Abstract
The endocannabinoid system (ECS) consists particularly of cannabinoid receptors 1 and 2 (CB1 and CB2), their endogenous ligands, and enzymes that synthesize and degrade their ligands. It acts in a variety of organs and disease states ranging from cancer progression over neuropathic pain to neurodegeneration. Protein components engaged in the signaling, trafficking, and homeostasis machinery of the G-protein coupled CB2, are however largely unknown. It is therefore important to identify further interaction partners to better understand CB2 receptor functions in physiology and pathophysiology. For this purpose, we used an affinity purification and mass spectrometry-based proteomics approach of Strep-HA-CB2 receptor in HEK293 cells. After subtraction of background interactions and protein frequency library assessment we could identify 83 proteins that were classified by the identification of minimally 2 unique peptides as highly probable interactors. A functional protein association network analysis obtained an interaction network with a significant enrichment of proteins functionally involved in protein metabolic process, in endoplasmic reticulum, response to stress but also in lipid metabolism and membrane organization. The network especially contains proteins involved in biosynthesis and trafficking like calnexin, Sec61A, tubulin chains TUBA1C and TUBB2B, TMED2, and TMED10. Six proteins that were only expressed in stable CB2 expressing cells were DHC24, DHRS7, GGT7, HECD3, KIAA2013, and PLS1. To exemplify the validity of our approach, we chose a candidate having a relatively low number of edges in the network to increase the likelihood of a direct protein interaction with CB2 and focused on the scaffold/phagosomal protein p62/SQSTM1. Indeed, we independently confirmed the interaction by co-immunoprecipitation and immunocytochemical colocalization studies. 3D reconstruction of confocal images furthermore showed CB2 localization in close proximity to p62 positive vesicles at the cell membrane. In summary, we provide a comprehensive repository of the CB2 interactome in HEK293 cells identified by a systematic unbiased approach, which can be used in future experiments to decipher the signaling and trafficking complex of this cannabinoid receptor. Future studies will have to analyze the exact mechanism of the p62-CB2 interaction as well as its putative role in disease pathophysiology.
Keywords: AP-MS, interactome, SQSTM1, protein–protein interaction, GPCR, endocannabinoid system
Introduction
Cellular processes are usually dependent on network dynamics and interactions of macromolecular protein complexes. Identification of interactomes of proteins and consequently connecting molecular processes are important steps for understanding complex functions such as cell signaling and receptor trafficking. Here we focused on the identification of the interactome of the CB2, which belongs to the potentially therapeutic GPCRs. GPCRs are implicated in the pathophysiology of numerous human diseases like hypertension (Brinks and Eckhart, 2010), Parkinson’s disease (Pinna et al., 2005; Gandia et al., 2013), various cancers (Lappano and Maggiolini, 2011; Xiang et al., 2018), and infectious diseases (Heng et al., 2013), thus reflecting their immense therapeutic and clinical relevance. Due to difficulties in preserving the complex three-dimensional GPCR structure during receptor solubilization the identification of their interacting partners is demanding (Daulat et al., 2009). In this context methods of tandem AP-MS and MYTH approaches have been optimized for robustness and reproducibility to obtain high-confidence protein complex information and to systematically find interactions of full-length integral membrane receptors (Daulat et al., 2009; Glatter et al., 2009; Sokolina et al., 2017). These methods have indeed been successfully used for studies on clinically relevant GPCRs such as serotonin 5-HT4d, adenosine ADORA2A receptors (Sokolina et al., 2017), and CB1 (Mattheus et al., 2016) but not for CB2.
The CB2 receptor has been shown to contribute to various pathological states in animal models especially in neurodegenerative, immunological, inflammatory, cardiovascular, hepatic, and bone disorders (Pertwee, 2012). CB2 acts as a modulator of immune suppression, induction of apoptosis, and cell migration (Basu and Dittel, 2011). In pathophysiological conditions CB2 is often upregulated in immune cells, where it inhibits proinflammatory cytokine production (Steffens et al., 2005). The pharmacological modulation of the transcription factor PPARalpha represents an important mechanism through which CB2 expression could be regulated (Guida et al., 2017). Of note, CB2 receptors are involved in the regulation of co-stimulatory factors, such as LPS and TNF-alpha, which signal via distinct membrane receptors (Toguri et al., 2014). Hence, it is of interest to identify interactors of CB2 that might act as signaling hubs for the crosstalk of different signaling events or that regulate the trafficking and degradation of the receptor.
By an AP-MS approach we have now identified p62 (also named sequestosome 1, SQSTM1) as an interacting partner of CB2 receptor. The scaffold protein p62 contains various domains that modulate protein–protein interactions; p62, e.g., associates with TRAF6 and with the death-domain kinase RIP1 and regulates NF-κB (nuclear factor- light-chain-enhancer of activated B cells) and MAP kinase signaling downstream of the TNF receptor, RANK receptor and nerve growth factor receptor (Moscat et al., 2007). Mutations in p62 are associated with neurodegenerative diseases and with PDB. The latter is characterized by increased bone remodeling in focal areas leading to bone pain, deformities and fractures (Laurin et al., 2002; Seitz et al., 2008). A P392L exchange found in the UBA of the C-terminal part of p62 protein is a prominent mutation present in patients with PDB and with amyotrophic lateral sclerosis and/or frontotemporal dementia (Laurin et al., 2002; Cavey et al., 2006; Moscat et al., 2007; Fecto et al., 2011). This UBA domain is also of importance for the interaction of p62 with polyubiquitinated proteins, destined for degradation by autophagy (Moscat and Diaz-Meco, 2011). Through its interaction with LC3B p62 recruits ubiquitinated cargo proteins to the autophagosomal degradation pathway (Pankiv et al., 2007). Defects in autophagy lead to an accumulation of p62 (Komatsu et al., 2007). P62 aggregates are highly accumulated in neurons and glial cells in patients with neurodegenerative diseases (Seilhean et al., 2009; Al-Sarraj et al., 2011). Interestingly, in a number of neurodegenerative disorders a CB2 upregulation especially in microglial cells has been described and neuroprotective effects of CB2 ligands were shown (Cassano et al., 2017).
In summary, with a systematic unbiased approach we identified the CB2 interactome. Our data will serve in future studies to decipher cannabinoid receptor trafficking and signaling complexes and receptor homeostasis. We could furthermore validate our approach by confirming an interaction of CB2 with p62. Interestingly, it has been shown that the dysfunction of both proteins alone is critically involved in related disorders affecting brain and bone (Pacher and Mechoulam, 2011; Duran et al., 2016; Sanchez-Martin and Komatsu, 2018), so that future studies will have to analyze the exact mechanism of this interaction and investigate its role in pathophysiological conditions.
Materials and Methods
Antibodies and Reagents
The following antibodies and chemicals were used: Rabbit anti-p62 (#P0067, Sigma), mouse anti-CB2 (#H00001269-M01, Abnova), secondary antibodies; Alexa Fluor®488 goat anti-mouse (#A21202, Thermo Fisher Scientific), Alexa Fluor®649 goat anti-rabbit (#111-496-144, JacksonImmuno Research), rabbit anti-FLAG (#F7425, Sigma). Lectin WGA conjugated with Texas Red®-X (#W7024, Thermo Fisher Scientific) HU-210 from Tocris (CAS No. 112830-95-2), poly-L-Lysine (#P6282-5mg, Sigma), bovine serum albumin (#A9418, Sigma), Triton X-100 (#A4975, AppliChem), ProLong® Gold antifade reagent with DAPI (#P36935, Thermo Fisher Scientific), HBSS (#H6648, Sigma).
Plasmids and Cell Culture
Human HEK293 cells (obtained from ATCC) were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific) supplemented with 10% FBS, penicillin (50 units/ml), and streptomycin (50 μg/ml) at 37°C, 5% CO2. Transient transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Flag-p62 and CB2 expression vectors were generated by inserting human cDNA in-frame into EcoRI and XhoI sites into the pcDNA3.1 vector (Thermo Fisher Scientific). The deletion constructs for p62 and CB2 were generated by site-directed mutagenesis using the QuikChange site directed mutagenesis kit (#200523, Agilent) according to the manufacturer’s protocol. The ΔC-CB2 construct was generated by PCR amplification introducing a stop codon after the amino acid position 300 of human CB2 protein sequence. CB2 deletion constructs of intracellular loops (Δi) and p62 deletion constructs via XL II Gold Site-Directed Mutagenesis Kit (#200521, Agilent) were generated using following primer pairs (Table 1).
TABLE 1.
Sequences of primer pairs used for the generation of CB2 deletion constructs of intracellular loops (Δi) and p62 deletion constructs.
| Flag-plasmids | Forward primer (5′–3′) | Reverse primer (5′–3′) |
| Δi1-CB2 | GTGCTCTATCTGATCCTGCTGTTCATTGGCAGCTTG | CAAGCTGCCAATGAACAGCAGGATCAGATAGAGCAC |
| Δi2-CB2 | CCTCCTGCTGACCGCCATTGCACTGGTGA | TCACCAGTGCAATGGCGGTCAGCAGGAGG |
| Δi3-CB2 | GGGCATGTTCTCTGGACCCTAGGGCTAGTG | CACTAGCCCTAGGGTCCAGAGAACATGCCC |
| p62-ΔZZ122-167 | AGGGGCTGGGGAACATGTTGCGGGGC | GCCCCGCAACATGTTCCCCAGCCCCT |
| P62-ΔPEST266-294 | CCCACCCGGCTTTCTTTTCCCTCCGTGCT | AGCACGGAGGGAAAAGAAAGCCGGGTGGG |
| P62-ΔUBA389-434 | CACATCTCCCGCCAAAGCATCCCCCGCC | GGCGGGGGATGCTTTGGCGGGAGATGTG |
Bait Cloning and Stable Cell Line (Dualsystems Biotech AG)
The human open reading frame of the CB2 receptor (NM_001841.3; corresponding to NP_001832) was inserted into the bait vector pN-TGSH (Dualsystems). Bait expression construct pN-TGSH/CB2 with N-terminal Strep-HA fusion was used for the generation of a stable cell line using the FRT-HEK293 cells (Thermo Fisher Scientific) containing a single genomic FRT site supporting the rapid generation of bait-expressing cell lines by Flp-mediated recombination (O’Gorman et al., 1991; Glatter et al., 2009). Stable cell line HEK293/CB2 was cultured in complete medium (D-MEM (high glucose), 10% FBS, 2 mM L-glutamine, 100 ug/ml Hygromycin) at 37°C, 5% CO2.
Affinity Purification and Mass Spectrometric Analysis
The Dualsystems CaptiVate approach including LC–MS/MS analysis and peptide assignment were described previously in detail (Glatter et al., 2009). For the analysis of stable cell line and bait expression protein extraction was performed with 0.5% DDM containing affinity purification lysis buffer (Dualsystems) followed by a 12% SDS-PAGE and Western blotting. A mouse monoclonal anti-HA antibody was used for bait detection. Calculated mass of the fusion protein was 50 kDa. Pulldown was performed with CB2 expressing and non-expressing control Flp-In HEK293 cell lysates and 100 μl Strep-Tactin Sepharose. Affinity purification and mass spectrometric analysis of Strep-HA CB2 was performed in three biological replicates for quantitative mass spectrometric analysis and monitored by Western blotting. The samples were analyzed on a Thermo LTQ Orbitrap XL spectrometer using a C18 column, ESI and a 60 min gradient. The variation of the biological replicates of untransfected (replicates 1–3 control cells) or Strep-HA CB2 expressing HEK293 cells (replicates 1–3 CB2 cells) was below 10% that was calculated as Pearson’s correlation coefficient of variation (r) by GraphPad Prism software. All steps of the CaptiVate approach were done by Dualsystems Biotech AG (Schlieren, Switzerland) and were described in details previously (Glatter et al., 2009). Results and experimental parameters for mass spectrometric analysis and protein identification are given in the Supplementary Tables 1–6.
Background Subtraction
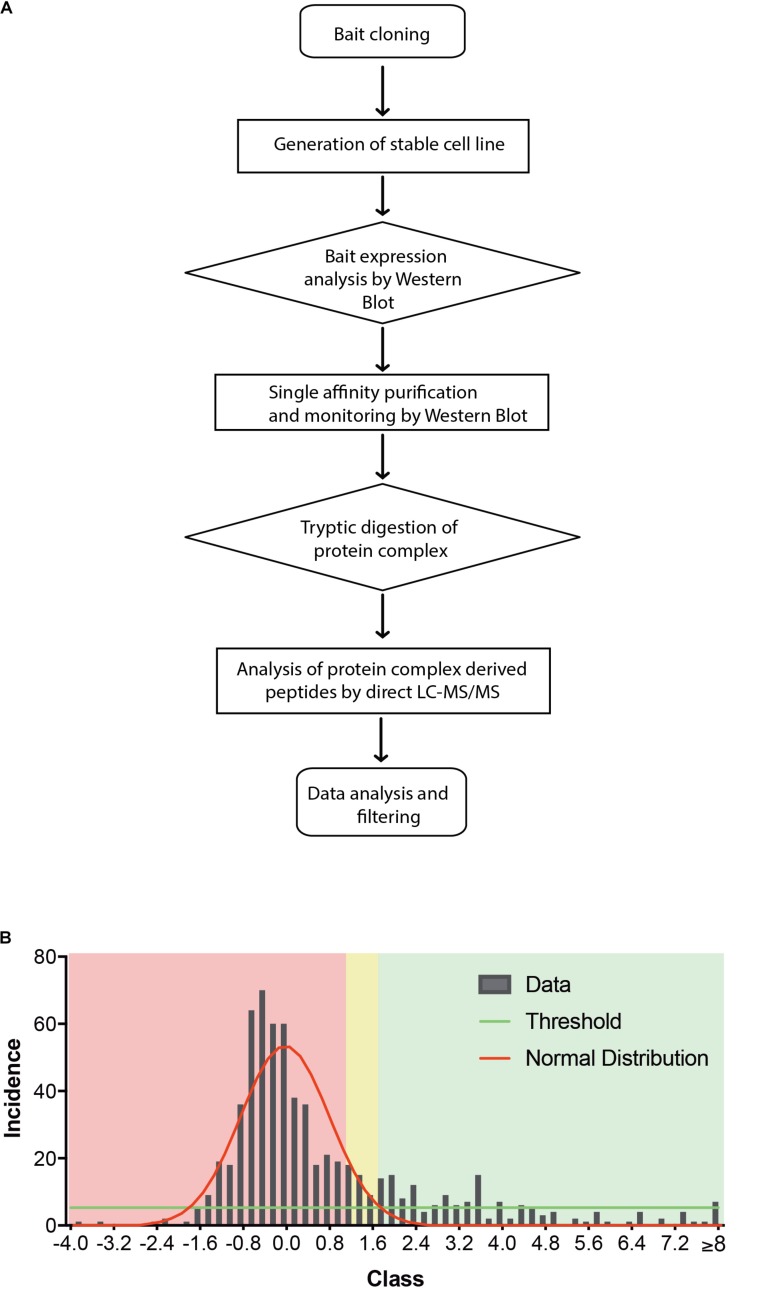
To define the background the affinity purified sample without CB2 (control sample) was used as a negative control experiment. Such a negative control should reproduce a maximum of unspecific bindings that can be subtracted to identify the interacting portion of the obtained peptides. Therefore, we calculated log2 of intensity of probe samples versus control samples (column B) with the range of >1,000 and analyzed the normal distribution (Gauss curve) by counting and distributing them on a bin histogram. For the whole range of log2 values defined bins with increments of 0.2 were set. The adjusted values were multiplicated by total number of values in column B (used for frequency distribution calculation). The threshold was set to 10% of the maximal peak height at 5.3152.
Protein Interaction Analysis
All identified proteins that were detected by more than one unique peptide were used for the generation of an interaction network using the STRING program on https://string-db.org/ (Szklarczyk et al., 2015). The top five results of GO analysis categorized into BP, MF, and CC were also obtained by STRING database. Further GO annotations were performed by the DAVID Bioinformatics Database1 (Huang da et al., 2009a, b). The enrichment score represents the overall enrichment for a group based on the EASE Score of each term members. EASE Score is a modified Fisher Exact p-Value, for gene-enrichment analysis. GraphPad PRISM software (Version 7) was used for linear regression and Gaussian distribution.
Immunocytochemistry
HEK293 cells were plated on poly-L-Lysine coated glass cover slips and transfected with CB2-plasmid (2 μg) with Lipofectamine 2000. After 24 h of transfection cells were washed with HBSS and fixed with Rotifix (4% PFA/PBS) for 15 min at 37°C. After washing the cells were stained with the lectin WGA conjugated with texas red (Texas Red®-X conjugate of WGA 1:200, 15 min, RT) which binds to sialic acid and N-acetylglucosaminyl residues that is used as a marker for plasma membranes before permeabilization. After three washing steps cells were permeabilized with 2% Triton X-100 in 2% BSA in PBS for 45 min at RT on a shaker followed by an incubation with primary antibodies for CB2 (Abnova, 1:300) and p62 (Sigma, 1:300) overnight at 4°C in 2% BSA-PBS. Secondary antibodies (1:1,000 in 2% BSA and 1% normal horse serum in PBS) were incubated for 1 h at RT, cells were washed and mounted with ProLong® Gold antifade reagent with DAPI.
Imaging and 3D Reconstruction Modeling
Four colors images were acquired by mean of a confocal laser scan microscope, i.e., Leica TCS SP5. As excitation four laser lines, 405, 488, 561, 633 nm were employed. For excitation and for detection of the fluorescence signal a 63x NA 1.4 oil objective lens was used. An AOBS separated the detected light from the excitation signal. The fluorescence signal, after being spectrally separated, was directed to a HyD. An estimation of the p62 co-localization with CB2 was performed by using the Manders coefficient. Therefore, we imaged a field of view including approximately 10 cells from eight coverslips in an area of approximately 125 μm2 with a pixel size of approximately 81 nm and a z-stack spacing of 300 nm. The laser light and detector gain were set in a way that the saturation of the detected signal was avoided. After repetitive measurements the background was estimated to be equal to 10/5% of the peak signal for p62/CB2 channels respectively and was subtracted in each channel accordingly. All the post-processing analysis and evaluation were performed with the software Bitplane Imaris. The mean values obtained per image were used for the calculation of mean ± SD. In all the three-dimensional acquisitions the lateral (xy) pixel size was set to be 100 nm (x, y) and the transversal pixel size was set to be 300 nm, i.e., the distance between image planes within a stack (z). Laser intensities and detection gain were to be the same in order to perform comparative studies between the acquired images. The software Bitplane Imaris was used to visualize and study the confocal pictures. As preliminary data visualization the signal offset was determined and subtracted from each image channel. Afterward, a segmentation algorithm was used for visualizing the spatial distribution of the samples under investigation. This algorithm uses a variable intensity threshold based on the local variation of signal to offset ratio to isolate a structure and it was the ideal choice for analyzing objects that, like the ones under investigation, are not continuous within the image volume. Finally, all signal above the local threshold belongs to the structure and all signal under the threshold are set to zero. For each fluorophore a threshold value was determined that represented an optimal coverage of the raw images. Segmented images were than displayed and image snapshots were taken at different visualization angles as well as different zoom factors.
Stimulation of HEK293 Cells With CB2- Synthetic Cannabinoids
Transiently transfected HEK293 cells were stimulated with 100 nM HU-210 for 10 min about 24 h after transfection. Therefore, medium was aspirated and 500 μl of DMEM containing 10% FCS and synthetic cannabinoid or solvent control (ethanol), respectively, were added. HEK293 cells were incubated in 5% CO2 atmosphere at 37°C. Afterward, cell lysates were prepared as described in the following paragraphs.
Co-immunoprecipitation
To immunoprecipitate Flag-tagged proteins transiently transfected HEK293 cells in 6-well cell culture plate were used. About 24 h after transfection the medium of the HEK293 cells was aspirated and the cells were washed with 800 μl ice-cold PBS from the plate and transferred to a 1.5 ml reaction vessel. After a centrifugation step at 8.000 × g at RT for 2 min supernatant was removed and the cell pellet was resuspended in 120 μl 0.2% DDM lysis buffer. Cell suspension was incubated by end-over-end rotation at 4°C for 30 min and centrifuged at 16.200 × g at 4°C for 15 min to remove cell debris. Cell lysates could be directly used for immunoprecipitation or stored at −80°C. 20 μl of cell lysates were used as input control, to this end it was mixed with 5 μl SDS loading buffer and stored on ice until SDS-PAGE. For each immunoprecipitation sample 8 μl anti-Flag® M2 affinity gel was prepared. For that purpose, the whole volume of anti-Flag® M2 affinity gel was washed three times with about 500 μl 0.2% DDM lysis buffer. For each sample 100 μl 0.2% DDM lysis buffer was added to the washed anti-Flag® M2 affinity gel. The remaining cell lysate was diluted with 200 μl 0.2% DDM lysis buffer and 100 μl anti-Flag® M2 affinity gel were added and incubated by end-over-end rotation at 4°C for 1 h. The immunoprecipitates were washed three times with 500 μl lysis buffer containing 0.2% DDM and one time with 500 μl ice-cold PBS. Remaining supernatant was removed with a syringe and cannula. After washing 18 μl SDS loading buffer were added to the immunoprecipitates which were incubated at 70°C for 10 min. The input samples were not heated. Immunoprecipitates and input control samples were analyzed by SDS-PAGE and Western blotting.
Western Blotting Analysis
HEK293 cell lysates were mixed with 6x Laemmli buffer [100 mM Tris-HCl (pH 6.8), 4% SDS, 60% glycerol, 0.2% bromophenol blue, and 10 mM DTT in dH2O]. Proteins of the cell lysate were separated via SDS-PAGE with 12% gels and transferred to Amersham Hybond ECL nitrocellulose membrane (#RPN2032D, GE Healthcare, United Kingdom). After blocking with 5% skimmed milk powder in TBS-T (150 mM NaCl, 10 mM Tris and 0.025% Tween® 20 in dH2O), the membrane was incubated with specific primary antibodies in TBS-T or blocking buffer over night at 4°C. Afterward, fluorescence dye-conjugated or HRP-coupled secondary antibodies were incubated for 1 h at RT. After washing the proteins were detected with the two-channel IR direct detection imaging system from Odyssey LI-COR, United States, or by chemiluminescence. Signal quantifications were done using the Image Studio Software 5.2.5 of LI-COR Bioscience.
Results
Analysis of Affinity Purified CB2 Receptor Complexes
Most biochemical investigations of GPCRs including CB2 are methodologically challenging due to their topology as multiple transmembrane proteins, that is vulnerable to common extraction procedures. We therefore used a method especially applicable for membrane receptors combining systematic affinity purification and mass spectrometric analysis to identify CB2 putative interaction partners (Figure 1A, CaptiVate Dualsystems, performed by Dualsystems Biotech AG, Switzerland) (Glatter et al., 2009). Due to the lack of specific and reliable antibodies for a detection of endogenous CB2 receptors (Cecyre et al., 2014), we were restricted in our studies to use heterologous expression systems. To this aim the bait CB2 was sub-cloned and stably expressed in HEK293 Flp-In cells. Affinity purification and quantitative mass spectrometric analysis (LC–MS/MS) of Strep-HA CB2 were performed in three biological replicates. The samples analyzed showed a variation below 10% of the biological replicates of untransfected or Strep-HA CB2 expressing HEK293 cells (Supplementary Figure 1).
FIGURE 1.
(A) Flow chart of CaptiVate screening procedure. The bait CB2 was amplified and sub-cloned into the expression vector pN-TGSH (Dualsystems). The bait sequence was verified by sequencing. For constitutive expression, the bait was transfected into HEK293 Flp-In cells and selected for stable integration. Bait expression and solubility was monitored by immunoblotting using anti-HA antibodies. The expression level was compared to an internal standard and judged suitable for affinity purification and mass spectrometric analysis. (B) Background filtering of CB2 affinity purification: histogram of enrichment factors (CB2/control) of identifications to determine threshold of enrichment for specific interactors. Red area represents Gaussian distribution of unspecific interactors, yellow area is in the border and in the green area are the specific interactors (threshold as 10% of maximum peak height).
Indeed, a total of 39.2% of the CB2 protein sequence was covered by mass spectrometric data. In total 698 proteins were identified. As expected for a GPCR guanine-nucleotide-binding protein G subunits alpha GNAI3 and GNAS1 have each been identified by one unique peptide. After subtraction of background interactions (Figure 1B) and protein frequency library assessment (Glatter et al., 2009) we could identify 83 putative interactors that were classified by the identification of minimally two unique peptides as highly probable interactors (Table 2).
TABLE 2.
CB2 interactors in HEK293 cells.
| Entry | Entry name | Protein names | Unique | Sequence | Molecular | Ratio |
| peptides | coverage [%] | weight [kDa] | CB2/ctrl | |||
| P34972 | CNR2_HUMAN | Cannabinoid receptor 2 | 16 | 39.2 | 39.68 | 845.0 |
| Q15392 | DHC24_HUMAN | Delta(24)-sterol reductase | 2 | 5.4 | 60.101 | Only in CB2 |
| Q9Y394 | DHRS7_HUMAN | Dehydrogenase/reductase SDR family member 7 | 2 | 14.2 | 38.298 | Only in CB2 |
| Q9UJ14 | GGT7_HUMAN | Glutathione hydrolase 7 | 4 | 11.2 | 70.466 | Only in CB2 |
| Q5T447 | HECD3_HUMAN | E3 ubiquitin-protein ligase HECTD3 | 2 | 3.8 | 97.112 | Only in CB2 |
| Q8IYS2 | K2013_HUMAN | Uncharacterized protein KIAA2013 | 4 | 11.1 | 72.68 | Only in CB2 |
| O15162 | PLS1_HUMAN | Phospholipid scramblase 1 | 4 | 14.8 | 35.049 | Only in CB2 |
| O14967 | CLGN_HUMAN | Calmegin | 19 | 45.7 | 70.038 | 12388.6 |
| Q06136 | KDSR_HUMAN | 3-Ketodihydrosphingosine reductase (KDS reductase) | 7 | 32.2 | 36.187 | 2214.8 |
| Q96JJ7 | TMX3_HUMAN | Protein disulfide-isomerase TMX3 | 13 | 39 | 51.871 | 551.6 |
| Q99720 | SGMR1_HUMAN | Sigma non-opioid intracellular receptor 1 | 6 | 40.4 | 25.127 | 370.9 |
| P16615 | AT2A2_HUMAN | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 (SERCA2) | 44 | 48.6 | 114.76 | 306.2 |
| P27824 | CALX_HUMAN | Calnexin (IP90) (Major histocompatibility complex class I antigen-binding protein p88) | 38 | 57.9 | 67.567 | 200.4 |
| Q8IWV7 | UBR1_HUMAN | E3 ubiquitin-protein ligase UBR1 | 3 | 2.8 | 200.21 | 170.0 |
| P37268 | FDFT_HUMAN | Squalene synthase | 8 | 24.5 | 48.115 | 168.0 |
| Q99442 | SEC62_HUMAN | Translocation protein SEC62 | 4 | 10 | 45.861 | 164.7 |
| O43505 | B4GA1_HUMAN | Beta-1.4-glucuronyltransferase 1 | 2 | 12.8 | 47.119 | 161.7 |
| Q9UBV2 | SE1L1_HUMAN | Protein sel-1 homolog 1 | 2 | 6.2 | 88.754 | 124.8 |
| P10586 | PTPRF_HUMAN | Receptor-type tyrosine-protein phosphatase F | 5 | 4.5 | 212.88 | 88.1 |
| O95260 | ATE1_HUMAN | Arginyl-tRNA–protein transferase 1 | 4 | 10.2 | 59.09 | 87.4 |
| Q96A33 | CCD47_HUMAN | Coiled-coil domain-containing protein 47 | 11 | 34.4 | 55.873 | 85.3 |
| O15269 | SPTC1_HUMAN | Serine palmitoyltransferase 1 | 4 | 11.8 | 52.743 | 77.3 |
| Q4ZIN3 | MBRL_HUMAN | Membralin (Transmembrane protein 259) | 3 | 7.3 | 67.888 | 62.9 |
| O43149 | ZZEF1_HUMAN | Zinc finger ZZ-type and EF-hand domain-containing protein 1 | 5 | 2.8 | 331.07 | 55.5 |
| Q13501 | SQSTM_HUMAN | Sequestosome-1 | 6 | 23.4 | 47.687 | 52.7 |
| O15258 | RER1_HUMAN | Protein RER1 | 2 | 15.3 | 22.958 | 51.5 |
| Q9H3N1 | TMX1_HUMAN | Thioredoxin-related transmembrane protein 1 | 3 | 12.9 | 31.791 | 42.2 |
| O00264 | PGRC1_HUMAN | Membrane-associated progesterone receptor component 1 | 8 | 57.4 | 21.671 | 40.5 |
| Q15043 | S39AE_HUMAN | Zinc transporter ZIP14 (LIV-1 subfamily of ZIP zinc transporter 4) | 2 | 6.7 | 54.212 | 31.2 |
| Q8NI60 | COQ8A_HUMAN | Atypical kinase COQ8A mitochondrial | 3 | 8.3 | 71.949 | 28.5 |
| P55084 | ECHB_HUMAN | Trifunctional enzyme subunit beta mitochondrial | 15 | 43.7 | 51.294 | 27.8 |
| P42356 | PI4KA_HUMAN | Phosphatidylinositol 4-kinase alpha | 2 | 1.6 | 231.32 | 26.3 |
| O75915 | PRAF3_HUMAN | PRA1 family protein 3 (ADP-ribosylation factor-like protein 6-interacting protein 5) | 2 | 16 | 21.614 | 25.6 |
| A1L0T0 | ILVBL_HUMAN | Acetolactate synthase-like protein | 4 | 10.9 | 67.867 | 23.1 |
| Q9Y4P3 | TBL2_HUMAN | Transducin beta-like protein 2 | 2 | 5.6 | 49.797 | 22.5 |
| O60884 | DNJA2_HUMAN | DnaJ homolog subfamily A member 2 | 2 | 6.8 | 45.745 | 22.2 |
| Q29963 | 1C06_HUMAN | HLA class I histocompatibility antigen. Cw-6 alpha chain (MHC class I antigen Cw∗6) | 5 | 21.9 | 40.968 | 21.5 |
| P20020 | AT2B1_HUMAN | Plasma membrane calcium-transporting ATPase 1 | 3 | 4.2 | 138.75 | 21.1 |
| P49755 | TMEDA_HUMAN | Transmembrane emp24 domain-containing protein 10 | 2 | 12.8 | 24.976 | 19.8 |
| P62341 | SELT_HUMAN | Thioredoxin reductase-like selenoprotein T (SelT) | 2 | 17.4 | 22.174 | 18.8 |
| P50395 | GDIB_HUMAN | Rab GDP dissociation inhibitor beta | 2 | 8.5 | 50.663 | 17.0 |
| P49411 | EFTU_HUMAN | Elongation factor Tu mitochondrial (EF-Tu) | 23 | 61.5 | 49.541 | 15.2 |
| Q8IXI1 | MIRO2_HUMAN | Mitochondrial Rho GTPase 2 (MIRO-2) (hMiro-2) | 2 | 4.4 | 68.117 | 15.2 |
| Q14257 | RCN2_HUMAN | Reticulocalbin-2 | 2 | 11 | 36.876 | 14.6 |
| Q9BQE3 | TBA1C_HUMAN | Tubulin alpha-1C chain (Alpha-tubulin 6) | 2 | 76.2 | 49.895 | 12.4 |
| Q9BXW9 | FACD2_HUMAN | Fanconi anemia group D2 protein (Protein FACD2) | 2 | 2.3 | 166.46 | 12.1 |
| P04844 | RPN2_HUMAN | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 2 | 11 | 37.9 | 69.283 | 12.1 |
| P04843 | RPN1_HUMAN | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase subunit 1 | 19 | 43 | 68.569 | 12.0 |
| P40939 | ECHA_HUMAN | Trifunctional enzyme subunit alpha mitochondrial | 27 | 53.1 | 82.999 | 12.0 |
| P39656 | OST48_HUMAN | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase 48 kDa subunit | 11 | 45.2 | 50.8 | 11.9 |
| Q9BVA1 | TBB2B_HUMAN | Tubulin beta-2B chain | 3 | 67.2 | 49.953 | 11.5 |
| Q9BQB6 | VKOR1_HUMAN | Vitamin K epoxide reductase complex subunit 1 | 2 | 19 | 18.234 | 11.1 |
| P13797 | PLST_HUMAN | Plastin-3 (T-plastin) | 2 | 6.3 | 70.81 | 11.0 |
| Q9HCU5 | PREB_HUMAN | Prolactin regulatory element-binding protein | 3 | 15.1 | 45.468 | 10.8 |
| P07237 | PDIA1_HUMAN | Protein disulfide-isomerase (PDI) | 2 | 4.3 | 57.116 | 10.3 |
| P51648 | AL3A2_HUMAN | Fatty aldehyde dehydrogenase | 6 | 14.4 | 57.669 | 9.9 |
| P13073 | COX41_HUMAN | Cytochrome c oxidase subunit 4 isoform 1 mitochondrial | 2 | 13.6 | 19.576 | 9.0 |
| P08195 | 4F2_HUMAN | 4F2 cell-surface antigen heavy chain (4F2hc) | 3 | 7 | 71.122 | 9.0 |
| Q15363 | TMED2_HUMAN | Transmembrane emp24 domain-containing protein 2 (Membrane protein p24A) | 2 | 10.9 | 22.761 | 8.8 |
| P61204 | ARF3_HUMAN | ADP-ribosylation factor 3 | 2 | 18.2 | 20.601 | 8.8 |
| O15173 | PGRC2_HUMAN | Membrane-associated progesterone receptor component 2 | 2 | 18.8 | 23.818 | 7.9 |
| O95831 | AIFM1_HUMAN | Apoptosis-inducing factor 1 mitochondrial | 5 | 11.1 | 66.9 | 6.8 |
| P00403 | COX2_HUMAN | Cytochrome c oxidase subunit 2 | 3 | 16.3 | 25.565 | 6.1 |
| P05109 | S10A8_HUMAN | Protein S100-A8 (Calgranulin-A) | 3 | 32.3 | 10.834 | 6.0 |
| Q12931 | TRAP1_HUMAN | Heat shock protein 75 kDa mitochondrial (HSP 75) (TNFR-associated protein 1) | 7 | 13.2 | 80.109 | 5.5 |
| P22102 | PUR2_HUMAN | Trifunctional purine biosynthetic protein adenosine-3 | 4 | 7.5 | 107.77 | 5.4 |
| O14975 | S27A2_HUMAN | Very long-chain acyl-CoA synthetase (VLACS) (VLCS) | 2 | 5.5 | 70.311 | 5.2 |
| P11586 | C1TC_HUMAN | C-1-Tetrahydrofolate synthase cytoplasmic (C1-THF synthase) | 2 | 2.7 | 101.56 | 5.1 |
| O75396 | SC22B_HUMAN | Vesicle-trafficking protein SEC22b (ER-Golgi SNARE of 24 kDa) | 2 | 10.2 | 24.593 | 5.1 |
| P30101 | PDIA3_HUMAN | Protein disulfide-isomerase A3 | 5 | 13.9 | 56.782 | 5.1 |
| P53618 | COPB_HUMAN | Coatomer subunit beta (Beta-coat protein) | 2 | 2.2 | 107.14 | 4.9 |
| P51571 | SSRD_HUMAN | Translocon-associated protein subunit delta (TRAP-delta) | 2 | 18.5 | 18.998 | 4.8 |
| P61619 | S61A1_HUMAN | Protein transport protein Sec61 subunit alpha isoform 1 (Sec61 alpha-1) | 2 | 9 | 52.264 | 4.6 |
| P30048 | PRDX3_HUMAN | Thioredoxin-dependent peroxide reductase mitochondrial | 2 | 9.8 | 27.692 | 4.5 |
| Q7Z6Z7 | HUWE1_HUMAN | E3 ubiquitin-protein ligase HUWE1 | 2 | 0.5 | 481.89 | 4.3 |
| P50402 | EMD_HUMAN | Emerin | 3 | 17.3 | 28.994 | 4.3 |
| P24534 | EF1B_HUMAN | Elongation factor 1-beta (EF-1-beta) | 3 | 26.7 | 24.763 | 4.1 |
| Q53GQ0 | DHB12_HUMAN | Very-long-chain 3-oxoacyl-CoA reductase | 4 | 21.2 | 34.324 | 4.0 |
| Q9P035 | HACD3_HUMAN | Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase 3 | 9 | 33.1 | 43.159 | 3.8 |
| Q00325 | MPCP_HUMAN | Phosphate carrier protein mitochondrial (Phosphate transport protein) (PTP) | 8 | 28.5 | 39.958 | 3.7 |
| O14980 | XPO1_HUMAN | Exportin-1 | 5 | 8.3 | 123.38 | 3.7 |
| P06702 | S10A9_HUMAN | Protein S100-A9 (Calgranulin-B) | 3 | 36 | 13.242 | 3.7 |
| Q86VU5 | CMTD1_HUMAN | Catechol O-methyltransferase domain-containing protein 1 | 4 | 25.2 | 28.808 | 3.6 |
| Q93008 | USP9X_HUMAN | Probable ubiquitin carboxyl-terminal hydrolase FAF-X | 3 | 1.7 | 292.28 | 3.6 |
All specific interactors are classified by the identification in mass spectrometry data of triplicates by at least two unique peptides. The Uniprot accession numbers with protein name, descriptions and molecular weights are given. Sequence coverage by identified peptides is presented in %. Additionally, the ratio of identification in CB2 expressing cells to control cells was calculated.
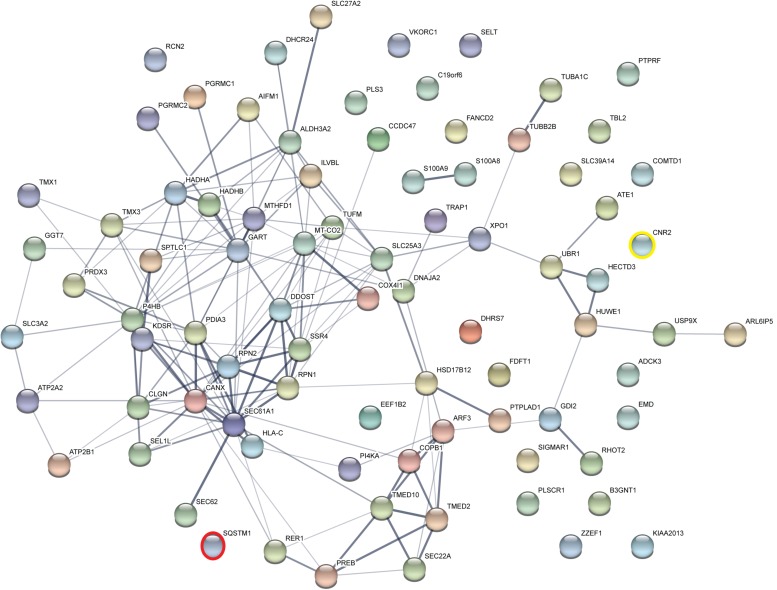
Taking those 83 putative interactors we processed a functional protein association network analysis using the STRING online tool (Szklarczyk et al., 2015) obtaining an interaction network with a protein–protein-interaction enrichment p-value < 1.0e-16. This network consists of 84 nodes (including CB2) and 163 edges with an average node degree of 3.8 and an average local clustering coefficient of 0.392. In the network a significant functional enrichment of proteins involved in protein metabolic process and in response to endoplasmic reticulum (ER) stress were obtained using the GO BP analysis (Table 3). These functions are in line with the findings of CC ontology revealing an annotation of ER in 35 CB2 interacting proteins (Table 3). Further, the MF gene ontology analysis unveiled an enrichment of proteins with catalytic activity and oxidoreductase activity (Table 3). Functional annotations that were used by the STRING database to illustrate interactions network were determined by: text mining, experiments, or databases with a minimum required interaction score of 0.400 (Figure 2). The nodes represent proteins whereas color saturation of the edges mark confidence of the interactions indicating the strength of data support. Several proteins were not connected by any known interactions to the other proteins and are displayed as nodes without any edges. In contrast, some proteins particularly located in the center of the network with many interactions are ER proteins like calnexin and Sec61A1. Also the cargo receptors for GPCR trafficking and resensitization TMED2 and TMED10 have been identified in this network.
TABLE 3.
Gene ontology (GO) analysis of 84 CB2 specific interactors (including CB2) using the STRING database (https://string-db.org/).
| Biological process (GO) | |||
| Pathway ID | Pathway description | Count in | False discovery |
| gene set | rate | ||
| GO:0019538 | Protein metabolic process | 37 | 2.8e-05 |
| GO:0034976 | Response to endoplasmic reticulum stress | 10 | 2.8e-05 |
| GO:0044267 | Cellular protein metabolic process | 34 | 2.8e-05 |
| GO:0006457 | Protein folding | 9 | 0.000337 |
| GO:0061024 | Membrane organization | 16 | 0.000342 |
| Molecular function (GO) | |||
| GO:0003824 | Catalytic activity | 41 | 0.000457 |
| GO:0016491 | Oxidoreductase activity | 14 | 0.000457 |
| GO:0003756 | Protein disulfide isomerase activity | 4 | 0.00114 |
| GO:0016408 | C-Acyltransferase activity | 3 | 0.0118 |
| GO:0016509 | Long-chain-3-hydroxyacyl-CoA dehydrogenase activity | 2 | 0.0118 |
| Cellular component (GO) | |||
| GO:0042175 | Nuclear outer membrane-endoplasmic reticulum membrane network | 31 | 3.40e-17 |
| GO:0044432 | Endoplasmic reticulum part | 29 | 1.17e-16 |
| GO:0005789 | Endoplasmic reticulum membrane | 31 | 1.17e-16 |
| GO:0005783 | Endoplasmic reticulum | 35 | 4.01e-16 |
| GO:0031090 | Organelle membrane | 42 | 4.50e-13 |
Displayed are the most significant top five GO pathways in the category Biological Process (BP), Molecular Function (MF), Cellular Component (CC).
FIGURE 2.
Functional Annotations were used to visualize protein–protein interaction network of 83 identified CB2 interactors by STRING. Illustrated interaction network was determined by text mining, experiments or databases with a minimum required interaction score of 0.400. In this view, the nodes represent proteins and the color saturation of the edges represents the confidence score of data support. CNR2 represents CB2 and SQSTM1 represents p62.
To identify further functional annotations, a systematic analysis of all 83 CB2 interactors was performed using the “DAVID Functional Annotation Tool” (Huang da et al., 2009a, b). The analysis identified 20 clusters with 10 clusters having an enrichment score higher than 2.0 (Table 4). The top five of these clusters with a highly significant enrichment (p < 0.0001) in the obtained CB2 interactome (Table 4) represent the ER (cluster 1), membrane (cluster 2), oxidoreductase (cluster 3), ER stress and cell redox homeostasis (cluster 4), and lipid metabolism (cluster 5). As the CB2 receptor is expected to interact with nucleotide binding G protein subunits, we screened the functional annotation list with the keyword “nucleotide binding” and identified 13 proteins belonging to this cluster (p-value = 5.4 e-2). Out of them five CB2 interactors were enriched for the GOTerm “GTP binding,” namely the ADP-ribosylation factor 3 (ARF3), the translation elongation factor TUFM, the member of the Rho family of GTPases RHOT2, and the tubulin chains TUBA1C and TUBB2B (p-value of 2.5 e-2).
TABLE 4.
Gene ontology (GO) functional annotation clustering.
| Count | p_Value | FDR | ||
| Cluster 1 | Enrichment score: 15.5 | |||
| UP_KEYWORDS | Endoplasmic reticulum | 34 | 2.7E-21 | 3.3E-18 |
| GOTERM_CC_DIRECT | Endoplasmic reticulum membrane | 27 | 3.2E-15 | 3.8E-12 |
| UP_SEQ_FEATURE | Topological domain:lumenal | 18 | 3.6E-12 | 4.9E-9 |
| Cluster 2 | Enrichment score: 9.61 | |||
| UP_KEYWORDS | Membrane | 63 | 1.3E-12 | 1.6E-9 |
| UP_KEYWORDS | Transmembrane helix | 54 | 4.0E-12 | 5.0E-9 |
| UP_KEYWORDS | Transmembrane | 54 | 4.6E-12 | 5.6E-9 |
| UP_SEQ_FEATURE | Transmembrane region | 48 | 1.1E-9 | 1.5E-6 |
| GOTERM_CC_DIRECT | Integral component of membrane | 50 | 2.7E-9 | 3.2E-6 |
| UP_SEQ_FEATURE | Topological domain:cytoplasmic | 33 | 3.1E-6 | 4.3E-3 |
| Cluster 3 | Enrichment score: 3.59 | |||
| UP_KEYWORDS | Oxidoreductase | 12 | 2.1E-5 | 2.6E-2 |
| GOTERM_BP_DIRECT | Oxidation–reduction process | 12 | 7.7E-5 | 1.1E-1 |
| GOTERM_MF_DIRECT | Oxidoreductase activity | 7 | 3.7E-4 | 4.7E-1 |
| UP_KEYWORDS | NADP | 5 | 6.8E-3 | 8.1E0 |
| Cluster 4 | Enrichment score: 3.17 | |||
| UP_KEYWORDS | Redox-active center | 7 | 4.4E-8 | 5.4E-5 |
| GOTERM_BP_DIRECT | Response to endoplasmic reticulum stress | 6 | 2.4E-5 | 3.4E-2 |
| GOTERM_BP_DIRECT | Cell redox homeostasis | 6 | 2.7E-5 | 3.9E-2 |
| INTERPRO | Thioredoxin domain | 5 | 3.0E-5 | 3.9E-2 |
| INTERPRO | Thioredoxin, conserved site | 4 | 5.2E-5 | 6.8E-2 |
| GOTERM_MF_DIRECT | Protein disulfide isomerase activity | 4 | 1.5E-4 | 1.9E-1 |
| INTERPRO | Thioredoxin-like fold | 6 | 2.6E-4 | 3.4E-1 |
| GOTERM_CC_DIRECT | Cell | 5 | 1.1E-3 | 1.3E0 |
| UP_SEQ_FEATURE | Short sequence motif:prevents secretion from ER | 4 | 2.1E-3 | 2.8E0 |
| GOTERM_MF_DIRECT | Isomerase activity | 3 | 4.4E-3 | 5.4E0 |
| UP_SEQ_FEATURE | Domain:thioredoxin | 3 | 8.2E-3 | 1.1E1 |
| UP_KEYWORDS | Isomerase | 3 | 8.7E-2 | 6.7E1 |
| UP_SEQ_FEATURE | Disulfide bond | 14 | 4.3E-1 | 1.0E2 |
| UP_KEYWORDS | Disulfide bond | 13 | 7.5E-1 | 1.0E2 |
| Cluster 5 | Enrichment score: 2.87 | |||
| GOTERM_MF_DIRECT | Long-chain-3-hydroxyacyl-CoA dehydrogenase activity | 3 | 6.6E-5 | 8.3E-2 |
| UP_KEYWORDS | Lipid metabolism | 9 | 3.6E-4 | 4.4E-1 |
| KEGG_PATHWAY | Fatty acid elongation | 4 | 5.4E-4 | 5.8E-1 |
| KEGG_PATHWAY | Fatty acid metabolism | 4 | 3.6E-3 | 3.8E0 |
| KEGG_PATHWAY | Biosynthesis of unsaturated fatty acids | 3 | 9.6E-3 | 9.9E0 |
| UP_KEYWORDS | Fatty acid metabolism | 4 | 1.4E-2 | 1.6E1 |
| Cluster 6 | Enrichment score: 2.56 | |||
| GOTERM_CC_DIRECT | Mitochondrial inner membrane | 9 | 8.2E-4 | 9.8E-1 |
| UP_SEQ_FEATURE | Transit peptide:mitochondrion | 9 | 8.5E-4 | 1.2E0 |
| UP_KEYWORDS | Transit peptide | 9 | 1.4E-3 | 1.7E0 |
| UP_KEYWORDS | Mitochondrion inner membrane | 6 | 4.7E-3 | 5.7E0 |
| GOTERM_CC_DIRECT | Mitochondrion | 14 | 6.3E-3 | 7.2E0 |
| UP_KEYWORDS | Mitochondrion | 11 | 1.4E-2 | 1.6E1 |
| Cluster 7 | Enrichment score: 2.54 | |||
| GOTERM_BP_DIRECT | Protein folding | 7 | 1.8E-4 | 2.5E-1 |
| UP_KEYWORDS | Chaperone | 5 | 8.9E-3 | 1.0E1 |
| GOTERM_MF_DIRECT | Unfolded protein binding | 4 | 1.5E-2 | 1.8E1 |
| Cluster 8 | Enrichment score: 2.38 | |||
| GOTERM_CC_DIRECT | Endoplasmic reticulum-Golgi intermediate compartment | 6 | 1.4E-5 | 1.6E-2 |
| GOTERM_BP_DIRECT | Retrograde vesicle-mediated transport, Golgi to ER | 6 | 3.7E-5 | 5.2E-2 |
| UP_KEYWORDS | ER-Golgi transport | 6 | 3.9E-5 | 4.8E-2 |
| GOTERM_CC_DIRECT | ER to Golgi transport vesicle membrane | 4 | 1.7E-3 | 2.0E0 |
| GOTERM_BP_DIRECT | COPII vesicle coating | 4 | 2.8E-3 | 3.9E0 |
| UP_KEYWORDS | Protein transport | 9 | 3.2E-3 | 3.8E0 |
| GOTERM_CC_DIRECT | Golgi membrane | 9 | 5.1E-3 | 5.9E0 |
| GOTERM_BP_DIRECT | ER to Golgi vesicle-mediated transport | 5 | 6.3E-3 | 8.6E0 |
| GOTERM_CC_DIRECT | Transport vesicle | 4 | 9.0E-3 | 1.0E1 |
| GOTERM_CC_DIRECT | Golgi apparatus | 10 | 1.5E-2 | 1.7E1 |
| GOTERM_CC_DIRECT | Endoplasmic reticulum-Golgi intermediate compartment membrane | 3 | 3.4E-2 | 3.4E1 |
| UP_KEYWORDS | Golgi apparatus | 8 | 4.5E-2 | 4.4E1 |
| GOTERM_BP_DIRECT | Transport | 5 | 7.6E-2 | 6.7E1 |
| GOTERM_BP_DIRECT | Intracellular protein transport | 4 | 9.4E-2 | 7.6E1 |
| UP_KEYWORDS | Cytoplasmic vesicle | 5 | 1.4E-1 | 8.3E1 |
| Cluster 9 | Enrichment score: 2.32 | |||
| UP_KEYWORDS | Lipid metabolism | 9 | 3.6E-4 | 4.4E-1 |
| UP_KEYWORDS | NADP | 5 | 6.8E-3 | 8.1E0 |
| UP_KEYWORDS | Steroid biosynthesis | 3 | 8.8E-3 | 1.0E1 |
| UP_KEYWORDS | Lipid biosynthesis | 4 | 2.5E-2 | 2.7E1 |
| Cluster 10 | Enrichment score: 2.11 | |||
| GOTERM_MF_DIRECT | Dolichyl-diphosphooligosaccharide-protein glycotransferase activity | 3 | 7.8E-4 | 9.8E-1 |
| GOTERM_CC_DIRECT | Oligosaccharyltransferase complex | 3 | 8.8E-4 | 1.0E0 |
| GOTERM_BP_DIRECT | Protein N-linked glycosylation via asparagine | 3 | 1.4E-2 | 1.9E1 |
| KEGG_PATHWAY | N-Glycan biosynthesis | 3 | 4.0E-2 | 3.6E1 |
| UP_KEYWORDS | Glycosyltransferase | 4 | 7.0E-2 | 5.9E1 |
After analyzing 84 CB2 specific interacting proteins (including CB2) with DAVID (https://david.ncifcrf.gov/home.jsp) 20 clusters were identified of which 10 were represented in this table with an enrichment score >2.
Furthermore, we identified six proteins that were only expressed in the stable Strep-HA-CB2-HEK293 cells; the delta(24)-sterol reductase (DHC24), dehydrogenase/reductase SDR family member 7 (DHRS7), glutathione hydrolase 7 (GGT7), E3 ubiquitin-protein ligase HECTD3 (HECD3), an uncharacterized protein KIAA2013 (K2013), and the phospholipid scramblase 1 (PLS1).
P62 Is an Interaction Partner of Cannabinoid Receptor CB2
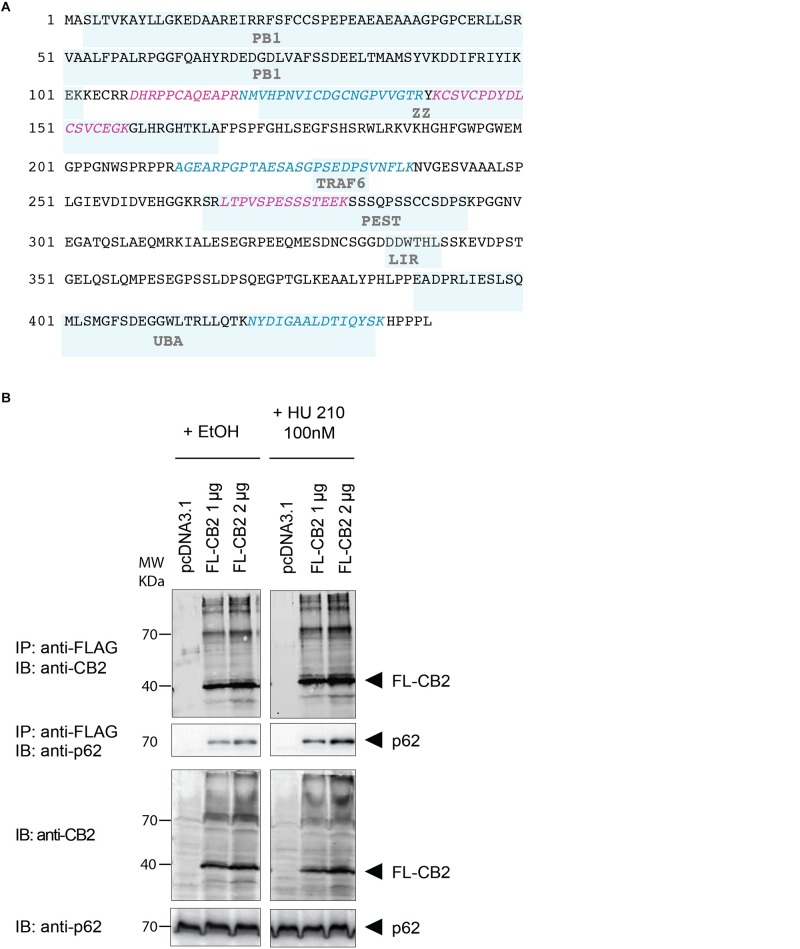
To verify the experimental approach and the obtained interactome list, we selected one specific protein with a high probability of interaction. To increase the likelihood of a direct protein interaction with CB2, this candidate should have a relatively low number of edges in our STRING network (Figure 2). Furthermore, the candidate should represent a signaling hub protein that might be involved in receptor and signaling crosstalk. Therefore, we chose p62/SQSTM1 for further experiments. The scaffold protein p62 is included in the 34 proteins of the functional annotation cluster 1 representing ER association. It is involved in TNF receptor signaling (Kim and Ozato, 2009) and in the development of PDB (Laurin et al., 2002) and functions as a signaling hub and as an autophagy adaptor (Katsuragi et al., 2015). The identification of p62 based on six specific tryptic peptides (i.e., AGEARPGPTAESASGPSEDPSVNFLK, CSVCPDYDLCSVCEGK, DHRPPCAQEA-PR, LTPVSPESSST EEK, NMVHPNVICDGCNGPVVGTR, NYDIGAALDTIQYSK) that represent 23.4% of the whole protein sequence (Figure 3A). Indeed, the quantification ratio of CB2 stable expressing versus control cells showed that p62 specific peptides were identified 52 times more in CB2-cells than in controls (Table 2).
FIGURE 3.
(A) Protein sequence of p62 with protein domains underlaid in light blue boxes (Phox and Bem1p domain (PB1), Zinc finger-type (ZZ), TNF receptor-associated factor 6 (TRAF6), LC3-interacting region (LIR), protein sequence rich in proline (P), glutamic acid (E), serine (S), and threonine (T) (PEST), UBA. The six peptides identified after tryptic digestion by mass spectrometry are highlighted in red and blue. (B) Verification of the interaction of CB2 receptors with p62 by co-immunoprecipitation (Co-IP) of transiently overexpressed flag tagged CB2 receptors (FL-CB2) with endogenously expressed p62 in HEK293 cells. The precipitated p62 protein intensity directly correlated with the amount of transfected CB2-receptor plasmid (either 1 or 2 μg) that was not influenced by 10 min stimulation with the CB2 agonist HU-210. Representative result of n > 3.
In the next step, we verified the physical interaction of CB2 receptors with p62 by co-immunoprecipitation (Figure 3B). To this end we generated a Flag-tagged CB2 receptor using the expression vector pcDNA3.1 and transiently transfected HEK293 cells. We precipitated CB2 using beads carrying FLAG M2 antibodies and detected endogenous p62 by using an antibody against the human p62 protein (Figure 3B). The endogenous p62 was co-precipitated in Flag-tagged CB2-transfected (FL-CB2) cell extracts in an intensity directly correlated with the amount of transfected CB2-receptor plasmid (either 1 or 2 μg). In all pre-IP extracts (input) p62 was detectable in comparable intensities whereas in control transfected protein extracts (pcDNA3.1) no p62 band was visible after immunoprecipitation. The stimulation with a CB2 receptor agonist (HU-210) did not alter the interaction (Figure 3B). These results strongly indicate that CB2 receptor interacts with p62 and that stimulation of CB2 does not interfere with this interaction.
CB2 Receptors Colocalize With p62 Vesicles Mainly in Plasma Membrane Associated Regions
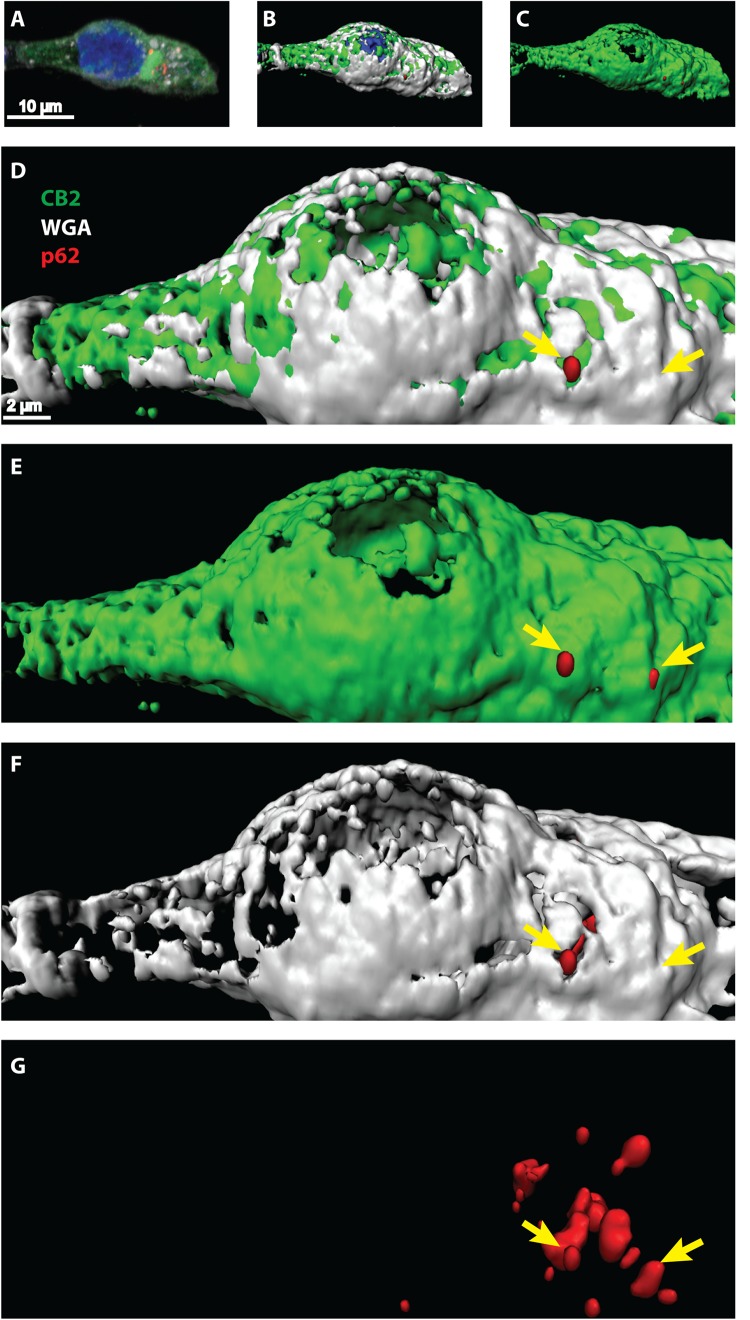
Once activated, GPCRs are typically internalized and either degraded or recycled to the membrane for another activation cycle. Degradation can occur via lysosome and autophagy pathways where the scaffolding protein p62 is involved in. Since we showed that CB2 interacts with p62 in whole cell lysates we wondered at which cellular compartment this interaction is taking place. We performed immunocytochemical studies of HEK293 cells expressing endogenously p62 and transiently CB2. For surface membrane staining we used WGA-texas red labeled probes. Confocal microscopy revealed a broad and predominant membrane localization of transiently expressed CB2 receptors (Figure 4 and Supplementary Figure 2) but a localization of p62 in dotted and vesicular like structures that are most probable lysosome and autophagosome areas. Because of the relatively strong heterologous expression of CB2 we selected the p62 positive areas and analyzed the co-localization with CB2 receptors using the Manders coefficient and obtained a mean value of 0.46 ± 0.07. Additionally, we aimed to better visualize and to clearly distinguish localizations of both proteins in co-localization with the membrane marker. Therefore, we performed a 3D reconstruction modeling of confocal images. With this approach we remodeled the p62 expression in red vesicular structures (Figures 4D–G). We used for our analysis a segmentation algorithm that eliminates signals below a threshold to visualize the p62 vesicles optimally. Thus, any signals of p62 that are weaker are not present in the 3D-reconstruction images. In white/gray we visualized the areas that were positively stained with membrane marker (Figures 4D,F) that intermingled with green CB2 receptor signals (Figures 4D,E). Our 3D reconstruction images indicated that those p62 vesicles were surrounded by CB2 positive areas that were adjacent to the surface membrane (Figure 4D). These results demonstrate that about half of the p62 positive areas co-expressed CB2 and that these p62-CB2 vesicles are at the cell membrane.
FIGURE 4.
(A–G) Confocal light microscopy of HEK293 cells transiently expressing CB2 receptors. (A) Immunohistochemical staining of CB2 (green), p62 (red), and DAPI (blue). The membrane was stained by Texas-red conjugated wheat-germ-agglutinin (white) and DAPI revealed the localization of the nucleus. (B–G) 3D reconstruction modeling of confocal images using a segmentation algorithm to distinguish single signal from different channels. (D–G) Higher zoom in of 3D reconstructed cell showing a strong overlap of CB2 positive areas with the membrane marker (white). P62 was found in vesicular like structures that intermingled with membranal CB2 positive areas (yellow arrows).
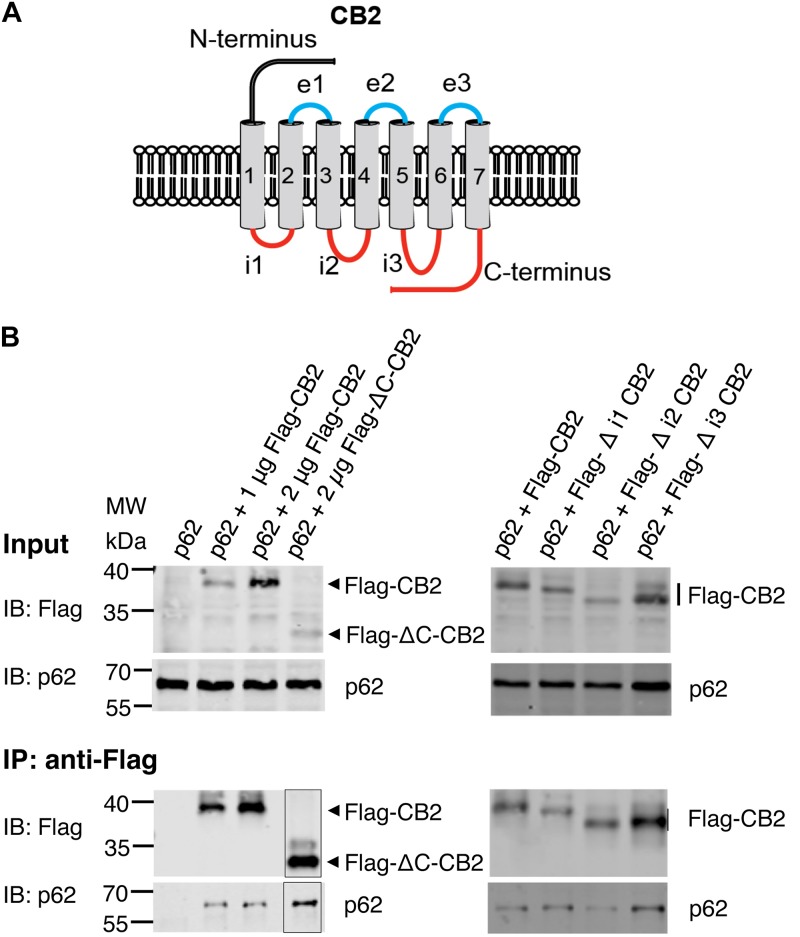
Intracellular Regions of CB2 Receptor Are Not Responsible for the Interaction With p62
Since p62 is localized intracellularly we wondered whether p62 interacts with intracellular domains of CB2. We designed expression constructs deleting the three intracellular loops and the C-terminus of the CB2 receptor (Figure 5A). Surprisingly none of these deletion constructs altered the interaction of CB2 with p62 in co-IP studies if correlated to the different expression levels in the input and the IPs (Figure 5B). We conclude from this experiment that it is not individual intracellular loops that are responsible for the interaction, but rather that multiple domains are involved in the interaction with p62. Because receptor structure is strongly dependent on the transmembrane domains, we have not investigated further deletion constructs of CB2 in this assay.
FIGURE 5.
(A) Schematic drawing of CB2 receptor protein with extracellular N-terminus and three extracellular loops (e1, e2, e3), seven transmembrane domains, and three intracellularly localized loops (i1, i2, i3) and the C-terminus. The red marked intracellular regions were deleted in CB2 expression plasmids and used for following Co-IP studies. (B) Representative Western blot results of CB2 precipitations using Flag antibody with lysates of HEK293 cells transiently transfected with p62-WT (1 μg) and Flag-CB2 constructs with the respective deletions (ΔC-CB2, Δ i1-CB2, Δ i2-CB2, Δ i3-CB2). Considering lower expression levels of deletion constructs compared to WT-full length Flag-CB2, the co-precipitated p62 protein levels were similar in all approaches. The IP figure was grouped from cropped parts of the same gels/blots indicated by the border line. This experiment has been performed at least two times independently.
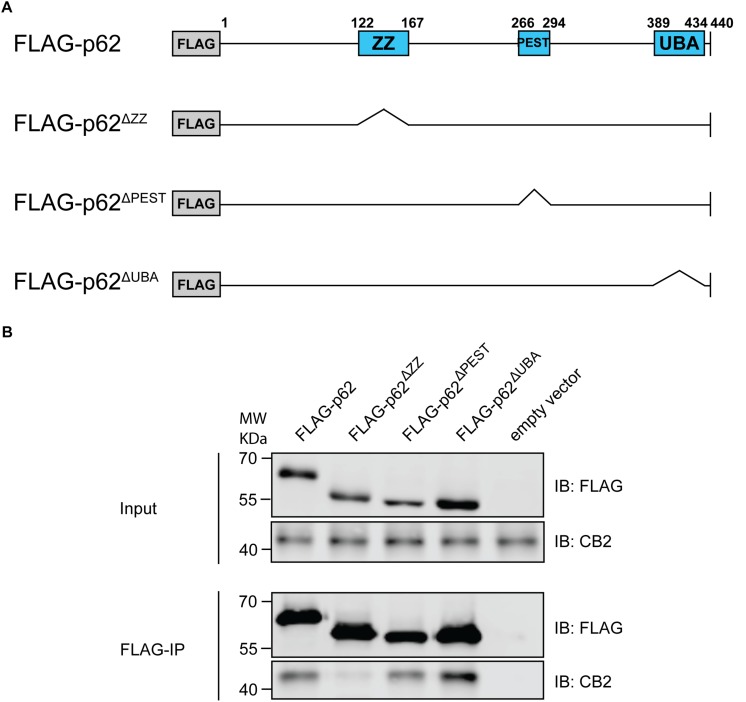
ZZ Domain of p62 Is Important for Interaction With CB2
P62 is a cargo and multifunctional protein carrying several motifs and domains responsible for the interaction with a variety of proteins (Moscat et al., 2007). Therefore, we aimed to identify the motif responsible for CB2 interaction and co-expressed CB2 with deletion constructs of p62 lacking the PB1 domain and the binding motifs ZZ and UBA respectively (Figure 6A). Only the ZZ deletion construct failed to co-precipitate CB2 indicating that this region is important for the interaction of p62 with CB2 (Figure 6B). All other investigated p62 constructs including the UBA-domain deletion protein resulted in CB2 receptor co-precipitations that were comparable to positive controls. We therefore showed that CB2 interacts with p62 via the ZZ-domain.
FIGURE 6.
(A) Schematic illustration of Flag-p62 deletion constructs for ZZ, PEST, and UBA motifs that were used for following co-immunoprecipitation experiments to identify interaction region. (B) Stably expressing HA-CB2 HEK293 cells were transiently transfected with different Flag-p62-deletion constructs, respectively. Protein extracts were used for precipitation of Flag-p62 using Flag antibody. The expression levels in input samples for CB2 and p62 were comparable in all constructs. Immunoprecipitation result of Flag-p62 ΔZZ revealed a significantly reduced coprecipitation of CB2 indicating that this domain in p62 is involved in the interaction with CB2. Representative results of three independent experiments were presented.
In summary, we identified the CB2 interactome in HEK293 cells consisting of 83 interacting proteins involved in, e.g., biosynthesis, trafficking, and signaling. The identified proteins are mainly localized at the ER and other membrane structures in the cell and in intracellular compartments. The autophagy receptor p62 was identified by six unique peptides as an interactor with a very high probability of interaction with CB2 receptor. In our investigations we focused on p62-CB2 complex formation and could verify that CB2 receptor interacts with the ZZ domain of p62 at membranous structures close to the cell surface. Future investigations will concentrate on the characterization of the interaction and its physiological impact in the cell.
Discussion
Investigations to identify GPCR interactomes are valuable approaches for the understanding of receptor function and methods like AP-MS and MYTH enabled first reproducible identifications of GPCR interactomes (Kittanakom et al., 2014; Sokolina et al., 2017). In the present work we applied tandem mass spectrometry and identified a CB2 receptor interactome consisting of 83 proteins and characterized its interaction with p62 in more detail. The identified interactome especially contains protein networks involved in signaling, biosynthesis and trafficking like calnexin, tubulin chains, and TMED10.
One major concern in the investigations of protein interactomes is the reduction of false-positive findings, which we minimized by the inclusion of only those proteins that were identified by at least two unique peptides. To further drastically reduce the false discovery rate a method established by Glatter et al. (2009) was applied and analyzed the CB2 receptor interactome in triplicates with a reproducibility of at least 90%. Thus, the identified interaction data in our study should represent data with a high level of robustness and reliability. The lack of specific CB2 antibodies (Cecyre et al., 2014) limited our experimental set up to cultured model cells reconstituted with an epitope- tagged CB2 receptor. But the Flp-mediated recombination enabled an insertion of CB2 receptor cDNA into a single genomic locus, which minimize the overexpression levels in this model system. Nevertheless, the relatively high biosynthesis of the CB2 protein could be represented in the interaction network, since GO annotation analysis showed that a relatively high portion of the CB2 interacting proteins were associated with ER and with proteins involved in biosynthesis and degradation pathways. Sec61A1, calnexin, and DNAJA2 are examples for interactors that are functionally relevant in translocation of newly synthesized polypeptides to the ER, correct folding and further trafficking of proteins. Previously, binding of calnexin to the GPCRs rhodopsin and delta-opioid receptor and an interaction of the chaperone of the J protein family DNAJ2B to rhodopsin had been demonstrated (Chapple and Cheetham, 2003; Rosenbaum et al., 2006; Tuusa et al., 2010) emphasizing the relevance of our investigations.
The identified GTP binding proteins are involved in a variety of functions ranging from recycling and trafficking to microtubule stability, cytokinesis, and mitochondrial biosynthesis. The signaling alpha subunits of guanine nucleotide-binding proteins (GNA proteins) were also identified in the list of interacting proteins. Interestingly, GNAS1 and the GNAI3 proteins were present by a single unique peptide but not GNAI1 or GNAI2. The CB2 receptor has mainly been associated with G alpha-i subunit coupled responses and signaling via G alpha s coupled subunits has, to our knowledge, not been reported for CB2 signaling. Possibly, our results hint to similar options of CB2 receptor signaling via GNAS and GNAI3 as has already been described for CB1 receptor (Mukhopadhyay et al., 2000; Finlay et al., 2017).
Findings for other GPCRs further support our newly identified interactions of CB2 receptors. For example, a direct interaction of the C-terminus of alpha2B-adrenergic receptor (alpha2B-AR) with tubulin has been demonstrated to mediate receptor trafficking from ER to cell surface (Duvernay et al., 2011). Our findings of an interaction of CB2 with the tubulin chains, TUBB2B and TUBA1C, might implicate a similar role for proper receptor transport. In line with this, the interactions with the trafficking proteins, TMED2 and TMED10, might hint at an endocytic transport of resensitized CB2 receptor via these proteins as has already been demonstrated for opioid receptor OPRM1 and purinergic nucleotide receptor P2RY4 (Luo et al., 2011). Our results open the path for future studies to investigate, e.g., if CB2 receptor trafficking and homeostasis is coordinated by its newly identified interacting proteins.
Those six proteins that were only identified in CB2-expressing cells DHC24, DHRS7, GGT7, HECD3, KIAA2013, and PLS1 are not forming an interaction network. Nevertheless, the interaction of CB2 with DHC24, a sterol intermediate catalyzing enzyme, and PLS1, an enzyme that flips phosphatidylserines between membrane leaflets, strongly argues for a functional importance in metabolism of lipid molecules connected to the cell membrane that might be of importance, e.g., for viral entry (Williams et al., 2014; Cheshenko et al., 2018).
The reliability of our data is supported by the fact that we have analyzed one protein by Co-IP and could verify the results obtained by the AP-MS approach. For the detailed interaction analysis, we focused on the scaffold protein p62. Our results show a localization of CB2 at p62 positive vesicles that are also WGA positive. WGA selectively binds to N-acetylglucosamine and N-acetylneuraminic acid (sialic acid) residues at the cell membrane (Wright, 1984). Because of the threshold that we used for 3D-reconstructions, the amount of p62 proteins cannot be compared between the models of confocal images and Western blots.
The protein p62 acts as a cargo protein in autophagy and proteasome targeting by its UBA and PB1 domain (Cohen-Kaplan et al., 2016) and regulates the activation of NF-κB (Sanz et al., 1999). We identified the ZZ domain as a main contributor to the interaction with CB2. Recently, binding of the ZZ domain of p62 to arginylated BiP (the ER-localized homolog of Hsp70) was demonstrated to be involved in p62 oligomerization and interaction with the autophagy marker LC3 leading to p62 targeting to autophagosomes and selective lysosomal co-degradation of R-BiP and p62 together with associated cargoes (Cha-Molstad et al., 2015). The enzyme responsible for the N-terminal arginylation is catalyzed by arginyltransferase 1 (ATE1)-encoded Arg-transfer RNA transferases (Cha-Molstad et al., 2015), which was also identified in the CB2 interactome. Therefore, the ZZ domain is particularly important for redirecting N-end rule substrates to the autophagy pathway (Kwon et al., 2018). Thus, biologically the p62-CB2-interaction could be of relevance in cells for an autophagosomal degradation and clearance of CB2 receptors. However, the interaction via the ZZ-type zinc finger motif of p62 also hints at a signaling complex formation as, e.g., the binding to RIP1 protein that links PKCz to form a signaling complex (Festjens et al., 2007; Lin et al., 2013). RIP1 kinase binds to TNF receptor influencing NF-κB and p38 MAPK signaling pathways (Lin et al., 2013), which are also activated by CB2 receptor expression or activation (Dhopeshwarkar and Mackie, 2014; Toguri et al., 2014). Generally, CB2 receptor functions are connected to co-stimulatory effects of, e.g., cytokines, which are then modulated by the GPCR action (Cabral et al., 2015a). Because dysfunction of p62 as well as CB2 receptors are involved in neurodegenerative diseases, inflammation, cancer and bone biology (Pacher and Mechoulam, 2011; Duran et al., 2016; Sanchez-Martin and Komatsu, 2018), it has to be clarified in future experiments if the complex formation of p62 and CB2 is involved in these pathophysiologic functions. Since p62 acts as a signaling platform for, e.g., TNF receptor 1, IL-1 receptor, and RANK receptor (Moscat et al., 2007) and is involved in Toll-like receptor 4 (TLR4) – mediated autophagy (Fujita et al., 2011), our findings might indicate that this platform could function as a signaling hub, which enables the crosstalk of different receptors with CB2 protein. Recently identified interaction between CB2 and TLR4 and their co-expression, co-precipitation and co-functioning in macrophage activation and cancer progression (Xiang et al., 2018) might be one possible example for these protein complexes.
In general, the CB2 receptor is emerging as an attractive therapeutic target for immunomodulation (Cabral et al., 2015b). The potential of CB2 antagonists as a target for treating cancer (Xiang et al., 2018) and liver fibrosis (Mallat et al., 2013) and of CB2 receptor agonists in the treatment of neuropathic pain, neuroinflammation, and neurodegenerative disorders (Guindon and Hohmann, 2008; Lunn et al., 2008; Racz et al., 2008a, b; Contino et al., 2017) hint to the need of molecular insights into the activation mechanism of CB2.
Recently, the crystal structure of the human CB1 and CB2 receptors have been revealed (Hua et al., 2016; Krishna Kumar et al., 2019; Li et al., 2019) and led to the discovery of an opposing functional profile of CB2 antagonism versus CB1 agonism (Li et al., 2019). The clarification of ligand selectivity or function will provide new insights into precise modulation of the ECS by structural analysis, molecular docking and mutagenesis studies (Li et al., 2019). A combination of crystal structure data with results from protein interactome screenings will contribute to facilitate rational drug design and understand mode of action of agonists or antagonists/inverse agonists. Our data add a first step toward this aim, but in future it would be of additional relevance to investigate the CB2 interactome by applying a broader spectrum of cannabinoids.
Despite the use of a heterologous expression system in HEK293 cells, the present work reveals for the first time a CB2 interactome and represent a basal and representative overview. HEK293 cells have been shown to express a variety of GPCR class A transcripts like histamine, dopamine, chemokine, and adenosine receptors (Atwood et al., 2011). Thus, the machinery for GPCR biosynthesis, signaling, and trafficking is present in these cells and is available for the over-expressed cannabinoid receptors that are functional in HEK293 cells (Nagler et al., 2016; Hunter et al., 2017). Nevertheless, investigations of native CB2 expressing cells like lymphocytes and macrophages might result into partially different interactomes. It will therefore be valuable to perform similar screening approaches in different cell types under stimulation with different ligands and systematically compare the interactomes to get a specific and detailed understanding of the CB2 receptor function.
In summary, with the identification of new protein interaction partners for CB2, our findings provide new molecular insights contributing for a better understanding of CB2 receptor biosynthesis, signaling and functions that might account to improve therapeutic strategies of this clinically relevant protein.
Data Availability
The datasets generated for this study can be found in the Supplementary Data to this manuscript.
Author Contributions
MK was responsible for research design. AS, LM, SR, CK, MS, LP, BM, and MK were involved in the generation of expression plasmids, IP experiments, the analysis of these experiments, and figure preparation. ND performed the IPs with deletion constructs of CB2 receptor loops and prepared the figures. AF, SR, and MK conducted the microscopic experiments including analysis, figure preparation, and interpretation. MK performed the bioinformatic analyzes that were approved by HK. MK, AS, and MR wrote or contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- AOBS
Acusto Optic Beam Splitter
- AP-MS
affinity purification coupled to mass spectrometry
- BP
biological process
- CB1
cannabinoid receptor 1
- CB2
cannabinoid receptor 2
- CC
cellular component
- DDM
dodecylmaltosid
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- FCS
fetal calf serum
- GPCRs
G-protein coupled receptors
- HyD
high sensitive hybrid detectors
- LPS
lipopolysaccharide
- MF
molecular function
- MYTH
modified membrane yeast to hybrid
- NF- κ B
nuclear factor- light-chain-enhancer of activated B cells
- PDB
Paget’s disease of bone
- RANK
receptor activator of nuclear factor-kappa B
- RANKL
receptor activator of nuclear factor-kappa B ligand
- RT
room temperature
- TLRs
toll like receptors
- TNF
tumor necrosis factor
- TRAF6
TNF-receptor associated factor 6
- TUBA1C
tubulin alpha-1C
- TUBB2B
tubulin beta-2B
- UBA
ubiquitin-associated domain
- WGA
wheat germ agglutinin.
Funding. This work was supported by the German Research Foundation to MK (DFG Grant Number KA2306/3-1).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00224/full#supplementary-material
References
- Al-Sarraj S., King A., Troakes C., Smith B., Maekawa S., Bodi I., et al. (2011). p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 122 691–702. 10.1007/s00401-011-0911-2 [DOI] [PubMed] [Google Scholar]
- Atwood B. K., Lopez J., Wager-Miller J., Mackie K., Straiker A. (2011). Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12:14. 10.1186/1471-2164-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., Dittel B. N. (2011). Unraveling the complexities of cannabinoid receptor 2 (CB2) immune regulation in health and disease. Immunol. Res. 51 26–38. 10.1007/s12026-011-8210-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinks H. L., Eckhart A. D. (2010). Regulation of GPCR signaling in hypertension. Biochim. Biophys Acta 1802 1268–1275. 10.1016/j.bbadis.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G. A., Ferreira G. A., Jamerson M. J. (2015a). Endocannabinoids and the immune system in health and disease. Handb. Exp. Pharmacol. 231 185–211. 10.1007/978-3-319-20825-1_6 [DOI] [PubMed] [Google Scholar]
- Cabral G. A., Rogers T. J., Lichtman A. H. (2015b). Turning over a new leaf: cannabinoid and endocannabinoid modulation of immune function. J. Neuroimmune Pharmacol. 10 193–203. 10.1007/s11481-015-9615-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassano T., Calcagnini S., Pace L., De Marco F., Romano A., Gaetani S. (2017). Cannabinoid receptor 2 signaling in neurodegenerative disorders: from pathogenesis to a promising therapeutic target. Front. Neurosci. 11:30. 10.3389/fnins.2017.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey J. R., Ralston S. H., Sheppard P. W., Ciani B., Gallagher T. R., Long J. E., et al. (2006). Loss of ubiquitin binding is a unifying mechanism by which mutations of SQSTM1 cause Paget’s disease of bone. Calcif. Tissue Int. 78 271–277. 10.1007/s00223-005-1299-6 [DOI] [PubMed] [Google Scholar]
- Cecyre B., Thomas S., Ptito M., Casanova C., Bouchard J. F. (2014). Evaluation of the specificity of antibodies raised against cannabinoid receptor type 2 in the mouse retina. Naunyn Schmiedebergs Arch. Pharmacol. 387 175–184. 10.1007/s00210-013-0930-8 [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H., Sung K. S., Hwang J., Kim K. A., Yu J. E., Yoo Y. D., et al. (2015). Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 17 917–929. 10.1038/ncb3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple J. P., Cheetham M. E. (2003). The chaperone environment at the cytoplasmic face of the endoplasmic reticulum can modulate rhodopsin processing and inclusion formation. J. Biol. Chem. 278 19087–19094. 10.1074/jbc.m212349200 [DOI] [PubMed] [Google Scholar]
- Cheshenko N., Pierce C., Herold B. C. (2018). Herpes simplex viruses activate phospholipid scramblase to redistribute phosphatidylserines and Akt to the outer leaflet of the plasma membrane and promote viral entry. PLoS Pathog. 14:e1006766. 10.1371/journal.ppat.1006766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Kaplan V., Livneh I., Avni N., Fabre B., Ziv T., Kwon Y. T., et al. (2016). p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. U.S.A. 113 E7490–E7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contino M., Capparelli E., Colabufo N. A., Bush A. I. (2017). Editorial: the CB2 cannabinoid system: a new strategy in neurodegenerative disorder and neuroinflammation. Front. Neurosci. 11:196 10.3389/fnins.2017.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulat A. M., Maurice P., Jockers R. (2009). Recent methodological advances in the discovery of GPCR-associated protein complexes. Trends Pharmacol. Sci. 30 72–78. 10.1016/j.tips.2008.10.009 [DOI] [PubMed] [Google Scholar]
- Dhopeshwarkar A., Mackie K. (2014). CB2 cannabinoid receptors as a therapeutic target-what does the future hold? Mol. Pharmacol. 86 430–437. 10.1124/mol.114.094649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A., Hernandez E. D., Reina-Campos M., Castilla E. A., Subramaniam S., Raghunandan S., et al. (2016). p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell 30 595–609. 10.1016/j.ccell.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernay M. T., Wang H., Dong C., Guidry J. J., Sackett D. L., Wu G. (2011). Alpha2B-adrenergic receptor interaction with tubulin controls its transport from the endoplasmic reticulum to the cell surface. J. Biol. Chem. 286 14080–14089. 10.1074/jbc.M111.222323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecto F., Yan J., Vemula S. P., Liu E., Yang Y., Chen W., et al. (2011). SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 68 1440–1446. 10.1001/archneurol.2011.250 [DOI] [PubMed] [Google Scholar]
- Festjens N., Vanden Berghe T., Cornelis S., Vandenabeele P. (2007). RIP1, a kinase on the crossroads of a cell’s decision to live or die. Cell Death Differ. 14 400–410. 10.1038/sj.cdd.4402085 [DOI] [PubMed] [Google Scholar]
- Finlay D. B., Cawston E. E., Grimsey N. L., Hunter M. R., Korde A., Vemuri V. K., et al. (2017). Galphas signalling of the CB1 receptor and the influence of receptor number. Br. J. Pharmacol. 174 2545–2562. 10.1111/bph.13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Maeda D., Xiao Q., Srinivasula S. M. (2011). Nrf2-mediated induction of p62 controls toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc. Natl. Acad. Sci. U.S.A. 108 1427–1432. 10.1073/pnas.1014156108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandia J., Fernandez-Duenas V., Morato X., Caltabiano G., Gonzalez-Muniz R., Pardo L., et al. (2013). The Parkinson’s disease-associated GPR37 receptor-mediated cytotoxicity is controlled by its intracellular cysteine-rich domain. J. Neurochem. 125 362–372. 10.1111/jnc.12196 [DOI] [PubMed] [Google Scholar]
- Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009). An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5:237. 10.1038/msb.2008.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida F., Luongo L., Boccella S., Giordano M. E., Romano R., Bellini G., et al. (2017). Palmitoylethanolamide induces microglia changes associated with increased migration and phagocytic activity: involvement of the CB2 receptor. Sci. Rep. 7:375. 10.1038/s41598-017-00342-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon J., Hohmann A. G. (2008). Cannabinoid CB2 receptors: a therapeutic target for the treatment of inflammatory and neuropathic pain. Br. J. Pharmacol. 153 319–334. 10.1038/sj.bjp.0707531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng B. C., Aubel D., Fussenegger M. (2013). An overview of the diverse roles of G-protein coupled receptors (GPCRs) in the pathophysiology of various human diseases. Biotechnol. Adv. 31 1676–1694. 10.1016/j.biotechadv.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Hua T., Vemuri K., Pu M., Qu L., Han G. W., Wu Y., et al. (2016). Crystal structure of the human cannabinoid receptor CB1. Cell 167 750.e14–762.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Hunter M. R., Finlay D. B., Macdonald C. E., Cawston E. E., Grimsey N. L., Glass M. (2017). Real-time measurement of cannabinoid receptor-mediated cAMP signaling. Methods Enzymol. 593 43–59. 10.1016/bs.mie.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Katsuragi Y., Ichimura Y., Komatsu M. (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282 4672–4678. 10.1111/febs.13540 [DOI] [PubMed] [Google Scholar]
- Kim J. Y., Ozato K. (2009). The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-kappaB activity. J. Immunol. 182 2131–2140. 10.4049/jimmunol.0802755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittanakom S., Barrios-Rodiles M., Petschnigg J., Arnoldo A., Wong V., Kotlyar M., et al. (2014). CHIP-MYTH: a novel interactive proteomics method for the assessment of agonist-dependent interactions of the human beta(2)-adrenergic receptor. Biochem. Biophys. Res. Commun. 445 746–756. 10.1016/j.bbrc.2014.02.033 [DOI] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131 1149–1163. 10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- Krishna Kumar K., Shalev-Benami M., Robertson M. J., Hu H., Banister S. D., Hollingsworth S. A., et al. (2019). Structure of a signaling cannabinoid receptor 1-G protein complex. Cell 176 448.e12–458.e12. 10.1016/j.cell.2018.11.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D. H., Park O. H., Kim L., Jung Y. O., Park Y., Jeong H., et al. (2018). Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat. Commun. 9:3291. 10.1038/s41467-018-05825-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappano R., Maggiolini M. (2011). G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 10 47–60. 10.1038/nrd3320 [DOI] [PubMed] [Google Scholar]
- Laurin N., Brown J. P., Morissette J., Raymond V. (2002). Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in paget disease of bone. Am. J. Hum. Genet. 70 1582–1588. 10.1086/340731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Hua T., Vemuri K., Ho J. H., Wu Y., Wu L., et al. (2019). Crystal structure of the human cannabinoid receptor CB2. Cell 176 459.e13–467.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Li S., Zhao Y., Ma X., Zhang K., He X., et al. (2013). Interaction domains of p62: a bridge between p62 and selective autophagy. DNA Cell Biol. 32 220–227. 10.1089/dna.2012.1915 [DOI] [PubMed] [Google Scholar]
- Lunn C. A., Reich E. P., Fine J. S., Lavey B., Kozlowski J. A., Hipkin R. W., et al. (2008). Biology and therapeutic potential of cannabinoid CB2 receptor inverse agonists. Br. J. Pharmacol. 153 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Wang Y., Reiser G. (2011). Proteinase-activated receptors, nucleotide P2Y receptors, and mu-opioid receptor-1B are under the control of the type I transmembrane proteins p23 and p24A in post-Golgi trafficking. J. Neurochem. 117 71–81. 10.1111/j.1471-4159.2011.07173.x [DOI] [PubMed] [Google Scholar]
- Mallat A., Teixeira-Clerc F., Lotersztajn S. (2013). Cannabinoid signaling and liver therapeutics. J. Hepatol. 59 891–896. 10.1016/j.jhep.2013.03.032 [DOI] [PubMed] [Google Scholar]
- Mattheus T., Kukla K., Zimmermann T., Tenzer S., Lutz B. (2016). Cell Type-specific tandem affinity purification of the mouse hippocampal CB1 receptor-associated proteome. J. Proteome Res. 15 3585–3601. 10.1021/acs.jproteome.6b00339 [DOI] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M. T. (2011). Feedback on fat: p62-mTORC1-autophagy connections. Cell 147 724–727. 10.1016/j.cell.2011.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M. T., Wooten M. W. (2007). Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32 95–100. 10.1016/j.tibs.2006.12.002 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Mcintosh H. H., Houston D. B., Howlett A. C. (2000). The CB(1) cannabinoid receptor juxtamembrane C-terminal peptide confers activation to specific G proteins in brain. Mol. Pharmacol. 57 162–170. [PubMed] [Google Scholar]
- Nagler M., Palkowitsch L., Rading S., Moepps B., Karsak M. (2016). Cannabinoid receptor 2 expression modulates Gbeta(1)gamma(2) protein interaction with the activator of G protein signalling 2/dynein light chain protein Tctex-1. Biochem. Pharmacol. 99 60–72. 10.1016/j.bcp.2015.09.017 [DOI] [PubMed] [Google Scholar]
- O’Gorman S., Fox D. T., Wahl G. M. (1991). Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251 1351–1355. 10.1126/science.1900642 [DOI] [PubMed] [Google Scholar]
- Pacher P., Mechoulam R. (2011). Is lipid signaling through cannabinoid 2 receptors part of a protective system? Prog. Lipid Res. 50 193–211. 10.1016/j.plipres.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., et al. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282 24131–24145. 10.1074/jbc.m702824200 [DOI] [PubMed] [Google Scholar]
- Pertwee R. G. (2012). Targeting the endocannabinoid system with cannabinoid receptor agonists: pharmacological strategies and therapeutic possibilities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367 3353–3363. 10.1098/rstb.2011.0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna A., Wardas J., Simola N., Morelli M. (2005). New therapies for the treatment of Parkinson’s disease: adenosine A2A receptor antagonists. Life Sci. 77 3259–3267. 10.1016/j.lfs.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Racz I., Nadal X., Alferink J., Banos J. E., Rehnelt J., Martin M., et al. (2008a). Crucial role of CB(2) cannabinoid receptor in the regulation of central immune responses during neuropathic pain. J. Neurosci. 28 12125–12135. 10.1523/JNEUROSCI.3400-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racz I., Nadal X., Alferink J., Banos J. E., Rehnelt J., Martin M., et al. (2008b). Interferon-gamma is a critical modulator of CB(2) cannabinoid receptor signaling during neuropathic pain. J. Neurosci. 28 12136–12145. 10.1523/JNEUROSCI.3402-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum E. E., Hardie R. C., Colley N. J. (2006). Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron 49 229–241. 10.1016/j.neuron.2005.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Martin P., Komatsu M. (2018). p62/SQSTM1 - steering the cell through health and disease. J. Cell Sci. 131:jcs222836. 10.1242/jcs.222836 [DOI] [PubMed] [Google Scholar]
- Sanz L., Sanchez P., Lallena M. J., Diaz-Meco M. T., Moscat J. (1999). The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 18 3044–3053. 10.1093/emboj/18.11.3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seilhean D., Cazeneuve C., Thuries V., Russaouen O., Millecamps S., Salachas F., et al. (2009). Accumulation of TDP-43 and alpha-actin in an amyotrophic lateral sclerosis patient with the K17I ANG mutation. Acta Neuropathol. 118 561–573. 10.1007/s00401-009-0545-9 [DOI] [PubMed] [Google Scholar]
- Seitz S., Priemel M., Von Domarus C., Beil F. T., Barvencik F., Amling M., et al. (2008). The second most common bone disease: a review on Paget’s disease of bone. Eur. J. Trauma Emerg. Surg. 34 549–553. 10.1007/s00068-008-8208-4 [DOI] [PubMed] [Google Scholar]
- Sokolina K., Kittanakom S., Snider J., Kotlyar M., Maurice P., Gandia J., et al. (2017). Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Mol. Syst. Biol. 13:918. 10.15252/msb.20167430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens S., Veillard N. R., Arnaud C., Pelli G., Burger F., Staub C., et al. (2005). Low dose oral cannabinoid therapy reduces progression of atherosclerosis in mice. Nature 434 782–786. 10.1038/nature03389 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguri J. T., Lehmann C., Laprairie R. B., Szczesniak A. M., Zhou J., Denovan-Wright E. M., et al. (2014). Anti-inflammatory effects of cannabinoid CB(2) receptor activation in endotoxin-induced uveitis. Br. J. Pharmacol. 171 1448–1461. 10.1111/bph.12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuusa J. T., Leskela T. T., Petaja-Repo U. E. (2010). Human delta opioid receptor biogenesis is regulated via interactions with SERCA2b and calnexin. FEBS J. 277 2815–2829. 10.1111/j.1742-4658.2010.07699.x [DOI] [PubMed] [Google Scholar]
- Williams J. C., Appelberg S., Goldberger B. A., Klein T. W., Sleasman J. W., Goodenow M. M. (2014). Delta(9)-Tetrahydrocannabinol treatment during human monocyte differentiation reduces macrophage susceptibility to HIV-1 infection. J. Neuroimmune Pharmacol. 9 369–379. 10.1007/s11481-014-9527-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S. (1984). Structural comparison of the two distinct sugar binding sites in wheat germ agglutinin isolectin II. J. Mol. Biol. 178 91–104. 10.1016/0022-2836(84)90232-8 [DOI] [PubMed] [Google Scholar]
- Xiang W., Shi R., Kang X., Zhang X., Chen P., Zhang L., et al. (2018). Monoacylglycerol lipase regulates cannabinoid receptor 2-dependent macrophage activation and cancer progression. Nat. Commun. 9:2574. 10.1038/s41467-018-04999-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Supplementary Data to this manuscript.