Abstract
Nontraditional glycemic biomarkers including fructosamine, glycated albumin and 1,5-anhydroglucitol (1,5-AG) are potential alternatives or complements to traditional measures of hyperglycemia. Genetic variants are associated with these biomarkers, but the heritability, or extent to which genetics control their variation, is not known. We estimated pedigree-based, SNP-based and bivariate heritabilities for traditional glycemic biomarkers (fasting glucose, HbA1c), and nontraditional biomarkers (fructosamine, glycated albumin, 1,5-AG) among white participants in the Atherosclerosis Risk in Communities (ARIC) Study (N=400 first-degree relatives from sibships, N=5,575 unrelated individuals). Pedigree-based heritabilities (representing heritability from the entire genome) for nontraditional biomarkers were substantial (0.44 – 0.55) and comparable to HbA1c (0.34); the fasting glucose estimate was nonsignificant. SNP-based heritabilities (representing heritability from common variants) were lower than pedigree-based heritabilities for all biomarkers. Bivariate heritabilities showed shared genetics between fructosamine and glycated albumin (0.46 pedigree-based, 1.00 SNP-based) and glycated albumin and 1,5-AG (0.50 pedigree-based, 0.47 SNP-based). Genetic factors contribute to a considerable proportion of the variance of fructosamine, glycated albumin and 1,5-AG and a portion of this heritability likely comes from common variants.
Keywords: fructosamine; glycated albumin; 1,5-AG; glycemic biomarkers; heritability
INTRODUCTION
Type 2 diabetes mellitus is a major public health problem that affects over 10% of the US adult population and is associated with substantially increased risks of mortality and serious clinical outcomes such as heart disease, stroke, chronic kidney disease and retinopathy (Selvin 2014, Marathe 2017). Diabetes is defined by hyperglycemia, or elevated glucose concentrations in the blood. Fasting glucose and hemoglobin A1c (HbA1c) are the most common biomarkers used for screening and diagnosis of diabetes, but have limitations. Fasting glucose requires substantial patient preparation (i.e., an eight-hour fast), has high pre-analytic variability with sample stability issues, is acutely affected by factors such as recent physical activity or illness, and has moderate intra-individual variability. HbA1c is less affected by these factors, but the interpretation of HbA1c can be problematic in the setting of altered red blood cell turnover or changes in hemoglobin, factors due to characteristics of the biomarker and unrelated to circulating glucose (Marathe 2017, Parrinello 2014, Sacks 2011, Sacks 2014). The limitations of traditional measures of hyperglycemia have led to a growing interest in nontraditional biomarkers including fructosamine, glycated albumin and 1,5-anhydroglucitol (1,5-AG) (Parrinello 2014, Goldstein 2004).
Fructosamine, glycated albumin and 1,5-AG are indirect measures of blood glucose levels. Fructosamine and glycated albumin are both biomarkers where glucose is bound to protein. Fructosamine is glucose bound to serum total protein, and glycated albumin is glucose bound to serum albumin. The majority of serum protein is comprised of albumin, thus there are expected similarities between these two biomarkers and they represent the average blood glucose over the previous ~2–3 weeks (Armbruster 1987).
1,5-AG is a molecule structurally similar to glucose and is consumed through food. During hyperglycemic conditions, when glucose exceeds the renal threshold, glucose is preferentially reabsorbed from urine by the kidney, leading to excretion of 1,5-AG in the urine and a reduction of serum 1,5-AG levels. Blood 1,5-AG concentrations represent glycemic excursions above the renal threshold over the previous 1–2 weeks (Buse 2003, Dungan 2008, Yamanouchi 1994).
Heritability, the proportion of variance in a phenotype that can be attributed to genetics, is population specific, and is affected by the relative genetic and environmental impacts on the phenotype. Previous studies in various populations have estimated the narrow-sense heritability using a pedigree-based approach of fasting glucose to range from 0.30 to 0.70, and HbA1c to range from 0.20 to 0.75 (Shin 2014, Meigs 2002, Pilia 2006, Watanabe 1999, Mills 2004, Hsueh 2000, Simonis-Bik 2008, Snieder 2001, Mitchell 1996). Recent studies evaluating hundreds of metabolites using non-targeted assays have estimated the heritability of 1,5-AG to be 0.61 to 0.63 (Shin 2014, Long 2017) in population-based studies. To date, no study has estimated the heritability of fructosamine or glycated albumin. Quantifying the genetic contribution of these biomarkers will inform the extent to which genetics may play a role in these non-traditional biomarkers, and determine if they are comparable to traditional diabetes biomarker (fasting glucose and HbA1c) heritabilities.
An underlying assumption of heritability is that if a trait is heritable, individuals who share more of their genetics (i.e., are more closely related) will have more similar phenotypes than those who share less of their genetics (i.e., are distantly related or unrelated). Traditional heritability methods use closely related individuals (first- and second-degree relatives) and infer the degree of shared genetics (shared identity by descent (IBD)) based on family structure. These pedigree-based methods provide estimates of narrow-sense heritability (), or the proportion of additive genetic variance in a phenotype passed down from parents to offspring. However, these estimates may be influenced by shared environments between related pairs. In newer SNP-based () heritability methods, the amount of shared genetics among unrelated individuals can be estimated using measured genotypes, taking advantage of the small amount of shared genetics across all humans from our common ancestor as a species (Speed 2012, Yang 2010). These methods are less likely to be influenced by shared environment. is commonly estimated using genome-wide association studies (GWAS) data, which target common or less frequent SNPs (minor allele frequency (MAF)>0.01) that are in linkage disequilibrium (LD) with causal variants, and thus generally represents the h2 due to common genetic variation. Thus comparing and h2 (representing the entire proportion of a phenotype due to genetics) can inform the genetic architecture of a trait, representing the proportion due to common variants.
In this analysis, both pedigree-based and SNP-based heritability were estimated for fructosamine, glycated albumin and 1,5-AG using the same participants from the Atherosclerosis Risk in Communities (ARIC) Study, and compared across the different glycemic biomarkers.
METHODS
Study population
The ARIC Study is a prospective cohort study initiated in 1987 to evaluate risk factors for cardiovascular disease in a community-based setting. Briefly, participants were recruited from four study sites: Forsyth, North Carolina; suburban Minneapolis, Minnesota; Jackson, Mississippi; and Washington County, Maryland. Overall, 15,792 middle-aged adults participated in the initial study visit (visit 1, 1987–1989), with 6 subsequent study visits (1990–2019). All study participants provided written informed consent, and the study protocols were approved by the relevant institutional review boards (ARIC Investigators 1989).
Glycemic biomarkers
Samples for all glycemic biomarkers were collected at ARIC visit 2 (1990–1992). Fructosamine (Roche Diagnostics, Indianapolis IN, USA), glycated albumin (GA-L Asashi Kasei Pharma Corporation, Tokyo, Japan) and 1,5-AG (GlycoMark, Winston-Salem, NC) were measured in 2012–2013 using a Roche Modular P800 system from samples stored at −70°C. Glucose was measured at visit 2 using the Roche Hitachi 911 analyzer using the hexokinase method (Roche Diagnostics). HbA1c was measured at visit 2 in stored whole blood samples using high performance liquid chromatography, using NGSP-certified assays standardized to the Diabetes Control and Complications Trial (Selvin 2010).
Genotyping and Quality Control
Genotyping was performed using the Affymetrix 6.0 array. Samples with sex mismatches, genetic outliers, failed concordance with Taqman genotypes, or missingness >98% were excluded. First-degree relatives were defined by a DST value>0.8 (DST = IBS distance (IBS2 + 0.5*IBS1) / (N SNP pairs)) generated from PLINK (Purcell 2007). Both members of each first-degree relative pair were included in the pedigree-based heritability estimation, and one member of a first-degree relative pair were excluded in SNP-based heritability estimation. SNPs were excluded if missingness was >5%, Hardy-Weinberg Equilibrium (HWE)<0.00001, low MAF <0.005. Imputation was pre-phased using ShapeIt (v1.r532) and then imputed using IMPUTE2 to 1,000 Genomes Phase I (March 2012) (1000 Genomes Project Consortium 2015).
From the 30,038,522 imputed SNPs, SNPs were excluded if they had bases other than G, C, T or A, had duplicate base pair positions, imputation quality info score<0.99 and minor allele frequency (MAF)<0.01 to obtain a dataset with 3,224,517 SNPs. Imputed scores were converted to hard calls for the SNP-based heritability analyses using PLINK (Purcell 2007).
Family-based study sample for pedigree-based heritability
Because pedigree data was not available for this data, we used genetic data to infer relatedness and took a conservative approach to our inclusion criteria. We chose to include only first-degree relatives, as the distinction between parent-offspring and sibling pairs can be reasonably estimated by age. Through genotyping, 384 first-degree relative pairs were identified (688 individuals, some were part of multiple pairs). Individuals were excluded if they met the following criteria: failed genetic quality control (N=50), did not attend visit 2 (N=29), did not fast for at least 8 hours (N=11) or missing fasting status (N=1), had diagnosed diabetes (N=32), or missing fasting glucose, HbA1c, fructosamine, glycated albumin, and 1,5-AG data (N=40). Individuals were further excluded if their related pair member did not pass quality control (N=91), potential parent-child relationships (N=20, defined as first-degree relative pairs with >15 year age difference), and likely monozygotic twins (first-degree relatives with the same age and sex, N=14), leaving 400 individuals who were members of sibling-pairs (Figure 1). With this sample size, we had 80% power to detect a heritability of 0.16 or greater as estimated by the h2power feature in the program SOLAR-Eclipse (Almasy 1998).
Figure 1.
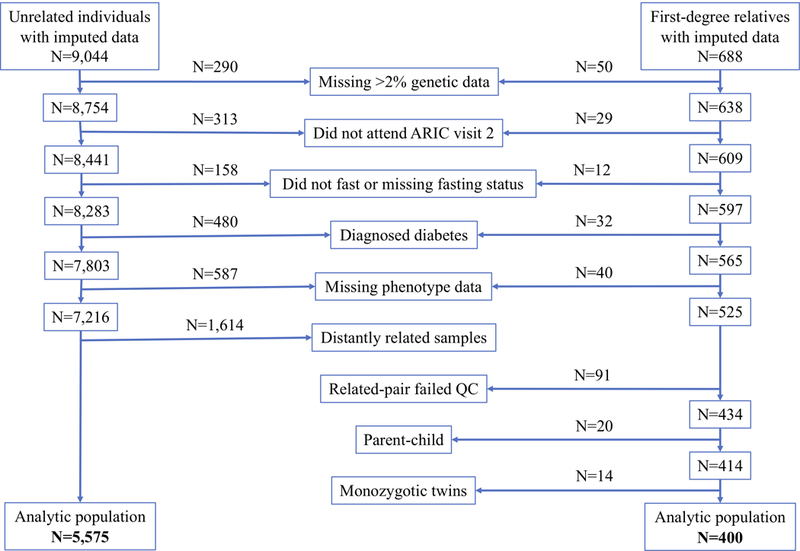
Study participant exclusions.
Pedigree-based heritability analysis
Pedigree-based heritability was estimated using the variance components method using the program SOLAR-Eclipse (Almasy 1998). This method uses a linear mixed model, with covariates (age, sex and ARIC study center) as fixed effects and genetics and environment as random effects. It partitions the variance between genetic and environmental effects and then heritability is calculated as the ratio of genetic variance to the total variance. The distributions of all glycemic biomarkers were skewed to the right among all participants, and were therefore inverse normal transformed for all analyses.
SNP-based heritability study sample
The present study was restricted to self-identified white individuals because of limited power due to the smaller sample size of self-identified black participants (N=1,483 after exclusions; recommended sample size for =4,000 (Speed 2012)). Of the 9,044 white ARIC participants with available genotyping data, participants with low quality genotype data (missingness>2%; N=290), did not attend ARIC visit 2 (N=313), did not fast for at least 8 hours (N=147) or missing fasting status (N=11), individuals with diagnosed diabetes (self-reported physician diagnosis or use of diabetes medications; N=480), or missing fasting glucose, HbA1c, fructosamine, glycated albumin, and 1,5-AG data (N=587), were excluded (Figure 1).
LDAK SNP-based heritability analysis
The method Linkage Disequilibrium Adjusted Kinships (LDAK) was used to analyze SNP-based heritability for fasting glucose, HbA1c, fructosamine, glycated albumin and 1,5-AG (Speed 2012, Speed 2017). This method employs a linear mixed model, with covariates such as age and sex as fixed effects and a genetic relationship matrix (GRM) calculated from genotyped SNPs for all pairs of individuals as random effects. The variance of the random effects is partitioned to isolate the variance due to genetics, and restriction maximum likelihood estimation is then used to estimate that variance. Heritability is then calculated as the proportion of total variance in the outcome due to genetics. We used recommended parameters for all analyses. The first step in LDAK is to calculate weights for each SNP, dividing the genome into approximately 1000kb sections and weighting SNPs based on the local linkage disequilibrium (LD) structure such that areas of high LD had lower weights than those with low LD. The total weight of these SNPs was 113,120, representing the approximate number of independent loci evaluated.
Closely related individuals may affect SNP heritability analysis due to their shared environment or shared regions of LD, and hence we excluded them from analysis. To determine relatedness, kinship was calculated based on a thinned set of SNPs (not within 1Mb of each other or in LD, with r2>0.2) using alpha = −0.25 (alpha is a parameter representing the relationship between heritability and MAF). Individuals were excluded so that no pair of individuals had a kinship value greater than the smallest observed kinship (−0.025, approximately no more related than cousins twice or thrice removed). Our analytic sample contained 5,575 individuals (Figure 1). The ARIC study included a large percentage of married participants (McAdams-DeMarco 2011) (N=4,500 spousal pairs, 57% of individuals who attended visit 1), which represents a form of shared environment. However, the biomarker correlations among married couples was low (<0.10).
In each analysis, age, sex, ARIC study center, and the top 20 principal components (PC) were included as covariates. Predictor loadings from 1000 genomes were projected onto our data and the top 10 loadings were controlled for as recommended by the LDAK developers (Speed 2017). For each biomarker, strongly associated SNPs (p<1×10−20) were evaluated using linear regression and excluded from heritability estimates to avoid biasing results (rs182549 for 1,5-AG). Inflation due to population substructure was determined by calculating heritability separately in four chunks of chromosomes (chromosomes 1–3, 4–7, 8–11, 12–22) and comparing the sum of the heritabilities from the chunks to the heritability calculated using all of the chromosomes. If population substructure was present, the chromosomes would be correlated and hence the sum of heritability from the four chunks would be greater than heritability from all of the chromosomes (because each chunk would be representing more than just the heritability from the chromosomes in that chunk). Sensitivity analyses were performed excluding undiagnosed diabetes cases (defined as fasting for at least 8 hours and glucose ≥126 mg/dL) from the heritability estimations.
GCTA SNP-based heritability analysis
Because there has been much debate but no consensus in the literature as to whether LDAK or the originally proposed SNP-based heritability method, Genome-wide Complex Trait Analysis (GCTA) (Yang 2010) provide more accurate SNP-based heritability estimates (Speed 2017, Yang 2017, Yang 2015, Lee 2013) both methods were used in this analysis. Due to sample size constraints, the most recent version of GCTA (GCTA-LDMS) did not run and therefore the original version of GCTA (GCTA-SC) was used. We ran GCTA using recommended parameter settings. Individuals with kinship>0.05 were removed, leaving 6,443 individuals. A genetic relationship matrix was calculated and SNP-based heritabilities were estimated controlling for age, sex, ARIC study center and the first 10 principal components.
Bivariate heritability analyses
To explore the shared heritability among the glycemic biomarkers, bivariate heritability was performed, which calculates the percentage of heritability shared across two traits. Bivariate heritability models two traits as the outcome and estimates the genetic correlation between the traits using the equation
Where ρp is the phenotypic correlation, ρg is the genetic correlation, ρe is the environmental correlation, and h21 and h22 are the heritabilities of trait 1 and trait 2.
A negative correlation (between −1 and 0) indicates that the same genes increase the values of one trait while decreasing the values of the other trait, and a positive correlation (between 0 and 1) indicates that the same genes increase the values of both traits. Pedigree-based bivariate heritability was estimated using SOLAR-Eclipse and SNP-based bivariate heritability using GCTA.
RESULTS
There were 5,575 unrelated individuals in the SNP-based heritability analytic sample and 400 first-degree relatives in the pedigree-based heritability analytic sample. Approximately half of the participants were female. Mean biomarker values were similar across both samples, and 5–7% of samples had undiagnosed diabetes (Table 1).
Table 1.
Demographic and clinical characteristics in unrelated and first-degree relative study participants†
| SNP-based heritability | Pedigree-based heritability |
|
|---|---|---|
| Unrelated Participants (N=5,575) |
First-degree relatives (N=400) |
|
| Female | 54% | 55% |
| Age (years) | 57 (5.7) | 58 (5.3) |
| Fructosamine (μmol/L) | 227 (23) | 227 (22) |
| Glycated albumin (%) | 12.6 (1.6) | 12.5 (1.5) |
| 1,5-AG (μg/mL) | 18.7 (5.7) | 19.9 (6.4) |
| HbA1c (mmol/mol) | 36 (5.5) | 37 (6.5) |
| (%) | 5.4 (0.5) | 5.5 (0.6) |
| Fasting glucose (mg/dL) | 104 (17) | 104 (16) |
| Undiagnosed diabetes‡ | 5% | 7% |
Continuous variables shown as mean (SD) and categorical variables shown as %
Undiagnosed diabetes defined as fasting and glucose≥ 126 or nonfasting and glucose≥ 200
Pedigree-based heritability
The pedigree-based heritability estimates using sibling-pairs for 1,5-AG (=0.55), glycated albumin (=0.45) and fructosamine (=0.44) were statistically significant (p<1.9×10−4) (Figure 2, Table 2) and comparable to HbA1c (= 0.34). The fasting glucose estimate was not significant (p=0.43), but analysis using visit 1 data (N=522) estimated heritability was 0.23 (p=0.03).
Figure 2.
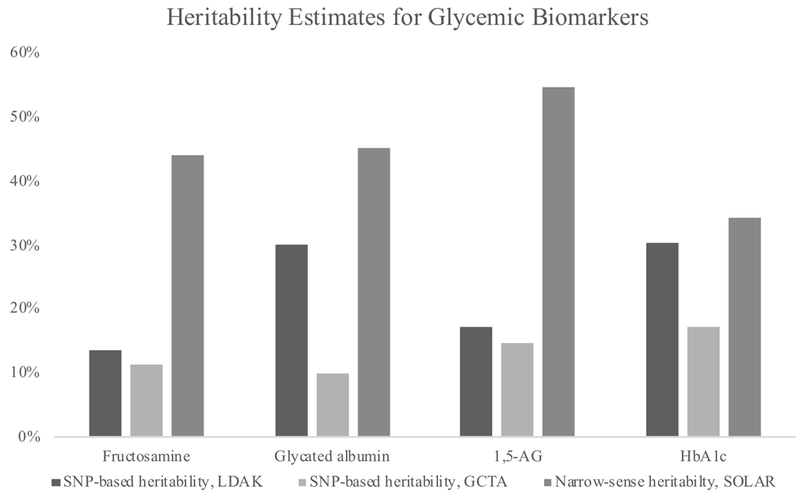
Heritability estimates for glycemic biomarkers by method†
†p<0.05 for all estimates
Table 2.
SNP-based ()and pedigree-based () heritability estimates for glycemic biomarkers†
| (SOLAR) | (LDAK‡) | (GCTA) | ||||
|---|---|---|---|---|---|---|
| Heritability (SE) |
P-value | Heritability (SD) |
P-value | Heritability (SE) |
P-value | |
| Fructosamine | 0.44 (0.13) | 2.9E–04 | 0.13 (0.06) | 0.01 | 0.11 (0.03) | 1.1E–03 |
| Glycated albumin | 0.45 (0.13) | 1.8E–04 | 0.30 (0.07) | 8.6E–07 | 0.10 (0.04) | 2.5E–03 |
| 1,5–AG | 0.55 (0.13) | 1.3E–05 | 0.17 (0.06) | 3.6E–03 | 0.15 (0.04) | 5.3E–05 |
| HbA1c | 0.34 (0.13) | 0.01 | 0.30 (0.07) | 2.6E–06 | 0.17 (0.04) | 1.1E–06 |
| Fasting glucose | 0.02 (0.03) | 0.43 | 0.08 (0.07) | 0.11 | 0.05 (0.04) | 0.08 |
LDAK N=5,575, relatedness cutoff=0.25; GCTA N=6443, relatedness cutoff=0.05; SOLAR N=400
LDAK inflation <3.3% for all heritability estimates
SNP-based heritability
The glycated albumin SNP-based heritability estimated using LDAK was (= 0.30), followed by 1,5-AG (= 0.17) and fructosamine (= 0.13) (Figure 2, Table 2). HbA1c had similar SNP-based heritability to the nontraditional biomarkers (= 0.30). The fasting glucose result was nonsignificant (p=0.11). Inflation for SNP-based heritability of the biomarkers was low (<3.3%). Sensitivity analyses using a relatedness cutoff of 0.05 consistent with the GCTA methods produced similar results (results not shown).
SNP-based heritabilities estimated by GCTA were lower than estimates using LDAK. Fructosamine (= 0.11), glycated albumin (= 0.10), 1,5-AG (= 0.15) and HbA1c (= 0.17) (Figure 2, Table 2). The fasting glucose estimate was not significant (p=0.08).
For all methods, excluding undiagnosed diabetes cases had little impact on heritability estimates (Table 3).
Table 3.
SNP-based ()and pedigree-based ()heritability estimates for glycemic biomarkers, excluding undiagnosed diabetes cases.†
| (SOLAR) | (LDAK‡) | (GCTA) | ||||
|---|---|---|---|---|---|---|
| Heritability (SE) |
P-value | Heritability (SD) |
P-value | Heritability (SE) |
P-value | |
| Fructosamine | 0.40 (0.14) | 2.7E–03 | 0.14 (0.07) | 0.01 | 0.12 (0.04) | 7.1E–04 |
| Glycated albumin | 0.48 (0.14) | 3.8E–04 | 0.27 (0.07) | 1.7E–05 | 0.09 (0.04) | 8.4E–03 |
| 1,5–AG | 0.52 (0.13) | 7.4E–05 | 0.20 (0.07) | 3.0E–03 | 0.17 (0.04) | 7.4E–06 |
| HbA1c | 0.42 (0.14) | 0.002 | 0.37 (0.07) | 1.1E–07 | 0.20 (0.04) | 9.8E–08 |
| Fasting glucose | 0.07 (0.15) | 0.31 | 0.13 (0.07) | 0.03 | 0.08 (0.04) | 0.02 |
LDAK N=5,281, relatedness cutoff=0.25; GCTA N=6099, relatedness cutoff=0.05; SOLAR N=372
LDAK inflation <3.3% for all heritability estimates
Bivariate heritability analyses
Bivariate heritability estimates for fructosamine and glycated albumin showed shared genetics using SOLAR-Eclipse (0.46) with nearly complete overlap using GCTA (1.00). Glycated albumin and 1,5-AG had shared genetics which influence these traits in opposite directions, consistent with the inverse correlation of these biomarkers (−0.50 in SOLAR, −0.47 in GCTA); Table 4). No other pairs of biomarkers had significantly shared heritability using GCTA or SOLAR-Eclipse.
Table 4.
Bivariate heritability results†
| Fructosamine | Glycated Albumin |
1,5-AG | HbA1c | ||
|---|---|---|---|---|---|
| Fructosamine | G: 0.46 (p=0.04) E: 0.67 (p=0.0001) |
G: −0.22 (P=0.25) E: −0.11 (p=0.54) |
G: 0.10 (p=0.70) E: 0.08 (p=0.58) |
SOLAR
heritability (P-value) |
|
| Glycated Albumin | 1.00 (0.12) | G: −0.50 (p=0.009) E: −0.03 (p=0.88) |
G: 0.03 (p=0.89) E: 0.31 (p=0.04) |
||
| 1,5-AG | −0.31 (0.21) | −0.47 (0.21) | G: −0.30 (p=0.18) E: −0.07 (p=0.66) |
||
| HbA1c | 0.24 (0.19) | 0.15 (0.20) | 0.13 (0.18) | ||
| GCTA heritability (SE) | |||||
p<0.05 are in bold
G=shared genetic heritability estimated with SOLAR
E=shared environmental heritability estimated with SOLAR
DISCUSSION
In this study, both pedigree-based and SNP-based heritabilities were estimated for nontraditional glycemic biomarkers. Because heritability is a population-specific measure that depends on relative genetic and environmental factors, it is important to estimate heritabilities in the same population in order to compare heritabilities across traits. This was done this for both traditional and nontraditional glycemic biomarkers, using the same population of white individuals participating in the ARIC Study.
Approximately half of the variation in fructosamine, glycated albumin and 1,5-AG was estimated to be controlled by genetic factors. Our results for 1,5-AG are consistent with previous estimations (0.55 in our study vs. 0.61 to 0.63 in previous studies) (Shin 2014, Long 2017). There are no published reports of the heritability of fructosamine and glycated albumin. Our results illustrate that genetics play an important role in nontraditional glycemic biomarkers, and may affect these markers in a similar manner to HbA1c. Given that 60 variants are associated with HbA1c (Wheeler 2017) (heritability = 0.20 to 0.75) (Meigs 2002, Pilia 2006, Mills 2004, Hsueh 2000, Simonis-Bik 2008, Snieder 2001), it is likely that more than the currently discovered (1 for fructosamine, 1 for glycated albumin, 7 for 1,5-AG) (Li 2017, Loomis 2018) variants are associated with nontraditional glycemic biomarkers and these low numbers of SNPs may reflect the limited sample sizes to date for GWAS and sequencing studies for these biomarkers. The nonsignificant heritability estimates for fasting glucose may be in part due to its pre-analytic and intra-individual variability. Although nonsignificant, SNP-based heritability was similar to that estimated in a previous study in ARIC using participants from visit 1 (=0.13 vs 0.08 in our LDAK analyses) (Vattikuti 2012). The significant and larger pedigree-based heritability estimated using visit 1 data (N=521, =0.23, p=0.03) indicates that the smaller sample from visit 2 likely reduced power to estimate fasting glucose heritability.
As expected, the SNP-based heritabilities were lower than the pedigree-based heritabilities. SNP-based methods estimate heritability based on the variants for which data is collected (genotyped or sequenced), while pedigree-based heritability among related individuals is based on the entire genome. Additionally, pedigree-based heritability may be influenced by shared environment among family members, which has much lower impact SNP-heritability estimates. The two SNP-based heritability methods give different estimates for some of the biomarkers, which may be due to differences in the methods. For instance, our sample size was too small to be adequately powered to run the most recent iteration of GCTA (GCTA-LDMS (Yang 2015)), which accounts for LD structure. While there is still debate as to which method of SNP-based heritability estimation is preferred (Speed 2017, Yang 2017, Yang 2015, Lee 2013), we presented results from multiple methods but refrain from quantitative comparison. Regardless of the estimated value, for all biomarkers, SNP-based heritabilities represented a portion of the pedigree-based heritability, indicating that the genetic influences on these traits is likely due to both common and rare variants.
Bivariate heritability was also estimated across all of the biomarkers, examining how much heritability is shared between them. The shared heritability between fructosamine and glycated albumin was significant, suggesting a substantial portion of overlapping genetics. However, this was expected due to the biological similarity of these two measures (80% of glycated proteins (i.e., fructosamine) are glycated albumin) (Anguizola 2013, Cohen 2013) and their strong phenotypic correlation (correlation coefficient = 0.76, Table 5). The SNP-based bivariate heritability of 1, however, is likely an overestimation and some of the phenotypic correlation is due to environmental correlation (environmental correlation among related =0.67, Table 4). It is possible that the shared variants between fructosamine and glycated albumin are more common and thus captured by SNP-based heritability. While the biomarkers in the present study all aim to capture hyperglycemia, some variability in these measures may be explained by non-glycemic factors (e.g. the known nonglycemic variants associated with HbA1c (Wheeler 2017), which may be particularly important in the non-diabetic range. Alternatively, it could simply indicate the biomarkers are under control of different genes for other reasons such as the different time frames each biomarker represents (2–3 weeks for fructosamine and glycated albumin, 1–2 weeks for 1,5-AG 2–3 months for HbA1c, instantaneous for fasting glucose) or the differences in variability of these measures. Unfortunately, the lack of significance across the other bivariate analyses limit our ability to draw other conclusions about the amount of shared genetics across the other biomarkers.
Table 5.
Phenotypic correlations across glycemic biomarkers (N=5,575).
| Fructosamine | Glycated Albumin | 1,5-AG | HbA1c | Fasting Glucose |
|
|---|---|---|---|---|---|
| Fructosamine | 1 | ||||
| Glycated albumin | 0.76 | 1 | |||
| 1,5-AG | −0.18 | −0.24 | 1 | ||
| HbA1c | 0.46 | 0.54 | −0.18 | 1 | |
| Fasting glucose | 0.53 | 0.56 | −0.17 | 0.71 | 1 |
The substantial heritability estimates for nontraditional biomarkers indicate a strong genetic component. Additional studies focused on the identification of genetic variants associated with fructosamine, glycated albumin and 1,5-AG will inform the biology of these biomarkers, and may identify limitations and implications for their clinical interpretation.
ACKNOWLDEGEMENTS
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I and HHSN268201700005I), R01HL087641, R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
The work of AK was funded by DFG KO 3598/3–1 and KO 3598/4–1. SL was supported by an institutional training grant from the NIH/NHLBI (T32 HL007024). This work was also supported by NIH/NIDDK grants K24DK106414 and R01DK089174 to Dr. Selvin. Reagents for the 1,5-anhydroglucitol assays were donated by GlycoMark, Inc. Reagents for the glycated albumin assays were donated by the Asahi Kasei Corporation. Reagents for the fructosamine assays were donated by Roche Diagnostics Corporation.
Footnotes
Data Availability Statement: The genetic data that support the findings of this study are openly available in dbGAP at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000090.v5.p1&phv=80479&phd=2057&pha=&pht=114&phvf=&phdf=&phaf=&phtf=&dssp=1&consent=&temp=1, dbGaP Study Accession: phs000090.v5.p1.
REFERENCES
- 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguizola J, Matsuda R, Barnaby OS, et al. Review: Glycation of human serum albumin. Clin Chim Acta. 2013;425:64–76. doi: 10.1016/j.cca.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARIC investigators. The atherosclerosis risk in communities (ARIC) study: Design and objectives. Am J Epidemiol. 1989;129(4):687–702. doi: 10.1093/oxfordjournals.aje.a115184. [DOI] [PubMed] [Google Scholar]
- Armbruster DA. Fructosamine: Structure, analysis, and clinical usefulness. Clin Chem. 1987;33(12):2153–2163. [PubMed] [Google Scholar]
- Buse JB, Freeman JLR, Edelman SV, Jovanovic L, McGill JB. Serum 1,5-anhydroglucitol (GlycoMark ): A short-term glycemic marker. Diabetes Technol Ther. 2003;5(3):355–363. doi: 10.1089/152091503765691839. [DOI] [PubMed] [Google Scholar]
- Cohen MP. Clinical, pathophysiological and structure/function consequences of modification of albumin by amadori-glucose adducts. Biochim Biophys Acta. 2013;1830(12):5480–5485. doi: 10.1016/j.bbagen.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Dungan KM. 1,5-anhydroglucitol (GlycoMark) as a marker of short-term glycemic control and glycemic excursions. Expert Rev Mol Diagn. 2008;8(1):9–19. doi: 10.1586/14737159.8.1.9. [DOI] [PubMed] [Google Scholar]
- Goldstein DE, Little RR, Lorenz RA, et al. Tests of glycemia in diabetes. Diabetes Care. 2004;27(7):1761–1773. doi: 10.2337/diacare.27.7.1761 [DOI] [PubMed] [Google Scholar]
- Hsueh WC, Mitchell BD, Aburomia R, et al. Diabetes in the old order Amish: Characterization and heritability analysis of the Amish family diabetes study. Diabetes Care. 2000;23(5):595–601. doi: 10.2337/diacare.23.5.595 [DOI] [PubMed] [Google Scholar]
- Lee SH, Yang J, Chen G, et al. Estimation of SNP heritability from dense genotype data. Am J Hum Genet. 2013;93(6):1151–1155. doi: 10.1016/j.ajhg.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Maruthur NM, Loomis SJ, et al. Genome-wide association study of 1,5-anhydroglucitol identifies novel genetic loci linked to glucose metabolism. Sci Rep. 2017;7(1):2812. doi: 10.1038/s41598-017-02287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Hicks M, Yu H, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet. 2017;49(4):568–578. doi: 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- Loomis SJ, Li M, Maruthur NM, et al. Genome-wide association study of serum fructosamine and glycated albumin in adults without diagnosed diabetes: results from the Atherosclerosis Risk in Communities Study. Diabetes. 2018. August;67(8):1684–1696. doi: 10.2337/db17-1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe PH, Gao HX, Close KL. American diabetes association standards of medical care in diabetes 2017. J Diabetes. 2017;9(4):320–324. doi: 10.1111/1753-0407.12524. [DOI] [PubMed] [Google Scholar]
- McAdams DeMarco M, Coresh J, Woodward M, et al. Hypertension status, treatment, and control among spousal pairs in a middle-aged adult cohort. Am J Epidemiol. 2011. October 1;174(7):790–6. doi: 10.1093/aje/kwr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs JB, Panhuysen CIM, Myers RH, Wilson PWF, Cupples LA. A genome-wide scan for loci linked to plasma levels of glucose and HbA(1c) in a community-based sample of caucasian pedigrees: The framingham offspring study. Diabetes. 2002;51(3):833–840. doi: 10.2337/diabetes.51.3.833 [DOI] [PubMed] [Google Scholar]
- Mills GW, Avery PJ, McCarthy MI, et al. Heritability estimates for beta cell function and features of the insulin resistance syndrome in UK families with an increased susceptibility to type 2 diabetes. Diabetologia. 2004;47(4):732–738. doi: 10.1007/s00125-004-1338-2. [DOI] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio family heart study. Circulation. 1996;94(9):2159–2170. doi: 10.1161/01.CIR.94.9.2159 [DOI] [PubMed] [Google Scholar]
- Parrinello CM, Selvin E. Beyond HbA1c and glucose: The role of nontraditional glycemic markers in diabetes diagnosis, prognosis, and management. Curr Diab Rep. 2014;14(11):548. doi: 10.1007/s11892-014-0548-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilia G, Chen W, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2(8):e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Tood-Brown K, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. AJHG. 2007. September;81(3):559–75. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DB. A1C versus glucose testing: A comparison. Diabetes Care. 2011;34(2):518–523. doi: 10.2337/dc10-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks DB, John WG. Interpretation of hemoglobin A1c values. JAMA. 2014;311(22):2271–2272. doi: 10.1001/jama.2014.6342. [DOI] [PubMed] [Google Scholar]
- Selvin E, Coresh J, Zhu H, Folsom A, Steffes MW. Measurement of HbA1c from stored whole blood samples in the atherosclerosis risk in communities study. J Diabetes. 2010;2(2):118–124. doi: 10.1111/j.1753-0407.2010.00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvin E, Parrinello CM, Sacks DB, Coresh J. Trends in prevalence and control of diabetes in the united states, 1988–1994 and 1999–2010. Ann Intern Med. 2014;160(8):517–525. doi: 10.7326/M13-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Fauman EB, Petersen A, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis-Bik AMC, Eekhoff EMW, Diamant M, et al. The heritability of HbA1c and fasting blood glucose in different measurement settings. Twin Res Hum Genet. 2008;11(6):597–602. doi: 10.1375/twin.11.6.597. [DOI] [PubMed] [Google Scholar]
- Snieder H, Sawtell PA, Ross L, Walker J, Spector TD, Leslie RD. HbA(1c) levels are genetically determined even in type 1 diabetes: Evidence from healthy and diabetic twins. Diabetes. 2001;50(12):2858–2863. doi: 10.2337/diabetes.50.12.2858 [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91(6):1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Cai N, Johnson MR, Nejentsev S, Balding DJ. Reevaluation of SNP heritability in complex human traits. Nat Genet. 2017;49(7):986–992. doi: 10.1038/ng.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattikuti S, Guo J, Chow CC. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012;8(3):e1002637. doi: 10.1371/journal.pgen.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe RM, Valle T, Hauser ER, et al. Familiality of quantitative metabolic traits in finnish families with non-insulin-dependent diabetes mellitus. Finland-United States investigation of NIDDM genetics (FUSION) study investigators. Hum Hered. 1999;49(3):159–168. doi: 10.1159/000022865. [DOI] [PubMed] [Google Scholar]
- Wheeler E, Leong A, Liu C, et al. Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14(9):e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanouchi T, Akanuma Y. Serum 1,5-anhydroglucitol (1,5 AG): New clinical marker for glycemic control. Diabetes Res Clin Pract. 1994;24 Suppl:261. doi: 10.1016/0168-8227(94)90259-3 [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bakshi A, Zhu Z, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zeng J, Goddard ME, Wray NR, Visscher PM. Concepts, estimation and interpretation of SNP-based heritability. Nat Genet. 2017;49(9):1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
- [dataset] GENEVA: The Atherosclerosis Risk in Communities (ARIC) Study, A sub-study of Atherosclerosis Risk in Communities (ARIC) Cohort; dbGAP; dbGaP Study Accession: phs000090.v5.p1


