Abstract
Break-Induced Replication (BIR) is an important pathway specializing in repair of one-ended double-strand DNA breaks (DSBs). This type of DSB break typically arises at collapsed replication forks or at eroded telomeres. BIR initiates by invasion of a broken DNA end into a homologous template followed by initiation of DNA synthesis that can proceed for hundreds of kilobases. This synthesis is drastically different from S-phase replication in that instead of a replication fork, BIR proceeds via a migrating bubble and is associated with conservative inheritance of newly synthesized DNA. This unusual mode of DNA replication is responsible for frequent genetic instabilities associated with BIR, including hyper-mutagenesis, which can lead to the formation of mutation clusters, extensive loss of heterozygosity, chromosomal translocations, copy-number variations, and complex genomic rearrangements. In addition to budding yeast experimental systems that were initially employed to investigate eukaryotic BIR, recent studies in different organisms including humans, have provided multiple examples of BIR initiated within different cellular contexts, including collapsed replication fork and telomere maintenance in the absence of telomerase. In addition, significant progress has been made towards understanding microhomology-mediated BIR (MMBIR) that can promote complex chromosomal rearrangements, including those associated with cancer and those leading to a number of neurological disorders in humans.
Keywords: break-induced replication, alternative lengthening of telomeres, Rad51-dependent BIR, Rad51-independent BIR, MMBIR, half-crossover, mutation cluster
Pathways of double-strand break repair
Double-strand breaks (DSBs) are dangerous and potentially lethal events that can result from many sources including, a collapse of DNA replication, problems in chromosome segregation, and exposure to DNA damaging agents. The repair of DSBs is essential for cell survival and for preventing genome instability. However, some DSB repair pathways can also be destabilizing, which makes the choice of repair very important. In general, DSB repair occurs by non-homologous end joining (NHEJ) or by homologous recombination (HR). NHEJ heals breaks by joining two broken DNA ends (see (Rodgers and McVey, 2016) for further details). NHEJ has traditionally been considered error-prone because it does not use a homologous template for repair and often results in the loss of genetic material. In contrast, repair by HR utilizes a sister chromatid, a homologous chromosome, or a homologous region of another chromosome as a template to heal the break. Usually HR results in accurate repair, however, there are many cases where it leads to problems, including mutations and chromosomal rearrangements making HR repair an important contributor of genome instability (reviewed in (Malkova and Haber, 2012, Rodgers and McVey, 2016). HR repair is further subdivided into several pathways, including: (i) gene conversion (GC), which can occur via gap repair or by synthesis-dependent strand annealing (SDSA), (ii) single-strand annealing (SSA), and (iii) break-induced replication (BIR).
For the initiation of all HR repair pathways, DSB resection is required. In the case of GC and BIR, resection is followed by Rad51-mediated strand invasion of the broken DNA end into a donor DNA template, which leads to initiation of repair synthesis that restores the broken DNA back into double-stranded DNA (dsDNA). GC is associated with short patch repair synthesis, and can be also associated with crossing-over, which results in the exchange of genetic material at flanking regions, and can yield such phenomena as sister chromatid exchange, loss of heterozygosity, meiotic crossing-over, and translocations. Importantly, GC occurs in situations when both sides of the break are homologous to the donor and participate in repair. In contrast, BIR is employed when only one end of a break can find a homologous template for repair. This scenario may arise at eroded telomeres, in situations when a replication fork encounters nicked DNA that subsequently is converted into one-ended DSBs, or in cases where one of two broken DSB ends fails to find a homologous partner. BIR may also occur when two ends of a break find homologous templates independently of each other, for example at different ectopic positions.
Experimental systems to study BIR
BIR was first described as a pathway responsible for late replication in bacteriophage T4, and was named Recombination-Dependent Replication (RDR) (Luder and Mosig, 1982, Kreuzer et al., 1995, Kreuzer and Brister, 2010). RDR was also found in Escherichia coli, as a process responsible for stable DNA replication (Asai et al., 1993, Asai et al., 1994), DSB repair (Asai et al., 1993, Asai et al., 1994, Kuzminov and Stahl, 1999), repair of collapsed replication forks (Heller and Marians, 2006, Gabbai and Marians, 2010), and for the formation of adaptive mutations (Cairns and Foster, 1991, Harris et al., 1994). Overall, the large body of information about RDR in bacteria and bacteriophage systems have been thoroughly summarized previously (Malkova and Haber, 2012, Sakofsky et al., 2012, Malkova and Ira, 2013) and will not be reviewed here. In this review we will focus on BIR in eukaryotes, with a special emphasis on data that has recently become available, as well as on existing questions in the field that require further investigation.
Initially, the only eukaryotic organism where BIR was investigated was the budding yeast, Saccharomyces cerevisiae. Several different approaches were developed to study BIR in this organism. First, transformation of yeast cells with a linearized DNA fragment was used to initiate BIR by strand invasion of the broken fragment end into a homologous chromosomal region (Morrow et al., 1997). It was demonstrated that BIR initiated in this way can proceed for hundreds of kilobases leading to the stabilization of the fragment. The second approach employed HO site-specific endonuclease to induce a chromosomal DSB in such a way that only one broken DNA end could find homology in the yeast genome for strand invasion, which was followed by initiation of BIR (Malkova et al., 1996, Morrow et al., 1997, Bosco and Haber, 1998). This approach allowed for both genetic and physical monitoring of BIR, which led to the discovery of Rad51-dependent (Davis and Symington, 2004, Malkova et al., 2005) and Rad51-independent BIR (Malkova et al., 1996, Davis and Symington, 2004). In addition, this approach allowed the mode of BIR synthesis to be determined (Donnianni and Symington, 2013, Saini et al., 2013), as well as enabled a number of genomic destabilizing events resulting from BIR to be characterized (Deem et al., 2008, Sakofsky et al., 2014, Vasan et al., 2014, Sakofsky et al., 2015). A third approach used yeast strains that lacked telomerase, which maintained their telomeres using the alternative lengthening of telomeres (ALT) pathway that proceeds via BIR (Le et al., 1999, Lydeard et al., 2007). Additional insight into BIR became possible by the development of new assays where a site-specific DNA nick was introduced and later converted to a one-ended DSB during replication that can initiate BIR (Cortes-Ledesma and Aguilera, 2006, Nielsen et al., 2009, Katz et al., 2014, Mayle et al., 2015). Investigation of DNA repair initiated by replication fork collapse in other studies has also provided important information about BIR, as well as about microhomology-mediated BIR (MMBIR), a variation of the BIR pathway (Lemoine et al., 2005, Narayanan et al., 2006, Payen et al., 2008, Kim et al., 2017).
Recently, the development of several new experimental systems has allowed the investigation of BIR in higher eukaryotes. First, a system in Xenopus laevis egg extracts has been used to study how replication restarts following replication-induced one-ended DSBs (Hashimoto and Costanzo, 2011). In this system, replication stress from aphidicolin resulted in the accumulation of single-stranded (ss) DNA which was cut by a single-strand specific nuclease (S1 or mung bean), thus generating a single-end break. The authors observed the restoration of the replisome by a Rad51-dependent BIR-like pathway.
Also, a reporter-based system employed in mammalian cell lines enabled the first genetic study of BIR to be conducted in mammalian cells (Costantino et al., 2014). Additional insight came from studies of long-tract gene conversion (LTGC) that is activated in BRCA1-defective mammalian cells and appears mechanistically similar to BIR (Chandramouly et al., 2013). Further studies will be needed however, to determine if the properties of BIR (e.g., long, conservative replication) observed in yeast are similar to what has been observed in higher eukaryotes. Also, a number of important findings has recently come from studies of ALT (proceeding via BIR) performed in human cancer cells lacking telomerase (Cho et al., 2014, Dilley et al., 2016, Garcia-Exposito et al., 2016, Roumelioti et al., 2016), and even in normal somatic mammalian cells (Neumann et al., 2013). Finally, investigation of repair DNA synthesis initiated by replication collapse at mammalian fragile sites has recently become an important source of information about BIR and MMBIR (Minocherhomji et al., 2015, Bhowmick et al., 2016).
Initiation of BIR.
Budding yeast was the first eukaryotic organism used to study initiation of BIR and therefore, we will begin summarizing important findings obtained in this organism. In particular, it was demonstrated that BIR starts by 5’−3’ DSB resection that can be extensive and leads to the formation of a 3’ single-stranded DNA initiating the search for a homologous DNA template (Figure 1A) (Chung et al., 2010). The interaction between the exposed ssDNA and homologous template requires Rad52 and can proceed via Rad51-dependent or Rad51-independent BIR pathways (Malkova et al., 1996, Davis and Symington, 2004, Malkova et al., 2005). The Rad51-dependent pathway is more efficient, requires significant (more than 200bp) regions of homology between recombining DNA molecules (Lydeard et al., 2007), and involves Rad55, Rad57, and Rad54 (Signon et al., 2001).
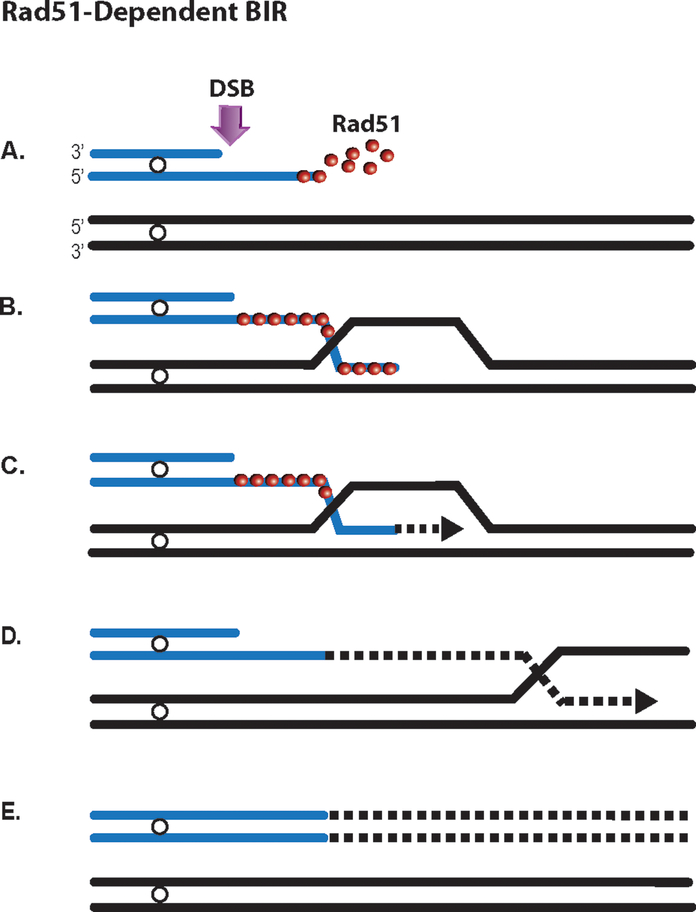
Figure 1:
Model of Rad51-dependent BIR. A. 5’ to 3’ resection of a one-ended DSB generating a 3’ ssDNA end (blue), where the assembly of Rad51 filaments takes place. B. Invasion of the 3’ end containing Rad51 nucleoprotein filament into a homologous chromosome (black), followed by D-loop formation. C. Initiation of BIR DNA synthesis using a homologous chromosome as a template. Nascent DNA represented by dotted lines. D. Progression of DNA synthesis with the migration of the BIR replication bubble, generating a long, persistent ssDNA tail behind the bubble. E. Completion of BIR associated with conservative inheritance of a newly synthesized strand (Donnianni and Symington, 2013, Saini et al., 2013). A color version of the figure is available online.
Rad51-independent BIR is a less efficient pathway, and this pathway, in addition to Rad52, also depends on Rad59 as well as on Mre11-Rad50-Xrs2 complex and on Rdh54 Swi2/Snf2 chromatin remodeler complex (Malkova et al., 1996, Signon et al., 2001). Rad51-indepenent Rad52-dependent BIR was first described in yeast diploids where an HO-induced DSB was introduced in the middle of chromosome III (Malkova et al., 1996). Despite the large regions of homology between homologous chromosomes, recombination induced in rad51Δ diploids almost never occurred at the break, likely due to the inability of efficient strand invasion in the absence of Rad51. Instead, the broken ends were resected back 30kb or more, until two closely spaced inverted Ty1 elements were exposed (Malkova et al., 2001, VanHulle et al., 2007, Downing et al., 2008). This promoted inter-sister single-strand annealing between inverted Ty1 repeats resulting in the formation of dicentric inverted dimers, which subsequently induced a cycle of chromosome breakage and recombination events, which allowed to maintain the broken chromosome through multiple cell generations leading eventually to a variety of chromosomal rearrangements. These rearrangements resulted from Rad51-independent BIR that proceeded via strand invasions into multiple positions of Ty1 elements dispersed throughout different regions of the genome.
Downing et al. proposed that Rad51-independent BIR proceeds via Rad52-mediated annealing of a broken DNA end to ssDNA regions that are homeologous or contain only small amounts of homology, and that are formed from secondary DNA structures, replication, or transcription (Figure 4A) (Downing et al., 2008). These authors observed that Rad51-independent BIR was suppressed in rad54Δ strains where Rad51 filaments could form, but was activated in both rad51Δ and rad51Δ rad54Δ double mutants. Based on these observations, the authors proposed that so-called Rad51-independent BIR is actually suppressed by Rad51 filaments, which preclude interactions between DNA containing only limited homology. BIR operating at short regions of homology that is also suppressed by Rad51 was proposed to mediate repair initiated by HO cleavage between inverted repeats that shared only 33-bp homology on plasmid DNA (Ira and Haber, 2002). The authors proposed that these events proceed via Rad51-independent BIR followed by SSA. The preference of Rad51-independent BIR for short regions of homology (or homeology) allowed Hastings et al. to propose that this pathway is similar or overlaps with another branch of BIR called microhomology- mediated BIR, or MMBIR (Hastings et al., 2009a). Rad51-independent MMBIR that was at least in part dependent on Rad52 was an underlying source for the formation of segmental duplications in yeast initiated by replication stress (Payen et al., 2008). In addition, recent data suggest that BIR often proceeds via a Rad52-dependent Rad51-independent mechanism in mammals when breaks occur at telomeres (ALT), or when cells experience replication stress from replication fork breakage at fragile sites or from overexpression of oncogenes (Costantino et al., 2014, Minocherhomji et al., 2015, Bhowmick et al., 2016, Dilley et al., 2016, Sotiriou et al., 2016). Importantly, BIR in mammals also may proceed via annealing at microhomologies, and the presence of Rad51 seems to suppress this Rad51-independent MMBIR repair (see below for more details). This pathway was proposed as a mechanism responsible for complex genomic rearrangements detected in patients with a number of neurological diseases (reviewed in (Carvalho and Lupski, 2016)). Hastings et al. proposed that such MMBIR events can be stimulated by temporal suppression of Rad51(Hastings et al., 2009b), while Carvahlo et al. proposed that initiation of BIR can lead to a switch to MMBIR that can proceed without dependence on Rad51 (Carvalho et al., 2013, Carvalho et al., 2015, Carvalho and Lupski, 2016). Overall, BIR appears to be especially dangerous when it is initiated in the absence of Rad51 because it proceeds by annealing at microhomologies mediated by Rad52. This pathway is likely stimulated by conditions that promote the formation of ssDNA templates. One recent example supporting this idea comes from the demonstration that an increased level of TERRA transcription (transcription of RNA encoded by telomeres) causes massive formation of R-loops, which are structures induced by formation of RNA-DNA hybrids that lead to exposure of ssDNA regions. The formation of these R-loops stimulates Rad51-independent BIR at telomeres (Yu et al., 2014) (see below for details). Overall, Rad51-independent BIR is much less efficient in yeast as compared to Rad-51-dependent BIR, which makes it difficult to study its mechanism at the molecular level.
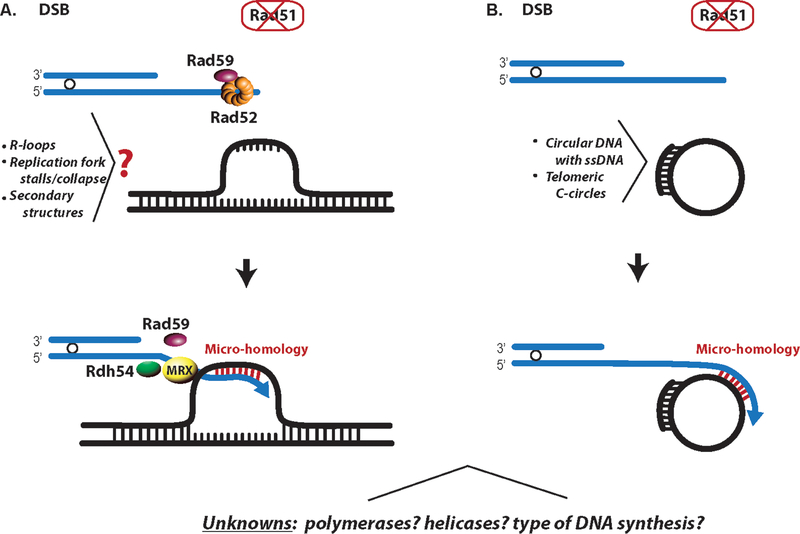
Figure 4:
Rad51-independent BIR. A. A model of Rad51-independent BIR in yeast mediated by Rad52, Rad59, Rdh54 and the MRX complex in the absence of Rad51 where the 3’ end of a one-ended broken chromosome anneals to regions of transient ssDNA formed as a result of secondary DNA structures, R-loops, or stalled/collapsed replication forks. Annealing to ssDNA likely occurs at positions of microhomology. B. Model of Rad51-independent BIR to explain alternative lengthening of telomeres in human cells (similar to Type II-ALT events in yeast). ALT is initiated by annealing of resected uncapped telomere end to ssDNA in telomeric C-circles (as described in (Cesare and Reddel, 2010, Dilley and Greenberg, 2015)). A color version of the figure is available online.
Unusual DNA synthesis associated with BIR
The delayed initiation of BIR synthesis
DNA synthesis has been studied in detail for only Rad51-dependent BIR, and therefore this is the pathway we will focus on here. The initiation of BIR synthesis can take several hours longer as compared to gene conversion, while the exact kinetics of BIR initiation and the extent of the delay has varied for different experimental systems (Malkova et al., 2005, Donnianni and Symington, 2013). The long delay in initiation of BIR was proposed to result from a recombination execution checkpoint (REC) expressed in response to the absence of the second DSB end capable of participating in repair (Jain et al., 2009). While the details of how this checkpoint functions remain unknown, it appears that two helicases, Sgs1 and Mph1, mediate its operation (Jain et al., 2009, Jain et al., 2016). When only one side of a break invades a homologous region, these helicases may promote multiple rounds of dissociation and reinvasion of the broken DNA end into its template. Importantly, the double deletion of MPH1 and SGS1 significantly accelerates the initiation of BIR (Jain et al., 2016). In addition, the absence of Mph1 can also shift the competition between GC and BIR in the direction of BIR (Mehta et al., 2016). The idea of multiple rounds of strand invasions was proved by the discovery of Symington and colleagues who analysed BIR using the chromosome fragmentation assay in diploid cells with polymorphic chromosome III homologues (Smith et al., 2007). They observed that more than 20% of BIR products were derived from both copies of chromosome III, which led them to conclude that BIR undergoes frequent cycles of strand invasions called template switching. Importantly, deletion of MPH1 eliminates template switching, consistent with a key role of this helicase in dismantling D-loops leading to an interruption of BIR. (Stafa et al., 2014). In addition, the overall increase of BIR efficiency in the absence of Mph1 protein, suggested that multiple cycles of strand invasions may occur even in situations when template switching was not detected (Stafa et al., 2014, Mehta et al., 2016). Another important observation was that template switching events only occurred over a 10-kb region of initial BIR synthesis (Smith et al., 2007). Based on this result, the authors proposed that stabilization of BIR synthesis occurs after approximately 10-kb of synthesis, however, the reasons for this stabilization remain unknown. Also, Anand et al. examined similar template switching using a yeast system where a single DSB was repaired by a two-step process that first involved BIR, which was then followed by a template switch that led to the formation of complex translocations that reconstituted a URA3 gene (Anand et al., 2014). In particular, they observed that template switching events required less homology between recombining molecules and was less dependent on Rad51 than the original strand invasion, but was instead dependent on Rdh54 protein.
The unusual mode of DNA synthesis.
From the time BIR was discovered, two major models of its DNA synthesis were postulated. According to the first model, the D-loop was resolved to form a normal replication fork associated with synchronous leading and lagging strand synthesis and semi-conservative inheritance of newly-synthesized strands. In the second model, BIR is carried out by a migrating D-loop (bubble), where branch migration of an unresolved Holliday junction (HJ) promotes the displacement of a newly synthesized strand and therefore leads to conservative inheritance of nascent DNA (Figure 1) (reviewed in (Anand et al., 2013, Malkova and Ira, 2013)). The second model was found to be true for BIR that was initiated in yeast by HO-induced DSBs and that proceeded by using another chromosome as a template during the G2 stage of the cell cycle (Donnianni and Symington, 2013, Saini et al., 2013, Wilson et al., 2013). The main molecular intermediate of this synthesis, a migrating bubble, was detected using two-dimensional gel electrophoresis, while the conservative mode of synthesis was demonstrated using molecular combing analysis and by a combination of Southern and Western hybridization techniques. Another important feature of BIR synthesis is the asynchrony between leading and lagging strand synthesis (Saini et al., 2013). In particular, only the leading strand synthesis is initiated by the extension of the 3’-OH broken DNA end, and is accumulated as ssDNA behind the BIR bubble. Later, this strand serves as a template for lagging strand synthesis, a process that remains uncharacterized in BIR. In particular, it remains unclear when lagging strand synthesis starts and how it proceeds. For example, it is unknown whether lagging strand synthesis proceeds via the formation of Okazaki fragments as discontinuous synthesis, or as continuous synthesis beginning from the terminal end of the leading strand followed by contiguous synthesis to its completion. It is likely that the HJ structure formed at the beginning of BIR remains unresolved and that bubble migration proceeds until the end of the chromosome. This suggestion came from the observation that a single deletion of a gene encoding a structure-specific nuclease, either MUS81 or YEN1, had a very small effect on BIR that was initiated by HO endonuclease at G2 stage of the cell cycle, and which proceeded by using another chromosome, while the deletion of both of these genes still only led to a small decrease in BIR efficiency (Wilson et al., 2013). The small decrease of BIR efficiency in the double mutant might reflect the need for these enzymes to resolve the intermediates of stalled BIR to enable BIR to re-start. In either case, however, it is the unusual mode of DNA synthesis during BIR that is the underlying source of many different BIR-associated genetic instabilities, including mutations and chromosomal rearrangements (see below). Interestingly, Mus81 is capable of resolving intermediates of BIR initiated during S-phase by a replication collapse through an ssDNA nick that initiates BIR progression using a sister chromatid as a template (Mayle et al., 2015). In this situation, the resolution promoted by Mus81 may interrupt BIR, which allows a colliding replication fork to reach the region of BIR collapse, or it may promote the establishment of a normal replication fork. The molecular mechanisms behind the differential resolution of BIR intermediates during G2 vs S-phase of the cell cycle remain unclear.
DNA polymerases
Studies in budding yeast demonstrated that all three replicative polymerases, Polα, Polδ and Polε participate in BIR (Lydeard et al., 2007). However, their exact roles and the distribution of labor between these enzymes in BIR remain unclear. Despite the gaps in our knowledge, at this point there has been a lot of evidence in support of Polδ playing a very important role in BIR (Lydeard et al., 2007, Deem et al., 2008, Smith et al., 2009). It likely performs the bulk of leading strand synthesis, and may participate in lagging strand synthesis. A specific feature of BIR is its dependence on Pol32, a subunit of Polδ which is not essential for S-phase DNA synthesis. The exact reason behind the requirement for Pol32 during the course of BIR remains unclear. During S-phase replication, Pol32 enables Polδ to carry out DNA synthesis via strand displacement (Stith et al., 2008), thus it possible that BIR leading strand synthesis executed by Polδ is mechanistically similar to this displacement mode of synthesis (reviewed in (McVey et al., 2016). Pol32 is also required for Rad51-independent BIR, including MMBIR (Payen et al., 2008), Rad51-indepenent BIR promoting alternative telomere maintenance in yeast and humans (Lydeard et al., 2007, Dilley et al., 2016), and BIR initiated in human cells by the collapse of a replication fork (Costantino et al., 2014, Minocherhomji et al., 2015, Bhowmick et al., 2016, Sotiriou et al., 2016). In general, the dependence on Pol32 has become a key factor allowing BIR to be implicated in various DNA repair and/ or chromosome rearrangement pathways. However, this criterion should be used with caution since Pol32 was also shown to impact other repair pathways, including gene conversion and MMEJ (Jain et al., 2009, Villarreal et al., 2012). The exact role of Polε in BIR remains unclear. Polε is not required for initial BIR synthesis, but is needed for efficient BIR to continue after the first several kilobases of synthesis even though at least 25% of the final BIR product can be synthesized in its absence (Lydeard et al., 2007). In addition, the non-essential subunits of Polε, Dpb3 and Dpb4, are not required for BIR (Lydeard et al., 2010).
BIR requires Polα-primase and two factors that help Polα remain processive, Mcm10 and Ctf4 (Lydeard et al., 2007, Lydeard et al., 2010). In addition, several other factors known as pre-replication origin-loading factors, such as Dpb11-Sld2, and Sld3 are also involved in BIR. Importantly, without Polα-primase and all of these other factors, even the initial extension of the 3’ end (the start of leading strand synthesis) was not observed. While the involvement of Polα-primase in lagging strand synthesis is expected, it remains unclear why it would be required to initiate the synthesis of the leading strand since this strand contains a 3’-OH end, which by itself is sufficient to prime DNA synthesis. To explain this puzzling result, Lydeard et al. proposed that leading and lagging strand synthesis are tightly coordinated, and therefore leading strand synthesis cannot proceed when lagging strand synthesis is halted due to the absence of Polα-primase. However, in later studies, it became clear that BIR lagging strand synthesis is dis-coordinated from the leading strand (Saini et al., 2013), and therefore other possibilities to explain the role of Polα in BIR should be considered. It is possible that the invading 3’-OH DNA does not make a good primer because it is either not well annealed to the template, it is degraded, or it just cannot serve as a good substrate for Polδ to initiate replication. It was also suggested that Polδ cannot efficiently initiate synthesis in the context of a D-loop (reviewed in (McVey et al., 2016). For example, in DT40 chicken cells, as well as in mammalian cells, synthesis inside the D-loop is started by translesion polymerase Polη, and only later continued by Polδ. Experiments in yeast found no BIR defects in the absence of Polη or Polζ (another translesion polymerase) (Deem et al., 2011), but it remains possible that these polymerases participate. Finally, it is possible that leading strand BIR synthesis can in fact be initiated without Polα, but the intermediates of this reaction (with only leading strand initiated) cannot be detected due to their instability.
DNA helicases and other replication proteins
Another important and not fully answered question concerns the identity and respective roles of helicases supporting the movement of the BIR bubble. Lydeard et al. proposed that BIR, similar to S-phase replication, uses the Mcm2–7 helicase complex as its main replicative helicase (Lydeard et al., 2010). In addition, they showed that initiation of BIR also requires Cdc45, Cdt1, and the GINS complex, which all participate in the initiation of S-phase replication at replication origins. However, later experiments performed by Ira and colleagues came to a different conclusion, possibly due to the utilization of a different assay. They found that the MCM complex is in fact not essential for BIR (Wilson et al., 2013). Instead, they determined that another helicase, Pif1, is essential for long-range BIR and in its absence BIR cannot proceed beyond 20kb of synthesis (see also in (Saini et al., 2013)). However, the specific role that Pif1 is playing during BIR is not fully understood. Several different and not mutually exclusive roles of Pif1 in BIR have been proposed, including operating ahead of the replication bubble to unwind duplex DNA, recruiting and stabilizing Polδ to the BIR bubble, and tracking behind the replication bubble to ‘pull’ the newly synthesized DNA strand in order to overt topological hindrance of the nascent strand with the template strand (Wilson et al., 2013). Overall, future studies will be needed to better understand how BIR helicase functions are distributed between MCM, Pif1, and/or other helicases.
Similar to normal DNA replication, PCNA plays an important role in BIR. Interestingly, Lydeard et al. identified that BIR-specific mutations in PCNA, pol30-FF248,249AA, suppress BIR, but not S-phase replication (Lydeard et al., 2010). Surprisingly, these mutations are dominant, suggesting that even one mutant subunit in a PCNA trimer can make BIR defective. Another important participant of BIR is the single-stranded DNA binding protein, RPA. The requirement of RPA stems from the need to protect single-strand DNA that accumulates and persists in large amounts during BIR due to extensive DSB resection and during leading strand synthesis. Symington and colleagues observed that hypomorphic alleles of RFA1, the gene encoding the large RPA subunit, caused a profound BIR defect, while S-phase DNA replication and gap repair were either mildly affected, or not at all (Ruff et al., 2016). The observed BIR defect was consistent with a problem at the beginning of BIR when strand invasion is occurring. The authors proposed that the deficit of RPA binding led to problems of Rad51 loading, thus compromising strand invasion and leading to the loss of the 3’ssDNA invading end, which lacks protection by RPA thus allowing degradation by the Sgs1/Dna2 complex.
Hyper-mutagenesis associated with BIR
DNA synthesis associated with BIR is extremely imprecise. The first implication that BIR was mutagenic came from the observation of frequent template switching events at the beginning of BIR synthesis that were explained by low processivity of BIR (Smith et al., 2007). These template switches were restricted to the first 10kb of BIR, after which a more processive BIR was established. Interestingly, despite even an apparently processive BIR, the level of mutagenesis still remains high. This was first demonstrated by Deem et al. using a disomic yeast experimental system wherein lys2 frameshift reporters were inserted in a chromosome that served as a template during BIR at different distances from the place of BIR strand invasion (Deem et al., 2011). Deem et al. observed that the frequency of frameshifts during BIR was approximately 1000-fold higher than in normal DNA replication, and this high increase of frameshifts was observed at all positions throughout the track of BIR (Deem et al., 2011). The majority of mutations were deletions of one nucleotide and were consistent with polymerase slippage (Figure 2A). Some of the mutations were likely formed as a result of jumps between quasi-palindromic sequences. Importantly, Polδ, the main replicative polymerase, was responsible for the majority of frameshift mutations associated with BIR. The likely reason for the reduced fidelity of Polδ is a bubble-migration mechanism that may lead to increased frequency of Polδ dissociations from the template. In addition, a bubble migration mechanism associated with rapid dissociation of newly synthesized DNA from its template likely interferes with mismatch repair (MMR) of mispairing errors resulting from BIR. Indeed, while MMR is able to correct some BIR errors, its efficiency is significantly lower as compared to MMR efficiency during S-phase replication (Deem et al., 2011). Additionally, another contributor to increased frameshifts during BIR was found to be increased nucleotide pools at the time of BIR (Deem et al., 2011).
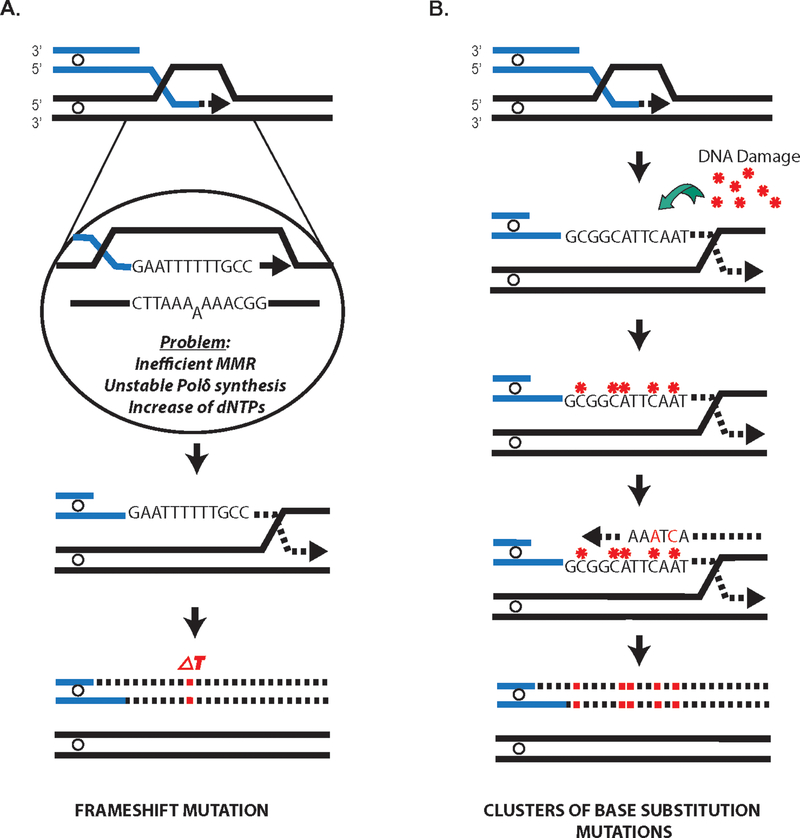
Figure 2:
Mechanisms of BIR-induced mutagenesis. A. Generation of frameshift mutations during BIR at the position of a homo-nucleotide run (Deem et al., 2011). Frameshift mutations result from decreased fidelity of Polδ, mismatch repair defect and increased dNTP levels (Deem et al., 2011). B. Formation of closely spaced (clusters) of base pair substitutions resulting from damage in ssDNA accumulating behind the BIR bubble followed by error-prone DNA lesion bypass during lagging strand BIR synthesis that leads to mutations (see (Sakofsky et al., 2014; Saini et al., 2013) for details). Red stars indicate ssDNA lesions, while red squares indicate mutated DNA bases. A color version of the figure is available online.
Importantly, base substitutions also increase during BIR. In particular, the rate of base substitutions in the ura3–29 reporter placed on the track of BIR, which is capable of reverting to Ura+ via C→T, C→G, and C→A base substitutions, increased more than 500 times as compared to reversions during S-phase DNA replication (Saini et al., 2013). The increase of base substitutions resulted from another unusual property of the BIR mechanism: the asynchrony between leading and lagging strand synthesis that results in long ssDNA regions accumulating during leading strand synthesis behind the BIR bubble (Figure 2B) (Saini et al., 2013). It is known that ssDNA is mutagenic, particularly in combination with DNA damaging agents (Yang et al., 2008). This is because lesions formed in ssDNA go unrepaired, since DNA repair pathways usually require another, undamaged DNA strand as a template for repair. Therefore, DNA damage tend to accumulate in ssDNA and can be bypassed by translesion synthesis polymerases that incorporate erroneous bases opposite the lesion (Yang et al., 2008, Roberts et al., 2012, Sakofsky et al., 2014). In the case of BIR, the long stretch of ssDNA accumulating behind the replication bubble is indeed highly vulnerable to DNA damage, and constitutes the major source of hyper-mutagenesis associated with BIR. In particular, using a ura3–29 reversion assay, Saini et al. demonstrated that the mutagenic effect of BIR synergized with the effect of a ssDNA specific mutagen, methyl methanesulfonate (MMS) that attacks cytidines and adenosines in ssDNA at the N3 and N1 positions, respectively, creating highly mutagenic DNA damage (Saini et al., 2013). Importantly, the synergistic effect (20-fold increase of Ura− to Ura+ reversions upon exposure to MMS as compared to cells undergoing BIR without MMS exposure) was specifically observed for one orientation of the ura3–29 reporter that placed the cytidine in the mutant position in ssDNA (leading strand) formed by BIR synthesis. These data provided convincing evidence that DNA damage in the context of ssDNA generated during BIR is a potent source of hyper-mutagenesis. In another study, the cells undergoing BIR were exposed to MMS, and the unselected BIR outcomes were analyzed by whole genome sequencing (Sakofsky et al., 2014). The authors observed that BIR was a potent source of mutation clusters that shared similar characteristics to mutation clusters found in several types of cancers that resulted from ssDNA damage introduced by APOBEC cytidine deaminases (Nik-Zainal et al., 2012, Roberts et al., 2012, Roberts et al., 2013). The mutation clusters observed in association with BIR were almost exclusively localized to the chromosome III region surrounding the HO-induced DSB that initiated BIR and extended up to 115kb in length (out of approximately ~ 120kbs that were potentially copied during BIR in the experimental assay). Also, as predicted from the established MMS mutation specificity, mutations in BIR clusters were located at positions of cytidines and also (less frequently) at the positions of adenosines, specifically in the leading strand which persists as ssDNA during BIR synthesis. In addition, mutation clusters detected in this study were frequently associated with chromosomal rearrangements that likely resulted from pausing of the BIR bubble at the positions of DNA damage in the BIR template. This was similar to APOBEC-induced clusters observed in cancer cells that are also frequently associated with chromosomal rearrangements (reviewed in (Roberts and Gordenin, 2014, Chan and Gordenin, 2015)). Overall, it is likely that BIR is a prominent source of mutation clusters not only in yeast, but also in cancer genomes where APOBEC enzymes could damage ssDNA accumulated during BIR.
BIR promotes half-crossovers and non-reciprocal translocations
The replication bubble established during BIR is highly unstable and is a potent contributor of chromosomal rearrangements. Two major types of chromosomal rearrangements associated with BIR include (i) non-reciprocal translocations (NRTs) resulting from strand invasion into a non-allelic position (Figure 3A) (Bosco and Haber, 1998, Lydeard et al., 2007), and (2) half-crossovers (HC), resulting from fusions between donor and recipient chromosomes participating in BIR (Figure 3B) (Deem et al., 2008, Smith et al., 2009). NRTs result from BIR that proceeds via invasion of repetitive DNA, typically abundant throughout the genome, such as transposable elements (Ty and delta elements in yeast). Excessive resection at a DSB prior to BIR initiation can expose repetitive elements that allow recombination with other repetitive elements located at ectopic positions resulting in BIR-induced NRT events (Figure 3A) (VanHulle et al., 2007, Deem et al., 2008, Vasan et al., 2014).
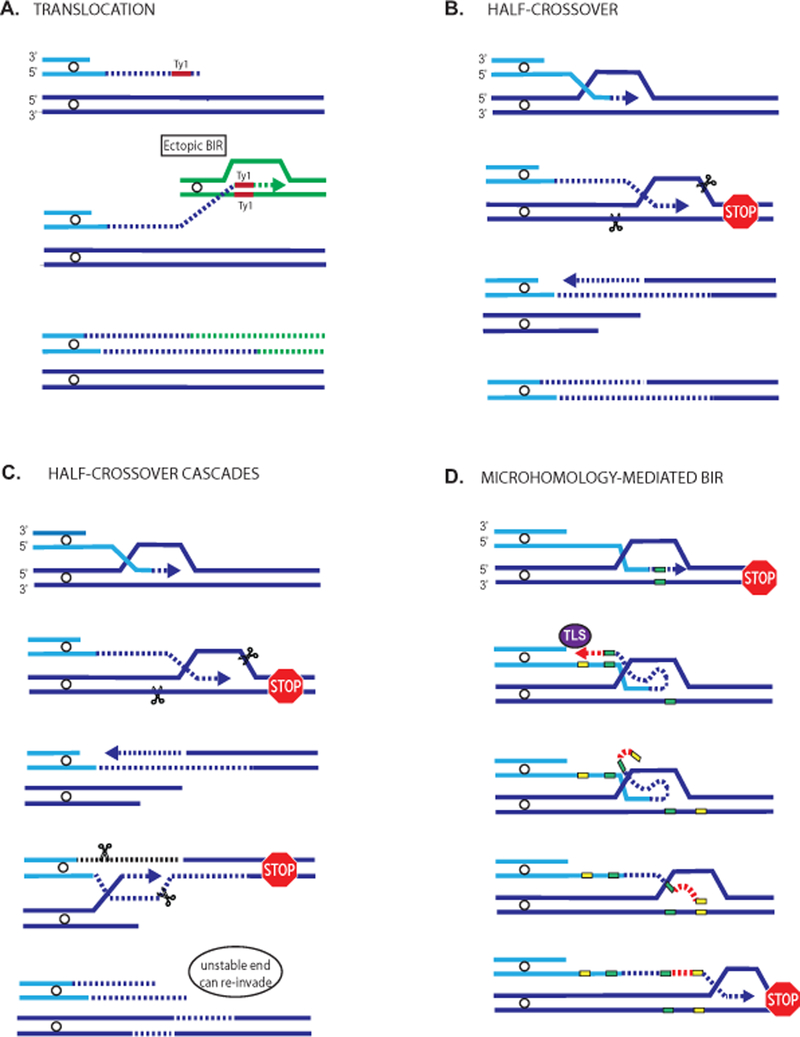
Figure 3:
Mechanisms of chromosomal rearrangements associated with BIR. A. A simple non-reciprocal translocation (NRT) resulting from long ssDNA resection at a DSB exposing repetitive DNA (Ty elements shown in red rectangles) that can initiate BIR at an ectopic position (non-homologous chromosome represented in green) through annealing of Ty elements. B. Formation of half-crossovers resulting from interruption of BIR that promotes a resolution of the migrating bubble, and resulting in fusion of the recombining portions of recipient and donor chromosome (shown in light blue and dark blue respectively). The remaining portion of the broken donor chromosome can either be lost as depicted in (B), or it can invade another donor at ectopic position (shown in green) leading to translocation (A). The invasion at ectopic position usually occurs at position of Ty or other repeated element. (C). The formation of half-crossover is followed by secondary BIR event resulting from the invasion of the broken donor into the half-crossover product, which can perpetuate breaking and re-invading cycles leading to half-crossover cascades (as described in (Vasan et al., 2014), or cycles of NRTs (not illustrated). D. Microhomology-mediated BIR resulting from a collapse of BIR synthesis resulting in dissociation of 3’ end from its template followed by annealing at microhomology in the region of ssDNA accumulated behind the BIR bubble. Using this annealing intermediate as a primer, TLS polymerases initiate DNA synthesis that eventually disrupts causing a second template switch by re-annealing to the original track of BIR through microhomology as described in (Sakofsky et al., 2015). Yellow and green boxes show regions of microhomology that can consist of 1–10bp. A color version of the figure is available online.
Formation of HCs results from interruption of BIR, leading to the resolution of BIR intermediates followed by the fusion between parts of recombining molecules while other parts remain broken or lost. HCs are formed in situations where initiation of BIR (strand invasion) occurs, but DNA synthesis is compromised (Figure 3B). For example, HCs were frequent in yeast strains containing Polδ mutations, including pol3-ct, pol3-Y708A, and pol32Δ that hindered initiation of DNA synthesis (Deem et al., 2008, Smith et al., 2009, Vasan et al., 2014). In addition, a pol3-t mutation, which is known to decrease processivity of Polδ, also promoted an increase in HCs (Vasan et al., 2014). An increase in the incidence of HCs have also been observed when BIR initiates in cells with defective or absent Pif1 helicase. Pif1 is essential for long-range BIR synthesis, and in its absence BIR synthesis is interrupted within the first 20kb, leading to rearrangements (Saini et al., 2013, Wilson et al., 2013, Sakofsky et al., 2015). Also, BIR pausing induced by DNA damage in the BIR template, can promote HC formation (Vasan et al., 2014). It was suggested that unrepaired damage in double-strand DNA ahead of BIR replication inhibits progression of the BIR replication bubble, thus promoting HCs. Finally, a premature onset of mitosis due to compromised checkpoint response in cells undergoing BIR also stimulated HC formation (Vasan et al., 2014).
Importantly, in addition to fusions between recombining molecules, HCs also lead to breakage of previously intact donors, which can initiate recurrent cycles of genetic instability. In particular, the broken fragment can invade another template located at an allelic or ectopic position, which starts an HC-initiated cascade (HCC) (Figure 3C). When yeast were exposed to high levels of alkylating damage from MMS, complex genetic events resulting from multiple chromosomal recombination gave rise to clusters of mutations associated with HCC events (Sakofsky et al., 2014). In addition, when strand invasion occurs at ectopic positions, HCC can lead to cycles of NRTs. These cycles resemble the cycles of NRTs in cancer cells (also called a cycles of telomere acquisition), where the breakage of one chromosome initiates recombination leading to perpetuating cycles of a breakage and re-invasion (Sabatier et al., 2005). In addition to cycles of NRTs, simple NRTs and even HCs have also been associated with rearrangement events in cancer and other diseases leading to the loss of heterozygosity (LOH) in cancer cells, or to the absence of heterozygosity (AOH) described in neurological diseases (see for details (Carvalho et al., 2015, Carvalho and Lupski, 2016)).
The role of BIR in recovering collapsed replication forks
BIR has always been implicated in the restart of collapsed replication forks. Fork collapse can occur when a replication fork encounters a single-stranded DNA nick, which is a major type of spontaneous DNA damage. This situation leads to the formation of a single-ended double strand break that needs to be repaired by HR. In E. coli, where replication involves two replication forks moving in opposite directions from a single replication origin and terminating at a single locus, broken replication forks are indeed recovered by a process similar to BIR called recombination-induced replication (RDR). During RDR, recovery proceeds with the invasion of a sister chromatid by the broken chromosome end followed by the assembly of a new replication fork (reviewed in (Sakofsky et al., 2012, Anand et al., 2013, Malkova and Ira, 2013)). In E.coli, RDR is an essential process since it is required for the recovery of broken forks in this organism.
BIR is also able to re-start broken replication forks in eukaryotes. For example, studies using Xenopus laevis egg extracts directly demonstrate that BIR can re-start collapsed replication forks in vertebrates (Hashimoto and Costanzo, 2011). However, eukaryotic cells are less dependent on BIR for the recovery of broken replication forks because they have multiple replication origins per chromosome, which eliminates the requirement for extensive DNA synthesis from a broken fork. Indeed, a direct experiment where replication was forced to proceed through a ssDNA nick initiated by a modified FLP nuclease, showed that mutagenic BIR was initiated, but rapidly terminated upon very short synthesis (Mayle et al., 2015). The authors demonstrated that two mechanisms contributed to the interruption of mutagenic BIR initiated by the collapse of a replication fork: (i) the convergence of a replication fork coming from the opposite direction of a BIR D-loop, and (ii) the cleavage of BIR intermediates by Mus81 structure-specific nuclease. The important role that converging replication forks and Mus81 cleavage play in the recovery of collapsed replication forks was consistent with reports from several other studies (Doe et al., 2002, Roseaulin et al., 2008, Munoz-Galvan et al., 2012, Nguyen et al., 2015). Mayle et al. concluded that mutagenic BIR is limited to only short-track synthesis, while long-track BIR is precluded (Mayle et al., 2015). However, long-track BIR was observed when replication collapse occurred in mus81Δ mutants in sub-telomeric chromosomal regions, with no replication forks coming from the opposing side. In this situation, the long-range BIR was similar to the one described in G2 phase by the fact that they both involved continuous DNA synthesis to the end of a chromosome, both were dependent on POL32 and PIF1, and both were associated with hyper-mutagenesis and template switching. Therefore, it appears that long-range BIR in S-phase is limited to certain conditions. Additionally, a number of reports suggested that in unusual circumstances, including the collapse of replication at positions of atypical DNA structures, changes in regulation of replication, or unusual timing of DNA synthesis can all trigger BIR. For example, BIR events have been reported to occur following replication collapse at fragile sites in yeast cells deficient in DNA polymerases (Lemoine et al., 2005), and in regions that can adopt unusual secondary structure promoting replication pausing and collapse (Narayanan et al., 2006). BIR events were reported in tumors and cancer cell lines following replication stress initiated by oncogene overexpression (Costantino et al., 2014, Sotiriou et al., 2016). These events depend on Rad52 and also require POLD3, a mammalian homolog of the BIR-specific Polδ subunit Pol32. Moreover, it was observed by Hickson and colleagues, that a replication collapse at human fragile sites initiates BIR-like DNA synthesis occurring during mitosis (Minocherhomji et al., 2015, Bhowmick et al., 2016). Similar to BIR, this synthesis is conservative and also depends on POLD3. Additionally, this synthesis also depends on Rad52, but not on Rad51, and therefore represents Rad51-independent BIR. Hickson and colleagues proposed that initiation of this BIR-like synthesis was promoted by cleavage of stalled replication forks at fragile sites by Slx4-associated Mus81-Eme1 complex. They demonstrated that initiation of this mitotic synthesis was critical for the prevention of mitotic chromosome segregation problems (Minocherhomji et al., 2015, Bhowmick et al., 2016). Similarly, BIR-like synthesis during mitosis was also induced by replication stress in mouse models, and results from mutations in MCM4 (Luebben et al., 2014). An additional example of Rad51-independent, but Rad52-dependent events promoted in cancer cells by replication stress resulting from p53-independent expression of p21 that could potentially result from BIR has been recently documented (Galanos et al., 2016).
Interestingly, BIR was also recently reported to initiate in yeast as a result of replication collapse at positions of (CAG) n /(CTG) n repeats, known for their propensity to adopt unusual DNA conformations that can stall DNA replication (Kim et al., 2017). Interestingly, the authors reported that these BIR events led to large-scale expansions of trinucleotide repeats, which were similar to expansions responsible for various neurodegenerative diseases in humans (reviewed in (Schmidt and Pearson, 2016)). The exact cause of these trinucleotide repeat expansions during BIR has remained unclear so far. In principle, they could result either from the out-of-register strand invasion of a broken DNA strand into the region containing trinucleotide repeats were already replicated from multiple template switching events commonly occurring at the beginning of BIR, or from the unusual mode of DNA synthesis during BIR. The latter is possible in light of a recent report that replication restart in DNA regions containing trinucleotide repeats might be a lengthy process that is completed at the nuclear pores (Schmidt and Pearson, 2016). This may suggest that BIR initiated at positions of unusual DNA structures may follow rules that are different from those established for S-phase BIR (Mayle et al., 2015), and is similar in its mechanism and genetic consequences to the BIR that occurs during the G2 phase of the cell cycle (Saini et al., 2013). In addition, the exact molecular details of BIR initiation at positions of trinucleotide repeats remain unclear.
Alternative lengthening of telomeres (ALT) and BIR
In proliferating human somatic cells, telomeres shorten every cell cycle, which leads to senescence, but can be counteracted by telomerase that is often re-activated in cancer cells, providing cells with unlimited replication potential. Another pathway of telomere maintenance employed by 5%−15% of cancers does not involve telomerase, but relies on recombination resembling BIR (reviewed in (Conomos et al., 2013)). This pathway, called alternative lengthening of telomeres (ALT), has been originally described in yeast mutants lacking functional telomerase (Lundblad and Blackburn, 1993). In these mutants, telomeres shorten causing a majority of cells to undergo senescence, however, survivors with elongated telomeres emerge. The formation of these survivors depends on the main yeast recombination protein, Rad52. Importantly, two Rad52-dependent ALT pathways have been described. They differ from each other by the structure of telomeres and by their genetic requirements. In the Type I pathway, telomeres are extended by acquiring copies of yeast transposon-like sequences called Y’-elements, which are normally located in sub-telomeric regions (Lundblad and Blackburn, 1993, Louis et al., 1994). This pathway depends on classic recombination proteins, including Rad51, Rad54, Rad55, Rad57, and proceed by recombination that involves either Y’ sequences themselves, or occurs between unprotected telomeres and TG1–3 repeats located between tandem Y’ elements (Le et al., 1999, Teng and Zakian, 1999). The telomeric repeats in Type I survivors are usually very short. The second ALT pathway (Type II) is independent of Rad51, but depends on Rad59 and the MRX complex (Lundblad and Blackburn, 1993, McEachern and Blackburn, 1996, Le et al., 1999, Teng and Zakian, 1999, Teng et al., 2000). In general, the genetic requirements of Type II ALT are similar to Rad51-independent BIR. Type II survivors, are characterized by the elongation of the terminal telomeric repeat tracts. Type II survivors arise more rarely than Type I, but grow faster and eventually come to dominate liquid-grown cultures. The mechanism of the formation of telomeres remains unclear, but they could either result from rolling circle replication or from DNA synthesis initiated by annealing of the broken ssDNA telomeric end to ssDNA regions formed at other telomeres (reviewed in (McEachern and Haber, 2006, Anand et al., 2013), see also (Dilley et al., 2016)). Importantly, both yeast ALT pathways require the main BIR-specific protein, Pol32, thus implicating BIR as an underlying mechanism of ALT (Lydeard et al., 2007).
In the case of human ALT, several observations support that it proceeds via a BIR-like telomere recombination mechanism (reviewed in (Cesare and Reddel, 2010, Dilley and Greenberg, 2015)). First, in ALT cells, a DNA tag inserted at a single telomere was frequently copied to other chromosomal ends (Dunham et al., 2000). Second, telomeres are highly heterogeneous and fluctuate in size, consistent with recombination (Murnane et al., 1994, Bryan et al., 1995). Third, a high level of inter-chromatid recombination has been associated with ALT (Londono-Vallejo et al., 2004). Fourth, ALT involves the accumulation of extrachromosomal telomeric repeat DNA (ECTR), which can be either linear or circular, as well as partially single-stranded and represent products of ALT, but interestingly may also serve as ALT templates (Cesare and Griffith, 2004, Nabetani and Ishikawa, 2009). Finally, telomere maintenance during ALT is associated with formation of special ALT-specific cellular centers termed ALT-associated promyelocytic leukemia (PML) bodies (Yeager et al., 1999). PMLs contain a number of recombination proteins that are involved in ALT, including, Rad51, BRCA2, and BLM, as well as telomeres and ECTRs (reviewed in (Dilley and Greenberg, 2015). Overall, it was proposed that ALT is initiated by recombination between broken chromosomal (telomere) ends and a homologous DNA template located (i) on a sister chromatid, (ii) on another chromosome, or (iii) on ECTR, and proceeds inside PMLs.
A series of recent studies used a telomere-specific endonuclease (TRF1-Fok1) to model initiation of ALT by induction of DSBs at telomeres. The observation was that DSBs at telomeres started a long-range homology search, manifested in large-scale telomere movements that required the main recombination protein Rad51 (Cho et al., 2014). In addition, these movements involved a meiosis-specific protein, Hop2, expressed in ALT cells. Based on this observation, the researchers proposed that ALT resembled meiosis in its preference for non-sister chromatid recombination. Also, induction of the telomere-specific DSBs initiated BIR-like DNA synthesis which generated a long-tract (up to 70-kb long) and required Polδ, and PCNA, loaded by replication factor C (RFC). Similar to BIR, this synthesis required POLD3 (a Pol32 homolog). Importantly, this synthesis was readily observed in the absence of Rad51, MCM complex, Polε and Polα-primase (Dilley et al., 2016). Overall, it appears that induction of telomere-specific DSBs led to initiation of two different ALT pathways proceeding via Rad51-dependent and Rad51-independent BIR, respectively. The absence of MCM-dependence observed here is consistent with the data reported by (Wilson et al., 2013), while independence of Polε and Polα-primase may reflect the difference between Rad51-dependent and Rad51-independent BIR. Importantly, investigation of naturally occurring ALT demonstrated that its associated DNA synthesis is conservative, similar to BIR (Roumelioti et al., 2016). Additionally, the proteomic analysis of ALT telomeres demonstrated participation of an additional polymerase, Polη, in initiation of ALT DNA synthesis (Garcia-Exposito et al., 2016). It was proposed that following strand invasion, Polη initiates DNA synthesis, which is continued by Polδ.
While modelling telomere DSBs has provided important characteristics of ALT DNA synthesis, the source of endogeneous DSBs initiating ALT and the details of such initiation remain unclear. It has always been assumed that a 3’-ssDNA end resulting from telomere erosion is the main initiating factor of ALT. However, in addition to 3’-G-rich overhangs, ALT cells also contain abundant 5’-C-rich telomere overhangs (Oganesian and Karlseder, 2011, Oganesian and Karlseder, 2013), which might also be used during ALT initiation. In addition, recent studies suggested that ALT is also promoted by a replication collapse at positions of R-loops formed by annealing of long non-coding TERRA RNA that is transcribed from the telomeres, which remains attached to the template (Azzalin et al., 2007, Schoeftner and Blasco, 2008, Flynn et al., 2011, Lovejoy et al., 2012, Arora et al., 2014, Episkopou et al., 2014, Flynn et al., 2015). Replication fragility at telomeres due to R-loop formation, or for other reasons, is likely promoted by significant changes of telomere chromatin structure in ALT cells resulting from mutations in chromatin factors ATRX, DAXX, and H3.3 that are enriched in ALT positive tumors (Heaphy et al., 2011, Schwartzentruber et al., 2012). Additionally, the disruption of Asf1, a histone chaperone, also promotes ALT (O’Sullivan et al., 2014). These changes of chromatin structure and upregulation of TERRA promote accumulation of RPA at ALT telomeres, which also promotes recombination. Finally, the same chromatin changes and formation of R-loops may also contribute to accumulation of telomere repeat ssDNA regions that can be used as templates for annealing to initiate Rad51-independent ALT. It is also possible that this type of ALT employs partially single-stranded C-circles as a template (Figure 4B).
BIR and complex genomic rearrangements
Several types of complex genomic rearrangements (CGRs) have been recently described in association with a number of human diseases. One of them, called chromothripsis, was discovered in cancer cells, and is characterized by a massive number of chromosomal rearrangements that are usually localized to a single chromosome (Berger et al., 2011, Kloosterman et al., 2011b, Stephens et al., 2011, Kloosterman et al., 2012, Molenaar et al., 2012, Malhotra et al., 2013, Zack et al., 2013). Another one, chromoanasynthesis, represents a combination of chromosomal rearrangements with copy number gains, and has been found in patients with various congenital disorders (Carvalho et al., 2011, Liu et al., 2011, Carvalho et al., 2013, Beck et al., 2015, Carvalho et al., 2015, Carvalho and Lupski, 2016). Chromoanasynthesis was explained by an unusual type of DNA synthesis called microhomology-mediated break-induced replication (MMBIR) (Hastings et al., 2009a). The idea of MMBIR came from the discovery that the CNV regions underlining several neurological disorders were not comprised of simple tandem duplications, but consisted of DNA regions containing interspersed segments that were duplicated, triplicated, and quadruplicated, with many containing microhomologies at their junctions (Lee et al., 2007, Carvalho et al., 2011, Carvalho et al., 2013, Beck et al., 2015). According to current models (Hastings et al., 2009a, Hastings et al., 2009b), MMBIR is initiated by DNA breakage generating a single DNA end, and proceeds with multiple template switches at positions of microhomologies that could be as short as 1–3bp (Lee et al., 2007, Carvalho et al., 2011, Liu et al., 2011, Carvalho et al., 2013, Beck et al., 2015). In addition, MMBIR-like events have been described in bacteria (Slack et al., 2006, Lin et al., 2011), Arabidopsis (Marechal et al., 2009, Kwon et al., 2010), Caenorhabditis elegans (Meier et al., 2014) and mouse embryonic stem cells (Arlt et al., 2012). Also, a recent study of chromothripsis that was experimentally induced by a micro-nuclear formation, revealed multiple templated insertions that likely resulted from MMBIR (Zhang et al., 2015), which linked MMBIR to chromothripsis. Also, the MMBIR pathway has been used to explain telomere healing (Lowden et al., 2011, Yatsenko et al., 2012) and several other types of CGRs in a number of diseases including cancer (Vissers et al., 2009, Lawson et al., 2011, Wang et al., 2015).
Hastings and colleagues proposed that MMBIR represents a type of Rad51-independent BIR, and is induced under specific cellular conditions, for example by hypoxia, which leads to Rad51 repression that stimulates Rad51-independent MMBIR proceeding at positions of microhomology (Hastings et al., 2009a). It remains to be directly tested whether MMBIR and Rad51-independent BIR represent the same pathway, since they were never studied in the same system, and their respective genetics has not been fully investigated. However, some experimental support has been obtained. First, studies in yeast have documented the existence of an MMBIR-like Rad51-independent pathway that is induced by replication stress, depends on the signature BIR protein Pol32, and leads to the formation of segmental duplications containing microhomologies at their junctions (Payen et al., 2008). In addition, the recently described Rad52-dependent mitotic synthesis initiated as a result of replication fork collapse at human fragile sites, was also proposed to represent MMBIR (Minocherhomji et al., 2015, Bhowmick et al., 2016).
An important mechanistic insight into MMBIR has come from the sequence analyses of CGRs found in patients with PMD and MECP2 duplication syndrome (Carvalho et al., 2011, Carvalho et al., 2013, Beck et al., 2015), which suggested that MMBIR is coupled to BIR. In particular, the authors proposed that initiation of MMBIR observed in these patients involved the following two steps. The first step involved breakage of a replication fork that led to homology-driven BIR involving two highly homologous inverted repeats, which generated an inverted segment and led to a copy number gain (Carvalho et al., 2011, Beck et al., 2015). In the second step, a switch from homology-mediated BIR to MMBIR was postulated. The involvement of MMBIR was supported by a series of template switching events mediated by microhomologies that resulted in genomic changes of various sizes from large-scale copy number variations (CNVs) to small templated insertions (Carvalho et al., 2013, Beck et al., 2015). The support for this idea came from two studies in yeast. First, Anand et al. reported that template switching initiated during BIR required less homology than the original BIR strand invasion (Anand et al., 2014). Second, Sakofsky et al. discovered that interruption of BIR due to deficiency of repair synthesis (modeled in their study by the lack of Pif1 helicase), leads to a switch from homology- to microhomology-driven BIR (Sakofsky et al., 2015). Interestingly, at least two template switching events lead to the formation of each DNA repair outcome (Figure 3D). Importantly, in Sakofsky et al. the switch from BIR to MMBIR was promoted by an exchange of DNA polymerase Polδ that was driving normal BIR, with the translesion polymerase Polζ that initiated DNA synthesis at 0–6 nucleotide microhomologies during MMBIR (Sakofsky et al., 2015).
In the future, it will be necessary to determine whether the switch from BIR to MMBIR may also initiate in other circumstances of BIR interruption, including other defects in BIR replication machinery, DNA damage accumulation in the template for BIR, or due to formation of secondary structures that impede progression of BIR leading-strand synthesis. The detection of these events may require the usage of new high-throughput screening approaches. This will likely require the development of new bioinformatics tools that can allow for robust detection of MMBIR events often overlooked by existing algorithms due to the intrinsic complexity of the MMBIR pattern. It will be also important to figure out whether MMBIR associated with neurological or other disorders in humans may also involve Polζ, or one of several other known human polymerases. Finally, it might be useful to revisit a number of CGRs that were previously explained by NHEJ – based on the lack a copy number increase and/or on the absence of homology at their junctions (see for example in (Carvalho et al., 2011, Kloosterman et al., 2011a, Kloosterman et al., 2011b, Arlt et al., 2012)). It is possible that they alternatively can result from MMBIR since these events often occur with no microhomology, and can be often linked to half-crossover events (Carvalho et al., 2015), which do not result in copy number increase. It is important to note however that NHEJ and microhomology end-joining (MMEJ) are highly prevalent in mammalian cells, and therefore it is difficult in many cases to distinguish between MMBIR, NHEJ and MMEJ events.
Conclusions
BIR remains one of the most interesting and least understood DNA repair pathways. The interest in understanding BIR is driven by its dual consequences for genomic stability: it stabilizes genomes by repairing DSBs, but it can also be destabilizing by its ability to generate high levels of mutations and complex chromosomal rearrangements. Remarkably, recent progress in BIR investigation has allowed scientists to characterize its molecular mechanism, to identify proteins participating in BIR, and to unravel the role of BIR and MMBIR in promoting complex genomic rearrangements and mutations. In addition, the development of several new experimental systems has allowed for the expansion of BIR studies to include, (i) different organisms (e.g., humans), (ii) the initiation of BIR at different stages of the cell cycle, and (iii) the induction of BIR by different types of DNA damage, including targeted DSBs, replication collapse, and telomere erosion.
As our understanding of BIR progresses, there is also a realization that the details of the molecular mechanisms and regulation differ between BIR events taking place in different organisms, at different stages of the cell cycle, and also in how initiation of BIR may be influenced by different types of damage. For example, BIR initiated by a replication fork collapse using a sister chromatid as a template is regulated more tightly than BIR initiated by targeted DSBs that use a non-sister chromatid as a template, such as BIR events reported at G2. The reasons behind this difference remain unclear, however contributing factors may include possible differences in the structure of BIR intermediates or cell-cycle specific modifications of structure-specific resolvases responsible for cleaving BIR intermediates.
In addition, many recent examples of BIR pathways in mammalian cells are genetically similar to Rad51-independent BIR that was previously discovered in yeast: they depend on Rad52, but not on Rad51. All these findings underscore the importance of a genetic and mechanistic characterization of Rad51-independent BIR pathway that has never thoroughly been investigated. In particular, it is critical to determine the proteins participating in this pathway, as well as to characterize the mode and intermediates of DNA synthesis associated with Rad51-independent BIR and to characterize its regulation. In addition, it remains unclear whether Rad51-independent BIR and MMBIR represent that same or different pathways. Also, it is becoming increasingly important to characterize the role of BIR and MMBIR in the onset of various human diseases, including cancer and neurological diseases.
Acknowledgments
We thank Grzegorz Ira for helpful comments on the manuscript.
Declaration of interest
The research in AM laboratory is funded by NIH grant GM084242
Footnotes
Disclosure statement
Authors declare no conflict of interest.
References
- Anand RP, Lovett ST & Haber JE, 2013. Break-induced DNA replication. Cold Spring Harb Perspect Biol, 5, a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand RP, Tsaponina O, Greenwell PW, Lee C-S, Du W, Petes TD & Haber JE, 2014. Chromosome rearrangements via template switching between diverged repeated sequences. Genes & Development, 28, 2394–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE & Glover TW, 2012. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end joining. PLoS Genet, 8, e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Lee Y, Wischnewski H, Brun CM, Schwarz T & Azzalin CM, 2014. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat Commun, 5, 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Bates DB & Kogoma T, 1994. DNA replication triggered by double-stranded breaks in E. coli: dependence on homologous recombination functions. Cell, 78, 1051–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Sommer S, Bailone A & Kogoma T, 1993. Homologous recombination-dependent initiation of DNA replication from DNA damage-inducible origins in Escherichia coli. EMBO J, 12, 3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E & Lingner J, 2007. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science, 318, 798–801. [DOI] [PubMed] [Google Scholar]
- Beck CR, Carvalho CM, Banser L, Gambin T, Stubbolo D, Yuan B, Sperle K, Mccahan SM, Henneke M, Seeman P, Garbern JY, Hobson GM & Lupski JR, 2015. Complex genomic rearrangements at the PLP1 locus include triplication and quadruplication. PLoS Genet, 11, e1005050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, Macdonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA & Garraway LA, 2011. The genomic complexity of primary human prostate cancer. Nature, 470, 214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S & Hickson ID, 2016. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell, 64, 1117–1126. [DOI] [PubMed] [Google Scholar]
- Bosco G & Haber JE, 1998. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics, 150, 1037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Englezou A, Gupta J, Bacchetti S & Reddel RR, 1995. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J, 14, 4240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J & Foster PL, 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics, 128, 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM & Lupski JR, 2016. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet, 17, 224–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, Belmont JW, Hastings PJ & Lupski JR, 2013. Replicative mechanisms for CNV formation are error prone. Nat Genet, 45, 1319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Pfundt R, King DA, Lindsay SJ, Zuccherato LW, Macville MV, Liu P, Johnson D, Stankiewicz P, Brown CW, Study DDD, Shaw CA, Hurles ME, Ira G, Hastings PJ, Brunner HG & Lupski JR, 2015. Absence of heterozygosity due to template switching during replicative rearrangements. Am J Hum Genet, 96, 555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, Mccall A, Pivnick EK, Hines-Dowell S, Seaver LH, Friehling L, Lee S, Smith R, Del Gaudio D, Withers M, Liu P, Cheung SW, Belmont JW, Zoghbi HY, Hastings PJ & Lupski JR, 2011. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet, 43, 1074–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ & Griffith JD, 2004. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol Cell Biol, 24, 9948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ & Reddel RR, 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet, 11, 319–30. [DOI] [PubMed] [Google Scholar]
- Chan K & Gordenin DA, 2015. Clusters of Multiple Mutations: Incidence and Molecular Mechanisms. Annu Rev Genet, 49, 243–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouly G, Kwok A, Huang B, Willis NA, Xie A & Scully R, 2013. BRCA1 and CtIP suppress long-tract gene conversion between sister chromatids. Nat Commun, 4, 2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho NW, Dilley RL, Lampson MA & Greenberg RA, 2014. Interchromosomal homology searches drive directional ALT telomere movement and synapsis. Cell, 159, 108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A & Ira G, 2010. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet, 6, e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos D, Pickett HA & Reddel RR, 2013. Alternative lengthening of telomeres: remodeling the telomere architecture. Front Oncol, 3, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ledesma F & Aguilera A, 2006. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep, 7, 919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP & Halazonetis TD, 2014. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science, 343, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP & Symington LS, 2004. RAD51-dependent break-induced replication in yeast. Mol Cell Biol, 24, 2344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Barker K, Vanhulle K, Downing B, Vayl A & Malkova A, 2008. Defective break-induced replication leads to half-crossovers in Saccharomyces cerevisiae. Genetics, 179, 1845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A & Malkova A, 2011. Break-induced replication is highly inaccurate. PLoS Biol, 9, e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL & Greenberg RA, 2015. ALTernative Telomere Maintenance and Cancer. Trends Cancer, 1, 145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR & Greenberg RA, 2016. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature, 539, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J & Whitby MC, 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem, 277, 32753–9. [DOI] [PubMed] [Google Scholar]
- Donnianni RA & Symington LS, 2013. Break-induced replication occurs by conservative DNA synthesis. Proc Natl Acad Sci U S A, 110, 13475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing B, Morgan R, Vanhulle K, Deem A & Malkova A, 2008. Large inverted repeats in the vicinity of a single double-strand break strongly affect repair in yeast diploids lacking Rad51. Mutat Res, 645, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MA, Neumann AA, Fasching CL & Reddel RR, 2000. Telomere maintenance by recombination in human cells. Nat Genet, 26, 447–50. [DOI] [PubMed] [Google Scholar]
- Episkopou H, Draskovic I, Van Beneden A, Tilman G, Mattiussi M, Gobin M, Arnoult N, Londono-Vallejo A & Decottignies A, 2014. Alternative Lengthening of Telomeres is characterized by reduced compaction of telomeric chromatin. Nucleic Acids Res, 42, 4391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J & Zou L, 2011. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature, 471, 532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, Bersani F, Pineda JR, Suva ML, Benes CH, Haber DA, Boussin FD & Zou L, 2015. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science, 347, 273–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbai CB & Marians KJ, 2010. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst), 9, 202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanos P, Vougas K, Walter D, Polyzos A, Maya-Mendoza A, Haagensen EJ, Kokkalis A, Roumelioti FM, Gagos S, Tzetis M, Canovas B, Igea A, Ahuja AK, Zellweger R, Havaki S, Kanavakis E, Kletsas D, Roninson IB, Garbis SD, Lopes M, Nebreda A, Thanos D, Blow JJ, Townsend P, Sorensen CS, Bartek J & Gorgoulis VG, 2016. Chronic p53-independent p21 expression causes genomic instability by deregulating replication licensing. Nat Cell Biol, 18, 777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Fernandez M, Saugar I, Ortiz-Bazan MA, Vazquez MV & Tercero JA, 2012. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res, 40, 8325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Exposito L, Bournique E, Bergoglio V, Bose A, Barroso-Gonzalez J, Zhang S, Roncaioli JL, Lee M, Wallace CT, Watkins SC, Opresko PL, Hoffmann JS & O’sullivan RJ, 2016. Proteomic Profiling Reveals a Specific Role for Translesion DNA Polymerase eta in the Alternative Lengthening of Telomeres. Cell Rep, 17, 1858–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Longerich S & Rosenberg SM, 1994. Recombination in adaptive mutation. Science, 264, 258–60. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y & Costanzo V, 2011. Studying DNA replication fork stability in Xenopus egg extract. Methods Mol Biol, 745, 437–45. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Ira G & Lupski JR, 2009a. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet, 5, e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM & Ira G, 2009b. Mechanisms of change in gene copy number. Nat Rev Genet, 10, 551–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, De Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, Offerhaus GJ, Mclendon R, Rasheed BA, He Y, Yan H, Bigner DD, Oba-Shinjo SM, Marie SK, Riggins GJ, Kinzler KW, Vogelstein B, Hruban RH, Maitra A, Papadopoulos N & Meeker AK, 2011. Altered telomeres in tumors with ATRX and DAXX mutations. Science, 333, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC & Marians KJ, 2006. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol, 7, 932–43. [DOI] [PubMed] [Google Scholar]
- Ira G & Haber JE, 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol, 22, 6384–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N & Haber JE, 2009. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev, 23, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Mehta A, Ryu T & Haber JE, 2016. Sgs1 and Mph1 Helicases Enforce the Recombination Execution Checkpoint During DNA Double-Strand Break Repair in Saccharomyces cerevisiae. Genetics, 203, 667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz SS, Gimble FS & Storici F, 2014. To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human cells. PLoS One, 9, e88840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JC, Harris ST, Dinter T, Shah KA & Mirkin SM, 2017. The role of break-induced replication in large-scale expansions of (CAG)n/(CTG)n repeats. Nat Struct Mol Biol, 24, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, Van Roosmalen M, Duran KJ, De Bruijn E, Bakker SC, Letteboer T, Van Nesselrooij B, Hochstenbach R, Poot M & Cuppen E, 2011a. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet, 20, 1916–24. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, Van Roosmalen MJ, Van Lieshout S, Nijman IJ, Roessingh W, Van ‘T Slot R, Van De Belt J, Guryev V, Koudijs M, Voest E & Cuppen E, 2011b. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol, 12, R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, Van Roosmalen MJ, Van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, Ruivenkamp CA, Jager M, Van Haeringen A, Ippel EF, Haaf T, Passarge E, Hochstenbach R, Menten B, Larizza L, Guryev V, Poot M & Cuppen E, 2012. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep, 1, 648–55. [DOI] [PubMed] [Google Scholar]
- Kreuzer KN & Brister JR, 2010. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J, 7, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN, Saunders M, Weislo LJ & Kreuzer HW, 1995. Recombination-dependent DNA replication stimulated by double-strand breaks in bacteriophage T4. J Bacteriol, 177, 6844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A & Stahl FW, 1999. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev, 13, 345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T, Huq E & Herrin DL, 2010. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci U S A, 107, 13954–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson AR, Hindley GF, Forshew T, Tatevossian RG, Jamie GA, Kelly GP, Neale GA, Ma J, Jones TA, Ellison DW & Sheer D, 2011. RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology. Genome Res, 21, 505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Moore JK, Haber JE & Greider CW, 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics, 152, 143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM & Lupski JR, 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell, 131, 1235–47. [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K & Petes TD, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell, 120, 587–98. [DOI] [PubMed] [Google Scholar]
- Lin D, Gibson IB, Moore JM, Thornton PC, Leal SM & Hastings PJ, 2011. Global chromosomal structural instability in a subpopulation of starving Escherichia coli cells. PLoS Genet, 7, e1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, Bacino CA, Campos-Acevedo LD, Delgado MR, Freedenberg D, Garnica A, Grebe TA, Hernandez-Almaguer D, Immken L, Lalani SR, Mclean SD, Northrup H, Scaglia F, Strathearn L, Trapane P, Kang SH, Patel A, Cheung SW, Hastings PJ, Stankiewicz P, Lupski JR & Bi W, 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell, 146, 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S & Reddel RR, 2004. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res, 64, 2324–7. [DOI] [PubMed] [Google Scholar]
- Louis EJ, Naumova ES, Lee A, Naumov G & Haber JE, 1994. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics, 136, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, De Lange T, De S, Petrini JH, Sung PA, Jasin M, Rosenbluh J, Zwang Y, Weir BA, Hatton C, Ivanova E, Macconaill L, Hanna M, Hahn WC, Lue NF, Reddel RR, Jiao Y, Kinzler K, Vogelstein B, Papadopoulos N, Meeker AK & Consortium ALTSC, 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet, 8, e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Flibotte S, Moerman DG & Ahmed S, 2011. DNA synthesis generates terminal duplications that seal end-to-end chromosome fusions. Science, 332, 468–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luder A & Mosig G, 1982. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc Natl Acad Sci U S A, 79, 1101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebben SW, Kawabata T, Johnson CS, O’sullivan MG & Shima N, 2014. A concomitant loss of dormant origins and FANCC exacerbates genome instability by impairing DNA replication fork progression. Nucleic Acids Res, 42, 5605–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V & Blackburn EH, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell, 73, 347–60. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M & Haber JE, 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature, 448, 820–3. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM & Haber JE, 2010. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev, 24, 1133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Quinlan AR & Hall IM, 2013. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res, 23, 762–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A & Haber JE, 2012. Mutations arising during repair of chromosome breaks. Annu Rev Genet, 46, 455–73. [DOI] [PubMed] [Google Scholar]
- Malkova A & Ira G, 2013. Break-induced replication: functions and molecular mechanism. Curr Opin Genet Dev, 23, 271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL & Haber JE, 1996. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci U S A, 93, 7131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G & Haber JE, 2005. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol, 25, 933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, Newlon CS & Haber JE, 2001. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev, 15, 1055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A, Parent JS, Veronneau-Lafortune F, Joyeux A, Lang BF & Brisson N, 2009. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci U S A, 106, 14693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM & West SC, 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell, 147, 158–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG & West SC, 2013. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep, 4, 76–86. [DOI] [PubMed] [Google Scholar]
- Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, Shaw CA, Bjergbaek L, Lupski JR & Ira G, 2015. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science, 349, 742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mceachern MJ & Blackburn EH, 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev, 10, 1822–34. [DOI] [PubMed] [Google Scholar]
- Mceachern MJ & Haber JE, 2006. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem, 75, 111–35. [DOI] [PubMed] [Google Scholar]
- Mcvey M, Khodaverdian VY, Meyer D, Cerqueira PG & Heyer WD, 2016. Eukaryotic DNA Polymerases in Homologous Recombination. Annu Rev Genet, 50, 393–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Beach A & Haber JE, 2016. Homology Requirements and Competition between Gene Conversion and Break-Induced Replication during Double-Strand Break Repair. Mol Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B, Cooke SL, Weiss J, Bailly AP, Alexandrov LB, Marshall J, Raine K, Maddison M, Anderson E, Stratton MR, Gartner A & Campbell PJ, 2014. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Res, 24, 1624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y & Hickson ID, 2015. Replication stress activates DNA repair synthesis in mitosis. Nature, 528, 286–90. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, Van Sluis P, Valentijn LJ, Van Der Ploeg I, Hamdi M, Van Nes J, Westerman BA, Van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, Van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM & Versteeg R, 2012. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature, 483, 589–93. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Connelly C & Hieter P, 1997. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics, 147, 371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Galvan S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD & Aguilera A, 2012. Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol, 32, 1592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L, Marder BA & Morgan WF, 1994. Telomere dynamics in an immortal human cell line. EMBO J, 13, 4953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani A & Ishikawa F, 2009. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol, 29, 703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD & Lobachev KS, 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell, 125, 1283–96. [DOI] [PubMed] [Google Scholar]
- Neumann AA, Watson CM, Noble JR, Pickett HA, Tam PP & Reddel RR, 2013. Alternative lengthening of telomeres in normal mammalian somatic cells. Genes Dev, 27, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MO, Jalan M, Morrow CA, Osman F & Whitby MC, 2015. Recombination occurs within minutes of replication blockage by RTS1 producing restarted forks that are prone to collapse. Elife, 4, e04539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen I, Bentsen IB, Lisby M, Hansen S, Mundbjerg K, Andersen AH & Bjergbaek L, 2009. A Flp-nick system to study repair of a single protein-bound nick in vivo. Nat Methods, 6, 753–7. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, Menzies A, Martin S, Leung K, Chen L, Leroy C, Ramakrishna M, Rance R, Lau KW, Mudie LJ, Varela I, Mcbride DJ, Bignell GR, Cooke SL, Shlien A, Gamble J, Whitmore I, Maddison M, Tarpey PS, Davies HR, Papaemmanuil E, Stephens PJ, Mclaren S, Butler AP, Teague JW, Jonsson G, Garber JE, Silver D, Miron P, Fatima A, Boyault S, Langerod A, Tutt A, Martens JW, Aparicio SA, Borg A, Salomon AV, Thomas G, Borresen-Dale AL, Richardson AL, Neuberger MS, Futreal PA, Campbell PJ, Stratton MR & Breast Cancer Working Group of the International Cancer Genome, C., 2012. Mutational processes molding the genomes of 21 breast cancers. Cell, 149, 979–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’sullivan RJ, Arnoult N, Lackner DH, Oganesian L, Haggblom C, Corpet A, Almouzni G & Karlseder J, 2014. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat Struct Mol Biol, 21, 167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L & Karlseder J, 2011. Mammalian 5’ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell, 42, 224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesian L & Karlseder J, 2013. 5’ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst), 12, 238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B & Fischer G, 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet, 4, e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA & Gordenin DA, 2014. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer, 14, 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G & Gordenin DA, 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet, 45, 970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA & Gordenin DA, 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell, 46, 424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers K & Mcvey M, 2016. Error-Prone Repair of DNA Double-Strand Breaks. J Cell Physiol, 231, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H & Arcangioli B, 2008. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J, 27, 1378–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumelioti FM, Sotiriou SK, Katsini V, Chiourea M, Halazonetis TD & Gagos S, 2016. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep, 17, 1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff P, Donnianni RA, Glancy E, Oh J & Symington LS, 2016. RPA Stabilization of Single-Stranded DNA Is Critical for Break-Induced Replication. Cell Rep, 17, 3359–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier L, Ricoul M, Pottier G & Murnane JP, 2005. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res, 3, 139–50. [DOI] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS & Malkova A, 2013. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature, 502, 389–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Ayyar S, Deem AK, Chung WH, Ira G & Malkova A, 2015. Translesion Polymerases Drive Microhomology-Mediated Break-Induced Replication Leading to Complex Chromosomal Rearrangements. Mol Cell, 60, 860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Ayyar S & Malkova A, 2012. Break-induced replication and genome stability. Biomolecules, 2, 483–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakofsky CJ, Roberts SA, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA & Malkova A, 2014. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep, 7, 1640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MH & Pearson CE, 2016. Disease-associated repeat instability and mismatch repair. DNA Repair (Amst), 38, 117–26. [DOI] [PubMed] [Google Scholar]
- Schoeftner S & Blasco MA, 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol, 10, 228–36. [DOI] [PubMed] [Google Scholar]
- Schwartz EK & Heyer WD, 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma, 120, 109–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, Von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM & Jabado N, 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature, 482, 226–31. [DOI] [PubMed] [Google Scholar]
- Signon L, Malkova A, Naylor ML, Klein H & Haber JE, 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol Cell Biol, 21, 2048–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM & Hastings PJ, 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet, 2, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Lam AF & Symington LS, 2009. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase delta complex. Mol Cell Biol, 29, 1432–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B & Symington LS, 2007. Template switching during break-induced replication. Nature, 447, 102–5. [DOI] [PubMed] [Google Scholar]
- Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, Ochs F, Lukas C, Lukas J, Gorgoulis VG, Scapozza L & Halazonetis TD, 2016. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell, 64, 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafa A, Donnianni RA, Timashev LA, Lam AF & Symington LS, 2014. Template switching during break-induced replication is promoted by the Mph1 helicase in Saccharomyces cerevisiae. Genetics, 196, 1017–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, Mclaren S, Lin ML, Mcbride DJ, Varela I, Nik-Zainal S, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Quail MA, Burton J, Swerdlow H, Carter NP, Morsberger LA, Iacobuzio-Donahue C, Follows GA, Green AR, Flanagan AM, Stratton MR, Futreal PA & Campbell PJ, 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stith CM, Sterling J, Resnick MA, Gordenin DA & Burgers PM, 2008. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J Biol Chem, 283, 34129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng SC, Chang J, Mccowan B & Zakian VA, 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol Cell, 6, 947–52. [DOI] [PubMed] [Google Scholar]
- Teng SC & Zakian VA, 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol, 19, 8083–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, Mccullough C, Bellinger M, Lobachev K, Petes TD & Malkova A, 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol, 27, 2601–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasan S, Deem A, Ramakrishnan S, Argueso JL & Malkova A, 2014. Cascades of genetic instability resulting from compromised break-induced replication. PLoS Genet, 10, e1004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal DD, Lee K, Deem A, Shim EY, Malkova A & Lee SE, 2012. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet, 8, e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LE, Bhatt SS, Janssen IM, Xia Z, Lalani SR, Pfundt R, Derwinska K, De Vries BB, Gilissen C, Hoischen A, Nesteruk M, Wisniowiecka-Kowalnik B, Smyk M, Brunner HG, Cheung SW, Van Kessel AG, Veltman JA & Stankiewicz P, 2009. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum Mol Genet, 18, 3579–93. [DOI] [PubMed] [Google Scholar]
- Wang Y, Su P, Hu B, Zhu W, Li Q, Yuan P, Li J, Guan X, Li F, Jing X, Li R, Zhang Y, Ferec C, Cooper DN, Wang J, Huang D, Chen JM & Wang Y, 2015. Characterization of 26 deletion CNVs reveals the frequent occurrence of micro-mutations within the breakpoint-flanking regions and frequent repair of double-strand breaks by templated insertions derived from remote genomic regions. Hum Genet, 134, 589–603. [DOI] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P & Ira G, 2013. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature, 502, 393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA & Gordenin DA, 2008. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet, 4, e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Hixson P, Roney EK, Scott DA, Schaaf CP, Ng YT, Palmer R, Fisher RB, Patel A, Cheung SW & Lupski JR, 2012. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Hum Genet, 131, 1895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR & Reddel RR, 1999. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res, 59, 4175–9. [PubMed] [Google Scholar]
- Yu TY, Kao YW & Lin JJ, 2014. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc Natl Acad Sci U S A, 111, 3377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M & Beroukhim R, 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet, 45, 1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M & Pellman D, 2015. Chromothripsis from DNA damage in micronuclei. Nature, 522, 179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]