Abstract
Meiosis is a conserved tenet of sexual reproduction in eukaryotes, yet this program is seemingly absent from many extant species. In the human fungal pathogen Candida albicans, mating of diploid cells generates tetraploid products that return to the diploid state via a non-meiotic process of depolyploidization known as concerted chromosome loss (CCL). Here, we report that recombination rates are more than three orders of magnitude higher during CCL than during normal mitotic growth. Furthermore, two conserved ‘meiosis-specific’ factors play central roles in CCL as SPO11 mediates DNA double-strand break formation while both SPO11 and REC8 regulate chromosome stability and promote inter-homolog recombination. Unexpectedly, SPO11 also promotes DNA repair and recombination during normal mitotic divisions. These results indicate that C. albicans CCL represents a ‘parameiosis’ that blurs the conventional boundaries between mitosis and meiosis. They also reveal parallels with depolyploidization in mammalian cells and provide potential insights into the evolution of meiosis.
Subject terms: Fungal genetics, Pathogens, DNA recombination
Mating of Candida albicans produces tetraploid products that return to the diploid state via a non-meiotic process known as concerted chromosome loss (CCL). Here, Anderson et al. show high recombination rates during CCL and identify factors that are essential for chromosome stability and recombination during CCL.
Introduction
Sexual reproduction arose early in the eukaryotic lineage and has been maintained by most extant species1,2. Sex involves gamete fusion and a doubling of cell ploidy, whereas ploidy reduction is typically achieved via meiosis in which two successive cell divisions halve the DNA content in the cell. Extensive recombination between chromosome homologs is characteristic of meiosis and promotes adaptation through the efficient joining of beneficial alleles and the removal of deleterious ones3–5. Meiosis presumably evolved from the apparatus responsible for mitotic cell divisions although its exact origins, as well as the benefits of sex, continue to be debated6,7. Additional insight is therefore required into meiosis and the emergence of sex to understand how these fundamental processes arose and have been retained in eukaryotes.
Key aspects of meiosis are conserved at both the cellular and molecular level8,9. Typically, segregation of homologous chromosomes occurs during Meiosis I and sister chromatids remain tethered until Meiosis II, which mechanistically resembles a mitotic cell division10. Hallmarks of Meiosis I include synapsis of homologous chromosomes followed by extensive recombination, which are both essential for faithful chromosome segregation11,12. Central to recombination is the directed formation of DNA double-strand breaks (DSBs) by the topoisomerase-like protein Spo11, which, with the exception of dictyostelids13, represents a conserved meiosis-specific protein8,14–17. Spo11-induced DNA breaks lead to the formation of chromosome crossovers via Holliday junction intermediates or the formation of noncrossovers via synthesis-dependent strand annealing9,18,19.
Cohesins also play a key role in meiosis including the meiosis-specific kleisin Rec8 that assembles on chromosomes prior to premeiotic DNA replication20–22. In the model yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, a protease cleaves cohesin attachments along the chromosome arms during the metaphase to anaphase transition in Meiosis I, and subsequently releases centromeric cohesion during Meiosis II. This mechanism ensures correct segregation of chromosome homologs in the first meiotic division, whereas sister chromatids remain attached until the second meiotic division23–26.
Both SPO11 and REC8 are conserved from yeast to humans and loss of either factor causes defects in synapsis and meiotic recombination9,27. However, both genes are also present in species that lack a defined meiosis, which could indicate the presence of uncharacterized sexual cycles or functions unrelated to sexual reproduction1,28–30. Moreover, several studies now indicate that ‘meiosis-specific’ genes can be expressed in tumor cells and are associated with ploidy reduction in polyploid (> 2 N) cells, a process known as depolyploidization31–33. There is, therefore, a pressing need to understand how ‘meiosis-specific’ factors promote ploidy change and genetic exchange during both mitotic and meiotic processes.
Fungal species have contributed extensively to our understanding of ploidy cycling, sexual reproduction, and recombination. Complete sexual cycles exist in many fungi yet atypical ploidy cycles have also been uncovered such as that in the pathogenic yeast Candida albicans. Here, efficient mating of diploid a and α cells generates tetraploid a/α cells, yet neither meiosis nor sexual sporulation has been reported34–37. Instead, tetraploid instability can be induced by growth on S. cerevisiae ‘PRE-SPO’ medium, upon which C. albicans cells undergo “concerted chromosome loss” (CCL)38,39. Mitotic non-disjunction events are thought to drive CCL and yet analysis of a limited number of products suggested that Spo11 may enhance recombination during CCL39. However, these studies did not address the degree of homologous recombination accompanying CCL, or the precise role of Spo11 or other meiosis-specific factors during this process.
The studies outlined here reveal the extent of genetic diversity produced via CCL in C. albicans and highlight roles for both Spo11 and Rec8 in this process. We demonstrate that Spo11 promotes high levels of DNA double-strand breaks and recombination during CCL, producing recombination frequencies orders of magnitude higher than during normal mitotic growth. Rec8 also enhances recombination frequencies during CCL, and yet Spo11 and Rec8 have opposing effects on chromosome loss as they increase and decrease chromosome stability, respectively. Interestingly, Spo11 also influences chromosome stability and recombination during normal mitotic passaging. We therefore show that two ‘meiosis-specific’ factors contribute to genome dynamics and genetic exchange during CCL, with additional roles for Spo11 during mitotic growth. These results suggest that C. albicans can undergo a ‘parameiosis’ with potential parallels to depolyploidization processes observed in higher eukaryotes.
Results
Recombination and DNA DSB formation during CCL
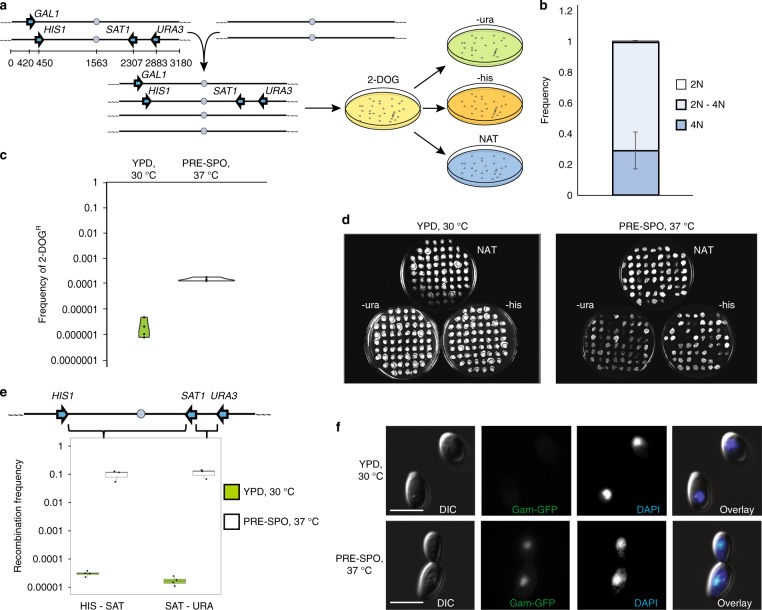
To precisely determine the frequency of inter-homolog recombination during CCL, a tetraploid C. albicans strain carrying four heterozygous markers (GAL1, URA3, HIS1, and SAT1) at different positions on chromosome (Chr) 1 was constructed (Fig. 1a and Supplementary Fig. 1). The genetically marked strain was grown under CCL-inducing conditions (PRE-SPO medium at 37 oC for 7 days) and cells transferred to 2-deoxygalactose (2-DOG) medium to select for gal1− cells. 2-DOGR colonies were subsequently evaluated for the presence or absence of HIS1, SAT1, and URA3 markers by growth on selective media (Fig. 1a). Using this approach, most 2-DOGR colonies (70.3%) had undergone a reduction in ploidy and were intermediate between diploid and tetraploid when analyzed by flow cytometry (Fig. 1b and Supplementary Fig. 2), consistent with the frequent formation of aneuploid CCL progeny38,39.
Fig. 1.
High rates of inter-homolog recombination during concerted chromosome loss (CCL). a Schematic showing the construction and screening of a genetically marked tetraploid strain for monitoring chromosome loss and recombination. b Frequency of different ploidy states in 2-DOGR (gal1−) progeny derived from tetraploid cells by CCL. Error bars signify standard deviation. n = 3 biologically independent experiments. c Frequency of 2-DOGR colonies following growth of the genetically marked tetraploid strain on solid agar under standard conditions (green; YPD, 30 °C) or under conditions that promote CCL (white; PRE-SPO medium, 37 °C) for 7 days. n = 4 biologically independent experiments. d Example images of selection plates indicating growth of progeny on selective media for different markers. e Recombination frequencies in HIS1-SAT1 and SAT1-URA3 intervals following growth on standard medium (green) or CCL-inducing medium (white) for 7 days. SAT1 encodes resistance to nourseothricin (NATR). Boxplots are presented as the 75th to 25th percentile with the thick line denoting the median. Whiskers indicate the largest and smallest values within 1.5 × of the interquartile range. f C. albicans tetraploid cells expressing the Gam-GFP construct following 24 h growth on PRE-SPO or YPD medium (CCL-inducing or standard growth conditions, respectively). Scale bar, 5 μm. Error bars indicate standard deviation
Growth under CCL-inducing conditions increased GAL1 loss in tetraploid cells by ~ 400-fold relative to culture on standard yeast extract peptone dextrose (YPD) medium at 30 °C (Fig. 1c). Furthermore, recombination frequencies within the HIS1-SAT1 and SAT1-URA3 genetic intervals in 2-DOGR progeny were ~ 2700-fold and ~ 8200-fold higher, respectively, when grown under CCL-inducing conditions compared with standard YPD growth (Fig. 1d, e). This corresponds to 1 in 10 cells, on average, having undergone recombination within the tested Chr1 intervals during CCL, indicative of high recombination frequencies.
We next examined whether DSB formation, a hallmark of a conventional meiosis, is observed during CCL. To test this, we introduced a GFP-tagged version of Gam protein from bacteriophage Mu into tetraploid C. albicans cells. Gam-GFP has been shown to bind to DSBs and can be used as a visual readout of DSB formation in live cells40. In control experiments, doxycycline-induced Gam-GFP expression generated a nuclear GFP signal in C. albicans cells treated with ionizing irradiation (Supplementary Fig. 3 and 4). A subset of tetraploid C. albicans cells grown under CCL-inducing conditions also displayed an intense Gam-GFP signal within the nucleus (Fig. 1f, Supplementary Fig. 5), whereas tetraploid cells cultured under standard growth conditions did not. Growth under CCL-inducing conditions similarly promoted the formation of DSBs based on the prevalence of cells positive for terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) staining (Supplementary Fig. 6). Together, these results establish that there is an increased prevalence of DSBs during CCL relative to normal mitotic growth.
Meiosis-specific factors regulate parasexual processes
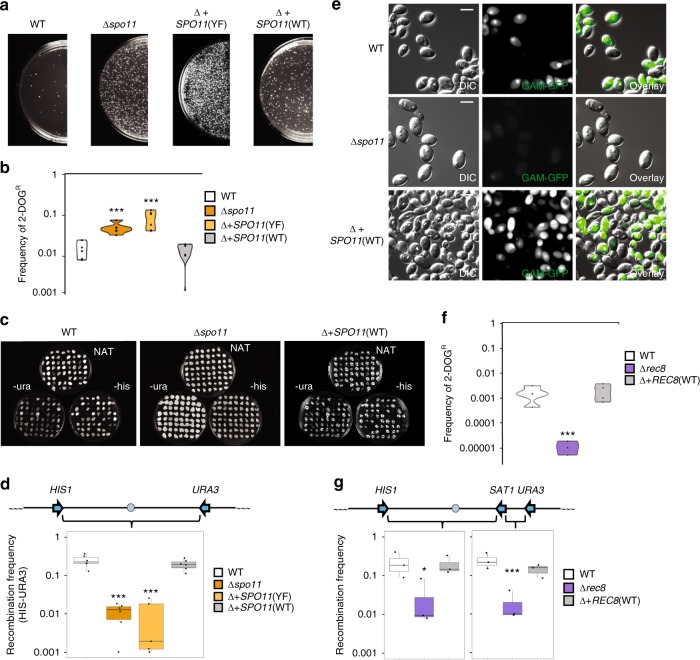
To evaluate the functions of ‘meiosis-specific’ genes in CCL, we focused on C. albicans SPO11 and REC8 homologs, given that these genes play critical and conserved roles in meiosis from yeast to mammals. Both genes were individually deleted in tetraploid C. albicans strains carrying genetic markers along Chr1 (Supplementary Fig. 1). Interestingly, deletion of SPO11 led to ~ 10-fold higher frequencies of 2-DOGR colonies reflecting increased GAL1 marker loss relative to the wildtype control (two-sample t (6.12) = − 4.31, p = 1.57E-3, Fig. 2a, b). Complementation of the Δspo11 mutant with a single copy of SPO11 was sufficient to restore normal levels of marker stability during CCL (Δ + SPO11(WT) vs. WT: two-sample t (9.49) = 0.73, p = 0.48). In contrast, integration of a mutated SPO11 allele lacking the putative catalytic residue (Y65F substitution, Supplementary Fig. 7) failed to rescue the mutant phenotype (Δ + SPO11(YF) vs. WT: two-sample t (5.13) = −3.75, p = 5.25E-3, Fig. 2a, b). Flow cytometry measurements of DNA content in cells 48 h after inducing CCL revealed that Δspo11 cells generally had ploidy levels that were lower than either the wildtype or SPO11-complemented strains (Supplementary Fig. 2). A slight, but not significant, reduction in DNA content was observed in CCL progeny from SPO11-complemented cells relative to those from wildtype cells. Overall, these results indicate that Spo11 promotes chromosome stability (or reduces chromosome loss) under CCL conditions.
Fig. 2.
Homologous recombination during CCL requires the ‘meiosis’ factors Spo11 and Rec8. a Images of plates illustrating 2-DOGR colonies produced by CCL in C. albicans wildtype and Δspo11 tetraploid strains, as well as in Δspo11 strains complemented with wildtype SPO11, SPO11(WT), or an active site mutant, SPO11(YF). b Quantification of the frequency of 2-DOGR colonies in different strain backgrounds following CCL. n = 6 biologically independent experiments. c Images of plates examining CCL progeny grown on different selective media. d Frequency of recombination in CCL progeny from different strain backgrounds. n = 6 biologically independent experiments. e Analysis of Gam-GFP signal in tetraploid strains with different SPO11 genotypes undergoing CCL. Images shown after 24 h on PRE-SPO medium with 50 μg/mL doxycycline at 37 °C. Scale bars = 5 μm. f Quantification of the frequency of 2-DOGR colonies in different REC8 strain backgrounds following CCL. n = 4 biologically independent experiments. g Frequency of recombination in CCL progeny derived from different REC8 strain backgrounds in HIS1-SAT1 and SAT1-URA3 intervals. n = 3 biologically independent experiments. * denotes p < 0.05. *** denotes p < 0.001 as calculated by two-sample t b, f or Wilcoxon d, g tests. Boxplots are presented as the 25th to 75th percentile with the thick line denoting the median. Whiskers indicate the largest and smallest values within 1.5 × of the interquartile range. White, orange, light orange, and gray denote WT, Δspo11, Δ+SPO11(YF), and Δ+SPO11(WT), respectively. White, purple, and gray denote WT, Δrec8, and Δ+REC8(WT), respectively
Deletion of SPO11 also led to marked changes in recombination frequencies during CCL. Thus, tetraploid strains lacking SPO11 showed a ~ 24-fold reduction in recombination rates in the HIS1-URA3 interval on Chr1 compared with the wildtype strain (Wilcoxon test (W(40)), p = 5.04E-5, Fig. 2c, d, and Supplementary Fig. 8). Reintegration of a wildtype SPO11 allele into the Δspo11 tetraploid strain restored recombination frequencies (Δ + SPO11(WT) vs. WT: Wilcoxon test (W(26)), p = 0.29, Fig. 2c, d). In contrast, complementation with the SPO11-YF allele failed to complement the recombination defect during CCL (Δ + SPO11(YF) vs WT: Wilcoxon test (W(34)), p = 1.75E-4, Fig. 2c, d). A Gam-GFP signal was present only at background levels in most Δspo11 cells during CCL indicative of reduced DSB formation, and this signal was restored in the WT SPO11-complemented strain (Δ + SPO11(WT) vs. Δspo11: two-sample t (1.12) = 9.59, p = 0.015; Fig. 2e and Supplementary Fig. 5). Examination of cells complemented with the SPO11-YF allele revealed fewer cells were Gam-GFP positive than those with the WT allele, but significantly more cells were positive than in Δspo11 cells (Supplementary Fig. 5). This suggests that the mutant Spo11 protein may contribute to DSB levels via a non-enzymatic mechanism. Together, these experiments establish that Spo11 promotes DSB formation and supports both chromosome stability and DNA recombination in polyploid cells undergoing CCL.
Next, we evaluated the role of a second meiosis-specific factor, REC8, in tetraploid cells. Interestingly, loss of REC8 had the opposite effect on chromosome stability during CCL to that of SPO11, as Δrec8 mutants showed a 48-fold reduction in the formation of 2-DOGR colonies relative to the wildtype control (two-sample t (6) = 3.73, p = 9.83E-3, Fig. 2f, Supplementary Fig. 9a and 10). Similar to loss of SPO11, however, deletion of REC8 reduced the recombination frequency (~ 12-fold) in the HIS1-SAT1 and SAT1-URA3 intervals compared with the wildtype strain (Fig. 2g, Supplementary Fig. 9b). Recombination frequencies were restored to wildtype levels by reintroduction of one copy of REC8 into the Δrec8 background (Fig. 2g). Both SPO11 and REC8 therefore promote high levels of recombination in polyploid cells undergoing CCL. However, SPO11 enhances chromosome stability during CCL whereas REC8 has a destabilizing effect on the genome under these conditions.
Genome-wide patterns of recombination following CCL
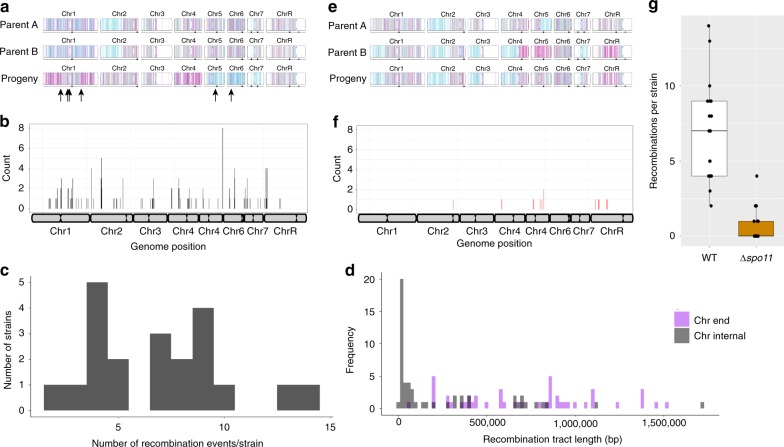
To investigate global patterns of homologous recombination, we genotyped CCL progeny using double digest restriction-site associated DNA sequencing (ddRAD-Seq), which selectively sequences a large number of marker positions across the genome (see Methods). Wildtype tetraploid cells were induced to undergo CCL and 21 2-DOGR progeny analyzed by ddRAD-Seq. Approximately half (10/21) of these CCL isolates were aneuploid, with 13.6% (20/147) of the chromosomes present as supernumerary copies. Trisomies of several smaller chromosomes (Chrs 4, 5, and 6) were the most common in CCL progeny (Supplementary Fig. 11a). Interrogation of ddRAD-Seq data revealed multiple recombination events in the 21 progeny (Fig. 3a, b). The number of detectable recombination events varied between 1 and 14 crossovers per strain and recombinant segments ranged from ~ 10 kb to 1.5 Mb (Fig. 3c). Recombination tracts extending to the ends of chromosome arms could represent break-induced replication (BIR) or crossover events, and these tracts were significantly longer than internal recombination tracts (Wilcoxon test (W(2114), p = 1.66E-7, Fig. 3d)). Recombination frequency correlated with chromosome size (Pearson, r = 0.78, p = 0.027, Supplementary Fig. 12) although breakpoints were not evenly distributed across all chromosomes (Fig. 3b). Mapping the 149 recombination events within the 21 progeny highlighted regions containing multiple recombination events near the centromere of Chr3, the left arm of Chr2, and telomere-proximal regions on ChrR and Chr4. These regions spanned 20–40 kb and represent potential hotspots for recombination, but were not obviously enriched for repetitive genomic elements that are associated with increased recombination frequencies41.
Fig. 3.
Genomic analysis of C. albicans CCL progeny. Parental and progeny genotypes are shown for three representative strains in WT a and Δspo11 e backgrounds. The parental genotypes are indicated on the top two lines with a representative progeny underneath. Arrows indicate recombination breakpoints. Homozygous alleles for homolog A or B are represented by cyan and magenta lines, respectively, with heterozygous positions denoted as gray. The recombination breakpoints were mapped to the genome across all 21 genotyped WT b and Δspo11 f progeny. c The number of recombination events within each WT progeny following CCL. d The length of each recombination tract in WT progeny was determined for chromosome internal tracts (gray) and tracts extending to the chromosome end (purple). n = 46 biologically independent data points. g The number of recombination breakpoints was compared for WT (white) and Δspo11 (orange) progeny. Boxplots are presented as the 25th to 75th percentile with the thick line denoting the median. Whiskers indicate the largest and smallest values within 1.5 × of the interquartile range. n = 20 biologically independent samples
We also used ddRAD-Seq to examine global recombination patterns in 21 CCL progeny from the Δspo11 tetraploid and found that recombination frequencies were significantly lower in Δspo11 progeny than in wildtype progeny (Mann–Whitney U test (W(456)), p = 2.23E-8, Fig. 3g). Only 13 recombination events were detected in the 21 Δspo11 progeny, with most progeny (15 out of 21) having no detectable recombination events (Fig. 3e, f). Recombination tracts that did not extend to the chromosome ends were, on average, sixfold shorter in Δspo11 cells than in wildtype cells (47 kb vs 260 kb, respectively), whereas those extending to the telomere were similar in size between backgrounds (535 kb vs 734 kb, respectively). This suggests that Spo11 selectively impacts the tract length of internal recombination events, although there was insufficient power to compare with the wildtype owing to the limited number of events in the mutant strain. We also note that only 4 of the 21 Δspo11 progeny were aneuploid owing to five imbalanced chromosomes (3.4% of 147 chromosomes; Supplementary Fig. 11b), lending support to the model that chromosome instability (and a return to the 2 N state) may be enhanced by SPO11 loss.
A functional role for SPO11 during mitotic growth
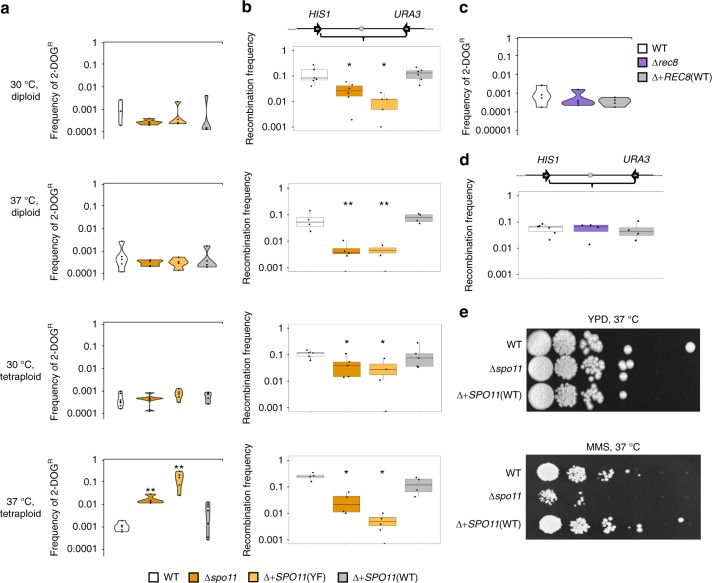
In addition to evaluating SPO11 and REC8 function during CCL, we examined their potential role(s) in genome stability and recombination during normal mitotic growth in diploid and tetraploid cells. Although SPO11 and REC8 are both expressed in mitotic cells39,42, a functional role for these genes during mitotic divisions has not been reported. Genetically marked diploid or tetraploid strains were passaged in YPD medium at 30 oC or 37 oC (serially diluted every 24 h for 7 days) and analyzed for the presence/absence of markers on Chr1 (Fig. 1a). In general, similar levels of GAL1 marker stability were observed in diploid and tetraploid cells grown at both 30 oC and 37 oC (Fig. 4a, Supplementary Fig. 13a). However, recombination frequencies varied markedly between conditions, with tetraploid cells exhibiting higher levels of recombination at 37 oC than at 30 oC, as well as higher recombination frequencies than diploid cells grown at either temperature (Fig. 4b, Supplementary Fig. 13b). Notably, loss of SPO11 activity resulted in a decrease in recombination in diploid and tetraploid cells at both temperatures, as recombination frequencies were lower in Δspo11 mutants (2–13-fold) and in cells expressing the SPO11-YF allele (5–54-fold) than in wildtype controls (Fig. 4b, Supplementary Fig. 13b). Reintegration of an intact SPO11 allele complemented the recombinational defect of the Δspo11 mutant in mitotic cells (Fig. 4b, Supplementary Fig. 13b).
Fig. 4.
Spo11, but not Rec8, promotes recombination in mitotically dividing C. albicans cells. a–d Genetically marked diploid and tetraploid C. albicans strains were serially passaged in YPD medium at 30 °C or 37 °C (cells diluted 1:100 every 24 h), and evaluated for loss of the GAL1 marker (2-DOGR frequency) and for the frequency of recombination in the HIS1-URA3 interval. a Frequency of 2-DOGR colonies in diploid/tetraploid wildtype and SPO11 mutant strains. n = 4, 5, 6, and 6 biologically independent experiments for 30 oC diploid, 37 oC diploid, 30 oC tetraploid, and 37 oC tetraploid, respectively. b Frequency of recombination between HIS1 and URA3 on Chr1 in diploid/tetraploid wildtype and SPO11 mutant strains. n = 4, 4, 5, and 5 biologically independent experiments for 30 oC diploid, 37 oC diploid, 30 oC tetraploid, and 37 oC tetraploid, respectively. c Frequency of 2-DOGR colonies in wildtype or REC8 mutant tetraploid strains passaged at 30 oC. n = 4 biologically independent experiments. d Analysis of recombination in the HIS1-URA3 interval for wildtype or REC8 mutant tetraploid strains passaged at 30 oC. n = 4 biologically independent experiments. e Tenfold spot dilution assays performed for wildtype, Δspo11, and SPO11-complemented tetraploid cells grown on YPD medium in the presence or absence of 0.01% methyl methanesulfonate (MMS) and imaged after 2 days at 37 oC. * denotes p < 0.05. ** denotes p < 0.01 as calculated by two-sample t (chromosome loss) or Wilcoxon (recombination frequency) tests. Boxplots are presented as the 25th to 75th percentile with the thick line denoting the median. Whiskers indicate the largest and smallest values within 1.5 × of the interquartile range. White, orange, light orange, and gray denote WT, Δspo11, Δ+SPO11(YF), and Δ+SPO11(WT), respectively. White, purple, and gray denote WT, Δrec8, and Δ+REC8(WT), respectively
We also note that SPO11 influenced the stability of the GAL1 marker during mitotic passaging, as tetraploid Δspo11 cells grown at 37 oC experienced a 16-fold increase in GAL1 marker loss relative to wildtype cells (two-sample t (3.69) = 5.50, p = 1.86E-3), whereas cells expressing the SPO11-YF variant showed a 135-fold increase in GAL1 loss compared with wildtype cells (two-sample t (3.01) = 6.53, p = 0.013) (Fig. 4a, Supplementary Fig. 13a and 14). In contrast to SPO11, cells lacking REC8 displayed no change in recombination frequency or GAL1 marker stability during standard mitotic growth (Fig. 4c, d).
To determine whether Spo11 also contributes to mitotic DNA repair phenotypes in response to genotoxic stress, we compared wildtype and Δspo11 diploid cells in the presence of the DNA damaging agent methyl methanesulfonate (MMS). We found that diploid Δspo11 cells showed a reproducible increase in MMS susceptibility relative to wildtype cells indicating a defect in DNA repair (Fig. 4e), whereas exposure to other cellular stresses did not produce detectable Δspo11 phenotypes (Supplementary Fig. 15). These findings establish that Spo11 can function in DNA recombination and repair in mitotically dividing C. albicans cells, in addition to its more specialized role during CCL.
Discussion
The current study details key aspects of CCL in C. albicans, during which tetraploid cells undergo extensive genome instability leading to a reduction in ploidy and experience high levels of homologous recombination. Given the apparent lack of a conventional meiosis, this mechanism provides an effective pathway by which C. albicans cells can complete a diploid–tetraploid–diploid parasexual cycle and generate diverse progeny. Understanding of the CCL process can therefore shed light on alternative mechanisms of sexual/parasexual reproduction, as well as detailing how parasex produces genetic diversity in this important human pathogen.
Our experiments establish that high recombination frequencies accompany CCL in C. albicans, with inter-homolog exchanges on Chr1 being more than three orders of magnitude higher than those during normal mitotic passaging. We also reveal prominent roles for the ‘meiosis-specific’ factors Spo11 and Rec8 in regulating genome dynamics during CCL, with parallels to their conventional roles during meiosis. In particular, it is now evident that Spo11 introduces extensive DSBs during both CCL and meiosis, and that Spo11 and Rec8 are similarly required for promoting high levels of inter-homolog recombination during both of these processes. We therefore propose that CCL represents a ‘parameiosis’43 in which both ploidy reduction and homologous recombination are dependent on key meiosis-specific factors. CCL clearly differs from a conventional fungal meiosis, however, in that it is highly uncoordinated and ploidy reduction is uncoupled from the process of sexual sporulation, as the latter has never been observed in C. albicans.
In meiosis, Spo11 function can affect chromosome pairing at two steps: first, Spo11 promotes homolog alignment independent of its ability to form DSBs, and second, a more precise pairing of homologs occurs during synapsis that is dependent on Spo11-mediated DSBs44–46. We found that chromosome instability in C. albicans cells undergoing CCL was impacted by Spo11 catalytic activity, as mutation of the active site increased genome instability, which may mirror its role in promoting chromosome pairing during meiosis. Furthermore, loss of Spo11 clearly diminished, but did not abrogate, DSB formation and recombination during CCL, indicating that additional factors contribute to these processes. Surprisingly, Spo11 also impacted DNA repair and recombination during mitotic growth of C. albicans diploid and tetraploid cells, revealing a general role for this factor in genome dynamics beyond its function in CCL.
In marked contrast to Spo11, C. albicans tetraploid cells lacking Rec8 exhibited reduced GAL1 marker loss during CCL. The cause of this phenotype and its relationship to the meiotic function of Rec8 is unclear, although Spo11 and Rec8 have opposing roles in promoting S-phase progression in S. cerevisiae meiosis45. Interestingly, recent experiments also revealed that ectopic overexpression of REC8 can increase chromosome loss in mitotic S. cerevisiae cells47. This result is in line with our current findings where Rec8 enhances chromosome instability in C. albicans cells, at least under CCL conditions.
It is possible that parameiosis represents an ancestral mechanism for ploidy cycling and thus reflects an intermediary step in the evolutionary path from mitosis to meiosis. The dual function of C. albicans Spo11 in both mitotic and parameiotic recombination could also indicate a general role in DNA repair prior to being co-opted for meiosis-specific purposes. However, given the ubiquity of meiosis in the eukaryotic kingdom48 and its presence in other extant Candida species48–50, a more parsimonious model is that the ancestor of C. albicans underwent meiosis and that this program then degenerated into the parameiotic program present today. In line with this, certain C. albicans genes (e.g., DLH1/DMC1) are able to complement for the loss of their meiotic orthologs in S. cerevisiae51, whereas others (e.g., NDT80) have been reprogrammed for alternative functions such as biofilm formation52.
Regardless of whether parameiosis is an ancestral program or more recently derived, this study highlights how mating and ploidy cycling could have evolved without the precise chromosome dynamics that characterize a conventional meiosis. Moreover, despite its rudimentary mechanism, CCL can still achieve several key functions of meiosis including (1) a reduction in ploidy, (2) generation of recombinant progeny, and (3) the purging of deleterious alleles/unmasking of advantageous recessive alleles43. The uncoupling of ploidy cycling from sexual sporulation may have been a key step in the emergence of parameiosis in C. albicans and conferred a fitness advantage to a commensal organism whose sexual spores would likely be antigenic to the host53.
Finally, we note that non-meiotic processes are now recognized as important drivers of ploidy change in multiple eukaryotic species. For example, the filamentous fungus Aspergillus nidulans can undergo a parasexual cycle in which diploid cells readily lose chromosomes to return to the haploid state, a process that can generate population diversity de novo54,55. Parasexuality has also been described in the parasitic protozoans Trypanosoma cruzi and Giardia intestinalis, in which genetic exchange can take place in the absence of meiosis but where ‘meiosis-specific’ genes such as SPO11 are expressed, at least in G. intestinalis56–58. Mechanistic parallels also extend to depolyploidization processes observed in mammalian cells. In particular, polyploid tumor cells can undergo depolyploidization that is accompanied by upregulation of SPO11 and REC8 genes32,33. Expression of such genes in several cancer types is thought to promote genome instability and drive the formation of aneuploid products31,59. These findings are analogous to the high rate of aneuploid formation arising in C. albicans cells as a result of CCL. Thus, diverse eukaryotic species appear to be able to use conserved meiosis genes for alternative mechanisms driving ploidy reduction. These studies establish that new roles for meiosis-specific factors are being uncovered that blur the boundaries between conventional mitotic and meiotic programs, as well as providing potential insights into how ploidy cycling and meiosis could have evolved in early eukaryotes.
Methods
Media used
Media was prepared by dissolving the appropriate nutrient into ddH2O and autoclaving as previously described60,61. YPD plates containing 200 μg/mL nourseothricin (NAT) were used for selection of strains that were resistant to nourseothricin (SATR strains)62. S. cerevisiae pre-sporulation (‘PRE-SPO’) medium contained 0.8% yeast extract, 0.3% peptone, 10% glucose (added prior to autoclaving), and 2% agar63. Medium with 2-deoxygalactose (2-DOG) consisted of Synthetic Complete medium supplemented with 2% glycerol, 2 mg/mL 2-DOG (Fluka #31050), and 2% agar. Maltose plates were prepared as YP media supplemented with 2% maltose and 2% agar.
Construction of genetically marked strains
All strains are listed in Table S1. The genetically marked chromosome strain was produced by multiple rounds of targeted integration in C. albicans strain SN78 obtained from the Noble lab (UCSF)64. To target GAL1, a plasmid was constructed in which the 5′ sequence flanking GAL1 was PCR amplified with primers gcggccatgggcccTCCCGACACCAAAATCATAATT and caggcgcgCTCGAGTCAAACGTAGGAACTGACATGG and the 3′ flank was PCR amplified with primers gcggccaCCGCGGCCATCTATGGGTAGTTGTATTG and caggcgcgGAGCTCGACTTGTATAAGCCTACTTTGC. These products were cut with enzymes ApaI/XhoI and SacII/SacI, respectively, and cloned into pSFS2A62. The resulting plasmid, pRB472, was treated with ApaI and SacI and the products used to delete GAL1 by transformation into SN78 followed by recycling of the SAT1 selectable marker by induction of FLP by plating to maltose. The allele deleted was phased to a specific chromosome homolog by sequencing of the remaining homolog. A single copy of HIS1 was amplified from the genome using primers ggcgccAAGCTTGGTTGGCTCTCTTAATACGAT and gccggcGGATCCTGGTAAACCTGAGTGAGATG that target HIS1 adjacent to SNP #23 in the genome65. Phasing of HIS1 relative to SNP #23 was performed using primers AGCCAACCATATTTCAGGATTGAC and GTGCCAACTAGTAATGGTTGTCAT and digest with HincII. Following HIS1 phasing, URA3 was amplified from the SC5314 genome using primers CAGAAAAAAAAATTTTGATGATGAGATGGTCTATTTATAGTGCGCGAAATTTAGAAGCATCTCATGGTTTTCTAGAAGGACCACCTTTGA and ATGAAAATTTATTTGATGTCTTGGGAGCCCTAATTATTTTTCTTCTTCATTAATCTTTGATCTCATCGCCGTTGCTGTAGTGCCATTGAT and introduced at SNP #4266. Phasing of URA3 was performed using primers GAAATCCACCGCATAAGAAATGGTT and CTGGTGAGGCATGAGTTTCT and digest with MluI. MTLa or MTLα were deleted using the previously described plasmids pRB101 and pRB102, respectively67. Introduction of SAT1 at SNP #30 was accomplished by amplifying the SAT1 marker from pSFS2a62 with primers AGTCATTAACTTGAAACTTCACCTTCCACTACAATCCTTTTCTCAAAGCAAAGCTAAAAAGTAATGCTACCCATCATAAAATGTCGAGCG and TAAGATTGATGTGTAATGGGAACGTTATCAAAGTAAATAAGTGTGGTGCGACAACAACGACAGCCAGAAACGCTCTAGAACTAGTGGATC. Phasing of SAT1 as SNP #30 was determined using primers ACAGCGATGTACTGGTACTG and GCATGGTTTGAACAAGTGATAGAGT and digest with AluI.
Deletion of SPO11 and REC8 (orf19.3589 and orf19.776, respectively) was performed by homologous recombination of PCR products amplified from pSFS2A62 with 70 bp of flanking sequence to the desired locus. Two successive rounds of PCR was performed to target each of the two SPO11 alleles with CTATAACAATATCTCATACTGAGTCCATGTATGTTGGAAACAATCATATCATCAATTTTCTGAAAACGGGAGGGAACAAAAGCTGGAGCT/ CCATCTTTTTCAATGTAGACTTTGTAGTCTAAAGTGTTGGCAGCACCAACTTGACGAGTAGAGTATGAAGACATCCTCGAGGAAGTTCCTATACT for deletion of the first allele and CTGAAAACGGGAAGACTAACTTTCTTTAAGAAACAAATTACAGGTGCTCCTAAATATTTGGGAACAAGTTGGGAACAAAAGCTGGAGCT/ TTTGGGAACTTTTGCTCGATTTTGTTGAGAACAAATGAAAATTGTTTGAATCTCCGCTTAACATAAATAGCTCGAGGAACTTCCTATACT for the second round. Deletion of REC8 was similarly performed with GAGGTCATATACATTGTTTGGCTAGTAGTAGTTGTGTGTGTGCTTGTTTGTACAATTTTTTAGCAACCACAGGGAACAAAAGCTGGAGCT/ AGCAAACACCTAGCAAAAGAAGTTGCGAAATAAACTGATCACATACAAATCGTTAACCTCATTCCAAAGGCCTCGAGGAAGTTCCTATACT for both rounds of transformation. The SAT1 gene was recycled by plating for 100 colonies on SC + maltose to induce expression of the FLP flippase62 and top spread with 20 µg/mL NAT. Small colonies were screened for loss of SAT1 by replating to 200 µg/mL NAT. Deletion of SPO11 or REC8 was performed in diploid strains prior to integration of the SAT1 marker at SNP #3066.
The function of each marker was assessed at each step of strain construction and markers were phased again at the conclusion of strain construction to insure they remained heterozygous and associated with the SNP variant. The MTLa/a marked strain and an unmarked MTLα/α sibling strain were switched to the opaque state, mated to one another, and the products screened by DNA-staining with Sytox Green followed by flow cytometry to identify tetraploid mating products as described previously68 and as detailed below.
Reintegration of an intact SPO11, SPO11(Y65F), or REC8 was performed in the corresponding Δspo11 or Δrec8 tetraploid background. The SPO11-YF plasmid was constructed by PCR amplification of the gene in two fragments using oligos GGCGCCGGGCCCCAAACGTTGATTTCTTGGCC and CAATGGTTGAAAACTTCCACATCTTGGAAATATATATCACGAATGGTTGTTATTTGAT in one reaction and oligos ATCAAATAACAACCATTCGTGATATATATTTCCAAGATGTGGAAGTTTTCAACCATTG and GCCGGCCTCGAGTGTGGGAAGCAGTTTCTGAC in a second reaction. The two PCR products were combined by fusion PCR and cloned into the pCR-Blunt II-TOPO vector (Invitrogen) and sequenced to confirm the presence of the Y65F mutation. The resulting vector was treated with enzymes ApaI/XhoI to release the SPO11(YF) gene and cloned into the corresponding restriction sites in plasmid pSFS2A. The resulting plasmid, pRB589, was linearized with BsgI and used to transform strain CAY7268/CAY7270 to generate strains CAY7701 and CAY7703. A plasmid containing the REC8 ORF was obtained by amplification of REC8 using primers 5′-AAGCGGCCGCGGGTTTGGTTTAATTGTGGC-3′ and 5′-GGTGGTGAGCTCCTTGAACACGAACACGGTGA-3′, digesting the resulting PCR product and pSFS2a with SacI and NotI, and ligating the two products. The integrity of the REC8 sequence was assessed by Sanger sequencing of the plasmid. The resulting plasmid, pRB543, was linearized with SacII and transformed into strain CAY7351/CAY7354 to generate strains MAY572/573).
Assays to determine chromosome instability and recombination
Cells were induced to undergo CCL by heavily inoculating cells in quadrants on S. cerevisiae PRE-SPO medium and culturing at 37 °C as previously described38. After 7 days on PRE-SPO medium, cells were scraped from the plates, counted via hemocytometer, and plated to 2-DOG (50 K, 200 K, and two million cells) and to YPD plates (100 cells). The frequency of 2-DOGR colonies was determined relative to the number of colonies present on YPD. Chromosome loss across all strains and conditions appeared to be less efficient across these experiments compared to previous studies38,39. 2-DOG plates were also replica plated to synthetic complete defined (SCD) media lacking histidine or uracil to select for HIS1 or URA3, respectively, or to YPD + NAT (200 µg/mL) to select for SAT1. The frequency of recombination was identified by the presence or absence of genetic markers (HIS1/URA3/SAT1) in colonies that were 2-DOGR (gal1−). Recombination frequency was calculated as the number of colonies observed to have recombined between two markers relative to the total number of 2-DOGR. Loss of all three markers (HIS3/URA3/SAT1) suggested the marked chromosome itself had been lost. All direct comparisons were made between data sets performed in parallel as considerable variation was observed between CCL experiments performed on different days, possibly owing to differences in plate humidity or other environmental conditions.
Selective genotyping of strains by ddRAD-Seq
Progeny from either wildtype or Δspo11 tetraploid strains that had undergone CCL were plated to 2-DOG and screened by flow cytometry for near-diploid DNA content. Twenty-one colonies from each genotype along with five colonies of each parental genotype were prepared for double digest restriction-site associated (ddRAD-Seq) sequencing essentially as described69. In brief, DNA from each sample was digested with MboI and MfeI restriction endonucleases (New England Biolabs, Ipswich, MA) to produce non-complementary overhangs. P1 and P2 adapters complementary to each overhang, respectively, were ligated to the DNA fragments. P1 adapters encode unique 7 bp barcodes and P2 adapters corresponded to the D701–D712 Illumina adapter set. Adapter-ligated pools of eight samples were gel excised between 200 and 500 bp69. The resulting pools were amplified using universal P5 and P7 Illumina amplification primers. Prepared libraries were sequenced on a HiSeq 2500 at the Genewiz Sequencing Facility (South Plainfield, NJ). Approximately two million reads were generated for each sample. Reads were demultiplexed using the barcode_splitter scripts70 and quality controlled using FastQC71. The ploidy and allelic composition of parental and progeny strains were determined using YMAP72. On average, 2169 marker positions were captured across each progeny genome (range: 1458–2513), resulting in an average spacing of ~ 7100 bp between markers.
Recombination breakpoints were determined by comparing the allelic patterns of each progeny to the appropriate parental genotypes. Progeny genotypes that could be inherited from whole parental chromosomes were not considered recombinant. A recombination bin was required to span a minimum of 10 kb as defined by at least four loci that did not match the genotype possible from inheritance of parental chromosomes.
Flow cytometry of DNA content
Tetraploid cells passaged or induced to undergo CCL were isolated as single colonies after 7 days. Single colonies were grown overnight in YPD in 96-well plates and prepared for flow cytometry as previously described68. In brief, cells were washed and resuspended in 50:50 TE (50 mm Tris, pH 8, 50 mm EDTA) solution. Cells were then treated with 1 mg/mL RNAse A for 4 hours at 37 oC followed by 5 mg/mL Proteinase K treatment at 37 oC for 45 min. Cells were washed with 50:50 TE and resuspended in SybrGreen (1:100 dilution in 50:50 TE) and incubated overnight at 4 oC. Stained cells were washed and resuspended in 50:50 TE and SybrGreen staining data was obtained for 50,000 events for each sample using a BD FACSCanto II. Ten independent known diploid and tetraploid samples served as controls.
Gam-GFP
The plasmid pRS11 contains a Gam-GFP cassette under the control of a TetON promoter and allows for visualization of DNA double-strand breaks. C. albicans codon-optimized Gam flanked by two SalI restriction sites was synthesized, digested with SalI and cloned into the pNIM1 vector73 at the SalI site to generate the tetO-Gam-GFP plasmid. For generation of Gam-GFP-marked strains, pRS10 was digested with KpnI/SacII to linearize the plasmid for targeting to the ADH1 locus and transformants selected on YPD + nourseothricin.
Strains containing the Gam-GFP construct were grown for 24 h on either PRE-SPO or SCD media in the presence or absence of 50 μg/mL doxycycline. Cells were taken from the plates and visualized across 10 fields of view. Nuclear staining was performed with 25 μg/mL 4′,6-diamidino-2-phenylindole.
For western blots, strains containing the Gam-GFP construct were grown for 24 h on YPD in the presence or absence of 50 μg/mL doxycycline. Cell lysates were prepared as previously described74. In brief, cells were spun down and resuspended in Thorner buffer (40 mm Tris, pH 8.0, 5% sodium dodecyl sulfate, 8 m urea, and 100 μm EDTA) and transferred to a 2 mL cryovial with 100 μL of 500 micron glass beads. The tubes were heated at 100 oC for 3 min and bead beat for four intervals of 1 min ON/1 min OFF. Western blots were performed with mouse anti-GFP (Sigma, St. Louis, MO) and mouse anti-PSTAIR (Abcam, Cambridge, MA).
TUNEL assay
Tetraploid cells grown on PRE-SPO solid medium for 48 h were collected and spheroplasted as previously described75. In briefly, 1 mL of cells (OD = 2) were fixed in 4% formaldehyde for 1 h, followed by quenching with 125 mm glycine for 10 min, and then spheroplasted using 5 U of zymolyase according to the manufacturer’s instructions (G-Biosciences, St. Louis, MO). Spheroplasting was assessed visually with ~ 90% of the cells appearing as ‘ghosts’. Spheroplasted cells were then incubated with TUNEL reagents according to the manufacturer’s instructions (Roche, Basel, Switzerland) to mark DNA double-strand breaks. Cells were visualized by microscopy for fluorescence indicating DNA breaks, and an unlabeled sample served as a negative control.
Stress resistance
Overnight cultures were grown in YPD liquid medium at 30 oC. Cell density was determined using OD600 and cultures were adjusted to an OD600 of 1.0 in 1 mL ddH20. These dilutions were used as a base for five sequential tenfold dilutions. For each stress condition, 5 μL of each dilution was spotted to the appropriate prewarmed agar plates including a SCD medium plate absent any stressor as a control for growth. Plates were then incubated at 37 oC and imaged at 48 h. Conditions assayed included 100 μg/mL calcofluor white, 2 mm hydroxyurea, 0.01% MMS, and control SCD media. Experiments were performed with a minimum of three biological replicates on separate wildtype, Δspo11, and SPO11-complemented isolates.
Statistics
Statistics are provided throughout the manuscript. All two-sample t tests were two-sided comparisons. All measurements were taken from distinct biological samples.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Peter Belenky, Rebecca Shapiro, and Suzanne Noble for gifting plasmids. We thank Judy Berman and Mason Clark for assistance with enumerating recombination assays. This work was supported by National Institutes of Health grants AI081704, AI122011, and AI141893 (to R.J.B.), a PATH award from the Burroughs Wellcome Fund (to R.J.B.), a Research Supplement to Promote Diversity in Health-Related Research award (to M.Z.A.), and a F31 DE023726 (to M.P.H.).
Author contributions
M.Z.A. and R.J.B. designed the study; M.Z.A. and G.J.T. collected the data; M.Z.A analyzed the data; M.Z.A. and M.P.H. constructed strains used in this study; M.Z.A. and R.J.B. drafted and edited the manuscript.
Data availability
Sequence data used for ddRAD-Seq analysis is available at the BioProject database under accession code PRJNA560397. Other data sets generated and/or analyzed during the current study are available in this article and its Supplementary Information files, or from the corresponding author on request.
Code availability
The code used to generate genotypes for each marker position within the ddRAD-Seq data can be obtained through the YMAP workflow (http://lovelace.cs.umn.edu/Ymap/)72 or from Dr. Judy Berman’s lab upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Matthew Z. Anderson, Email: Anderson.3196@osu.edu
Richard J. Bennett, Email: richard_bennett@brown.edu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41467-019-12376-2.
References
- 1.Schurko AM, Logsdon JM., Jr Using a meiosis detection toolkit to investigate ancient asexual “scandals” and the evolution of sex. Bioessays. 2008;30:579–589. doi: 10.1002/bies.20764. [DOI] [PubMed] [Google Scholar]
- 2.Speijer D, Lukes J, Elias M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl Acad. Sci. USA. 2015;112:8827–8834. doi: 10.1073/pnas.1501725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goddard MR. Molecular evolution: sex accelerates adaptation. Nature. 2016;531:176–177. doi: 10.1038/nature17304. [DOI] [PubMed] [Google Scholar]
- 4.Goddard MR, Godfray HC, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- 5.McDonald MJ, Rice DP, Desai MM. Sex speeds adaptation by altering the dynamics of molecular evolution. Nature. 2016;531:233–236. doi: 10.1038/nature17143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenormand, T., Engelstadter, J., Johnston, S. E., Wijnker, E. & Haag, C. R. Evolutionary mysteries in meiosis. Philos. Trans. R Soc. Lond B Biol. Sci.371, pii: 20160001 (2016). [DOI] [PMC free article] [PubMed]
- 7.Wilkins AS, Holliday R. The evolution of meiosis from mitosis. Genetics. 2009;181:3–12. doi: 10.1534/genetics.108.099762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramesh MA, Malik SB, Logsdon JM., Jr A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr. Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 2014;7:a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/S0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 11.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 12.Szekvolgyi L, Nicolas A. From meiosis to postmeiotic events: homologous recombination is obligatory but flexible. FEBS J. 2010;277:571–589. doi: 10.1111/j.1742-4658.2009.07502.x. [DOI] [PubMed] [Google Scholar]
- 13.Bloomfield G. Spo11-independent meiosis in social amoebae. Annu. Rev. Microbiol. 2018;72:293–307. doi: 10.1146/annurev-micro-090817-062232. [DOI] [PubMed] [Google Scholar]
- 14.Bergerat A, et al. An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 15.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/S0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 16.Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr. Opin. Genet. Dev. 2013;23:147–155. doi: 10.1016/j.gde.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Keeney, S. Spo11 and the formation of DNA double-strand breaks in meiosis. In Recombination and Meiosis, Vol. 2 (ed. Lankenau, D.) (Springer, Berlin/Heidelberg, 2007).
- 18.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/S0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 19.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2007;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 21.Klein F, et al. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 22.Mehta GD, Rizvi SM, Ghosh SK. Cohesin: a guardian of genome integrity. Biochim. Biophys. Acta. 2012;1823:1324–1342. doi: 10.1016/j.bbamcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
- 23.Buonomo SB, et al. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/S0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 24.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/S0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 25.Tomonaga T, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitajima TS, Miyazaki Y, Yamamoto M, Watanabe Y. Rec8 cleavage by separase is required for meiotic nuclear divisions in fission yeast. EMBO J. 2003;22:5643–5653. doi: 10.1093/emboj/cdg527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe Y. Modifying sister chromatid cohesion for meiosis. J. Cell Sci. 2004;117:4017–4023. doi: 10.1242/jcs.01352. [DOI] [PubMed] [Google Scholar]
- 28.Bloomfield G. Atypical ploidy cycles, Spo11, and the evolution of meiosis. Semin Cell Dev. Biol. 2016;54:158–164. doi: 10.1016/j.semcdb.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 29.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzung KW, et al. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl Acad. Sci. USA. 2001;98:3249–3253. doi: 10.1073/pnas.061628798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, A. Y. & Gjerstorff, M. F. Ectopic expression of testis germ cell proteins in cancer and its potential role in genomic instability. Int. J. Mol. Sci.17, pii: E890 (2016). [DOI] [PMC free article] [PubMed]
- 32.Ianzini F, et al. Activation of meiosis-specific genes is associated with depolyploidization of human tumor cells following radiation-induced mitotic catastrophe. Cancer Res. 2009;69:2296–2304. doi: 10.1158/0008-5472.CAN-08-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalejs M, et al. Upregulation of meiosis-specific genes in lymphoma cell lines following genotoxic insult and induction of mitotic catastrophe. BMC Cancer. 2006;6:6. doi: 10.1186/1471-2407-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hull CM, Raisner RM, Johnson AD. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science. 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- 35.Bennett RJ, Johnson AD. Mating in Candida albicans and the search for a sexual cycle. Annu Rev. Microbiol. 2005;59:233–255. doi: 10.1146/annurev.micro.59.030804.121310. [DOI] [PubMed] [Google Scholar]
- 36.Magee BB, Magee PT. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science. 2000;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- 37.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/S0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 38.Bennett RJ, Johnson AD. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 2003;22:2505–2515. doi: 10.1093/emboj/cdg235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forche A, et al. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shee C, et al. Engineered proteins detect spontaneous DNA breakage in human and bacterial cells. Elife. 2013;2:e01222. doi: 10.7554/eLife.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ene IV, et al. Global analysis of mutations driving microevolution of a heterozygous diploid fungal pathogen. Proc. Natl Acad. Sci. USA. 2018;115:E8688–E8697. doi: 10.1073/pnas.1806002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuch BB, et al. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodenough, U. & Heitman, J. Origins of eukaryotic sexual reproduction. Cold Spring Harb. Perspect. Biol.6, pii: a016154 (2014). [DOI] [PMC free article] [PubMed]
- 44.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding SPO11-mediated double-strand breaks in mice. Dev. Cell. 2013;24:196–205. doi: 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- 46.Loidl J. The hidden talents of SPO11. Dev. Cell. 2013;24:123–124. doi: 10.1016/j.devcel.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Tutaj, H., Pogoda, E., Tomala, K. & Korona, R. Gene overexpression screen for chromosome instability in yeast primarily identifies cell cycle progression genes. Curr. Genet.65, 483–492 (2018). [DOI] [PMC free article] [PubMed]
- 48.Butler G, et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 2009;459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherwood RK, Scaduto CM, Torres SE, Bennett RJ. Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature. 2014;506:387–390. doi: 10.1038/nature12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reedy JL, Floyd AM, Heitman J. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr. Biol. 2009;19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diener AC, Fink GR. DLH1 is a functional Candida albicans homologue of the meiosis-specific gene DMC1. Genetics. 1996;143:769–776. doi: 10.1093/genetics/143.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nocedal, I., Mancera, E. & Johnson, A. D. Gene regulatory network plasticity predates a switch in function of a conserved transcription regulator. Elife6, pii: e23250 (2017). [DOI] [PMC free article] [PubMed]
- 53.Nielsen K, Heitman J. Sex and virulence of human pathogenic fungi. Adv. Genet. 2007;57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- 54.Schoustra SE, Debets AJ, Slakhorst M, Hoekstra RF. Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans. PLoS Genet. 2007;3:e68. doi: 10.1371/journal.pgen.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD, Bufton AW. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/S0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 56.Poxleitner MK, et al. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 57.Carpenter ML, Assaf ZJ, Gourguechon S, Cande WZ. Nuclear inheritance and genetic exchange without meiosis in the binucleate parasite Giardia intestinalis. J. Cell Sci. 2012;125:2523–2532. doi: 10.1242/jcs.103879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaunt MW, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 59.Rivera M, et al. Acquisition of meiotic DNA repair regulators maintain genome stability in glioblastoma. Cell Death Dis. 2015;6:e1732. doi: 10.1038/cddis.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 61.Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 62.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 63.Codon AC, Gasent-Ramirez JM, Benitez T. Factors which affect the frequency of asporulation and tetrad formation in Saccharomyces cerevisiae baker’s yeasts. Appl Environ. Microbiol. 1995;61:1677. doi: 10.1128/aem.61.4.1677-1677b.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forche A, Steinbach M, Berman J. Efficient and rapid identification of Candida albicans allelic status using SNP-RFLP. FEMS Yeast Res. 2009;9:1061–1069. doi: 10.1111/j.1567-1364.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forche A, Magee PT, Magee BB, May G. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell. 2004;3:705–714. doi: 10.1128/EC.3.3.705-714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alby K, Bennett RJ. Stress-induced phenotypic switching in Candida albicans. Mol. Biol. Cell. 2009;20:3178–3191. doi: 10.1091/mbc.e09-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hickman MA, Paulson C, Dudley A, Berman J. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics. 2015;200:781–794. doi: 10.1534/genetics.115.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parsons, R.L.a.L. Barcode Splitter, version 0.18.0. (2017).
- 71.Andrews, S. FastQC: a quality control tool for high throughput sequence data. (2010).
- 72.Abbey DA, et al. YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Med. 2014;6:100. doi: 10.1186/s13073-014-0100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ketel C, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data used for ddRAD-Seq analysis is available at the BioProject database under accession code PRJNA560397. Other data sets generated and/or analyzed during the current study are available in this article and its Supplementary Information files, or from the corresponding author on request.
The code used to generate genotypes for each marker position within the ddRAD-Seq data can be obtained through the YMAP workflow (http://lovelace.cs.umn.edu/Ymap/)72 or from Dr. Judy Berman’s lab upon request.