inflammatory signals. We further corroborated reports from other groups by demonstrating enhanced gene expression of prominent cytokines, including IL6 and IL32 (62, 63). The strong signals of cell adhesion genes may be attributed to the massive infiltration of immune cells into VAT, as shown in previous studies (28, 37), and higher complexity in cellular composition than SAT. Additionally, we identified high gene expression levels of the secretory adipokines SAA1 and IGF1 in VAT. As proteins potentially secreted from VAT, these factors may impact systemic energy homeostasis (64) or function as growth factors (65). The data presented in the present study substantiate the unique role of VAT as a highly active endocrine organ that may constitute a crucial modifier of the obesity–cancer link.
Exploratory survival analyses showed no significant association between expression of selected genes that were upregulated in VAT compared with SAT and overall survival of patients with CRC 2 years after primary surgery. We assume that our analyses had insufficient statistical power to detect significant differences. Emerging evidence proposes that body fat distribution, rather than BMI, is a prognostic marker for poor cancer-related outcome (66, 67). With improving anticancer treatment and a rising number of CRC survivors, there is an urgent need to understand the biological mechanisms that link body fat distribution to poor metabolic health and prognosis of CRC patients. However, to our knowledge no study has systematically investigated molecular perturbations within VAT that may underlie this link and affect disease outcome. We have uncovered a VAT-specific metabolic gene expression signature among patients with CRC that implies upregulation of critical enzymes involved in arachidonic acid metabolism and potentially secreted cytokines, chemokines, and growth factors with known prognostic relevance (17, 68, 69). These data provide an initial hint of potential mechanistic relationships and glean important insight that may inform future interventional studies of cancer survivorship. Prospective studies with larger sample sizes are required to comprehensively assess these relationships.
Although gene expression profiles strongly differed between VAT and SAT, in both compartments, an increase in depot-specific fat mass was associated with similar transcriptomic alterations. We confirm previously described downregulation of metabolic processes and upregulation of immune response–related gene expression in SAT and VAT (36). We further observed that gene expression of key enzymes of lipid metabolism, AGPAT9 and PPAP2B, was downregulated in both compartments, suggesting that triglyceride synthesis and storage were impaired with increasing fat mass. Moreover, our data could validate the strong correlation between fat mass and gene expression of LEP, HSD11B1, TNMD, and NQO1, regulators of energy homeostasis, angiogenesis, and oxidative stress defense, respectively, in both compartments (8, 11, 16, 70). However, only in SAT was an increase of SFA associated with transcriptional upregulation of two integrins and the cytokine CXCL16. Interestingly, prominent markers of adipose tissue inflammation, such as IL6 or IL1B, did not show obesity-related gene expression levels in VAT or SAT. Rather, inflammatory gene expression patterns were only detectable when analyzing whole gene sets in GSEA, which reduces the number of parallel testing and thereby enables measurement of modest signals. In line with these observations, there is emerging evidence that macrophages, identified in obese adipose tissue without features of classical activation, may be “metabolically activated” and characterized by a modest increase in proinflammatory cytokine expression (71, 72). Based on these published data and our observations, we suggest that in human adipose tissue, obesity-related transcriptional upregulation of inflammatory cytokines is rather weak.
Notably, comparative analyses of paired samples provided evidence that the association between fat area and adipose tissue gene expression was more pronounced in SAT than in VAT. LEP, genes of lipid metabolism, integrins, as well as CXCL16 were uniquely altered in SAT in these analyses, suggesting a stronger association with fat area in SAT compared with VAT. Our observations support data from a recent study that reported that macrophage counts in SAT, but not in VAT, were associated with obesity in healthy individuals (73). Based on these data, we propose a higher transcriptomic adaptability of SAT in response to adipose mass expansion. Importantly, our analyses were based on relative gene expression measurements that do not allow drawing conclusions about the absolute level of inflammation or metabolic activity within distinct adipose tissue depots or the entire organism. It is conceivable that inflammatory and metabolic factors that are highly expressed in VAT may not be transcriptionally upregulated but quantitatively highly expressed in patients with great absolute VAT mass.
In the present cohort, self-reported BMI was a weak indicator for VFA among patients with visceral obesity and was poorly associated with transcriptomic alterations within VAT. In contrast, BMI was tightly correlated with SFA among normal-weight patients as well as patients with obesity and resembled SAT transcriptome shifts that were associated with SFA. There is growing consensus that BMI fails to sufficiently describe distinct body fat distribution patterns, in particular the central obesity phenotype (74–77), which is strongly related to enhanced VAT mass, as well as the increased incidence of metabolic diseases (78–81). In the present study, we provide a biological rationale for observed disparities between obesity measures regarding their associations with risk profiles of metabolic diseases and CRC in particular.
Importantly, the development of colorectal tumors potentially induces molecular as well as morphological changes in the surrounding adipose tissue, including lipolysis, inflammation, and adipose tissue browning (82–84). Although there is only limited knowledge on adipose tissue alterations related to chemotherapy and/or radiotherapy, neoadjuvant therapy prior to adipose tissue collection may have also affected adipose tissue physiology, including its lipid storage capacity and fibrosis (85, 86). We have accounted for this treatment effect by adjusting all analyses for neoadjuvant therapy. Moreover, patients who underwent surgery several weeks after diagnosis may have undergone short-term changes in lifestyle habits, including diet and physical activity, that could have further influenced adipose tissue physiology (87–89). Still, we were able to validate findings from other studies that investigated cancer-free individuals, indicating that in many aspects the correlation between obesity and adipose tissue gene expression surpasses the potential impact of CRC, cancer therapy, and short-term lifestyle changes on adipose tissue gene expression. Moreover, we did not identify any significant associations of weight change or weight loss prior to surgery with adipose tissue gene expression (data not shown), indicating that the impact of weight change on adipose gene expression is rather modest in the analyzed cohort.
Future confirmatory and functional studies are required to validate the identified gene expression signatures and to mechanistically confirm our findings. Moreover, the gene expression patterns presented in this study relate to whole adipose tissue. It will be interesting to investigate the contribution of distinct cell populations to these patterns in future studies.
To our knowledge, the presented results are based on the largest study characterizing human mesenteric VAT and abdominal SAT gene expression in combination with absolute quantification of distinct fat areas among patients with CRC. These comprehensive data refine our understanding of molecular disparities between distinct adipose tissue compartments and their metabolic transcriptome profiles in obesity, and they illustrate that the BMI is a poor indicator for fat mass–associated metabolic gene expression alterations in VAT. We thereby provide a biological rationale for the necessity to differentiate between distinct adipose tissue compartments and body fat distribution patterns. Given the rising number of CRC survivors, there is an instant need for introducing direct measurements of depot-specific adipose tissue mass and defining VAT mass scores to correctly classify CRC patients at high risk for metabolic disturbances that may fuel cancer recurrence and poor prognosis.
Acknowledgments
The authors thank the National Center for Tumor Diseases Tissue Biobank for sample handling and storage, and the Microarray Unit of the DKFZ Genomics and Proteomics Core Facility for providing the Illumina Whole-Genome Expression BeadChips and related services. We are grateful to all of the study staff who have made this study possible, especially Torsten Kölsch, Susanne Jakob, Stefanie Skender, Werner Diehl, Rifraz Farook, Lin Zielske, Anett Brendel, Marita Wenzel, and Renate Skatula.
Financial Support: The Heidelberg ColoCare Study was supported by the National Institutes of Health Grant R01 CA189184, the Matthias Lackas Foundation, the Claussen-Simon Stiftung, the German Consortium of Translational Cancer Research (DKTK), institutional funding from the German Cancer Research Center (DKFZ) Heidelberg, and by the Helmholtz Association (portfolio theme “Metabolic Dysfunction”). C.M.U. was supported by the Huntsman Cancer Foundation and by National Institutes of Health Grants U01 CA206110, R01 CA189184, R01 CA207371, and P30 CA042014. A.N.H. was supported by the National Institutes of Health under Ruth L. Kirschstein National Research Service Award T32 HG008962 from the National Human Genome Research Institute.
Clinical Trial Information: ClinicalTrials.gov no. NCT02328677 (registered 31 December 2014).
Author Contributions: M.H., M.K., C.M.U., D.S. and H.G. conceived the study. M.H. collected biological samples and clinical data. C.M.U., H.G., M.S., A.U., E.H., P.S., B.K.S., J.N., H.-U.K., and H.B. provided resources. P.S.-K., J.B. and N.H. coordinated the Heidelberg ColoCare Study. B.G. maintained the ColoCare Study databases. M.H., R.T., A.B. and T.L. analyzed the data and performed statistical analysis. M.H., M.K., C.M.U., C.R.B., H.G. and D.S. interpreted the results. M.H., D.S., A.N.H. and C.M.U. wrote the manuscript. All authors read and approved the manuscript.
Additional Information
Disclosure Summary: C.M.U. has as cancer center director oversight over research funded by several pharmaceutical companies, but has not received funding directly and has no conflicts in relationship to the current work. The remaining authors have nothing to disclose.
Data Availability: All data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
Glossary
Abbreviations:
- BMI
body mass index
- CRC
colorectal cancer
- GSEA
gene set enrichment analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- SAT
subcutaneous adipose tissue
- SFA
subcutaneous fat area
- VAT
visceral adipose tissue
- VFA
visceral fat area
References and Notes
- 1. Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K; International Agency for Research on Cancer Handbook Working Group. Body fatness and cancer—viewpoint of the IARC Working Group. N Engl J Med. 2016;375(8):794–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Long MT, Fox CS. The Framingham Heart Study—67 years of discovery in metabolic disease. Nat Rev Endocrinol. 2016;12(3):177–183. [DOI] [PubMed] [Google Scholar]
- 3. Ulrich CM, Himbert C, Holowatyj AN, Hursting SD. Energy balance and gastrointestinal cancer: risk, interventions, outcomes and mechanisms. Nat Rev Gastroenterol Hepatol. 2018;15(11):683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murphy N, Jenab M, Gunter MJ. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol. 2018;15(11):659–670. [DOI] [PubMed] [Google Scholar]
- 5. Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW Jr. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sartipy P, Loskutoff DJ. Monocyte chemoattractant protein 1 in obesity and insulin resistance. Proc Natl Acad Sci USA. 2003;100(12):7265–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. [DOI] [PubMed] [Google Scholar]
- 8. Saiki A, Olsson M, Jernås M, Gummesson A, McTernan PG, Andersson J, Jacobson P, Sjöholm K, Olsson B, Yamamura S, Walley A, Froguel P, Carlsson B, Sjöström L, Svensson PA, Carlsson LM. Tenomodulin is highly expressed in adipose tissue, increased in obesity, and down-regulated during diet-induced weight loss. J Clin Endocrinol Metab. 2009;94(10):3987–3994. [DOI] [PubMed] [Google Scholar]
- 9. Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol. 2017;13(11):633–643. [DOI] [PubMed] [Google Scholar]
- 10. Huber J, Kiefer FW, Zeyda M, Ludvik B, Silberhumer GR, Prager G, Zlabinger GJ, Stulnig TM. CC chemokine and CC chemokine receptor profiles in visceral and subcutaneous adipose tissue are altered in human obesity. J Clin Endocrinol Metab. 2008;93(8):3215–3221. [DOI] [PubMed] [Google Scholar]
- 11. Palming J, Sjöholm K, Jernås M, Lystig TC, Gummesson A, Romeo S, Lönn L, Lönn M, Carlsson B, Carlsson LMS. The expression of NAD(P)H:quinone oxidoreductase 1 is high in human adipose tissue, reduced by weight loss, and correlates with adiposity, insulin sensitivity, and markers of liver dysfunction. J Clin Endocrinol Metab. 2007;92(6):2346–2352. [DOI] [PubMed] [Google Scholar]
- 12. Himbert C, Delphan M, Scherer D, Bowers LW, Hursting S, Ulrich CM. Signals from the adipose microenvironment and the obesity–cancer link—a systematic review. Cancer Prev Res (Phila). 2017;10(9):494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng T, Lyon CJ, Bergin S, Caligiuri MA, Hsueh WA. Obesity, inflammation, and cancer. Annu Rev Pathol. 2016;11(1):421–449. [DOI] [PubMed] [Google Scholar]
- 14. Glad CAM, Svensson PA, Nystrom FH, Jacobson P, Carlsson LM, Johannsson G, Andersson-Assarsson JC. Expression of GHR and downstream signaling genes in human adipose tissue—relation to obesity and weight-change. J Clin Endocrinol Metab. J Clin Endocrinol Metab. 2019;104(5):1459–1470. [DOI] [PubMed] [Google Scholar]
- 15. Aguilar-Salinas CA, García EG, Robles L, Riaño D, Ruiz-Gomez DG, García-Ulloa AC, Melgarejo MA, Zamora M, Guillen-Pineda LE, Mehta R, Canizales-Quinteros S, Tusie Luna MT, Gómez-Pérez FJ. High adiponectin concentrations are associated with the metabolically healthy obese phenotype. J Clin Endocrinol Metab. 2008;93(10):4075–4079. [DOI] [PubMed] [Google Scholar]
- 16. Mojiminiyi OA, Abdella NA, Al Arouj M, Ben Nakhi A. Adiponectin, insulin resistance and clinical expression of the metabolic syndrome in patients with type 2 diabetes. Int J Obes (Lond). 2007;31(2):213–220. [DOI] [PubMed] [Google Scholar]
- 17. Major JM, Laughlin GA, Kritz-Silverstein D, Wingard DL, Barrett-Connor E. Insulin-like growth factor-I and cancer mortality in older men. J Clin Endocrinol Metab. 2010;95(3):1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stattin P, Lukanova A, Biessy C, Söderberg S, Palmqvist R, Kaaks R, Olsson T, Jellum E. Obesity and colon cancer: does leptin provide a link? Int J Cancer. 2004;109(1):149–152. [DOI] [PubMed] [Google Scholar]
- 19. Greenlee H, Unger JM, LeBlanc M, Ramsey S, Hershman DL. Association between body mass index and cancer survival in a pooled analysis of 22 clinical trials. Cancer Epidemiol Biomarkers Prev. 2017;26(1):21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schlesinger S, Siegert S, Koch M, Walter J, Heits N, Hinz S, Jacobs G, Hampe J, Schafmayer C, Nöthlings U. Postdiagnosis body mass index and risk of mortality in colorectal cancer survivors: a prospective study and meta-analysis. Cancer Causes Control. 2014;25(10):1407–1418. [DOI] [PubMed] [Google Scholar]
- 21. Calle EE, Rodriguez C, Walker-Thurmond K, Thun MJ. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–1638. [DOI] [PubMed] [Google Scholar]
- 22. Caan BJ, Cespedes Feliciano EM, Kroenke CH. The importance of body composition in explaining the overweight paradox in cancer—counterpoint. Cancer Res. 2018;78(8):1906–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park Y, Peterson LL, Colditz GA. The plausibility of obesity paradox in cancer—point. Cancer Res. 2018;78(8):1898–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organization. Obesity and overweight. Available at: http://www.who.int/mediacentre/factsheets/fs311/en/. Accessed 1 February 2019.
- 25. Stevens J, McClain JE, Truesdale KP. Selection of measures in epidemiologic studies of the consequences of obesity. Int J Obes. 2008;32(Suppl 3):S60–S66. [DOI] [PubMed] [Google Scholar]
- 26. Rosenquist KJ, Massaro JM, Pedley A, Long MT, Kreger BE, Vasan RS, Murabito JM, Hoffmann U, Fox CS. Fat quality and incident cardiovascular disease, all-cause mortality, and cancer mortality. J Clin Endocrinol Metab. 2015;100(1):227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Britton KA, Massaro JM, Murabito JM, Kreger BE, Hoffmann U, Fox CS. Body fat distribution, incident cardiovascular disease, cancer, and all-cause mortality. J Am Coll Cardiol. 2013;62(10):921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harman-Boehm I, Blüher M, Redel H, Sion-Vardy N, Ovadia S, Avinoach E, Shai I, Klöting N, Stumvoll M, Bashan N, Rudich A. Macrophage infiltration into omental versus subcutaneous fat across different populations: effect of regional adiposity and the comorbidities of obesity. J Clin Endocrinol Metab. 2007;92(6):2240–2247. [DOI] [PubMed] [Google Scholar]
- 29. Bruun JM, Lihn AS, Pedersen SB, Richelsen B. Monocyte chemoattractant protein-1 release is higher in visceral than subcutaneous human adipose tissue (AT): implication of macrophages resident in the AT. J Clin Endocrinol Metab. 2005;90(4):2282–2289. [DOI] [PubMed] [Google Scholar]
- 30. Lozano-Bartolomé J, Llauradó G, Portero-Otin M, Altuna-Coy A, Rojo-Martínez G, Vendrell J, Jorba R, Rodríguez-Gallego E, Chacón MR. Altered Expression of miR-181a-5p and miR-23a-3p is associated with obesity and TNFα-induced insulin resistance. J Clin Endocrinol Metab. 2018;103(4):1447–1458. [DOI] [PubMed] [Google Scholar]
- 31. Moreno-Navarrete JM, Ortega F, Serrano M, Pérez-Pérez R, Sabater M, Ricart W, Tinahones F, Peral B, Fernández-Real JM. Decreased STAMP2 expression in association with visceral adipose tissue dysfunction. J Clin Endocrinol Metab. 2011;96(11):E1816–E1825. [DOI] [PubMed] [Google Scholar]
- 32. Shimobayashi M, Albert V, Woelnerhanssen B, Frei IC, Weissenberger D, Meyer-Gerspach AC, Clement N, Moes S, Colombi M, Meier JA, Swierczynska MM, Jenö P, Beglinger C, Peterli R, Hall MN. Insulin resistance causes inflammation in adipose tissue. J Clin Invest. 2018;128(4):1538–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alba DL, Farooq JA, Lin MYC, Schafer AL, Shepherd J, Koliwad SK. Subcutaneous fat fibrosis links obesity to insulin resistance in Chinese Americans. J Clin Endocrinol Metab. 2018;103(9):3194–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hetemäki N, Savolainen-Peltonen H, Tikkanen MJ, Wang F, Paatela H, Hämäläinen E, Turpeinen U, Haanpää M, Vihma V, Mikkola TS. Estrogen metabolism in abdominal subcutaneous and visceral adipose tissue in postmenopausal women. J Clin Endocrinol Metab. 2017;102(12):4588–4595. [DOI] [PubMed] [Google Scholar]
- 35. Gauthier MS, Pérusse JR, Lavoie ME, Sladek R, Madiraju SR, Ruderman NB, Coulombe B, Prentki M, Rabasa-Lhoret R. Increased subcutaneous adipose tissue expression of genes involved in glycerolipid-fatty acid cycling in obese insulin-resistant versus -sensitive individuals. J Clin Endocrinol Metab. 2014;99(12):E2518–E2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klimcáková E, Roussel B, Márquez-Quiñones A, Kovácová Z, Kováciková M, Combes M, Siklová-Vítková M, Hejnová J, Srámková P, Bouloumié A, Viguerie N, Stich V, Langin D. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J Clin Endocrinol Metab. 2011;96(1):E73–E82. [DOI] [PubMed] [Google Scholar]
- 37. O’Rourke RW, Metcalf MD, White AE, Madala A, Winters BR, Maizlin II, Jobe BA, Roberts CT, Slifka MK, Marks DL. Depot-specific differences in inflammatory mediators and a role for NK cells and IFN-γ in inflammation in human adipose tissue. Int J Obes (Lond). 2009;33(9):978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maury E, Ehala-Aleksejev K, Guiot Y, Detry R, Vandenhooft A, Brichard SM. Adipokines oversecreted by omental adipose tissue in human obesity. Am J Physiol Endocrinol Metab. 2007;293(3):E656–E665. [DOI] [PubMed] [Google Scholar]
- 39. Zhang Y, Bossé Y, Marceau P, Biron S, Lebel S, Richard D, Vohl M-C, Tchernof A. Gene expression variability in subcutaneous and omental adipose tissue of obese men. Gene Expr. 2007;14(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vohl MC, Sladek R, Robitaille J, Gurd S, Marceau P, Richard D, Hudson TJ, Tchernof A. A survey of genes differentially expressed in subcutaneous and visceral adipose tissue in men. Obes Res. 2004;12(8):1217–1222. [DOI] [PubMed] [Google Scholar]
- 41. Ulrich CM, Gigic B, Böhm J, Ose J, Viskochil R, Schneider M, Colditz GA, Figueiredo JC, Grady WM, Li CI, Shibata D, Siegel EM, Toriola AT, Ulrich A. The ColoCare Study: a paradigm of transdisciplinary science in colorectal cancer outcomes. Cancer Epidemiol Biomarkers Prev. 2019;28(3):591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gigic B, Boeing H, Toth R, Böhm J, Habermann N, Scherer D, Schrotz-King P, Abbenhardt-Martin C, Skender S, Brenner H, Chang-Claude J, Hoffmeister M, Syrjala K, Jacobsen PB, Schneider M, Ulrich A, Ulrich CM. Associations between dietary patterns and longitudinal quality of life changes in colorectal cancer patients: the ColoCare Study. Nutr Cancer. 2018;70(1):51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Himbert C, Ose J, Lin T, Warby CA, Gigic B, Steindorf K, Schrotz-King P, Abbenhardt-Martin C, Zielske L, Boehm J, Ulrich CM. Inflammation- and angiogenesis-related biomarkers are correlated with cancer-related fatigue in colorectal cancer patients: results from the ColoCare Study. Eur J Cancer Care (Engl). 2019;28(4):e13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Himbert C, Ose J, Nattenmüller J, Warby CA, Holowatyj AN, Böhm J, Lin T, Haffa M, Gigic B, Hardikar S, Scherer D, Zielske L, Schrotz-King P, Kölsch T, Siegel EM, Shibata D, Ulrich A, Schneider M, Hursting SD, Kauczor HU, Ulrich CM. Body fatness, adipose tissue compartments, and biomarkers of inflammation and angiogenesis in colorectal cancer: the ColoCare Study. Cancer Epidemiol Biomarkers Prev. 2019;28(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Irlbeck T, Massaro JM, Bamberg F, O’Donnell CJ, Hoffmann U, Fox CS. Association between single-slice measurements of visceral and abdominal subcutaneous adipose tissue with volumetric measurements: the Framingham Heart Study. Int J Obes (Lond). 2010;34(4):781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nattenmueller J, Hoegenauer H, Boehm J, Scherer D, Paskow M, Gigic B, Schrotz-King P, Grenacher L, Ulrich C, Kauczor HU. CT-based compartmental quantification of adipose tissue versus body metrics in colorectal cancer patients. Eur Radiol. 2016;26(11):4131–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liesenfeld DB, Grapov D, Fahrmann JF, Salou M, Scherer D, Toth R, Habermann N, Böhm J, Schrotz-King P, Gigic B, Schneider M, Ulrich A, Herpel E, Schirmacher P, Fiehn O, Lampe JW, Ulrich CM. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare Study. Am J Clin Nutr. 2015;102(2):433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 49. R Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2016. Available at: https://www.R-project.org/.
- 50. Schmid R, Baum P, Ittrich C, Fundel-Clemens K, Huber W, Brors B, Eils R, Weith A, Mennerich D, Quast K. Comparison of normalization methods for Illumina BeadChip HumanHT-12 v3. BMC Genomics. 2010;11(1):349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ritchie ME, Dunning MJ, Smith ML, Shi W, Lynch AG. BeadArray expression analysis using Bioconductor. PLOS Comput Biol. 2011;7(12):e1002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fong Y, Huang Y, Lemos MP, Mcelrath MJ. Rank-based two-sample tests for paired data with missing values. Biostatistics. 2018;19(3):281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 54. Wang X, Terfve C, Rose JC, Markowetz F. HTSanalyzeR: an R/Bioconductor package for integrated network analysis of high-throughput screens. Bioinformatics. 2011;27(6):879–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6(2):65–70. [Google Scholar]
- 57. Pounds S, Cheng C. Sample size determination for the false discovery rate. Bioinformatics. 2005;21(23):4263–4271. [DOI] [PubMed] [Google Scholar]
- 58. Greulich F, Hemmer MC, Rollins DA, Rogatsky I, Uhlenhaut NH. There goes the neighborhood: assembly of transcriptional complexes during the regulation of metabolism and inflammation by the glucocorticoid receptor. Steroids. 2016;114:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hübner N, Evans RM. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49(1):158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kleiman A, Hübner S, Rodriguez Parkitna JM, Neumann A, Hofer S, Weigand MA, Bauer M, Schmid W, Schütz G, Libert C, Reichardt HM, Tuckermann JP. Glucocorticoid receptor dimerization is required for survival in septic shock via suppression of interleukin-1 in macrophages. FASEB J. 2012;26(2):722–729. [DOI] [PubMed] [Google Scholar]
- 61. Haffa M, Holowatyj AN, Kratz M, Toth R, Benner A, Gigic B, Habermann N, Schrotz-King P, Böhm J, Brenner H, Schneider M, Ulrich A, Herpel E, Schirmacher P, Straub BK, Nattenmüller J, Kauczor H-U, Lin T, Ball CR, Ulrich CM, Glimm H, Scherer D. Data from: Transcriptome profiling of adipose tissue reveals depot-specific metabolic alterations among patients with colorectal cancer. Zenodo Repository 2019. Deposited 3 June 2019. 10.5281/zenodo.3237786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lysaght J, van der Stok EP, Allott EH, Casey R, Donohoe CL, Howard JM, McGarrigle SA, Ravi N, Reynolds JV, Pidgeon GP. Pro-inflammatory and tumour proliferative properties of excess visceral adipose tissue. Cancer Lett. 2011;312(1):62–72. [DOI] [PubMed] [Google Scholar]
- 63. Catalán V, Gómez-Ambrosi J, Rodríguez A, Ramírez B, Ortega VA, Hernández-Lizoain JL, Baixauli J, Becerril S, Rotellar F, Valentí V, Moncada R, Silva C, Salvador J, Frühbeck G. IL-32α-induced inflammation constitutes a link between obesity and colon cancer. OncoImmunology. 2017;6(7):e1328338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thomas MJ, Sorci-Thomas MG. SAA: a link between cholesterol efflux capacity and inflammation? J Lipid Res. 2015;56(8):1383–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–591. [DOI] [PubMed] [Google Scholar]
- 66. Xiao J, Mazurak VC, Olobatuyi TA, Caan BJ, Prado CM. Visceral adiposity and cancer survival: a review of imaging studies. Eur J Cancer Care (Engl). 2018;27(2):e12611. [DOI] [PubMed] [Google Scholar]
- 67. Malietzis G, Aziz O, Bagnall NM, Johns N, Fearon KC, Jenkins JT. The role of body composition evaluation by computerized tomography in determining colorectal cancer treatment outcomes: a systematic review. Eur J Surg Oncol. 2015;41(2):186–196. [DOI] [PubMed] [Google Scholar]
- 68. Lippitz BE, Harris RA. Cytokine patterns in cancer patients: a review of the correlation between interleukin 6 and prognosis. OncoImmunology. 2016;5(5):e1093722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, Baumgartner KB, Gilliland FD, Sorensen BE, McTiernan A, Ulrich CM. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol. 2009;27(21):3437–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Paulsen SK, Pedersen SB, Fisker S, Richelsen B. 11β-HSD type 1 expression in human adipose tissue: impact of gender, obesity, and fat localization. Obesity (Silver Spring). 2007;15(8):1954–1960. [DOI] [PubMed] [Google Scholar]
- 71. Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, Landerholm RW, Crouthamel M, Gozal D, Hwang S, Singh PK, Becker L. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20(4):614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, Ferrante AW Jr. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18(6):816–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lesna IK, Cejkova S, Kralova A, Fronek J, Petras M, Sekerkova A, Thieme F, Janousek L, Poledne R. Human adipose tissue accumulation is associated with pro-inflammatory changes in subcutaneous rather than visceral adipose tissue. Nutr Diabetes. 2017;7(4):e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ross R, Léger L, Morris D, de Guise J, Guardo R. Quantification of adipose tissue by MRI: relationship with anthropometric variables. J Appl Physiol (1985). 1992;72(2):787–795. [DOI] [PubMed] [Google Scholar]
- 75. Camhi SM, Bray GA, Bouchard C, Greenway FL, Johnson WD, Newton RL, Ravussin E, Ryan DH, Smith SR, Katzmarzyk PT. The relationship of waist circumference and BMI to visceral, subcutaneous, and total body fat: sex and race differences. Obesity (Silver Spring). 2011;19(2):402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Neamat-Allah J, Wald D, Hüsing A, Teucher B, Wendt A, Delorme S, Dinkel J, Vigl M, Bergmann MM, Feller S, Hierholzer J, Boeing H, Kaaks R. Validation of anthropometric indices of adiposity against whole-body magnetic resonance imaging—a study within the German European Prospective Investigation into Cancer and Nutrition (EPIC) cohorts. PLoS One. 2014;9(3):e91586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abraham TM, Pedley A, Massaro JM, Hoffmann U, Fox CS. Association between visceral and subcutaneous adipose depots and incident cardiovascular disease risk factors. Circulation. 2015;132(17):1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Katzmarzyk PT, Heymsfield SB, Bouchard C. Clinical utility of visceral adipose tissue for the identification of cardiometabolic risk in white and African American adults. Am J Clin Nutr. 2013;97(3):480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McLaughlin T, Lamendola C, Liu A, Abbasi F. Preferential fat deposition in subcutaneous versus visceral depots is associated with insulin sensitivity. J Clin Endocrinol Metab. 2011;96(11):E1756–E1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Després J-P, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444(7121):881–887. [DOI] [PubMed] [Google Scholar]
- 81. Vague J. The degree of masculine differentiation of obesities: a factor determining predisposition to diabetes, atherosclerosis, gout, and uric calculous disease. Am J Clin Nutr. 1956;4(1):20–34. [DOI] [PubMed] [Google Scholar]
- 82. Neto NI, Murari AS, Oyama LM, Otoch JP, Alcântara PS, Tokeshi F, Figuerêdo RG, Alves MJ, Lima JD, Matos-Neto EM, Seelaender M, Oller do Nascimento CM. Peritumoural adipose tissue pro-inflammatory cytokines are associated with tumoural growth factors in cancer cachexia patients. J Cachexia Sarcopenia Muscle. 2018;9(6):1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zoico E, Rizzatti V, Darra E, Budui SL, Franceschetti G, Vinante F, Pedrazzani C, Guglielmi A, De Manzoni G, Mazzali G, Rossi AP, Fantin F, Zamboni M. Morphological and functional changes in the peritumoral adipose tissue of colorectal cancer patients. Obesity (Silver Spring). 2017;25(Suppl 2):S87–S94. [DOI] [PubMed] [Google Scholar]
- 84. Petruzzelli M, Schweiger M, Schreiber R, Campos-Olivas R, Tsoli M, Allen J, Swarbrick M, Rose-John S, Rincon M, Robertson G, Zechner R, Wagner EF. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metab. 2014;20(3):433–447. [DOI] [PubMed] [Google Scholar]
- 85. Ebadi M, Field CJ, Lehner R, Mazurak VC. Chemotherapy diminishes lipid storage capacity of adipose tissue in a preclinical model of colon cancer. Lipids Health Dis. 2017;16(1):247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Reggiani Bonetti L, Domati F, Farinetti A, Migaldi M, Manenti A. Radiotherapy-induced mesorectum alterations: histological evaluation of 90 consecutive cases. Scand J Gastroenterol. 2015;50(2):197–203. [DOI] [PubMed] [Google Scholar]
- 87. Schübel R, Nattenmüller J, Sookthai D, Nonnenmacher T, Graf ME, Riedl L, Schlett CL, von Stackelberg O, Johnson T, Nabers D, Kirsten R, Kratz M, Kauczor H-U, Ulrich CM, Kaaks R, Kühn T. Effects of intermittent and continuous calorie restriction on body weight and metabolism over 50 wk: a randomized controlled trial. Am J Clin Nutr. 2018;108(5):933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Campbell KL, Landells CE, Fan J, Brenner DR. A systematic review of the effect of lifestyle interventions on adipose tissue gene expression: implications for carcinogenesis. Obesity (Silver Spring). 2017;25(Suppl 2):S40–S51. [DOI] [PubMed] [Google Scholar]
- 89. Campbell KL, Foster-Schubert KE, Makar KW, Kratz M, Hagman D, Schur EA, Habermann N, Horton M, Abbenhardt C, Kuan LY, Xiao L, Davison J, Morgan M, Wang CY, Duggan C, McTiernan A, Ulrich CM. Gene expression changes in adipose tissue with diet- and/or exercise-induced weight loss. Cancer Prev Res (Phila). 2013;6(3):217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]