Abstract
Phenylketonuria (PKU) is considered to be a paradigm for a monogenic metabolic disorder but was never thought to be a primary application for human gene therapy due to established alternative treatment. However, somewhat unanticipated improvement in neuropsychiatric outcome upon long-term treatment of adults with PKU with enzyme substitution therapy might slowly change this assumption. In parallel, PKU was for a long time considered to be an excellent test system for experimental gene therapy of a Mendelian autosomal recessive defect of the liver due to an outstanding mouse model and the easy to analyze and well-defined therapeutic end point, that is, blood l-phenylalanine concentration. Lifelong treatment by targeting the mouse liver (or skeletal muscle) was achieved using different approaches, including (1) recombinant adeno-associated viral (rAAV) or nonviral naked DNA vector-based gene addition, (2) genome editing using base editors delivered by rAAV vectors, and (3) by delivering rAAVs for promoter-less insertion of the PAH-cDNA into the Pah locus. In this article we summarize the gene therapeutic attempts of correcting a mouse model for PKU and discuss the future implications for human gene therapy.
Keywords: rAAV, nonviral minicircle vector, gene delivery, base editing, liver gene therapy
Introduction
Hyperphenylalaninemia (HPA) also termed phenylketonuria (PKU; OMIM 261600) is an inborn error of metabolism defined as blood l-phenylalanine (l-Phe) levels >120 μM (or 2 mg/dL). The primary cause is autosomal recessively inherited mutations in the phenylalanine hydroxylase gene (PAH).1,2 The hepatic enzyme PAH (EC 1.14.16.1) is responsible for converting l-Phe into tyrosine, using molecular oxygen and the cofactor tetrahydrobiopterin (BH4) for its catalytic activity.3 Besides “classical” PAH deficiency, rare variants of HPA can be caused by either BH4 cofactor deficiency due to defects in its biosynthesis or regeneration or autosomal recessive variants in DNAJC12, encoding a cochaperone and folding aid of the PAH enzyme.4 All these deficiencies lead to systemic accumulation of l-Phe within the whole body, including the brain. Although the liver itself remains unaffected, HPA is neurotoxic and impairs postnatal development, leading to intellectual disability, seizures, and irreversible neuropsychiatric and neurocognitive impairments.2 In many countries, newborns are detected for HPA by measuring blood l-Phe levels through newborn screening programs, which were initiated more than 5 decades ago.5 The incidence of PKU in Caucasian populations is around 1 in 10,000, and >1,000 individual variants are tabulated in the PAH locus-specific database.*
Current treatments for PKU
HPA caused by PAH deficiency can be treated by a low l-Phe diet or, more recently, by enzyme substitution therapy.6 A lifelong dietary treatment consisting of an l-Phe-restricted diet supplemented with medical food has been the mainstay of contemporary therapy for PKU with a goal of maintaining blood l-Phe levels in the range of 120–360 μM (2–6 mg/dL) in all ages recommended by American College of Medical Genetics and Genomics (ACMG) practice guidelines7 or 120–600 μM (2–10 mg/dL) for individuals older than 12 years according to European Society for Phenylketonuria and Allied Disorders treated as Phenylketonuria (ESPKU).8,9
Although lowering l-Phe levels is associated with improved neurological performance,10 problems in executive functioning, motor ability, social skill, and behavior can still occur despite early treatment intervention.2,11–13 Adults with PKU who discontinued the low-Phe diet during adolescence as the dietary treatment is burdensome have been reported to have increased incidence in mental disorders, headache, hyperactivity and hypoactivity, and poorer intellectual ability.9,14 Pregnancy presents another major problem for women with PKU, as high levels of l-Phe are toxic to the brain of the developing fetus, causing mental retardation. Subsequently, microcephaly, congenital heart disease, and intrauterine growth retardation can also be found in the non-phenylketonuric offspring, resulting in a defined maternal PKU syndrome.15 Therefore, blood l-Phe concentrations should ideally be kept between 120 and 360 μM (2–6 mg/dL) for women with PKU before conception and during pregnancy for normal fetal development.7,8
An enzyme substitution therapy, pegvaliase (PALYNZIQ™; BioMarin, San Rafael, CA), has been approved by U.S. Food and Drug Administration (FDA) in May, 2018, for use in adults with PKU whose blood l-Phe levels are ≥600 μM (10 mg/dL); more recently (April, 2019) this treatment approach was approved for adolescent adults >16 years age by the European Medicines Agency.
Pegvaliase is a PEGylated recombinant Anabaena variabilis phenylalanine ammonia lyase (PAL) administrated by continuous subcutaneous injections that lowers blood l-Phe independently of PAH and its BH4 cofactor.16 PAL catalyzes a reaction converting l-Phe to ammonia and trans-cinnamic acid, which are readily metabolized by the liver and excreted in the urine.17 Results from the phase 3 clinical trial program (PRISM) demonstrated that 60.7% of patients were able to achieve blood l-Phe concentrations <360 μM (6 mg/dL) at 24 months18 without restricting dietary protein intake. Reduction of l-Phe in blood was accompanied with improvements in neuropsychiatric outcomes that were sustained with long-term treatment. However, immune-mediated adverse reactions and anaphylaxis have been reported to be associated with this therapy.19
Several other groups have investigated in experimental setups alternatives for delivery of PAL. The group from Leuzzi and coworkers has shown to successfully normalize the blood l-Phe levels by infusing autologous erythrocytes loaded with PAL in early age PKU mice (15 postnatal days).20 Similar results were achieved by feeding mice and primates synthetic live bacteria expressing PAL (from Photorhabdus luminescens) and l-amino acid deaminase, a membrane-associated enzyme that converts l-Phe to phenylpyruvate.21
Pahenu2/2 mice: a model of human PKU
To explore the potential of novel therapies for PKU, an appropriate animal model that accurately recapitulates the human disease is necessary. The most commonly used mouse model for PKU is the Pahenu2/2 mouse that was first described by McDonald almost 30 years ago.22 It was generated by random chemical mutagenesis in a Black and Tan Brachyury-mouse strain background using the alkylating agent N-ethyl-N-nitrososurea, which resulted in a missense mutation in exon 7 (p.Phe263Ser), a region that encodes the active site of the PAH enzyme.23 Although protein levels of mutant PAH are reduced compared with wild type, it can be detected by Western blot in liver and kidney from Pahenu2/2 mice. PAH enzyme activity, nevertheless, is completely abolished.24 Homozygous Pahenu2/2 animals exhibit blood l-Phe levels >1,500 μM (25 mg/dL), are phenotypically hypopigmented, mildly growth retarded25 and cognitively impaired,26 have abnormal low serotonin and dopamine neurotransmitter metabolites in the brain,27 and exhibit osteopenia28 that is also observed in some of the PKU patients.29,30 Affected PKU female mice are fertile but with significantly increased spontaneous abortions.31 Furthermore, maternal PKU syndrome is also observed in this mouse model.31,32 The PKU mouse is not only a valuable model for human PKU, it is also an outstanding validated genetic model to study hepatic gene transfer approaches in vivo, as it offers a direct readout for therapeutic efficacy, that is, lowering of systemic high blood levels of l-Phe.
Experimental restoration of PAH enzyme activity by gene addition or correction in the homozygous Pahenu2/2 “PKU” mouse model has been the goal of many investigations. Several reviews have previously summarized different strategies for gene therapy in the PKU mouse.33–38 In this review, we compiled all the approaches with a focus on the more recent achievements using recombinant adeno-associated viral (rAAV) vectors as well as nonviral naked DNA vectors, genome editing by base editors, and integration of promoter-less PAH–mRNA into the Pah locus (for an overview of all successful approaches thus far see Table 1). Finally, the plans for clinical gene therapy trials in humans are discussed.
Table 1.
Overview of experimental gene therapy for phenylketonuria using the Pahenu2/2 mouse model
| Vector System | Transgene(s)a | Promoterb | Target Organ | Injection Methodc | Documented Duration of Treatment | References |
|---|---|---|---|---|---|---|
| Adenovirus | hPAH | RSV-LTR | Liver | i.p | 1 week | Fang et al.42 |
| hPAH | CAG | Liver | i.p or i.v. | 11 days | Nagasaki et al.43 | |
| rAAV2/2 | hPAH | EF | Liver | i.p | Up to 25 weeks | Oh et al.44 |
| rAAV2/5 | mPah | CBA | Liver | i.p | Up to 40 weeks | Mochizuki et al.45 |
| rAAV2/8 | mPah | CBA | Liver | i.v. or i.p. | Up to 42 weeks | Ding et al.49 |
| rAAV2/8 | mPah | LPS | Liver | i.p. | Up to 17 weeks | Harding et al.48 |
| rAAV2/1 | Pah, Gch1, Pts | CMV | Muscle | i.m | Up to 70 weeks | Ding et al.55 |
| rAAV2/1, rAAV2/2, rAAV2/8 | mPah | CBA | Liver | i.m. | Up to 53 weeks | Rebuffat et al.52 |
| scAAV2/8 | mPah | LP1 | Liver | i.p. | Up to 52 weeks | Yagi et al.50 |
| Naked DNA (minicircle) | mPah | P3 | Liver | HTV | Up to 52 weeks | Viecelli et al.57 |
| mcoPah | P3, endogenous | Liver | HTV | Up to 60 weeks | Grisch-Chan et al.62 | |
| rAAV2/8 intein | APOBEC-nSaCas9 sgRNA |
P3, U6 | Liver | i.v. | Up to 26 weeks | Villiger et al.80 |
| AAVHSC15 | hPAH | Endogenous | Liver | i.v. | Up to 8 weeks | Wright et al.54 |
Note that all Pahenu2/2 mice were treated as adults.
hPAH, human Pah gene; mPah, murine Pah gene; mcoPah, murine, codon-optimized Pah gene; Gch1, guanosine triphosphate cyclohydrolase I; Pts, 6-pyruvoyltetraphydrobiopterin synthase.
RSV-LTR, Rous sarcoma virus long-terminal repeat; CBA, CMV (cytomegalovirus) early enhancer/chicken β actin promoter; EF, human elongation factor 1-α promoter; CMV, cytomegalovirus promoter; LPS; liver-specific promoter is a combination of two copies of a human a1-microglobulin/bikunin enhancer and the promoter from the human thyroid hormone-binding globulin gene48; LP1 promoter consists of the human apolipoprotein E/C-I hepatic control region and the human α1-antitrypsin promoter50; P3, synthetic liver-specific promoter61; U6, RNA polymerase III promoter.
i,p., intraportal vein infusion; i.v., intravenous tail vein infusion; i.m., intramuscular (M. gastrognemius); i.p, intraperitoneal injection; HTV, hydrodynamic tail vein injection.
Physiologic Requirements of Successful Liver-Directed Gene Therapy in PKU
As already mentioned, the liver is not affected by high l-Phe levels and at the same time it is the exclusive site for PAH for regulated degradation of l-Phe to physiological levels. This makes the liver the ideal target organ for a gene therapy approach. What are the physiologic requirements for successful liver-directed gene therapy for PKU? The vector of course will need to express in the liver sufficient PAH protein to fully metabolize the daily load of l-Phe from dietary and endogenous sources. PAH requires BH4 cofactor for catalytic activity, but this pterin cofactor is abundantly synthesized and rapidly recycled in hepatocytes so its supply is not a limitation to the success of liver-directed gene therapy. Individuals who are heterozygous for pathogenic PAH mutations are not hyperphenylalaninemic despite expressing reduced liver PAH activity; this fact suggests that it will not be necessary to fully restore cellular PAH activity to 100% after liver-directed gene transfer to effect blood l-Phe concentrations. However, cell transplantation experiments demonstrate that PAH activity must be distributed across a minimum number of hepatocytes, otherwise l-Phe flux will be limited.39,40 Repopulation of PAH-deficient Pahenu2/2 mouse liver with >10% wild type hepatocytes expressing 100% PAH activity completely corrects blood l-Phe. In contrast, blood l-Phe remains mildly elevated (300–700 μM or 5–12 mg/dL) even when the animals had 5–10% repopulation.39 Furthermore, identical results are achieved after repopulation with hepatocytes from heterozygous Pahenu2/+ mice, which express <50% PAH activity per cell.40 This result suggests that at a low frequency of PAH expressing hepatocytes, the absolute number of cells is primarily limiting l-Phe clearance rather than total PAH enzyme activity.
Liver Gene Therapy with Viral Vectors
An early approach of using a recombinant retrovirus vector was able to induce PAH activity in PAH-deficient mouse hepatocytes in vitro, but no in vivo studies have been reported thereafter.41 Two independent studies showed that liver-directed gene therapy using an adenovirus vector corrected serum l-Phe transiently in vivo.42,43 However, this approach was discontinued as the vector provoked a profound host immune response against the recombinant virus.
Recombinant adeno-associated virus vector for PKU
rAAV is currently the favored vector system for safe and effective liver-directed gene addition, with active clinical trials ongoing for adults with Hemophilia A or B, ornithine transcarbamylase deficiency, glycogen storage disease type 1A, acute intermittent porphyria, Crigler–Najjar syndrome, and others.* rAAV-mediated liver-directed gene therapy for PKU has been explored preclinically by several investigators using the Pahenu2/2 mouse model for ∼20 years. A complete review of the history of rAAV liver gene therapy is beyond the scope of this review, so we will necessarily focus upon the development of this treatment approach specifically for PKU.
Attempts at liver-directed gene therapy using early rAAV vectors were likely impaired by the problem of poor transduction frequency and consequently incomplete correction of HPA. Administration of a PAH-expressing rAAV2 serotype 244 or serotype 545 yielded correction of blood l-Phe in male mice but only after administration of very high doses (1012–1014 vector genome [vg]/mouse). Female mice responded only at extremely high vector doses (1014 vg/mouse) and the duration of efficacy was longer in male mice than in female mice. In the study of the Kume laboratory in Japan, blood l-Phe concentrations had gone back up to pretreatment levels by 40 weeks after rAAV2/5 injection in both sexes.45 In both studies, the vectors had been delivered by portal vein injection. Delivery through tail vein or intramuscular injection yielded no change in blood l-Phe concentrations.44 Improved efficacy at a lower AAV vector dose was seen when a Woodchuck Posttranscriptional Regulatory Element (WPRE) was included in the vector genome, but administration of WPRE-containing vectors was associated with the development of hepatocellular dysplasia in the livers of treated animals.46
Pseudotyping of rAAV2 genomes with capsid proteins from alternative rAAV serotypes alters tissue tropism and enhances therapeutic gene expression.47 The discovery and implementation of rAAV2 serotype 8 vectors were a major breakthrough for AAV-mediated liver-directed gene therapy as the AAV8 capsid shows the greatest affinity for and yields the best transduction frequency in rodent liver of any available AAV serotype. rAAV2/8 vectors expressing either murine or human PAH-cDNAs have been administered by either tail vein, portal vein, or intraperitoneal injection to successfully treat murine PKU by 3 different laboratories.48–50 Administration of as few as 5 × 1011 vg rAAV2/8 expressing the murine (m) Pah-cDNA under the transcriptional control of a strong liver-specific promoter through portal vein injection yielded complete correction of blood l-Phe concentrations in both male and female mice.48 At this dose, 8–25 vgs were detected per haploid liver genome in animals euthanized 17 weeks after AAV treatment and this level of transduction was associated with restoration of liver PAH activity to approximately 11–15.1% of wild type liver PAH activity. Although this effect was stable up to 17 weeks in the original report, PAH expression has been maintained up to at least 1 year after AAV treatment49). Nevertheless, blood l-Phe levels began to slowly increase, primarily in female mice, and this reemergence of HPA is associated with a gradual dilution of vector genome copy number in liver. Larger initial vector doses (5 × 1012 vg/mouse) administered by portal vein yielded ∼1,000 vgs per diploid genome in liver and restoration of liver PAH activity to almost 100% of wild type activity with sustained correction of blood l-Phe concentrations to beyond 42 weeks in experiments carried out in the Thöny laboratory.49 No difference in efficacy between male and female mice was seen. Similar results were obtained if the rAAV2/8 vectors were delivered by tail vein injection, but a higher dose was required (2 × 1013 vg/mouse). No major toxicity was seen after rAAV2/8 treatment, and no anti-PAH antibodies were detected.49
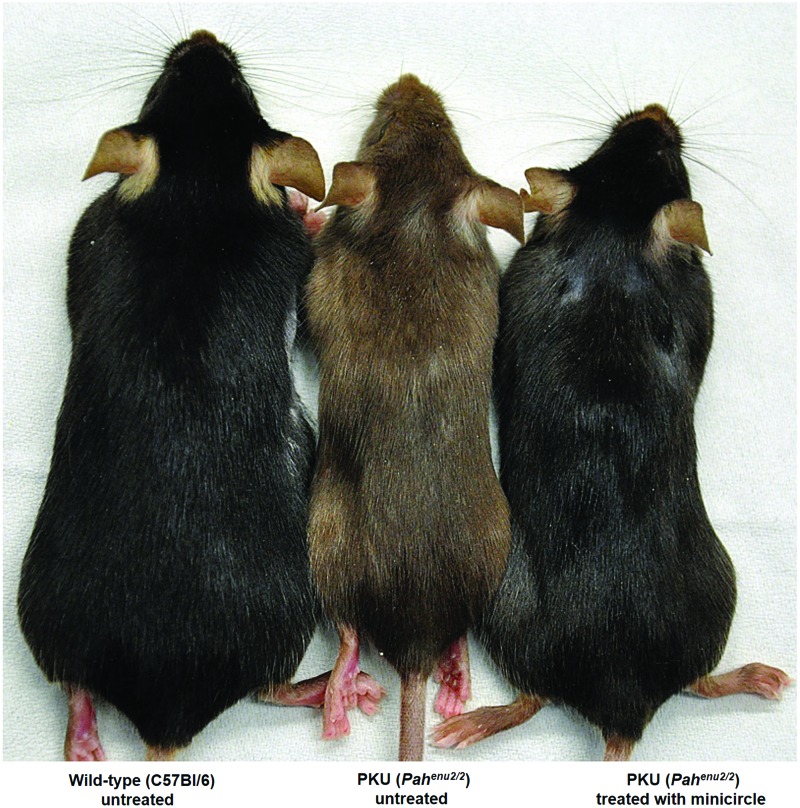
In all reported successful trials, the hypopigmentation associated with chronic HPA in the mice has been fully reversed after rAAV2/8-mediated liver-directed gene therapy (see Fig. 1).51 Later, direct comparison of serotypes 1, 2, and 8 vectors demonstrated the unequivocal superiority of serotype 8 vector for the treatment of Pahenu2/2 model through liver-directed gene therapy and, furthermore, it was demonstrated that even simple intramuscular injection of rAAV2/8 vector can lead to trafficking of vector to and transduction of liver with robust PAH expression and correction of HPA.52
Figure 1.
Phenotype reversion of fair hair upon gene therapy of the PKU mice. Reversion from brown to black coat of treated (C57Bl/6) PKU mice as depicted in the figure is a phenomenon that can be seen from gene therapy or any other treatment, including dietary or pegvaliase treatment. Not only mice but also patients with untreated PKU exhibit blond hair and fair skin. Tyrosine that is needed to make melanin, the pigment that gives skin and hair its coloring, is limited under untreated PKU conditions and the corresponding enzyme, tyrosinase, is competitively inhibited by elevated phenylalanine levels. The figure is reproduced with permission from Viecelli et al.57 wherein mice were treated with nonviral naked DNA vector expressing phenylalanine hydroxylase on the right, wild type mice untreated on the left, and PKU mice untreated in the middle. PKU, phenylketonuria. Color images are available online.
The Kume laboratory constructed a self-complementary (sc) AAV2/8 vector that expressed the mPah-cDNA using a compact enhancer/promoter cassette (a human apolipoprotein E/C-I hepatic control region fused to the human alpha-1-antitrypsin promoter) and demonstrated improved efficacy at lower doses than a comparable single-stranded AAV2/8 vector that included the cytomegalovirus (CMV) immediate–early enhancer/chicken β-actin promoter.50 Blood l-Phe was corrected to normal after administration of as little at 1 × 1011 vg scAAV2/8 injected through the intraperitoneal route even in female mice. In addition to measuring blood l-Phe, efficacy was also assessed through the measurement of 13C-phenylalanine oxidation to 13CO2 in collected breath. The portion of 13C-phenylalanine dose oxidized to 13CO2 was directly related to the vector dose administered and the number of vector genomes detected in total liver DNA at euthanasia and had continued to increase even after blood l-Phe had normalized. Correction of HPA was sustained to beyond 1 year after treatment and was associated with marked improvement of the brain dopamine, norepinephrine, and serotonin deficiency associated with chronic HPA in the Pahenu2/2 mouse model.53
A novel set of proprietary AAV vectors derived from human CD34+ hematopoietic stem cells (AAVHSCs) from Homology Medicines, Inc. (Bedford, MA) containing a promoter-less human (h) PAH-cDNA has recently been reported to correct HPA in Pahenu2/2 mice after intravenous tail vein injection.54 The hPAH-cDNA was integrated into the mPah locus through homologous recombination. As a result, blood l-Phe level was maintained below therapeutic level (≤360 μM or 6 mg/dL) for up to 8 weeks. Further animal studies confirmed that genome editing upon delivering AAVHSC15 containing the promoter-less cDNA encoding hPAH targeted precisely the PAH allele in a mouse repopulated with human hepatocytes, whereas no measurable editing in mouse hepatocytes was detectable.
An entirely different approach is the targeting of skeletal muscle for PKU gene therapy. The systemically elevated l-Phe that is responsible for the symptoms of PKU could be degraded in muscle tissue as it comprises 30–40% of body mass, is highly vascularized and supplied with blood, moderately regenerating, and easily accessible for percutaneous injection. The limiting factor turned out the BH4 cofactor that is not present in muscle tissue. Therefore, the PAH protein was coexpressed from a CMV promoter along with the enzymes necessary to synthesize BH4. A corresponding triple-cistronic rAAV serotype 1 vector that was directly injected into the gastrocnemius muscle of Pahenu2/2 mice led to cofactor biosynthesis, normalization of blood l-Phe levels, and long-term treatment of PKU.55
Liver Gene Therapy with Nonviral Vectors
The development of nonviral gene delivery systems as an alternative to viral vectors has gradually gained attention due to the potentially better safety profile and low production costs. The recent FDA approval of patisiran (Onpattro™; Alnylam Pharmaceuticals, Cambridge, MA), the first siRNA drug to use RNA interference to downregulate protein as a treatment for polyneuropathy in adults with hereditary transthyretin-mediated amyloidosis, is a breakthrough in the drug discover field. This drug is encapsulated in lipid nanoparticles and N-acetylgalactosamine for efficient and specific delivery to the liver.56 DNA-mediated gene transfer into tissues or organs is more preferable for long-term treatment for monogenic metabolic diseases but generally suffers from lower rates of cell transduction and transient therapeutic responses.
The Thöny laboratory has reported on persistent and efficacious treatment of PKU mice through hydrodynamic delivery of nonviral naked minicircle (MC)-DNA vectors without adverse effects.57 MC-DNA vectors are a form of supercoiled DNA for use in nonviral gene transfer that contain a minimal expression cassette and from which all bacterial DNA is removed by intravector recombination.58 Studies have shown that bacterial DNA contributes to biological safety problems and transgene silencing.59,60 MC-DNA vectors (3 × 1013 vg/mouse) expressing murine PAH from a synthetic liver-specific promoter (designated as P361) yielded therapeutic PAH activity accompanied by complete revision of hypopigmentation in the PKU mouse model (Fig. 1) for 1 year after liver gene transfer by a single hydrodynamic tail vein injection.57 At the same time, there was no reduction of l-Phe in blood in the mice treated with the (parental) plasmid, indicating a rapid loss of either vector DNA or gene expression or both due to the bacterial backbone.
Since there is no limitation of package capacity of the DNA insert, the expression cassette consisting of a 3.6-kb native endogenous Pah promoter/enhancer sequence, codon-optimized mPah–cDNA, and a truncated intron was included in the MC-DNA vector.62 The efficacy of this improved expression cassette of MC-DNA vectors resulted in a significant reduction in vector dosage (6 × 1012 vg/mouse) required for successful treatment. Analysis of treated mice upon sacrifice revealed that statistically <1 copy of MC vector was present per diploid hepatocyte genome in a whole liver. The loss of MC-DNA vectors and PAH activity after partial hepatectomy and liver regeneration confirmed that MC-DNA vectors do not integrate into the genome and remain episomal. However, a limiting factor for human usage and treatment of PKU remains delivery of naked DNA vectors to liver (and other tissues).
In summary, naked DNA-based therapeutic tools are still in its infancy compared with the much more advanced AAV vector field, but nonviral alternatives for gene delivery are receiving growing attention and it is anticipated that this trend will continue in the future.
Liver Gene Therapy by Gene Editing
In recent years, a number of studies have implemented genome editing for the treatment of animal models of genetic liver diseases.63 It has the advantage compared with gene addition approaches that endogenous loci can be corrected. Thus, temporary intervention enables permanent repair and expression of the functional gene under its endogenous promoter.
The most widespread genome editing tool is the CRISPR/Cas9 system, where a programmable chimeric guide RNA targets the Cas9 endonuclease to the locus of interest.64,65 If generated DNA double-stranded breaks (DSBs) are restored by homology-directed repair (HDR) from an exogenous template DNA, mutations are repaired with single base precision. However, although precise repair is essential for correcting autosomal recessive disorders such as PKU, hepatocytes in the adult liver predominantly repair DSBs through nonhomologous end-joining pathways, resulting in deletion or insertion (indel) mutations.66,67 Thus, when CRISPR/Cas9 was applied to adult mouse models for ornithine transcarbamylase deficiency, hereditary tyrosinemia, and hemophilia B, repair rates were extremely low, with 2%, 0.4%, and 1%, respectively.68–70 Unfortunately, such low repair rates would likely not be sufficient to reduce blood l-Phe in PKU patients below pathological levels.39,40 In addition, recent reports suggest that CRISPR/Cas9-induced DSBs could create serious safety concerns to patients, as they often cause unpredictable, complex, and potentially harmful genetic alterations,71,72 and activate the tumor suppressor TP53.73,74
Recently a novel CRISPR-based genome editing tool that enables direct conversion of single bases has been developed. These so-called base editors are chimeric proteins that consist of nuclease-impaired Cas9 fused to catalytic domains that enable DNA deamination. Cytidine base editors enable single C·G to T·A base pair conversions through uracil intermediates, and adenine base editors enable single A·T to G·C base pair conversions through inosine intermediates.75,76 Since base editors enable precise exchange of single base pairs independently of DSBs formation and HDR, they also enable efficient base editing in postmitotic cells such as adult hepatocytes.77–79
In a recent study, cytidine base editors have been employed to correct the G to A missense mutation in the Pahenu2/2 mouse model.80 rAAV-mediated delivery of base editors into the liver resulted in repair rates of the target base >20%, and a reduction of blood l-Phe to physiological levels. These data suggest that the base editors have great potential for treatment of a subset of PKU patients with targetable T to C, A to G, G to A, and C to T mutations. Nevertheless, before clinical application, safety studies that closely characterize potentially deleterious effects from off-target editing are still needed.81–83 In addition, current base editor studies in the liver use viral delivery vectors that cause prolonged expression of the Cas9-deaminase, and for application in patients, the establishment of temporally active base editor approaches would be desired.
Trends in the Pharmaceutical Industry for Clinical Gene Therapy Trials in PKU Patients
As of this writing (May, 2019), no gene transfer into humans for the purpose of treating PKU has yet been attempted, but several pharmaceutical companies have publicly announced their intentions to do so in the very near future employing either rAAV- or lentivirus-mediated gene addition strategies.
Homology Medicines, Inc. has presented preclinical data in abstract form demonstrating sustained correction of blood l-Phe concentrations in the Pahenu2/2 model after a single IV injection of their proprietary AAVHSC15 vector expressing human PAH (HMI-102).84 They have completed the necessary preclinical pharmacology/toxicology studies and have recently been granted clearance of their Investigational New Drug (IND) application by the US FDA. They are planning to initiate a human Phase 1/2 clinical trial (pheNIX, NCT03952156) in adults with PKU within the next few months.
BioMarin Pharmaceutical, Inc. has publicly announced its plans to investigate gene addition in adults with PKU using an rAAV2/5 vector expressing human PAH. BioMarin scientists are completing the preclinical validation of their PAH-expressing rAAV2/5 vector and will be applying for an IND soon.
In any rAAV-mediated gene addition trial, the majority of therapeutic gene expression will be from episomal vector genomes residing within hepatocytes. Any stimulus to hepatocyte regeneration would threaten the long-term stability of expression, and truthfully the ultimate duration of therapeutic gene expression after a single IV infusion of a liver-directed rAAV2 vector to an adult human is unknown, although robust Factor IX expression has been measured in scAAV2/8-treated individuals >10 years from their initial injection. In addition, the efficacy of this gene addition approach is only temporary in juvenile animals during rapid hepatocyte proliferation due to episomal dilution.85–87
As detailed elsewhere, direct gene editing or the use of integrating vectors avoids this issue, but these approaches all struggle to achieve editing or integration frequencies (in the absence of a selective growth advantage for PAH expressing hepatocytes) sufficient to yield a physiologically relevant PAH positive cell population. However, American Gene Technologies, Inc. (Rockville, MD) has publicly announced that it is developing a liver-directed gene therapy strategy based upon a proprietary integrating lentivirus-based vector system. Its vector has been granted Orphan Drug status by the FDA (no. DRU-2018-6572), but the company has not released any preclinical data or other information regarding its progress toward a clinical trial.
Outlook
Gene addition is for some rare monogenic disorders where no good alternative treatment is available, becoming a reality for patients with severe genetic defects. Furthermore, for in vivo approaches to target, for example, the liver, AAV vectors are currently the most effective choice. Owing to the long history of successful dietary treatment, if started early in life, PKU was not considered to be a primary target for a genetic treatment. On top of that, newborn screening programs implemented today in most countries for PKU guarantee early diagnosis and intervention. It is acknowledged today that treatment from early in life with dietary therapy has protected adults with PKU from permanent neuropathologic damage associated with chronic HPA. Adherence to diet to keep blood l-Phe levels low during pregnancy protects also against the maternal PKU syndrome. Some adults, however, struggle to maintain necessary dietary control of blood l-Phe and consequently suffer cognitive, behavioral, and psychiatric symptoms associated with HPA (for a discussion see for instance88).
Since most of these symptoms should be reversible upon successful correction of blood l-Phe concentrations to normal, these adult patients may benefit from new treatment options such as for instance gene addition therapies. In this regard, the U.S.7 guidelines state the following in the future direction section: “New therapies, including gene therapy, and hepatocyte transplant, have shown some efficacy in animal or limited human trials but require further development and validation for routine clinical use.” However, the more recent European guidelines8,9 for PKU mention dietary and enzyme replacement treatment but not gene therapy as an option. Nevertheless, and as already mentioned, treating adults with PKU with AAV vector approaches is currently pursued by several companies in the United States.
Whether gene therapy for PKU with the current technology and experience available is also an option in other countries such as in Europe or in Asia remains to be seen. Intravenous administration of AAV vectors has proven safe in several clinical trials and evidence of efficacy has also been seen for many. A critical question to be answered is which serotype will be most efficacious at which minimally effective dose, and whether the treatment effect will be permanent. The goal of course is sustained disease correction over many years without the need for redosing. However, in children with PKU, this approach will likely not be successful, as episomal AAV genomes are known to not be stably maintained in growing liver of juvenile mammals.85,86
Solutions for permanent cure in, for example, pediatric patients with a growing liver are either integration or addition of replication-competent gene copy, or correction of the disease-causing mutation. Such future approaches may include integration of a PAH-cDNA through AAV-directed homologous recombination,54 random integration of PAH-cDNA through liver-directed lentiviral vectors,89–91 hepatocyte transfection with replicating nonviral vectors,92–94 or genome editing with base editors through liver-directed AAV vectors or lipid-nanoparticles for mRNA delivery.80,94 Although some of these technologies have proven therapeutic in the PKU mouse model, they are still at its developing level and need to be improved for efficacy and/or assessed for safety profiles. We look forward to a series of robust clinical trials, which hopefully will address pitfalls of current PKU therapies.
Footnotes
Author Disclosure
C.O.H. has received consulting fees or funds in support of clinical or preclinical research from BioMarin Pharmaceutical, Inc., Ultragenyx, Inc., Horizon Pharmaceutical, Synlogic, Inc., Rubius Therapeutics, Cydan, Inc., StrideBio, Inc., Voyager Therapeutics, and Pfizer, Inc. All other authors have no conflict of interest.
Funding Information
Our study was funded by Forschungszentrum für das Kind (to H.M.G-C.), the Swiss National Science Foundation (3100A0-105250 and CRSII5_180257/1 to B.T., 31003A_160230 to G.S.), Wolfermann Nägeli Stiftung (to B.T.), and NIH (R01NS080866 to C.O.H.).
References
- 1. Donlon J, Sarkissian C, Levy H, et al. In: Beaudet AL, Vogelstein B, Kinzler KW, et al., eds. The Online Metabolic and Molecular Bases of Inherited Disease. New York, NY: The McGraw-Hill Companies, Inc., 2014 [Google Scholar]
- 2. Blau N, van Spronsen FJ, Levy HL. Phenylketonuria. Lancet 2010;376:1417–1427 [DOI] [PubMed] [Google Scholar]
- 3. Thöny B, Auerbach G, Blau N, Tetrahydrobiopterin biosynthesis, regeneration and functions. Biochem J 2000;347:1–16 [PMC free article] [PubMed] [Google Scholar]
- 4. Blau N, Martinez A, Hoffmann GF, et al. . DNAJC12 deficiency: a new strategy in the diagnosis of hyperphenylalaninemias. Mol Genet Metab 2018;123:1–5 [DOI] [PubMed] [Google Scholar]
- 5. Guthrie R, Susi A. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 1963;32:338–343 [PubMed] [Google Scholar]
- 6. Levy HL, Sarkissian CN, Scriver CR. Phenylalanine ammonia lyase (PAL): from discovery to enzyme substitution therapy for phenylketonuria. Mol Genet Metab 2018:124:223–229 [DOI] [PubMed] [Google Scholar]
- 7. Vockley J, Andersson HC, Antshel KM, et al. . Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet Med 2014;16:188–200 [DOI] [PubMed] [Google Scholar]
- 8. van Spronsen FJ, van Wegberg AM, Ahring K, et al. . Key European guidelines for the diagnosis and management of patients with phenylketonuria. Lancet Diabetes Endocrinol 2017;5:743–756 [DOI] [PubMed] [Google Scholar]
- 9. van Wegberg AMJ, MacDonald A, Ahring K, et al. . The complete European guidelines on phenylketonuria: diagnosis and treatment. Orphanet J Rare Dis 2017;12:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bilder DA, Noel JK, Baker ER, et al. . Systematic review and meta-analysis of neuropsychiatric symptoms and executive functioning in adults with phenylketonuria. Dev Neuropsychol 2016;41:245–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DeRoche K, Welsh M. Twenty-five years of research on neurocognitive outcomes in early-treated phenylketonuria: intelligence and executive function. Dev Neuropsychol 2008;33:474–504 [DOI] [PubMed] [Google Scholar]
- 12. Christ SE, Huijbregts SC, de Sonneville LM, et al. . Executive function in early-treated phenylketonuria: profile and underlying mechanisms. Mol Genet Metab 2010;99:S22–S32 [DOI] [PubMed] [Google Scholar]
- 13. Jahja R, van Spronsen FJ, de Sonneville LMJ, et al. . Social-cognitive functioning and social skills in patients with early treated phenylketonuria: a PKU-COBESO study. J Inherit Metab Dis 2016;39:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koch R, Burton B, Hoganson G, et al. . Phenylketonuria in adulthood: a collaborative study. J Inherit Metab Dis 2002;25:333–346 [DOI] [PubMed] [Google Scholar]
- 15. Lenke RR, Levy HL. Maternal phenylketonuria and hyperphenylalaninemia. An international survey of the outcome of untreated and treated pregnancies. N Engl J Med 1980;303:1202–1208 [DOI] [PubMed] [Google Scholar]
- 16. Longo N, Dimmock D, Levy H, et al. . Evidence- and consensus-based recommendations for the use of pegvaliase in adults with phenylketonuria. Genet Med 2018. [Epub ahead of print]; DOI: 10.1038/s41436-018-0403-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mahan KC, Gandhi MA, Anand S. Pegvaliase: a novel treatment option for adults with phenylketonuria. Curr Med Res Opin 2019;35:647–651 [DOI] [PubMed] [Google Scholar]
- 18. Thomas J, Levy H, Amato S, et al. . Pegvaliase for the treatment of phenylketonuria: results of a long-term phase 3 clinical trial program (PRISM). Mol Genet Metab 2018;124:27–38 [DOI] [PubMed] [Google Scholar]
- 19. Gupta S, Lau K, Harding CO, et al. . Association of immune response with efficacy and safety outcomes in adults with phenylketonuria administered pegvaliase in phase 3 clinical trials. EBioMedicine 2018;37:366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pascucci T, Rossi L, Colamartino M, et al. . A new therapy prevents intellectual disability in mouse with phenylketonuria. Mol Genet Metab 2018;124:39–49 [DOI] [PubMed] [Google Scholar]
- 21. Isabella VM, Ha BN, Castillo MJ, et al. . Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol 2018;36:857–864 [DOI] [PubMed] [Google Scholar]
- 22. McDonald JD, Bode VC, Dove WF, et al. . Pahhph-5: a mouse mutant deficient in phenylalanine hydroxylase. Proc Natl Acad Sci U S A 1990;87:1965–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDonald JD, Charlton CK. Characterization of mutations at the mouse phenylalanine hydroxylase locus. Genomics 1997;39:402–405 [DOI] [PubMed] [Google Scholar]
- 24. Heintz C, Troxler H, Martinez A, et al. . Quantification of phenylalanine hydroxylase activity by isotope-dilution liquid chromatography-electrospray ionization tandem mass spectrometry. Mol Genet Metab 2012;105:559–565 [DOI] [PubMed] [Google Scholar]
- 25. McDonald JD. Postnatal growth in a mouse genetic model of classical phenylketonuria. Contemp Top Lab Anim Sci 2000;39:54–56 [PubMed] [Google Scholar]
- 26. Zagreda L, Goodman J, Druin DP, et al. . Cognitive deficits in a genetic mouse model of the most common biochemical cause of human mental retardation. J Neurosci 1999;19:6175–6182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winn SR, Scherer T, Thony B, et al. . Blood phenylalanine reduction corrects CNS dopamine and serotonin deficiencies and partially improves behavioral performance in adult phenylketonuric mice. Mol Genet Metab 2018;123:6–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dobrowolski SF, Tourkova IL, Robinson LJ, et al. . A bone mineralization defect in the Pah(enu2) model of classical phenylketonuria involves compromised mesenchymal stem cell differentiation. Mol Genet Metab 2018;125:193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perez-Duenas B, Cambra FJ, Vilaseca MA, et al. . New approach to osteopenia in phenylketonuric patients. Acta Paediatr 2002;91:899–904 [DOI] [PubMed] [Google Scholar]
- 30. van Spronsen FJ, Burgard P. The truth of treating patients with phenylketonuria after childhood: the need for a new guideline. J Inherit Metab Dis 2008;31:673–679 [DOI] [PubMed] [Google Scholar]
- 31. Cho S, McDonald JD. Effect of maternal blood phenylalanine level on mouse maternal phenylketonuria offspring. Mol Genet Metab 2001;74:420–425 [DOI] [PubMed] [Google Scholar]
- 32. McDonald JD, Dyer CA, Gailis L, et al. . Cardiovascular defects among the progeny of mouse phenylketonuria females. Pediatr Res 1997;42:103–107 [DOI] [PubMed] [Google Scholar]
- 33. Eisensmith RC, Woo SL. Gene therapy for phenylketonuria. Acta Paediatr Suppl 1994;407:124–129 [DOI] [PubMed] [Google Scholar]
- 34. Ding Z, Harding CO, Thöny B. State-of-the-art 2003 on PKU gene therapy. Mol Genet Metab 2004;81:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harding CO, Ding Z, Thöny B. Gene and cell therapies for phenylketonuria (PKU). In: Blau N, ed. PKU and BH4: Advances in Phenylketonuria and Tetrahydrobiopterin. Heilbronn: SPS Publications, 2006 [Google Scholar]
- 36. Sarkissian CN, Gamez A, Scriver CR. What we know that could influence future treatment of phenylketonuria. J Inherit Metab Dis 2009;32:3–9 [DOI] [PubMed] [Google Scholar]
- 37. Thony B. Long-term correction of murine phenylketonuria by viral gene transfer: liver versus muscle. J Inherit Metab Dis 2010;33:677–680 [DOI] [PubMed] [Google Scholar]
- 38. Al Hafid N, Christodoulou J. Phenylketonuria: a review of current and future treatments. Transl Pediatr 2015;4:304–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hamman K, Clark H, Montini E, et al. . Low therapeutic threshold for hepatocyte replacement in murine phenylketonuria. Mol Ther 2005;12:337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hamman KJ, Winn SR, Harding CO. Hepatocytes from wild-type or heterozygous donors are equally effective in achieving successful therapeutic liver repopulation in murine phenylketonuria (PKU). Mol Genet Metab 2011;104:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin CM, Tan Y, Lee YM, et al. . Expression of human phenylalanine hydroxylase activity in T lymphocytes of classical phenylketonuria children by retroviral-mediated gene transfer. J Inherit Metab Dis 1997;20:742–754 [DOI] [PubMed] [Google Scholar]
- 42. Fang B, Eisensmith RC, Li XH, et al. . Gene therapy for phenylketonuria: phenotypic correction in a genetically deficient mouse model by adenovirus-mediated hepatic gene transfer. Gene Ther 1994;1:247–254 [PubMed] [Google Scholar]
- 43. Nagasaki Y, Matsubara Y, Takano H, et al. . Reversal of hypopigmentation in phenylketonuria mice by adenovirus-mediated gene transfer. Pediatr Res 1999;45:465–473 [DOI] [PubMed] [Google Scholar]
- 44. Oh HJ, Park ES, Kang S, et al. . Long-term enzymatic and phenotypic correction in the phenylketonuria mouse model by adeno-associated virus vector-mediated gene transfer. Pediatr Res 2004;56:278–284 [DOI] [PubMed] [Google Scholar]
- 45. Mochizuki S, Mizukami H, Ogura T, et al. . Long-term correction of hyperphenylalaninemia by AAV-mediated gene transfer leads to behavioral recovery in phenylketonuria mice. Gene Ther 2004;11:1081–1086 [DOI] [PubMed] [Google Scholar]
- 46. Embury J, Frost S, Charron CE, et al. . Hepatitis virus protein X-phenylalanine hydroxylase fusion proteins identified in PKU mice treated with AAV-WPRE vectors. Gene Ther Mol Biol 2008;12:69–76 [Google Scholar]
- 47. Gao GP, Alvira MR, Wang L, et al. . Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci U S A 2002;99:11854–11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harding CO, Gillingham MB, Hamman K, et al. . Complete correction of hyperphenylalaninemia following liver-directed, recombinant AAV2/8 vector-mediated gene therapy in murine phenylketonuria. Gene Ther 2006;13:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ding Z, Georgiev P, Thony B. Administration-route and gender-independent long-term therapeutic correction of phenylketonuria (PKU) in a mouse model by recombinant adeno-associated virus 8 pseudotyped vector-mediated gene transfer. Gene Ther 2006;13:587–593 [DOI] [PubMed] [Google Scholar]
- 50. Yagi H, Ogura T, Mizukami H, et al. . Complete restoration of phenylalanine oxidation in phenylketonuria mouse by a self-complementary adeno-associated virus vector. J Gene Med 2011;13:114–122 [DOI] [PubMed] [Google Scholar]
- 51. Thony B, Ding Z, Rebuffat A, et al. . Phenotypic reversion of fair hair upon gene therapy of the phenylketonuria mice. Hum Gene Ther 2014;25;573–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rebuffat A, Harding CO, Ding Z, et al. . Comparison of adeno-associated virus pseudotype 1, 2, and 8 vectors administered by intramuscular injection in the treatment of murine phenylketonuria. Hum Gene Ther 2010;21:463–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yagi H, Sanechika S, Ichinose H, et al. . Recovery of neurogenic amines in phenylketonuria mice after liver-targeted gene therapy. Neuroreport 2012;23:30–34 [DOI] [PubMed] [Google Scholar]
- 54. Wright J, Ellsworth JF, St. Martin T, et al. . Nuclease-free and promoter-less AAVHSC-mediated genome editing in vivo corrects the disease phenotype in a mouse model of phenylketonuria. Mol Ther 2019;27:462 [Google Scholar]
- 55. Ding Z, Harding CO, Rebuffat A, et al. . Correction of murine PKU following AAV-mediated intramuscular expression of a complete phenylalanine hydroxylating system. Mol Ther 2008;16:673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Al Shaer D, Al Musaimi O, Albericio F, et al. . 2018 FDA tides harvest. Pharmaceuticals (Basel) 2019;12:E52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Viecelli HM, Harbottle RP, Wong SP, et al. . Treatment of phenylketonuria using minicircle-based naked-DNA gene transfer to murine liver. Hepatology 2014;60:1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gaspar V, de Melo-Diogo D, Costa E, et al. . Minicircle DNA vectors for gene therapy: advances and applications. Expert Opin Biol Ther 2015;15:353–379 [DOI] [PubMed] [Google Scholar]
- 59. Gill DR, Pringle IA, Hyde SC. Progress and prospects: the design and production of plasmid vectors. Gene Ther 2009;16:165–171 [DOI] [PubMed] [Google Scholar]
- 60. Hyde SC, Pringle IA, Abdullah S, et al. . CpG-free plasmids confer reduced inflammation and sustained pulmonary gene expression. Nat Biotechnol 2008;26:549–551 [DOI] [PubMed] [Google Scholar]
- 61. Nair N, Rincon MY, Evens H, et al. . Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood 2014;123:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Grisch-Chan HM, Schlegel A, Scherer T, et al. . Low-dose gene therapy for murine PKU using episomal naked DNA vectors expressing PAH from its endogenous liver promoter. Mol Ther Nucleic Acids 2017;7:339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aravalli RN, Steer CJ. CRISPR/Cas9 therapeutics for liver diseases. J Cell Biochem 2018;119:4265–4278 [DOI] [PubMed] [Google Scholar]
- 64. Cong L, Ran FA, Cox D, et al. . Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mali P, Yang L, Esvelt KM, et al. . RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Barnes DE. Non-homologous end joining as a mechanism of DNA repair. Curr Biol 2001;11:R455–R457 [DOI] [PubMed] [Google Scholar]
- 67. van den Bosch M, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem 2002;383:873–892 [DOI] [PubMed] [Google Scholar]
- 68. Huai C, Jia C, Sun R, et al. . CRISPR/Cas9-mediated somatic and germline gene correction to restore hemostasis in hemophilia B mice. Hum Genet 2017;136:875–883 [DOI] [PubMed] [Google Scholar]
- 69. Yin H, Xue W, Chen S, et al. . Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ohmori T, Nagao Y, Mizukami H, et al. . CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep 2017;7:4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Adikusuma F, Piltz S, Corbett MA, et al. . Large deletions induced by Cas9 cleavage. Nature 2018;560:E8–E9 [DOI] [PubMed] [Google Scholar]
- 72. Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018;36:765–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Haapaniemi E, Botla S, Persson J, et al. . CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018;24:927–930 [DOI] [PubMed] [Google Scholar]
- 74. Ihry RJ, Worringer KA, Salick MR, et al. . p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 2018;24:939–946 [DOI] [PubMed] [Google Scholar]
- 75. Gaudelli NM, Komor AC, Rees HA, et al. . Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017;551:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Komor AC, Kim YB, Packer MS, et al. . Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rees HA, Komor AC, Yeh WH, et al. . Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat Commun 2017;8:15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Song C-Q, Jiang T, Richter M, et al. . Adenine base editing in an adult mouse model of tyrosinaemia. Nat Biomed Eng 2019; DOI: 10.1038/s41551-019-0357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yeh WH, Chiang H, Rees HA, et al. . In vivo base editing of post-mitotic sensory cells. Nat Commun 2018;9:2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Villiger L, Grisch-Chan HM, Lindsay H, et al. . Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med 2018;24:1519–1525 [DOI] [PubMed] [Google Scholar]
- 81. Grunewald J, Zhou R, Garcia SP, et al. . Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019;569; DOI: 10.1038/s41586-019-1161-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Jin S, Zong Y, Gao Q, et al. . Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019;364:292–295 [DOI] [PubMed] [Google Scholar]
- 83. Zuo E, Sun Y, Wei W, et al. . Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019;364:289–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wright TL, Ellsworth JF, Ahmed SS, et al. . A novel AAVHSC15 encoding human phenylalanine hydroxylase, in cynomolgus monkeys. Mol Ther 2019;27:67 [Google Scholar]
- 85. Cunningham SC, Spinoulas A, Carpenter KH, et al. . AAV2/8-mediated correction of OTC deficiency is robust in adult but not neonatal Spfash mice. Mol Ther 2009;17:1340–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang L, Wang H, Bell P, et al. . Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther 2012;23:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wang L, Bell P, Lin J, et al. . AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta). Mol Ther 2011;19:2012–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Brown CS. Family reflections on phenylketonuria. Pediatr Res 2018;84:797–798 [DOI] [PubMed] [Google Scholar]
- 89. Milani M, Starinieri F, Canepari C, et al. . Liver-directed gene therapy with lentiviral vectors in newborn mice and dog puppies. Mol Ther 2019;27:323 [Google Scholar]
- 90. Vonada A, Nygaard S, Grompe M. Selective expansion of gene-targeted hepatocytes using acetaminophen leads to reproducible long-term liver repopulation. Mol Ther 2019;27:32630660488 [Google Scholar]
- 91. Merlin S, Follenzi A. Transcriptional targeting and microRNA regulation of lentiviral vectors. Mol Ther Methods Clin Dev 2019;12:223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Argyros O, Wong SP, Harbottle RP. Non-viral episomal modification of cells using S/MAR elements. Expert Opin Biol Ther 2011;11:1177–1191 [DOI] [PubMed] [Google Scholar]
- 93. Jackson DA, Juranek S, Lipps HJ. Designing nonviral vectors for efficient gene transfer and long-term gene expression. Mol Ther 2006;14:613–626 [DOI] [PubMed] [Google Scholar]
- 94. Finn JD, Smith AR, Patel MC, et al. . A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep 2018;22:2227–2235 [DOI] [PubMed] [Google Scholar]