Abstract
Rice bran supplementation provides nutrients, prebiotics and phytochemicals that enhance gut immunity, reduce enteric pathogens and diarrhea, and warrants attention for improvement of environmental enteric dysfunction (EED) in children. EED is a subclinical condition associated with stunting due to impaired nutrient absorption. This study investigated the effects of rice bran supplementation on weight for age and length for age z-scores (WAZ, LAZ), EED stool biomarkers, as well as microbiota and metabolome signatures in weaning infants from 6 to 12 months old that reside in Nicaragua and Mali. Healthy infants were randomized to a control (no intervention) or a rice bran group that received daily supplementation with increasing doses at each month (1–5 g/day). Stool microbiota were characterized using 16S rDNA amplicon sequencing. Stool metabolomes were analyzed using ultra-high-performance liquid-chromatography tandem mass-spectrometry. Statistical comparisons were completed at 6, 8, and 12 months of age. Daily consumption of rice bran was safe and feasible to support changes in LAZ from 6–8 and 8–12 months of age in Nicaragua and Mali infants when compared to control. WAZ was significantly improved only for Mali infants at 8 and 12 months. Mali and Nicaraguan infants showed major differences in the overall gut microbiota and metabolome composition and structure at baseline, and thus each country cohort demonstrated distinct microbial and metabolite profile responses to rice bran supplementation when compared to control. Rice bran is a practical dietary intervention strategy that merits development in rice-growing regions that have a high prevalence of growth stunting due to malnutrition and diarrheal diseases. Rice is grown as a staple food, and the bran is used as animal feed or wasted in many low- and middle-income countries where EED and stunting is prevalent.
Subject terms: Microbiome, Randomized controlled trials, Paediatric research
Introduction
The prevalence of malnutrition in low and middle-income countries (LMIC) has negative consequences on growth of children during the first five years of life and has lifelong health consequences1,2. There is an increased risk of death among children under 5 years of age due to being underweight and stunted2,3. Risk factors for undernutrition may include, but are not limited to: low birth weight, inadequate breastfeeding, improper complementary feeding, and recurrent infections3,4. Diarrheal diseases are some of the primary causes of undernutrition in children under five years of age1,3,4.
Environmental enteric dysfunction (EED) is an acquired subclinical condition of the small intestine among LMIC children5–10. Chronic exposure to enteric pathogens early in life is one likely contributor to EED11. The altered gastrointestinal functions in EED include intestinal nutrient malabsorption and increased intestinal permeability that leads to protein loss6,7. Infant weaning has been identified as a critical window for intervention12. Previous intervention efforts in young children have targeted micronutrient deficiencies, such as Vitamin A, Zn and Fe13–16, oral rehydration salts for treating diarrhea17, antimicrobial use18,19, and community hygiene improvements20. Given the worldwide trends and variable timing of growth faltering that has been reported, assessing interventions in diverse populations merits further attention21.
Rice bran is a nutrient dense food with a unique profile and ratio of bioactive phytochemicals such as gamma oryzanol, tocotrienols, ferulic acid, vitamin B, beta-sitosterol and many others. The metabolome of rice bran from 17 countries was analyzed and revealed a varied suite of bioactive molecules that span multiple chemical classes22. Rice bran has shown chronic disease fighting properties in both animal and human studies, and the promising results from feeding neonatal pigs provided translational support for investigation of dietary feasibility in weaning infants. Many rice bran components, such as phenolics, fatty acids and soluble fibers work together to prevent enteric pathogens and diarrheal disease in mice and pigs23–26, and to favorably promote gut health in adults27,28. The effect of rice bran supplementation on host resistance to enteric infections and enhanced gut mucosal immunity was demonstrated for Salmonella enterica Typhimurium25,26, rotavirus29–31, and norovirus23. Rice bran merits attention because it has widespread scalability for consumption globally32, and particularly in LMIC regions where EED is prevalent33.
Stool EED biomarkers, gut microbiota34 and metabolite profiling analysis15,35 are important surrogate markers for analysis as intestinal tissue from infants is not easily accessible to evaluate. Stool myeloperoxidase (MPO)36, calprotectin (CAL)37, and neopterin (NEO)38 are indicators of inflammation; and alpha-1-antitrypsin (AAT)36 is an indicator of barrier lumen disruption. Chronic, elevated concentrations of all four biomarkers have been associated with poor linear growth in infants up to 24 months old10,36–38, and as the gut microbiota is maturing over the first 3 years of life21,39. Gut microbiota composition and metabolism is influenced by delivery mode, environment, and nutrition40,41. Recent studies have demonstrated that malnutrition and immature microbiota of infants are only partially, and temporarily improved by some nutritional interventions15,42. The primary objective of this study was to investigate safety and tolerability of dietary rice bran supplementation during infant weaning, including effects on growth, EED biomarkers, gut microbiota and metabolome from six to twelve months of age in Nicaragua and Mali. Nicaragua and Mali have agricultural rice production systems, yet they currently do not use the rice bran for human consumption after it is polished from the whole grain. Our study hypothesis was that daily consumption of rice bran for six months is tolerable, safe and feasible for children during weaning and that daily intake will be associated with improved growth and decreased gut permeability alongside favorable modulation of the gut microbiota and metabolome.
Results
Safety and feasibility
Daily rice bran consumption was completed in a randomized controlled trial with infants from 6 to 12 months of age. While the types of weaning foods differ between Mali and Nicaragua, there were no differences detected in the types of weaning foods consumed between rice bran and control group infants within each country site. Rice bran supplementation was palatable and safe for weaning infants based on a high-level of compliance and no adverse events related to rice bran intake at the increasing doses over time. Dietary compliance to rice bran was averaged per month during the 6-month intervention and was 90% in Nicaragua and 99% in Mali. The feasibility and tolerability for infants to consume 1–5 g of rice bran/day over the six-month study period was determined by how mothers fed the rice bran powder directly or as reported by consumption with drinking water, staple grain porridges (i.e. millet, sorghum, and white rice), soups, milk, fruits, juices, eggs, and fish when available. The nutritional profile of rice bran provided in this study is shown in Table 1.
Table 1.
Nutrient composition of Rice Bran (as provided by USDA National Nutrient Database).
| Nutrient | Unit | Value per 1 g | Value per 5 g | Value per 540 g |
|---|---|---|---|---|
| Minimum daily consumption | Maximum daily consumption | Total consumption over 6 months | ||
| Water | g | 0.0613 | 0.3065 | 33.102 |
| Energy | kcal | 3.16 | 15.8 | 1706.4 |
| Protein | g | 0.1335 | 0.6675 | 72.09 |
| Total lipid (fat) | g | 0.2085 | 1.0425 | 112.59 |
| Carbohydrate, by difference | g | 0.4969 | 2.4845 | 268.326 |
| Fiber, total dietary | g | 0.21 | 1.05 | 113.4 |
| Sugars, total | g | 0.009 | 0.045 | 4.86 |
| Minerals | ||||
| Calcium, Ca | mg | 0.57 | 2.85 | 307.8 |
| Iron, Fe | mg | 0.1854 | 0.927 | 100.116 |
| Magnesium, Mg | mg | 7.81 | 39.05 | 4217.4 |
| Phosphorus, P | mg | 16.77 | 83.85 | 9055.8 |
| Potassium, K | mg | 14.85 | 74.25 | 8019 |
| Sodium, Na | mg | 0.05 | 0.25 | 27 |
| Zinc, Zn | mg | 0.0604 | 0.302 | 32.616 |
| Vitamins | ||||
| Vitamin C, ascorbic acid | mg | 0 | 0 | 0 |
| Thiamin | mg | 0.02753 | 0.13765 | 14.8662 |
| Riboflavin | mg | 0.00284 | 0.0142 | 1.5336 |
| Niacin | mg | 0.33995 | 1.69975 | 183.573 |
| Vitamin B-6 | mg | 0.0407 | 0.2035 | 21.978 |
| Folate, DFE | µg | 0.63 | 3.15 | 340.2 |
| Vitamin B-12 | µg | 0 | 0 | 0 |
| Vitamin A, RAE | µg | 0 | 0 | 0 |
| Vitamin A, IU | IU | 0 | 0 | 0 |
| Vitamin E (alpha-tocopherol) | mg | 0.0492 | 0.246 | 26.568 |
| Vitamin D (D2 + D3) | µg | 0 | 0 | 0 |
| Vitamin D | IU | 0 | 0 | 0 |
| Vitamin K (phylloquinone) | µg | 0.019 | 0.095 | 10.26 |
| Lipids | ||||
| Fatty acids, saturated | g | 0.04171 | 0.20855 | 22.5234 |
| Fatty acids, monounsaturated | g | 0.07549 | 0.37745 | 40.7646 |
| Fatty acids, polyunsaturated | g | 0.07459 | 0.37295 | 40.2786 |
The minimum daily dose, maximum daily dose, and total dose of rice bran provided to infants in the Nicaragua and Mali randomized controlled trials.
Study participant characteristics
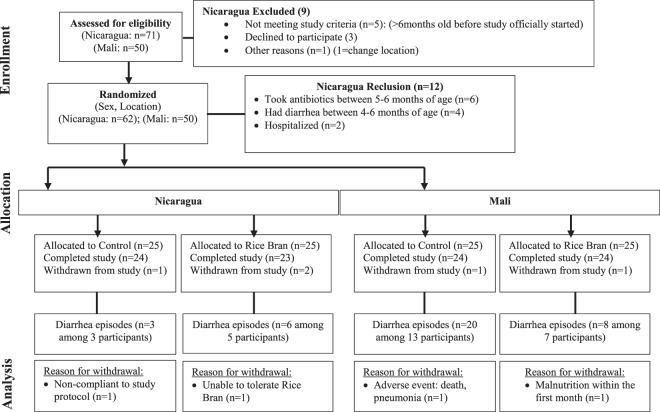
To study the effect of daily rice bran supplementation, monthly stool samples from 47 Nicaraguan and 48 Malian children were collected (average of 7 samples per child, total of 567 samples). The flow and number of infants from study recruitment to study completion is shown in Fig. 1. Baseline participant characteristics for Nicaragua and Mali are shown in Table 2. We collected information on demographic factors and infants’ household characteristics. In Nicaragua, 54.2% of infants were born via caesarean section in the control group and 30.4% in the rice bran group. All participants from Mali were delivered vaginally. For breastfeeding status, 96% of the control group and 83% in the rice bran group were consuming breast milk at six months old in Nicaragua and all children in the Mali group were consuming breast milk at the beginning and throughout the study. Of the 95 infants enrolled, 52 received antibiotics with 87 total antibiotic courses in the six-month period. Most courses consisted of systemic antibiotics given orally, with some delivered by injection for respiratory, skin, ear or diarrheal infections.
Figure 1.
Study recruitment and participation based on CONSORT statement guidelines for clinical trials conducted in Nicaragua and Mali (NCT02615886, NCT0255737315). 95 infants from León, Nicaragua and Dioro, Mali enrolled after meeting eligibility criteria, randomized by sex and location to one of two study arms. The number of diarrhea episodes and reasons for withdrawal were reported for each child.
Table 2.
Baseline infant participant characteristics from Nicaragua and Mali.
| Variable | Nicaragua | Mali | ||||
|---|---|---|---|---|---|---|
| Control (n = 24) | Rice bran (n = 23) | p-valuec | Control (n = 24) | Rice bran (n = 24) | p-valuec | |
| Sex (%) | ||||||
| Male | 14 (58.0) | 12 (52.0) | 0.6711 | 12 (50) | 12 (50) | 1 |
| Female | 10 (42.0) | 11 (48.0) | 12 (50) | 12 (50) | ||
| Water source (%) | ||||||
| Indoor municipal | 24 (100) | 23 (100) | — | 0 (0) | 0 (0) | — |
| Untreated ground water | 0 (0) | 0 (0) | — | 24 (100) | 24 (100) | — |
| Delivery type (%) | ||||||
| Vagina | 11 (45.8) | 16 (69.6) | 0.099 | 100 (100) | 100 (100) | — |
| Caesarean | 13 (54.2) | 7 (30.4) | 0 (0) | 0 (0) | ||
| Sanitation System | ||||||
| None | 0 (0) | 1 (4.3) | — | 0 (0) | 0 (0) | — |
| Community latrine | 0 (0) | 0 (0) | — | 21 (87.5) | 19 (79.2) | — |
| Latrine | 4 (16.7) | 9 (39.1) | — | 3 (12.5) | 5 (20.8) | — |
| Indoor toilet | 20 (83.3) | 13 (56.5) | — | 0 (0) | 0 (0) | — |
| Mother education (%) | ||||||
| None | 1 (4.2) | 0 (0) | — | 12 (50) | 11 (46) | — |
| Some primary | 3 (12.5) | 7 (30.4) | — | 4 (17) | 7 (29) | — |
| Completed primary | 3 (12.5) | 2 (8.7) | — | 6 (25) | 1 (4) | — |
| Some secondary | 8 (33.3) | 5 (21.7) | — | 1 (4) | 2 (8) | — |
| Completed secondary | 4 (16.7) | 5 (21.7) | — | 1 (4) | 3 (13) | — |
| University | 5 (20.8) | 4 (17.4) | — | 0 (0) | 0 (0) | — |
| Breastfeeding Status (%) | ||||||
| 6 months | 23 (95.8) | 19 (82.6) | 0.1415 | 24 (100) | 24 (100) | — |
| Antibiotic Use (6–12 months) | ||||||
| # Infants antibiotic use | 14 (58.3) | 11 (47.8) | — | 14 (58.3) | 13 (54.2) | — |
| Antibiotic courses | 21 (58.3) | 15 (41.6) | — | 26 (51.0) | 25 (49.0) | — |
| Household Animals a | ||||||
| Poultry | 3 (12.5) | 9 (37.5) | 0.3583 | 21 (88) | 21 (88) | 0.4091 |
| Livestock | 2 (8.3) | 2 (8.7) | 21 (88) | 17 (71) | ||
| Domesticated pets | 17 (70.8) | 16 (69.6) | 5 (21) | 1 (4) | ||
| None | 7 (29.2) | 5 (21.7) | 1 (4) | 2 (8) | ||
| Anthropometry b, d | ||||||
| Weight at Birth (kg) | 3.17 ± 0.39 | 2.94 ± 0.38 | 0.0505 | 3.07 ± 0.45 | 3.29 ± 0.49 | 0.1146 |
| Weight 6 months (kg) | 8.09 ± 1.10 | 7.93 ± 0.89 | 0.5833 | 7.02 ± 0.88 | 7.14 ± 0.99 | 0.4832 |
| Length Birth (cm) | 50.67 ± 1.93 | 49.55 ± 3.03 | 0.1472 | 49.77 ± 1.56 | 50.50 ± 2.04 | 0.1789 |
| Length 6 months (cm) | 66.38 ± 2.10 | 66.26 ± 2.90 | 0.8787 | 65.57 ± 2.56 | 66.56 ± 3.12 | 0.2960 |
| LAZ 0 months (cm) | 0.87 ± 0.93 | 0.37 ± 1.60 | 0.2045 | 0.12 ± 0.88 | 0.45 ± 1.18 | 0.3289 |
| LAZ 6 months (cm) | −0.03 ± 0.82 | 0.07 ± 1.29 | 0.7325 | −0.30 ± 1.70 | −0.15 ± 1.46 | 0.7427 |
| WAZ 0 months (cm) | −0.30 ± 0.85 | −0.82 ± 0.89 | 0.0497 | −0.48 ± 1.04 | 0.02 ± 1.06 | 0.1389 |
| WAZ 6 months (cm) | 0.33 ± 1.09 | 0.27 ± 0.98 | 0.8412 | −0.65 ± 1.29 | −0.64 ± 1.09 | 0.9827 |
| WLZ 0 months (cm) | −1.54 ± 1.41 | −2.07 ± 1.63 | 0.2585 | −0.92 ± 1.31 | −0.39 ± 1.49 | 0.2571 |
| WLZ 6 months (cm) | 0.53 ± 1.25 | 0.41 ± 0.96 | 0.7107 | −0.51 ± 0.95 | −0.58 ± 1.05 | 0.7963 |
aMore than one category may be represented per household. bMean ± standard deviation. cp-value:Chi-squared test. dAnthropometric p-values calculated by two-sample t-test.
Anthropometric growth measurements
Anthropometric data were collected using standardized procedures across study sites at 6, 8, and 12 months of age for each child and included length-for-age Z-score (LAZ), weight-for-age Z-score (WAZ) and weight-for-length Z-score (WLZ). Table 3 reports the z-scores analyzed for each country by repeated measures and adjusted by treatment (rice bran) and the rate of growth was significantly changed by age groups 6–8 months and 8–12 months. Significant differences were observed for anthropometric measures between treatment and ages in both cohorts. In Nicaraguan infants who consumed rice bran, LAZ was significantly changed over time (p-value = 0.0002) and for WLZ at 6–8 months (p-value < 0.0001). The Malian infants who consumed rice bran had significant growth results for WAZ (6–8 months, p-value = 0.0001, 8–12 months, p-value = 0.0175) and WLZ (6–8 months, p-value = 0.0141, 8–12 months, p-value = 0.0134). Figure 2 displays LAZ, WAZ and WLZ for each age group and by country. The significant changes observed in LAZ at 8 months and 12 months in Nicaraguan infants who consumed rice bran was compared to control group (Fig. 2A, p < 0.01). No significant differences were observed for WAZ and WLZ with this control group comparison (Fig. 2B,C).
Table 3.
Anthropometric measures adjusted by treatment and age in Nicaraguan and Malian Infants.
| Indicator | Nicaragua | Mali | ||||||
|---|---|---|---|---|---|---|---|---|
| Control | Rice Bran | Control | Rice Bran | |||||
| n = 24a | p-valueb | n = 23a | p-valueb | n = 24a | p-valueb | n = 24a | p-valueb | |
| Length-for-age Z-score | ||||||||
| Months | ||||||||
| 6 | −0.03 (0.17) | 0.8689 | 0.07 (0.27) | <0.0001 | −0.30 (0.35) | 0.9165 | −0.15 (0.30) | 0.1111 |
| 8 | −0.13 (0.14) | 1.18 (0.26) | −0.20 (0.23) | 0.19 (0.25) | ||||
| 0.0098 | 0.0002 | 0.1609 | 0.6229 | |||||
| 12 | −0.73 (0.18) | 0.35 (0.21) | −0.58 (0.22) | 0.01 (0.22) | ||||
| Weight-for-age Z-score | ||||||||
| Months | ||||||||
| 6 | 0.33 (0.22) | 0.5648 | 0.27 (0.20) | 0.3335 | −0.65 (0.26) | 0.4575 | −0.64 (0.22) | <0.0001 |
| 8 | 0.22 (0.23) | 0.11 (0.20) | −0.44 (0.23) | −0.02 (0.21) | ||||
| 0.0558 | 0.9919 | <0.0001 | 0.0175 | |||||
| 12 | −0.03 (0.20) | 0.11 (0.18) | −1.10 (0.24) | −0.44 (0.21) | ||||
| Weight-for-length Z-score | ||||||||
| Months | ||||||||
| 6 | 0.53 (0.26) | 0.8997 | 0.41 (0.20) | <0.0001 | −0.51 (0.19) | 0.8419 | −0.58 (0.21) | 0.0141 |
| 8 | 0.44 (0.28) | −0.54 (0.22) | −0.36 (0.21) | −0.05 (0.20) | ||||
| 0.9892 | 0.0569 | <0.0001 | 0.0134 | |||||
| 12 | 0.41 (0.22) | −0.06 (0.18) | −1.18 (0.24) | −0.59 (0.22) | ||||
aMean (SEM). bAjusted p-value by repeated measures for each treatment and time point (6–8 and 8–12 months) LAZ: Length for Age Z-score, WAZ: Weight for Age Z-score, WLZ: Weight for Length Z-score.
Figure 2.
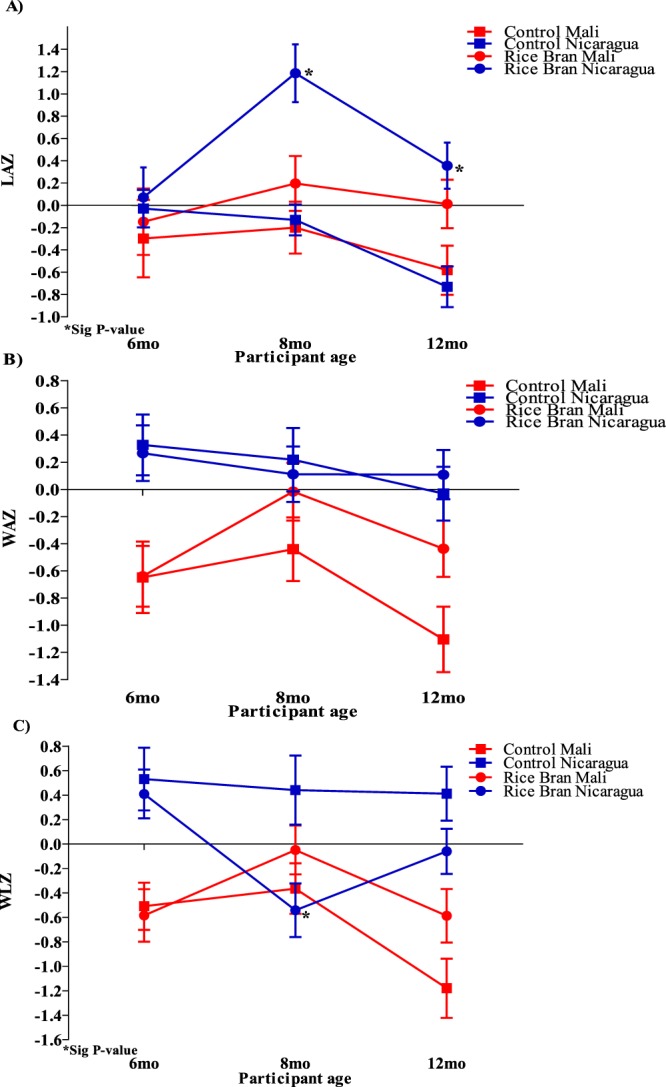
Anthropometric Z-scores for Nicaraguan and Malian infants in rice bran and control groups at 6, 8 and 12 months. (A) Significant LAZ (p < 0.05) at 8 and 12 months in the rice bran group compared to control for Nicaraguan infants. (B) No WAZ significant changes between rice bran and control group in Nicaraguan and Malian infants. (C) WLZ at 8 months was significantly lower for the rice bran group compared to control in Nicaragua.
Diarrheal episodes and stool environmental enteric dysfunction (EED) biomarkers
For the Mali cohort, the diarrheal disease incidence was 33% in the rice bran intervention group compared to 79% in the control group over the 6-month study period. There was one rice bran intervention participant who experienced a single repeat episode of diarrhea, and 5 control group participants experienced between 2 and 4 repeat episodes of diarrhea during the 6-month trial. There were a higher number of diarrheal episodes in the Mali control group infants when compared to rice bran intervention group. No differences were detected for diarrheal episodes for the Nicaragua rice bran and control groups. We next evaluated four EED stool biomarkers at 6, 8 and 12 months of age using ELISA (see materials and methods). A significant decrease in AAT was observed at 12 months of age (p = 0.0368) in Nicaragua infants who consumed rice bran compared to control (Table 4). No significant differences were detected for the three other surrogate markers NEO, MPO and CAL between treatment groups. There was, however, a slight decrease observed in the median concentration of all stool EED biomarkers in the rice bran group compared to control for both countries.
Table 4.
Environmental enteric dysfunction (EED) biomarkers in stool at 6, 8, and 12 months of age for Nicaraguan and Malian infants.
| EED Biomarker | Nicaragua | Mali | ||||
|---|---|---|---|---|---|---|
| Controla (n = 24) | Rice Brana (n = 23) | p-valueb | Controla (n = 24) | Rice Brana (n = 24) | p-valueb | |
| Neopterin (nmol/L) | ||||||
| 6 | 150.8 (182.2) | 208.6 (131.7) | 0.7220 | 20.2 (31.0) | 34.6 (28.0) | 0.5929 |
| 8 | 222.3 (241.1) | 144.8 (196.9) | 0.5771 | 36.2 (27.6) | 28.9 (28.5) | 0.4314 |
| 12 | 137.5 (285.2) | 182.4 (230.4) | 0.8727 | 12.0 (33.0) | 36.0 (32.6) | 0.9880 |
| Myeloperoxidase (ng/ml) | ||||||
| 6 | 277.0 (374.5) | 237.5 (376.5) | 0.0847 | 3970.6 (17776.6) | 5400.9 (20794.8) | 0.6139 |
| 8 | 331.1 (312.3) | 266.3 (236.4) | 0.8454 | 15838.7 (20511.9) | 7451.0 (12972.7) | 0.3763 |
| 12 | 182.0 (324.8) | 158.5 (376.7) | 0.3345 | 4846.4 (11266.9) | 3153.9 (14095.2) | 0.5437 |
| Calprotectin (µg/g) | ||||||
| 6 | 32.2 (108.7) | 88.0 (281.1) | 0.2394 | 51.7 (101.2) | 53.3 (106.4) | 0.7136 |
| 8 | 24.7 (87.5) | 58.2 (213.2) | 0.8023 | 130.0 (769.7) | 68.7 (126.8) | 0.0639 |
| 12 | 24.0 (74.4) | 50.5 (133.9) | 0.2629 | 35.0 (70.2) | 20.9 (56.0) | 0.2103 |
| Alpha-1 Antitrypsin (ng/ml) | ||||||
| 6 | 130.1 (177.7) | 109.5 (217.4) | 0.7199 | 247.5 (499.6) | 463.1 (891.8) | 0.0999 |
| 8 | 152.0 (115.4) | 73.5 (122.4) | 0.1221 | 619.2 (759.0) | 579.9 (899.9) | 0.2237 |
| 12 | 130.9 (129.8) | 70.8 (87.8) | 0.0368 | 663.7 (580.5) | 453.2 (807.5) | 0.4796 |
aMedian (IQR).
bp-value by repeated measures comparing treatments at each time point.
Infant gut microbiota
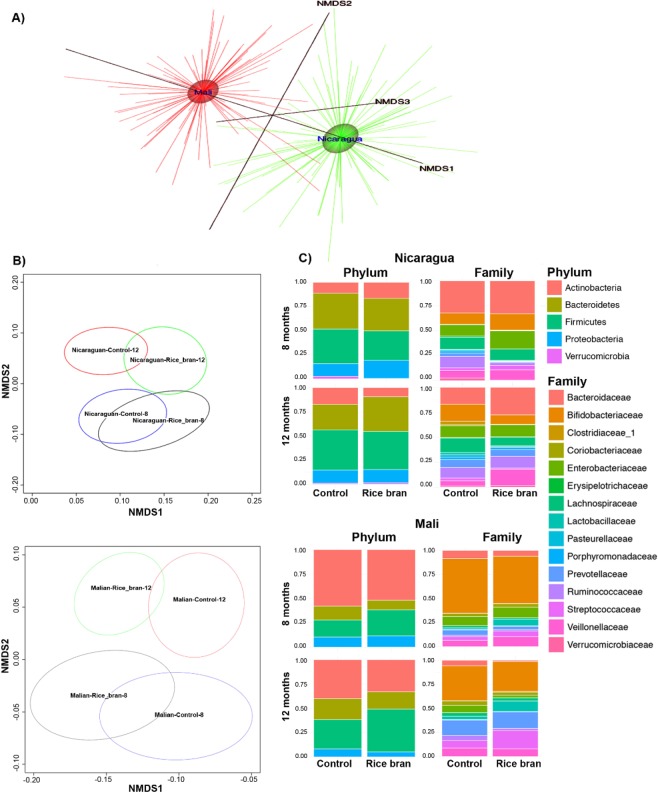
The microbiota were characterized and compared for 48 Malian and 47 Nicaraguan infants at 8 and 12 months of age in the rice bran and control study groups. DNA was isolated from stool samples and the V4 hypervariable region of the 16S rRNA gene was sequenced utilizing the Earth Microbiome Project protocol43–47. Sequences were preprocessed for quality assurance and classified into operational taxonomic units (OTUs) (see materials and methods), and the results were integrated to construct family and genus-level composition profiles for all 192 samples from both countries. No major differences were detected in alpha diversity indices (Observed, Shannon, InvSimpson and Richness) calculated for rice bran group and control group at 8 and 12 months (Table S1). A complete country-level separation in the overall gut microbial community composition was depicted for beta diversity using Nonmetric Multidimensional Scaling (NMDS) plot based on the Bray-Curtis distance measure (Fig. 3A). The magnitude of age and country-level differences in infant gut microbiota composition was expected, as differences were previously reported that compared these geographical regions21. The differences in microbiota composition at baseline provided compelling rationale for separately evaluating the control and rice bran responsive microbial communities in infants at each age and for each country.
Figure 3.
Rice bran and control infant stool microbiota at 8 and 12 months of age in Nicaragua and Mali. Nonmetric Multidimensional Scaling (NMDS) for (A). Nicaragua and Mali all samples (B). Control groups and rice bran groups at 8 and 12 months. NMDS was used on the OTU level to assess possible trends and clustering in the microbial community structure per treatment and time point. (C) Bacterial taxa at phylum and family level in Nicaragua (top) and Mali (bottom). Bar-graphs show phylum and family relative abundance based on the resulting OTU table generated using the ggplot2 package in R. These plots were generated for the data at the phylum and the family levels and meant to describe the microbial community structure per sampled group and per time point (8 months and 12 months) under each of the treatment levels (control and rice bran).
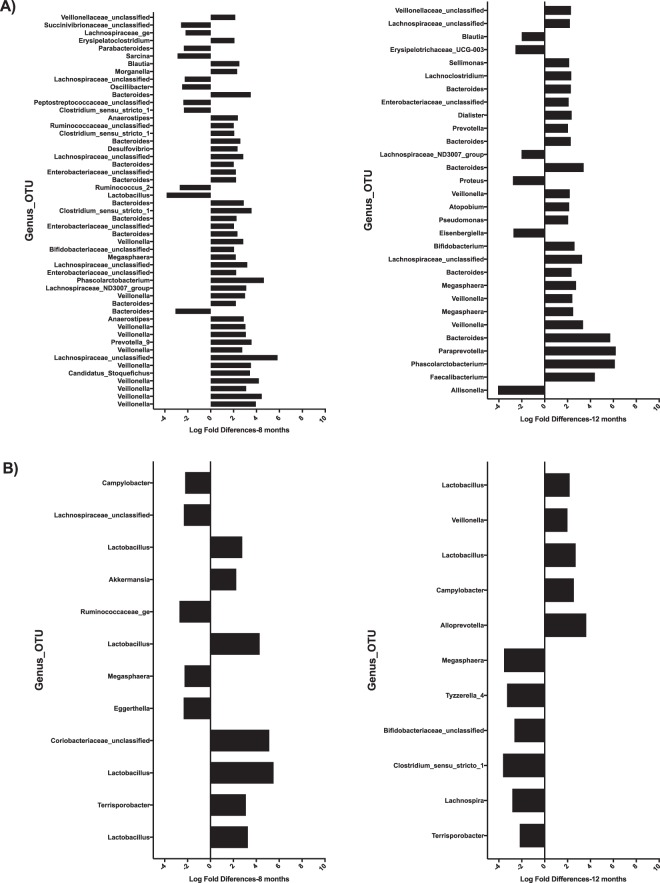
Figure 3B shows the NMDS plot separated by country. This figure highlights differences in the microbiota between two time periods, 8 and 12 months, which is more pronounced in the Malian samples. There is overlap of microbiota within each time period that indicates putative similarity between microbial community structures during growth. The overlap was observed to a greater level at 8 months of age as compared to 12 months that may illustrate microbial adaptation to new exposures33. Figure 4 illustrates the OTU with at least 2 log-fold change between the rice bran and control groups per country and per age group. These OTU were ordered based on statistical significance (FDR-adjusted p-value, from bottom (lowest p-value) to top). See additional details provided in Table S2 for Nicaragua and Table S3 for Mali. The effects of daily dietary rice bran supplementation on the infant microbiota were evaluated at 8 months of age for each country because this was the time point in which measurable growth differences (LAZ) were already observed for the rice bran group compared to control.
Figure 4.
Microbiota differences between Nicaragua and Mali at 8 and 12 months between rice bran and control groups. Fold differences in relative percentage of OTUs different between control and rice bran groups at 8 months and 12 months. (A) Nicaragua, and (B) Mali. OTUs with fold difference more than 2 are shown for infants at 8 months (left) and 12 months (right). Fold difference for OTUs with FDR less than 0.05 is shown with the most significant OTUs on the bottom of each graph.
In Nicaragua, differential abundance testing led to the identification of 145 OTUs that were significantly different between control and rice bran (Table S2). There were 74 OTUs that showed greater than or equal to 2 log fold differences between rice bran and control groups at 8 or 12 months (Fig. 4A, adjusted p-value < 0.05). Seven of these 74 OTUs overlapped between the ages 8 and 12 months. In Mali, 42 bacterial OTUs were identified to significantly differ between control and rice bran across samples, and 19 showed more than 2 log fold changes between rice bran and control at 8 or 12 months (Fig. 4B, adjusted p-value < 0.05) with only three overlapping between the two age groups. Next, we explored the genus level assignments that were responsive to rice bran intake for each country. For Nicaraguan infants (Table S2), the notable rice bran responsive taxa which changed at 8 months of age compared to control group were Lachnospiraceae-unclassified-Otu0280 (log-FC 5.84, adjusted p-value 3.95E-08) Bifidobacterium-unclassified-Otu0314 (log-FC 2.04, adjusted p-value 1.38E-6), Ruminococcaceae-unclassified-Otu0238 (log-FC 2.01, adjusted p-value 0.00097), Veillonella (11different OTUs, log-FC > 2.0, adjusted p-value < 0.05), and Bacteroides (log-FC > 2.0, adjusted p-value < 0.05). The fold difference for genus level taxa that were lower in relative percent abundance for rice bran group were Bacteroides-Otu0192 (log-FC −3.08, adjusted p-value 2.34E-07), Parabacteroides-Otu0086 (log-FC −2.34, adjusted p-value 0.0074), Lachnospiraceae-unclassified-Otu0174 (log-FC −2.27, adjusted p-value 0.0033), Lactobacillus-Out0053 (log-FC −3.85, adjusted p-value 1.09E-05), Oscillibacter (log-FC −2.49, adjusted p-value 0.0029) and Ruminococcaceae_2 (log-FC −2.69, adjusted p-value 1.98E-05).
In Mali infants, (Table S3), there were 2-fold increased differences observed for rice bran fed infants in Lactobacillus-Out0356 (log-FC 3.2, adjusted p-value 1.35E-09) and decreased for Lachnospiraceae-unclassified-Otu0010 (log-FC −2.3, adjusted p-value 0.016). The infant gut microbiota showed changes that were observed by age and for each country with respect to rice bran intake. At 12 months of age, the Nicaragua rice bran group had increased Paraprevotella (log-FC 6.2, adjusted p-value 4.27E-08), Phascolarctobacterium (log-FC 6.12, adjusted p-value 1.60E-08) Veillonella (log-FC 3.35, adjusted p-value 3.30E-07) and Bifidobacterium (log-FC 2.6, adjusted p-value 1.44E-05). Lower abundant taxa in rice bran infants at 12 months of age from Nicaragua were Lachnospiraceae_ND3007_group (log-FC −2.0, adjusted p-value 0.00029) and Alisonella (log-FC −4.0, adjusted p-value 1.60E-08). Malian rice bran fed infants at 12 months of age showed increased Lactobacillus_Otu0053 (log-FC 2.7, adjusted p-value 0.0098) and Alloprevotella (log-FC 3.6, adjusted p-value 0.00034). The significant changes observed for Malian infants between rice bran and control groups at 12 months were Bifidobacteriaceae_unclassified_Otu0265 (log-FC −2.6, adjusted p-value 3.95E-05), Clostridium_sensu_stricto_1_Otu0076, and Terrisoporobacter (log-FC −2.1, adjusted p-value 7.03E-06).
Gut microbiota changes that were reflective of a dietary response to rice bran in both the Mali and Nicaragua cohorts involved 12 OTUs at either 8 or 12 months of age when compared to control. The highest area of overlap occurred for both Lactobacillaceae_Lactobacillus_Otu0024 and the Lactobacillus_Otu0053 (Table S4). There was additional overlap in both countries with respect to microbiota changes following rice bran intake that included Bifidobacterium, Faecalibacterium, and Lachnospiriaceae.
Infant stool metabolomes
A total of 309 stool samples were analyzed for metabolomics from this 6-month prospective study to evaluate the effects of rice bran supplementation compared to control. Stool metabolite analysis at 8 months of age in Nicaraguan and Malian infants resulted in the detection of 1449 biochemicals, of which 1016 metabolites had confirmed names and 433 compounds were of unknown structural identity (see Table S5). ANOVA contrasts and Welch’s two-sample t-test were used to identify biochemicals that significantly differed between experimental groups at 8-months of age. Table 5 lists the fold differences in the stool metabolite relative abundance between study diet groups at 8 months of age. There are 39 (Nicaragua) and 44 (Mali) stool metabolites with significant fold differences between children consuming rice bran for two months when compared to control. There were also 33 unknown metabolites for Nicaragua and 31 unknown metabolites for Mali that showed significant differences between groups (data shown in Table S5). Significant fold differences occurred for 15 amino acids, 2 peptides, 3 carbohydrates, 9 lipids, 1 cofactor and vitamin, and 9 xenobiotics (six of these metabolites were considered to be food components/plant-derived) in Nicaraguan children who consumed rice bran compared to control. At 8 months of age, the stool from Mali infants had 6 amino acids, 1 energy metabolite, 14 lipids, 6 cofactor and vitamins, 5 nucleotides and 12 xenobiotics (7 classified as food components/ plant-derived) that were significantly different between rice bran and control (Table 5).
Table 5.
Stool metabolites significantly modulated by rice bran supplementation compared to control for Nicaragua & Mali infants at 8 months of age.
| Metabolic Pathway | Metabolitea | HMDBb | Nicaragua | Mali | ||
|---|---|---|---|---|---|---|
| Fold Diffc | p-value | Fold Diffc | p-value | |||
| Cofactors and Vitamins | ||||||
| Nicotinate and Nicotinamide Metabolism | Quinolinate | HMDB00232 | 0.88 | 0.7272 | 0.44 | 0.0313 |
| Nicotinate | HMDB01488 | 1.15 | 0.3958 | 1.6 | 0.0053 | |
| Tocopherol Metabolism | alpha-tocotrienol | HMDB06327 | 0.69 | 0.3922 | 3.01 | 0.0168 |
| gamma-tocotrienol | HMDB12958 | 0.67 | 0.1909 | 2.53 | 0.0044 | |
| gamma-CEHC glucuronide* | 0.41 | 0.0035 | 0.96 | 0.8862 | ||
| Vitamin B6 Metabolism | pyridoxine (Vitamin B6) | HMDB02075 | 1.58 | 0.3051 | 4.65 | 0.0011 |
| Pyridoxate | HMDB00017 | 0.86 | 0.5441 | 2.34 | 0.0014 | |
| Xenobiotics | ||||||
| Benzoate Metabolism | 4-hydroxybenzoate | HMDB00500 | 0.88 | 0.6260 | 1.8 | 0.0272 |
| methyl-4-hydroxybenzoate | HMDB32572 | 0.57 | 0.0299 | 0.9 | 0.7061 | |
| 3-(4-hydroxyphenyl)propionate | HMDB02199 | 1.08 | 0.8898 | 6.94 | 0.0007 | |
| Xanthine Metabolism | Theophylline | HMDB01889 | 0.86 | 0.6266 | 0.44 | 0.0091 |
| Food Component/Plant | Indoleacetylaspartate | HMDB38666 | 1.1 | 0.7543 | 2.14 | 0.0134 |
| Vanillate | HMDB00484 | 0.82 | 0.5775 | 2.27 | 0.0246 | |
| deoxymugineic acid | 0.6 | 0.2733 | 4.5 | 0.0021 | ||
| dihydroferulic acid | 0.63 | 0.4522 | 6.99 | 0.0032 | ||
| Ferulate | HMDB00954 | 1.24 | 0.6403 | 3.5 | 0.0100 | |
| ferulic acid 4-sulfate | HMDB29200 | 1.14 | 0.8494 | 4.88 | 0.0274 | |
| ferulylglycine (1) | 0.4 | 0.0256 | 2.16 | 0.0665 | ||
| Rosmarinate | HMDB03572 | 0.57 | 0.0206 | 1.28 | 0.3008 | |
| Tyrosol | HMDB04284 | 1.01 | 0.9832 | 1.98 | 0.0412 | |
| Diosmetin | HMDB29676 | 0.29 | 0.0192 | 1.25 | 0.6872 | |
| daidzein sulfate (2) | 0.39 | 0.0123 | 1.38 | 0.4129 | ||
| daidzein sulfate (1) | 0.35 | 0.0023 | 1.04 | 0.9083 | ||
| Salicylate | HMDB01895 | 1.78 | 0.0970 | 4.67 | 0.0000 | |
| N-propionylmethionine | 1.09 | 0.8631 | 3.6 | 0.0111 | ||
| malonylgenistin | 0.51 | 0.0023 | 0.99 | 0.9670 | ||
| Drug - Analgesics, Anesthetics | 4-acetamidophenylglucuronide | HMDB10316 | 0.99 | 0.0484 | 1 | 1.0000 |
| 2-methoxyacetaminophen glucuronide* | 0.77 | 0.0049 | 1 | 1.0000 | ||
| Amino Acid | ||||||
| Glycine, Serine and Threonine Metabolism | Glycine | HMDB00123 | 0.66 | 0.0062 | 1.2 | 0.2384 |
| dimethylglycine | HMDB00092 | 0.69 | 0.2880 | 0.42 | 0.0153 | |
| Lysine Metabolism | N6-formyllysine | 0.6 | 0.1944 | 2.37 | 0.0385 | |
| Phenylalanine Metabolism | phenylpyruvate | HMDB00205 | 0.59 | 0.0332 | 1.18 | 0.5305 |
| phenyllactate (PLA) | HMDB00779 | 0.44 | 0.0223 | 1.49 | 0.2855 | |
| Tyrosine Metabolism | 4-hydroxyphenylpyruvate | HMDB00707 | 0.55 | 0.0210 | 0.92 | 0.7455 |
| vanillic alcohol sulfate | 0.7 | 0.2572 | 2.01 | 0.0378 | ||
| Tryptophan Metabolism | kynurenate | HMDB00715 | 0.48 | 0.0079 | 1.13 | 0.6565 |
| N-formylanthranilic acid | HMDB04089 | 1.2 | 0.5416 | 0.51 | 0.0321 | |
| indolepropionate | HMDB02302 | 4.67 | 0.0189 | 1.33 | 0.6727 | |
| Leucine, Isoleucine and Valine Metabolism | alpha-hydroxyisocaproate | HMDB00746 | 0.45 | 0.0325 | 1.05 | 0.8904 |
| alpha-hydroxyisovalerate | HMDB00407 | 0.47 | 0.0335 | 1.08 | 0.8323 | |
| 3-methyl-2-oxobutyrate | HMDB00019 | 0.52 | 0.0299 | 1.4 | 0.2751 | |
| 2-hydroxy-3-methylvalerate | HMDB00317 | 0.46 | 0.0224 | 1.14 | 0.7078 | |
| Methionine, Cysteine, SAM and Taurine Metabolism | N-acetylmethionine | HMDB11745 | 0.89 | 0.8154 | 3.24 | 0.0214 |
| N-formylmethionine | HMDB01015 | 0.56 | 0.2035 | 2.84 | 0.0294 | |
| cysteine | HMDB00574 | 0.66 | 0.0498 | 1.29 | 0.2445 | |
| hypotaurine | HMDB00965 | 0.52 | 0.0439 | 0.9 | 0.7512 | |
| Urea cycle; Arginine and Proline Metabolism | dimethylarginine (SDMA + ADMA) | HMDB01539 | 0.53 | 0.0064 | 0.92 | 0.7445 |
| Glutathione Metabolism | 5-oxoproline | HMDB00267 | 0.5 | 0.0085 | 1.27 | 0.3819 |
| 2-hydroxybutyrate/2-hydroxyisobutyrate | 0.45 | 0.0153 | 0.73 | 0.3554 | ||
| Peptide | ||||||
| Gamma-glutamyl Amino Acid | gamma-glutamylglutamine | HMDB11738 | 0.49 | 0.0278 | 1.05 | 0.8844 |
| gamma-glutamyl-epsilon-lysine | HMDB03869 | 0.49 | 0.0137 | 0.84 | 0.5553 | |
| Carbohydrate | ||||||
| Glycolysis, Gluconeogenesis, and Pyruvate Metabolism | pyruvate | HMDB00243 | 0.46 | 0.0237 | 1.26 | 0.5202 |
| Disaccharides and Oligosaccharides | 3-sialyllactose | HMDB00825 | 0.8 | 0.0484 | 1 | 1.0000 |
| Lewis a trisaccharide | 1.36 | 0.0443 | 0.99 | 0.9520 | ||
| Energy | ||||||
| TCA Cycle | alpha-ketoglutarate | HMDB00208 | 0.65 | 0.1785 | 1.96 | 0.0484 |
| Lipid | ||||||
| Fatty Acid, Dicarboxylate | pimelate (C7-DC) | HMDB00857 | 0.93 | 0.7811 | 2.13 | 0.0078 |
| Fatty Acid Metabolism (Acyl Choline) | palmitoloelycholine | 1.17 | 0.1301 | 0.74 | 0.0070 | |
| linoleoylcholine* | 1.85 | 0.0459 | 1.5 | 0.2118 | ||
| Fatty Acid, Monohydroxy | 8-hydroxyoctanoate | HMDB61914 | 0.75 | 0.1576 | 1.61 | 0.0224 |
| Fatty Acid, Dihydroxy | 12,13-DiHOME | HMDB04705 | 0.89 | 0.7839 | 2.78 | 0.0246 |
| 9,10-DiHOME | HMDB04704 | 0.9 | 0.7816 | 3.75 | 0.0012 | |
| Monoacylglycerol | 1-linolenoylglycerol (18:3) | HMDB11569 | 2.36 | 0.0457 | 1.01 | 0.9871 |
| Diacylglycerol | linoleoyl-linolenoyl-glycerol (18:2/18:3) [1]* | HMDB07249 | 2.04 | 0.0285 | 1.18 | 0.6201 |
| linolenoyl-linolenoyl-glycerol (18:3/18:3) [2]* | HMDB07278 | 2.29 | 0.0404 | 0.76 | 0.5121 | |
| linoleoyl-docosahexaenoyl-glycerol (18:2/22:6) [1]* | 1.03 | 0.8045 | 0.66 | 0.0006 | ||
| linoleoyl-docosahexaenoyl-glycerol (18:2/22:6) [2]* | HMDB07266 | 0.98 | 0.9291 | 0.59 | 0.0165 | |
| Sphingolipid Metabolism | N-acetylsphingosine | HMDB04950 | 1.11 | 0.7987 | 2.44 | 0.0387 |
| Mevalonate Metabolism | 3-hydroxy-3-methylglutarate | HMDB00355 | 0.74 | 0.4206 | 2.77 | 0.0098 |
| Sterol | beta-sitosterol | HMDB00852 | 0.77 | 0.3699 | 2.89 | 0.0006 |
| stigmasterol | HMDB00937 | 1.14 | 0.6191 | 2.01 | 0.0159 | |
| campesterol | HMDB02869 | 0.86 | 0.6689 | 2.1 | 0.0415 | |
| Androgenic Steroids | 5alpha-androstan-3alpha,17alpha-diol disulfate | 0.56 | 0.0199 | 0.63 | 0.0625 | |
| androstenediol (3beta,17beta) disulfate (2) | HMDB03818 | 0.71 | 0.2185 | 0.54 | 0.0324 | |
| Primary Bile Acid Metabolism | glycocholate | HMDB00138 | 0.39 | 0.0483 | 1.37 | 0.5237 |
| glycochenodeoxycholate | HMDB00637 | 0.43 | 0.0434 | 0.64 | 0.3052 | |
| glycochenodeoxycholate glucuronide (2) | 0.37 | 0.0195 | 0.78 | 0.5717 | ||
| glycochenodeoxycholate sulfate | 0.57 | 0.2283 | 0.35 | 0.0284 | ||
| Secondary Bile Acid Metabolism | 7alpha-hydroxycholestenone | HMDB01993 | 0.64 | 0.0411 | 0.88 | 0.5716 |
| Nucleotide | ||||||
| Purine Metabolism, Adenine containing | N6-dimethylallyladenine | 0.94 | 0.8249 | 0.28 | 0.0000 | |
| Purine Metabolism, Guanine containing | Guanine | HMDB00132 | 0.47 | 0.0876 | 3.22 | 0.0112 |
| Pyrimidine Metabolism, Orotate containing | N-carbamoylaspartate | HMDB00828 | 1.14 | 0.5814 | 0.59 | 0.0326 |
| Pyrimidine Metabolism, Uracil containing | uridine-2′,3′-cyclic monophosphate | HMDB11640 | 1.27 | 0.3081 | 1.75 | 0.0243 |
| Pyrimidine Metabolism, Thymine containing | 3-aminoisobutyrate | HMDB03911 | 1.23 | 0.6961 | 0.27 | 0.0175 |
aTable displays metabolites with statistically significant differences between rice bran and control group in stool. Bold metabolites are present in the rice bran (Calrose) that the children consumed.
bHMDB refers to the Human Metabolome Database.
cFold differences (Fold Diff) between study groups was calculated by dividing the scaled relative abundance of rice bran vs control.
There were 62 stool metabolites from the Nicaraguan cohort at 8 months that had lower relative abundances, and there were 10 stool metabolites with significantly increased fold differences in abundance between groups. The stool metabolites of food and nutritional relevance that were associated with increasing intake of rice bran were classified to the tryptophan metabolism pathway (e.g. indolepropionate), as well as monoacylglycerol (1-linolenoylglycerol) and diacylglycerol metabolic (linoleoyl-linolenoyl-glycerol) pathways.
In contrast to Nicaragua, there were 54 distinct stool metabolites from the Mali cohort that increased with significant fold differences at 8 months between rice bran and control infants. Other stool metabolites with gut health and nutritional importance that originated from rice bran food intake, included alpha-tocotrienol (vitamin E component), pyridoxine (vitamin B6), ferulic acid 4-sulfate, tyrosol, and N-acetyl sphingosine (Table 5). An estimated false discovery rate (q-value) was calculated to account for the multiple comparisons across metabolites that are typical of metabolomic-based studies.
Discussion
This study demonstrated that rice bran supplementation was feasible and safe for weaning infants with strong compliance in two distinct LMIC countries. Rice bran was well tolerated in a dose-escalation, supplementation trial regimen during the first 6 months of weaning, and was without side effects or adverse interactions. The distinct agro-ecological climates, dietary patterns, and birth methods between infants in West Africa and Latin America were considered highly relevant sources of variation that added scientific rigor to the generalizability of these clinical trial results with respect to safety, feasibility, and tolerability. However, when evaluating the infant gut microbiome and stool metabolite responses to increasing rice bran intake, this study showed distinct country and age specific variability in weaning infants. Rice bran supplementation in the diet supported the healthy growth of Nicaraguan and Malian infants with changes detected in LAZ between 6–8 months and 8–12 months of age. Nicaraguan infants had reduced gut permeability as measured by AAT at 12 months. Malian infants also showed changes over time for WAZ. The 95 healthy infants enrolled in this study had slightly higher WAZ, LAZ and WLZ scores than the documented country-wide averages that use the WHO scoring index12.
There is a growing body of scientific evidence for a strong relationship between EED and growth deficits in children7,10,48. EED biomarkers were selected in this study because high concentrations of stool AAT and MPO were associated with decreased growth in children48, Naylor et al. found that elevated AAT levels were associated with decreased oral rotavirus vaccine response49, and were associated with seroconversion to rotavirus vaccination in Nicaraguan infants37. Rice bran was shown to reduce AAT in neonatal pigs challenged with human Rotavirus infection24 and was a finding of translational importance to this clinical trial because rice bran intake improved AAT levels in Nicaraguan infants (Table 4). The lack of any statistically significant differences in EED stool biomarkers for Mali infants may be due to the higher number of overall diarrheal episodes that this cohort experienced, the level of variability within individuals, and breastfeeding status for all infants throughout the study period. Our findings did concur with EED biomarker concentrations reported in the MAL-ED cohort36. EED biomarkers merit continuous review for relevance with anthropometric measures due to extensive global variability in concentrations reported across studies6,50–52.
A study limitation and possible confounder of rice bran effects to modulate the infant gut microbiota was the percentage of exclusively breastfed infants and the number of vaginal deliveries being lower in the Nicaragua cohort. This was in contrast to the Mali cohort that had 100% of children who were breastfed and had vaginal delivery. These are key considerations to evaluating microbiota that has been previously characterized as less mature53, and with varied structure by geographical location54, diet deficiencies55–59, environmental exposures54,60,61 and host factors60,62. Infants from each country showed favorable changes in both the stool microbiota and metabolome over time (with increasing age and growth), and between rice bran and control groups at 8 months of age.
Given that dietary rice bran intake has been previously shown to promote beneficial stool microbial communities, such as native gut probiotics in mice25,63, pigs23,24,31 and adults27,64,65, differential abundance testing at the genus level assignment between rice bran and control fed infants was conducted at 8 months and 12 months of age. In Nicaragua, the Lactobacillus, Lachnospiraceae, Bifidobacterium, Ruminococcaceae and Veillonella were identified as responsive following rice bran consumption compared to an age and control matched group. These taxa have recognized saccharolytic mechanisms of action66–68, are known to produce and promote crossfeeding of short chain fatty acids69,70, as well as provide competitive inhibition of pathogen colonization71,72. The microbial enrichment of these communities has important implications for assessing how gut microbes metabolize rice bran that can support changes to infant LAZ growth outcomes. In Mali, the observed increase in the relative abundance of Lactobacillus in rice bran fed infants was consistent with prior studies of rice bran feeding to young animals. This finding was considered alongside evidence for human milk oligosaccharides that also promote Lactobacillus in breastfed infants73. Rice bran metabolism by Lactobacillus spp. was shown to produce a suite of small molecules that may be absorbed by the host or detected in stool. Stool metabolites from infants fed rice bran showed significant fold differences amongst several essential amino acids, cofactors and vitamins, lipids, phytochemicals, and in energy metabolism pathways when compared to the control. The stool metabolites that were highly likely to have originated from the rice bran consumed by both Nicaragua and Malian infants provided additional confirmation of compliance to the dietary intervention74. As predicted, we observed and reported exceptionally distinct profiles for both the stool microbiota and metabolome composition of infants between Mali and Nicaragua at all ages41,54,75 and therefore separately discussed the response to rice bran supplementation by each region. For example, the nearly 5-fold increase of stool indolepropionate in rice bran infants compared to control from Nicaragua represents a tryptophan metabolite produced by the gut microbiota that may influence the developing immune system and intestinal homeostasis76,77. Increased levels of N-acetylmethionine (nutritionally and metabolically equal to L-methionine) and N-formylmethionine in Malian infants also represented rice bran derived amino acids required for normal growth and development78. There are several cofactors and vitamins from rice bran supplementation, such as alpha and gamma-tocotrienol and pyridoxine (vitamin B6) that merit attention for demonstrating multiple health benefits such as synthesis of amino acids and neurotransmitter precursors, as well as preventing anemia and skin problems74,79,80. Additional microbial digested food components in the stool metabolome that come from rice bran were ferulic acid 4-sulfate, indoleacetylaspartate, and Tyrosol. This trial had only supplemented the rice bran for a 6 month window and according to WHO, growth assessment should be standardized and compared globally over the first two years of life81. We put forth that rice bran metabolism by host and gut microbes between 12–36 months of age should be captured in future assessments of early influences on growth velocity81,82. The focus on changes between 8 and 12 months in this study showed modifications in gut microbial communities and metabolites by rice bran intake, and suggests there will be long-term impact on the overall microbiota composition as it continues to develop and mature83.
This was the first randomized controlled trial of rice bran supplementation in LMIC infants and provides compelling rationale for continued follow-up investigation of rice bran supplementation for reducing risk of malnutrition, as well as for eliciting changes during child growth that protect against enteric pathogens and diarrheal diseases. The dose and feasibility outcomes from this study support development of rice bran based complementary weaning foods. Incorporating rice bran from local rice production and processing facilities should be a priority in subsequent trial designs. The development of sustainable, local and affordable food products for consumption by weaning infants and throughout childhood is particularly critical in regions where political instability continuously threaten the regions food and nutritional security.
Materials and Methods
Study design
A 6-month, prospective, randomized-controlled, phase one dose escalation dietary intervention was conducted in a cohort of weaning infants residing in León, Nicaragua and in the community of Dioro, Mali, West Africa. Nicaraguan infants were recruited from public health rosters provided by the local Health Ministry from Perla Maria and Sutiava health sectors, and Malian infants were recruited from the Dioro Community Health Center. To be eligible, male and female infants were screened between 4–5 months of age for health status, and then followed weekly for diarrhea episodes. Participants were excluded if they had experienced diarrhea or received antibiotic treatment within the previous month; had known allergies, or immune-compromising conditions (e.g. parasitic or malarial infections); had previously been hospitalized; and/or enrolled in a malnutrition treatment program. In Nicaragua, all eligible participants received 3 doses of the rotavirus vaccine per regular administration through the Immunization Program82. Rotavirus vaccination was not yet administrated to the Mali cohort. All Malian participants received vitamin A supplementation upon enrollment. Dietary intervention with rice bran started when infants were 6 months of age because WHO guidelines promote exclusive breastfeeding for the first six months of life84,85.
The required ethical board reviews and approvals were completed for Mali and Nicaragua as provided by the Internal Review Board (IRB) of the Colorado State University Research Integrity and the Compliance Review office (protocol ID# 14-5233 H Nicaragua, 15-5744 H Mali). In Mali, the Institut National de Recherche en Santé Publique (National Institute of Research in Public Health, FWA 00000892) approved the intervention, which occurred between October 2015 and May of 2016 and registered at clinicaltrial.gov as (NCT02557373) on 23 September 2015. Ethical review and approvals for the Nicaraguan intervention that occurred between March 2015 and October 2015 were provided by the IRBs of the Universidad Nacional Autónoma de Nicaragua – León, University of North Carolina at Chapel Hill, and Virginia Polytechnic Institute and State University and registered at clinicaltrial.gov on 26 November 2105 as (NCT02615886). Written informed consent was obtained from the infant’s parent or responsible guardian prior to any data collection. Infant participants that met the eligibility criteria were randomized within each health sector, and sex (Nicaragua) and geographic location of household and sex (Mali) to either rice bran or control group (see Fig. S1 for enrollment details). Randomization was completed using sequential enrollment for each site independently. Participants were randomized by CSU, enrolled and assigned to groups by study coordinators in each country. The participants were not blinded. Complete study protocol is available online (http://csu-cvmbs.colostate.edu/academics/erhs/Pages/elizabeth-ryan-lab-global-health.aspx).
Rice bran packaging for consumption
The United States Department of Agriculture-Agricultural Research Service (USDA-ARS) Dale Bumpers National Rice Research Center provided rice bran that was polished from the U.S. variety, Calrose. Rice bran is prone to fat oxidation and heat-stabilization was performed to increase shelf-life by heating the bran at 100 degrees Celsius for five minutes to inactivate the lipase/lipoxygenase enzymes that cause rancidity86. The rice bran was sifted to remove any debris (rice husk, rice grain). Packaging of the rice bran was completed by Western Innovations, Inc. (Denver, CO) where 22 kg of rice bran was weighed into 1 g increments, separated into water-proof sachets, and heat-sealed to ensure the rice bran would be administered with accurate doses to infants.
Fourteen sachets (1 g/sachet) were filled into a 4″ × 3″ × 2″ box that was labeled for study participants and included nutrient information. These boxes were stored in a cool, dark, dry place until they were provided to study participants.
Nicaragua and Mali intervention
The study team (doctor, nurse and study coordinator) in Nicaragua and the community health workers (CHWs) in Mali provided a 2-week supply of rice bran at each routine home visit and instructed the participant’s parent or guardian to add the daily amount of rice bran to the participant’s food. At 6–7 months of age, participants in the rice bran group consumed 1 g of rice bran/day (1 sachet). Between the ages of 7–8 months, participants consumed 2 g of rice bran/day (2 sachets). At 8–10 months of age, participants consumed 3 g of rice bran/day (3 sachets). The amount increased to 4 g of rice bran/day (4 sachets) from 10–11 months, and 5 g of rice bran/day (5 sachets) from 11–12 months of age, respectively. The rice bran was added to appropriate weaning foods, such as rice cereal, yogurt, fruit and natural juices, vegetables, and soups. At the beginning of the intervention (six months of age), infant’s parents or guardians were instructed and monitored daily for one week by study personnel to ensure that guardians knew how to administer and record the amount of rice bran consumed. Compliance to the rice bran intervention was calculated from records that had the dose/amount of rice bran consumed circled in daily increments of none (0%), half (50%), or all (100%). The study team also collected any unused boxes or sachets during these visits. Participants in the control group did not receive any rice bran and there were no reports of brown rice intake during the 6-month study duration.
In Nicaragua, study personnel visited all infants weekly. In Mali, the CHWs visited each participant’s household daily for the duration of the 6-month study to assess compliance and diarrhea episodes. If a participant had a diarrhea episode, the study team would collect a stool sample, and collect information that included the diarrhea onset date, how long the episode lasted, numbers of bowel movements within 24 hours, any associated signs and symptoms (e.g. nausea, vomiting, fever), if any other family members had diarrhea, and if any treatment was provided (e.g. antibiotics, rehydration).
The study team in Nicaragua collected data for control group participants at 6, 8, and 12 months old, and rice bran group every month. The anthropometric measures (weight and length) were collected via a portable stadiometer and weighing balance. Mali participants visited the Community Health Clinic every month. Length was measured in supine position using a reclining length-board. Length was collected to the nearest centimeter and weight to the nearest 0.1 kg. Anthropometric measures were calculated for LAZ, WAZ, and WLZ scores following the World Health Organizations (WHO) child growth standards using the WHO Anthro software (version 3.2.2)87.
Diapers were provided to all study participants. Stool was collected directly from soiled diapers. Freshly collected stool was diluted 20-fold and homogenized in a sterile pre-reduced anaerobic saline −0.1 M potassium phosphate buffer (pH 7.2) containing 20% glycerol (vol/vol). Four aliquot suspensions were prepared in 15 mL falcon tubes, transported on dry ice to the UNAN-León-Center of Infectious Diseases Laboratories (and liquid nitrogen in Bamako,Mali), immediately transferred to a −80 °C freezer, shipped in a liquid nitrogen chilled dry shipping dewar to Colorado State University, where they were relocated into a −80 °C freezer prior to analysis.
A study questionnaire was completed by the participant’s caretaker (e.g. mother, father, or grandparent) to assess for duration of breastfeeding, types of and timing of introductions to complementary foods, as well as antibiotic use. The breastfeeding questions included whether or not the child was receiving breast milk, and/or had the child been receiving received formula. The complementary feeding history included a list of common Nicaraguan and Malian foods that are normally introduced to infants during weaning. Infant’s parents or guardians recorded how often the infant consumed each of the eleven foods. The questionnaire also recorded if a participant had received treatment with antibiotics since the last visit, the reason for taking the antibiotic, the name of the antibiotic, as well as the length of time the participant had been taking the antibiotic. A household survey was also completed at the beginning of the trial to collect mother’s education level, drinking water source, household flooring type, and animals present in the household. Analysis of breastfeeding and formula feeding patterns, complementary feeding practices, and associations with nutritional status at 6-months old (i.e. baseline) were previously reported for Nicaragua88. Monthly visits to the Mali community health clinic provided monitoring for malnutrition and severe adverse events; no adverse events were reported in the rice bran intervention group. There was one participant death reported in the control group (respiratory infection) and another withdrew to receive malnutrition treatment in the second month of the study. Diarrheal episodes were recorded, and a sample was collected in both countries using the same protocol.
Stool analysis for EED markers
Stool biomarkers were selected to report gut inflammation and epithelial integrity as indicators of EED. These included neopterin (NEO), myeloperoxidase (MPO), calprotectin, (CAL) and alpha-1 antitrypsin (AAT)89. Suspended stool samples from 6, 8, and 12-month collections were centrifuged at 3,000 RPM to remove debris, following agitation, and the remaining supernatant was used for Enzyme-Linked-Immunosorbant-Assay (ELISA) determination of EED biomarker concentrations. Laboratory analysis protocols included in commercial kits were followed. Concentrations of CAL were determined at a 1:360 final dilution factor (Eagle Biosciences- Nashua, NH. Ref: CAL35-K01). Samples were diluted to 1:100 for determination of NEO concentrations (GenWay Biotech Inc- San Diego, CA, USA). MPO concentrations were determined at a 1:500 dilution factor (Immundiagnostik AG- Bensheim, Germany). Samples were diluted to 1:12,500 for determination of AAT concentrations (Immuchrom GMBH- Heppenheim, Germany), and dilution factors accounted for stool suspension ratios (20-fold). Final concentrations were determined from averages of three replicate assays and duplicate optical density readings, and interpolated using Graphpad 6.0 according to standards measured on each 96-well plate.
Stool microbiota analysis for Nicaragua and Mali
The infant stool was collected at 6, 8 and 12 months of age from diapers and placed in a 1:19 ratio with Phosphate Buffered Saline + Glycerol solution. Diarrhea samples were collected using the same protocol. Suspended stool samples were vortexed before centrifuging at 3000 RPM to separate the stool debris. The remaining supernatant was used for Enzyme-Linked-Immunosorbant-Assay (ELISA) determination of EED biomarkers whereas; DNA was extracted for 16S microbial analysis from the stool pellet. DNA extraction was conducted using MoBio PowerSoil Kit (Reference number 12888, MoBio Laboratories Inc., Solana Beach, CA). PCR amplification of 390 bp amplicons was done in 50 µl reaction using Fischer Hot Start Master Mix and EMP standard protocols43–47. SPRI magnetic beads were used to purify DNA, and flourimetric quantification of Sybr Green tags was used to confirm adequate concentration of DNA. The pooled library was created with 50 ng DNA per sample and quantified using Kapa Kit (Kapa Biosystems). The pooled library was run on Illumina-MiSeq with 15% PhiX mock library to reduce discrepancies in read clustering, using the Illumina V2 500 cycle kit (2 × 250/250 paired-end reads).
Microbiota data processing and analysis
Sequence data were processed using mothur90 version 1.39.5 and using a custom pipeline that provides an adjustment on the developers’ standard operating procedure (SOP) for OTU calling and taxonomic classification of MiSeq data first presented in Kozich, et al.91. For alignment and classification within this SOP we used the SILVA database92 version 128. Clustering, for OTU identification, was performed using VSEARCH using the distance based greedy clustering (DGC) option as implanted in mothur and utilizing 0.97 sequence similarity cutoff. We also used a cutoff of one read that was subtracted from all OTU read counts to guard against overestimation of sample richness. Rarefaction curves were generated using the package vegan93 as implemented in R version 3.4.494 to assess diversity and suitability of depth of coverage per sample. The resulting OTU table was utilized in further data analyses as follows.
Exploring the data: Bar-graphs for relative abundance data based on the resulting OTU table were generated using the ggplot295 package in R. These plots were generated for the data at the genus and the family levels and meant to describe the microbial community structure per sampled infant and per time point under each of the treatment levels.
Data were normalized using cumulative sum scaling (CSS)96 prior to beta diversity and log-fold change analyses. Nonmetric Multidimensional Scaling (NMDS)97 was used on the OTU level to assess possible trends and clustering in the microbial community structure comparing the two countries, the treatment conditions and the two time points, using the vegan package and utilizing Bray-Curtis dissimilarity97. Data were separated per country and the metagenomeSeq.98 package in R94 was used to fit a zero inflated normal (ZIN) model to test for log-fold change differences between the rice bran treatment and control per age group. Benjamini and Hochberg’s99 false discovery rate (FDR) method was used to correct for multiple testing and compute the adjusted p-values used to determine significance of differences in the log-fold change of OTU abundance.
Stool metabolomics analysis
Stool samples were sent to Metabolon Inc. (Durham, NC, USA) for non-targeted metabolite profiling via ultrahigh-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS). All samples were accessioned into the Metabolon Library Information Management Systems (LIMS) and prepared using the automated MicroLab Star® system (Hamilton Company, Switzerland). Eight to ten recovery standards were added prior to the first step in the extraction process for quality control purposes. Extraction was performed using 80% ice-cold methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation to remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix. Each stool extract was divided into five fractions: two for analysis by two separate reverse phase UPLC-MS/MS methods with positive ion mode electrospray ionization, 1 for analysis by reverse phase UPLC-MS/MS methods with negative ion mode electrospray ionization, 1 for hydrophilic interaction liquid chromatography UPLC-MS/MS with negative ion mode electrospray ionization, and 1 sample for backup. All samples were placed briefly on Concentration Evaporator (TurboVap® Zymark) to remove organic solvent. UPLC-MS/MS methods utilized a Waters ACQUITY ultra-performance liquid chromatography and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. Raw data was extracted, peak-identified and processed for quality control using Metabolon’s hardware and software.
Statistical analysis
Statistical analyses for anthropometric measures (length, weight, LAZ, WAZ, and WLZ) and stool EED biomarkers were completed using SAS 9.4 (Cary, NC, USA). The sample size was calculated for achieving greater than 85% power and based on expected changes in selected stool metabolites following dietary rice bran consumption for one month27. Normality was evaluated by visual inspection. For anthropometric variables, two-sample t-tests were used to compare means for the 2 treatment groups (rice bran and control) separately at birth and 6 months (prior to start of treatment). A repeated measures analysis was performed for each response variable separately using SAS Proc Mixed. Specifically, treatment (rice bran or control) and age (6, 8 or 12 months), and treatment-age interaction were included in the model as fixed effects. The participant was included as a random effect to account for repeated measures. At each age, treatment groups were compared using contrasts of the model. A similar repeated measures analysis was used EED biomarkers, but log transformation was used to satisfy model assumptions. For stool metabolites, Welch’s two-sample t-test was used to analyze statistical significance between groups’ stool metabolites, after participating in the 6-month dietary trial. A p-value of ≤ 0.05 was used for statistical significance. An estimated false discovery rate (q-value) was calculated to account for the multiple comparisons across metabolites that are typical of metabolomic-based studies.
One sentence summary
Dietary rice bran supplementation during infant weaning from 6–12 months of age improved growth outcomes and supported metabolism by the gut microbiota.
Supplementary information
Acknowledgements
The authors thank the study participants, community health workers, and local clinical staff for their assistance in the conduct of the clinical trials. This study was supported by the Grand Challenges Explorations in Global Health award from the Bill and Melinda Gates Foundation (OPP1043255) and the Fulbright Faculty Development scholarship award.
Author Contributions
For Nicaragua were as follows: S.V., S.B.D., L.Y. and E.P.R. designed research and maintained study oversight; E.C.B. and L.Z. conducted research and sample collection; I.Z., S.M., J.P. and C.P. analyzed stool and serum samples; A.H. and L.Z. data analysis; L.Z., E.C.B. and E.P.R. wrote paper; E.P.R. had primary responsibility for the final product. For Mali E.P.R., O.K. and A.M. drafted the research protocols, and designed the rice bran intervention with assistance from A.B., A.C., E.C.B. and L.S. A.B. and A.C. coordinated the study in the community, and L.S. oversaw sample collection and field laboratory operations. L.D., S.D. and K.K. provided community and participant engagement, collected samples and data, and carried out laboratory analyses in Mali. S.A.M., L.E.Z., E.C.B., H.I., Z.A. and L.Z. carried out further laboratory and data analyses in the USA. A.H. and Z.A. contributed to the analytical approach and carried out the statistical analysis. S.A.M., H.I., Z.A., O.K. and E.P.R. collectively interpreted data, carried out the literature search, constructed tables and figures, and contributed to manuscript preparation. All authors read and approved the final manuscript.
Data Availability
16S sequence data were submitted to the National Center for Biotechnology Information SRA under accession no. (SRP159269) and Bio-project (PRJNA488807).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ousmane Koita, Email: okoita@icermali.org.
Samuel Vilchez, Email: samuelvilchez@gmail.com.
Elizabeth P. Ryan, Email: E.P.Ryan@colostate.edu
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50344-4.
References
- 1.WHO. Diarrhoeal disease, http://www.who.int/mediacentre/factsheets/fs330/en/ (2017).
- 2.Black RE, et al. Maternal and child undernutrition: global and regional exposures and health consequences. Lancet. 2008;371:243–260. doi: 10.1016/S0140-6736(07)61690-0. [DOI] [PubMed] [Google Scholar]
- 3.Bhutta ZA, Salam RA. Global nutrition epidemiology and trends. Ann Nutr Metab. 2012;61(Suppl 1):19–27. doi: 10.1159/000345167. [DOI] [PubMed] [Google Scholar]
- 4.Joseph SA, et al. Risk Factors Associated with Malnutrition in One-Year-Old Children Living in the Peruvian Amazon. PLOS Neglected Tropical Diseases. 2014;8:e3369. doi: 10.1371/journal.pntd.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keusch GT, et al. Implications of acquired environmental enteric dysfunction for growth and stunting in infants and children living in low- and middle-income countries. Food Nutr Bull. 2013;34:357–364. doi: 10.1177/156482651303400308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keusch GT, et al. Environmental Enteric Dysfunction: Pathogenesis, Diagnosis, and Clinical Consequences. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2014;59:S207–S212. doi: 10.1093/cid/ciu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Syed SAA, Duggan C. Environmental Enteric Dysfunction in Children: A Review. J Pediatr Gastroenterol Nutr. 2016;63:9. doi: 10.1097/MPG.0000000000001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lunn PGN-CCA, Downes RM. Intestinal permeability, mucosal injury, and growth faltering in Gambian infants. The Lancet. 1991;338:4. doi: 10.1016/0140-6736(91)91772-M. [DOI] [PubMed] [Google Scholar]
- 9.Campbell DIEMALPG. Growth Faltering in Rural Gambian Infants Is Associated with Impaired Small Intestinal Barrier Function, Leading to Endotoxemia and Systemic Inflammation. J. Nutr. 2003;133:7. doi: 10.1093/jn/133.4.7e. [DOI] [PubMed] [Google Scholar]
- 10.Kosek M, et al. Fecal markers of intestinal inflammation and permeability associated with the subsequent acquisition of linear growth deficits in infants. The American journal of tropical medicine and hygiene. 2013;88:390–396. doi: 10.4269/ajtmh.2012.12-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korpe PS, Petri WA., Jr. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol Med. 2012;18:328–336. doi: 10.1016/j.molmed.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Victora CG, de Onis M, Hallal PC, Blossner M, Shrimpton R. Worldwide timing of growth faltering: revisiting implications for interventions. Pediatrics. 2010;125:e473–480. doi: 10.1542/peds.2009-1519. [DOI] [PubMed] [Google Scholar]
- 13.Caulfield ELHLS, Piwoz GE. Interventions to improve intake of complementary foods by infants 6 to 12 months of age in developing countries: Impact on growth and on the prevalence of malnutrition and potential contribution to child survival. Food and Nutrition Bulletin. 1999;20:18. doi: 10.1177/156482659902000203. [DOI] [Google Scholar]
- 14.Dewey KG, Cohen RJ, Brown KH, Rivera LL. Age of introduction of complementary foods and growth of term, low-birth-weight, breast-fed infants: a randomized intervention study in Honduras. Am J Clin Nutr. 1999;69:679–686. doi: 10.1093/ajcn/69.4.679. [DOI] [PubMed] [Google Scholar]
- 15.Smith HE, et al. Multiple micronutrient supplementation transiently ameliorates environmental enteropathy in Malawian children aged 12-35 months in a randomized controlled clinical trial. J Nutr. 2014;144:2059–2065. doi: 10.3945/jn.114.201673. [DOI] [PubMed] [Google Scholar]
- 16.Wang AZ, et al. A Combined Intervention of Zinc, Multiple Micronutrients, and Albendazole Does Not Ameliorate Environmental Enteric Dysfunction or Stunting in Rural Malawian Children in a Double-Blind Randomized Controlled Trial. J Nutr. 2017;147:97–103. doi: 10.3945/jn.116.237735. [DOI] [PubMed] [Google Scholar]
- 17.Madhi SA, et al. Effect of human rotavirus vaccine on severe diarrhea in African infants. The New England journal of medicine. 2010;362:289–298. doi: 10.1056/NEJMoa0904797. [DOI] [PubMed] [Google Scholar]
- 18.Jones KD, Thitiri J, Ngari M, Berkley JA. Childhood malnutrition: toward an understanding of infections, inflammation, and antimicrobials. Food Nutr Bull. 2014;35:S64–70. doi: 10.1177/15648265140352S110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keenan JD, et al. Azithromycin to Reduce Childhood Mortality in Sub-Saharan Africa. New England Journal of Medicine. 2018;378:1583–1592. doi: 10.1056/NEJMoa1715474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pickering AJ, Djebbari H, Lopez C, Coulibaly M, Alzua ML. Effect of a community-led sanitation intervention on child diarrhoea and child growth in rural Mali: a cluster-randomised controlled trial. Lancet. Glob Health. 2015;3:e701–711. doi: 10.1016/S2214-109X(15)00144-8. [DOI] [PubMed] [Google Scholar]
- 21.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarei Iman, Luna Emily, Leach Jan, McClung Anna, Vilchez Samuel, Koita Ousmane, Ryan Elizabeth. Comparative Rice Bran Metabolomics across Diverse Cultivars and Functional Rice Gene–Bran Metabolite Relationships. Metabolites. 2018;8(4):63. doi: 10.3390/metabo8040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei S, et al. High Protective Efficacy of Probiotics and Rice Bran against Human Norovirus Infection and Diarrhea in Gnotobiotic Pigs. Frontiers in microbiology. 2016;7:1699. doi: 10.3389/fmicb.2016.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, et al. High protective efficacy of rice bran against human rotavirus diarrhea via enhancing probiotic growth, gut barrier function, and innate immunity. Scientific reports. 2015;5:15004. doi: 10.1038/srep15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar A, et al. Dietary rice bran promotes resistance to Salmonella enterica serovar Typhimurium colonization in mice. BMC microbiology. 2012;12:71. doi: 10.1186/1471-2180-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodyear A, et al. Dietary rice bran supplementation prevents Salmonella colonization differentially across varieties and by priming intestinal immunity. Journal of Functional Foods. 2015;18:653–664. doi: 10.1016/j.jff.2015.08.027. [DOI] [Google Scholar]
- 27.Sheflin AM, et al. Pilot dietary intervention with heat-stabilized rice bran modulates stool microbiota and metabolites in healthy adults. Nutrients. 2015;7:1282–1300. doi: 10.3390/nu7021282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarei I, Brown DG, Nealon NJ, Ryan EP. Rice Bran Metabolome Contains Amino Acids, Vitamins & Cofactors, and Phytochemicals with Medicinal and Nutritional Properties. Rice. 2017;10:24. doi: 10.1186/s12284-017-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Twitchell EL, et al. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut pathogens. 2016;8:51. doi: 10.1186/s13099-016-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, et al. Dietary rice bran protects against rotavirus diarrhea and promotes Th1-type immune responses to human rotavirus vaccine in gnotobiotic pigs. Clinical and vaccine immunology: CVI. 2014;21:1396–1403. doi: 10.1128/cvi.00210-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nealon NJ, Yuan L, Yang X, Ryan EP. Rice Bran and Probiotics Alter the Porcine Large Intestine and Serum Metabolomes for Protection against Human Rotavirus Diarrhea. Frontiers in microbiology. 2017;8:653. doi: 10.3389/fmicb.2017.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. An overview of global rice production, supply, trade, and consumption. Annals of the New York Academy of Sciences. 2014;1324:7–14. doi: 10.1111/nyas.12540. [DOI] [PubMed] [Google Scholar]
- 33.Bokulich NA, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Science translational medicine. 2016;8:343ra382–343ra382. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ordiz MI, et al. Environmental Enteric Dysfunction and the Fecal Microbiota in Malawian Children. The American journal of tropical medicine and hygiene. 2017;96:473–476. doi: 10.4269/ajtmh.16-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semba RD, et al. Environmental Enteric Dysfunction is Associated with Carnitine Deficiency and Altered Fatty Acid Oxidation. EBioMedicine. 2017;17:57–66. doi: 10.1016/j.ebiom.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCormick BJ, et al. Dynamics and Trends in Fecal Biomarkers of Gut Function in Children from 1-24 Months in the MAL-ED Study. The American journal of tropical medicine and hygiene. 2017;96:465–472. doi: 10.4269/ajtmh.16-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker-Dreps S, et al. The Association Between Fecal Biomarkers of Environmental Enteropathy and Rotavirus Vaccine Response in Nicaraguan Infants. The Pediatric infectious disease journal. 2017;36:412–416. doi: 10.1097/INF.0000000000001457. [DOI] [PubMed] [Google Scholar]
- 38.Campbell DI, McPhail G, Lunn PG, Elia M, Jeffries DJ. Intestinal inflammation measured by fecal neopterin in Gambian children with enteropathy: association with growth failure, Giardia lamblia, and intestinal permeability. J Pediatr Gastroenterol Nutr. 2004;39:153–157. doi: 10.1097/00005176-200408000-00005. [DOI] [PubMed] [Google Scholar]
- 39.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell host & microbe. 2015;17:852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Subramanian S, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Apprill, A., McNally, S., Parsons, R. & Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. 75 (2015).
- 44.Caporaso J Gregory, Lauber Christian L, Walters William A, Berg-Lyons Donna, Huntley James, Fierer Noah, Owens Sarah M, Betley Jason, Fraser Louise, Bauer Markus, Gormley Niall, Gilbert Jack A, Smith Geoff, Knight Rob. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 47.Walters, W. et al. Improved Bacterial 16S rRNA Gene (V4 and V4-5) and Fungal Internal Transcribed Spacer Marker Gene Primers for Microbial Community Surveys. mSystems1, 10.1128/mSystems.00009-15 (2016). [DOI] [PMC free article] [PubMed]
- 48.Guerrant RL, et al. Biomarkers of Environmental Enteropathy, Inflammation,Stunting,and Impaired Growth in Children in Northeast Brazil. journal.pone. 2016;11:20. doi: 10.1371/journal.pone.0158772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naylor C, et al. Environmental Enteropathy, Oral Vaccine Failure and Growth Faltering in Infants in Bangladesh. EBioMedicine. 2015;2:1759–1766. doi: 10.1016/j.ebiom.2015.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harper KM, Mutasa M, Prendergast AJ, Humphrey J, Manges AR. Environmental enteric dysfunction pathways and child stunting: A systematic review. PLOS Neglected Tropical Diseases. 2018;12:e0006205. doi: 10.1371/journal.pntd.0006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, et al. Environmental Enteric Dysfunction Includes a Broad Spectrum of Inflammatory Responses and Epithelial Repair Processes. Cell Mol Gastroenterol Hepatol. 2016;2:158–174 e151. doi: 10.1016/j.jcmgh.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petri WA, Jr., Naylor C, Haque R. Environmental enteropathy and malnutrition: do we know enough to intervene? BMC Med. 2014;12:187. doi: 10.1186/s12916-014-0187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown EM, et al. Diet and specific microbial exposure trigger features of environmental enteropathy in a novel murine model. Nature communications. 2015;6:7806. doi: 10.1038/ncomms8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayeni FA, et al. Infant and Adult Gut Microbiome and Metabolome in Rural Bassa and Urban Settlers from Nigeria. Cell reports. 2018;23:3056–3067. doi: 10.1016/j.celrep.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 55.Mayneris-Perxachs J, et al. Protein- and zinc-deficient diets modulate the murine microbiome and metabolic phenotype. Am J Clin Nutr. 2016;104:1253–1262. doi: 10.3945/ajcn.116.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma N, Tian Y, Wu Y, Ma X. Contributions of the Interaction Between Dietary Protein and Gut Microbiota to Intestinal Health. Current protein & peptide science. 2017;18:795–808. doi: 10.2174/1389203718666170216153505. [DOI] [PubMed] [Google Scholar]
- 57.Lin H, An Y, Hao F, Wang Y, Tang H. Correlations of Fecal Metabonomic and Microbiomic Changes Induced by High-fat Diet in the Pre-Obesity State. Scientific reports. 2016;6:21618. doi: 10.1038/srep21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parra-Llorca A, et al. Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk. Frontiers in microbiology. 2018;9:1376. doi: 10.3389/fmicb.2018.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velly H, Britton RA, Preidis GA. Mechanisms of cross-talk between the diet, the intestinal microbiome, and the undernourished host. Gut microbes. 2017;8:98–112. doi: 10.1080/19490976.2016.1267888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merrifield CA, et al. Neonatal environment exerts a sustained influence on the development of the intestinal microbiota and metabolic phenotype. The ISME journal. 2016;10:145–157. doi: 10.1038/ismej.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blekhman R, et al. Host genetic variation impacts microbiome composition across human body sites. Genome biology. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henderson AJ, Kumar A, Barnett B, Dow SW, Ryan EP. Consumption of rice bran increases mucosal immunoglobulin A concentrations and numbers of intestinal Lactobacillus spp. Journal of medicinal food. 2012;15:469–475. doi: 10.1089/jmf.2011.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown DG, Borresen EC, Brown RJ, Ryan EP. Heat-stabilised rice bran consumption by colorectal cancer survivors modulates stool metabolite profiles and metabolic networks: a randomised controlled trial. British Journal of Nutrition. 2017;117:1244–1256. doi: 10.1017/S0007114517001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.So WKW, Law BMH, Law PTW, Chan CWH, Chair SY. Current Hypothesis for the Relationship between Dietary Rice Bran Intake, the Intestinal Microbiota and Colorectal Cancer Prevention. Nutrients. 2016;8:569. doi: 10.3390/nu8090569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hamer HM, Preter VD, Windey K, Verbeke K. Functional analysis of colonic bacterial metabolism: relevant to health? American Journal of Physiology-Gastrointestinal and Liver Physiology. 2012;302:G1–G9. doi: 10.1152/ajpgi.00048.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton DF, Macfarlane S, Macfarlane GT. Effects of Antibiotics on Bacterial Species Composition and Metabolic Activities in Chemostats Containing Defined Populations of Human Gut Microorganisms. Antimicrobial Agents and Chemotherapy. 2013;57:2016–2025. doi: 10.1128/aac.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: lights and shadows. Frontiers in Cellular and Infection Microbiology2, 10.3389/fcimb.2012.00086 (2012). [DOI] [PMC free article] [PubMed]
- 69.Leone V, et al. Effects of Diurnal Variation of Gut Microbes and High-Fat Feeding on Host Circadian Clock Function and Metabolism. Cell host & microbe. 2015;17:681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tahara Y, et al. Gut Microbiota-Derived Short Chain Fatty Acids Induce Circadian Clock Entrainment in Mouse Peripheral Tissue. Scientific Reports. 2018;8:1395. doi: 10.1038/s41598-018-19836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunological Reviews. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ubeda C, Djukovic A, Isaac S. Roles of the intestinal microbiota in pathogen protection. Clinical & Translational Immunology. 2017;6:e128. doi: 10.1038/cti.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davis, J. C. C. et al. Growth and Morbidity of Gambian Infants are Influenced by Maternal Milk Oligosaccharides and Infant Gut Microbiota. Scientific Reports7, 40466, 10.1038/srep40466, https://www.nature.com/articles/srep40466#supplementary-information (2017). [DOI] [PMC free article] [PubMed]
- 74.Li, K. J., Borresen, E. C., Jenkins-Puccetti, N., Luckasen, G. & Ryan, E. P. Navy Bean and Rice Bran Intake Alters the Plasma Metabolome of Children at Risk for Cardiovascular Disease. Frontiers in Nutrition4, 10.3389/fnut.2017.00071 (2018). [DOI] [PMC free article] [PubMed]
- 75.Gupta, V. K., Paul, S. & Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Frontiers in microbiology8, 10.3389/fmicb.2017.01162 (2017). [DOI] [PMC free article] [PubMed]
- 76.Alexeev EE, et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. The American journal of pathology. 2018;188:1183–1194. doi: 10.1016/j.ajpath.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao J, et al. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front Cell Infect Microbiol. 2018;8:13. doi: 10.3389/fcimb.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McBreairty LE, Bertolo RF. The dynamics of methionine supply and demand during early development. Applied Physiology, Nutrition & Metabolism. 2016;41:581–587. doi: 10.1139/apnm-2015-0577. [DOI] [PubMed] [Google Scholar]
- 79.Baj, T. & Sieniawska, E. In Pharmacognosy (eds Simone Badal & Rupika Delgoda) 281–292 (Academic Press, 2017).
- 80.Borresen EC, et al. A Pilot Randomized Controlled Clinical Trial to Assess Tolerance and Efficacy of Navy Bean and Rice Bran Supplementation for Lowering Cholesterol in Children. Glob Pediatr Health. 2017;4:2333794X17694231. doi: 10.1177/2333794X17694231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.WHO. Training course on child growth assessment. (WHO, Geneva, 2008).
- 82.MINSA. Normativa 121 (ed Programa Ampliado de Inmunizaciones) 246 (Managua, Nicaragua, 2013).
- 83.Putignani L, et al. Gut Microbiota Dysbiosis as Risk and Premorbid Factors of IBD and IBS Along the Childhood-Adulthood Transition. Inflammatory bowel diseases. 2016;22:487–504. doi: 10.1097/mib.0000000000000602. [DOI] [PubMed] [Google Scholar]
- 84.MINSA (2012).
- 85.WHO. Global Database on Child Growth and Malnutrition. 74 (WHO, Geneva, 1997).
- 86.Lakkakula NR, Lima M, Walker T. Rice bran stabilization and rice bran oil extraction using ohmic heating. Bioresource Technology. 2004;92:157–161. doi: 10.1016/j.biortech.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 87.WHO Anthro and macros v. version 3.2.2 (2011).
- 88.Borresen EC et al. Association between Infant Feeding Practices and Nutritional Status in Healthy Nicaraguan Infants. J Food Nutri Diete1, 6, 14000110 (2016).
- 89.George CM, et al. Geophagy is associated with environmental enteropathy and stunting in children in rural Bangladesh. The American journal of tropical medicine and hygiene. 2015;92:1117–1124. doi: 10.4269/ajtmh.14-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–7541. doi: 10.1128/aem.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology. 2013;79:5112–5120. doi: 10.1128/aem.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oksanen, J. et al. vegan: Community Ecology Package (2014).
- 94.R Core Team. R: A language and environment for statistical computing (2017).
- 95.Wickham, H. ggplot2: Elegant graphics for data analysis. Springer Science+Business Media (2009).
- 96.Paulson Joseph N, Stine O Colin, Bravo Héctor Corrada, Pop Mihai. Differential abundance analysis for microbial marker-gene surveys. Nature Methods. 2013;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Legendre, P. L. L. Numerical Ecology. Elsevier B.V20 (1998).
- 98.Paulson, J. N. metagenomeSeq: Statistical analysis for sparse high-throughput sequencing. Bioconductor. Package 1 (2014).
- 99.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S sequence data were submitted to the National Center for Biotechnology Information SRA under accession no. (SRP159269) and Bio-project (PRJNA488807).