Abstract
Nervous system function relies on complex assemblies of distinct neuronal cell types with unique anatomical and functional properties instructed by molecular programs. Alternative splicing is a key mechanism for the expansion of molecular repertoires and protein splice isoforms shape neuronal cell surface recognition and function. However, the logic of how alternative splicing programs are arrayed across neuronal cells types is poorly understood. We systematically mapped ribosome-associated transcript isoforms in genetically-defined neuron types of the mouse forebrain. Our dataset provides an extensive resource of transcript diversity across major neuron classes. We find that neuronal transcript isoform profiles reliably distinguish even closely-related classes of pyramidal cells and inhibitory interneurons in the mouse hippocampus and neocortex. These highly specific alternative splicing programs selectively control synaptic proteins and intrinsic neuronal properties. Thus, transcript diversification by alternative splicing is a central mechanism for the functional specification of neuronal cell types and circuits.
Introduction
The mammalian brain contains hundreds of cell types with unique anatomical and functional properties. Cell type characteristics are fundamental underpinnings of neuronal circuit function and – ultimately – the control of behaviors. Many of the distinctive neuronal morphologies were recognized one hundred years ago 1. More recent studies uncovered electrophysiological properties and characteristic gene expression profiles that are associated with specific neuron types 2,3. Yet, we still lack comprehensive knowledge of how the multitude of neuronal properties is encoded by a limited number of genes. Evolutionary comparisons revealed a significant increase in alternative splicing heterogeneity in more complex organisms. Within those, the nervous system exhibits the most extensive usage of alternative transcript isoforms 4,5. Single gene studies provided evidence that individual protein variants generated through alternative splicing can exhibit unique isoform-specific functions 6–10. However, in many molecular studies on neuronal connectivity and function the identity of splice isoforms endogenous to the cell type of interest are unknown. This is a significant bottleneck for interpretation of gain and loss-of-function studies. While loss of RNA binding proteins that regulate alternative splicing and cell type-specific knock-outs exhibit severe impacts on neuronal function and synaptic transmission 11–16, most of these proteins are commonly expressed in all neuronal cell types 17–20. Thus, the general logic of how alternative splicing programs relate to brain complexity is poorly understood.
Previous bulk-sequencing analyses contrasted neuronal and non-neuronal splicing regulation 21. Developmental analysis of mouse neocortex uncovered a series of temporally controlled coordinated splicing switches in brain tissues. These developmental switches were pan-neuronal and occurred across all neuronal populations 21–24. Only very recent studies are beginning to probe whether well-defined neuronal cell types rely on alternative splicing for the regulation of specific biological functions 16,20. However, it remains debated to what extent transcript isoforms detected by RNA-sequencing are indeed recruited for translation to produce protein isoforms 25,26. To address these questions, we generated genome-wide maps of transcript isoforms that are recruited for translation in genetically-defined neuronal cell populations. Our analysis identified hundreds of differentially regulated splicing events across distinct neuron types. Moreover, we demonstrate that cell type-specific splice isoforms define neuronal cell populations and shape intrinsic properties and synaptic protein complexes. The dataset provides a rich resource for selecting endogenously expressed splice isoforms to be used in functional studies, for interpreting impact of gene mutations in disease states, and for the dissection of enhancers and promoters that drive cell type-specific transcripts from alternative transcription start sites.
Results
Deep mapping of actively translated transcript isoforms in cortical and hippocampal neuron populations
In order to obtain a comprehensive mapping of transcript isoforms in the mouse forebrain we conducted large-scale tagged-ribosomal affinity purification (RiboTRAP) of ribosome-associated mRNAs from genetically-defined neurons (Fig. 1a). The endogenous ribosomal protein Rpl22 was conditionally HA-tagged in glutamatergic neurons (using CamK2-cre for most neocortical pyramidal cells and Scnn1a-cre for spiny stellate and star pyramid layer 4 cells), and GABAergic interneurons [with somatostatin-cre (SST), parvalbumin-cre (PV) and vasointestinal peptide-cre (VIP)]. Within the hippocampus, we further targeted Cornu ammonis 1 (CA1) neurons (CamK2-cre), CA3 neurons (Grik4-cre), and SST-positive interneurons (SST-cre) (Fig. 1b, Supplementary Fig. 1). Using an optimized affinity-isolation protocol and strict quality control measures followed by deep RNA-sequencing (paired-end, read length 100bp, >100 Million reads per biological replicate) we detected > 12’000 genes per sample with full-length coverage across transcripts (Supplementary Fig. 2, 3a) and low variance between biological replicates (Fig. 1c). Transcriptome analysis confirmed appropriate enrichment and de-enrichment of known and newly discovered markers. Widely expressed non-neuronal genes such as astrocyte markers were either not detected or showed low level background in some of these isolates (Supplementary Fig. 1b, 3b-d, 4, Supplementary Table 1). Thus, this deep dataset enables reliable dissection of transcript isoforms translated in specific cell types.
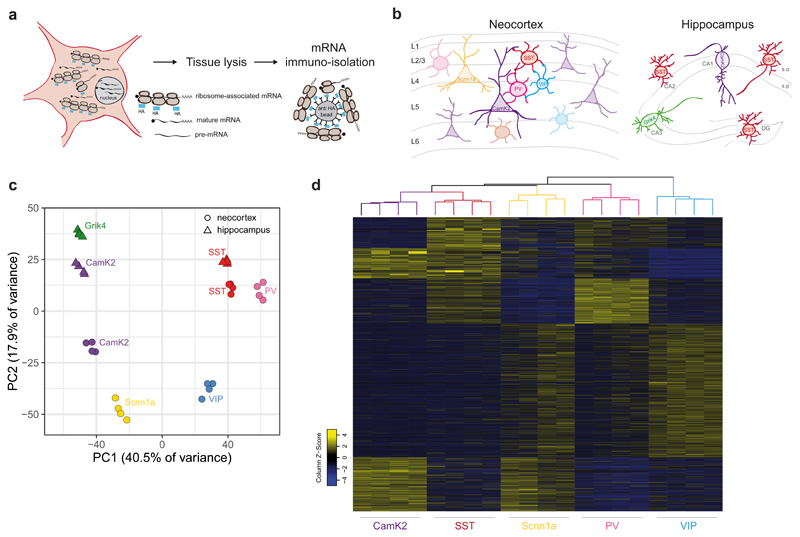
Fig. 1. Extensive alternative exon usage defines classes of forebrain neurons.
a, Schematic representation of RiboTRAP pulldowns. HA-tagged ribosomes in the cytoplasm of genetically defined cell populations load fully mature mRNAs. Following whole tissue lysis, ribosome-associated mRNAs are immuno-isolated using anti-HA beads and subsequently purified. b, Cartoons representing neuronal cell populations isolated from mouse neocortex (left panel) and hippocampus (right panel). c, Principal component analysis of genes expressed in each neocortical and hippocampal sample (n=4 biologically independent samples). Variance explained by the principal components 1 and 2 (PC1 and PC2) is indicated. Gene expression values were normalized by Variance Stabilizing Transformation (VST) d, Heatmap of SI values obtained from EXON analysis for each neocortical cell class (see methods for details) of all 2898 differentially regulated exons involved in alternative splicing events (log2(FC) ≥ 1 and ≤ -1, p-value ≤ 0.01, unpaired Student’s t-test. Base mean includes all neocortical samples). Alternative exon inclusion identifies sub-groups of exons that define distinct neocortical populations.
Alternative transcript repertoires define neuronal populations
To map transcript repertoires across neuronal cell types we quantified alternative isoforms using two complementary computational methods. First, we analyzed differential exon usage by quantifying reads mapping onto individual exons relative to the number of reads on constitutive exons derived from the same gene (constitutive exons are defined in the annotated transcript database FAST DB27, EXON analysis, Supplementary Fig.5, see methods for details). Second, the differential usage of splicing patterns was assessed using exonic and junctional reads mapping to transcript isoforms annotated in FAST DB (PATTERN analysis, Supplementary Fig. 5, see methods for details). This quantitative mapping of alternative transcript isoforms uncovered hundreds of highly differentially regulated transcript isoforms in neocortical and hippocampal cell populations [log2 fold-change (log2FC) in splicing index (SI) ≥ 1 or ≤ -1, p-value ≤ 0.01]. Independent experimental validations with semi-quantitative RT-PCR confirmed the accuracy of the computational pipeline (validation rate > 90%, Supplementary Fig. 6). Therefore, this validated dataset represents a comprehensive resource for alternative transcripts in the major forebrain neuron populations (Supplementary Table 2, 3 and https://scheiffele-splice.scicore.unibas.ch for a web-based look-up tool to query isoforms for individual genes). Divergent transcript isoforms may arise from alternative splicing but also alternative transcription start sites (TSS) 28,29. The PATTERN analysis enabled us to separate transcript isoforms arising from these mechanistically different forms of transcript diversification. Remarkably, the exons differentially regulated by alternative splicing reliably segregated neuronal cell classes (Fig. 1d). Thus, Scnn1a-defined layer 4 cells are characterized by 310 exons included in 214 different genes. Similarly, the two medial ganglionic eminence (MGE)-derived interneuron classes (PV and SST populations) are distinguished by 628 and 719 exons from 407 and 486 genes, respectively. Moreover, alternative transcript isoforms in the CGE (Caudal Ganglionic Eminence)-derived VIP interneurons were distinct from PV and SST-populations, with 609 exons differentially included in 407 genes. Overall, we did not observe a correlation of changes in splicing indices and gene expression level, indicating that our analysis captures differentially regulated exons across a broad spectrum of transcript expression levels (Supplementary Fig. 7). In previous studies, microexons (defined as exons 3-27 nucleotides long) were shown to preferentially contribute to transcript diversification in the nervous system30. Amongst all exons differentially regulated (DR) across neuronal cell classes, we find 3.8-5.3% to be microexons. These percentages are slightly higher compared to the percentage of total microexons detected in the neocortex (2.8%, see methods for details). Thus, differential alternative splicing across cell types is substantial for microexons but also other types of splicing events. In summary, this analysis demonstrates that extensive alternative splicing regulation distinguishes major neuronal cell classes in the mouse neocortex.
Wide use of alternative transcription start sites across cortical neuron sub-classes
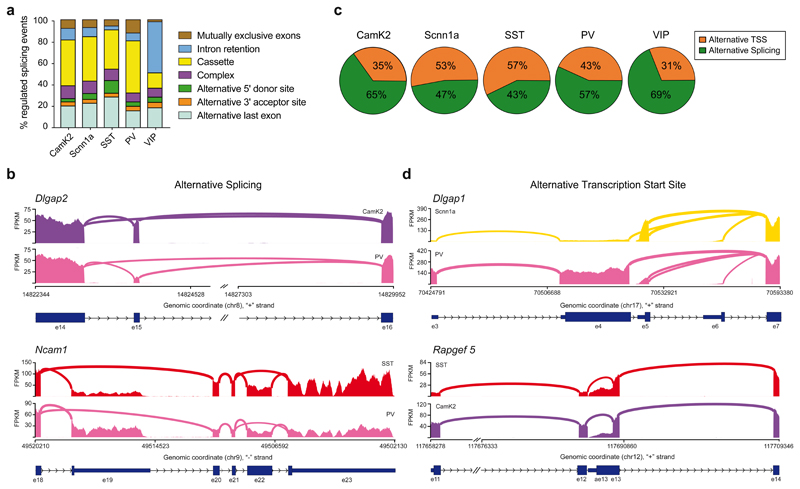
We quantified the frequency of distinct patterns underlying the differentially regulated splicing events in neocortical cell populations and found that they distributed over multiple categories (Figure 2a, Supplementary Table 2). Usage of cassette exons was the most frequent differentially regulated alternative splicing event across cell populations (Fig. 2a, 2b for Dlgap2 as example). Interestingly, alternative last exons (ALE), which result in a modification of the 3’UTR of transcripts, represented about 20% of events (Fig 2a, 2b Ncam1 as example). This is notable considering that alternative last exons can impose cell type-specific protein expression as well as subcellular localization of mRNAs 10,31. In addition to transcript diversification by alternative splicing we found that across all neocortical cell classes, ~30-60% of differentially regulated transcript isoforms arose from alternative TSS (Fig. 2c, Supplementary Table 2). This implies a frequent action of cell type-specific enhancers and promoters. An example for alternative TSS regulation is Dlgap1, which encodes a major glutamatergic scaffolding protein. In neocortical PV-positive cells, we identified an alternative TSS in exon3 of Dlgap1 which switches to exon 5 in the Scnn1a-positive layer 4 cells (Fig. 2d). This differential regulation results in transcripts that differ in the 5’UTR and the N-terminal amino acids (see Fig. 2d Rapgef5 for an additional example). When comparing the segregation of differential TSSs and ALEs we find that either of these types of events efficiently segregated neocortical excitatory and inhibitory cell classes, including MGE- and CGE-derived interneurons (Supplementary Fig. S8 a and b, respectively. Exon numbers involved are indicated in the figure legend). Thus, our analysis demonstrates that not only alternative splicing but also alternative TSS are major drivers of neuronal cell type-specific transcript isoform expression in the mouse forebrain.
Fig. 2. Alternative splicing and alternative transcription start site usage drive transcript diversification in neocortical neurons.
a, Histogram representing the relative percentage of differentially regulated (DR) alternative splicing event types (log2(FC) ≥ 1, p-value ≤ 0.01. Base mean includes all neocortical samples). The distinct pattern categories (mutually exclusive exon, cassette exon, intron retention, alternative 5’ and 3’ donor and acceptor site, alternative last exon, complex) are indicated. Total number of DR alternative splicing events are: 261 for CamK2, 165 for Scnn1a, 85 for SST, 316 for PV and 373 for VIP. Note that VIP interneurons show higher number of intron retention events. Otherwise, overall all neocortical populations show similar rates of splicing pattern types usage. b, Sashimi plots illustrating read distribution and splice junctions of Dlgap2 (upper panel) and Ncam1 (lower panel). One representative replicate for each cell population is shown. Genomic coordinates, chromosome number, strand and exon number are indicated below. Coding regions are indicated as thicker boxes, non-coding regions as thinner boxes). The alternative cassette exon 15 of Dlgap2 is preferentially included in CamK2-positive neurons, vice versa excluded in PV-positive interneurons. On the other hand, Ncam1 shows differential usage of exon 19 or exon 23 as alternative last exon, even between two GABAergic populations, with SST-positive cells preferentially using e23 compared to PV-positive neurons. c, Pie charts indicating the relative percentage of alternative transcription start sites (TSS) and alternative splicing of differentially regulated events (log2(FC) ≥ 1, p-value ≤ 0.01. Base mean includes all neocortical samples) identified by the PATTERN analysis (see methods for details) in neocortical populations. Total number of DR events are: 402 for CamK2, 349 for Scnn1a, 199 for SST, 557 for PV and 540 for VIP. d, Example sashimi plots for alternative TSS usage in Dlgap1 (upper panel) or Rapgef5 (lower panel). For Dlgap1, transcripts preferentially start with exon 5 in Scnn1a-positive cells and with exon 3 in PV-positive interneurons (note that coding region starts in exon 4). For Rapgef5, SST- and CamK2-positive cells show differential usage of exon 11 or exon 13 as first exon, with SST preferentially including exon 13.
Divergent alternative splicing programs across closely related cells in different anatomical positions
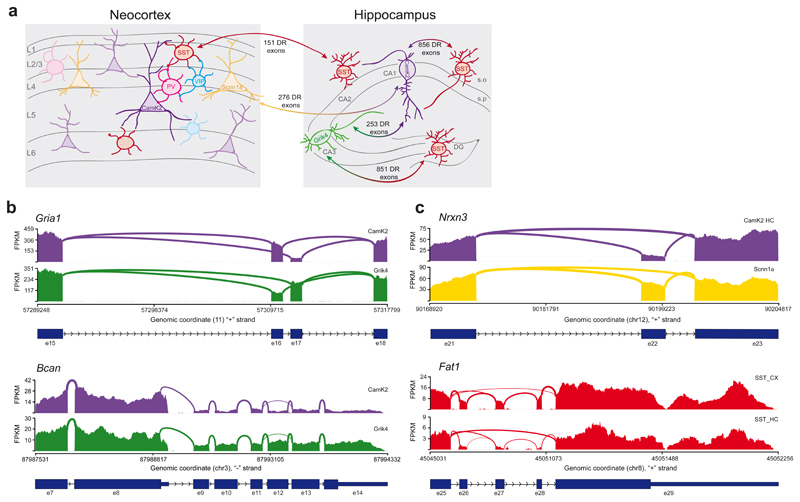
Hippocampal CA1 and CA3 pyramidal neurons exhibit certain unique functional properties and overall similar transcriptomes 32. Thus, we explored whether there are differential splicing programs specific for these closely-related glutamatergic cell classes. We identified hundreds of differentially expressed transcript isoforms arising from different patterns of alternative splicing, as well as alternative TSS between CA1 and CA3 cell preparations (253 DR exons, log2FC ≥ 1 or ≤ -1, p-value ≤ 0.01, Fig. 3a, Supplementary Fig. 9, Supplementary Table 3). These include key isoforms with well-characterized functional properties such as the mutually exclusive exons which regulate flip/flop variants of the Gria1 AMPAR subunit 33, or alternative last exons in Brevican, which control expression of a secreted Brevican isoform implicated in neuronal adhesion 34 (Fig. 3b). Similarly, cortical L4 excitatory neurons and hippocampal CA1 pyramidal neurons exhibited 276 differentially regulated exons (Figure 3a) and transcript isoforms derived from multiple patterns of alternative splicing (Figure 3c Nrxn3 as example, Supplementary Fig. 9, Supplementary Table 4). These comparisons highlight vastly divergent transcript isoform content between different classes of glutamatergic neurons, including closely related pyramidal cells from hippocampal sub-fields. By comparison, splicing programs were much more similar between hippocampal versus neocortical SST-positive interneurons (only 151 highly differentially regulated exons Fig. 3a, 3c cassette exon regulation in Fat1 as example, Supplementary Fig. 9, Supplementary Table 4, Supplementary Table 5 for an overview of differentially expressed genes and alternatively regulated splicing events). In sum, we conclude that alternative splicing plays a major role in diversifying molecular repertoires at the level of neuronal sub-classes and cell types within and across anatomical positions.
Fig. 3. Alternative splicing programs distinguish closely related neuronal populations within the hippocampus and across brain regions.
a, Cartoon illustrating the schematic representation of neuronal classes in the neocortex and hippocampus. The numbers shown indicate differentially regulated (DR) exons (log2(FC) ≥ 1 and ≤ -1, p-value ≤ 0.01) in pairwise comparisons between CamK2-positive CA1 and Grik4-positive CA3 pyramidal neurons (253 exons from 177 genes), between CA1 or CA3 neurons vs SST-positive hippocampal neurons and between neocortical (856 exons from 551 genes and 851 exons from 517 genes) and hippocampal SST-positive interneurons (151 exons from 103 genes) or layer 4 vs CA1 pyramidal neurons (276 exons from 209 genes). Note that only exons involved in AS events are indicated. Hippocampal SST-positive neurons show high diversity of alternative isoform expression compared to pyramidal cells. Also closely related glutamatergic populations (CA1 vs CA3 and CA1 vs Scnn1a) exhibit highly differential exon usage. On the other hand, SST-positive neurons isolated from distinct anatomical brain regions (hippocampus vs neocortex) present fewer but still significant differences in alternative isoform expression. s.o.=stratum oriens, s.r.=stratum radiatum, DG=dentate gyrus, CA=cornu ammonis. Note that this panel only displays select pairwise comparisons. Analysis for hippocampal SST interneurons considered all SST neurons without sub-regional distinction. b-c, Sashimi plots illustrating read distribution and splice junctions of transcripts differentially spliced between CA1 and CA3 pyramidal neurons (panel b) or between related cell classes across distinct brain regions (SSTHc vs SSTCx, CA1 vs Scnn1a glutamatergic neurons, panel c). Genomic coordinates, chromosome number, strand and exon number are indicated below. Coding regions are indicated as thicker boxes, non-coding as thinner boxes.In b, CamK2- and Grik4-positive glutamatergic neurons show differential usage of the mutually exclusive exons 16 and 17 in Gria1 (upper panel) and the alternative last exons 8 and 14 in Bcan (lower panel). In c, Nrxn3 shows differential rate of inclusion of cassette exon 22 in the hippocampal CA1 vs cortical Scnn1a-positive pyramidal neurons (upper panel). Neocortical SST-positive interneurons show preferential inclusion of cassette exons 26, 27 and 28 in Fat1, while hippocampal SST-positive cells exclude them (the lower panel).
Identification of neuronal subclass-specific splicing factors in neocortical and hippocampal cells
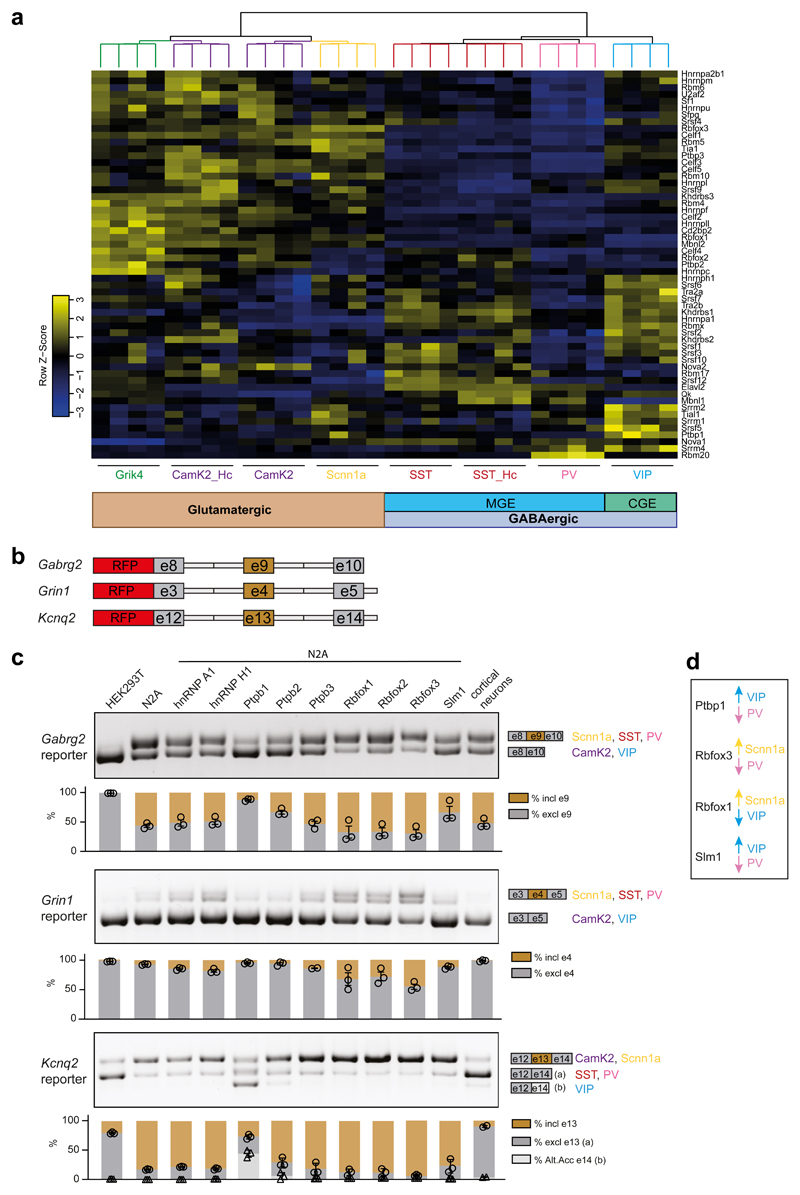
Most neuronal RNA binding proteins (RBPs) studied thus far are pan-neuronally expressed. Given the extensive differential alternative transcript regulation in neuronal sub-classes, we sought to identify RBPs that might regulate alternative splicing in a cell type-specific manner. We generated a hand-curated list of 57 bona fide splicing regulators based on databases and previous publications and evaluated their expression across neuronal populations (Supplementary Table 6). As expected, several splicing factor transcripts exhibited broad expression with little difference across neocortical and hippocampal cell classes (e.g., Hnrnpa2b1, Hnrnpl, Srrm1, Fig. 4a, Supplementary Table 6). By contrast, other splicing factors showed highly selective expression with some segregating between glutamatergic and GABAergic neuron groups and others highly enriched in certain neuron classes (Fig. 4a). For example, the Rbm20 transcript is preferentially expressed in PV-interneurons, Ptbp1 in VIP-interneurons, and Rbfox3 – also called NeuN – is preferentially expressed in glutamatergic cells (Fig. 4a, Supplementary Table 6). Fluorescent in situ hybridizations for select RBPs confirmed the differential expression patterns extracted from RiboTRAP sequencing data (Supplementary Fig. S10, Supplementary Table 6 for statistical analysis). To investigate whether some of the differentially expressed splicing factors represent candidates that drive cell type-specific alternative splicing choices, we employed splice reporter assays in Neuroblastoma 2A (N2A) cells (see Methods for details). We generated reporter constructs for exons that we found to be differentially regulated across neocortical neurons (alternative cassette exons in neurotransmitter receptors Gabrg2 and Grin1, and the voltage-gated potassium channel Kcnq2, Fig. 4b). Co-expression in N2A cells of several RBPs (e.g., hnRNP A1, hnRNP H1, Ptbp3) did not shift splicing patterns in vitro. On the other hand, co-expression of Ptbp1 (which is preferentially expressed in VIP-interneurons Fig. 4a,d) shifted splicing of Gabrg2 and Kcnq2 reporters to the pattern observed for endogenous mRNAs in VIP-interneurons (Fig 4c). Similarly, Rbfox3 shifted the Grin1 reporter splicing to the pattern enriched in Scnn1a cells (which express high levels of Rbfox3) (Fig. 4c,d). Thus, these differentially expressed splicing regulators represent possible candidates for the regulation of the respective splicing events in vivo. In summary, this analysis identifies candidate neuronal cell class-specific splicing regulators for the differential regulation of transcripts.
Fig. 4. Differential splicing factors expression for cell class-specific splicing programs in mature neurons.
a, Gene expression heatmap of a hand curated list of splicing factors in neocortical and hippocampal neuron classes. Sub-groups of splicing factors with highly significant or modest changes in gene expression across cell populations can be identified. Overall, expression rates of splicing factors across forebrain neurons populations can segregate cells according to their neurotransmitter phenotype and their developmental origin (glutamatergic vs GABAergic, Medial Ganglionic Eminence (MGE)-derived vs Caudal Ganglionic Eminence (CGE)-derived). For the complete list of enrichments and significance values for splicing factors across neocortical and hippocampal neurons, see Supplementary table 6. b, Cartoon illustrating the design of splicing reporters for Gabrg2, Grin1 and Kcnq2 (see methods for details). c, RT-PCR for splicing reporters overexpressed in HEK293T cells, Neuro2A (N2A) cells and cultured cortical neurons, or in combination with overexpression of several splicing factors (indicated above) in N2A cells. On the right, schematic representation of reporter exons amplified and cell types in which a given splicing pattern is enriched are indicated. For each sample, three PCR reactions were performed and band intensity was quantified. Representative images are shown. Below, histograms represent the percentage of inclusion (in brown) or exclusion (in light and dark gray) band intensity relative to the sum intensity of all bands. n=2-3 RT-PCRs, single data points and SEM are indicated. Circles represent quantification of exclusion values for all reporters, triangles, only for Kcnq2, the quantification of the alternative acceptor e14. Top PCR panel: Expression of the splicing reporter for exon 9 of Gabrg2 leads to differential exon inclusion in non-neuronal (HEK293T, excl. e9) versus neuronal (N2A or cortical neurons, 50% excl. e9) cells. Co-expression of Ptbp1 and, to lower extents, Ptbp2 and Slm1 (enriched in VIP) lead to higher exclusion rates, a pattern significantly enriched in VIP neurons. Co-expression of Rbfox1/2/3 slightly reduces exon exclusion rates, consistent with the splicing pattern observed in purifications of Scnn1a. Middle PCR panel: Similar effects of the same splicing factors can be observed for the splicing pattern of exon 4 of Grin1. Note that the amplification of Grin1 isoform including e4 generates a doublet band. Lower PCR panel: Exon 13 of Kcnq2 splice reporter is preferentially included in N2A cells. Addition of Ptbp1 and Ptbp2 induces the additional alternative acceptor site usage found in VIP neurons. Overall, these experiments indicate a correlation between splicing factor expression and alternative isoform usage. d, Highest or lowest expression levels in different cortical populations for Ptbp1, Rbofox1, Rbfox3 and Slm1 (Khdrbs2) are indicated by arrows.
Alternative splicing programs are highly dedicated to controlling synaptic interactions and neuronal architecture
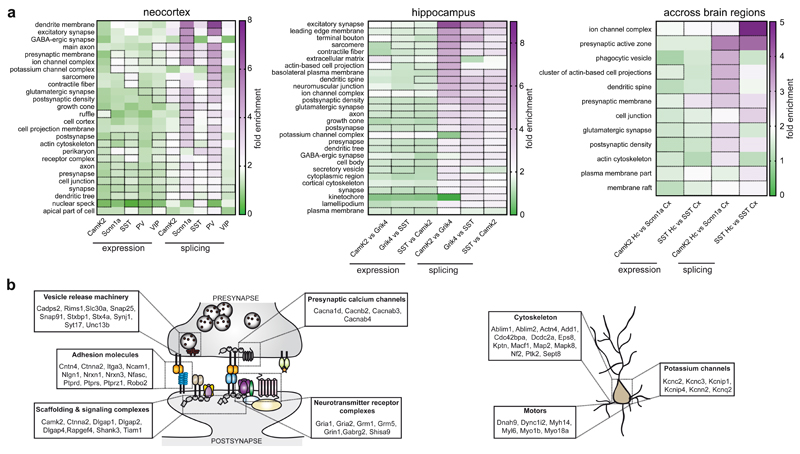
To probe which cellular properties are regulated by alternative splicing, we assessed the enrichment of Gene Ontology (GO) terms for transcripts regulated across all neocortical cell classes, hippocampal comparisons, and across brain regions. Given that alternative first exons result from transcriptional regulation, we excluded them from this analysis. Remarkably, the differential alternative splicing regulation almost exclusively targets transcripts encoding for synaptic proteins and intrinsic neuronal properties. Enrichment of the top GO terms significant for genes differentially regulated by splicing was 2 - 4 fold higher as compared to genes differentially expressed (Fig. 5a, Supplementary Fig. 11a,b for enriched categories of differentially expressed genes, Supplementary Table 7 for all GO terms). Note that the enrichment of genes encoding synaptic proteins was not simply a consequence of such genes containing larger numbers of exons (Supplementary Figure 11c). Specifically, the enriched GO terms map onto five key categories, which fundamentally shape synapse function and intrinsic neuronal properties: Adhesion complexes (e.g. Cntn4, Cntnap2, Ncam1, Nlgn1, Nrxn3, Nfasc, Ptprs, Robo2) implicated in formation and specification of neuronal synapses 35–37, voltage-gated calcium channels (e.g. Cacnb2, Cacnab4, Cacna1g, Cacna1d), presynaptic release machinery (e.g. Rims, Synj1, Stxbp1, Syt17, Unc13b), postsynaptic neurotransmitter receptor complexes (e.g. Grm1, Grm5, Gria1, Gria2, Shisa9) and associated scaffolding proteins (e.g. Camk2, Dlgap1, Rapgef4, Shank3, Tiam1) (Fig. 5b). All of these genes encode key regulators of synaptic function and plasticity. Three further categories highly targeted by cell type-specific alternative splicing are potassium channels, motor proteins, and regulators of cytoskeletal rearrangements – elements central for the control of intrinsic neuronal properties (Fig. 5c). In particular, potassium channels are key determinants of neuronal excitability at the level of after-hyperpolarization upon action potential firing (Kcnn2), at the level of M-currents (Kcnq2), or through calcium-dependent regulation of A-currents (Kcnip1,4) 38. Thus, neuronal cell type-specific alternative splicing programs specifically encode intrinsic neuronal properties and synapse specification.
Fig. 5. Alternative splicing programs are highly dedicated to the control of synaptic interactions and neuronal architecture.
a, Heatmap representing the fold enrichment of Gene Ontology (GO) terms for transcripts differentially regulated at gene expression level or by alternative splicing identified by the Panther Classification System (see methods for details). Terms listed were selected based on the splicing analysis and had to be significant in at least one neocortical population (left panel), hippocampal comparison (middle panel) or comparison across brain regions (right panel). Corresponding values from analysis of differentially expressed genes were included on the left. Fields for the statistically significant enrichments (Fisher’s exact test with Benjamini-Hochberg false discovery rate correction, p-value ≤ 0.05) are highlighted by a dashed outline. Overall, transcripts undergoing differential alternative splicing show higher fold enrichments compared to differentially expressed genes. Splicing-dependent transcript isoforms in VIP interneurons present lower, whereas Scnn1a and PV exhibit higher fold enrichment of GO categories. See Supplementary Table 7 for the raw output from the GO analysis. b, Cartoon illustrating the main categories of genes whose alternative splicing is differentially regulated between cell populations (log2(FC) ≥ 1 and ≤ -1, p-value ≤ 0.01). Among the most enriched categories, we find genes encoding presynaptic proteins modulating calcium influx or vesicle fusion, pre- and postsynaptic adhesion molecules and postsynaptic scaffolding molecules. c, Cartoon illustrating examples of differentially expressed transcript isoforms encoding for proteins which modulate intrinsic properties of neurons (e.g., potassium channels, proteins involved in cytoskeletal remodeling and cellular transport along neurites).
Discussion
Previous studies highlighted an expansion of splicing complexity across vertebrate species with a particular increase in alternative exon usage in the brain 4. This increase in alternative splicing may relate to neuronal cell types and functions in multiple ways. Single gene studies illustrated stochastic splice isoform choices at the single cell level 39,40 but also reproducible splicing patterns linked to cell types 16,20,41–44. Here, we demonstrate that complex alternative splicing programs define sub-classes of cortical and hippocampal neuron types. Thus, the selection of cell type-specific transcript variants is not an exceptional feature for individual protein families but a fundamental program of highly differentiated cell types in a complex organism. During embryonic development, the lineage decisions for interneuron and pyramidal cell differentiation are mainly driven by transcription factor codes 45,46. We propose that cell type-specific expression of RNA-binding proteins imposes splicing-dependent regulation for terminal differentiation of these neuron classes. Consistent with this notion, several of the candidate splicing specificity factors that we mapped here are already detected in interneurons at embryonic stages of development 46,47.
The RiboTrap approach used in our study facilitates interrogation of splice isoforms with excellent coverage across the entire transcript. Low level background contamination for some cell classes may influence detected splicing differences, particularly for widely expressed non-neuronal genes. However, the RiboTrap approach benefits from deep coverage of splice junctions and rare transcript isoforms that go undetected or cannot reliably be quantified from single cell sequencing data. Moreover, mapping ribosome-associated mRNAs focuses the analysis on transcript isoforms that are recruited by the translational machinery.
An unexpected finding in our study was the highly divergent usage of alternative transcription start sites across neuronal populations. This suggests prominent roles for cell type-specific enhancers and promoters in generating transcript isoforms. We propose that this complex transcript regulation evolved not only to modify protein isoforms but also to afford unique spatio-temporal modulation of neuronal gene expression. Complex alternative splicing programs control diverse biological processes from chromatin and RNA regulators, to ion homeostasis and mitochondrial function. Considering this broad range of splicing-regulated processes, it is remarkable that the neuronal cell type-specific splicing programs are selectively geared to the control of synaptic and intrinsic neuronal properties. Splice isoforms of neuronal receptors, ion channels, synaptic adhesion and scaffolding proteins exhibit fundamentally divergent functions 19,48 and significant splicing disruptions have been linked to neurodevelopmental disorders 30. In humans, more than 90% of gene products are modified by alternative splicing. A major impediment for exploring the functional relevance of transcript isoforms in neuronal wiring has been the lack of knowledge of how splice isoforms are arrayed over neuronal cell types. Our comprehensive genome-wide analysis uncovers hundreds of cell class-specific transcript isoforms encoding key regulators of synaptic function and intrinsic neuronal properties. To maximize the accessibility of this large dataset and to simplify the identification of differentially expressed transcript isoforms, we established a web-based “splicecode database” where users can retrieve differential isoform expression data for any gene of interest (https://scheiffele-splice.scicore.unibas.ch). In the future, targeted manipulation of cell type-specific splicing events may open the door for a new class of therapeutic interventions in disease states.
Methods
Mice
All procedures involving animals were approved by and performed in accordance with the guidelines of the Kantonales Veterinäramt Basel-Stadt. Male and female mice were used in this study.
Rpl22-HA (RiboTag) mice 49, Pvalb-cre mice 50, SST-cre mice 51, CamK2-cre mice 52, Grik4-cre mice 53, VIP-cre mice 51 and Scnn1a-cre mice 54 were obtained from Jackson Laboratories (Jax stock no: 011029, 017320, 013044, 005359, 006474, 031628, 009613, respectively). All lines were maintained on a C57Bl6/J background. The specificity of cre-lines for recombination of the Rpl22-allele was confirmed by immunohistochemistry and matched previous reports in the literature.
Immunohistochemistry and imaging
Animals (males and females) from postnatal day 25 to 42 were transcardially perfused with fixative (4% paraformaldehyde in 100mM phosphate buffer, pH 7.4). The brains were post-fixed overnight in same fixative at 4°C. Coronal brain slices were cut between Bregma -1.43 and -2.15 (including somatosensory cortex and dorsal hippocampus) at 50 µm with a vibratome (Leica Microsystems VT1000). For immunohistochemistry, brain sections were kept in PBS before incubation for 1 hour with blocking solution containing 0.05% Triton X-100 and 10% normal donkey serum. Slices were incubated with primary antibodies in blocking solution at 4°C overnight and washed three times in PBS containing 0.05% Triton X-100, followed by incubation for 2 hours at room temperature with a secondary antibody. Sections were washed three times in PBS before mounting onto microscope slides with Fluoromount-G (SouthernBiotech, 0100-01). The following primary antibody was used in this study: rat anti-HA (Roche, 11867431001, 1:1000); Secondary antibody was: donkey anti-rat IgG-Cy3 (Jackson ImmunoResearch, 712-165-153, 1:1000). Hoechst dye was co-applied with primary antibody at a final concentration of 0.5 µg/ml. Images for assessing the Rpl22-HA expression were acquired at room temperature on a Slidescanner AxioScan.Z1 (Zeiss) using 10X objective. Stacks of 24 µm width (4 µm interval) were acquired and were then processed by doing maximum projection. Images were assembled using Fiji and Illustrator Software.
RNA isolation by RiboTRAP pulldowns
For Ribotag purifications, the procedure of Heiman and colleagues for affinity-purification of polysomes 55 was modified as follows: Neoortices and hippocampi from mice between postnatal day 25 and 28 (males and females) were dissected in ice-cold PBS. For CamK2-cre and Grik4-cre mice, given the later onset of cre-recombinase expression, hippocampi between postnatal day 39 and 42 were used. Moreover, control samples from animals negative for either Rpl22 or cre-recombinase were used to ensure for specificity of the pulldown. For interneuron pulldowns, four hippocampi or cortical hemispheres (2 animals per condition) were lysed in respectively 1 mL or 14 mL (1:20 weight per volume) of homogenization buffer containing 100mM KCl, 50mM Tris-HCl pH 7.4, 12mM MgCl2, 100µg/mL cycloheximide (Sigma-Aldrich), 1mg/mL heparin (Sigma-Aldrich), 1x complete mini, EDTA-free protease inhibitor cocktail (Roche), 200 units/mL RNasin© plus inhibitor (Promega) and 1mM DTT (Sigma-Aldrich). Excitatory neuron pulldowns were performed on single animals and hippocampi or neocortices were homogenized in 0.5mL or 7mL of homogenization buffer, respectively. The lysate was centrifuged at 2’000 x g for 10 minutes. Igepal-CA630 (Sigma-Aldrich) was then added to the supernatant to a final concentration of 1%. After 5 minutes incubation on ice, the lysate was centrifuged at 12’000 x g for 10 minutes. Before incubation with beads, 1% of the supernatant was taken (Input) and resuspended in 350 µL of RLT plus buffer from RNeasy Plus Micro Kit (Qiagen, 74034) supplemented with 2-Mercaptoethanol (Sigma-Aldrich) as suggested by manufacturer’s instructions. Anti-HA coupled magnetic beads (Pierce, 88837) were added to the supernatant: 140 µL of beads for all neocortical samples, 20 µL for CamK2-cre and Grik4-cre hippocampal samples and 15 µL for SST-cre hippocampal samples. Incubation was performed under gentle rotation at 4°C for 3-4 hours. After incubation, beads were quickly washed 3-4 times in washing buffer containing 300mM KCl, 1% Igepal-CA630 (Sigma-Aldrich), 50mM Tris-HCl, pH7,4, 12mM MgCl2, 100 µg/mL Cycloheximide (Sigma-Aldrich) and 1mM DTT (Sigma-Aldrich). Beads were than eluted in 350 µL of RLT plus buffer from RNeasy Plus Micro Kit (Qiagen) supplemented with 2-Mercaptoethanol (Sigma-Aldrich).
RNA purification, quantification, quality check and RT-qPCRs
RNA purification (for both input and immunoprecipitated RNA) was performed using RNeasy Plus Micro Kit (Qiagen), following the manufacturers’ instructions. RNA was quality-checked on the Bioanalyzer instrument (Agilent Technologies) using the RNA 6000 Pico Chip (Agilent, 5067-1513). Only RNA samples with RNA integrity number (RIN) higher than 7.5 were used for the following steps. RNA concentration was quantified by Fluorometry using the QuantiFluor RNA System (Promega, E3310). 90 ng and 20 ng of RNA was reverse transcribed from neocortical and hippocampal samples, respectively, using random hexamers (Promega) and Superscript III Reverse Transcriptase (Invitrogen,18080093).
To determine the fold-enrichment of respective marker genes in immunoprecipitated RNA compared to input purifications, DNA oligonucleotides were used with FastStart Universal SYBR Green Master (Roche, 4913914001) and comparative CT method. Samples were considered to be specific if immunoprecipitated RNA exhibited correct de- or enrichments of respective marker genes and if RNA of control samples did not show any selectivity for marker genes. For each assay, two technical replicates were performed and the mean was calculated. The mRNA levels were normalized to gapdh mRNA. RT-qPCR assays were analyzed with the StepOne software.
The assessment of fold-enrichments for RNA obtained from VIP-cre was performed using commercial Taqman probes from Applied Biosystems: Vip (Mm00660234_m1), vGat (Mm00494138_m1), vGlut1 (Mm00812886_m1), Gfap (Mm01253033_m1).
DNA Oligonucleotides used with SYBR Green-based real-time PCR (name and sequence 5’->3’ are indicated):
| CamK2-F: | AGAAGTTCAATGCCAGGAG |
| CamK2-R: | CAGAAGATTCCTTCACACCA |
| Gad67-F: | GTACTTCCCAGAAGTGAAGAC |
| Gad67-R: | GAATAGTGACTGTGTTCTGAGG |
| Gapdh-F: | GCTTGTCATCAACGGGAAG |
| Gapdh-R: | TTGTCATATTTCTCGTGGTTCA |
| Gfap-F: | CTCGTGTGGATTTGGAGAG |
| Gfap-R: | AGTTCTCGAACTTCCTCCT |
| Pvalb-F: | CATTGAGGAGGATGAGCTG |
| Pvalb-R: | AGTGGAGAATTCTTCAACCC |
| Pvrl3-F: | TGACTGTGTTAGTTGAACCCA |
| Pvrl3-R: | CTGCTACTGTCTCATTCCCT |
| Rgs14-F: | ATGGATTTGGAGAATCCAGTG |
| Rgs14-R: | TTCACTCATCTTTGCATCCG |
| Scnn1a-F: | AAAGAGAAGCGGGAGTCAGC |
| Scnn1a-R: | CGGTGAGTTGGAGACGTCAA |
| SST-F: | CGTCAGTTTCTGCAGAAGTC |
| SST-R: | AGTACTTGGCCAGTTCCTG |
| Tdo2-F: | ATGAGTGGGTGCCCGTTTG |
| Tdo2-R: | GGCTCTGTTTACACCAGTTTGAG |
| Vgat-F: | CGTGACAAATGCCATTCAG |
| Vgat-R: | AAGATGATGAGGAACAACCC |
| Vglut1-F: | ACCCTGTTACGAAGTTTAACAC |
| Vglut1-R: | CAGGTAGAAGGTCCAGCTG |
| Wsf1-F: | CATCATTCCCACCAACCTG |
| Wsf1-R: | TACTTCACCACCTTCTGGC |
Library preparation and Illumina sequencing
For all five neocortical and three hippocampal neuronal populations, 4 biological replicates with RIN > 7.5 were analyzed, resulting in a total of 32 individual samples. Library preparation for all samples was performed with 50 ng of RNA using the TruSeq PolyA+ Stranded mRNA Library Prep Kit High Throughput (Illumina, RS-122-2103). Libraries were quality-checked on a Fragment Analyzer (Advanced Analytical) using the Standard Sensitivity NGS Fragment Analysis Kit (Advanced Analytica, DNF-473), revealing excellent quality of libraries (average concentration was 49±14 nmol/L and average library size was 329±8 base pairs). The 32 samples were pooled to equal molarity and the pool was quantified by PicoGreen Fluorometric measurement. The pool was adjusted to 10pM for clustering on C-Bot (Illumina) and then sequenced Paired-End 101 bases using the HiSeq SBS Kit v4 (Illumina, FC-401-4003) on a HiSeq 2500 system. Primary data analysis was performed with the Illumina RTA version 1.18.66.3 and bcl2fastq-v2.20.0.422.
QC and RNA-seq pre-processing
The splicing analysis of RNA-Seq data were performed by GenoSplice technology (www.genosplice.com). Data quality, reads repartition (e.g., for potential ribosomal contamination), and insert size estimation were performed using FastQC, Picard-Tools, Samtools and rseqc. Reads were mapped using STARv2.4.0 56 against the exons defined in the proprietary Mouse FAST DB v2016_1 database27, using a mismatch cutoff of 2 and discarding reads with 10 or more alignments. The minimum chimeric segment length was 15. Read counts were summarized using featureCounts 57 in two stages. First, unique reads per exon were counted. In the second stage, multimapping reads were fractionally allocated to exons based on the distribution of unique counts of exons within a gene. Total counts were then calculated based on three constitutivity classes defined in FAST DB: class 2 includes exons present in more than 75% of annotated transcripts for a gene (“constitutive”), class 1 includes exons present in 50-75% of transcripts (“semi-constitutive”), and class 0 includes exons present in less than 50% of transcripts (“alternative”). Total counts per gene were summed from constitutivity class 2 exons if their FPKM values exceed 96% of the background FPKM based on intergenic regions. If counts from class 2 exons were insufficient to exceed the detection threshold, class 1 and eventually class 0 exon counts were included to reach the detection threshold.
Differential gene expression analysis
The analysis of differential expression was conducted using DESeq2 v1.22.2 58. The input read count matrix was the same as used for the splicing analysis. Neocortical samples were modeled as a group with a shared base mean, and each sample set was contrasted against all neocortical samples. The hippocampal samples were contrasted in all pairwise combinations. A series of additional pairwise contrasts were conducted for comparisons across anatomical brain regions. Results for these contrasts using the Ward test and Benjamini Hochberg for p-value adjustment as implemented in DESeq2 are compiled in an Excel workbook (Supplementary Table1). For the principal component analysis, counts were normalized using the variance stabilizing transform (vst) as implemented in DESeq2. For heatmaps and the web app plots, the internal normalization factors of DESeq2 were used to normalize the counts.
Alternative splicing analysis
Analysis at the splicing level is first performed taking into account reads mapping to exonic regions and to exon-exon junctions (EXON analysis) in order to potentially detect new alternative events that could be differentially regulated (i.e., without taking into account known alternative events). When mapping to exon-exon junctions, reads were assigned to both exons, therefore counted twice (the minimum number of nucleotides for a read to be considered mapped to an exon is 7).
In order to consider an exon expressed, FPKM values for exons must be greater than 96% of the background FPKM value based on intergenic regions. Only exons expressed in at least 3 of the 4 biological replicates of each condition and in at least one of the compared experimental conditions were further analyzed.
For EXON analysis illustrative cartoon, refer to Supplementary Figure S5. Briefly, for every expressed exon from expressed genes, a Splicing Index [SI, defined as the ratio between read density on the exon of interest (= row number of reads on the exon/exon length in nucleotides) and read density on constitutive exons of the gene; “class 2”] are generated, as well as fold-change (log2(FC), calculated by comparing the SI value in one condition to the mean SI value in all conditions considered) and p-value (unpaired Student’s t-test). Results are considered statistically significant for p-values ≤ 0.01 and log2(FC) ≥ 1 or ≤ -1.
Analysis at the splicing level is also performed by taking into account known splicing patterns (PATTERN analysis) annotated in the FAST DB database 27 (i.e., for each gene, all annotated splicing patterns are defined, and a Splicing Index (SI) is generated from the comparison of normalized read density on the alternative annotated patterns. For PATTERN analysis illustrative cartoon refer to Supplementary Figure S5). All types of alternative events can be analyzed: Alternative transcription start site, alternative last exons, cassette exon, mutually exclusive exons, alternative 5’ donor splice site, alternative 3’ acceptor splice site, intron retention, internal exon deletion and complex events (corresponding to mix of several alternative event categories). In Figure 2 and S8 we have merged intron retention and internal exon deletion events to one single category (“intron retention”).
Pattern analysis is performed for every condition; log2(fold-change) of SI against all conditions considered and p-value (unpaired Student’s t-test) are generated. Results are considered statistically significant for p-values ≤ 0.01 and log2(FC) ≥ 1.
FAST DB database includes annotations of 4965 microexons (defined as exons 3-27 nucleotides long), out of the 268827 total exons annotated (1.8%). In the neuronal populations analyzed, we identified 4140 (in the neocortex) and 3889 (in the hippocampus) microexons reaching the cutoff for detection (2.8% and 2.7% of the total exons detected in these forebrain regions, respectively). Specifically, amongst the total DR exons within a neuronal class we find 5.1% of microexons in Camk2, 3.8% in Scnn1a, 5.3% in SST, 4.7% in both PV and VIP.
Validation of regulated alternative splicing events
We used RT-PCR for experimental validation of differentially regulated splicing events detected by RNA-Seq. The cDNA amounts and PCR cycle numbers were carefully titrated to ensure linear amplification range and avoid signal saturation that would interfere with quantification. Standard PCR reactions were performed using 5X Firepol Master mix (Solis BioDyne, 04-11-00125) and designed exon-flanking DNA oligonucleotides. PCR products were resolved by gel electrophoresis and images acquired on a Biorad analyzer and processed in Fiji. Supplementary Figure 12 shows uncropped gel images. DNA Oligonucleotides used for RT-PCR are listed below (name and sequence 5’->3’ are indicated).
| Anks1b-exon21-F: | GCGATGCAAGGAGGAGAAGA |
| Anks1b-exon23-R: | GAGGTCGCAAGGTGATGGAA |
| Cadps-exon18-F: | CACGCCTATCTGAGTATGCCA |
| Cadps-exon21-R: | GTCTTCGAGCTTTTTGGCGG |
| Ctnna2-exon17-F: | CAAAGTGAAGGCCGAGGTTC |
| Ctnna2-exon19-R: | TTTTGGCTGCCTGGATGAGT |
| Cacna1d-exon32-F: | GAACATGGTCTTCACAGGGGT |
| Cacna1d-exon33-R: | CTGCCGATTACGATGAGGGA |
| Cacna1d-exon32-F: | ACATGGTCTTCACAGGGGTC |
| Cacna1d-exon34-R: | CAACGACGCTACCAACAACA |
| Nlgn1-exon4-F: | ACCATGGCACTTCCCAGATG |
| Nlgn1-exon6-R: | ACCTTCCATGTAAGAGCCGC |
| Stxbp1-exon19-F: | AGATGCGCTGTGCTTACGAA |
| Stxbp1-exon21-R: | ACCCTAAGCCTCCCCCATAG |
| Kcnq2-exon9-F: | CTAACCTCTCACGCACCGAC |
| Kcnq2-exon11-R: | CTCCAGCTGGTTCAGAGGTG |
| Cacna1d-exon8-F: | TGCGTTCTCAGGGAATGGAC |
| Cacna1d-exon9-R: | TGACAAAATACACCCAGGGCA |
| Cacna1d-exon8-F: | TGGCACGGAATGTAGGAGTG |
| Cacna1d-exon10-R: | ATACACCCATGGCCATTCCC |
| Gria1-exon15-F: | ACCGTCTGTGTTTGTTCGGA |
| Gria1-exon16-R: | TTTGTCCAAAAGCCCCTGCT |
| Gria1-exon15-F: | ACCGTCTGTGTTTGTTCGGA |
| Gria1-exon17-R: | TCCTTGCTTCCACATTCCCC |
| Shisa9-exon5-F: | AGCTCAACAAGTACGCCTCC |
| Shisa9-exon7-R: | TTGGTGCGAGACTTCTGTCC |
| Kcnq2-exon12-F: | CAGCCAGAGCCATCACCAAG |
| Kcnq2-exon14-R: | TCGGGCTGTCATCAAGACTC |
| Dlgap4-exon10-F: | CCTCCACGAACCACATCGAA |
| Dlgap4-exon11-R: | TGGGAACCGCTCCTTCTAGT |
| Dlgap4-exon12-F: | CCAGTCCATCGGGATTCAGG |
| Dlgap4-exon13-R: | AGGAACCAGTAGCCATCCCT |
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed adapting the ViewRNA ISH Cell Assay kit (Invitrogen, QVC0001) for tissue sections. P25 wild-type mouse brains (C57BL/6j background) were snap frozen in liquid nitrogen and 18 μm coronal sections were cut between Bregma -1.43 and -2.15 (including somatosensory cortex and dorsal hippocampus) on a cryostat. Sections were fixed at 4°C overnight with 4% paraformaldehyde in 100mM phosphate buffered saline, pH 7.4. The procedure followed the manufacturers’ instructions, except for protease treatment, which was performed at a dilution of 1:100.
Transcripts for splicing factors and cell type-specific markers were detected with the following commercial probes (Invitrogen): Celf4 (VB1-3044679), Elavl2 (VB1-3030263), Ptpb3 (VB1-3047128), Rbfox3 (VB1-13443), Camk2a (VB4-3112005), Pvalb (VB4-19638), Rorb (VB4-3131885), Sst (VB4-3112424) and Vip (VB4-3112423).
Images were acquired at room temperature with an upright LSM700 confocal microscope (Zeiss) using 40X Apochromat objectives. Stacks of 10-13 µm width (0.6 µm interval between stacks) were acquired from layer 2-3 (L2-3) of primary somatosensory area (S1) for CamK2-, SST-, PV- and VIP-positive neocortical cells and from layer4 (L4) of S1 for Rorb-positive cells. For CA1- or CA3-specific pyramidal cells, images were acquired from CamK2-positive cells in cornu ammonis 1 and 3 regions of hippocampus, while for hippocampal SST-positive interneurons images were taken from cells residing in stratum oriens. Cell types were identified based on the presence of the corresponding marker transcript. A region of interest (ROI) was drawn to define the area of the cell and dots in the ROI were manually counted throughout the stacks. The number of dots in the ROI were then normalized to 100 μm2 area. Images were assembled using Fiji and Illustrator Software.
Heatmaps and sashimi plots
For clustering analysis of gene expression, Normalized Feature Counts values were used and data were scaled by row. For clustering analysis of Splicing Index (SI) values obtained from EXON analysis, data were scaled by row and by column. Exons with NA values (not assigned, when no reads are mapping to constitutive parts of the gene) or Inf values (infinite, when no reads are mapping to the exon) were excluded, in order to not be biased by very lowly expressed genes or exons. In all cases, distance was calculated by Pearson correlation and the resulting distance matrix was clustered using Ward.D2. Heatmaps were generated in R using the heatmap.2 function of gplots package. Sashimi plots were generated using the MISO software package 59.
Plasmids
Splicing reporters for Gabrg2, Grin1 and Kcnq2 were assembled from synthetic DNA fragments ordered as gBlocks (Integrated DNA Technologies); for Grin1, an AT-rich intronic region that could not be synthesized was amplified from genomic mouse DNA using the following primers (name and sequence 5’->3’ are indicated):
| Grin1_i4_F: | GTATATATGCATGGACGTGCACGC |
| Grin1_i4_R: | CTGTGGTATGAGCAGGAGCGTTAGC |
Reporters are composed of two flanking constitutive exons, the alternative exon (exon 9, 4 or 13 for Gabrg2, Grin1 and Kcnq2, respectively) and the first and last 500 or 300 nucleotides of intronic regions (which contains regulatory elements for splicing reaction). A translational stop codon was included at the end of the last constitutive exons of Grin1 and Kcnq2 reporters. In the case of Gabrg2, exon 10 represents the last exon.
Chromosomal coordinates of splicing reporters are:
| Gabrg2: | 41915784 – 41916489, 41913470 – 41914493, 41912319 - 41913091 |
| Grin1: | 25312928 – 25313604, 25310375 - 25311905 |
| Kcnq2: | 181096650 – 181096979, 181091939 – 181092592, 181088347 – 181088870 |
Splicing reporters were then cloned into pmRFP-c1 vector cutting with SacI and SalI. All reporter plasmids are available on request.
The plasmids encoding splicing factors used are: pCMV-Ptbp1-His, pCMV-Ptbp2-His, pCMV-Ptbp3-His, pCMV-hnRNPA1-YFP, pCMV-hnRNPH1-YFP, pCS3-myc6-Rbfox1 (A016), pCS3-myc6-Rbfox2 (F011), pCS3-myc6-Rbfox3 (S).
Cell cultures and transfection
Cortical neuron cultures were prepared from E16.5 mouse embryos. Neocortices were dissociated by addition of papain (Worthington Biochemical, LK003176) for 30 min at 37 °C. 250,000 cells/wells were plated in 12-well plates and they were maintained in Neurobasal Medium (Gibco, 21103-049) containing 2% B27 supplement (Gibco, 17504-044), 1% GlutaMAX supplement (Gibco, 35050-038), and 1% penicillin/streptomycin (Sigma, P4333). At DIV7, cortical cultures were transfected with 400 ng/well of splicing reporters and Lipofectamine 3000 reagent (ThermoFisher Scientific, L3000008) diluted in opti-MEM medium (Gibco, 31985-062) using a 1:1.5 DNA-Lipofectamine ratio.
20,000 Neuroblastoma 2a (Neuro2a) or HEK293T cells (obtained from ATCC) were plated in 96 well plates and were kept in DMEM (Sigma, D5796) supplemented with 10% FBS (Gibco, 10270106) and 1% penicillin/streptomycin at 37°C. After 24h, cells were transfected using FuGENE HD Transfection reagent (Promega, E2691) with 50 ng of splicing reporter DNA alone or in combination with 50 ng of splicing factor DNA.
RNA isolation, Reverse transcription and RT-PCR
24 h (for Neuro2a and HEK293T cells) or 48h (for cortical neurons) post-transfection, cells were lysed with 100 or 600 μl, respectively, of RLT buffer from RNeasy Plus Micro Kit (Qiagen, 74034) supplemented with 2-Mercaptoethanol (Sigma-Aldrich) and RNA was purified according to the manufacturer’s instructions. 400 ng of RNA was reverse transcribed from all samples using random primers (Promega, C118A) and ImProm-II™ Reverse Transcriptase (Promega, A3802).
For evaluation of differential reporter processing in different cells and conditions, cDNA amounts and PCR cycle numbers were carefully titrated to ensure linear amplification range. Standard PCR reactions were performed using 5X Firepol Master mix (Solis BioDyne, 04-11-00125) and DNA oligonucleotides targeting the RFP sequence (to avoid detection of endogenous transcripts) and the last flanking exons.
DNA Oligonucleotides used for standard PCR (name and sequence 5’->3’ are indicated):
| RFP_F: | AAGCTGGACATCACCTCCCA |
| Gabrg2_e10_R: | ATGGTTGCTGATCTGGGACG |
| Grin1_e5_R: | ATCAGCAGAGCCGTCACATT |
| Kcnq2_e14_R: | TCGGGCTGTCATCAAGACTC |
Gene ontology
Analysis of GO terms both for neocortical and hippocampal samples was performed using the statistical overrepresentation test 60 of the PANTHER classification system (PANTHER14.1, released 2019-03-08 and 2019-04-17), available on http://pantherdb.org. Genes showing significant differential expression (log2(FC) ≥ 0.6 and ≤ -0.6, p-value ≤ 0.05; base mean for neocortex: all neocortical samples) and genes with significant alternative splicing events (log2(FC) ≥ 1 & ≤ -1; p-value ≤ 0.01 from either EXON or PATTERN analysis) were analyzed using the GO cellular component annotation data set and Fisher's Exact test with Benjamini-Hochberg false discovery rate correction for multiple testing. Alternative first exon events were excluded to analyze the functional role of alternative splicing programs only. In order to be considered significant, GO terms must have a minimum number of 10 genes, fold-enrichment ≥ 2 and False Discovery Rate (FDR) ≤ 0.05. As background reference list, all genes expressed (see methods for details of gene expression) in the neocortex for neocortical comparisons, or in either cell class of the pairwise comparisons were used. Panther output list GO terms in a hierarchical organization, enabling identification of super-categories which were further analyzed. Moreover, only terms significant in at least one neocortical population, hippocampal comparison or across anatomical region were used for heatmap visualization. In Fig. 5a, corresponding fold changes of the gene expression analysis were incorporated. Redundant term categories were excluded.
General statistical methods
Sample sizes were chosen based on previous experiments and literature surveys. No statistical methods were used to pre-determine sample sizes. Exclusion criteria used throughout this manuscript were pre-defined. There are detailed descriptions in the respective sections of the methods. Group assignment was defined by genotype, thus, no randomization was necessary. Knowledge of experimental conditions was needed for proper execution of experiments. Therefore, investigators were not blinded during data collection and/or analysis. Appropriate statistical tests were chosen based on sample size. Due to “n” in the analysis normal distribution and equal variances of measures were not formally tested. Thus, individual data points or measures are presented in the manuscript and data distribution was assumed to be normal. Sequencing analysis was performed on four animals per genotype exhibiting similar variances. N-numbers for in situ hybridizations and RT-PCRs are indicated in the figures. P-value calculations have been performed using the student t-test, Ward test, or one way ANOVA with Tukey’s multiple comparison test. FDR calculations were performed with the Benjamini Hochberg correction.
Supplementary Material
Acknowledgements
We are grateful to F. Doetsch, O. Mauger and L. Xiao for constructive comments on the manuscript and to members of the Scheiffele Lab for discussions, in particular to T.M. Nguyen for setting up the protocol for RiboTRAP purifications and C. Bornmann for expert help with fluorescent in situ hybridizations. We are grateful to S. Hrvatin and M. E. Greenberg for sharing detailed data from single cell sequencing studies for comparison to our dataset and to B.Rico for sharing data at an early stage of this project. We thank P. de la Grange, N. Robil and A. Jolly at Genosplice for help with analysis, F. Ambrosetti for occasional support with coding. Part of calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing center at the University of Basel, with support by the SIB - Swiss Institute of Bioinformatics. Sequencing and library preparations were performed with support from the Life Science Training Facility and the Quantitative Genomics Facility Basel. L.T. was supported by a Fellowship from the Boehringer Ingelheim PhD Fonds and the Doris Dietschy Stiftung. This work was supported by funds to P.S. from the Swiss National Science Foundation, a European Research Council Advanced Grant (SPLICECODE), and the Kanton Basel-Stadt.
Footnotes
Data availability
Detailed analyzed data is included as supplementary material. Raw sequencing data was deposited at GEO (GSE133291). Differential gene expression and splicing data for individual genes is provided on the freely available SpliceCode web-site (https://scheiffele-splice.scicore.unibas.ch). All renewable reagents and detailed protocols will be made available on request.
Accession codes
Code availability
Data analysis used standard software packages cited in the methods section. The FastDB database for quantitative splicing analysis is a proprietary database accessible through Genosplice Technology (http://www.genosplice.com/).
Author contributions
This work was jointly conceived by E.F., L.T. and P.S., all wet lab procedures were performed by E.F. and L.T., data analysis was conducted by E.F., L.T. and G.F., website design was performed by G.F., and the manuscript was jointly written by E.F., L.T. and P.S., with editing provided by G.F.
Competing interests
The authors have no competing interests.
References
- 1.Ramón y Cajal S. Histology of the nervous system of man and vertebrates. Oxford University Press; 1995. [Google Scholar]
- 2.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017;18:530–546. doi: 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa-Morais NL, Irimia M, Pan Q, et al. Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science (New York, N.Y. 2012;338:1587–1594. doi: 10.1111/aman.12147. [DOI] [PubMed] [Google Scholar]
- 5.Tapial J, et al. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome research. 2017;27:1759–1768. doi: 10.1101/gr.220962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154:75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traunmüller L, Gomez AM, Nguyen T-M, Scheiffele P. Control of neuronal synapse specification by highly dedicated alternative splicing program. Science (New York, N.Y. 2016;352:982–986. doi: 10.1126/science.aaf2397. [DOI] [PubMed] [Google Scholar]
- 8.Beffert U, et al. Modulation of synaptic plasticity and memory by Reelin involves differential splicing of the lipoprotein receptor Apoer2. Neuron. 2005;47:567–579. doi: 10.1016/j.neuron.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Yap K, Xiao Y, Friedman BA, Je HS, Makeyev EV. Polarizing the Neuron through Sustained Co-expression of Alternatively Spliced Isoforms. Cell reports. 2016;15:1316–1328. doi: 10.1016/j.celrep.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taliaferro JM, et al. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol Cell. 2016;61:821–833. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calarco JA, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Wang ET, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quesnel-Vallieres M, et al. Misregulation of an Activity-Dependent Splicing Network as a Common Mechanism Underlying Autism Spectrum Disorders. Mol Cell. 2016;64:1023–1034. doi: 10.1016/j.molcel.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 14.Vuong CK, et al. Rbfox1 Regulates Synaptic Transmission through the Inhibitory Neuron-Specific vSNARE Vamp1. Neuron. 2018;98:127–141. doi: 10.1016/j.neuron.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JA, et al. Cytoplasmic Rbfox1 Regulates the Expression of Synaptic and Autism-Related Genes. Neuron. 2016;89:113–128. doi: 10.1016/j.neuron.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wamsley B, et al. Rbfox1 Mediates Cell-type-Specific Splicing in Cortical Interneurons. Neuron. 2018 doi: 10.1016/j.neuron.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Black DL. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet. 2013;29:442–448. doi: 10.1016/j.tig.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlanis E, Scheiffele P. Regulation of neuronal differentiation, function, and plasticity by alternative splicing. Annual Review of Cell and Developmental Biology. 2018;34:451–469. doi: 10.1146/annurev-cellbio-100617-062826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito Y, et al. Differential NOVA2-Mediated Splicing in Excitatory and Inhibitory Neurons Regulates Cortical Development and Cerebellar Function. Neuron. 2019;101:707–720 e705. doi: 10.1016/j.neuron.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weyn-Vanhentenryck SM, et al. Precise temporal regulation of alternative splicing during neural development. Nat Commun. 2018;9:2189. doi: 10.1038/s41467-018-04559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell. 2016;166:1147–1162 e1115. doi: 10.1016/j.cell.2016.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, et al. Axonogenesis Is Coordinated by Neuron-Specific Alternative Splicing Programming and Splicing Regulator PTBP2. Neuron. 2019;101:690–706 e610. doi: 10.1016/j.neuron.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tress ML, Abascal F, Valencia A. Alternative Splicing May Not Be the Key to Proteome Complexity. Trends in biochemical sciences. 2017;42:98–110. doi: 10.1016/j.tibs.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weatheritt RJ, Sterne-Weiler T, Blencowe BJ. The ribosome-engaged landscape of alternative splicing. Nat Struct Mol Biol. 2016;23:1117–1123. doi: 10.1038/nsmb.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC bioinformatics. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal S, et al. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome research. 2011;21:1260–1272. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes A, Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic acids research. 2018;46:582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irimia M, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tushev G, et al. Alternative 3' UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Neuron. 2018;98:495–511 e496. doi: 10.1016/j.neuron.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Cembrowski MS, et al. Spatial Gene-Expression Gradients Underlie Prominent Heterogeneity of CA1 Pyramidal Neurons. Neuron. 2016;89:351–368. doi: 10.1016/j.neuron.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Sommer B, et al. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science (New York, N.Y. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 34.Brakebusch C, et al. Brevican-deficient mice display impaired hippocampal CA1 long-term potentiation but show no obvious deficits in learning and memory. Mol Cell Biol. 2002;22:7417–7427. doi: 10.1128/MCB.22.21.7417-7427.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Craig AM. Protein tyrosine phosphatases PTPdelta, PTPsigma, and LAR: presynaptic hubs for synapse organization. Trends in neurosciences. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- 38.Coetzee WA, et al. Molecular diversity of K+ channels. Annals of the New York Academy of Sciences. 1999;868:233–285. doi: 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 39.Miura SK, Martins A, Zhang KX, Graveley BR, Zipursky SL. Probabilistic splicing of Dscam1 establishes identity at the level of single neurons. Cell. 2013;155:1166–1177. doi: 10.1016/j.cell.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kise Y, Schmucker D. Role of self-avoidance in neuronal wiring. Curr Opin Neurobiol. 2013;23:983–989. doi: 10.1016/j.conb.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Iijima T, Iijima Y, Witte H, Scheiffele P. Neuronal cell type-specific alternative splicing is regulated by the KH domain protein SLM1. The Journal of cell biology. 2014;204:331–342. doi: 10.1083/jcb.201310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen TM, et al. An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. eLife. 2016;5 doi: 10.7554/eLife.22757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schreiner D, et al. Targeted Combinatorial Alternative Splicing Generates Brain Region-Specific Repertoires of Neurexins. Neuron. 2014 doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 44.Fuccillo MV, et al. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer C, et al. Developmental diversification of cortical inhibitory interneurons. Nature. 2018;555:457–462. doi: 10.1038/nature25999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mi D, et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science (New York, N.Y. 2018 doi: 10.1126/science.aar6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim L, et al. Optimization of interneuron function by direct coupling of cell migration and axonal targeting. Nature neuroscience. 2018;21:920–931. doi: 10.1038/s41593-018-016-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nature reviews. Neuroscience. 2016;17:265–281. doi: 10.1038/nrn.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanz E, et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A. 2009;106:13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsien JZ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 53.Nakazawa K, et al. Hippocampal CA3 NMDA receptors are crucial for memory acquisition of one-time experience. Neuron. 2003;38:305–315. doi: 10.1016/s0896-6273(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 54.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nature protocols. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nature protocols. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.