SUMMARY
Sleep is crucial for our survival, and many diseases are linked to long-term poor sleep quality. Before we can use sleep to enhance our health and performance and to alleviate diseases associated with poor sleep, a greater understanding of sleep regulation is necessary. We have identified a mutation in the β1-adrenergic receptor gene in humans who require fewer hours of sleep than most. In vitro, this mutation leads to decreased protein stability and dampened signaling in response to agonist treatment. In vivo, the mice carrying the same mutation demonstrated short sleep behavior. We found that this receptor is highly expressed in the dorsal pons and that these ADRB1+ neurons are active during REM sleep and wakefulness. Activating these neurons can lead to wakefulness and the activity of these neurons are affected by the mutation. These results highlight the important role of β1-adrenergic receptors in sleep/wake regulation.
Keywords: ADRB1, short sleep, Dorsal Pons
Blurb
A mutation in ADRB1 was found in humans who sleep fewer hours than most. Using mouse modeling and calcium imaging experiments, Shi et al demonstrated that dorsal pons ADRB1+ neurons activity is increased in the mutant, correlating with increased wakefulness.
INTRODUCTION
An understanding of the regulatory mechanism for sleep lays at the foundation for healthy living and aging. Sleep behavior has long been thought to be regulated by the interactions of circadian clock and sleep homeostasis pathways (Borbély, 1982; Borbély et al., 2016; Daan et al., 1984). In humans, variations of genetically inherited sleep features in the population have been recognized for a long time (Sehgal and Mignot, 2011; Shi et al., 2017). Importantly, human sleep has unique features that are different from that of animal models. For example, human sleep is usually consolidated, whereas mice sleep throughout the 24-hour day (though more in the light phase than in the dark phase). Drosophila sleep-like behavior is consolidated into one long period, but the level of similarity between the Drosophila and human molecular regulatory mechanisms remains unclear. Previously, we identified a series of genetic variations that influence the timing of sleep in humans, and mouse models of these mutations mostly recapitulate the phenotypes (Shi et al., 2017). Timing of sleep is heavily influenced by the circadian clock which has been intensely studied, and we now have a large and growing body of knowledge on how the clock is regulated at the molecular level. On the other hand, our understanding of sleep homeostasis regulation for human lags behind. We reported a mutation in the human DEC2 gene that causes mutation carriers to sleep 6 hours nightly for their entire lives without apparent negative effects (He et al., 2009). Another mutation in DEC2 was later reported in a single individual who is a short sleeper and resistant to sleep deprivation (Pellegrino et al., 2014). Identification of additional genes participating in modulation of human sleep duration provides a unique way to expand our knowledge of genes and pathways critical for human sleep homeostasis regulation.
Noradrenergic signaling in the CNS has long been known to regulate sleep (Berridge, 2008; Berridge et al., 2012; Szabadi, 2013). The network involving the noradrenergic neurons has been extensively studied and most of the receptor subtypes have been genetically defined. In contrast to α1 and α2 adrenergic receptors (AR), relatively little is known about the function of β receptors in the CNS (Berridge et al., 2012; Szabadi, 2013). β-ARs within the brain were previously suggested to mediate the effect of norepinephrine (NE) for alert waking and REM sleep (Berridge et al., 2012). Clinically, β-blockers are widely used and can be associated with difficulty falling asleep and staying asleep, possibly due to reduced production and release of melatonin (Helfand et al., 2009; Scheer et al., 2012; Stoschitzky et al., 1999; Yilmaz et al., 2008). We report here a rare mutation in the β1-adrenergic receptor (β1-AR) gene (ADRB1) found in humans with natural short sleep. Engineering the human mutation into mice resulted in a sleep phenotype similar to that seen in familial natural short sleepers. We show that β1AR is expressed at high levels in the dorsal pons (DP). Neuronal activity measured by calcium imaging in this region demonstrated that ADRB1+ neurons in DP are wake- and REM sleep-active. Manipulating the activity of these ADRB1+ neurons changes sleep/wake patterns. Also, the activity of these neurons was altered in mice harboring the mutation. Together, these results not only support the causative role of this ADRB1 mutation in the human subjects but also provide a mechanism for investigating noradrenaline and β1-AR in sleep regulation at the circuit level.
RESULTS
A mutation found in humans with familial natural short sleep
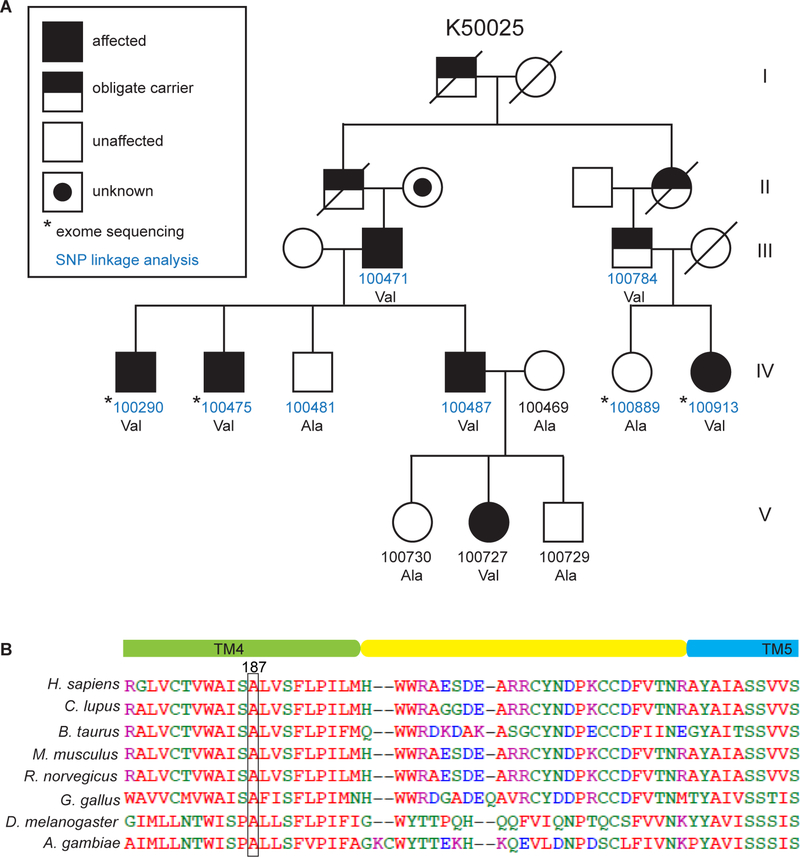
We searched for genes that are important in sleep regulation by screening for mutations in human individuals who exhibit unusual sleep patterns. ‘Natural short sleep’ (NSS) refers to individuals who have a life-long tendency to sleep only 4–6 hours/night and to feel well rested (He et al., 2009). Kindred 50025 is a family segregating an autosomal dominant allele for ‘Familial natural short sleep’ (FNSS) (Figure 1A and Table S1). SNP-based linkage analysis followed by whole-exome sequencing identified a very rare variant in the ADRB1 gene, located on chromosome 10q25.3. The mutation co-segregates with FNSS in the family and involves a C➔G change in the coding sequence that is predicted to cause an alanine➔valine alteration at amino acid position 187 of the β1-AR. This change was not found in the unaffected members of the same family. In the human population, this is a rare mutation with an incidence of 4.028/100,000 according to the Exome Aggregation Consortium database. The alanine at position 187 of β1-AR (A187) is highly conserved in vertebrates and invertebrates (Figure 1B). Notably, β1-AR is a G protein-coupled receptor (GPCR) that has seven transmembrane domains. The A187 residue is located in the 4th transmembrane domain.
Figure 1. An ADRB1 mutation was identified in a natural short sleep family.
(A) Pedigree of a family (K50025) carrying the ADRB1 mutation (A187V). See Table S1 for detailed sleep schedules.
(B) The β1AR A187V mutation is located in transmembrane domain 4 and is highly conserved among invertebrate and mammalian species.
Altered function of mutant β1-AR protein
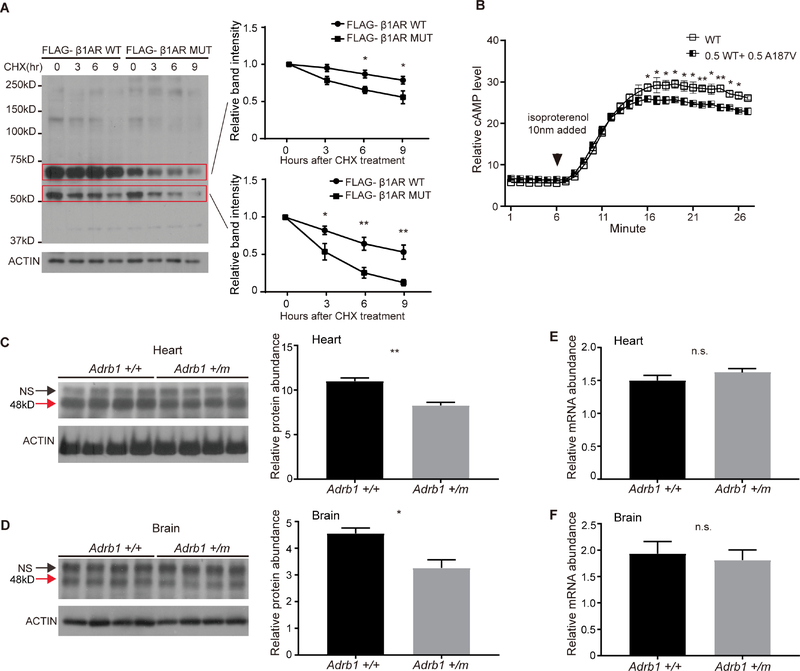
To gain insight into whether this mutation has functional consequences, we first compared the mutant vs. wild-type (WT) protein in cultured cells. As shown in Figure 2A, the mutant protein was less stable than the WT protein in a cycloheximide (CHX) assay. Because β1-AR mediates the catecholamine-induced activation of adenylatecyclase (thus cAMP-mediated signaling), we next examined the effect of the mutation on the synthesis of cAMP. To mimic the situation of human carriers, we transfected the cells with a mixture (1:1) of WT and mutant β1-AR. We found that heterozygous expression of the receptors led to decreased cAMP production in response to treatment with the non-selective agonist isoproterenol compared to expression of WT receptor alone (Figure 2B). This result indicates that the mutant protein likely has altered function. We thus generated an Adrb1-A187V knock-in mouse model using CRISPR/Cas9. Endogenous β1-AR levels were decreased significantly in the mutant (Adrb1 +/m) mice (Figures 2C and 2D), whereas mRNA levels remained unchanged (Figures 2E and 2F), suggesting the decreased protein levels were caused by post-transcriptional events. This is consistent with the finding that the mutant protein is less stable than the WT protein in the CHX assay in cultured cells (Figure 2A).
Figure 2. The β1AR A187V mutation alters protein stability and cAMP production.
(A) Degradation assay of β1AR in transfected HEK293 cells. Twenty-four hours after transfection, cells were treated with 100 μg/ml CHX and harvested at indicated time points. Bands inside the two red boxes indicate β1AR protein of different sizes in the SDS gel. Quantified results are shown on the right. β1AR protein levels at the starting point (t=0 hours) were normalized to 1.
(B) The β1AR A187V mutation confers altered downstream signaling output in response to isoproterenol in cultured cells. Heterozygous expression of β1AR A187V and WT leads to a reduction in cAMP production.
(C and D) Western blotting results of endogenous β1AR protein from the heart (C) and brain (D) lysates of Adrb1+/+, and Adrb1+/m animals. N=4 mice per group. NS, non-specific band. Quantified results are shown on the right.
(E and F) q-RTPCR results of Adrb1 mRNA normalized to Actin mRNA from the heart (E) and brain (F) tissues of Adrb1+/+ and +/m animals. N=4 mice per group.
* P<0.05, **P<0.01, n.s.= not significant. Two way RM ANOVA, post-hoc Sidak’s multiple comparisons test for (A) and (B). Two-tailed Student’s t-test for (C)-(F). Error bars represent ± SEM.
Mice with the Adrb1-A187V mutation exhibit a short sleep phenotype
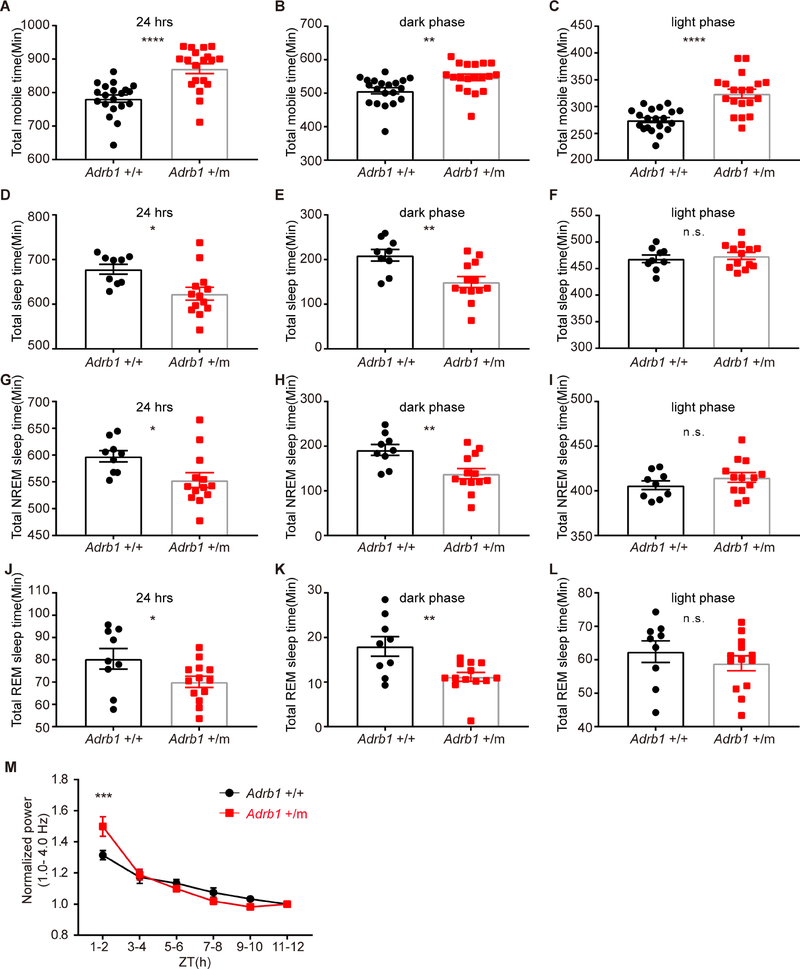
To determine whether this mutation has any influence on sleep related behaviors, we measured the sleep/wake behavior by two independent methods: ANY-maze (locomotor activity monitored by infrared cameras) and electroencephalogram/electromyogram (EEG/EMG). The mutant (Adrb1 +/m) mice showed increased activity by ANY-maze as reflected by more mobile time during the 24 hour day (Figure 3A). The increased mobile time in the mutant mice was observed in both the light and dark phases (Figures 3B and 3C). In parallel, total sleep time within 24 hours was ~55 minutes shorter in the mutant mice when measured by EEG/EMG (Figure 3D), and this decrease was only seen in the dark phase (Figures 3E, 3F and S1A). The short-sleep phenotype of these mice is also demonstrated by the significant shortening of both non-rapid eye movement (NREM) and rapid eye movement (REM) sleep in the dark phase for Adrb1 +/m mice when compared to WT mice. NREM sleep was ~53 minutes less and REM sleep was ~7 minutes less in the Adrb1 +/m vs. WT mice during the dark phase (Figures 3G - 3L, S1B and S1C). The decreased NREM and REM sleep were due to the reduction of sleep bouts rather than episode duration (Figures S1D–S1G). These results indicated that the Adrb1 A187V mutation can lead to short sleep and increased mobile time, similar to what we observe in the human subjects.
Figure 3. The Adrb1-A187V mutation alters sleep/wake related behaviors in the FNSS mouse model.
(A-C) Total mobile time by ANY-maze within 24 hours (A), dark phase (B) and light phase (C) were calculated in Adrb1 +/+ (N=20) and +/m (N=19) mice.
(D-F) Total sleep time by EEG/EMG within 24 hours (D), dark phase (E) and light phase (F) were calculated in Adrb1 +/+ (N=9) and +/m (N=13) mice.
(G-I) Total NREM sleep time within 24 hours (G), dark phase (H) and light phase (I) were calculated in Adrb1 +/+ (N=9) and +/m (N=13) mice.
(J-L) Total REM sleep time during a 24 hour day (J), dark phase (K) and light phase (L) were calculated in Adrb1 +/+ (N=9) and +/m (N=13) mice.
(M) Time course of EEG delta power (1.0–4.0 Hz) during NREM sleep across the light phase for Adrb1 +/+ (N=9) and +/m (N=9) mice.
* P<0.05, **P<0.01, *** P<0.001, **** P<0.0001, n.s.=not significant. Two-tailed Student’s t-test for (A)-(L).
Two way RM ANOVA, post-hoc Sidak’s multiple comparisons test for (M). Error bars represent ± SEM.
See Figures S1 for NREM and REM sleep bouts and duration calculation.
Since sleep pressure positively correlates with delta power (the 1–4.0 Hz frequency range of the EEG during NREM sleep) (Franken et al., 2001), we examined changes in the NREM EEG delta power during the light phase (see ‘Methods’). The delta power of the mutant mice was significantly higher at the beginning (ZT1–2) and decreased rapidly throughout the light phase when compared to WT mice (Figure 3M). These data suggested that the Adrb1 mutant mice accumulate more sleep pressure, probably due to the shorter sleep in the dark phase.
Expression of β1-AR in brain
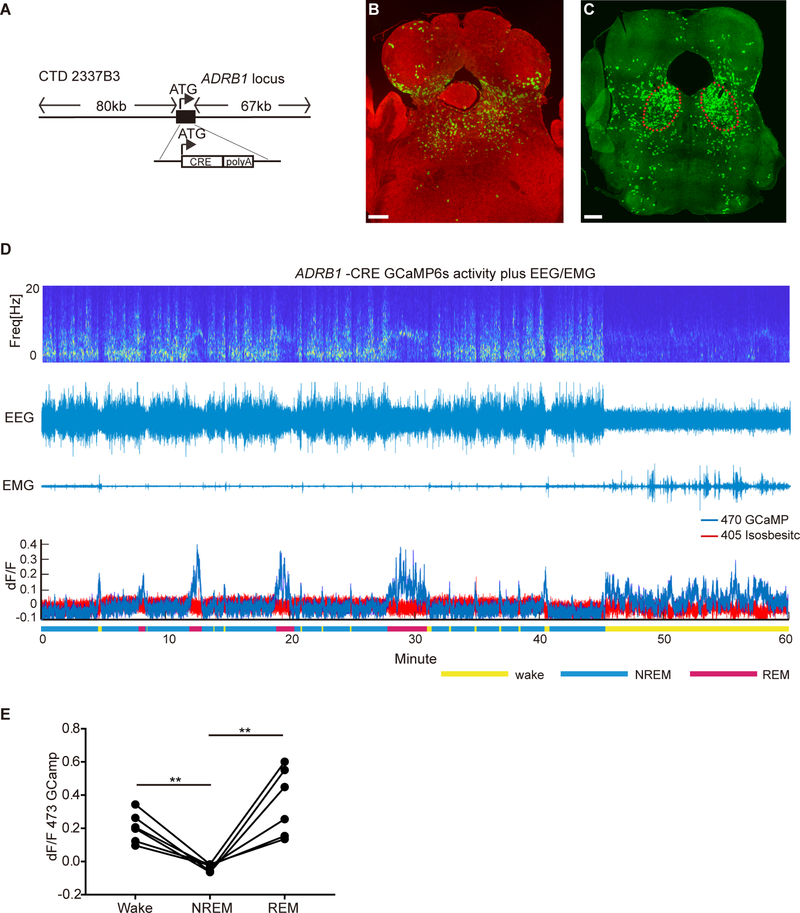
We next generated ADRB1-Cre BAC transgenic mice in order to study neural activity of ADRB1+ cells and their possible role in sleep regulation. A large BAC construct (150kb) containing the entire ADRB1 locus was modified so that the coding sequence of ADRB1 was replaced by that of Cre recombinase (Figure 4A) (Gong et al., 2003). The transgenic mice were crossed with two different reporter lines, ROSA mT/mG (Muzumdar et al., 2007) and Ai32 (loxP-flanked-ChR2–eYFP). We sectioned the whole brain to identify cells positive for the reporters. There was good agreement between the 2 reporter lines and several regions were found to have high CRE activity including hippocampus, lateral septal nucleus, medial prefrontal cortex, dorsal pons (DP) (Figures 4B and 4C), and various structures in the medulla (Figures S2). The DP region contains the major portions of the laterodorsal tegmental nucleus (LDTg), laterodorsal tegmental nucleus ventral (LDTgV), and part of parabrachial nucleus (PB), regions that are known to be involved in regulation of sleep behaviors (Figure S3A) (Boissard et al., 2002; Cox et al., 2016; Fuller et al., 2011). We thus focused our attention on the DP. We injected the Cre-dependent adeno-associated virus (AAV) encoding enhanced green fluorescent protein (EGFP) (AAV2/EF1a-DIO-EGFP-L10a) into this area and stained the brain sections with probes against endogenous Adrb1 and gfp. Approximately 80% of the GFP-positive cells also stained positively for Adrb1 (Figure S3B). Translating ribosome affinity purification (TRAP) of mRNA populations in CRE-expressing cells in DP also showed 3~4 fold enrichment of endogenous Adrb1 (Figures S3C and S3D) compared to control genes. Taken together, these results indicate that the Cre activity largely represents Adrb1 expression in this region.
Figure 4. Population activity of ADRB1+ neurons in DP is correlated with sleep/wake states.
(A) Tg (ADRB1-Cre) mice generated using BAC technology.
(B and C) Representative brain sections from ADRB1-Cre;ROSAmT/mG (B) and ADRB1-Cre;loxP-flanked-ChR2–eYFP (C) mice show high CRE activity in the DP area. Scale bar, 300 μm. See also Figures S2, S3 and S4 for further characterization.
(D) Representative EEG power spectrogram, EEG, EMG and fluorescence trace across spontaneous sleep–wake states. See Figures S5A–S5C for the fiber photometry setup and Figure S5D for fluorescence during state transitions.
(E) Quantified fluorescence signal across different sleep–wake states from N=6 mice. **P<0.01, two-tailed paired Student’s t-test.
Since DP contains different populations of neurons (Kohlmeier and Kristiansen, 2010; Wang and Morales, 2009), we then labeled the DP ADRB1+ neurons by injecting the virus AAV8-hSyn-DIO-mCherry (‘mCherry’ hereafter). Co-staining revealed that the ADRB1+ cells in the DP were glutamatergic (~37%, 221/600) or GABAergic (~25%, 150/600) based on the expression of vesicular glutamate transporter 2 (Vglut2) or glutamate decarboxylase 1 (Gad1), respectively (Figures S4A–S4F). There were very few cholinergic or noradrenergic cells based on the expression of choline acetyltransferase (ChAT) or tyrosine hydroxylase (TH) (Figures S4G–S4L).
Activity of dorsal pons ADRB1+ neurons in sleep/wake states
To assess the population activity of the ADRB1+ cells in DP across spontaneous sleep-wake states, we used fiber photometry to record calcium signals from ADRB1+ neurons in freely moving animals (Figure S5A) (Chen et al., 2013; Gunaydin et al., 2014). We injected a Cre-dependent AAV encoding the fluorescent calcium indicator GCaMP6s (AAV1/Syn-Flex-GCaMP6s-WPRE-SV40) into the DP of ADRB1-Cre mice. We implanted both a fiber optic probe (for subsequent delivery of excitation light and collection of fluorescent emission) and EEG/EMG electrodes (for simultaneous sleep-wake recordings) (Figures S5B and S5C) (Eban-Rothschild et al., 2016). When aligning the fluorescence signals of calcium activity across different sleep states, robust alterations in the population activity of the ADRB1+ neurons revealed that these cells are active during wakefulness and REM sleep and remain quiescent during NREM sleep (Figures 4D and 4E). Notably, these neurons began to increase their activity before NREM-to-REM and NREM-to-wake transitions and decrease their activity before wake-to-NREM and REM-to-wake transitions (Figure S5D). These findings demonstrate that ADRB1+ neurons in DP change their population activity across sleep–wake states and offer a mechanistic framework for their participation in the regulation of sleep and wakefulness.
Manipulation of dorsal pons ADRB1+ neurons changes sleep/wake states
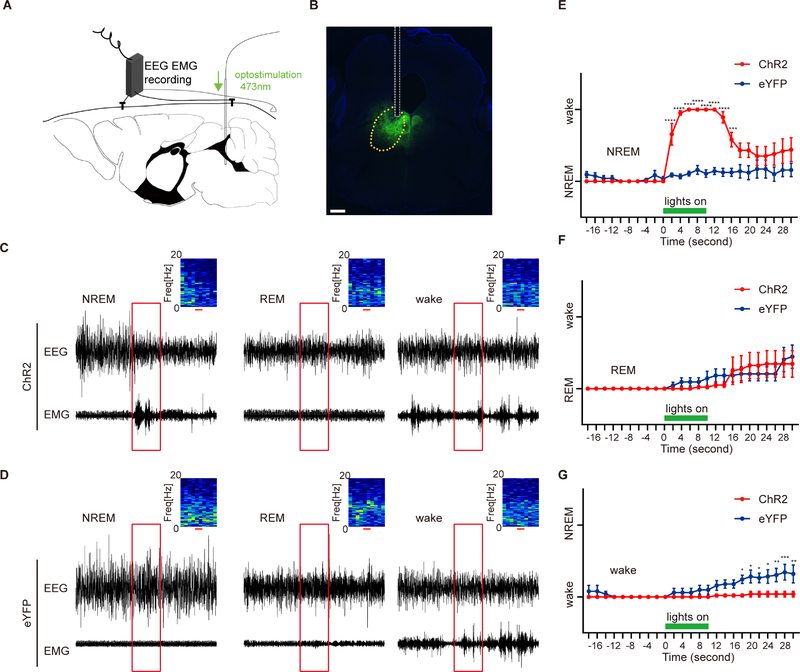
We next applied an optogenetic approach to activate the neurons in the DP. We unilaterally injected a Cre-inducible AAV expressing channel rhodopsin 2 fused with enhanced yellow fluorescent protein (AAV5-DIO-hChR2(H134R)-eYFP, ‘ChR2’ hereafter) or the control virus AAV5-EF1α-DIO-eYFP, ‘eYFP’ hereafter) into the DP of ADRB1-Cre mice and implanted the optical fiber and EEG-EMG electrodes (Figures 5A and 5B). The 10-s trains of light trials were randomly applied during EEG/EMG recording in the light phase. Based on the post hoc analysis of EEG/EMG, the trials were then categorized to be starting from NREM, REM and wakefulness. Interestingly, light stimulation during NREM sleep elicited immediate NREM-to-wake transitions in the ChR2-infected mice but not in the eYFP-infected mice (Figures 5C, 5D and 5E). However, similar stimulation could not trigger REM-to-wake transitions (Figures 5C, 5D and 5F). Stimulation during the wake phase produced no significant effect on state transitions although it may play a role in maintaining the wakefulness afterwards (Figures 5C, 5D and 5G). This is consistent with the fact that these neurons are wake and REM-active under physiological condition (Figures 4C and 4D). During wakefulness and REM sleep states, additional activation of these neurons cannot change the states because the neurons are already active. Nonetheless, the strong effect of NREM-to-wake transition demonstrates that ADRB1+ cells in the DP region are primarily wake-promoting.
Figure 5. ADRB1+ neurons in DP are wake-promoting.
(A) Schematic of simultaneous optogenetic stimulation/EEG /EMG set up for recording of the sleep/wake states with stimulation of DP ADRB1+ neurons.
(B) Representative slice showing viral expression and the placement of the fiber tip above the DP. Scale bar, 300μm.
(C and D) Representative EEG/EMG recordings of optogenetic trials (red box, 10 seconds) initiating from NREM, REM and wake states for ChR2-eYFP(C) and eYFP (D) mice. EEG power spectrograms are shown in the upper right. Red scales and boxes indicate 10 seconds light stimulation.
(E-G) Quantified results of NREM (E), REM (F) and wake (G) states for (C and D) from ChR2-eYFP (N=5) and eYFP (N=5) mice.
*P<0.05, **P<0.01, ***P<0.01, **** P<0.0001. Two way RM ANOVA, post-hoc Sidak’s multiple comparisons test for (E-G). Error bars represent ± SEM.
The Adrb1-A187V mutation alters the population activity of DP ADRB1+ neurons
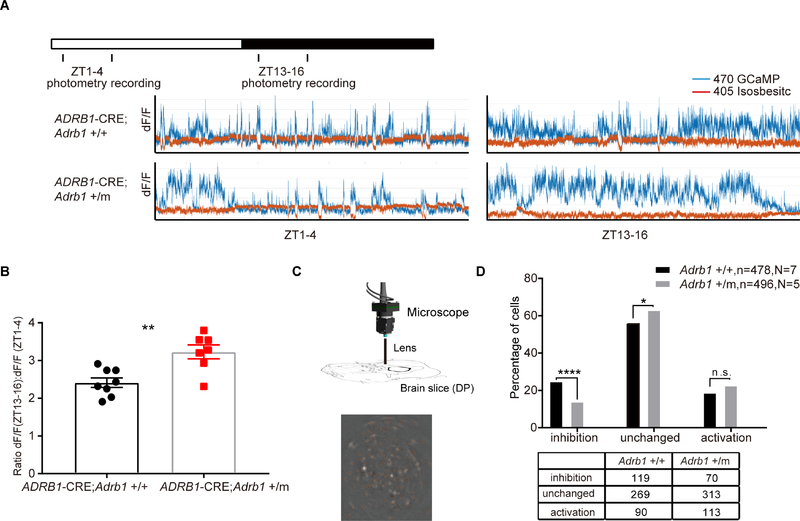
Given that the activity of ADRB1+ neurons of the DP region modulates the wakeful state and that the Adrb1-A187V mutant mice showed altered sleep/wake behavior, we next tested whether the Adrb1-A187V mutation affects the activity of ADRB1+ neurons in the DP. We crossed the transgenic ADRB1-Cre mice with the Adrb1-A187V mice, and a photometry strategy similar to that described above was applied to ADRB1-Cre; Adrb1+/+ and ADRB1-Cre; Adrb1+/m mice. The absolute photometry signals depend on the expression level of the virus around the tip of the optical fibers, which makes it difficult to analyze the neural activity between different mice directly. Thus, we compared the relative GCaMP signal amplitude from the active phase (ZT13–16) to the sleep phase (ZT1–4) in the same mouse. The relative amplitude was significantly greater in the Adrb1+/m mice than in the Adrb1+/+ mice (Figures 6A and 6B). These results suggest that activity of ADRB1+ neurons in the DP is altered by the Adrb1 A187V mutation. Notably, these neurons are wake-active and the Adrb1+/m mice spend 9% more time in wakefulness at ZT13–16. However, mutant mice exhibited 34% increase in the fluorescence signal shown in Figure 6B, suggesting the change of fluorescence here could not be solely attributed to the altered behavior.
Figure 6. Adrb1-A187V mutation alters DP neural activity.
(A) Representative photometry fluorescence traces at ZT1–4 and ZT13–16 from ADRB1-Cre; Adrb1+/+ (N=8) and ADRB1-Cre; Adrb1+/m (N=7) mice.
(B) Quantified ratio for (A).
(C) Schematic of calcium imaging set up for recording the activity of DP ADRB1+ neurons in brain slices (upper panel). One example of calcium imaging of DP in the slice (lower panel).
(D) Percentage of ADRB1+cells that respond differentially to dobutamine treatment in both Adrb1+/+ (N=7) and Adrb1+/m (N=5) brain slices. The bottom table shows the original cell numbers in different categories. See Figure S6 for the representative fluorescence traces of the cells in different categories.
**P<0.01, *** P<0.001. Two-tailed Student’s t-test for (B). Chi-square test for (D). Error bars represent ± SEM.
To further explore whether the changes of GCaMP signals are neuron-autonomous effects and not secondary to circuitry or behavior changes, we performed single cell imaging experiments on acutely isolated DP explants from ADRB1-Cre; Adrb1+/+ and ADRB1-Cre; Adrb1+/m mice (Figure 6C). We measured the percentage of ADRB1+ neurons responding with changes in GCaMP signal during treatment with dobutamine (a selective β1 agonist). Neurons were categorized into three groups based on their response to the dobutamine treatment: inhibition, no change, and activation (Figures S6A–S6C). This is consistent with the previous observations that β-ARs in CNS could be either excitatory or inhibitory (Szabadi, 2013). As shown in Figure 6D, the percentage of DP ADRB1+ neurons that were inhibited by dobutamine was significantly lower (56.7%) in the Adrb1+/m slice. Meanwhile, the percentages of DP ADRB1+ neurons that were activated by dobutamine were comparable between the two groups. These results indicate that the mutation affects ADRB1+ neurons to various extents. However, as a whole, the portion of neurons inhibited is much smaller in mutant than in WT, consistent with an overall increase in ADRB1+ neuron population activity in mutant mice (Figures 6A and 6B).
The Adrb1-A187V mutation changes the electrophysiological properties of DP ADRB1+ neurons
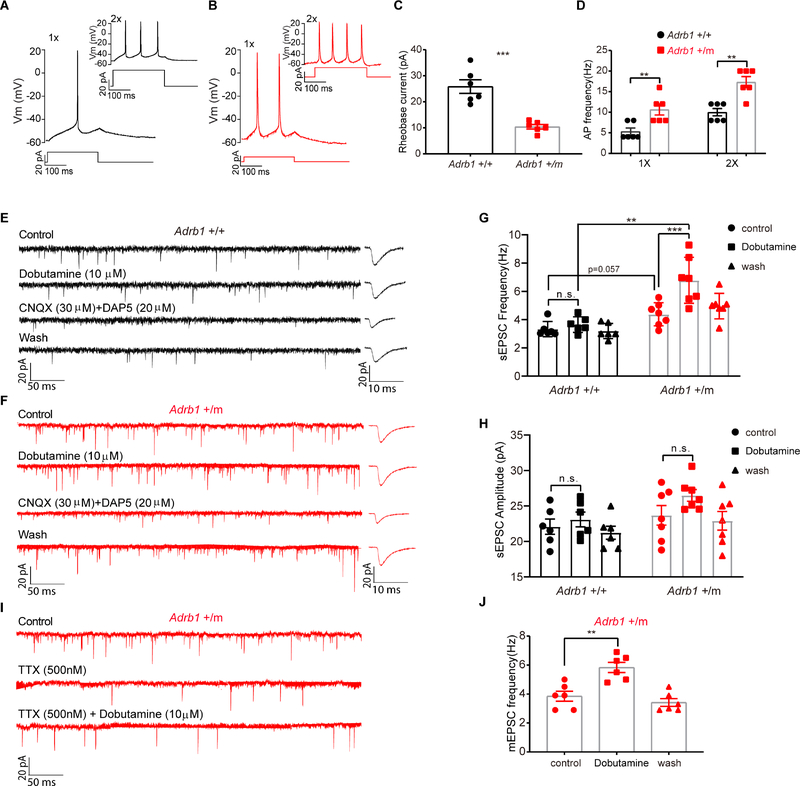
The increased population activity of ADRB1+ neurons from Adrb1 mutant mice could also be caused by enhanced excitability. To test this, we injected the Cre-dependent adeno-associated virus (AAV) encoding enhanced green fluorescent protein (EGFP) (AAV2/EF1a-DIO-EGFP-L10a) into the DP area to label the ADRB1+ cells with GFP. Using current clamp recordings of GFP-labeled ADRB1+ neurons from brain slices from either ADRB1-Cre; Adrb1+/+ (n=6, N=6 out of 23 brain slices from 11 animals) or ADRB1-Cre; Adrb1+/m mice (n=6, N=6 out of 15 brain slices from 15 animals), we obtained action potentials (Figures 7A and 7B). These action potentials were blocked by 500nM TTX (data not shown). We found that mutant neurons had a significantly lower rheobase current compared with that of neurons from Adrb1+/+ mice (Figure 7C) without any significant differences in the resting membrane potential (Adrb1 +/+, −60.14 ± 1.28 mV; Adrb1 +/m, −58.11 ± 1.32 mV), voltage threshold (Adrb1 +/+, −43.83 ± 3.00 mV; Adrb1 +/m, − 45.50 ± 2.48 mV or action potential overshoot (Adrb1 +/+, 21 ± 0.85 mV; Adrb1 +/m, 20.17 ± 0.14 mV). Also, a significant increase in firing frequencies was noted in Adrb1+/m neurons compared with those from Adrb1+/+ neurons (Figure 7D). These electrophysiological properties imply that Adrb1+/m neurons are more excitable.
Figure 7. The Adrb1-A187V mutation changes the electrophysiological properties of DP ADRB1+ neurons.
(A) Representative trace of a single action potential recorded from a DP Adrb1 +/+ neuron in response to a 250ms, 20pA (1X) pulse from the resting membrane potential (RMP). The same neuron fired three action potentials in response to a 40pA (2X) pulse (inset).
(B) Representative trace of two action potentials recorded from a DP Adrb1 +/m neuron in response to a 10 pA (1X) pulse from RMP. A 20pA (2X) pulse in the same neuron produced four action potentials (inset).
(C and D) Rheobase currents (C) and action potential frequency (D) were calculated for Adrb1 +/+ (n=6) and +/m (n=6) neurons.
(E and F) Representative traces of spontaneous EPSCs recorded under voltage clamp conditions (at −60 mV) with different treatments for Adrb1 +/+ (E) and Adrb1 +/m (F) neurons.
(G and H) Frequency (G) and amplitude (H) before and after dobutamine (10μM) treatment were calculated for Adrb1 +/+ (n=6) and +/m (n=7) neurons.
(I) Representative traces of mEPSCs recorded in one Adrb1 +/m neuron with different treatments.
(J) mEPSCs frequency were calculated for Adrb1 +/m (n=6) neurons before and after dobutamine (10μM) treatment in the presence of TTX.
**P<0.01, *** P<0.001, n.s. =not significant. Two-tailed Student’s t-test for (C) and (D). Two way RM ANOVA, post-hoc Sidak’s (genotype) and Tukey’s (dobutamine treatment) multiple comparisons test for (G) and (H). One way RM ANOVA, post-hoc Tukey’s multiple comparisons test for (J). Error bars represent ± SEM.
We also recorded spontaneous excitatory postsynaptic currents (sEPSCs) in slices from ADRB1-Cre; Adrb1+/+ or ADRB1-Cre; Adrb1+/m mice in the presence of the GABAergic antagonist, bicuculline (BIC, 50 μM). Because cells in the DP region are heterogeneous (Kohlmeier and Kristiansen, 2010; Wang and Morales, 2009), we analyzed sEPSCs that were blocked by post hoc treatment of glutamatergic antagonists CNQX and DAP5 (6/15 cells for Adrb1+/+, 40%; 7/22 cells for Adrb1+/m, 32%) (Figures 7E and 7F). Basal sEPSC frequency was slightly higher in Adrb1+/m than Adrb1+/+ neurons (p=0.057) (Figure 7G). Interestingly, dobutamine treatment increased sEPSC frequency in the Adrb1+/m but not Adrb1+/+ neurons (Figure 7G). Moreover, no significant change in sEPSC amplitude was found for either the genotype or treatment of dobutamine (Figure 7H). We further analyzed the miniature EPSCs (mEPSCs) in the presence of tetrodotoxin (TTX) from Adrb1+/m slices (Figure 7I). We found that the increase of EPSC frequencies were preserved in the presence of TTX, indicating that the increase in spontaneous glutamate release from the presynaptic terminals was independent of action potential generation (Figure 7J). This significant increase of the sEPSC (mEPSC) frequency suggests that presynaptic neurons (of the patched ADRB+ neuron) are more excitable as a population in response to dobutamine and that glutamatergic transmission is more active in the ADRB1+ neurons in mutant brain slices.
The dominant effects of Adrb1-A187V mutation
We analyzed the Adrb1 homozygous mutant mice (Adrb1 m/m) together with the heterozygous mice to clarify the mechanism. Although the protein level in homozygotes is even less than heterozygotes (Figures S7A and S7B), homozygous mutant mice have a similar mobile/sleep phenotype with heterozygotes (Figures S7C and S7D). In addition, in vivo and in vitro calcium imaging results were also similar between heterozygotes and homozygotes mice (Figures S7E and S7F). Together, these data further demonstrate the causative role of ADRB1 A187V mutation and imply that this mutation exerts dominant effects.
DISCUSSION
Sleep per se is thought to be a conserved behavior from vertebrates to invertebrates, including flies and nematodes (Cirelli et al., 2005; Koh et al., 2008; Raizen et al., 2008). Cutting-edge technologies have been applied to animal models and revealed mechanistic and circuit-level information (Artiushin and Sehgal, 2017; Chung et al., 2017; Guo et al., 2016; Niwa et al., 2018; Weber and Dan, 2016) for these organisms. In parallel, large-scale mutagenesis screening with animal models have been used to identify potential genes involved in sleep homeostasis regulation in these animals (Cirelli et al., 2005; Franken et al., 2001; Funato et al., 2016; Koh et al., 2008; Tatsuki et al., 2016). However, it remains unclear to what extent the genes and systems identified in model organisms are conserved in humans.
While NSS has long been recognized in sporadic cases from the general population, familial NSS was only first reported in 2009 (He et al., 2009), thus enabling the use of human genetics to identify novel sleep genes. The initial report was in a small nuclear family and thus, when a candidate mutation was identified in DEC2, in vitro and in vivo experiments were necessary to prove causality. We’ve continued to collect small and moderate sized families (2- and 3-generation) segregating FNSS alleles. It is likely that “required sleep time” is a genetic trait resulting from contributions of many variants in many genes. The >50 FNSS families identified to date have a sufficiently strong phenotype (lifelong requirement of <6.5 hours per/night) so that we have enriched for single mutations of large effect. Still, there is variability in the expression of FNSS among affected individuals, even within the same family. Taken together, these observations suggest that FNSS is caused by alleles of strong effect and that the phenotype is modified by the genetic background of each mutation carrier. In the moderate sized family reported here, there is clear transmission of an autosomal dominant allele in affected members of the family. We used a staged approach based on whole-exome sequencing, but using the genetic linkage data to take advantage of the information contained in the family structure to interpret the rare variants identified. This led to identification of only one coding variant from this locus that co-segregated with the FNSS phenotype. The ADRB1 allele is present in all ‘affected’ individuals but also in a carrier (#100784) with a total sleep time of 7 ½ hours. This is still one hour shorter than the population mean but does not meet our strict criteria to be classified as ‘affected’ (we classified this individual as ‘unknown’ and an obligate carrier since he has a definitely ‘affected’ daughter). Collectively, these data suggest that the ADRB1 mutation is likely to be causative for FNSS in this family. Variable expressivity is likely due to the genetic background of each mutation carrier.
Our mutant mice showed increased mobile time and a decreased sleep time of 55 minutes every 24 hours, whereas the human mutation carriers, on average, sleep 2 hours/day less than non-mutation carriers. It is not uncommon that animal models only partially recapitulate a human phenotype. This may be due to differences in physiology of mice vs. humans. Mice are nocturnal and have much more fragmented sleep than humans. Thus, it is possible that mice are less dependent on sleep duration, consolidation, timing, and other variables for sleep than humans. Also, the phenotype of mice modeling human mutations is often more subtle than those obtained in forward mutagenesis screens. Mutations identified in FNSS families exist in humans who have survived on this planet. It is not surprising that the phenotypes are not as strong as those seen in mutagenized flies and mice under laboratory conditions. Our mouse model resembles features of human FNSS, further strengthening the genetic data in support of a causative role of this ADRB1 mutation in sleep. Interestingly, the delta power of Adrb1 mutant mice is higher at the beginning of the sleep phase indicating a higher sleep pressure accumulated at the end of the active phase than the control mice. Further, the delta power decreased to the basal level during the light phase suggesting these mice can sustain a higher sleep pressure than control mice. More work is needed to test whether human mutation carriers have a similar alteration in delta power.
We found a heterogeneous group of neurons in dorsal pons that express ADRB1 at high levels. Using fiber photometry, we showed that these DP ADRB1+ neurons are either wake- or REM sleep-active under physiological conditions (Figure 4). Consistently, optogenetic studies indicated that these neurons are primarily wake-promoting (Figure 5). Thus, these sleep-relevant neurons provided an opportunity to investigate the neural behavior that might be affected by the Adrb1 A187V mutation. In mutant brain slices, the portion of ADRB1+ neurons that are potentially inhibited by agonist was significantly dampened while ADRB1+ neurons that are potentially excited by agonist remained unchanged (Figure 6D). Based on our finding in Figures 2, we speculated that the inhibitory function of β1-AR is more sensitive to the reduction of its protein abundance while the excitatory function is more tolerant to decreased protein levels. With fewer neurons being inhibited in the presence of natural ligand, the DP ADRB1+ neurons (as a population) are more active. This is consistent with the increased neuron activity in vivo in the mutant mice (Figure 6B) and the increased excitability and spontaneous excitatory glutamatergic neurotransmission in the mutant slices (Figure 7). Together, these results imply that the increased activity of the wake-promoting DP ADRB1+ neurons may contribute to the short sleep phenotype. Of note, although we chose to study ADRB1+ neurons in the DP, it is possible that ADRB1+ neurons (or glia) in other brain regions (Berridge et al., 2012; Paschalis et al., 2009) (Figures S2) may also play important roles in sleep regulation and contribute to the short sleep phenotype. Further investigation is needed to address this possibility.
In summary, we present human genetic data in FNSS, mouse modeling, in vitro, and in vivo functional data. Collectively, these diverse approaches support the role of the Adrb1-A187V as a causative mutation in FNSS and of DP β-ARs in regulation of sleep/wake behavior. This, in turn, provides an opportunity to further explore the mechanisms and potential drug targets of β1-AR for treatment of sleep related disorders. Much more work is needed to dissect the complex circuitry underlying sleep/wake regulation.
STAR⋆METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ying-Hui Fu (Ying-Hui.Fu@ucsf.edu). All reagents, cell lines and mouse models used in this manuscript are available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human studies
Human research subjects for this study are voluntary participants. The proband (first participant of this family) came to us because of an interest in his/her own sleep behavior and helped to recruit other family members. All human participants signed a consent form approved by the Institutional Review Boards at the University of Utah and the University of California, San Francisco (IRB# 10–03952). Self-reported habitual sleep-wake schedules were obtained during structured interviews by one of the authors (C.R.J., L.J.P, and A.K.). Blood sample collection and DNA preparation were performed as previously described (He et al., 2009).
Animal Sources
Adrb1-A187V knock-in mice were generated by CRISPR/Cas9. Briefly, DNA template for sgRNA was amplified with primers containing T7 promoter and the sgRNA-targeting sequence. The sequence for primers were as follows; forward, ‘TTAATACGACTCACTATAGGAAGGACACCAACGCCGAGAGTTTTAGAGCTAGAAATAGC’; reverse, ‘AAAAGCACCGACTCGGTGCC’. The sgRNA was then transcribed (MEGAshortscript T7 kit, Life Technologies) and purified (MEGAclear kit, Life Technologies) in RNase-free water.
Super-ovulated female FVB/N mice were mated to FVB/N males, and fertilized zygotes were collected from oviducts. Cas9 protein (50 ng/mL), sgRNA (20 ng/mL) and targeting oligo DNA (20 ng/mL) were mixed and injected into pronucleus of fertilized zygotes. Injected zygotes were implanted into oviducts of pseudopregnant CD1 female mice. Founders were genotyped by PCR and sequencing. Out of 24 pups genotyped, 18 were positive for the knockin. Mice were then backcrossed onto a C57Bl/6J background for at least six generations before phenotyping to dilute potential off-target effects. Two independent lines were chosen for experimentation, which gave similar results in all the tests. The oligo DNA sequence for recombination was as follows, ‘cgccctttcgctaccagagtttgctgacgcgcgcgcgagcgcgggccctcgtgtgcacagtgtgggccatctcggtcttggtgtccttcctgcccatcctcatgcactggtggcgggccgagagcgacgaagcgcgccgctgctacaacg’.
ADRB1-Cre BAC transgenic mice were generated as previously described (He et al., 2009; Xu et al., 2007). Human BAC: CTD 2337B3, containing the entire ADRB1 gene on a 150-kb genomic insert, was obtained from the CalTech human BAC library. The coding sequence of ADRB1 in the BAC was replaced by that of Cre through two steps of recombination. This BAC clone includes 80 kb of sequence upstream from the start codon and is expected to contain all cis-acting regulatory elements needed to accurately reproduce an endogenous expression pattern. Detailed mapping and sequencing were carried out for the modified BAC before injected into the mouse zygotes. This transgenic stain was maintained on a C57Bl/6J background.
ROSA mT/mG (stock 007676), Ai32 (loxP-flanked-ChR2–eYFP, stock 012569) reporter mice and Adrb1tm1Bkk Adrb2 tm1Bkk /J (stock 003810) mice were purchased from The Jackson Laboratory.
Animal Studies
All experimental animals were singly housed on a LD 12:12 cycle and given ad libitum access to food and water. Male mice were used for all behavioral experiments. Mice were at least eight weeks old at the time of surgery. For the studies of DREADDs/fiber photometry, ADRB1-Cre mice were randomly assigned to experimental or control group by receiving different virus injections. Littermates were used for the studies for the comparisons between WT and mutant mice. We noticed that the mouse sleep behaviors were significantly affected by light intensity. For the experiments that compared the sleep time and locomotor activity between WT and mutant mice, light intensity of the room was strictly controlled between 70–90lux.
All experimental protocols were approved by the University of California, San Francisco IACUC following NIH guidelines for the Care and Use of Laboratory Animals.
Cell Lines
HEK293 cells were obtained from the ATCC Human Primary Cell collection (https://www.atcc.org). They were grown in DMEM supplemented with 10% of heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin. All cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Transfection of the cells was performed using Lipofectamine 3000. Cell lines were tested for mycoplasma contamination every three months.
METHOD DETAILS
Nomenclature
For humans—gene (ADRB1), protein (ADRB1). For mouse—gene (Adrb1), protein (ADRB1). +/+ refers to wild type animals or unaffected human subjects and +/m refers to heterozygous mutant animals or affected human subjects.
SNP Genotyping
Genomic DNAs from 8 individuals (100290, 100471, 100475, 100481, 100487, 100784, 100889, 100913) were genotyped using the Illumina HumanCytoSNP-12 array. A total of 299,671 markers were called for each individual. SNP pruning was then performed using PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml). The genotype calls were filtered for the following criteria, in sequential order: genotype missingness, with a cutoff of 30% (289,305 SNPs remaining); Mendelian errors (289,305 SNPs remaining); and absence of linkage disequilibrium using pairwise genotypic correlation, with r2 cutoff of 0.1, and a sliding window of 50 SNPs (4,782 SNPs remaining). The filtered set of SNPs was used to calculate linkage, using the parametric linkage capabilities of the software package MERLIN (http://www.sph.umich.edu/csg/abecasis/Merlin/index.html). Map positions of each marker were calculated using interpolation of the Rutgers Combined Linkage-Physical Map Build 36/37. The parametric model assumed an autosomal dominant mode of inheritance with 90% penetrance.
Exome sequencing
Exome sequencing was performed on 4 individuals (100290, 100475, 100889, and 100913). Omicia Opal 0.10.0 software was used to annotate the potentially causative genes/variants in the exomes of each of the affected individuals (after filtering out common variants, dbSNP MAF > 1%) using the HGMD and OMIM databases. Variant prioritization (using Ingenuity Variant Analysis) included generating a list of all variants, filtered for 1) minor allele frequency < 1% according to 1000 Genomes Project, Complete Genomics public genomes, and NHLBI ESP exomes; 2) nonsynonymous, near a splice site/promoter, or a structural variant; 3) occur in all 3 affected and not in the control sample; and 4) within a region determined by parametric linkage to have LOD > 0.
Protein Extraction and Immunoblot Analysis
Proteins from mouse tissues and cultured HEK293 cells were prepared with the RIPA Lysis Buffer (Sigma). SDS-PAGE and immunoblot analysis were performed according to standard protocols. Antibodies used were mouse anti-FLAG (Sigma; FLAG M2), rabbit anti-ADRB1 (Santa Cruz; sc568) and mouse anti-actin (Abcam; ab6276). Band intensities were determined using Image J software (NIH). Actin was used as a loading control.
RNA Isolation and Real-Time PCR
Total RNA was isolated from frozen tissues (heart and brain) with Trizol reagent (Thermo Fisher Scientific). A total of 5ug total RNA was reverse transcribed using the Superscript IV Kit (Thermo Fisher Scientific). cDNA was then quantified using SYBR green real-time PCR analysis with the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). The real-time PCR data were normalized to Actb. See Key Resources Table for primer sequences.
Luminescence-based rapid cAMP assay
HEK293 cells were plated in 96-well dishes in 100 μl of growth medium. When cells reached 70–80% confluence, they were transfected with 50 ng β1AR-WT or β1AR-WT + β1AR-Mut (25 ng:25 ng) plasmids together with 50ng pGloSensor-22F (Promega) which produce rapid and reversible cAMP-dependent activation of luciferase activity. After incubation for an additional 20–24 hours, growth medium was replaced with 100μl equilibration medium (88% CO2-independent medium, 10% FBS, 2% GloSensor cAMP Reagent stock solution (Promega)). Cells were then equilibrated for 2 hours at room temperature. Baseline luminescence was measured every min for 10 min (synergy H4, Bio Tek). 10nM isoproterenol was then added to the medium, and Luminescence was immediately measured every min for 30 min (synergy H4, Bio Tek).
ANY-maze monitoring
Mice were kept in individual cages with free access to food and water. Mice were monitored by infrared camera and tracked by an automatic video tracking system (Storlting, Wood Dale, IL; RRID SCR_014289). Mice were entrained to LD 12:12 for 1 week and then locomotor activity was recorded for 3 or 4 days. Walking distance, rears and mobile times were calculated using ANY-maze software and data were averaged. Samples with over 500-meter walking distance or below 10,000-sec immobility time each day were excluded from the statistical analysis due to the failure of automatic tracking.
Stereotaxic Viral Injection, Cannulization, and EEG Implantation
Animals were anesthetized with 2% isoflurane and placed in a stereotaxic head frame on a heat pad. Opthalmic ointment was applied to the eyes to prevent drying. A midline incision was made down the scalp and a craniotomy was made using a dental drill. A 10 μl Nanofil Hamilton syringe (WPI, Sarasota, FL) with a pulled glass needle was used to infuse virus with a microsyringe pump (UMP3; WPI, Sarasota, FL) and its controller (Micro4; WPI, Sarasota, FL). Virus was infused at a rate of 50 nl/min. Following infusion, the needle was kept at the injection site for 10 min and then slowly withdrawn at 0.01 mm/sec. All stereotaxic coordinates are relative to bregma. ADRB1-Cre was injected into the DP (−5.1 mm anteroposterior (AP); 0.6 mm mediolateral (ML); −3.7 mm dorsoventral (DV)) with a total of 300 nl of virus.
Optical cannulas for optogenetics. Mice were implanted with a 200 μm diameter 0.22 NA mono fiber-optic cannula (ThorLabs) directly above the DP at −5.1 mm (AP); +0.6 mm (ML); −3.55 mm (DV). Cannulas were secured to the skull using a base layer of SuperGlue (LocTite) followed by a second layer of dental cement (Ortho-Jet; Lang, Wheeling, IL).
Optical cannulas for Fiber Photometry. Mice were implanted with a 400 μm diameter 0.22 NA mono fiber-optic cannula (Doric Lenses) directly above the DP at −5.1 mm (AP); +0.6 mm (ML); −3.55 mm (DV). Cannulas were secured to the skull using a base layer of SuperGlue (LocTite) followed by a second layer of dental cement (Ortho-Jet; Lang, Wheeling, IL).
Installation of EEG/EMG Headsets. After installation of the appropriate cannula, mice were also implanted with EEG/EMG headsets. Four guide holes were made using a 23-gauge surgical needle epidurally over the frontal cortical area (1 mm anterior to bregma, 1 mm lateral to the midline) and over the parietal area (3 mm posterior to bregma, 2.5 mm lateral to midline). One ground screw and three screws with leads were placed into the skull through the holes. The screws with leads were then soldered onto a 6-pin connector EEG/EMG headset (Pinnacle Technologies, Lawrence, Kansas). For EMG recordings, EMG leads from the headset were placed into the neck muscle. The headset was then covered with black dental cement to form a solid cap atop the mouse’s head. The incision was then closed with VetBond (3M, Santa Cruz Biotech) and animals were given a subcutaneous injection of marcaine (0.05 mg/kg) prior to recovery on a heating pad. Behavioral experiments were conducted 3 weeks later to allow for sufficient recovery and for viral expression.
All mice were subjected to histological analyses to confirm the area of AAV transfection and placement of fibers at the end of experiment.
EEG/EMG Recording and Scoring
For EEG/EMG recording, mice were singly-housed and habituated to the recording cable for 7 days in LD 12:12 conditions. Tethered pre-amplifiers were attached to the headset of the mice. The signals were relayed through the commutators which allowed the animal to move freely. Data was acquired through the Sirenia software package (Pinnacle Technologies, Lawrence, KS) (Anaclet et al., 2014). All the data were sampled at 500 Hz.
Sleep was scored semi-automatically by Sirenia Sleep Pro software in 10-s epochs for wakefulness, NREM, and REM sleep, and then subsequently hand-scored by researchers blinded to genotype with the assistance of spectral analysis using FFT. In general, wakefulness was defined as desynchronized low-amplitude EEG and heightened tonic EMG activity with phasic bursts. NREM sleep was defined as synchronized, high-amplitude, low-frequency (0.5–4 Hz) EEG and substantially reduced EMG activity compared with wakefulness. REM sleep was defined as having a pronounced theta rhythm (4–9 Hz) with no EMG activity. REM-like behavior was defined as having a pronounced theta rhythm (4–9 Hz) with tonic intermediate level of EMG activity. Under our experiment condition, this unnatural state could last from a few minutes to hours after CNO injection. This REM-like state was not seen in wild type animals. For calculation of total sleep time, total NREM sleep time and total REM sleep time, data were averaged from two consecutive days.
For spectral analysis, artifacts and state transition epochs were excluded. EEG power spectra and frequency bands for each vigilance state and condition were standardized by expressing each frequency bin as a percentage of the total power (0.5–25.0 Hz). The time course of delta power (1.0–4.0 Hz) in NREM sleep was computed as previously described (Cirelli, 2005; Franken et al., 2006; Vassalli and Franken, 2017; Wang et al., 2018). To investigate the temporal progression of delta power, values were computed relative to the last 4 hours of the light phase (i.e., ZT 8 to 12 defined as 1) in baseline. This interval corresponds to the period with lowest delta power (Franken et al., 2001; Mang and Franken, 2012).These analyses were restricted to the light phase, because in agreement with previous mouse studies (Franken et al., 2001; Halassa et al., 2009), there was too little NREM sleep in most mice during the active (dark) phase for accurate assessments.
TRAP
Custom recombinant cre-dependent AAV expressing GFP-tagged ribosomal subunit L10a (AAV2-EF1a-DIO-EGFP-L10a) was obtained from a previous report (Tan et al., 2016) and then packaged by the UNC vector core. This virus (300 nl) was infused bilaterally into the DP of ADRB1-Cre mice using the coordinates described above.
Animals were sacrificed for TRAP 2–3 weeks after injection. TRAP followed a detailed protocol supplied by the Heintz laboratory at the Rockefeller University (New York, New York, USA) (Doyle et al., 2008; Heiman et al., 2008). The small brain region including DP from 3–4 brains was pooled for each experiment, and a total of four replicate experiments were performed. The pooled tissues were then added to ice-cold polysome extraction buffer for homogenization. Monoclonal anti-GFP antibodies (Monoclonal Antibody Core Facility, Memorial Sloan-Kettering Cancer Center, New York, New York, USA) were coupled to Dynabeads (Thermo Fisher Scientific; 10004D) and added to a postmitochondrial preparation of tissue extract. Beads and extract were incubated at 4°C, with agitation, for 30 minutes. The beads were then washed with a large volume of wash buffer, and resuspended in lysis buffer. Total RNA was isolated with a column (QIAGEN RNAeasy Mini Kit). Equal amounts of RNA were used for the subsequent RT-qPCR assays using the strategy described above. Fold-enrichment was calculated using the ΔCt method with normalization to Actb.
Histology and Immunohistochemistry
Mice were transcardially perfused with PBS followed by 4% paraformaldehyde (PFA);PBS. Brains were postfixed overnight in 4% PFA;PBS and placed in 30% sucrose for another 24 hours. Free-floating sections (40 μm) were prepared with a cryostat (Leica). For GCaMP6s and DREADDs verification, slices were immediately mounted onto glass slides with DAPI mounting media, coverslipped, and imaged.
For colocalization of ChAT/TH and mCherry detection, sections were washed three times in TBST and then blocked for 1 hour in superblock buffer (Thermo Fisher Scientific). After block, the sections were incubated with a primary antibody for mCherry (1/1,000 rabbit anti-DsRed (Clontech; 632496)) together with another antibody for either ChAT (1/100 goat anti-ChAT (MilliPore; Ab144P)) or TH (1/1,000 mouse anti-Tyrosine Hydroxylase (MilliPore; MAB318)) at 4°C overnight. After washing three times in TBST, the sections were incubated with corresponding secondary antibodies (Jackson ImmunoResearch) for another 2 hours at room temperature before being mounted onto glass slides.
In-situ hybridization (ISH) of Vglut2 or Gad1 was performed using digoxigenin (DIG)-labeled RNA probes. The protocol was modified from previous reports(Hayashi et al., 2015; Xiu et al., 2014). Briefly, the RNA probes labeled by digoxygenin-UTP (Roche) were generated by in vitro transcription and dissolved at 1 μg/ml in the hybridization solution (50% formamide (vol/ vol), 5× SSC, 0.3 mg/ml yeast tRNA, 100 μg/ ml heparin, 1× Denhardt’s solution, 0.1% Tween 20 (vol/vol), 0.1% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS, wt/vol) and 5 mM EDTA). Free-floating sections were washed with PBS for 10 min and treated with 2% H2O2 (vol/vol) in 0.01 M PBS for 10 min, followed by another round of wash with PBS for 10 min at 25~28°C. They were then treated with 0.3% Triton X-100 (vol/vol) in PBS for 20 min and acetylated by 0.25% acetic anhydride (vol/vol) in 0.1 M triethanolamine (pH 8.0) for 10 min, followed by two rounds of washing with PBS. Afterward, the sections were placed in the hybridization solution without probes for 1 h and incubated in the hybridization solution with the corresponding probes for 16–18 h at 60°C. After hybridization, the sections were rinsed once and washed twice in 2× SSC for 15 min at 60°C, followed by treatment with 2 μg/ml RNase A in 2× SSC at 37°C for 30 min. They were further rinsed once and washed twice in 0.2× SSC for 30 min at 60°C, followed by three rounds of wash in PBS containing 0.05% Tween 20 (PBT) for 10 min at 25~28°C. The sections were blocked with 10% normal sheep serum (vol/vol) in PBT for 1 h at 25~28°C and incubated in the same solution with sheep anti-digoxygenin antibody (1:500, Roche 11207733910) at 4°C overnight. On the second day, the sections were washed three times in PBT for 10 min and incubated in the amplification solution with cyanine 3 tyramide (PerkinElmer NEL744B001KT) for 20 min at 25~28°C, followed by three rounds of washing in 0.01 M PBS for 10 min. Subsequently, immunohistochemistry (IHC) for mCherry was performed. Sections were incubated with primary antibody for mCherry (1/1,000 rabbit anti-DsRed (Clontech; 632496)) at 4°C overnight, followed by secondary antibody staining using Cy2-conjugated antibodies (Jackson Immuno Research). We confirmed that the ISH procedure eliminates the native fluorescence of mCherry. Anti-sense and sense probes were used in parallel in the preliminary experiments to confirm that only anti-sense probes gave rise to the positive signals. Images were captured with a laser confocal microscope. The Vglut2 probe covered the open reading frame (ORF) and the Gad1 probe shared identical sequences with the one used by Allen brain atlas (http://mouse.brain-map.org/).
ISH of endogenous Adrb1 was performed using the RNAscope assay (Cat# 320293/320513, Advanced Cell Diagnostics, Manual Fluorescent Multiplex kits). Fresh whole brain was dissected from EGFP virus (rAAV2/EF1a-DIO-EGFP-L10a) infused ADRB1-Cre animals and rapidly frozen in liquid nitrogen, embedded in cryo-embedding medium and sectioned on a cryostat. 20 μm sections were mounted on slides at −20°C. All subsequent steps were performed according to manufacturer’s instructions. Probes used: EGFP (400281-C3), Mm-Adrb1 (449761).
Fiber Photometry
ADRB1-Cre mice were infused with AAV1/Syn-Flex-GCaMP6s-WPRE-SV40 virus (300nl) unilaterally into the DP using the coordinates described above. Optical cannulas and EEG/EMG headsets were installed as described above. Control mice were infused with eYFP expressing virus and installed with the same cannulas and headsets. All animals were allowed to recover after patch cord attachment for at least 3 days.
A rig for performing fiber photometry recordings was constructed following basic specifications previously described with modifications (Chen et al., 2015; Gunaydin et al., 2014). Briefly, a 473 nm LED (ThorLabs) was used as the GCaMP6s excitation source. A 405 nm LED was used as isosbestic control. Both LEDs were fed into a fluorescence mini-cube (Doric Lenses, Quebec) which reflected the LED signal through two patchcords into the mouse cannula. Commutators were placed between the patchcords which allows the animals to move freely during recording. Fluorescence signals were then reflected back from the mini-cube onto a photoreceiver (Newport 2151). The signal was output into a RZ5P Processor (Tucker-Davis Technologies), which allowed for real-time modulation and demodulation of the signal.
All fiber photometry data were subjected to the following minimal processing steps in MATLAB. dF/F indicates (recorded fluorescence – F0)/ F0. For calculation of dF/F in the spontaneous sleep-wake state, trials were randomly started at the day time and lasted for 2–3 hours. F0 was defined as the median fluorescence over the trial. Different states were determined based on EEG/EMG scoring. dF/F value for each state was then calculated by averaging the value (dF/F 473-dF/F 405) from the first 10 episodes of that state. For calculation of the ratio dF/F (ZT13–16):dF/F (ZT1–4), photometry recording was carried out continuously from ZT12 (day 1) to ZT5 (day 2) in each trial. F0 was then defined as the median fluorescence from (ZT1–5 in day 2) of the same trial. dF/F (time window) is the summarization of all the values of (dF/F 473-dF/F 405 ) within the indicated time window.
Calcium imaging in DP explants
ADRB1-Cre mice (either WT or mutant, 8~12 weeks) were infused with AAV1/Syn-Flex- GCaMP6s -WPRE-SV40 virus (300nl) unilaterally into the DP using the coordinates described above 2–4 weeks before slice preparation. Slice were prepared following the previously reported protocol (Ting et al., 2014). Briefly, animals were anesthetized under isoflurane and briefly perfused intracardially with 10 ml of ice-cold NMDG solution (92mM NMDG, 30mM NaHCO3, 25 mM glucose, 20mM HEPES, 10mM MgSO4, 5mM sodium ascorbate, 3mM sodium pyruvate, 2.5mM KCl, 2mM thiourea, 1.25mM NaH2PO4, 0.5mM CaCl2 (pH 7.3, 300 mOsm, bubbled with 95% O2 and 5% CO2)). The brains were then quickly removed and placed into additional ice-cold NMDG solution for slicing. Coronal slices were cut using a Leica VT1200S vibratome at 300 μm thickness, and warmed to 36.5°C for 10 min. Slices containing DP were transferred to room temperature (22–24°C) HEPES holding solution containing 92mM NaCl, 30mM NaHCO3, 25mM glucose, 20mM HEPES, 5mM sodium ascorbate, 3mM sodium pyruvate, 2.5mM KCl, 2mM thiourea, 2mM MgSO4, 2mM CaCl2, and 1.25mM NaH2PO4, (pH 7.3, 300 mOsm, bubbled with 95% O2 and 5% CO2) for 1 – 2 hr.
After incubation, the slice were transferred to the recording chamber and constantly perfused with room-temperature (22–25 °C) recording solution containing 119mM NaCl, 2.5mM KCl, 1.25mM NaH2PO4, 24mM NaHCO3, 12.5mM glucose, 2mM CaCl2, and 2mM MgSO4 (pH 7.3, 300 mOsm, bubbled with 95% O2 and 5% CO2) at a rate of 4 ml per minute.
An integrated microscope (nVista HD, Inscopix) was used to image the GCaMP signal from the slice. We chose this small microscope because the lens that collects the light can go directly into the perfusion buffer from the top to image the surface-layer cells, which are usually healthier and more accessible to the drug. In addition, the recording and data processing software are commercially available and user-friendly. Although the resolution of the images might be lower than those gathered by some confocal microscopes, it is sufficient for our purpose in this scenario. We used the data acquisition software (nVista, Inscopix) to acquire the images (a range of 6%–10% of LED intensity, Gain 2–3, 2.5–5fps). During each recording, the recording solution containing dobutamine (10μM) was turned on and off at the indicated time point.
The video was then analyzed by the Inscopix Data Processing (Inscopix) software. Each video was processed with spatial crop, spatial filter, motion correction and dF/F calculation. Region of interests (ROIs, considered as a single cell) were manually selected based on the shape and dF/F changes throughout the recording. ROIs that exhibited short bursts of dF/F changes or fluctuations during the recording were analyzed. ROIs that showed steady decreases, increases or maintained constant ΔF/F were excluded from further analysis. The dF/F changes were then aligned with the time window of dobutamine treatment and the cells were categorized into three groups (inhibition, unchanged and activation) based on the alignment (Figures S6A–S6C). Researchers were blinded to the genotype of the slice when processing and analyzing the data to ensure the same criteria applied to calculate both the WT and mutant cells.
Electrophysiological recording in DP explants
ADRB1-Cre mice (either WT or mutant, 3~4 weeks) were infused with AAV2/EF1a-DIO-EGFP-L10a virus (250nl) unilaterally into the DP one week before slice preparation. On the experiment day, animals were deeply anesthetized and then decapitated. The brains were removed quickly and placed into ice-cold cutting solution of the following composition: 234 mM sucrose, 28 mM NaHCO3, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, 7 mM MgCl2, 7 mM glucose, 1 mM ascorbic acid, and 3 mM pyruvic acid, bubbled with 95% O2/5% CO2 at pH 7.4. For each experiment, coronal sections (250 μm) were obtained with a vibratome and kept for 30 min at 37°C in oxygenated standard artificial CSF (ACSF): 130 mM NaCl, 24 mM NaHCO3, 3.5 mM KCl, 1.25 mM NaH2PO4, 1.5 mM CaCl2, 1 mM MgSO4, and 10 mM glucose, bubbled with 95% O2/5% CO2 at pH 7.4. Slices were stored in modified interface chamber for 30–40 min at 37°C and then maintained at room temperature until being transferred to the recording chamber in oxygenated standard ACSF:130 NaCl mM, 24 mM NaHCO3, 3.5 mM KCl, 1.25 mM NaH2PO4, 1.5 mM CaCl2, 1 mM MgSO4, and 10 mM glucose, bubbled with 95% O2 and 5% CO2 at pH 7.4. The GFP+ cells were observed with the aid of a fluorescence microscope (BX-50WI; Olympus) and visualized with a chilled charge-coupled device video camera (Dage-MTI) with a 40× water-immersion differential interference contrast objective.
Whole cell current clamp and voltage clamp recordings were performed from GFP+ cells located LDT region. Patch pipettes with a resistance of 5–7 Mohm were pulled from glass capillaries (Sutter Instruments). For the detection of Action Potentials and spontaneous post synaptic currents (sEPSCs) pipettes were filled with (in mM) 144 K-gluconate, 0.2 EGTA, 3 MgCl2, 10 HEPES, 0.3 NaGTP and 4 Na2ATP (310 mOsm). RMP and input resistance was measured from each cell. Whole cell current and voltage clamp were obtained as it was described previously (Ardelt et al., 2013; Jayaraj et al., 2018). The input resistances of these cells were Adrb1+/+ (489.3 ± 3.87, n= 42) and Adrb1 +/m (502.6 ± 4.37, n=44). The rheobase was the minimum current injected that generated an action potential. Detection and measurement of spontaneous events were recorded for 2 to 3 min under the appropriate experimental configuration (baseline control, drug application). Event frequency and amplitude were determined by Clampfit 10.0. DL-2-Amino-5-phosphonopentanoic acid (DAP5, 50 μM, Tocris), and 6-cyano-7-nitroquinoxaline-2, 3-dione (CNQX, 10 μM, Tocris) were used to block NMDA and AMPA ionotrophic glutamate receptors, respectively. Tetrodotoxin with citrate (TTX; Alomone) was used to inhibit voltage-gated Na+ channels and applied at a final concentration of 500 nM.
Optical Stimulation
ADRB1-Cre mice were infused with rAAV5/EF1a-DIO-hCHR2(H134R)-eYFP virus (300nl) unilaterally into the DP using the above coordinates. Optical cannulas and EEG/EMG headsets were installed as described above. Control mice received eYFP virus injection and similar cannulas/headsets installation. During the stimulation experiment, the implanted cannulated fiber was attached to the LED light source integrated in the EEG/EMG preamplifier on the top of the skull (Pinnacle Technologies). This design enabled the animals to move freely. LED pulse width and frequency was controlled by external microcontroller (Arduino) and Sirenia Acquisition Software (Pinnacle Technologies). The current for LED was set at 300mA. LED pulses lasting for 10s at 10Hz with 10ms pulse width were generated every 5/10 minutes throughout the experiments. EEG/EMG was recorded simultaneously.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical parameters including sample sizes (N = number of animals per group), the statistical test used and statistical significance are reported in the Figures and Figure legends. All behavior trials involved age-matched littermates as controls where possible. Mice were randomly assigned before surgery to either Optogentics/GCaMP or control groups. No outliers were excluded from analysis unless specified. Data is judged to be statistically significant when p < 0.05. In figures, asterisks denote statistical significance *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All statistical analysis was performed using GraphPad PRISM 7 software.
DATA AND CODE AVAILABILITY
All data and custom software that support the current findings are available upon request from the Lead Contact.
Supplementary Material
Highlight.
A mutation in ADRB1 leads to natural short sleep trait in humans
Mice engineered with same mutation have similar short sleep behavior as humans
Activity of dorsal pons ADRB1+ neurons associates with REM sleep and wakefulness
Mutation increases the population activity of dorsal pons ADRB1+ neurons
ACKNOWLEDGEMENTS
We thank Dr. Y. Dan’s lab at the University of California, Berkeley for the help with our EEG setup; Y. Chen and D. Kuo for the help with photometry setup; Mr. S. Deverasetty and Mr. A. Huang for assistance with exome data processing; the University of North Carolina Virus Core for supplying AAV; J. Webb for comments on the manuscript; and all other members in Ptáček and Fu labs for helpful discussions. This work was supported by NINDS Informatics Center for Neurogenetics and Neurogenomics P30 NS062691 to G.C. and D.H.G.; by NIH grant NS099333 to L.J.P., NS072360 and NS104782 to Y-H.F. and by the William Bowes Neurogenetics Fund to L.J.P. and Y.H.F. The generation of mouse models was also supported by NIH P30 DK063720 to the Diabetes Center at UCSF.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information includes six figures and one table and can be found with this article online at:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anaclet C, Ferrari L, Arrigoni E, Bass CE, Saper CB, Lu J, and Fuller PM (2014). The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat. Neurosci. 17, 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardelt AA, Bhattacharyya BJ, Belmadani A, Ren D, and Miller RJ (2013). Stromal derived growth factor-1 (CXCL12) modulates synaptic transmission to immature neurons during post-ischemic cerebral repair. Exp. Neurol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiushin G, and Sehgal A (2017). The Drosophila circuitry of sleep–wake regulation. Curr. Opin. Neurobiol 44, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge CW (2008). Noradrenergic modulation of arousal. Brain Res. Rev 58, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge CW, Schmeichel BE, and España RA (2012). Noradrenergic modulation of wakefulness/arousal. Sleep Med. Rev 16, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissard R, Gervasoni D, Schmidt MH, Barbagli B, Fort P, and Luppi PH (2002). The rat ponto-medullary network responsible for paradoxical sleep onset and maintenance: A combined microinjection and functional neuroanatomical study. Eur. J. Neurosci 16, 1959–1973. [DOI] [PubMed] [Google Scholar]
- Borbély AA (1982). A two process model of sleep regulation. Hum. Neurobiol 1, 195–204. [PubMed] [Google Scholar]
- Borbély AA, Daan S, Wirz-Justice A, and Deboer T (2016). The two-process model of sleep regulation: A reappraisal. J. Sleep Res 25, 131–143. [DOI] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin YC, Kuo TW, and Knight ZA (2015). Sensory Detection of Food Rapidly Modulates Arcuate Feeding Circuits. Cell 160, 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Weber F, Zhong P, Tan CL, Nguyen TN, Beier KT, Hörmann N, Chang WC, Zhang Z, Do JP, et al. (2017). Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545, 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C (2005). Locus Ceruleus Control of Slow-Wave Homeostasis. J. Neurosci 25, 4503–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, and Tononi G (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092. [DOI] [PubMed] [Google Scholar]
- Cox J, Pinto L, and Dan Y (2016). Calcium imaging of sleep-wake related neuronal activity in the dorsal pons. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Beersma DG, and Borbely AA (1984). Timing of human sleep: recovery process gated by a circadian pacemaker. Am. J. Physiol. Integr. Comp. Physiol 246, R161–R183. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. (2008). Application of a Translational Profiling Approach for the Comparative Analysis of CNS Cell Types. Cell 135, 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eban-Rothschild A, Rothschild G, Giardino WJ, Jones JR, and De Lecea L (2016). VTA dopaminergic neurons regulate ethologically relevant sleep-wake behaviors. Nat. Neurosci 19, 1356–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Chollet D, and Tafti M (2001). The homeostatic regulation of sleep need is under genetic control. J. Neurosci 21, 2610–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Dudley CA, Estill SJ, Barakat M, Thomason R, O’Hara BF, and McKnight SL (2006). NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc. Natl. Acad. Sci 103, 7118–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller P, Sherman D, Pedersen NP, Saper CB, and Lu J (2011). Reassessment of the structural basis of the ascending arousal system. J. Comp. Neurol 519, 933–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, Ma J, Nakane S, Tomita J, Ikkyu A, et al. (2016). Forward-genetics analysis of sleep in randomly mutagenized mice. Nature 539, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, et al. (2014). Natural neural projection dynamics underlying social behavior. Cell 157, 1535–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, and Rosbash M (2016). Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature 536, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa MM, Florian C, Fellin T, Munoz JR, Lee SY, Abel T, Haydon PG, and Frank MG (2009). Astrocytic Modulation of Sleep Homeostasis and Cognitive Consequences of Sleep Loss. Neuron 61, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Kashiwagi M, Yasuda K, Ando R, Kanuka M, Sakai K, and Itohara S (2015). Cells of a common developmental origin regulate REM/non-REM sleep and wakefulness in mice. Science (80-. ). 350, 957–961. [DOI] [PubMed] [Google Scholar]
- He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Rossner MJ, Nishino S, and Fu YH (2009). The transcriptional repressor DEC2 regulates sleep length in mammals. Science (80-. ). 325, 866–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. (2008). A Translational Profiling Approach for the Molecular Characterization of CNS Cell Types. Cell 135, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand M, Peterson K, Christensen V, Dana T, and Thakurata S (2009). Drug Class Review Beta Adrenergic Blockers. Oregon Heal. Sci. Univ 1–616. [PubMed] [Google Scholar]
- Jayaraj ND, Bhattacharyya BJ, Belmadani AA, Ren D, Rathwell CA, Hackelberg S, Hopkins BE, Gupta HR, Miller RJ, and Menichella DM (2018). Reducing CXCR4-mediated nociceptor hyperexcitability reverses painful diabetic neuropathy. J. Clin. Invest [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, and Sehgal A (2008). Identification of SLEEPLESS, a sleep-promoting factor. Science (80-. ). 321, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier KA, and Kristiansen U (2010). GABAergic actions on cholinergic laterodorsal tegmental neurons: Implications for control of behavioral state. Neuroscience. [DOI] [PubMed] [Google Scholar]
- Mang GM, and Franken P (2012). Sleep and EEG Phenotyping in Mice. In Current Protocols in Mouse Biology, p. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li N, and Luo L (2007). A global double-fluorescent cre reporter mouse. Genesis 45, 593–605. [DOI] [PubMed] [Google Scholar]
- Niwa Y, Kanda GN, Yamada RG, Shi S, Sunagawa GA, Ukai-Tadenuma M, Fujishima H, Matsumoto N, Masumoto K hei, Nagano M, et al. (2018). Muscarinic Acetylcholine Receptors Chrm1 and Chrm3 Are Essential for REM Sleep. Cell Rep. [DOI] [PubMed] [Google Scholar]
- Paschalis A, Churchill L, Marina N, Kasymov V, Gourine A, and Ackland G (2009). beta(1)-Adrenoceptor distribution in the rat brain: An immunohistochemical study. Neurosci. Lett. 458, 84–88. [DOI] [PubMed] [Google Scholar]
- Pellegrino R, Kavakli IH, Goel N, Cardinale CJ, Dinges DF, Kuna ST, Maislin G, Van Dongen H.P. a., Tufik S, Hogenesch JB, et al. (2014). A Novel BHLHE41 Variant is Associated with Short Sleep and Resistance to Sleep Deprivation in Humans. Sleep 37, 327–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, and Pack AI (2008). Lethargus is a Caenorhabditis elegans sleep-like state. Nature 451, 569–572. [DOI] [PubMed] [Google Scholar]
- Scheer FAJL, Morris CJ, Garcia JI, Smales C, Kelly EE, Marks J, Malhotra A, and Shea SA (2012). Repeated Melatonin Supplementation Improves Sleep in Hypertensive Patients Treated with Beta-Blockers: A Randomized Controlled Trial. Sleep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, and Mignot E (2011). Genetics of sleep and sleep disorders. Cell 146, 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G, Wu D, Ptáček LJ, and Fu Y-H (2017). Human genetics and sleep behavior. Curr. Opin. Neurobiol 44, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoschitzky K, Sakotnik A, Lercher P, Zweiker R, Maier R, Liebmann P, and Lindner W (1999). Influence of beta-blockers on melatonin release. Eur. J. Clin. Pharmacol 55, 111–115. [DOI] [PubMed] [Google Scholar]
- Szabadi E (2013). Functional neuroanatomy of the central noradrenergic system. J. Psychopharmacol 27, 659–693. [DOI] [PubMed] [Google Scholar]
- Tan CL, Cooke EK, Leib DE, Lin YC, Daly GE, Zimmerman CA, and Knight ZA (2016). Warm-Sensitive Neurons that Control Body Temperature. Cell 167, 47–59.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuki F, Sunagawa GAA, Shi S, Susaki EAA, Yukinaga H, Perrin D, Sumiyama K, Ukai-Tadenuma M, Fujishima H, Ohno R ichiro, et al. (2016). Involvement of Ca2+-Dependent Hyperpolarization in Sleep Duration in Mammals. Neuron 90, 70–85. [DOI] [PubMed] [Google Scholar]
- Ting JT, Daigle TL, Chen Q, and Feng G (2014). Acute brain slice methods for adult and aging animals: Application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassalli A, and Franken P (2017). Hypocretin (orexin) is critical in sustaining theta/gamma-rich waking behaviors that drive sleep need. Proc. Natl. Acad. Sci 114, E5464–E5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, and Morales M (2009). Pedunculopontine and laterodorsal tegmental nuclei contain distinct populations of cholinergic, glutamatergic and GABAergic neurons in the rat. Eur. J. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma J, Miyoshi C, Li Y, Sato M, Ogawa Y, Lou T, Ma C, Gao X, Lee C, et al. (2018). Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nature. Weber F, and Dan Y (2016). Circuit-based interrogation of sleep control. Nature 538, 51–59. [DOI] [PubMed] [Google Scholar]
- Xiu J, Zhang Q, Zhou T, Zhou TT, Chen Y, and Hu H (2014). Visualizing an emotional valence map in the limbic forebrain by TAI-FISH. Nat. Neurosci. 17, 1552–1559. [DOI] [PubMed] [Google Scholar]
- Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, and Ptáček LJ (2007). Modeling of a Human Circadian Mutation Yields Insights into Clock Regulation by PER2. Cell 128, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz MB, Erdem A, Yalta K, Turgut OO, Yilmaz A, and Tandogan I (2008). Impact of beta-blockers on sleep in patients with mild hypertension: A randomized trial between nebivolol and metoprolol. Adv. Ther 25, 871–883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and custom software that support the current findings are available upon request from the Lead Contact.