Abstract
We advance two-photon microscopy for near diffraction-limited imaging down to 850 μm below the pia in awake mice. Our approach combines direct wavefront sensing of descanned fluorescence from Cy5.5-dextran in brain microvessels, which forms a guide star, and adaptive optics to compensate for tissue-induced aberrations of the wavefront. We achieve high signal-to-noise records of glutamate release from thalamocortical axons and calcium transients in spines of layer 5b basal dendrites during active sensing.
Two-photon laser scanning microscopy is indispensable for imaging through the mammalian brain with subcellular resolution [1]. However, the resolution and efficiency decreases with depth as a result of scattering and optical aberrations. To mitigate such effects, the use of high-energy excitation pulses [2] trades increased depth for the risk of nonlinear photodamage, while the use of an under-filled objective aperture [3] sacrifices spatial resolution for increased depth. Alternatively, adaptive optics (AO) [4] can improve multi-photon imaging by synthesizing a distortion to the wavefront of the excitatory beam that compensates for aberrations in the wavefront that are created by the tissue. The desired excitation wavefront can be determined by direct [5–7] or indirect [8–10] wavefront sensing methods. Among these two approaches, direct sensing of the wavefront from a descanned guide star signal leads to wavefront correction with high photon and time efficiency [6, 7]. This approach, with exogenous dye injected into cortex to form the guide star, enabled the determination of spine geometry down to 600 μm below the pia and functional imaging of spines down to 500 μm below the pia [7] (Supplementary Fig. 1), i.e., in granular but not infragranular layers, in an acute preparation. Thus an unmet challenge is to develop a robust imaging paradigm to resolve axons and spines throughout infragranular layers, which includes all output neurons.
We developed an approach that generates a guide star from two-photon excitation of microvessels in which the blood plasma is labeled with Cy5.5-conjugated-dextran (Fig. 1 and Supplementary Figs. 2 and 3). Cy5.5 has a strong two-photon absorbance at 1.25 μm and emission that peaks at 710 nm. Cy5.5-conjugated-dextran is delivered by retro-orbital injection and spreads into all vessels in the body. Unlike dye injected into a region of brain [7], the Cy5.5 is constantly replenished in brain vasculature, is nontoxic, and is comparable with use in chronic studies with closed cranial windows [11] or thinned-skull transcranial imaging [12] (Fig. 1a and Supplementary Video 1). We use descanned direct wavefront sensing (Fig. 1b) to achieve robust aberration measurements across the target region, typically a square of 50 to 100 μm on edge, exciting labeled microvessels within the target region. The wavefront of the descanned, ballistic light from the guide star is analyzed by a Shack-Hartmann wavefront sensor (SHWS) which produces a pattern of spots whose individual positions demarcate the tilt of a subregion of the wavefront (Fig. 1b,c and Supplementary Fig. 4a,b). This pattern serves as input to a reconstruction algorithm that weights the input according to the signal-to-background of each spot (Supplementary Fig. 4c,d) and then specifies the shape of the deformable mirror (DM). The DM alters the wavefront of the excitation beam to approximate a diffraction-limited focus inside the sample (Fig. 1c). We use 1.25 μm light to excite the guide star, as well as to measure blood flow, and use shorter wavelengths to excite subcellular structural and functional indicators. This avoids photobleaching of nonrenewable indicators during calibration. Emission from the indicators is detected with multi-pixel photon counters (MPPCs; Supplementary Fig. 3).
Figure 1. Aberration measurements up to 800 μm below the pia in parietal cortex of awake mouse via wavefront sensing from a microvascular-based guide star.
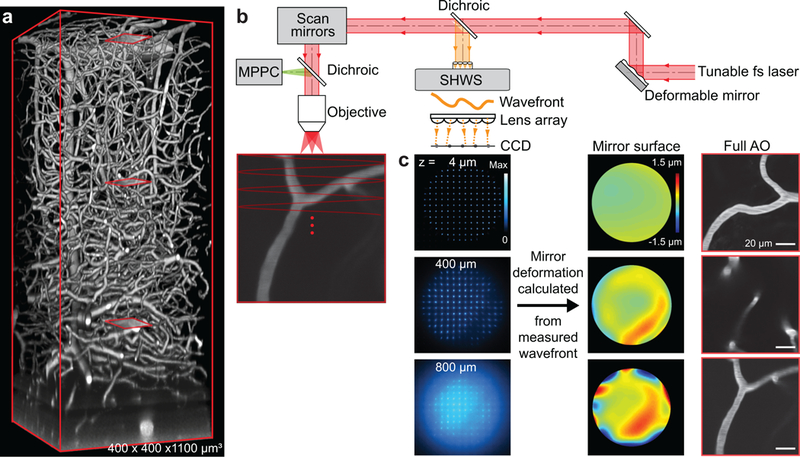
(a) In vivo two-photon imaging of vasculature in a 400×400×1100 μm3 volume of cortex with the blood plasma labeled with Cy5.5 conjugated to 2MD-dextran. The excitation wavelength was λ = 1.25 μm. Red boxes show three example sub-regions used for wavefront sensing in panel c.
(b) Adaptive optical two-photon microscopy based on direct wavefront sensing from the descanned signal of the guide star, formed by two-photon excitation of Cy5.5 in the vasculature lumen, with a Shack-Hartmann wavefront sensor. The wavefront is corrected by a deformable mirror. The signal for brain imaging is detected by the multi-pixel photon counters.
(c) Spot pattern (left) formed by the wavefront sensor from the descanned guide star signal; the position of each spot away from its center determined the tilt of the wavefront. Wavefront maps (center) reconstructed from the wavefront sensor spots patterns at different depths. Microvessels (right) at 14, 400, and 800 μm below the pia (red boxes in panel a) form the guide stars by two-photon excitation.
We evaluated our approach using the adult mouse primary vibrissa sensory (vS1) cortex with a closed cranial window and a two-photon microscope of local design (Supplementary Figs. 2 and 3). Aberration induced by imperfections in the microscope and the cranial window is nulled by a gradient descent method. The attenuation of the ballistic light leads to a decrease of the signal-to-background ratio of the SHWS pattern of spots and limits the depth for effective wavefront measurement to 850 μm below the pia (Supplementary Fig. 4). We refer to correction of the aberration within the microscope and cover glass as “System AO” and correction for all aberrations, including brain tissue, as “Full AO”.
We first consider in vivo morphological imaging. Dense labeling of projection neurons with yellow fluorescent protein (YFP) occurs with the Thy1-YFP SLICK-A mouse. Images obtained with Full AO contain fine dendritic structures in deep cortical layers that cannot be resolved with System AO alone, either at a depth of 690 μm or 770 μm below the pia (Fig. 2a,b and Supplementary Video 2). The improvement is highlighted by line profiles through the fine processes (Fig. 2c,d) and line scans of the shadows of red blood cells flowing in capillaries (Fig. 2e,f). Maps of the spectral power density show that higher spatial frequencies, k, are significantly restored by Full AO correction (Fig. 2g). A quantitative assessment of the depth limits for AO correction is achieved by comparing the power in the spatial frequency distribution between the Full AO and System AO correction at the level of the pia to that at 690, 770, and 850 μm deep (3 mice, Fig. 2h). All told, we achieve near diffraction-limited resolution with Full AO down to 700 μm below the pia–and enhanced imaging of fine structures down to 850 μm below the pia (Supplementary Figs. 5–7).
Figure 2. Adaptive optics correction with wavefront sensing from microvessels enable imaging of dendritic spines and blood velocity at layer 5 in mouse vS1 cortex in vivo.
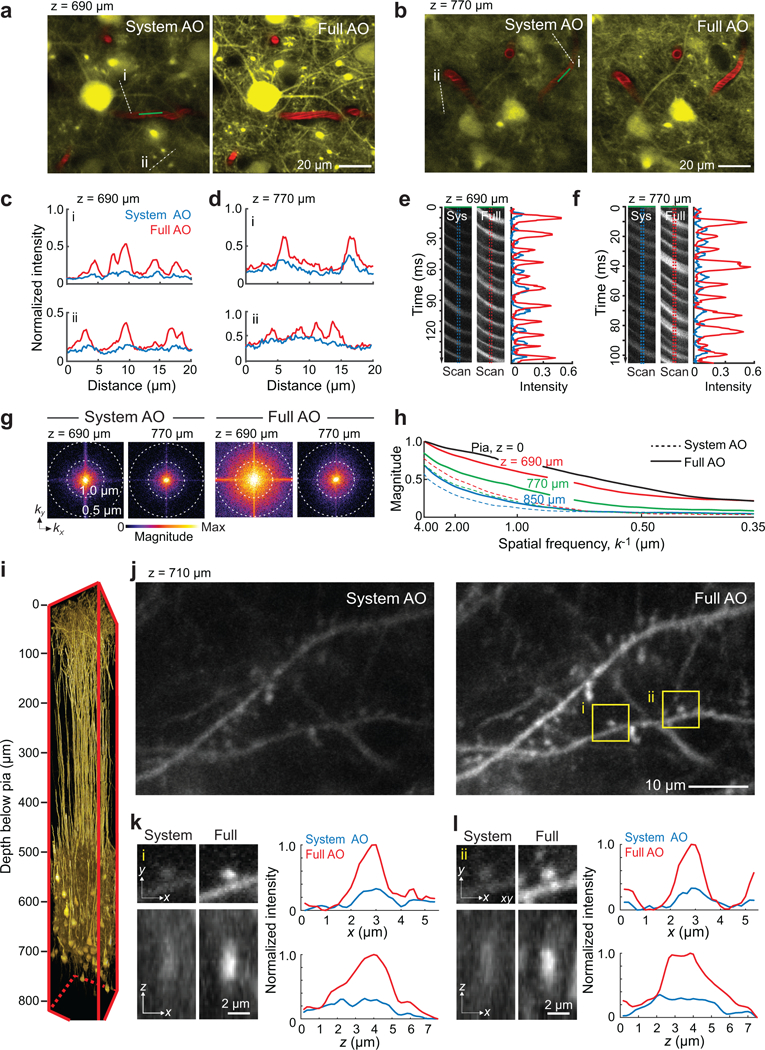
(a,b) Imaging at 690 μm (panel a) and 770 μm (panel b) below the pia in vibrissa primary sensory (vS1) cortex of a Thy1-YFP SLICK A mouse with at λ = 1.04 μm excitation. Image with System (left) and Full AO (right) correction.
(c,d) Signal profiles along the white dashed lines in panels a and b to compare between System AO (blue line) and Full AO (red line).
(e,f) Line scan signals along the green line through the blood vessels in panels a and b compared between System (left) and Full (right) AO, along with signal profiles
(g) Spectral power as a function of spatial frequency, k, for the images in panels a and d.
(h) |k|-space plot of the spatial frequency from images at the depth of the pia at 690 μm, 770 μm, and 850 μm below the pia with System AO (dashed lines) and Full AO (solid lines). Data from panels a and d and Supplementary Fig. 5b–d.
(i) In vivo imaging of mRuby2 labeled layer 5 pyramidal neurons in a 150×150×810 μm3 volume of vS1 cortex at an excitation wavelength of λ = 1.07 μm. The virus AAV-Flex-mRuby2 was injected into layer 5 of Tg(Rbp4-cre)KL100Gsat mice. Full AO corrections were measured and applied every 50 μm of depth.
(j-l) Imaging of dendritic spines at 710 μm below the pia in the same mouse in panel i with System (left) and Full (right) AO correction. Higher magnification views of post-synaptic spines, which correspond to the boxed regions in panel j, are shown in lateral and axial views in panels k and l along with the corresponding x and z profiles of the intensity.
Specific labeling of projection neurons in deep cortical layers is achieved with Rbp4-cre KL100 mice labeled with mRuby2 (Fig. 2i). Basal dendritic spines are clearly resolved at 710 μm below the pia (Fig. 2j). Intensity and resolution in both lateral and axial directions are significantly enhanced and approach the diffraction-limit after Full AO correction (Fig. 2k,l). Further, by the axial and lateral tiling of contiguous sub-regions in the cortex, Full AO correction for volumes that are up to 810 μm in depth and 300 μm on edge (Supplementary Figs. 6a and 8) are achieved.
We now consider the application of direct wavefront sensing to study pre- and postsynaptic responses in layer 5b of vS1 cortex during active sensing. We imaged calcium transients in spines of basal dendrites of layer 5b pyramidals using Rbp4-Cre KL100 mice that expressed jRGECO1a [13] (Fig. 3a,b and Supplementary Video 3) and concurrent measurements of whisking in response to air-puff stimulation of the vibrissa (Fig. 3c). Basal dendritic spines produced either robust calcium transients in response to the onset of stimulation (Fig. 3d) or tonic responses concurrent with changes in the midpoint of whisking (Fig. 3e). These results imply that infragranular layers in vS1 receive both input from vibrissa touch, via the lemniscal pathway, along with efferent copy input of self-motion.
Figure 3. In vivo imaging of calcium and glutamate signaling in dendritic spines and thalamocortical axons in deep layers of vS1 cortex with Full AO correction.
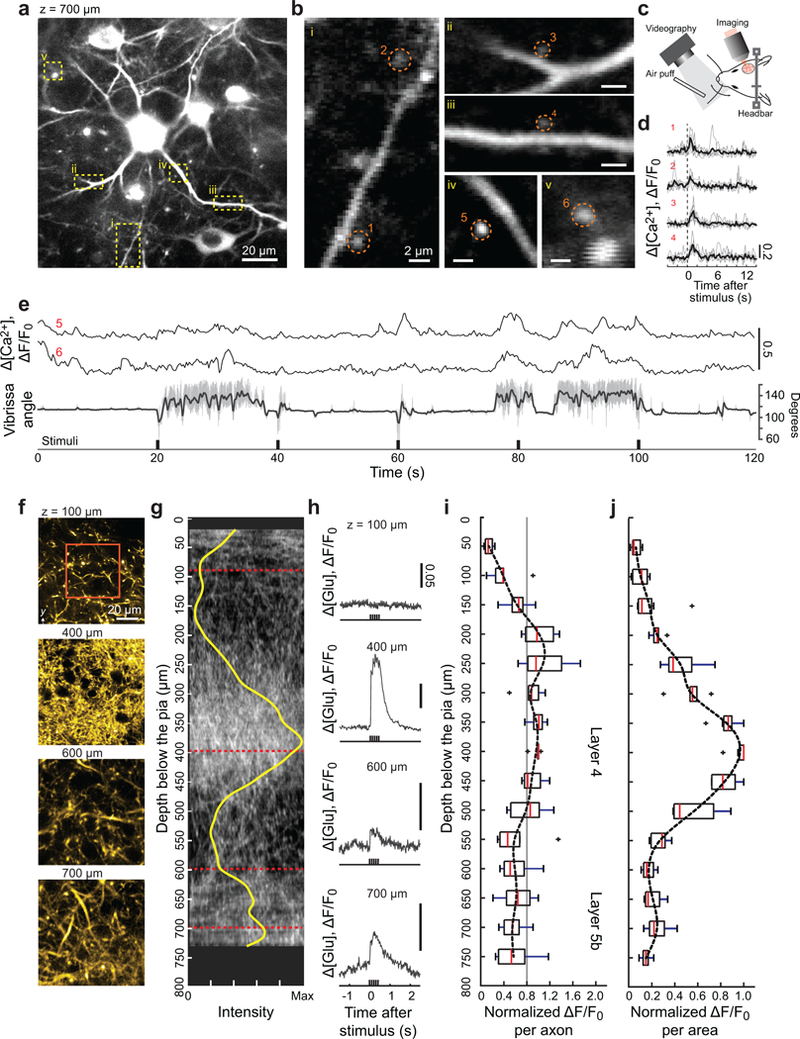
(a) Calcium imaging of L5b basal dendrites and spines at 700 μm below the pia in vS1 cortex at an excitation wavelength of λ = 1.07 μm. The virus AAV-Flex-jRGECO1a was injected into layer 5 of Tg(Rbp4-cre)KL100Gsat mice.
(b) Expanded views of the areas enclosed by the yellow dashed boxes in panel a.
(c) Vibrissa tracking with high-speed camera and air puff stimulation during imaging.
(d,e) Intracellular calcium response of dendritic spines enclosed by red dashed circles in panel b to air-puff stimulation of the C2 vibrissa. The vibrissa is held without whisking for the data of panel d and exhibited free whisking in the data of panel e. For the latter case, the concurrent vibrissa angle (grey) and set-point during rhythmic whisking (dark) are shown. The experiments were repeated in three mice with similar results.
(f-j) Imaging of thalamocortical axons to measure the activation of SF-Venus-iGluSnFR.A184S by glutamate release in response to air-puff stimulation of the vibrissa. Thalamocortical axons were labeled via injection of SF-Venus-iGluSnFR into the barreloids of ventral posterior medial thalamus and imaged at an excitation wavelength of λ = 1.03 μm. The glutamate response was measured every 50 μm from 50 to 730 – 750 μm below the pia in the cortical columns for vibrissae in vS1 cortex. Panel f shows example axons images for the B2 vibrissa cortical column (also see Supplementary Fig. 9b), while panel g shows a 175 μm thick projection in the x-z plane for one animals and the density (yellow) found as an average over all images. Panel h shows the corresponding time-dependent signal of glutamate release, averaged over the region demarked by the red box in panel f. The glutamate responses throughout the measurable depth of the cortical columns for the C1, C2, B2, or B3 vibrissae are shown as a population average of signal per axon in panel i. The gray line serves as a guide to a possible mean response. The same data, recalculated as signal per area, is shown in panel j. Box plots in panels i and j are shown as maximum, minimum, third quartile, first quartile and median (n = 7). Experiments were carried out on seven cortical columns across four mice.
We imaged glutamate release from presynaptic thalamocortical axons, using SF-Venus-iGluSnFR.A184S [14] expressed in thalamocortical neurons, to quantify the nature of input to L5b neurons in vS1 cortex (Fig. 3f,g and Supplementary Fig. 9). The SF-iGluSnFR basal fluorescence was measured every 50 μm of depth from the surface to a depth of 730–750 μm below the pia and showed peaks of fluorescence at the depths of layers 4 and 5b (Fig. 3g), consistent with morphological studies in rat [15]. In response to vibrissa stimulation, we observed reliable signals of glutamate release in granular and infragranular layers (Fig. 3h and Supplementary Fig. 9d) with a near constant release per axon throughout the depth of cortex (7 columns across 4 mice) (Fig. 3i). The greater release per area in layer 4 (Fig. 3j) is consistent with the higher density of axonal boutons in layer 4. These functional data support the consideration of layer 5b as a second thalamocortical projection hub in vS1 cortex [16].
We now consider the use of a thinned skull preparation for chronic, transcranial imaging. Despite the large aberration, we reliably imaged the vasculature and red blood cell flux down to a depth of 600 μm below the pia with improved spatial resolution and increased brightness with Full AO versus System AO (3 mice, Supplementary Fig. 10). Full AO restores the spatial frequency of neuronal images over wide field (Supplementary Fig. 11a–h) and to depth (3 mice, Supplementary Fig. 11i) which allowed us to detect spontaneous calcium spikes from bright processes down to 520 μm below the pia in Thy1-jRGECO1a mice (Supplementary Fig. 12).
A final issue concerns the potential phototoxicity on physiological function during long-term, deep imaging studies with Full AO [17–19]. We first consider the use of typical power at the focus, i.e., ~ 5 mW. For continuous imaging of glutamate in axons or calcium increases in dendrites and spines, we observe no change in functional response over 30 minutes (Supplementary Figs. 13 and 14). Nor did we observe degradation of the response on subsequent days (Supplementary Fig. 13). The dependence of phototoxicity with power was evaluated for the case of red blood cell flow in vessels with the plasma labeled with Cy5.5. We observed a stable flow rate at powers at or below 10 mW and degradation in the form of stalls [20] only for powers near or above 20 mW (Supplementary Fig. 15). Neither tissue-damage nor photobleaching was observed. We conclude that, under Full AO, we achieve long-term, chronic recording.
Direct wavefront sensing enables functional imaging of subcellular components down to 850 μm below the pia in the adult mouse brain. Significantly deeper imaging will require the use of longer excitation wavelengths and thus three- rather than two-photon excitation [21]. Here, greater optical penetration is traded against the lower cross-section for three-photon absorption and the need for higher pulse energies, albeit with a risk of photodamage [19].
Online Methods
Microscope design
A femtosecond laser beam tunable from 680 nm to 1300 nm wavelength (Coherent, Chameleon Discovery) passed through a Pockels Cell (Conoptics, 350–105-02 cell and 302 dirver) for power modulation (Supplementary Fig. 2a). Group delay dispersion was precompensated by the laser’s built-in dispersion compensation unit at wavelength of 1030 nm, 1040 nm, 1070 nm and 1250 nm. The beam was expanded 10-fold by lens pair L1 and L2 (Newport, f = 100 mm, KPX094AR.18 and f = 1000 mm, KPX124AR.18) to slightly overfill the aperture of the deformable mirror (Alpao, DM97–15). Excitation beam reflected by the DM was compressed 3-fold by a 4f telescope formed by L3 and L4 (Newport, f = 600 mm, KPX119AR.18 and f = 200 mm, KPX106AR.18), the DM and Galvo y were conjugated. Translation stage (TS) TS1, TS2 and TS3 carry TS2 and M1; DM and M2; M3 and M4 respectively. M3 and M4 can be adjusted to ensure the alignment of the 4f telescope between DM and Galvo y. Galvos x and y (Cambridge Technology, 5 mm mirror 6215H) were conjugated by a 4f relay formed by L5 and L6, both of which consist of two achromatic doublets (f = 150 mm, Thorlabs, AC508–150-B). The back focal plane of the objective (Olympus, XLPLN25XSVMP2, 25X, 1.0 NA and 4 mm working distance) and Galvo x were conjugated by a 4f telescope formed by scan lens L7 and tube lens L8. L7 consists of a plano-concave lens (f = −200 mm, Newport, KPC070AR.16), a bi-convex lens (f = 200 mm, Thorlabs, LB1199-B) and two achromatic doublets (f = 150 mm, Thorlabs, AC508–150-B). Tube lens L8 was formed by two achromatic doublets (f = 500 mm, Thorlabs, AC508–500-B). Beam reflected by Galvo x was expanded 3.25-fold before incident into the objective.
The fluorescent emission signal for imaging was reflected by primary dichroic D1 (Semrock, FF665-Di02–35×37, FF775-Di01–25×36 will be switched for Cy5.5 imaging) to three different channels, which were spectrally isolated by dichroic mirrors D2 (Semrock, FF552-Di02–25×36), D3 (Semrock, FF662-Di01–25×36) and emission filters F1 (Semrock, FF01–530/55–25) for green/yellow channel, F2 (Semrock, FF01–593/46–25) for red channel and F3 (Semrock, FF01–708/75–25) for far red channel. A fluorescence collection system built with off-the-shelf lenses (Thorlabs, LA1765-A, LA1805-A, LA1027-A and A240-A, Supplementary Fig. 3) coupled with the multi-pixel photon counters (Hamamatsu, C13366–3050GA) allowed the imaging signal to be detected with high efficiency.
In wavefront sensing mode, the emission signal from guide star transmitted through D1 and was descanned by Galvos x and y. The guide star emission signal was separated from excitation beam by dichroic mirror D4 (Semrock, FF775-Di01–25×36) and expanded 1.5-fold by a 4f relay formed with L9 and L10 (Thorlabs, f = 100 mm, AC254–100-A and f = 150 mm, AC254–150-A) before arriving the microlens array (Edmund, #64–483) of the Shack-Hartmann wavefront sensor (SHWS). The SHWS camera (Andor, iXon Ultra 888) was placed at the focal plane of the microlens array to acquire a guide star spots image. Note that the DM, Galvos, back focal plane of the objective and the SHWS microlens array were all located in the same conjugate plane.
The imaging system was controlled by Scanlmage (Vidrio Technologies) running on Matlab (Mathworks). Signals from MPPCs were amplified twice by a voltage preamplifier (Stanford Research Systems, SR445A) and anti-aliased filtered before digitization (National Instruments, PCI-6110). A custom Matlab GUI integrated with Scanlmage was built to control the DM and SHWS camera. Custom Matlab code was made for System AO, calibration, system aberration calibration and sample aberration measuring.
Detectors
The MPPCs used in this study are solid-state single-photon-sensitive detectors implemented by 60×60 Geiger-mode avalanche photodiode (APD) pixels with a pitch of 50 μm. The effective photosensitive area is 3 mm by 3 mm with a spectral response up to 900 nm. The MPPCs provides extremely low multiplicative noise compared to photomutiplier tubes [22], high sensitivity, and linear response for the typical number of photons emitted per laser pulse [23]. In this study, the MPPCs operates in the analog mode after passing the anti-aliasing filter.
System AO calibration
The DM was pre-calibrated in the factory so that commands for a flat mirror and ones to drive each of the first 60 Noll’s Zernike modes [24] at 1-μm root-mean-square amplitude are provided with the instrument. These transform each of the 60 Zernike modes into command voltages for each of 97 actuators which is denoted Z2C, where
The SHWS and the DM are calibrated with respect to each other after the system is well aligned. First, the objective is replaced by a flat mirror in the back focal plane. The calibration beam is incident on the DM, is then reflected by the pupil mirror, and finally projects into SHWS. The DM flat command is applied to obtain a wavefront spot pattern for calibration reference. Then each of the 60 individual Zernike modes is displayed by the DM and the deflection of the result spots on the SHWS, which correspond to changes on the wavefront of DM, are recorded. The vector of spots center location of the flat reference pattern SCref, where
and the 60 Zernike modes patterns, SCz, where
along with are determined by a Gaussian fit centroid algorithm [25] to achieve subpixel precision; there are 60×N elements in the array, where N is the spots number in each waveform pattern, and each elements has two entries, i.e., the x and y shift from the center. The spot shift array of the flat reference is subtracted from each of the 60 spot shift arrays for the different Zernike modes. This yields a 60×N calibration matrix SB that stores the spots shifts on the SH sensor that correspond to phase shifts for each Zernike mode at 1-μm root-mean-square amplitude on the DM, with elements
System aberration calibration
System aberration refers to imperfections of the microscope as well as the cranial window coverslip induced aberration. These are calibrated at the wavelength of 1070 nm and 1250 nm by imaging the central 100 μm by 100 μm field of a 2 to 5 % aqueous fluorescent solution of Sulforhodamine B or Sulfo-Cy5.5 with no. 1 coverslip on top. We use a gradient descent algorithm to optimize the amplitude of the different Zernike modes that are displayed by the DM to maximize the average intensity (Supplementary Figs. 2b,c). In each optimization cycle, the increments of Zernike mode coefficients were computed based on the gradient of mean intensity changes so that current Zernike modes kept on updating. Usually after 20 cycles of optimization the coefficients of the Zernike mode became stable. Tip, tilt and defocus were excluded in this calibration. This leads to a point spread function with the FWHM of 0.5 μm and 2 μm in lateral and axial direction respectively at 1070-nm wavelength, measured using 200-nm fluorescent beads (Supplementary Fig. 2d). We found that this procedure also leads to a near minimum point spread function in off-center regions, e.g., across the entire 700 μm by 700 μm field, albeit with some field curvature of the focal surface. The result of this procedure is a single, stable DM command CSy for system aberration correction, where
Sample aberration measurement
Sample aberrations are caused by the brain tissue. A reference spots pattern on SH sensor is recorded using the aqueous solution of Sulfo-Cy 5.5. Then we measure the wavefront from any location along blood vessels labeling with Cy5.5-dextran in the sample that fluoresces, using the descanned voxels as a guide star. The spots location of the sample aberration, SM, where as well as the reference, are determined using the above centroid algorithm. The relative location between the sample spots and reference spots are calculated which represents the sample aberration, Ss, where The sample spot shift array is decomposed into the calibration basis SB obtained previously to generate the 60 modes Zernike coefficient, when a weighting matrix W is calculated based on the signal-to-background ratio of each spot in the SHWS image (Supplementary Fig. 4e), where (·)+ indicates the generalized matrix inverse. For aberration correction, tip and tilt were excluded. The DM command for correcting sample aberration is then calculated as Cs, where To display the wavefront that we measured from the sample, modal wavefront reconstruction with Zernike polynomials was performed [26].
Full AO correction
The DM command for the system and sample corrections are added for the final correction. As a practical matter, the sample correction is applied to isoplanatic regions that vary between 50 μm and 100 μm of side length. The sample correction, denoted
Animal preparation
All experiments were conducted on adult male (> 8 weeks old) mice. Wild-type mice (C57BL/6J) and transgenic mice (Tg(Rbp4-cre)KL100Gsat [27], Tg(Thy1-jRGECO1a)GP8.31Dkim/J [28] and B6.Cg-Tg(Thy1-YFP-SLICK A)AGfng/J) [29] were used. All experimental procedures on our animals were accordance with Guide for the Care and Use of Laboratory Animals and have been approved by Institutional Animal Care and Use Committee at University of California, San Diego.
Mice were anesthetized with isoflurane using a precision vaporizer, 3 % (v/v) in oxygen for induction and 1 – 2 % (v/v) for maintenance. Body temperature was maintained at 37°C with a heating pad with feedback monitor during anesthesia. The animal was then given analgesic buprenorphine (s.c., 0.1 μg per g body weight) and placed in a stereotaxic frame. The scalp was sterilely removed, the periosteum on the parietal and occipital plates was removed gently.
Chronic cranial window.
A 4-mm diameter craniotomy was made over the right primary sensory vibrissa (vS1) cortex (centroid at 1.5 mm posterior to Bregma and 3.4 mm lateral from midline). Dura was left intact. An imaging window [11, 30] was constructed by a single 4-mm round coverslip (no. 1, 170-μm thick) that was embedded in the craniotomy and sealed around the edge with cyanoacrylate glue (Loctite, no. 401). Dental acrylic (Lang Dental) was further applied around the edge of the coverslip to reinforce the stability. A titanium headbar was glued onto the skull with Meta-bond (Parkell) for head-fixation. The remaining exposed bone and the headbar on top of the region were covered with dental acrylic to increase stability.
Chronic thinned-skull window.
A 3-mm diameter region of skull over the right vS1 cortex was thinned down to 50-μm to 100-μm thick approximately with a 250 μm dill bur [12, 31]. The thinned bone was dried and covered with cyanoacrylate glue and a 4-mm round coverslip. All other procedures are the same as cranial window surgery.
Virus injection.
Virus injection is conducted before the circular craniotomy was made. To label the pyramid neurons of layer 5 in vS1 cortex, virus containing jRGECO1a (AAV2/1.Syn.Flex.NES-jRGECO1a.WPRE.SV40, Penn Vector Core, genomic titer, 4.48×1013, dilution factor 6:1) or mRuby2 [32] (AAV2/1.hSyn.DIO.mRuby2, UCSD, genomic titer, 8.8×1012) was injected (50 nL, 10 nL per minute) at 45° angle into the target coordinates of the Rbp4-cre mice cortex: 1.5 mm posterior to Bregma, 3.4 mm lateral from midline and 0.7 mm depth. To label the thalamocortical projections in vS1 cortex, virus expressing SF-Venus-iGluSnFR ( AAV2/1.hSyn.Flex.SF-Venus-iGluSnFR.A184S, Janelia Research Campus, genomic titer, 1×1013) were mixed with AAV-Cre (AAV2/1.hSyn.Cre.WPRE.hGH, Addgene, genomic titer, 1×1013, dilution factor 1:100) at the ratio 1:1 and injected (50 nL, 10 nL per minute) perpendicular into the barreloids of ventral posteromedial nucleus in wide-type mice: 1.7 mm posterior to Bregma, 1.7 mm lateral from midline and 3.0 mm depth. For all injections glass pipettes (Drummond) were pulled and beveled to a sharp tip (30 μm outer diameter) and a syringe pump (Kd Science, Legato 185) was used to control the infusion.
In vivo imaging
Imaging was performed 7 days after the cranial window surgery or 14 to 21 days after cranial window surgery with virus injection. Animals with thinned-skull window were imaged 3 to 5 days after the surgery. Animals were handled and trained to head fixation for an hour daily to get habituated for three days ahead of imaging. Cyanine5.5 NHS ester (Lumiprobe, #27020) is conjugated with Amino-dextran 2000 kDa (Finabio, AD2000×200) to finally obtain a 5 % (w/v) solution in phosphate-buffered saline of Cyanine5.5-dextran (Cy5.5-dextran). Before imaging, mice were briefly anesthetized with isoflurane to label the lumen of blood vessels via a retro-orbital intravenous injection of 50-μL Cy5.5-dextran PBS solution [33]. The cranial window was carefully aligned to perpendicular to objective axis using a goniometer stage (Thorlabs, GNL20) based mice positioner.
During wavefront sensing the laser was tuned to 1250 nm and the average incident power through the objective ranged from 4 to 135 mW depending on depths in all experiments (Supplementary Table 1). Within a isoplanatic region, frame scanning over the whole field or multi-ROI scanning only on top of the microvessel are alternated to excite the guide star signal. To image vasculature, the primary dichroic is switch to FF775-Di01–25×36. Post-objective power was < 75 mW in all cranial window experiments within 700 μm depth and a maximum power of 135 mW was used for imaging depth over 800 μm or in the thinned skull window experiments (Supplementary Table 2).
For neuronal imaging, the laser was tuned to 1030, 1040, and 1070 nm to excited SF-Venus-iGluSnFR, eYFP and jRGECO1a/mRuby2 respectively. The imaging power post-objective was < 165 mW in most experiments within 700 μm depth. Maximum imaging power was limited to 200 mW for imaging over 760 μm depth or in several mice expressing SF-Venus-iGluSnFR at depth (Supplementary Table 3). Calcium imaging experiments with vibrissa stimulation and tracking were performed in awake animals. To image the glutamate response of thalamocortical axons to vibrissa stimulation, animals were under light anesthesia maintained by 1 % (v/v) isoflurane in oxygen.
Vibrissa tracking and stimulation
One day before behavioral experiments, mice were pre-imaged under the two-photon microscope to check the virus expressing in the vibrissa cortex. Locations in vS1 cortex were verified by intrinsic optical signal imaging [34]. Then the animals were anesthetized and vibrissae were trimmed, leaving only one or two vibrissae, i.e., C1, C2, B2 or B3, for about 10–15 mm long whose cortical column has the optimal jRGECOIa or SF-Venus-iGluSnFR expression. The vibrissa was painted lightly with a white fabric paint. The experiments were carried out in the dark and the vibrissa was illuminated by an infrared LED source (Thorlabs, M940L3). A high-speed camera (Basler, acA1300–200um) was used to capture the whisking at 500 fps.
Air puff deflection was used for vibrissae stimulation [35]. During the stimulation sessions, pulse controlled compressed air, 5 psi at the source, was delivered through a fine tube, which was placed parallel to the side of the mouse snout and 10 mm away from the targeted vibrissa. Pulse width was 20 ms. The frequency of the air puffs is 10 Hz and the stimulation last for 500 ms each trail.
Image analysis
All comparisons of the System AO and Full AO are performed at the same conditions including excitation laser power, imaging field, image size, pixel dwell time, frame average number (Supplementary Tables 1–3), as well as post motion correction process.
Images were processed using MATLAB or ImageJ [36]. Motion corrections were performed on mRuby2 or SF-Venus-iGluSnFR images using a Fourier transform-based algorithm [37]. Non-rigid motion corrections were performed on jRGECO1a calcium images and YFP images using NoRMCore [38], and on 3D stack of mRuby2 images using non-parametric image registration [39]. 3D data were rendered using 3D viewer in ImageJ [40].
For spine calcium imaging data (Fig. 3a,b) circular regions of interest were placed over individual dendrite spines to measure fluorescence. For the analysis of glutamate release per axons (Fig. 3i), the regions of axons were extracted using intensity threshold.
To compute the signal enhancement ratio in neuronal morphological images (Supplementary Figs. 5e and 11i), the line profiles were first extracted from selected fine processes and then the ratio of contrast, where contrast is defined as the difference of the peak and valley amplitudes of the signal, were calculated for both the System AO and Full AO. A similar analysis was performed for the red blood cell flux (Supplementary Figs. 7e and 10h).
Statistics
Box plots were used to show the signal enhancement ratio of fine processes in neuronal morphological images as well as of the red blood cell flux between System AO and Full AO (Supplementary Figs. 5e, 7e, 10h and 11i).
To compare glutamate responses of thalamocortical axons at different cortical layers (Fig. 3i,j), we use box plot to show the amplitudes of glutamate responses across samples. A fitting curve using the mean response amplitude at each depth is also used to shown the distribution trend.
Code availability
The system aberration calculation, DM and SHWS calibration and sample induced wavefront reconstruction and AO correction code are available in the Supplementary Software.
Data availability
The data that support the findings of this study are available from the corresponding author. Source data for Figs. 2 and 3 and Supplementary Figs. 5, 7, 10 and 11 are available online.
Supplementary Material
Acknowledgements
We gratefully acknowledge Na Ji, Chris Xu, and the late Joel Kubby for detailed advice on deep imaging. We further thank Takaki Komiyama for the gift of transgenic mice, Byungkook Lim for the gift of viruses, Martin Thunemann for instruction on synthesis of Cy5.5-dextran, Conrad Foo for assistance with the motion correction algorithm, Adrian Lozada and Celine Mateo for advice on surgery, Martin Deschênes, Michael Häusser, Benjamin Strowbridge and Karel Svoboda for discussions, and Beth Friedman for comments on an early version of the manuscript. This work was supported by the National Science Foundation (MRI grant PHY153264) and the National Institutes of Health (NINDS grant R35 NS097265).
References
- 1.Svoboda K, Denk W, Kleinfeld D, and Tank DW, In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature, 1997. 385: p. 161–165. [DOI] [PubMed] [Google Scholar]
- 2.Mittmann W, Wallace DJ, Czubayko U, Herb JT, Schaefer AT, Looger LL, Denk W, and Kerr JN, Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nature Neuroscience, 2011. 14: p. 1089–1093. [DOI] [PubMed] [Google Scholar]
- 3.Kondo M, Kobayashi K, Ohkura M, Nakai J, and Matsuzaki M, Two-photon calcium imaging of the medial prefrontal cortex and hippocampus without cortical invasion. Elife, 2017. 6: p. e26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji N, Adaptive optical fluorescence microscopy. Nature Methods, 2017. 14: p. 374–380. [DOI] [PubMed] [Google Scholar]
- 5.Tao X, Fernandez B, Azucena O, Fu M, Garcia D, Zuo Y, Chen DC, and Kubby J, Adaptive optics confocal microscopy using direct wavefront sensing. Optics letters, 2011. 36: p. 1062–1064. [DOI] [PubMed] [Google Scholar]
- 6.Wang K, Milkie DE, Saxena A, Engerer P, Misgeld T, Bronner ME, Mumm J, and Betzig E, Rapid adaptive optical recovery of optimal resolution over large volumes. Nature Methods, 2014. 11: p. 625–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Sun W, Richie CT, Harvey BK, Betzig E, and Ji N, Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nature Communications, 2015. 6: p. 7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji N, Milkie DE, and Betzig E, Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nature Methods, 2010. 7: p. 141–147. [DOI] [PubMed] [Google Scholar]
- 9.Débarre D, Botcherby EJ, Watanabe T, Srinivas S, Booth MJ, and Wilson T, Image-based adaptive optics for two-photon microscopy. Optics Letters, 2009. 34: p. 2495–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang J, Germain RN, and Cui M, Superpenetration optical microscopy by iterative multiphoton adaptive compensation technique. Proceedings of the National Academy of Sciences USA, 2012. 109: p. 8434–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee WC, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, and Wilbrecht L, Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nature Protocols, 2009. 4: p. 1128–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drew PJ, Shih AY, Driscoll JD, Knutsen PM, Davalos D, Blinder P, Akassoglou K, Tsai PS, and Kleinfeld D, Chronic optical access through a polished and reinforced thinned skull. Nature Methods, 2010. 7: p. 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D,., Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, and Kim DS, Sensitive red protein calcium indicators for imaging neural activity. eLIFE, 2016. 5: p. e12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marvin JS, Scholl B, Wilson DE, Podgorski K, Kazemipour A, Müller JA, Schoch S, Quiroz FJU, Rebola N, Bao H, Little JP, Tkachuk AN, Cai E, Hantman AW, Wang SSH, DePiero VJ, Borghuis BG, Chapman ER, Dietrich D, DiGregorio DA, Fitzpatrick D, and Looger LL, Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nature Methods, 2018. 15: p. 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oberlaender M, de Kock CPJ, Bruno RM, Ramirez A, Meyer HS, Dercksen VJ, Helmstaedter M, and Sakmann B, Cell type-specific three-dimensional structure of thalamocortical networks in a barrel column in rat vibrissal cortex. Cerebral Cortex, 2011. 22: p. 2375–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro-Alamancos MA and Connors BW, Thalamocortical synapses. Progress in Neurobiology, 1997. 51: p. 581–606. [DOI] [PubMed] [Google Scholar]
- 17.Hopt A and Neher E, Highly nonlinear photodamage in the two-photon fluorescence microscopy. Biophysical Journal, 2001. 80: p. 2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG, and Shroff H, Assessing phototoxicity in live fluorescence imaging. Nature Methods, 2017. 14: p. 657–661. [DOI] [PubMed] [Google Scholar]
- 19.Yildirim M, Sugihara H, So PTC, and Sur M, Functional imaging of visual cortical layers and subplate in awake mice with optimized three-photon microscopy. Nature Communications, 2019. 11: p. 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleinfeld D, Mitra PP, Helmchen F, and Denk W, Fluctuations and stimulus-induced changes in blood flow observed in individual capillaries in layers 2 through 4 of rat neocortex. Proceedings of the National Academy of Sciences USA, 1998. 95: p. 15741–15746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouzounov DG, Wang T, Wang M. Mengran, Feng DD, Horton NG, Cruz-Hernández JC, Cheng Y-T, Reimer J, Tolias AS, Nishimura N, and Xu C, In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nature Methods, 2017. 14: p. 388–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driscoll JD, Shih AY, Iyengar S, Field JJ, White GA, Squier JA, Cauwenberghs G, and Kleinfeld D, Photon counting, censor corrections, and lifetime imaging for improved detection in two-photon microscopy. Journal of Neurophysiology, 2011. 104: p. 1803–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai PS and Kleinfeld D, In vivo two-photon laser scanning microscopy with concurrent plasma-mediated ablation: Principles and hardware realization, in Methods for In Vivo Optical Imaging, 2nd edition, Frostig RD, Editor. 2009, CRC Press: Boca Raton: p. 59–115. [PubMed] [Google Scholar]
- 24.Noll RJ, Zernike polynomials and atmospheric-turbulence. Journal of the Optical Society of America, 1979. 66: p. 207–211. [Google Scholar]
- 25.Thompson RE, Larson DR, and Webb WW, Precise nanometer localization analysis for individual fluorescent probes. Biophysical Journal, 2002. 82: p. 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cubalchini R, Modal wavefront estimation from phase derivative measurements. Journal of the Optical Society of America, 1979. 69: p. 973–977. [Google Scholar]
- 27.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, and Heintz N, A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature, 2003. 425: p. 917–925. [DOI] [PubMed] [Google Scholar]
- 28.Dana H, Novak O, Guardado-Montesino M, Fransen JW, Hu A, Borghuis BG, Guo C, Kim DS, and Svoboda K, Thy1 transgenic mice expressing the red fluorescent calcium indicator jRGECO1a for neuronal population imaging in vivo. PLoS One, 2018. 13: p. e0205444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young P, Qiu L, Wang D, Zhao S, Gross J, and Feng G, Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nature Neuroscience, 2008. 11: p. 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morii S, Ngai AC, and Winn HR, Reactivity of rat pial arterioles and venules to adenosine and carbon dioxide: With detailed description of the closed cranial window technique in rats. Journal of Cerebral Blood Flow & Metabolism, 1986. 6: p. 34–41. [DOI] [PubMed] [Google Scholar]
- 31.Shih AY, Mateo C, Drew PJ, Tsai PS, and Kleinfeld D, A polished and reinforced thinned skull window for long-term imaging and optical manipulation of the mouse cortex. Journal of Visualized Experiments, 2012: p. http://www.jove.com/video/3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam AJ, St-Pierre F, Yiyang Gong Y., Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, D.M. W., Schnitzer MJ, Tsien RY, and Lin MZ, Improving FRET dynamic range with bright green and red fluorescent proteins. Nature Methods, 2012. 9: p. 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih AY, Driscoll JD, Drew PJ, Nishimura N, Schaffer CB, and Kleinfeld D, Two-photon microscopy as a tool to study blood flow and neurovascular coupling in the rodent brain. Journal of Cerebral Blood Flow & Metabolism, 2012. 32: p. 1277–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knutsen PM, Mateo C, and Kleinfeld D, Precision mapping of vibrissa representation within murine primary somatosensory cortex. Philosophical Transactions of the Royal Society B, 2016. 371: p. 20150351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleinfeld D, Sachdev RNS, Merchant LM, Jarvis MR, and Ebner FF, Adaptive filtering of vibrissa input in motor cortex of rat. Neuron, 2002. 34: p. 1021–1034. [DOI] [PubMed] [Google Scholar]
- 36.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, and Cardona A, Fiji: An open-source platform for biological-image analysis. Nature Methods, 2012. 9: p. 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guizar-Sicairos M, Thurman ST, and Fienup JR, Efficient subpixel image registration algorithms. Optics Letters, 2008. 33: p. 156–158. [DOI] [PubMed] [Google Scholar]
- 38.Pnevmatikakis EA and Giovannucci A, NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. Journal of Neuroscience Methods, 2017. 291: p. 83–94. [DOI] [PubMed] [Google Scholar]
- 39.Vercauteren T, Pennec X, Perchant A, and Ayache N, Diffeomorphic demons: Efficient non-parametric image registration. Neuroimage, 2009. 45: p. S61–72. [DOI] [PubMed] [Google Scholar]
- 40.Schmid B, Schindelin J, Cardona A, Longair M, and Heisenberg M, A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics, 2010. 11: p. 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author. Source data for Figs. 2 and 3 and Supplementary Figs. 5, 7, 10 and 11 are available online.


