Abstract
Musashi1 (Msi1) is a mammalian neural RNA-binding protein highly enriched in neural precursor cells that are capable of generating both neurons and glia during embryonic and postnatal CNS development. Here, we identified Musashi2 (Msi2), a novel mammalian RNA-binding protein that exhibits high sequence similarity to Msi1. The Msi2 transcript appeared to be distributed ubiquitously in a wide variety of tissues, consistent with the mRNA distribution of its Xenopushomolog, xrp1. However, the present study revealed cell type-specific and developmentally regulated expression of Msi2 in the mammalian CNS. Interestingly, Msi2 was expressed prominently in precursor cells in the ventricular zone and subventricular zone with the same pattern as Msi1 throughout CNS development. In the postnatal and adult CNS, this concurrent expression of Msi2 and Msi1 was seen in cells of the astrocyte lineage, including ependymal cells, a possible source for postnatal CNS stem cells. During neurogenesis, the expression of both Msi2 and Msi1 was lost in most postmitotic neurons, whereas Msi2 expression persisted in a subset of neuronal lineage cells, such as parvalbumin-containing GABA neurons in the neocortex and neurons in several nuclei of the basal ganglia. Msi2 may have a unique role that is required for the generation and/or maintenance of specific neuronal lineages. Furthermore, in vitro studies showed that Msi2 and Msi1 have similar RNA-binding specificity. These two RNA-binding proteins may exert common functions in neural precursor cells by regulating gene expression at the post-transcriptional level.
Keywords: RNA-binding protein, Musashi2, Musashi1, CNS development, neural precursor cells, CNS stem cell, astrocyte, parvalbumin, GABAergic neuron
A number of transcription factors that function in the proliferation and differentiation of neural precursor cells have been identified. However, the recent discovery of neural-specific RNA-binding proteins raises the possibility that the development of neural cells from the precursors may also be regulated at the post-transcriptional level, including by mRNA stabilization or translational control. We believe that two gene families are represented by the neural RNA-binding proteins found in both invertebrates and vertebrates (Okano, 1995). One, the Elav family, possesses three sets of ribonucleoprotein (RNP)-type RNA recognition motifs (RRMs) and includes the Drosophila elav gene (Yao et al., 1993) and the vertebrate Hu genes (Okano and Darnell, 1997). Members of this family are expressed in postmitotic neurons and are thought to function in the survival or differentiation of neurons (Akamatsu et al., 1999). The other family, the Musashi (Msi) family, is characterized by two RRMs and is conserved in ascidian (Kawashima et al., 2000), nematode (Yoda et al., 2000), fruit fly (Nakamura et al., 1994), frog (Richter et al., 1990), chick (Kaneko et al., 2000), mouse (Sakakibara et al., 1996), and human (Good et al., 1998). In contrast to the Elav family, the members of the Msi family are predominantly expressed in neural precursor cells (Sakakibara et al., 1996; Pincus et al., 1998; Kaneko et al., 2000). In Drosophila, Musashi (d-Msi) is expressed in numerous precursor cells of the embryonic CNS and in external sensory organ precursor cells (SOPs) (Nakamura et al., 1994) and is required for the two successive asymmetric cell divisions of the SOPs (Nakamura et al., 1994; Okabe et al., 2001). Although the molecular functions of the Msi and Elav families remain obscure, their different expression patterns suggest that the two families may play distinct roles in the development and maintenance of neural cells.
During mammalian CNS development, neurons and glial cells are thought to be generated from common neural precursor cells (CNS stem cells) in the embryonic ventricular zone (VZ) (Davis and Temple, 1994; Johe et al., 1996). We found previously that Msi1 is expressed predominantly in proliferating multipotent neural precursors but not in newly generated postmitotic neurons (Sakakibara et al., 1996; Kaneko et al., 2000). In the perinatal stage, the VZ shrinks and becomes a single epithelial layer, the ependyma. A second proliferative subventricular zone (SVZ) then appears at the late embryonic stage and persists into adulthood, in a considerably reduced form. It was believed for a long time that mammalian cortical neurogenesis occurs mainly in the embryonic VZ; however, several recent studies demonstrated that the SVZ astrocytes and/or ependymal cells correspond to a population of CNS stem cells throughout postnatal and adult life (Scheffler et al., 1999). Accordingly, substantial Msi1 protein expression persists in these putative precursor cells into adulthood (Sakakibara and Okano, 1997).
In the present study we identified Msi2, a novel member of the Msi family. Given their remarkable similarities in primary structure, RNA binding specificity, and expression profile during CNS development, Msi1 and Msi2 may play similar roles in neural precursor cells via post-transcriptional regulation.
MATERIALS AND METHODS
Cloning of mouse msi2 cDNA. A λgt 11 cDNA library obtained from adult mouse cerebellum (Sakakibara et al., 1996) was screened with both a 1.1 kb EcoRI fragment of the mousemsi1 coding region (GenBank accession no. D4965) (Sakakibara et al., 1996) and a 387 bp BamHI–NdeI fragment containing the C terminus of the coding region of Xenopus xrp1 cDNA (GenBank accession no. L02953) (Good et al., 1993). Hybridization was performed against 1 × 107 plaques in a buffer (1 mNaCl, 1% SDS, 10% dextran sulfate, 0.1 mg/ml salmon sperm DNA) containing 5 × 105 cpm/ml of32P-labeled random-primed probe for 18–24 hr at 60°C with the msi1 probe or at 55°C with thexrp1 probe. The hybridized filters were washed twice for 20 min each at room temperature in 2× SSC and 0.1% SDS (low stringency). Thirty-two positive clones that hybridized to both the msi1and xpr1 probes were obtained. Among them, nine that strongly hybridized to the xpr1 cDNA were selected, subcloned into pBluescriptII (Stratagene, La Jolla, CA), and sequenced by standard dideoxynucleotide sequencing using a dye-primer kit (Amersham Pharmacia Biotech, Buckinghamshire, UK). Sequencing analysis revealed multiple overlapping clones spanning a 0.5 kb 5′-UTR, a 0.8 kb 3′-UTR, and the 1.0 kb predicted open reading frame (ORF), which we designated mouse musashi2 (msi2). A putative alternatively spliced exon was found at the C terminus of the predicted coding region with a 54 bp insertion, producing short- and long-form ORFs of msi2 cDNA. Expression of the short and long forms ofmsi2 transcripts in vivo was confirmed by RT-PCR analysis of RNA isolated from embryonic day (E) 12 and adult brains. Homology searches and alignments were performed with BLAST and FASTA algorithms on the National Center for Biotechnology Information server. The GenBank accession numbers of the proteins used for phylogenetic tree analysis (clustalW program on the DNA Data Bank of Japan www server) are as follows: human (Hu) U1snRNP70K (A25707), mouse (Mus) heterogeneous nuclear ribonucleoprotein (hnRNP) A1 (NP 034577), Hu hnRNP A0 (Q13151), Mus hnRNP A2/B1 (O88569), Hu hnRNP A3 (P51991), Hu hnRNP type A/B (AAA36575), Rat AUF1 (BAB03468, BAB03466, BAB03467), Mus hnRNP C1/C2 (AAD03717), Hu hnRNP F (S43484), Mus hnRNP G (O35479), Hu hnRNP H (I39358), Mus PTB (hnRNP I) (P17225), Mus brPTB (NP 062423), Hu hnRNP L (P14866), Hu hnRNP M (P52272), Hu hnRNP R (T02673), Mus hnRNP U (NP 058085), Mus TIA-1 (P52912), Mus TIAR (S72436), Mus HuR (NP 034615), Mus HuB (AAC52644), Mus HuC (Q60900), Mus HuD (JC2298), Hu Brunol3 (AAB09040), Mus Lark (NP 033058), and Rat La/SS-B (JC1494).
Animals and tissue preparation. ICR (CD-1) mice, used for the preparation of tissue protein extracts, RNA, or tissue sections, were obtained from Charles River Japan Inc. (Kanagawa, Japan). The date of conception was established by the presence of a vaginal plug and recorded as E0, and the day of birth was designated as postnatal day (P) 0.
Northern blot analysis. Total RNA (20 μg) from each of the mouse tissues and embryos was isolated using Trizol (Life Technologies-BRL, Grand Island, NY) according to the manufacturer's instructions, fractionated by electrophoresis on 1% agarose-formaldehyde gels, and transferred onto Biodyne B nylon membranes (Pall, Port Washington, NY).32P-labeled probe was prepared using a random-primed DNA labeling kit (Roche Diagnostics, Mannheim, Germany) of a 750 bp 3′-UTR fragment of the mouse msi2 cDNA and hybridized in 50% formamide, 6× SSPE, 5× Denhardt's solution, 0.5% SDS, and 200 μg/ml salmon sperm DNA at 42°C for 16 hr. The incubation was followed by stringent washes in 0.1× SSC, 0.1% SDS at 50°C, and filters were exposed for 48 hr on Kodak X-OMAT film (Kodak, Rochester, NY). The integrity of the transferred RNA samples was verified by reprobing each blot with a radiolabeled β-actin cDNA fragment (Clontech, Palo Alto, CA).
Production and purification of anti-Msi2 antibodies. A peptide corresponding to the 14 amino acid terminal sequence of the Msi2 protein (MEANGSPGTSGSAN) followed by a cysteinamide residue was synthesized for the purpose of antibody production. This peptide sequence does not overlap with that of the RNA-binding domain, RRM1 (see Fig. 1) and does not show any similarities to the corresponding N-terminal region of the Msi1 protein. Approximately 15 mg of the peptide was coupled through the cysteinamide residue tom-bromosuccinimide-treated keyhole limpet hemocyanin and used to immunize New Zealand White rabbits. For the affinity purification of the anti-Msi2 antisera, the synthesized peptide (5 mg) was coupled covalently to activated 2-fluoro-1-methylpyridinium toluene-4-sulfonate (FMP) Cellulofine (Seikagaku Kogyo, Tokyo, Japan), according to the manufacturer's instructions. Ten milliliters of the filtered (0.45 μm) whole antisera were incubated with 3 ml of the peptide-FMP Cellulofine affinity resin, pre-equilibrated with TBS buffer (0.15 m NaCl, 20 mm Tris-HCl, pH 7.5) overnight at 4°C. The resin was then washed with 50 ml of 1 m NaCl, 1% Triton X-100, 20 mm Tris-HCl, pH 7.5, followed by 20 ml of 0.15 m NaCl, eluted with 4 ml of 100 mm glycine-HCl, pH 2.0, at 4°C, and immediately neutralized with 0.2 ml of 1 m Tris.
Fig. 1.
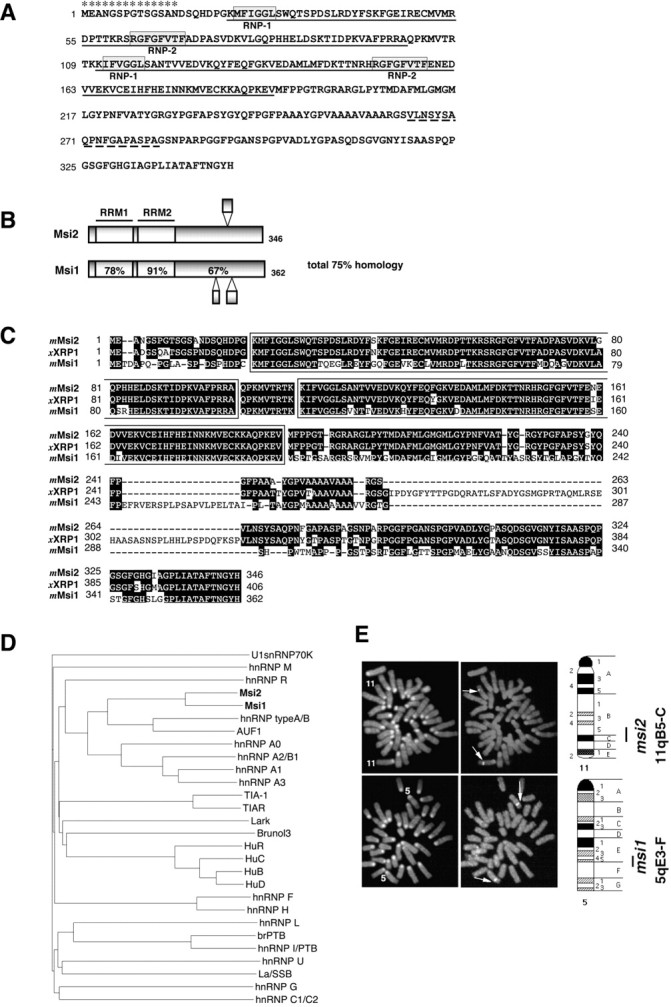
Primary structure and chromosomal localization of Msi2 and Msi1. A, The deduced amino acid sequence of mouse Msi2 protein. The two RNA recognition motifs areunderlined. Each RRM includes two highly conserved sequences designated RNP-1 and RNP-2(boxed sequences). The broken underlinesindicate the regions that are missing in the form arising from alternative mRNA splicing. Asterisks denote the peptide sequence used as an immunogen to generate anti-Msi2 antibody.B, Domain structures of the mouse Msi2 and Msi1 proteins. The percentages of amino acid identity between Msi1 and Msi2 within each RRM and in the C-terminal half are shown. Bulged small boxes represent the regions arising from alternative splicing. C, Amino acid sequence alignment of mouse Msi2 with its known homologs, Xenopus RNA-binding protein (XRP1), and mouse Msi1. The two RRMs are boxed, and gaps in the alignment are indicated by dashes. Amino acid residues conserved in more than two proteins are shaded.D, Dendrogram showing a multiple sequence comparison of various mammalian RRM-containing RNA-binding proteins. Sequences derived from selected proteins were compared using the clustalW program on the DDBJ www server. The branch lengths are proportional to the differences between sequences. The accession numbers of each protein are given in Materials and Methods. E, Chromosomal localization of the mouse msi1 and msi2genes by FISH. The left panels show metaphase chromosomes stained with DAPI for the identification of individual mouse chromosomes. Chromosomes 11 and 5are numbered. The right panels represent the same metaphase chromosomes with the hybridization signals from themsi2 (top panels) and msi1(bottom panels) genes (arrows indicate each signal). The closed bars on the right side of each ideogram indicate the possible location of the msi2 andmsi1 genes.
Recombinant Msi1 and Msi2 proteins. The 983 bp and 1072 bpBamHI–EcoRI fragments corresponding to the short- and long-form ORFs of the Msi2 protein were isolated by RT-PCR of the RNAs of E12 embryonic and adult brains and subcloned in-frame into the expression vector pRSET-A (Invitrogen, Carlsbad, CA) to construct pRSET-Msi2S (short form) and pRSET-Msi2L (long form) to generate fusion proteins containing six histidine residues at their amino termini. Expression vectors pRSET-Msi1 (Sakakibara et al., 1996), pRSET-Msi2S, and pRSET-Msi2L were then introduced intoEscherichia coli strain BL21(DE3) pLysS, and the fusion proteins were induced by incubation with 1 mmisopropyl-β-d-thiogalactopyranoside for 6 hr at 30°C. Recombinant fusion proteins (His6-Msi2S, His6-Msi2L, and His6-Msi1) were affinity purified using a Probond resin (Invitrogen) column as described by the supplier. The purity and concentration of the fusion proteins were verified by Coomassie Brilliant blue (Sigma, St. Louis, MO) staining of an SDS-polyacrylamide (SDS-PAGE) gel of the eluent, and by the Bradford assay (Bio-Rad, Hercules, CA).
Protein phosphatase treatment and immunoblotting. Tissue lysate was prepared by homogenization in Buffer A [50 mm Tris-HCl, pH 7.6, 1 mmpotassium acetate, 1.5 mm magnesium acetate, 2 mm dithiothreitol (DTT), 100 μg/ml phenylmethylsulfonyl fluoride (PMSF), 5 μg/ml aprotinin, 5 μg/ml leupeptin], followed by centrifugation at 10,000 × gfor 10 min. Bacterially expressed, purified recombinant proteins (50 ng per lane) or tissue lysate (30 μg of protein per lane) were resolved on 10% SDS-PAGE gels that were then electroblotted onto Immobilon-P membranes (Millipore, Bedford, MA) with a semidry transfer apparatus. Equal loading of total protein from each tissue was determined by standard Bradford assay and verified by Coomassie blue staining of a duplicate gel. The chemiluminescent signals were detected by ECL (Amersham Pharmacia Biotech) with Kodak X-OMAT film according to the manufacturer's instructions. For the protein dephosphorylation assay, endogenous Msi2 protein was partially purified from embryonic brain lysate (E12.5). E12.5 brains (1.0 gm of wet weight) were homogenized in 5 ml of Buffer A and spun to pellet the nuclei (12,000 ×g, 10 min, 4°C). The supernatant was removed and layered gently over a 1.5 ml sucrose (30% w/v in Buffer A) cushion and spun at 130,000 × g, 4°C for 2 hr, in a Beckman SW55Ti rotor. After removal of the S130 supernatant and sucrose cushion, the pelleted polysome fraction was rinsed and resuspended in 500 μl of Buffer A. A significant amount of Msi2 protein was recovered in this polysome fraction. For protein phosphatase treatment, 10 μg of protein from the cleared polysome fraction was incubated with 800 U of recombinant λ protein phosphatase (λPPase) (New England BioLabs, Beverly, MA) in 25 μl of 50 mm Tris-HCl, pH 7.5, 0.1 mm EDTA, 5 mm DTT, 0.01% polyoxyethylene (23) lauryl ether (Brij35), 2 mm MnCl2, 10 μg/ml PMSF, 5 μg/ml aprotinin, and 5 μg/ml leupeptin, at 30°C for 1 hr. The λPPase dephosphorylates phosphorylated serine, threonine, and tyrosine residues. Control samples lacking λPPase were incubated as described above. Reactions were stopped with SDS-PAGE sample buffer, and the samples were resolved on 10% SDS-PAGE gels for immunoblotting.
In vitro transcription/translation and RNA-binding assays. The cDNA fragments corresponding to the coding region of the Msi2 long form (nucleotides 524–1564; GenBank accession no.AB056103) and Msi1 (nucleotides 64–1152; GenBank accession no. D49654) were isolated by PCR, using primers encoding a FLAG tag at the amino termini, and subcloned into pCDNA3 (Invitrogen) to construct the expression vectors pCDNA-msi2 and pCDNA-msi1. These plasmids were transcribed and translated in rabbit reticulocyte lysate (TNT T7 quick-coupled transcription/translation system; Promega, Madison, WI) in the presence of 0.4 mCi/ml of35S-methionine (Amersham Pharmacia Biotech), according to the manufacturer's suggested conditions. Luciferase T7 control vector (Promega), which encodes a 61 kDa Luciferase protein, was also translated in vitro as described above. Binding of in vitro-translated proteins to RNA-homopolymers was performed essentially as described previously (Swanson and Dreyfuss, 1988), with minor modifications. Briefly, 20 μl of each ribohomopolymer-agarose bead (Sigma), equilibrated with the binding buffer (10 mm Tris-HCl, pH 7.4, 2.5 mm MgCl2, 0.5% Triton X-100, 2 mg/ml pepstatin, 2 mg/ml leupeptin, 0.5% aprotinin, 1 mg/ml heparin), was incubated with the35S-labeled proteins (1 × 105 cpm) in 500 μl of binding buffer containing either 100 or 250 mm NaCl for 15 min on a rocking platform at 4°C. The beads were pelleted with a brief spin and washed five times with 500 μl of binding buffer, before resuspension in 50 μl of SDS-PAGE loading buffer. Bound proteins were eluted by boiling, resolved on a 10% SDS-PAGE gel, and visualized by fluorography.
Immunohistochemistry. Pups and 8-week-old adults (anesthetized by ether inhalation) were perfused through the left ventricle with 4% paraformaldehyde (PFA) in 0.1m PBS, pH 7.4. The embryos were removed by cesarean section and immersed in the same fixative. Brains and other tissues were dissected and post-fixed overnight at 4°C, cryoprotected in 30% sucrose in PBS overnight at 4°C, and embedded in O.C.T. compound (Tissue Tek; Miles, Elkhart, IN). Cryostat sections (12 μm) were cut and affixed to 3-aminopropyltriethoxysilane-coated glass slides (Matsunami Glass, Osaka, Japan). Sections were permeabilized in 0.4% Triton X-100 in PBS for 30 min, followed by blocking with 10% normal serum of the species in which the secondary antibody was raised, 1% skim milk, and 0.1% Triton X-100 in PBS for 1 hr at room temperature; subsequently, the sections were incubated in a mixture of primary antibodies overnight at 4°C. The mixture always included the affinity-purified rabbit anti-Msi2 polyclonal antibody, diluted 1:300 in 10% normal serum and 0.1% Triton X-100 in PBS, and one of the following antibodies: anti-Msi1 (14H1, rat monoclonal IgG2b, ascites) (Kaneko et al., 2000), diluted 1:500; anti-Hu proteins (mouse monoclonal IgG2b, clone 16A11, which binds to Hu proteins including HuD, HuC, and HuB) (Marusich et al., 1994), diluted 1:500; anti-nestin (Rat401, mouse monoclonal IgG, Developmental Studies Hybridoma Bank of Iowa University), diluted 1:250; anti-Ki67 (mouse monoclonal IgG, Novocastra, Newcastle upon Tyne, UK), diluted 1:500; anti-glial fibrillary acidic protein (GFAP) (mouse monoclonal IgG1, Sigma), diluted 1:400; anti- 2′, 3′-cyclic nucleotide-3′-phosphohydrolase (CNPase) (mouse monoclonal IgG1, Sigma), diluted 1:100; anti-microtuble-associated protein 2 (MAP2) (mouse monoclonal IgG1, Sigma), diluted 1:500; anti-βIII tublin (TuJ1) (Berkeley Antibody; mouse monoclonal IgG2a/k), diluted 1:200; anti-parvalbumin (PV) (mouse monoclonal IgG1, Chemicon, Temecula, CA), diluted 1:500; anti-GABA (guinea pig polyclonal, Chemicon), diluted 1:400; anti-acetylcholine transferase (ChAT) (goat polyclonal antibody,Chemicon), diluted 1:100; anti-Calbindin (CB) D28K (mouse monoclonal IgG1, Sigma), diluted 1:1000; anti-tyrosine hydroxylase (TH) (mouse monoclonal IgG1, provided by Dr. H. Hatanaka, Osaka University), diluted 1:1000; anti-Mac1 (mouse monoclonal IgG1, Roche Diagnostics), diluted 1:100; or anti-bromodeoxyuridine (BrdU) (mouse monoclonal IgG1, Sigma), diluted 1:5000. After four washes with 0.1% Triton X-100 in PBS, the sections were incubated for 1 hr in a mixture containing the following secondary antibodies diluted 1:500 in the same solution as the primary antibodies: Alexa Fluor 568-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR) and Alexa Fluor 488-conjugated anti-mouse, rat, guinea pig, or goat IgG (Molecular Probes). After being rinsed in PBS, the sections were mounted and examined under a fluorescence microscope (Zeiss Axioplan 2) equipped with the appropriate epifluorescent filters. Optical sections were viewed using a scanning laser confocal imaging system (Zeiss LSM510). For control sections, the anti-Msi2 antibody was omitted or replaced with preimmune rabbit serum. The specificity of the anti-Msi2 antibody was examined by its preadsorption with the peptide (50 μg/ml) used for immunization, before its use for immunostaining tissue sections.
Cell culture. Neurosphere cultures were prepared as described previously (Nakamura et al., 2000). Briefly, cells from the frontal halves of E14.5 telencephalons were used for primary sphere formation (5 × 105 cells/5 ml per well, six-well plate) with the basic culture medium, DMEM/F-12 (1:1) (Life Technologies-BRL) containing 25 μg/ml insulin, 100 μg/ml transferrin, 20 nm progesterone, 60 μm putrescine, 30 nm sodium selenite, 20 ng/ml epidermal growth factor (EGF), and 10 ng/ml basic fibroblast growth factor (bFGF). After cultivation for 10 d, the neurospheres were fixed in 4% PFA for 30 min at 4°C and embedded in O.C.T. compound for cryosectioning. NG108–15, a hybrid cell line of mouse neuroblastoma and rat glioma (Amano et al., 1974), was grown in DMEM (Life Technologies-BRL) with 10% (v/v) fetal bovine serum. Cells were grown to a density of 5 × 105cells/cm2 on coverslips precoated with 0.01% poly-l-lysine (Sigma), fixed 4% PFA for 30 min at 4°C, and permeabilized in 0.05% Triton X-100 in PBS for 10 min. Immunoreactivity to Msi1 or Msi2 was visualized using rhodamine-conjugated donkey anti-rabbit IgG or anti-rat IgG secondary antibodies (Chemicon; 1:250 dilution). Cells were counterstained with 10 μm of Hoechst 33342 dye (Sigma) to identify nuclei. Optical images were obtained using a Zeiss LSM410 scanning laser confocal system.
Surgical procedures for brain lesions and bromodeoxyuridine incorporation. Brain injury experiments were performed as described previously (Sakakibara and Okano, 1997). Adult ICR mice, 2 months old, were anesthetized with Nembutal injections (10 mg/kg) (Abbott Laboratories, North Chicago, IL), and a midline skin incision was made. A unilateral linear craniectomy was performed on the skull using a drill, 3 mm left of the midline. The wound was made in the left cerebral hemisphere; a 27 gauge stainless-steel needle was placed 3 mm lateral to the sagittal fissure and 2 mm anterior to the lambda line, and inserted through the pia to a depth of 1.5 mm. The brain was cut sagittally, 4 mm anterior to the lambda line. The breaks in the skull were filled with Bonewax, and the wound was closed with sutures. These lesioned animals showed no obvious behavioral or motor deficits. At 4 d after surgery, the lesioned animals were given two intraperitoneal injections of BrdU (100 mg/kg of body weight; Sigma) over a 2 hr period and were again anesthetized and killed by perfusion 2 hr after the second injection. Immunostaining with anti-BrdU antibody was performed as described previously (Sakakibara and Okano, 1997).
Chromosomal mapping of mouse msi1 and msi2. Mouse 129 sv/J genomic DNA fragments of mouse msi1 (11.5 kb) and mousemsi2 (11 kb), each containing the ATG translation initiation codon, were subcloned into the pBluescriptII (Stratagene) and pZErO-1 (Invitrogen) plasmid vectors, respectively, and used for fluorescencein situ hybridization (FISH) as described previously (Satoh et al., 1993). Each of the plasmids was labeled with biotin-16-dUTP or digoxygenin-11-dUTP using a nick translation kit (Roche Diagnostics) and hybridized to metaphase chromosomes from mouse 129 embryonic stem cells. On each slide, 1 μg of labeled DNA was applied with a 5- to 10-fold excess amount of mouse Cot-1 DNA (Life Technologies-BRL) to block nonspecific hybridization to the repetitive sequences. The hybridization signals were visualized with FITC-avidin (Roche Diagnostics) or rhodamine-conjugated anti-digoxigenin (Vector Labs, Burlingame, CA). Chromosomes were counterstained with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) to identify individual chromosomes. Photographs were taken of the FISH signals superimposed on the images of DAPI-banded chromosomes.
GenBank accession numbers. The nucleotide sequence data reported in this paper have been deposited with the GSDB, DDBJ, EMBL, and NCBI under accession numbers AB056102 formsi2S and AB056103 for msi2L.
RESULTS
Identification of msi2 and characterization of its primary structure
To isolate the cDNA of a novel mammalian RNA-binding protein that was homologous to Msi1, a mouse nervous system cDNA library was screened using mouse msi1 and Xenopus xrp1 cDNA probes at reduced stringency. The Xenopus xrp1 gene (Good et al., 1993) encodes a protein that is related by sequence to the NRP1 protein, a Xenopus homolog of Msi1 (Sakakibara et al., 1996). The largest DNA insert obtained was used as a probe to rescreen the cDNA library at high stringency to obtain full-length cDNA. The longest and single open reading frame, encoding a 346 amino acid protein with a predicted molecular mass of 37 kDa, was identified from nine overlapping cDNAs (Fig.1A). Sequence analysis indicated that the gene product encoded by the cDNAs was a novel RNA-binding protein. We designated this Msi1-related gene as mousemusashi2. Multiple cDNA clones obtained from the library screening and RT-PCR analysis of msi2 transcripts from RNA isolated from E12 and adult mouse brains (data not shown) indicated that there were two alternatively spliced transcripts depending on the presence or absence of a short segment (18 amino acids) within the C-terminal half of Msi2 (Fig. 1A, broken underline). This alternative splicing could generate two Msi2 protein isoforms with predicted molecular masses of 36.9 and 35.7 kDa (termed Msi2L and Msi2S, respectively).
Many RNA-binding proteins contain a conserved sequence of 80–90 amino acids, termed an RRM, that includes two short, highly conserved core domains called RNP-1 and RNP-2 (Burd and Dreyfuss, 1994) (Fig.1A). Msi2 contains two RRMs (RRM-1 and -2) that contain well conserved RNP-1 consensus sequences, (K/R)G(F/Y)(G/A)FVX(F/Y) (Burd and Dreyfuss, 1994). RNP-2 is less conserved than RNP-1 but is characterized by its aliphatic and aromatic nature and its relative position to RNP-1 (Burd and Dreyfuss, 1994). Among a number of known RNA-binding proteins, a homology search against the expressed sequence tags (EST) and GenBank databases revealed that Msi2 shared remarkable sequence similarity with Xenopus XRP1 and mouse Msi1, a neural RNA-binding protein (Fig.1B–D). Msi2 and Msi1 showed significant sequence similarity (75% amino acid identity in the overall structure). In particular, the RRM regions of Msi2 showed 85 and 95% identity to those of Msi1 and XRP1, respectively (Fig.1B). The amino acid sequences of RNP-1 and RNP-2, which are critical for binding to RNA substrates, were completely conserved among these three proteins (Fig. 1C). This raises the possibility that Msi2 may bind the same RNA molecules that are recognized by Msi1 or XRP1. On the other hand, Msi2 did not show extensive similarity to the RRMs of any other RNA-binding proteins, including Drosophila Elav (Yao et al., 1993) and the vertebrate Hu proteins (HuB, HuC, HuD, and HuR) (Okano and Darnell, 1997). In the C-terminal tail of the protein, which does not contain the RRMs, Msi2 showed 67% amino acid identity with Msi1 (Fig.1B). The major difference between Msi2 and XRP1 was a 58 amino acid segment in the C-terminal half of XRP1, resulting in a length of 346 amino acids for Msi2 and 406 amino acids for XRP1 (Fig.1C). Except for the two RRM domains, no other functional domains or consensus sequences were identified in the putative Msi2 protein, as is also the case with Msi1 and XRP1. A short alanine-rich stretch, the function of which is not known, is present in the C-terminal tail of Msi2, Msi1, and XRP1 (Fig. 1C).
The relationship of the Msi2 and Msi1 proteins to other mammalian RRM-containing proteins was demonstrated by the pairwise comparisons presented in Figure 1D. Msi2 and Msi1 formed a unique subgroup within the hnRNP A/B class of proteins, the heterogeneous nuclear ribonucleoproteins that contain two copies of RRMs in addition to auxiliary domains. Among this class, hnRNP type A/B protein (Khan et al., 1991) and AUF1 (hnRNP D) (Zhang et al., 1993) were found to be somewhat related to this Msi subgroup (Fig. 1D). AUF1 binds the U-rich elements of the 3′-UTR found in many proto-oncogene and cytokine mRNAs and regulates the stability of these mRNAs (DeMaria and Brewer, 1996; Blaxall et al., 2000).
To determine whether msi2 mRNA is expressed in a tissue-specific manner, we performed Northern blotting of the RNA from various tissues from adult mice, probing with a 750 bp 3′-UTR fragment of mouse msi2 cDNA (Fig.2A). Substantial levels of a major msi2 transcript of 7.1 kb was detected in all tissues tested, with the highest expression observed in the cerebellum and smaller amounts in the liver and small intestine. A 1.5 kb short form of the msi2 transcript was detected only in the testis. Northern blots of RNA from different developmental stages indicated the continuous expression of the msi2 transcript in the CNS throughout development (Fig. 2B). This tissue distribution and developmental profile of the msi2transcripts paralleled the distribution profile reported forXenopus xrp1 mRNA (Good et al., 1993).
Fig. 2.
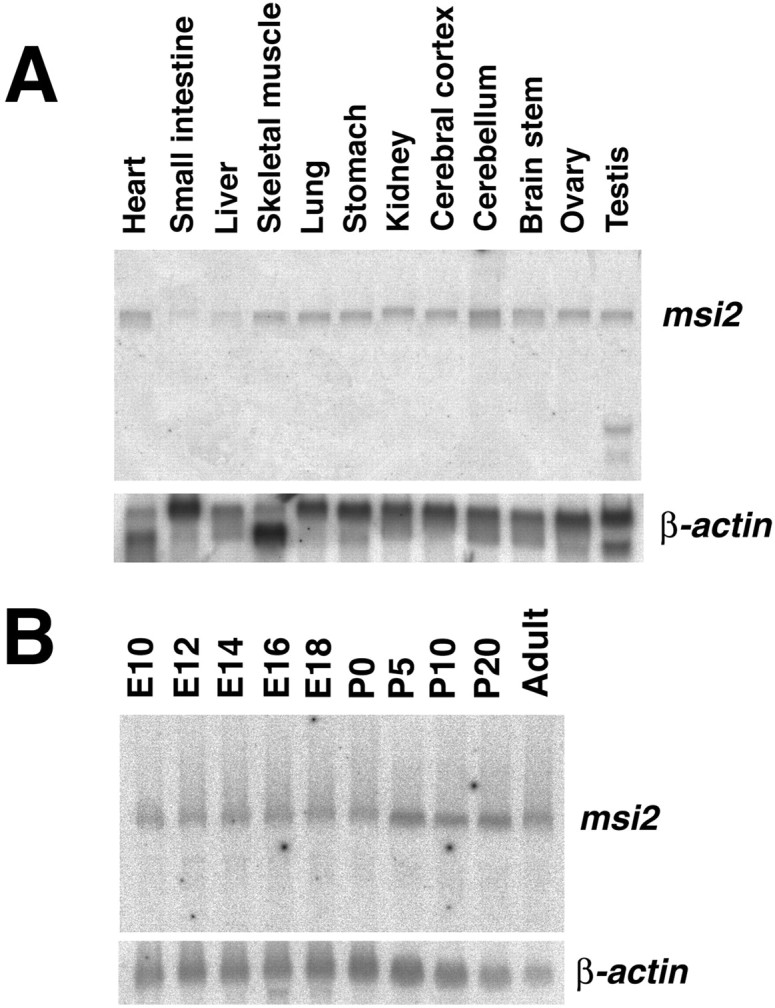
Tissue distribution and developmental expression of the msi2 transcripts. Total RNA (20 μg) from each mouse tissue was prepared, and the levels of msi2 mRNA were determined by Northern blot analysis. A, Tissue distribution of msi2 mRNA in adults. Expression of a 7.0 kb transcript can be seen in most lanes. A short transcript of ∼1.5 kb is observed only in the testis. B, Changes inmsi2 mRNA expression in the whole brain associated with development. Msi2 mRNA expression can be seen at E10, the earliest time point tested. The equal loading and quality of the RNAs were tested by performing the hybridization with a β-actin probe, shown in each bottom panel.
On the basis of the striking similarity of both the predicted protein sequence and the expression pattern, we concluded that themsi2 gene was a mouse homolog of Xenopus xrp1.Msi1 and Msi2 seem to constitute a novel RNA-binding protein family (Msi family) in rodents, although they show tissue distributions that are different from each other.
Chromosomal localization of the msi1 and msi2genes
Genomic Southern blot analysis using probes from msi1and msi2 genomic fragments yielded the predicted single bands for several appropriate restriction enzymes, indicating that both are likely to be single-copy genes. The cytogenetic locations of themsi1 and msi2 genes were determined by FISH. As shown in Figure 1E, the msi1 andmsi2 genes were mapped to chromosome 5qE3-F and 11qB5-C, respectively, by DAPI bands. Our previous FISH and somatic cell hybrid panel analyses (Good et al., 1998) localized the human msi1gene to chromosome 12q24.1–24.31, which corresponds to the syntenic region of the mouse msi1 locus. The msi2 gene was predicted to exist on chromosome 17 q11–21 in humans by the comparative map (Mouse Genomic Informatics database of the Jackson Laboratory). Indeed, search of the human genome database revealed a partial nucleotide sequence of the human msi2 gene on chromosome 17 (covered by a 165 kb BAC clone; GenBank accession no. AC005325), indicating the conservation of the msi2 andmsi1 genes across species, including Xenopus, mouse, and human. In human, this msi2 syntenic region contained several loci of hereditary neurological disorders: frontotemporal dementia with parkinsonism (FTDP17) (Spillantini and Geodert, 1998), Pick disease (Munoz-Garcia and Ludwin, 1984), familial progressive subcortical gliosis (GPSC) (Petersen et al., 1997), and neurofibromatosis type 1 (von Recklinghausen disease, NF1) (Mukonoweshuro et al., 1999).
RNA-binding properties of Msi2 protein
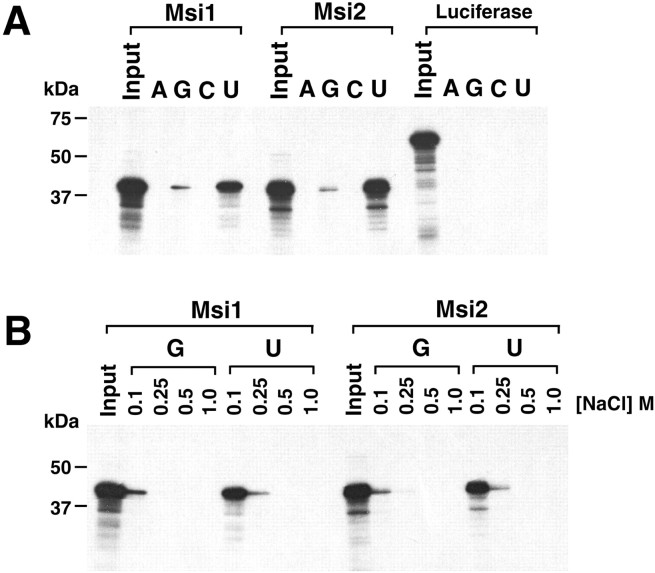
To test whether Msi2 could actually bind RNA, and if so, whether Msi1 and Msi2 recognize common RNA targets, we performed RNA-binding analysis in vitro. The full-length Msi2 and Msi1 proteins were prepared by in vitro transcription/translation, and their ability to bind RNA-homopolymers was determined. Both the Msi2 and Msi1 proteins showed strong binding to poly(U), weaker binding to poly(G), and no binding to poly(A) or poly(C), under normal stringency conditions in a buffer containing 100 mm NaCl (Fig.3A). A 61 kDa luciferase polypeptide showed no binding to any of the homopolymers, confirming the RNA-binding activity of the Msi2 and Msi1 proteins. Furthermore, binding experiments performed under various degrees of stringency, up to 1.0 m NaCl, revealed that the Msi1 and Msi2 proteins both bound poly(U) and poly(G). The binding to poly(U) was stable in 250 mm NaCl, although the binding to poly(G) was only seen at 100 mm NaCl (Fig. 3B). The binding properties of Msi1 observed in this study were slightly different from our previous report, in which we found, using a Northwestern assay, that Msi1 binds poly(G) more efficiently than poly(U) (Sakakibara et al., 1996). This discrepancy might be attributable to the different binding assays used in the two studies. Our previous Northwestern binding assay used a bacterially expressed, denatured, and immobilized Msi1 polypeptide. We believe that the binding properties observed in this report, in which soluble Msi1 protein produced fromin vitro transcription/translation was used, are more representative of the in vivo binding capacity of Msi1. Taken together, these observations indicated that Msi2 is an RNA-binding protein with in vitro RNA-binding specificity similar to that of Msi1. The Msi family proteins may share the same or similar RNA(s) characterized by uridine-rich sequences as their downstream target molecules in vivo.
Fig. 3.
Similar RNA-binding properties of Msi2 and Msi1 proteins. A, Binding of the full-length Msi2 and Msi1 proteins to ribonucleotide homopolymers.35S-methionine-labeled, in vitro-translated Msi2, Msi1, and Luciferase (as a control protein) were incubated with the indicated ribonucleotide homopolymer beads in buffer containing 100 mm NaCl. Bound proteins were visualized by SDS-PAGE and fluorography as described in Materials and Methods. Ribonucleotide homopolymers and proteins used in the assays are indicated at the top of each lane. Lanes marked Input contain each 35S-labeled protein used for binding reactions. The molecular sizes of markers are indicated on the left. Both Msi2 and Msi1 bound strongly to poly(U) and weakly to poly(G). Luciferase showed no binding to RNA homopolymers. B, Binding profile of Msi2 and Msi1 to RNA homopolymers under stringent conditions. Binding experiments ofin vitro-translated Msi2 and Msi1 with poly(U) and poly(G) beads were performed as described above at the indicated NaCl concentrations. In the buffer containing 0.25 m NaCl, Msi2 and Msi1 binding to poly(G) RNA was greatly diminished, whereas that to poly(U) remained significant. Note that the binding properties of Msi2 to poly(U) and poly(G) in the given salt concentrations were almost identical to those of Msi1.
Specificity of the Msi2 antibody
To localize the Msi2 protein in tissues, polyclonal antiserum (anti-Msi2) was generated in rabbits using a peptide corresponding to the amino-terminal 14 amino acids of mouse Msi2, in which no obvious homology to Msi1 could be seen (Fig.1A,C). As shown in Figure4A, the anti-Msi2 antiserum recognized both recombinant full-length Msi2L and Msi2S proteins but did not cross-react with Msi1 protein. Conversely, the anti-Msi1 monoclonal antibody (14H1) (Kaneko et al., 2000) detected Msi1 protein exclusively and not Msi2. In Western blots of embryonic (E12) and postnatal (P3) brains, the anti-Msi1 antibody recognized a doublet of ∼37–40 kDa (Figs. 1B,4B), which reflects alternative splicing products (Sakakibara et al., 1996). The Msi2 protein was also detected as two major polypeptides, with electrophoretic mobilities of 35 and 37 kDa (Fig. 4B, two arrows), that were likely to correspond to Msi2S and Msi2L, respectively. Both the 35 and 37 kDa proteins appeared to be composed of a closely spaced doublet (Fig.4B). These doublets were probably caused by the phosphorylation of Msi2L and Msi2S in vivo, because treatment of the partially purified endogenous Msi2 with λ protein phosphatase collapsed these doublets into single, faster-migrating bands (Fig. 4C). The Msi2 sequence is rich in serine and threonine residues (12%), many of which are potential phosphorylation sites for casein kinase I and II, GSK3, or PKC. Immunoblot analysis ofmsi2 knock-out mice verified that all these bands were derived from the msi2 gene (our unpublished results). Together, these results established the specificity of the anti-Msi1 and anti-Msi2 antibodies.
Fig. 4.
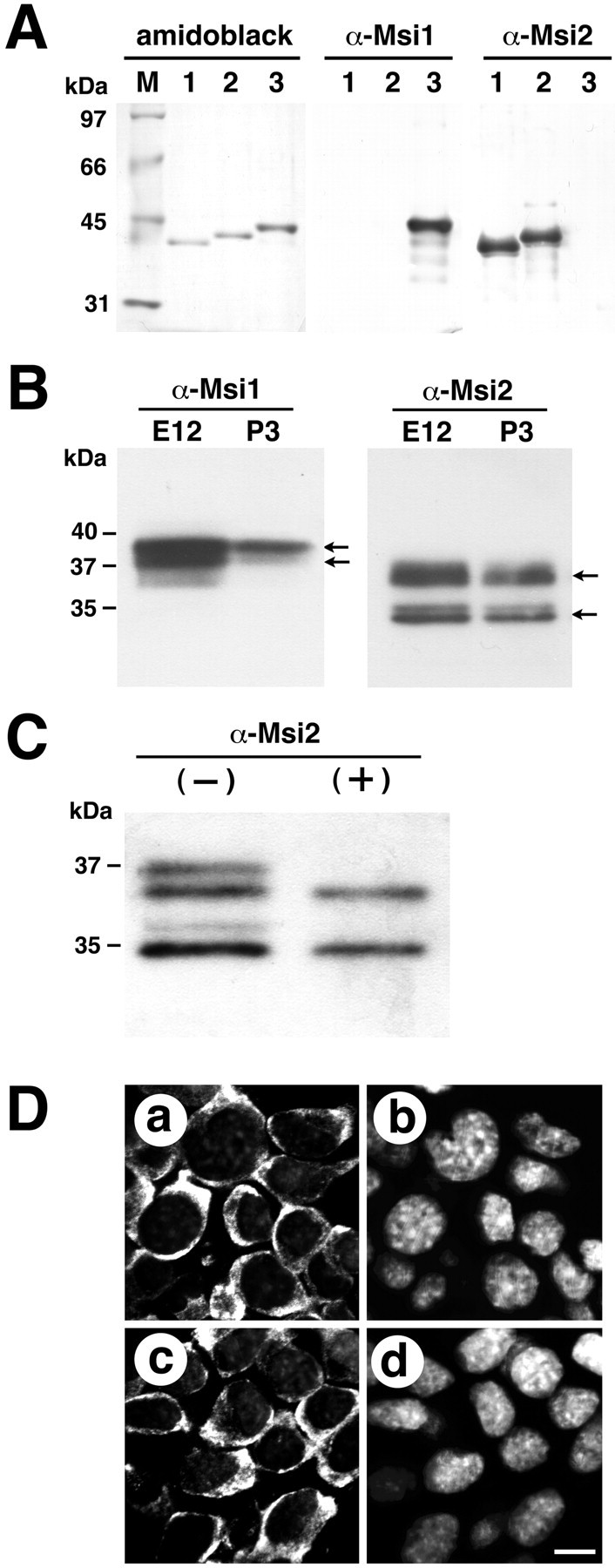
Western blot analysis and cytoplasmic localization of Msi1 and Msi2 proteins. A, Specificity of the anti-Msi2 and anti-Msi1 antibodies. Bacterially expressed full-length Msi2S (lane 1), Msi2L (lane2), and Msi1 (lane 3) proteins were subjected to immunoblotting. Membranes were probed with either affinity-purified rabbit polyclonal anti-Msi2 or rat monoclonal anti-Msi1 (14H1) antibodies. The anti-Msi2 antibody specifically detected Msi2L and Msi2S and did not cross-react with Msi1 protein. Conversely, the anti-Msi1 antibody recognized only the Msi1 protein. The left panel shows the blot stained with Amido black dye, confirming the integrity and equal loading of each fusion protein. Lane M indicates protein size markers. B, Changes in Msi2 and Msi1 protein expression during brain development. A Western blot of protein extracts from E12 and P3 brains (30 μg protein per lane) was probed with anti-Msi1 or anti-Msi2 antibodies. The anti-Msi1 antibody recognized a doublet at 37–40 kDa (arrows). Msi2 was detected as two major proteins of 35 and 37 kDa (arrows), and each protein consisted of a closely spaced doublet. C, Msi2 is a phosphoprotein. Western blot analysis for Msi2 in the polysome fraction from E12.5 brain was treated with (+) or without (−) protein phosphatase. Incubation with protein phosphatase produced a downward shift of the slower migrating form in each doublet. D, Confocal images of the neural cell line NG108 stained with anti-Msi1 (a) or anti-Msi2 (c), showing their cytoplasmic localization. b, d, The same fields, counterstained with Hoechst33342 dye to identify the individual nuclei, are shown. Scale bar (shown in d), 8 μm.
Subcellular distribution of the Msi2 and Msi1 proteins
The Msi2 and Msi1 proteins were localized exclusively to the cytoplasm of several neural cell lines, including NG108 (Fig.4D) and PC12 (data not shown) cells. Cytoplasmic expression of Msi2 and Msi1 was also observed in E10 neuroepithelial cells (Figs. 5G,H,insets, 6C). In addition, Msi2 was present in the cytoplasm of cells in non-neural tissues (data not shown) and in differentiated neurons, in which Msi2 protein was found in proximal regions of the somata rather than in the dendritic arborizations or axons (see Figs. 8G,9G). During subcellular fractionation of E12.5 brain lysate, a significant amount of Msi2 was recovered in the polysome fraction (Fig. 4C). Consistently, our observations by subcellular fractionation (Imai et al., 2001) as well as immunoelectron microscopy (our unpublished results) showed Msi1 protein to be associated with the free and membrane-bound polysomes in the embryonic VZ cells (our unpublished results), also supporting the notion that the Msi family plays a role in regulating RNA metabolism in the cytoplasm. However, it remains possible that Msi family proteins also participate in nuclear events, such as mRNA splicing. In fact, we previously observed the nuclear localization of Msi1 in a small population in cultured astrocytes, although the significance of this observation remains unclear (Kaneko et al., 2000).
Fig. 5.
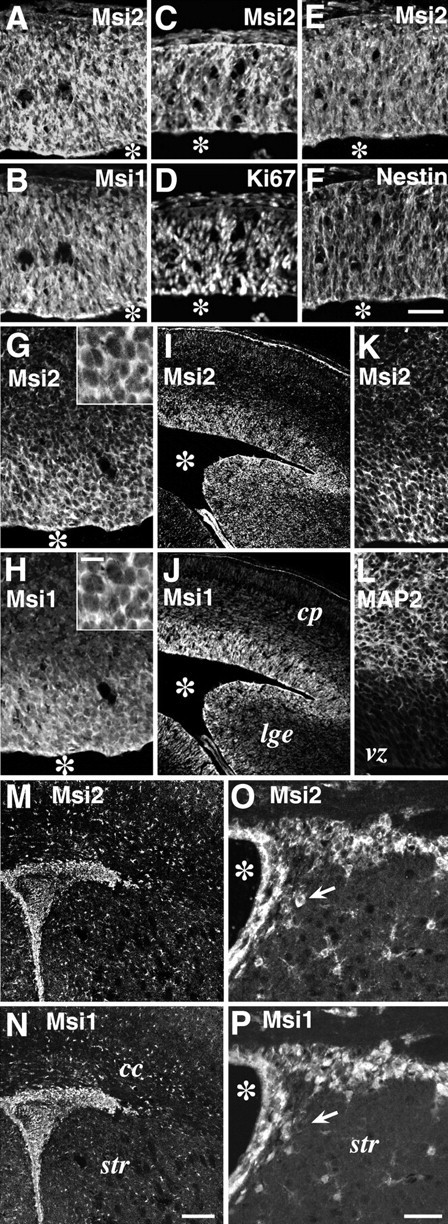
Msi2 expression in neural precursor cells residing in the embryonic VZ and postnatal SVZ. Double-immunofluorescence labeling of coronal sections through embryonic, postnatal, and adult brains, with antibodies against Msi2 and Msi1, Ki67, Nestin, or MAP2.A/B, C/D,E/F, G/H,I/J, K/L,M/N, andO/P represent each pair of double-stained photomicrographs. The primary antibodies used are indicated in each panel. A–F, Telencephalon at E10. Msi2 and Msi1 are uniformly distributed in the Ki67- and Nestin-positive neuroepithelium. G/H, Cerebral cortex at E12.Insets indicate higher magnifications of the VZ cells.I–L, Cerebral cortex at E14. Coincidental expression of Msi2 and Msi1 is seen in the VZ of the telencephalon and ganglionic eminences. Note the absence of Msi2 expression in the intermediate zone and cortical plate, which are occupied by MAP2-positive immature neurons.M/N, The SVZ region surrounding the lateral ventricle in P7 postnatal forebrain. O/P, Adult ependyma and SVZ lining the lateral ventricle. Lateral is to the right, and dorsal is up. Many densely packed cells that are brightly immunostained with both Msi2 and Msi1 are observed in the P7 SVZ, adult SVZ, and ependymal layer. Small scattered astrocytes in the parenchyma are also labeled with anti-Msi2 and anti-Msi1 antibodies. The arrow in O/P indicates a Msi2-positive but Msi1-negative neuron-like cell. Scale bars:A–H, K, L, 50 μm; I, J, M,N, 90 μm; O, P, 25 μm;insets in G and H, 8 μm.Asterisks indicate lateral ventricle. cp, Cortical plate; vz, ventricular zone;lge, lateral ganglionic eminence; cc, corpus callosum; str, striatum.
Fig. 6.
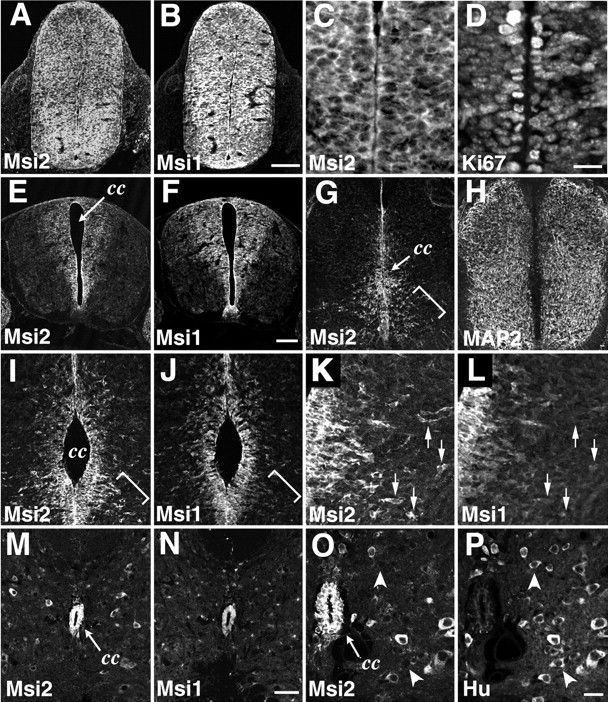
Msi2 expression in the spinal cord. Double-immunofluorescence labeling of horizontal sections through the embryonic and adult lumbar spinal cord, with antibodies against Msi2 and Msi1, MAP2, or Hu. A/B,C/D, E/F,G/H, I/J,K/L, M/N, andO/P represent each pair of double-stained photomicrographs. The primary antibodies used are indicated in each panel. A/B, E10 spinal cord. Msi2 and Msi1 are uniformly distributed within the neural tube. C/D, Higher magnification of the ventricular zone of E10 spinal cord, showing the Msi2 expression in the Ki67-positive proliferating neuroepithelial cells. E/F, E12 spinal cord.G–L, E14 spinal cord.K/L, Higher magnification of the bracketed area inI/J. Msi2 and Msi1 are simultaneously expressed in the embryonic VZ lining the central canal. Outside the VZ, most of the MAP2-positive postmitotic neurons do not show immunoreactivity for Msi2, whereas a subset of migrating neurons that are sparsely distributed in the ventrolateral area of the mantle layer coexpress Msi2 and MAP2 (bracketed area in G). These Msi2-positive cells are negative for Msi1 and have a few processes, indicating that they are migrating out of the VZ (bracketed portion in I/J, andarrows in K/L). M/N, Adult spinal cord. O/P, Higher magnification ofM/N. In the adult spinal cord, Msi2 and Msi1 are highly expressed in ependymal cells surrounding the central canal. Note the Msi2 expression in some, but not all, populations of neurons scattered within the parenchyma. Arrowheads in Oand P indicate Hu-positive but Msi2-negative neurons.cc, Central canal. Scale bars: A,B, 100 μm; C, D, 12 μm;E–H, 50 μm; I, J, M, N, 45 μm; K, L, O, P, 10 μm.
Fig. 8.
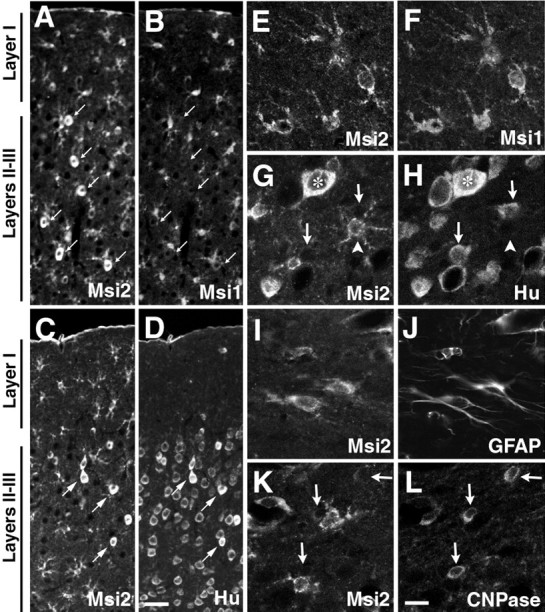
Distribution and cell type of Msi2-positive cells in adult cerebrum. Double immunofluorescence labeling of the somatosensory area of the adult neocortex, with antibodies against Msi2 and Msi1, or cell type-specific markers.A/B, C/D,E/F, G/H,I/J, andK/L represent each pair of double-stained photomicrographs. Primary antibodies used are indicated in each panel.A–D showing the expression of Msi2 in astrocytes scattered throughout the gray (layers II–III) and white matter (layer I) of the neocortex. Many small Msi2- and Msi1-positive astrocytes with small cell bodies are seen throughout the cortex (A, B). There is also a subset of neurons labeled with Msi2 (A/B, arrows) with larger cell bodies, which are found sparsely in layers II–III, and coexpress Hu antigens (C/D, arrows) but are never labeled with Msi1 (compareA/B with C/D). The pial surface is at thetop. E–L, High-power photomicrographs of Msi2-expressing cells in the gray matter (layers II–III, E-H) and white matter (molecular layer,I/J; subcortical white matter,K/L). In the gray matter, concurrent expression of Msi2 and Hu is observed in a few round neuronal cells (asterisks in G/H), whereas numerous other Hu-positive neurons lack immunoreactivity for Msi2 (arrows in G/H). There is also a considerable number of Hu-negative but Msi2-positive cells, which have small oval cell bodies with elaborated multiple short processes (arrowhead in G/H). These cells may represent the population of GFAP-negative protoplasmic astrocytes that are frequently observed in deep gray matter and are positive for Msi1 (E/F). Fibrous astrocytes in the superficial molecular layer and near the pial surface show colocalization of Msi2 and GFAP (I/J), whereas CNPase-positive oligodendroglial cell bodies predominant in the subcortical white matter are Msi2 negative (arrows in K/L). Scale bars: A–D, 25 μm;E–L, 8 μm.
Fig. 9.
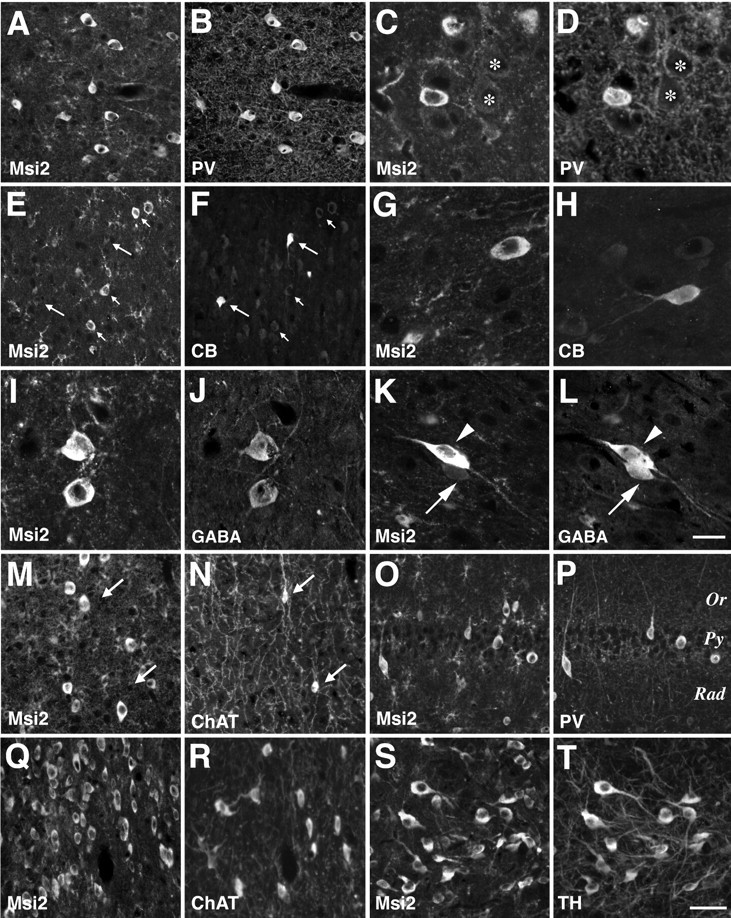
Msi2 expression in parvalbumin-positive GABAergic neurons in the adult neocortex. Double-immunofluorescence labeling with antibodies against Msi2 and parvalbumin (PV), calbindin-D28K (CB), GABA, choline acetyltransferase (ChAT), or tyrosine hydroxylase (TH) was performed on coronal sections of adult cerebral cortex, basal ganglia, and hippocampus.A/B, C/D,E/F, G/H,I/J, K/L,M/N, O/P,Q/R, andS/T represent each pair of double-stained photomicrographs. Primary antibodies used are indicated in each panel.A/B, Neocortex layers II–III, showing the coincidental expression of Msi2 and PV in GABAergic nonpyramidal neurons. Faint staining of Msi2 is also seen in the fine-branched processes and small cell bodies of astrocytes. C/D, Higher magnification of the Msi2- and PV-positive neurons in the deeper layer V. The somata of large pyramidal neurons surrounded by parvalbumin-positive multi-terminal endings express neither Msi2 nor PV (asterisks). E/F, Neocortex layers II–III stained with Msi2 and CB. Note that these two populations of neurons are intermingled but do not overlap with each other.Long arrows indicate Msi2-negative CB-positive neurons.Short arrows indicate the neurons that are Msi2 positive but CB negative. G/H, Higher magnification of neurons that express CB or Msi2 in layers II–III. I/J,K/L, Layers II–III stained with antibodies to Msi2 and GABA, showing the Msi2 expression in GABA-containing neurons. (I/J and arrowhead inK/L). Arrow in K/L points to a GABA-containing but Msi2-negative neuron. M/N, Gray matter of the neocortex, showing the absence of Msi2 expression from ChAT-positive cholinergic neurons (arrows).O/P, Hippocampal formation, showing the coexpression of Msi2 and PV in a small number of interneurons in the CA1 subfield. Numerous CA1 pyramidal neurons aligned in rows are not labeled with either Msi2 or PV. Q/R, S/T, Nuclei of the vertical limb of the diagonal band and the zona incerta, respectively. In the basal ganglia, Msi2 immunoreactivity was found in several ChAT-positive cholinergic neurons in the nuclei of diagonal bands (Ch2 and Ch3), in addition to the PV-positive neurons. Msi2 is also expressed in TH-positive dopaminergic neurons (A13) within the zona incerta in the subthalamic region. Scale bars:A, B, E, F,M–T, 25 μm; C,D, G, H,I–L, 8 μm. Rad, Stratum radiatum; Py, stratum pyramidale; Or, stratum oriens.
Developmental expression of Msi2 in the CNS
Although the Msi1 protein was highly enriched in the embryonic CNS, the Msi2 protein level appeared to stay at a relatively constant level during brain development (Fig. 4B) in parallel with its mRNA levels (Fig. 2B). However, the following immunohistochemical results clearly demonstrated that the Msi2 expression was not ubiquitous, at least in various regions of the embryonic and postnatal CNS. Rather, Msi2 was expressed in a cell type-specific manner, and its expression appeared to be strongly regulated spatially and temporally during the development of the mouse brain and spinal cord. Notably, in the developing CNS, Msi2 was mainly detected in proliferating precursor cells in the VZ and SVZ with a pattern equivalent to that of Msi1.
VZ and SVZ
First, we examined the sites of Msi2 expression in the CNS at the time when active neurogenesis and cell differentiation are known to occur. The neural tube of the E10 mouse embryo is uniformly occupied with mitotic and undifferentiated neuroepithelial cells. At this stage, Msi2 expression was intense throughout the telencephalon and spinal cord (Figs. 5, 6). Most of these Msi2-positive cells were Msi1-positive (Figs. 5A,B,6A,B), Ki67-positive (Figs.5C,D, 6C,D), and nestin-positive (Fig. 5E,F) neuroepithelial cells. Ki67 is a marker for proliferating cells (Nakamura et al., 2000), and nestin, an intermediate filament protein, is a selective marker for neural progenitor cells, including CNS stem cells (Lendahl et al., 1990). By E12–14, as CNS neurogenesis proceeds, Msi1-positive mitotic neural precursors become restricted to the VZ in the telencephalon and spinal cord; at this time, differentiated neurons occupy the peripheral locations (Sakakibara et al., 1996, Kaneko et al., 2000). At the same time, simultaneous expression of Msi2 and Msi1 was detected in the VZ cells of most CNS regions, including the telencephalon (Fig. 5G–L), spinal cord (Fig.6C–L), and ganglionic eminence (Fig.5I,J). Msi2 expression disappeared in most of the MAP2-positive postmitotic neurons migrating in the intermediate zone and cortical plate of the forebrain (Fig.5I–L) and in the spinal cord (Fig.6G,H).
In postnatal and adult CNS, cells in the ependyma and SVZ regions showed significant levels of Msi2 and Msi1 expression. At P7, during active gliogenesis, a large number of densely packed Msi2-positive cells were found in the dorsolateral corner of the SVZ bordered by the striatum and overlying the corpus callosum (Fig. 5M). This region is known to be proliferative and to contain not only glial precursors for astrocytes and oligodendrocytes (Levison and Goldman, 1993; Sakakibara and Okano, 1997) but also neuronal precursors (in the anterior part of the SVZ) for the interneurons of the olfactory bulb (Lois and Alvarez-Buylla, 1993; Luskin, 1993). As gliogenesis proceeds during postnatal development, the SVZ becomes thinner. However, it persists throughout adulthood as a mitotically active layer (Lewis, 1968). Many Msi2-positive cells were found in the ependyma and SVZ cells in the adult forebrain (Fig. 5O). In the adult spinal cord where the SVZ component had already disappeared, a high expression of Msi2 remained in the ependymal cells lining the central canal (Fig. 6M,O). Double immunofluorescence revealed the coexpression of Msi2 and Msi1 in the cells of the ependyma and SVZ throughout postnatal life. Recentin vivo and in vitro studies have provided evidence that the ependyma and/or SVZ of the postnatal mammalian forebrain may be a source of CNS stem cells, which have the potential to proliferate and differentiate into both neurons and glia (Morshead et al., 1994, 1998; Johe et al., 1996; Doetsch et al., 1999; Johansson et al., 1999).
To examine the expression of Msi proteins in a CNS stem cell population, we performed further immunostaining experiments using neurospheres. In the presence of mitogens such as EGF and bFGF, dissociated neural cells proliferate and form floating multicellular structures called “neurospheres' (Reynolds and Weiss, 1996;Nakamura et al., 2000). Most of the cells in a neurosphere are clonally derived from a single CNS stem cell/progenitor and are thought to possess the characteristics of CNS stem cells; i.e., they have self-renewing activity and are pluripotent, able to differentiate into either neurons or glia (Reynolds and Weiss, 1996; Nakamura et al., 2000). As shown in Figure7A–C, Msi2 and Msi1 were intensely coexpressed in cells of neurospheres derived from E14.5 telencephalon. Coexpression of Msi2 and Ki67 (Fig. 7D,inset) indicated that many Msi2-positive cells were endowed with the proliferative activity in neurospheres. These neurospheres were also uniformly immunolabeled with antibodies to nestin, and their pluripotency was confirmed by differentiation assays using sister neurospheres (data not shown).
Fig. 7.
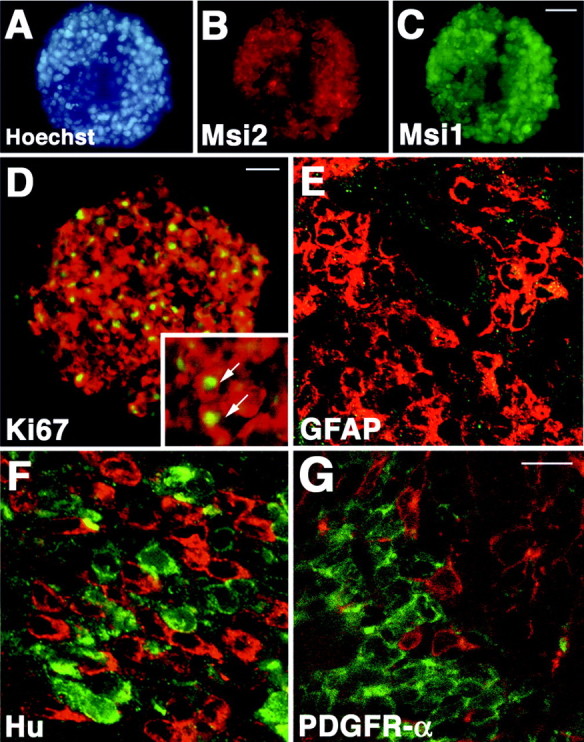
Msi2 expression in neurospheres and absence of Msi2 in the immature neurons or oligodendrocyte precursor cells lying in the SVZ. A–C, Double immunostaining of neurosphere from E14.5 embryonic telencephalon, showing the coexpression of Msi2 (B, red) and Msi1 (C, green). Nuclei are counterstained with Hoechst dye (A, blue). D, Msi2 expression (red) in Ki67-positive proliferating cells (green) in a neurosphere. Inset shows the higher view of the neurosphere. Arrows point to the Msi2 and Ki67 double-positive cells. Note that Msi2 is mainly localized in the cytoplasm of the cells in neurosphere, whereas the antibody to Ki67 only stains the nuclei. E–G, Scanning confocal images of the coronal sections through the SVZ surrounding the lateral ventricle of the P2 mouse forebrain. E, GFAP (green) and Msi2 (red). Msi2 is expressed in the GFAP-negative densely packed SVZ cells.F, Hu (red) and Msi2 (green). Hu-positive immature neurons and Msi2-positive cells distribute intermingled within the dorsolateral area of the SVZ, but these two populations of cells hardly overlap with each other. G, PDGFR-α (red) and Msi2 (green). Msi2 expression is never found in oligodendrocyte precursor cells that are positive for PDGFR-α and lie scattered in the SVZ. Scale bars: C, 50 μm;D, 20 μm; E–G, 10 μm.
To address whether Msi2 expression is restricted to the neural precursor cells/CNS stem cells in vivo, we double-stained the sections through the SVZ regions with the antibodies to GFAP, Hu, TuJ1, or PDGFR-α. In the postnatal SVZ surrounding the lateral ventricles, the prominent expression of Msi2 was observed in the densely packed cells as described above and as shown in Figure7E. We could not detect any cells reactive for GFAP within the SVZ (Fig. 7E), whereas a few Msi2- and GFAP-positive astrocytes that have multiple processes were present in the developing corpus callosum (data not shown). Hu belongs to another RNA-binding protein Elav family that is expressed in most immature and differentiated neurons (Barami et al., 1995; Okano and Darnell, 1997;Sakakibara and Okano 1997). The dorsolateral part of the SVZ region contained many Hu-positive immature neurons that were generated from the SVZ cells and appeared to be migrating into the lateral parenchyma. Although Msi2- and Hu-positive cells intermingled within these SVZ regions, Msi2-positive cells were exclusively immunonegative for Hu, and vice versa (Fig. 7F). Similarly, TuJ1-positive immature neurons and their tangled neurites in these SVZ regions were never labeled with Msi2 (data not shown). In oligodendrocyte lineage, PDGFR-α-positive cells residing in the postnatal SVZ were proposed to be the earliest form of oligodendrocyte progenitor cells (or O-2A progenitor cells), which divide in the SVZ to give rise to slightly more differentiated cells of the oligodendrocyte lineage in the nascent parenchyma, such as the corpus callosum and the neocortex (Nishiyama et al., 1996). Double immunofluorescence labeling demonstrated that these PDGFR-α-positive oligodendrocyte precursor cells showed no immunoreactivity to Msi2 (Fig. 7G).
Taken together, these results suggested that Msi2 expression was mainly associated with the proliferating precursor cells/CNS stem cells in the VZ and SVZ and disappeared from the oligodendrocyte lineage and most neurons early in their differentiation. This expression profile of Msi2 was quite similar to that of Msi1 during CNS development. In a subset(s) of differentiated neurons, however, sustained Msi2 expression was evident on the basis of the following immunohistochemical results.
Cerebrum
Neurons and glial cells, which originate in the embryonic VZ or the postnatal SVZ, respectively, migrate into appropriate layers through formative white matter, including the corpus callosum, to form the stratified adult cerebral cortex. Msi1 is known to be expressed in cells of the astrocyte lineage, which include both migrating GFAP-negative (protoplasmic) and GFAP-positive differentiated astrocytes (Miller and Raff, 1984; Sakakibara and Okano, 1997). Msi2-expressing cells were found in the parenchymal regions of the cerebrum throughout life, including the cerebral cortex, corpus callosum, and striatum, although a lower immunoreactivity and sparser distribution of Msi2 was seen in these regions than in the cells of the SVZ. To identify the cell type that expressed Msi2 in the mature CNS, double-label indirect immunostaining was performed using antibodies against Msi2 and one of the following marker proteins: Hu for neurons, CNPase for differentiated oligodendrocytes (Sprinkle 1989), and GFAP and Msi1 for astrocytes. We observed the accumulation of Msi2-positive cells throughout the cerebral cortex (Fig.8A,B). Some of these coexpressed Msi1 and possessed relatively thick and branched processes (Fig.8E,F), a feature typical of differentiated astrocytes. Double labeling with antibodies to Msi2 and GFAP confirmed the expression of Msi2 in astrocytes, as expected (Fig.8I,J). Cells that were immunoreactive for both Msi2 and GFAP were predominantly distributed in the molecular layer (layer I) (Fig.8I,J) and near the pial surface. In addition, the deeper layers (layers II–VI) of the cortex contained a considerable number of Msi2-positive but Hu-negative astrocytic cells (Fig. 8G,H,arrowhead). These cells coexpressed Msi1 (Fig.8E,F) and thus might represent protoplasmic astrocytes, which are known to be abundant in cortical layers II–VI but are sparse in layer I (Sakakibara and Okano, 1997). Many of these astrocytes are not stained by GFAP immunohistochemistry (Miller and Raff, 1984). In addition, astrocytes that were closely associated with the endothelial cells of vascular blood vessels were also immunoreactive for Msi2 (data not shown). In oligodendrocytes, although Msi2- and CNPase-positive cells were found commingled throughout the gray and white matter, CNPase-positive cells were exclusively immunonegative for Msi2, and vice versa (Fig.8K,L). This was consistent with the observation indicating the absence of Msi2 from the oligodendrocyte precursor cells residing in the postnatal SVZ, as described above.
On the other hand, the Msi2 protein was also expressed in a population of large, round cells, distinct from the small, Msi1-positive astrocytes (Fig. 8A,B,arrows). These Msi2-positive cells were found predominantly within layers II–VI and overlapped with Hu immunoreactivity, indicating that they were neurons (Fig. 8C,D,arrows, G,H, asterisk). However, a comparison of the Hu and Msi2 expression clearly revealed that the vast majority of Hu-positive neurons were negative for Msi2 (Fig. 8C,D, and arrows inG,H). Consistent with the expression of Msi2 in a subset of neurons in the cerebrum, Msi2-positive neuron-like cells were seen in the embryonic spinal cord (bracketed area in Fig. 6G, H, arrows in Fig. 6K, L) and adult spinal cord (Fig. 6O,P) and striatum (Fig. 5O,P), as mentioned above. These data indicated that Msi2 expression was retained in a subset of cells of the neuronal lineage, accompanied by its expression in the astrocytes. Msi2 expression appeared to be absent or primarily downregulated by the very early stage of the oligodendrocyte lineage, as is the case for Msi1 (Sakakibara and Okano, 1997).
Msi2 expression in a subpopulation of GABA neurons
To determine the neuronal cell types that express Msi2 in the adult cerebral cortex, we performed double immunofluorescence staining with several markers for cholinergic (ChAT), dopaminergic (TH), and GABAergic neurons (parvalbumin, calbindin-D28K, GABA). The cerebral cortex of the adult mouse is composed of many types of neurons, including large pyramidal neurons within layers II, III, IV, and V and medium-sized nonpyramidal neurons distributed throughout the cortex. Two calcium-binding proteins, PV and CB, are known to be expressed in a small number of nonpyramidal neurons that contain GABA as a neurotransmitter (Hendry et al., 1989; Celio 1990). Previous studies have demonstrated that PV and CB localize to two almost entirely distinct populations among these GABAergic interneurons (Hendry et al., 1989; DeFelipe, 1997): PV is expressed in chandelier and basket cells, whereas CB is found in double-bouquet cells (DeFelipe, 1997). As shown in Figure9A–D, expression of Msi2 and PV coincided in a large number of cells, regardless of the cortical area (i.e., parietal, frontal, or cingulate cortex) examined. All of the PV-positive cells intensely expressed Msi2 and were concentrated in layers II–V. We found very few or no cells that stained positively for both Msi2 and CB (Fig.9E–H). Consistent with these observations, double immunofluorescence labeling with antibodies to Msi2 and GABA indicated the expression of Msi2 in the subpopulations of GABAergic neurons. Although many Msi2-positive cells were overlapped with GABA-containing medium-sized neurons in layers II–III (Fig.9I,J), there was also a population of cells that were GABA positive but Msi2 negative (Fig.9K,L, arrow). In addition, Msi2 expression was absent from all the cortical pyramidal neurons that could be easily distinguished from other cells by their large cell body and typical morphology. In layer V, there were many somata and proximal dendrites of large pyramidal neurons (presumptive Betz cells) that were surrounded by PV-positive multi-terminal endings typical of basket cell axon terminals (Fig. 9D,asterisks), as observed previously (Hendry et al., 1989). Msi2 expression was absent from these PV-negative somata (Fig.9C, asterisks). As shown in Figure 9,M and N, ChAT-positive cholinergic cells with small oval cell bodies and fine ascending processes were found sparsely throughout the cortex, and they showed no Msi2 expression (Fig.9M,N, arrows). These observations indicated that during the development of the cerebral cortex, Msi2 expression is maintained in the lineage of PV-containing GABAergic neurons.
Cerebellum
A dynamic pattern of Msi2 expression was also observed in the developing cerebellum. In the cerebellar primordium at E14.5, Msi2 was expressed in the small tightly packed cells of the external granule cell layer (EGL) migrating out of the rhombic lip of the lateral recess and also in the VZ of the fourth ventricle (data not shown). During early postnatal development, the EGL covers the surface of the developing cerebellum and is composed exclusively of proliferating neuronal precursors. By P3, the expression of Msi2 could be detected in the EGL, although to a lesser degree than that of Msi1 (Fig.10A,B). As neurogenesis progresses, two populations of cells appear in the EGL: cells proliferating in the upper portion of the EGL (EGLa) and cells undergoing the initial step of neuronal differentiation deeper in the EGL (EGLb) (Altman, 1972; Kuhar et al., 1993). At P7, Msi1 expression was predominantly observed within EGLa cells, as reported previously (Sakakibara and Okano, 1997), whereas Hu was expressed in the differentiating granule neurons within EGLb (Fig.10E,G). At this time, only faint Msi2 expression was detected in EGLa (Fig.10F,H).
Fig. 10.
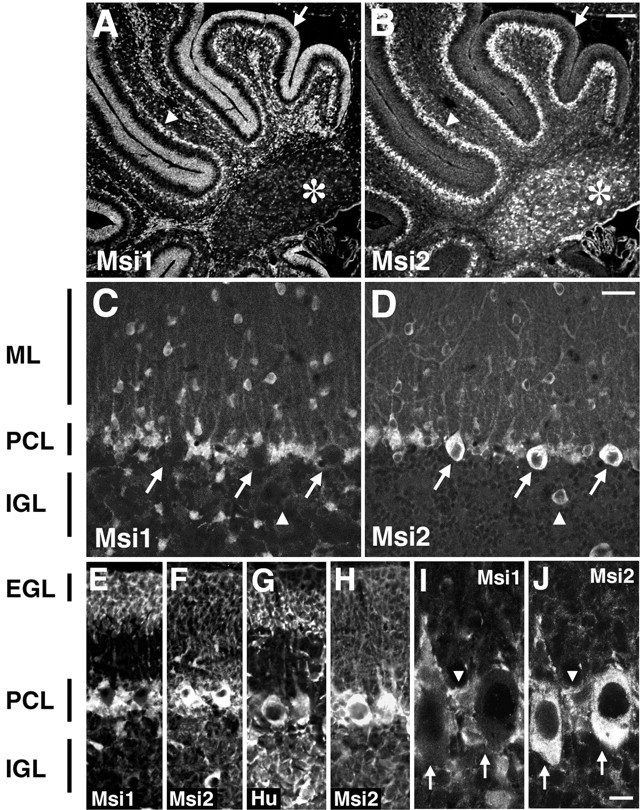
Msi2 expression in the cerebellum. Double-immunofluorescence labeling of sagittal sections through the early postnatal and adult cerebellum, with antibodies against Msi2 and Msi1 or Hu. A/B,C/D, E/F,G/H, andI/J represent each pair of double-stained photomicrographs. Primary antibodies used are indicated in each panel.A/B, P3 cerebellum. Intense Msi1 staining is observed in neuronal precursor cells in the EGL (indicated byarrow), in addition to the cells in the PCL (arrowhead) and numerous glial precursor cells in the folia white matter. Msi2 expression is prominent in the PCL and is faint in EGL cells and glial precursor cells in the white matter. Deep cerebellar nuclei (asterisk) are labeled only with Msi2.E–H, P7 cerebellum. The EGL is toward the top part of the panels. At P7, a low level of Msi2 expression is seen in the EGL (E/F). In the PCL, Purkinje cells forming a single row show immunoreactivity for both Msi2 and Hu but not for Msi1, whereas many small cells residing in the vicinity of the Purkinje cells express both Msi2 and Msi1 (compareE/F with G/H). These cells are likely to be the developing Bergmann glia. Msi2 expression is absent from Hu-positive granule neurons in the IGL. C/D, Adult cerebellum. In adults, Msi2 is predominantly expressed in the Purkinje cells (arrows in C/D) and Bergmann glial cells, which have cell bodies located adjacent to the large cell bodies of Purkinje cells and extend their tangential processes into the ML.Arrowhead in C/D represents the Golgi neuron in the IGL. I/J, Higher magnifications of the adult PCL. Arrows in I/J indicate the Purkinje cells, and arrowhead inI/J points to the soma of a Bergmann glial cell. ML, Molecular layer; PCL, Purkinje cell layer; IGL, internal granule layer;EGL, external granule cell layer. Scale bars:A, B, 100 μm; C–H, 30 μm; I, J, 8 μm.
The most striking difference between the Msi2 and Msi1 expression was seen in the neuronal population within the Purkinje cell layer (PCL) and deep cerebellar nuclei (Fig.10A,B). In the developing and adult PCL, Msi2 expression was conspicuous in the large Purkinje neurons, which absolutely lacked Msi1 immunoreactivity (Fig.10C,D,I,J,arrows). Msi2 expression in differentiated neurons was also observed within the deep cerebellar nuclei (Fig. 10B,asterisk) and in interneurons such as the stellate and basket cells residing in the molecular layer (Fig.10D). We noticed, however, that Msi2 was not expressed in numerous granule neurons in the internal granule cell layer (IGL), which were positive for Hu (Fig. 10G). In the IGL, Msi2 was detectable only in Golgi interneurons (Fig.10D, arrowhead). The sustained expression of Msi2 in the GABA neurons was also observed in the adult cerebellar cortex. Msi2 expression was confined to almost all the inhibitory GABA neurons, such as the stellate and basket cells in the molecular layer, the Purkinje cells, and the Golgi cells in the IGL (Fig. 10). Correspondingly, PV is known to be expressed in these cells (Celio, 1990).
In glial cells, Msi2 expression was detected in astrocytes within the central white matter tracts and the IGL, albeit at a greatly reduced level (Fig. 10B,D), compared with Msi1, which is highly expressed in astrocytes in these regions (Fig.10C) (Sakakibara and Okano, 1997). In the PCL, Msi2 and Msi1 expression were concomitantly observed in the radially aligned Bergmann glial cells and their fibers coursing from the PCL through the molecular layer to the pial surface (Fig.10C–J). Indirect double labeling on tissue sections of adult cerebellum with antibodies to GFAP and CNPase indicated the colocalization of Msi2 and GFAP in various regions, including the PCL (Bergmann glia) and foliar white matter tracts (data not shown). CNPase was never coexpressed with Msi2 in these regions (data not shown). These results indicated the expression of Msi2 in certain subpopulations of neurons and astrocytes in the cerebellum.
Other CNS regions
The persistent expression of Msi2 in certain subpopulations of neurons was also clearly observed in other brain regions. In the adult hippocampal formation, small, scattered Msi2-positive neurons were detected in the stratum pyramidale, stratum oriens, radiatum, and lacunosum-moleculare. A high-power view of the CA1 region showed intense expression of Msi2 in a small number of neurons along with diffuse expression in the astrocytes, which are uniformly distributed within this region (Fig. 9O,P). In the CA1 and CA3 subfields, ∼20% of all the GABAergic neurons contain PV (Kosaka et al., 1987). This PV-positive subpopulation of GABA neurons is thought to represent fast-spiking cells, which are electrically and metabolically more active than other local circuit neurons (Kawaguchi et al., 1987). Msi2 was mainly localized to these PV-containing GABAergic neurons in the CA1 region, although a few Msi2-positive neurons were negative for PV (Fig. 9O,P). In contrast, Msi2 immunoreactivity was absent from the CA1 pyramidal neurons (Fig. 9O), aligned in the stratum pyramidale, where CB is known to be expressed (Celio, 1990). Many granule neurons of the dentate gyrus did not express Msi2 either, whereas the anti-Msi2 antibody stained occasional cells in the subgranular region, the innermost region of the granular layer of the dentate gyrus (data not shown). Our previous study reported that Msi1 shows the same expression profile in this subgranular region (Kaneko et al., 2000). Neuron production in most hippocampal regions is completed before birth in the mouse; however, in the dentate gyrus, granule neurons continue to be generated into adulthood (Eriksson et al., 1998). Given that the subgranular region contains neural progenitor cells that are capable of postnatal neurogenesis (Palmer et al., 1997), the Msi2-positive cells in this region may represent a functionally distinct subset, i.e., a neural progenitor population in the postnatal hippocampus.
On the other hand, in other restricted brain regions including the basal ganglia and brainstem, Msi2 immunoreactivity was found in some non-GABA neurons, which contained neither PV nor CB. Within the basal ganglia, Msi2 was detected in subpopulations of neurons sparsely distributed in the ventral pallidum, globus pallidus, and caudate putamen (data not shown). Msi2 expression was seen in several ChAT-positive cholinergic projection neurons in the striatum, nuclei of diagonal bands (Ch2 and Ch3) (Fig. 9Q,R), medial septal nucleus (Ch1), and medial habenular nucleus (Ch7) (data not shown), in addition to the PV-positive neurons. Some dopaminergic neurons, identified by the presence of TH, also expressed Msi2. Figure9, S and T, shows the colocalization of Msi2 and TH in zona incerta dopaminergic neurons (A13) in the thalamic area surrounding the third ventricle. In the thalamus, epithalamus, and hypothalamic regions, the neuronal expression of Msi2 was identified in the medial preoptic nucleus, supraoptic nucleus, periventricular nucleus, and medial and lateral habenular nuclei, whereas Msi1 was expressed exclusively in astrocytes in these regions (data not shown).
Taken together, these observations substantially support the notion that during the development of several different regions of the brain (the cerebral cortex, hippocampus, and cerebellar cortex), Msi2 expression is maintained in the lineage of PV-containing GABAergic neurons ranging from the precursor stage to the postmitotic and differentiated stage, as well as in the astrocyte lineage. However, the Msi2 expression observed in cholinergic and dopaminergic cells in other brain regions (basal ganglia, brainstem) cannot be explained in this context.
Changes in Msi2 expression after injury
A previous report (Sakakibara and Okano, 1997) showed that Msi1 expression is drastically upregulated in reactive astrocytes after brain injury. The fact that Msi2 was also expressed in cells of the astrocyte lineage allowed us to examine the changes in Msi2 expression during the formation of glial scars after CNS injury. Experimentally induced damage to the CNS leads to the formation of a glial scar at the site of injury and to multiple changes in the astrocytes surrounding the damaged tissue. Numerous hypertrophic and hyperplastic reactive astrocytes display increased levels of GFAP and become mitotically active after the injury (Miyake et al., 1992). Immediately after the injury (day 0), the expression levels of Msi2 and GFAP were unaffected in the neighborhood of the injury site (cerebral cortex layers II–III) (Fig.11A,B). By day 2 after lesion, the number of Msi2-positive cells around the lesion site was not significantly different from the number in undamaged areas of the ipsilateral and contralateral hemispheres (data not shown). At 4 d after lesion, there was a dramatic increase in the number of Msi2-positive cells in the vicinity of the injury site (the region within 100–200 μm from the injury site) (Fig. 11). Immunohistochemical analysis of the area during glial scar formation showed dramatic and coincident induction of the Msi2 and Msi1 proteins at 3–4 d after lesion (Fig.11E,F). In this region, numerous GFAP-positive astrocytes were present. These cells had long and wavy processes extending in all directions and appeared to form a thick bundle of glia; these are typical characteristics of reactive astrocytes. These GFAP-positive cells were entirely immunopositive for Msi2 (Fig. 11G,H, arrows). BrdU administration experiments showed that these Msi2-positive reactive astrocytes continued to proliferate even 4 d after lesion (∼35% of the BrdU-incorporated cells expressed Msi2) (Fig.11C,D, arrows). This was consistent with previous studies showing the occurrence of cell division among GFAP-expressing astrocytes induced by brain injury (Miyake et al., 1992). On the other hand, there were many BrdU-positive but Msi2-negative proliferating cells at the site of injury. Most of these cells seemed to be activated ameboid microglia or infiltrated macrophages, which proliferated in response to the injury. The lack of Msi2 induction in the microglia and macrophages was verified by the double-staining with anti-Msi2 and anti-MacI antibodies (Thomas, 1992) (data not shown).
Fig. 11.
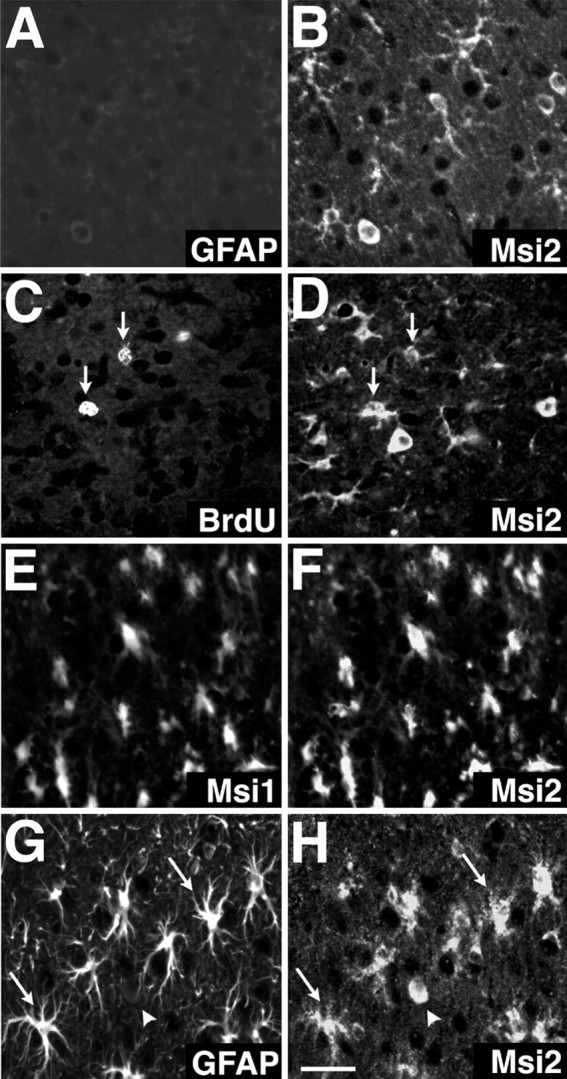
Induced expression of Msi2 and Msi1 in reactive astrocytes. Double-label fluorescence localization of Msi2 and Msi1, BrdU, or GFAP in the injured region of adult cerebral cortex immediately (day 0) (A, B) or 4 d (C–H) after lesioning. All photomicrographs correspond to cortex layers II–III, and the lesioned sites are toward the top in all panels. A,B, GFAP and Msi2 at day 0. The vicinity of the site of injury shows no immunoreactivity for GFAP. C,D, BrdU and Msi2 at 4 d after lesion. Msi2 is expressed in a population of BrdU-positive proliferating cells (arrows) that lie close to the injury site.E, F, Msi1 and Msi2 at 4 d after lesion. Simultaneous expression of Msi1 and Msi2 is evident in many enlarged reactive cells. G, H, GFAP and Msi2 at 4 d after lesion. Intense immunoreactivity of Msi2 is observed in an increased number of GFAP-positive reactive astrocytes; these cells exhibit enlarged, elongated cell bodies with multiple processes (arrows). Arrowhead indicates a rare Msi2-positive neuron-like cell that has an oval cell body and is immunonegative for GFAP. Scale bar (shown in H):A–H, 18 μm.
These observations demonstrated that Msi2 expression is induced at high levels in reactive astrocytes in the course of scar formation, with a profile similar to the expression of Msi1 (Sakakibara and Okano, 1997). Expression of the msi2 and msi1 genes therefore might be controlled by a common regulatory mechanism in reactive astrocytes.
DISCUSSION
Msi2 is an evolutionarily conserved RNA-binding protein found in different species of vertebrates (Xenopus, mouse, and human). On the basis of their striking sequence homology and similar RNA-binding preferences in vitro, Msi2 and Msi1 are likely to constitute a unique RNA-binding protein family (the Msi family). These two proteins are likely to play similar roles in the RNA metabolism that occurs in the cytoplasm, such as in the localization or stabilization of specific mRNAs, polyadenylation, or translational regulation (for review, see MacCarthy and Kollmus, 1995).
Role of the Msi family in neural precursor cells/CNS stem cells
In the present study we demonstrated the cell type-specific and developmentally regulated expression of Msi2 in the mammalian CNS. In particular, proliferative cells in the embryonic VZ and SVZ showed a high level of Msi2 expression. As neurogenesis proceeded, Msi2 expression disappeared in most of the postmitotic or migrating neurons within the intermediate zone or cortical plate. Interestingly, our double-labeling results indicated that Msi2 expression was coincident with the expression of Msi1 in proliferative cells. Previous in vivo and in vitro analyses have shown that Msi1 is predominantly expressed in neural precursor cells/multipotent CNS stem cells (Sakakibara et al., 1996; Kaneko et al., 2000); therefore, Msi2 and Msi1 are likely to be coexpressed in CNS stem cells during embryonic CNS development. Marked Msi2 expression was seen in growing neurospheres, further supporting its expression in CNS stem cells. Postnatally, the concurrent expression of Msi2 and Msi1 was maintained in proliferating neuronal and/or glial precursors in the SVZ (Sakakibara and Okano, 1997; present study). The postnatal SVZ mainly generates two glial lineages of cells: astrocytes and oligodendrocytes (Levison and Goldman, 1993). Although the Msi2 and Msi1 proteins disappeared from oligodendrocytes early in their differentiation, the proteins continued to be expressed in cells of the astrocyte lineage, including ependymal cells. The adult SVZ regions contain a population of CNS stem cells (Morshead et al., 1994; Johe et al., 1996). However, the SVZ might not be the only source of adult brain neurogenesis.Johansson et al. (1999) presented evidence that ependymal cells might be stem cells of the adult brain and spinal cord, whereas Doetsch et al. (1999) provided compelling evidence that SVZ astrocytes, referred to as “Type-B” cells, are precursors for neurogenesis and possibly stem cells. Type-B cells exhibit all of the characteristics of astrocytes, including GFAP expression. Consistent with this, recent heterotopic transplantation experiments (Leavitt et al., 1999) showed that mature neocortical astrocytes retain the capacity to dedifferentiate into radial glial-like cells, which are considered to be the neuronal and glial precursor cells in embryonic corticogenesis (Malatesta et al., 2000; Noctor et al., 2001). So far, the cell type that serves as the source of postnatal CNS stem cells in vivo is unresolved (Scheffler et al., 1999). In any case, the occurrence of Msi1 and Msi2 expression in the SVZ, ependymal cells, and astrocytes during postnatal life as well as in embryonic neuroepithelial cells of the VZ implies that Msi family proteins may function in cells harboring stem cell characteristics. The upregulation of Msi2 and Msi1 in proliferating reactive astrocytes may also reflect that these cells are endowed with the characteristics of glial precursors or of neural stem cells capable of generating astrocytes.
In accordance with this idea, we have obtained results from preliminaryin vitro experiments using neurospheres lacking expression of both the msi1 and msi2 genes that indicate that Msi1 and Msi2 are essential for the maintenance and/or proliferation of CNS stem cells (S. Sakakibara, unpublished results). The molecular basis for the function of the Msi family in CNS stem cells might be elucidated by identifying target mRNAs of the Msi1 and Msi2 proteins. We recently reported that Msi1 protein binds to the 3′-UTR of mouse numb mRNA, leading to repression of Numb expression (Imai et al., 2001). The mouse Numb protein is known to inhibit the Notch-signaling cascade (Wakamatsu et al., 1999), which is required for the self-renewing activity of CNS stem cells (Ohtsuka et al., 1999; Gaiano et al., 2000; Nakamura et al., 2000). The present sequence analysis and in vitro RNA-binding assay revealed a significant conservation of the RRM sequences and the RNA-binding specificity between Msi1 and Msi2, supporting the idea that Msi family proteins may share the same or similar target RNAs, which are characterized by uridine-rich sequences, as their downstream target molecules in vivo. Thus, Msi2 and Msi1 may exert common important functions in CNS stem cells by regulating the processing of the same specialized mRNAs. Loss-of-function experiments may be able to unequivocally reveal the function of the Msi proteins in embryonic and postnatal mammalian CNS development. Phenotypic analyses ofmsi2 and msi1 knock-out mice are currently in progress in our laboratory.
Msi2 in a subset of GABA neurons
During the development of the cerebral cortex, Msi2 expression is primarily downregulated in most of the cortical neurons except the GABA interneurons. Interneurons containing the GABA neurotransmitter are the major type of inhibitory neurons in the mammalian neocortex. These cells comprise 25% of all the cortical neurons, and exert powerful control over the local neuronal network by mediating both feedback and feedforward inhibition. Are there any significant differences between the developmental process of GABA interneurons and those of other types of neurons? It is well known that most differentiating cortical projection neurons migrate radially from the cortical VZ toward the pial surface and form the cortical plate, where they temporarily settle according to an inside-out gradient of positioning (Bayer and Altman, 1991). However, recent experiments using retroviral labeling or chimeric embryos suggest that cortical projection neurons and GABA interneurons are derived from distinct proliferative zones. Most, if not all, cortical interneurons (GABAergic neurons) are born in the VZ/SVZ of the lateral ganglionic eminence (LGE), the primordium of the striatum, and migrate tangentially from the LGE to the neocortex through the striatal–neocortical pathway (Tamamaki et al., 1997; Tan et al., 1998). Expression of Msi2 in the embryonic LGE is compatible with the idea that Msi2 is maintained by the GABAergic lineage undergoing a tangential mode of cell movement. Furthermore, it is possible that Msi2 expression is rapidly decreased in neurons that are fated to migrate radially into the neocortex.
In adult neocortex, GABA interneurons are classified into two separate subpopulations: the CB-positive double-bouquet cells and the PV-positive chandelier and basket cells (Hendry et al., 1989; DeFelipe, 1997). Msi2 expression is retained in a subset of PV-containing GABAergic neurons in the neocortex, hippocampus, and cerebellar cortex. In these regions, other CB-positive GABAergic neurons or pyramidal projection neurons showed little or no expression of Msi2. PV-containing GABA interneurons show electrophysiological fast-firing behavior and form a unique dense network connected by dendrodendritic gap junctions (electrical synapses), presumably enabling the generation of synchronous oscillations and the processing of spatiotemporal information within brain regions (Fukuda and Kosaka, 2000). Proper functioning of PV-positive GABA interneurons has been shown to require selective expression or rapid turnover of metabotropic glutamate receptor protein (mGluR5) (Whittington et al., 1995; Stinehelfer et al., 2000; van Hooft et al., 2000) or the highly localized dendritic expression of voltage-gated sodium and potassium ion channels (Martina et al., 2000). Therefore, Msi2 may participate in the generation and/or maintenance of these specific neuronal lineages by controlling the local translation or stability of the mRNAs for these receptors and channels, in addition to its role in neural precursor cells.
Intriguingly, the neuronal expression of Msi is not only a characteristic of vertebrates but occurs in invertebrates as well. InDrosophila, d-Msi is expressed in a small population of differentiated neurons, including the photoreceptor cells (Hirota et al., 1999), and in the precursor cells for the sensory organs and embryonic CNS (Nakamura et al., 1994). In Caenorhabditis elegans, Msi is expressed in certain populations of neurons, including the GABAergic neurons in the ventral cord, although its molecular significance in this organism remains unclear (Yoda et al., 2000). Because we have failed to find any other Msi-related genes in invertebrates, it is possible that mouse Msi2 has succeeded to the more restricted neuronal expression of invertebrate Msi protein during evolution.
Msi2 and neurological diseases
Previous immunohistochemical studies have indicated a selective reduction in or functional impairment of PV-containing GABA neurons in the neocortex under certain pathological conditions, which include schizophrenia (Beasley and Reynolds, 1997), hypothyroidism in rats (Berbel et al., 1996), and an animal model of cortical dysplasia (Roper et al., 1999). Msi2 may affect the pathogenesis of such diseases through its function in PV-containing neurons. On the other hand, themsi2 syntenic region contains several loci of hereditary human neurological disorders: FTDP17, Pick disease, GPSC, and NF1. FTDP17, GPSC, and Pick disease may be allelic and are accompanied by varying degrees of dementia and atrophic degeneration of frontal lobe (Lynch et al., 1994). NF1 is characterized by cafe-au-lait spots and fibromatous tumors of the skin, but it is also frequently associated with CNS tumors, including astrocytomas and ependymomas (Mukonoweshuro et al., 1999). Although inherited FTDP17 and NF1 are caused by mutations in the tau (Hutton et al., 1998) andneurofibromin (Streubel et al., 1999) genes, respectively, the complexities of their clinical and pathologic symptoms raise the possibility that additional loci may be involved in potentiating or modifying each syndrome (Lynch et al., 1994; Streubel et al., 1999). Indeed, a recent somatic mosaicism analysis of a patient with a complex clinical phenotype of NF1 and a large deletion of the contiguous genomic regions flanking the neurofibromin gene provides evidence for the involvement of adjacent loci in the clinical manifestations of this syndrome (Streubel et al., 1999). It will be interesting to determine whether the msi2 gene is implicated in these neurological disorders as a modifier.
Footnotes
This work was supported by grants from the Japanese Ministry of Education, Japan Science and Technology Corporation (Core Research for Evolutional Science and Technology).
Correspondence should be addressed to Dr. Hideyuki Okano, Department of Physiology, Keio University School of Medicine, 35 Shinanomachi, Shinjuku-ku, Tokyo 160-8582, Japan. E-mail:hidokano@sc.itc.keio.ac.jp.
S. Sakakibara's present address: Department of Histology and Neurobiology, Dokkyo University School of Medicine, Mibu, Tochigi 321-0293, Japan.
REFERENCES
- 1.Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and peripheral nervous systems. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altman J. Postnatal development of the cerebellar cortex in the rat. J Comp Neurol. 1972;145:353–398. doi: 10.1002/cne.901450305. [DOI] [PubMed] [Google Scholar]
- 3.Amano T, Hamprecht B, Kemper W. High activity of choline acetyltransferase induced in neuroblastoma × glioma hybrid cells. Exp Cell Res. 1974;85:399–408. doi: 10.1016/0014-4827(74)90142-6. [DOI] [PubMed] [Google Scholar]
- 4.Barami K, Iversen K, Furneaux H, Goldman SA. Early expression of Hu proteins by newly generated neurons of the adult avian forebrain. J Neurobiol. 1995;28:82–101. doi: 10.1002/neu.480280108. [DOI] [PubMed] [Google Scholar]
- 5.Bayer SA, Altman J. Neocortical development. Raven; New York: 1991. [Google Scholar]
- 6.Beasley CL, Reynolds GP. Parvalbumin-immunoreactive neurons are reduced in the prefrontal cortex of schizophrenics. Schizophrenia Res. 1997;24:349–355. doi: 10.1016/s0920-9964(96)00122-3. [DOI] [PubMed] [Google Scholar]
- 7.Berbel P, Marco P, Cerezo JR, DeFelipe J. Distribution of parvalbumin immunoreactivity in the neocortex of hypothyroid adult rats. Neurosci Lett. 1996;204:65–68. doi: 10.1016/0304-3940(96)12318-1. [DOI] [PubMed] [Google Scholar]
- 8.Blaxall BC, Pellett AC, Wu SC, Pende A, Port JD. Purification and characterization of β-adrenergic receptor mRNA-binding proteins. J Biol Chem. 2000;274:4290–4297. doi: 10.1074/jbc.275.6.4290. [DOI] [PubMed] [Google Scholar]
- 9.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 10.Celio MR. Calbindin D-28k and parvalbumin in the rat nervous system. Neuroscience. 1990;35:375–475. doi: 10.1016/0306-4522(90)90091-h. [DOI] [PubMed] [Google Scholar]
- 11.Davis AA, Temple S. A self-renewing multipotential stem cell in embryonic rat cerebral cortex. Nature. 1994;372:263–266. doi: 10.1038/372263a0. [DOI] [PubMed] [Google Scholar]
- 12.DeFelipe J. Types of neurons, synaptic connections and chemical characteristics of cells immunoreactive for calbindin-D28K, parvalbumin and calretinin in the neocortex. J Chem Neuroanat. 1997;14:1–19. doi: 10.1016/s0891-0618(97)10013-8. [DOI] [PubMed] [Google Scholar]
- 13.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 14.Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda T, Kosaka T. The dual network of GABAergic interneurons linked by both chemical and electrical synapses: a possible infrastructure of the cerebral cortex. Neurosci Res. 2000;38:123–130. doi: 10.1016/s0168-0102(00)00163-2. [DOI] [PubMed] [Google Scholar]
- 17.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 18.Good PJ, Rebbert ML, Dawid IB. Three new members of the RNP protein family in Xenopus. Nucleic Acids Res. 1993;21:999–1006. doi: 10.1093/nar/21.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Good PJ, Yoda A, Sakakibara S, Yamamoto A, Imai T, Sawa H, Ikeuchi T, Tsuji S, Satoh H, Okano H. The human Musashi homolog 1 (MSI1) gene encoding the homologue of Musashi/Nrp-1, a neural RNA-binding protein putatively expressed in CNS stem cells and neural progenitor cells. Genomics. 1998;52:382–384. doi: 10.1006/geno.1998.5456. [DOI] [PubMed] [Google Scholar]
- 20.Hendry SHC, Jones EG, Emson PC, Lawson DEM, Heizmann CW, Streit P. Two classes of cortical GABA neurons defined by differential calcium protein immunoreactivities. Exp Brain Res. 1989;76:467–472. doi: 10.1007/BF00247904. [DOI] [PubMed] [Google Scholar]
- 21.Hirota Y, Okabe M, Imai T, Kurusu M, Yamamoto A, Miyao S, Nakamura M, Sawamoto K, Okano H. Musashi and Seven in absentia downregulate tramtrack through distinct mechanisms in Drosophila eye development. Mech Dev. 1999;87:93–101. doi: 10.1016/s0925-4773(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 22.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J. Association of misssense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 23.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates the m-numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- 25.Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10:3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 27.Kawaguchi Y, Katsumaru H, Kosaka T, Heizmann CW, Hama K. Fast spiking cells in rat hippocampus (CA1 region) contain the calcium-binding protein parvalbumin. Brain Res. 1987;416:369–374. doi: 10.1016/0006-8993(87)90921-8. [DOI] [PubMed] [Google Scholar]
- 28.Kawashima T, Murakami AR, Ogasawara M, Tanaka KJ, Isoda R, Sasakura Y, Nishikata T, Okano H, Makabe KW. Expression patterns of musashi homologues of the ascidians, Halocynthia roretzi and Ciona intestinalis. Dev Gene Evol. 2000;210:162–165. doi: 10.1007/s004270050024. [DOI] [PubMed] [Google Scholar]
- 29.Khan F, Jaiswal AK, Szer W. Cloning and sequence analysis of a human type A/B hnRNP protein. FEBS Lett. 1991;290:159–161. doi: 10.1016/0014-5793(91)81249-8. [DOI] [PubMed] [Google Scholar]
- 30.Kosaka T, Katsumaru H, Hama K, Wu J-Y, Heizmann CW. GABAergic neurons containing the Ca2+-binding protein parvalbumin in the rat hippocampus and dentate gyrus. Brain Res. 1987;419:119–130. doi: 10.1016/0006-8993(87)90575-0. [DOI] [PubMed] [Google Scholar]
- 31.Kuhar SG, Feng L, Vidan S, Ross ME, Hatten ME, Heinz N. Changing patterns of gene expression define four stages of cerebellar granule neuron differentiation. Development. 1993;117:97–104. doi: 10.1242/dev.117.1.97. [DOI] [PubMed] [Google Scholar]
- 32.Leavitt BR, Hernit-Grant CS, Macklis JD. Mature astrocytes transform into transitional radial glia within adult mouse neocortex that supports directed migration of transplanted immature neurons. Exp Neurol. 1999;157:43–57. doi: 10.1006/exnr.1999.6982. [DOI] [PubMed] [Google Scholar]
- 33.Lendahl U, Zimmerman LB, McKay R. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 34.Levison SW, Goldman JE. Both oligodendrocytes and astrocytes develop from progenitors in the subventricular zone of postnatal rat forebrain. Neuron. 1993;10:201–212. doi: 10.1016/0896-6273(93)90311-e. [DOI] [PubMed] [Google Scholar]
- 35.Lewis PD. A quantitative study of cell proliferation in the subependymal layer of the adult rat brain. Exp Neurol. 1968;20:203–207. doi: 10.1016/0014-4886(68)90093-9. [DOI] [PubMed] [Google Scholar]
- 36.Lois C, Alvarez-Buylla A. Proliferating subventricular zone cells in the adult mammalian forebrain can differentiate into neurons and glia. Proc Natl Acad Sci USA. 1993;9:2073–2077. doi: 10.1073/pnas.90.5.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 38.Lynch T, Sano M, Marder KS, Bell KL, Foster NL, Defendini RF, Sima AAF, Keohane C, Nygaard TG, Fahn S, Mayeux R, Rowland LP, Wilhelmsen KC. Clinical characteristics of a family with chromosome 17-linked disinhibition-dementia-parkinsonism-amyotrophy complex. Neurology. 1994;44:1878–1884. doi: 10.1212/wnl.44.10.1878. [DOI] [PubMed] [Google Scholar]
- 39.Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 40.Martina M, Vida I, Jonas P. Distal initiation and active propagation of action potentials in interneuron dendrites. Science. 2000;287:295–300. doi: 10.1126/science.287.5451.295. [DOI] [PubMed] [Google Scholar]
- 41.Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy JE, Kollmus H. Cytoplasmic mRNA-protein interactions in eukaryotic gene expression. Trends Biochem Sci. 1995;20:191–197. doi: 10.1016/s0968-0004(00)89006-4. [DOI] [PubMed] [Google Scholar]
- 43.Miller RH, Raff MC. Fibrous and protoplasmic astrocytes are biochemically and developmentally distinct. J Neurosci. 1984;4:585–592. doi: 10.1523/JNEUROSCI.04-02-00585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyake T, Okada M, Kitamura T. Reactive proliferation of astrocytes studied by immunohistochemistry for proliferating cell nuclear antigen. Brain Res. 1992;590:300–302. doi: 10.1016/0006-8993(92)91109-r. [DOI] [PubMed] [Google Scholar]
- 45.Morshead CM, Reynolds BA, Craig CG, McBurney MW, Staines WA, Morassutti D, Weiss S, van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 46.Morshead CM, Craig CG, van der Kooy D. In vivo clonal analyses reveal the properties of endogenous neural stem cell proliferation in the adult mammalian forebrain. Development. 1998;125:2251–2261. doi: 10.1242/dev.125.12.2251. [DOI] [PubMed] [Google Scholar]
- 47.Mukonoweshuro W, Griffiths PD, Blaser S. Neurofibromatosis type 1: the role of neurology. Neuropediatrics. 1999;30:111–119. doi: 10.1055/s-2007-973474. [DOI] [PubMed] [Google Scholar]
- 48.Munoz-Garcia D, Ludwin SK. Classic and generalized variants of Pick's disease: a clinicopathological, ultrastructural, and immunocytochemical comparative study. Ann Neurol. 1984;16:467–480. doi: 10.1002/ana.410160408. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura M, Okano H, Blendy J, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura Y, Sakakibara S, Miyata T, Ogawa M, Shimazaki T, Weiss S, Kageyama R, Okano H. The bHLH gene Hes1 as a repressor of the neuronal commitment of CNS stem cells. J Neurosci. 2000;20:283–293. doi: 10.1523/JNEUROSCI.20-01-00283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishiyama A, Lin XH, Giese N, Helden CH, Stallcup WB. Co-localization of NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43:299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 52.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 53.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 55.Okano H. Two major mechanisms regulating cell-fate decisions in the developing nervous system. Dev Growth Differ. 1995;37:619–629. doi: 10.1046/j.1440-169X.1995.t01-5-00001.x. [DOI] [PubMed] [Google Scholar]
- 56.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol Cell Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- 58.Petersen RB, Tabaton M, Chen SG, Monari L, Richardson SL, Lynches T, Manetto V, Lanska DJ, Markesbery WR, Currier RD, Autilio-Gambetti L, Wilhelmsen KC, Bambetti P. Familial progressive subcortical gliosis: presence of prions and linkage to chromosome 17. Neurology. 1997;45:1062–1067. doi: 10.1212/wnl.45.6.1062. [DOI] [PubMed] [Google Scholar]
- 59.Pincus DW, Keyoung H, Restelli C, Goodman RR, Fraser RAR, Edgar M, Sakakibara S, Okano H, Nedergaad M, Goldman SA. FGF2/BDNF-responsive neuronal progenitor cells in the adult human subependyma. Ann Neurol. 1998;43:576–585. doi: 10.1002/ana.410430505. [DOI] [PubMed] [Google Scholar]
- 60.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 61.Richter K, Good PJ, Dawid IB. A developmentally regulated, nervous system-specific gene in Xenopus encodes a putative RNA-binding protein. New Biol. 1990;2:556–565. [PubMed] [Google Scholar]
- 62.Roper SN, Eisenschenk S, King MA. Reduced density of parvalbumin- and calbindin D28k-immunoreactive neurons in experimental cortical dysplasia. Epilepsy Res. 1999;37:63–71. doi: 10.1016/s0920-1211(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 63.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi Y, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 65.Satoh H, Nagai F, Homma H, Mori S, Matsui M. Regional assignment of rat androsterone UDP-glucuronosyltransferase gene (UGT2B2) to chromosome 14p21.2-p22. Cytogenet Cell Genet. 1993;62:49–51. doi: 10.1159/000133443. [DOI] [PubMed] [Google Scholar]
- 66.Scheffler B, Horn M, Blumcke I, Laywell ED, Coomes D, Kukekov VG, Steindler DA. Marrow-mindedness: a perspective on neuropoiesis. Trends Neurosci. 1999;22:348–357. doi: 10.1016/s0166-2236(99)01416-2. [DOI] [PubMed] [Google Scholar]
- 67.Spillantini MG, Geodert M. Tau protein pathology in neurodegenerative diseases. Trends Neurosci. 1998;21:428–433. doi: 10.1016/s0166-2236(98)01337-x. [DOI] [PubMed] [Google Scholar]
- 68.Sprinkle TJ. 2′ 3′-Cyclic nucleotide 3′-phosphodiesterase: an oligodendrocyte-Schwann cell and myelin-associated enzyme of the nervous system. Crit Rev Neurobiol. 1989;4:235–301. [PubMed] [Google Scholar]
- 69.Stinehelfer S, Vruwink M, Burette A. Immunolocalization of mGluR1α in specific populations of local circuit neurons in the cerebral cortex. Brain Res. 2000;861:37–44. doi: 10.1016/s0006-8993(00)01952-1. [DOI] [PubMed] [Google Scholar]
- 70.Streubel B, Latta E, Kehrer-Sawatzki H, Hoffmann GF, Fonatsch C, Rehder H. Somatic mosaicism of a greater than 1.7-Mb deletion of genomic DNA involving the entire NF1 gene as verified by FISH: further evidence for a contiguous gene syndrome in 17q11.2. Am J Med Genet. 1999;87:12–16. [PubMed] [Google Scholar]
- 71.Swanson MS, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA binding proteins. Mol Cell Biol. 1988;3:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamamaki N, Fujimori KE, Takauji R. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci. 1997;17:8313–8323. doi: 10.1523/JNEUROSCI.17-21-08313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan SS, Kalloniatis M, Sturm K, Tam PPL, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21:295–304. doi: 10.1016/s0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 74.Thomas WE. Brain macrophages: evaluation of microglia and their functions. Brain Res Rev. 1992;17:61–74. doi: 10.1016/0165-0173(92)90007-9. [DOI] [PubMed] [Google Scholar]
- 75.van Hooft JA, Giuffrida R, Blatow M, Monyer H (2000) Differential expression of group I metabotropic glutamate receptors in functionally distinct hippocampal interneurons. 20:3544–3551. [DOI] [PMC free article] [PubMed]
- 76.Wakamatsu Y, Maynard TM, Jones SU, Weston JA. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 77.Whittington MA, Traub RD, Jefferys JG. Synchronized oscillations in interneuron networks driven by metabotropic glutamate receptor activation. Nature. 1995;373:612–615. doi: 10.1038/373612a0. [DOI] [PubMed] [Google Scholar]
- 78.Yao K-M, Samson M-L, Reeves R, White K. Gene elav of Drosophila melanogaster: a prototype for neuron-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- 79.Yoda A, Sawa H, Okano H. MSI-1, a neural RNA-binding protein, is involved in male mating behavior in Caenorhabditis elegans. Genes Cells. 2000;5:1–12. doi: 10.1046/j.1365-2443.2000.00378.x. [DOI] [PubMed] [Google Scholar]
- 80.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]