Key Points
Question
Is the genetic liability to psychotic experiences shared with schizophrenia and/or other neuropsychiatric disorders and traits?
Findings
In this cohort study, genetic correlation, polygenic risk score, and copy number variation analyses indicated a shared genetic liability between psychotic experiences and major depressive disorder, schizophrenia, bipolar disorder, and neurodevelopmental disorders. Genome-wide association studies identified 4 genetic loci associated with psychotic experiences including loci in ANK3 and CNR2.
Meaning
Findings suggest that the genetic liability of psychotic experiences is shared with several psychiatric disorders, which include, but is not specific to, schizophrenia.
Abstract
Importance
Psychotic experiences, such as hallucinations and delusions, are reported by approximately 5% to 10% of the general population, although only a small proportion develop psychotic disorders such as schizophrenia. Studying the genetic causes of psychotic experiences in the general population, and its association with the genetic causes of other disorders, may increase the understanding of their pathologic significance.
Objectives
To determine whether genetic liability to psychotic experiences is shared with schizophrenia and/or other neuropsychiatric disorders and traits and to identify genetic loci associated with psychotic experiences.
Design, Setting and Participants
Analyses of genetic correlation, polygenic risk scores, and copy number variation were performed using data from participants in the UK Biobank from April 1, 2018, to March 20, 2019, to assess whether genetic liability to psychotic experiences is shared with schizophrenia and/or other neuropsychiatric disorders and traits. Genome-wide association studies of psychotic experience phenotypes were conducted to identify novel genetic loci. Participants in the final analyses after exclusions included 6123 individuals reporting any psychotic experience, 2143 individuals reporting distressing psychotic experiences, and 3337 individuals reporting multiple occurrences of psychotic experiences. A total of 121 843 individuals who did not report a psychotic experience formed the comparator group. Individuals with a psychotic disorder were excluded from all analyses.
Main Outcomes and Measures
Genetic associations with psychotic experience phenotypes.
Results
The study included a total of 127 966 participants (56.0% women and 44.0% men; mean [SD] age, 64.0 [7.6] years). Psychotic experiences were genetically correlated with major depressive disorder, schizophrenia, autism spectrum disorder, and attention-deficit/hyperactivity disorder. Analyses of polygenic risk scores identified associations between psychotic experiences and genetic liability for major depressive disorder, schizophrenia, bipolar disorder, autism spectrum disorder, and attention-deficit/hyperactivity disorder. Individuals reporting psychotic experiences had an increased burden of copy number variations previously associated with schizophrenia (odds ratio [OR], 2.04; 95% CI, 1.39-2.98; P = 2.49 × 10−4) and neurodevelopmental disorders more widely (OR, 1.75; 95% CI, 1.24-2.48; P = 1.41 × 10−3). Genome-wide association studies identified 4 significantly associated loci, including a locus in Ankyrin-3 (ANK3 [GenBank NM_020987]) (OR, 1.16; 95% CI, 1.10-1.23; P = 3.06 × 10−8) with any psychotic experience, and a locus in cannabinoid receptor 2 gene (CNR2 [GenBank NM_001841]) (OR, 0.66; 95% CI, 0.56-0.78; P = 3.78 × 10−8) with distressing psychotic experiences. The genome-wide association study of any psychotic experience had a low single-nucleotide polymorphism–based heritability estimate (h2 = 1.71%; 95% CI, 1.02%-2.40%).
Conclusions and Relevance
A large genetic association study of psychotic experiences from the population-based UK Biobank sample found support for a shared genetic liability between psychotic experiences and schizophrenia, major depressive disorder, bipolar disorder, and neurodevelopmental disorders.
This cohort study uses data from the UK Biobank to examine whether genetic liability to psychotic experiences is shared with schizophrenia and/or other neuropsychiatric disorders and traits and identify genetic loci associated with psychotic experiences.
Introduction
Psychotic experiences, such as hallucinations and delusions, are features of psychiatric disorders (eg, schizophrenia), but they are also reported by approximately 5% to 10% of the general population.1,2 Psychotic experiences are considered to be symptoms of psychiatric illness only if they co-occur with other features of that disorder, including some aspect of psychosocial impairment. It is currently unclear whether psychotic experiences in the general population are: (1) on a spectrum that, at the extreme, is associated specifically with schizophrenia, (2) largely unassociated with the psychotic symptoms experienced in schizophrenia and other major mental disorders, or (3) associated with liability to major mental disorders more generally.
Twin studies and genome-wide association studies (GWASs) have provided evidence that psychotic experiences are heritable (30%-50% from twin studies,3,4,5 and 3%-17% for single-nucleotide polymorphism [SNP]–heritability estimates6,7), indicating that common genetic variants play a role in their liability. There have been 3 GWASs of psychotic experiences to date in adolescent samples7,8,9 and no reported genome-wide significant findings. Although there was an initial assumption that psychotic experiences in adolescence would specifically increase the risk for schizophrenia in later life, epidemiologic evidence suggests a nonspecific increased risk for broader psychopathologic characteristics.10 However, to date, no study has found strong evidence for an association between genetic liabilities for schizophrenia or any other mental disorder and psychotic experiences.7,8,11,12,13
Although many individuals with a lifetime history of psychotic experiences have their first experience in adolescence, nearly one-quarter of first-onset psychotic experiences occur after 40 years of age.14 Our aims were to use data from the UK Biobank to identify genetic loci associated with psychotic experiences reported by adults in a population-based study, and to determine whether genetic liability to psychotic experiences is shared with schizophrenia and/or other neuropsychiatric disorders and traits.
Methods
Sample
Study individuals were from the UK Biobank, a large prospective population-based cohort study of approximately 500 000 individuals between 40 and 69 years of age who were recruited from across the UK between 2006 and 2010.15 The North West Multi-Centre Ethics Committee granted ethical approval to UK Biobank and all participants provided written informed consent. This study was conducted under UK Biobank project numbers 13310 and 14421.
Psychotic Experiences Phenotypes
A Mental Health Questionnaire (MHQ) was sent to all participants who provided an email address from July 13, 2016, to July 27, 2017, and was completed by 157 387 individuals (46.4% of those emailed; 31.4% of the total UK Biobank sample). For psychotic experiences, participants were asked about previous experience of visual hallucinations, auditory hallucinations, delusions of reference, delusions of persecution, as well as how often these experiences occurred and how distressing they found them (eAppendix in the Supplement). Individuals with a diagnosis of schizophrenia, bipolar disorder, or any other psychotic disorder were identified using all available sources (hospital records, death records, or self-report at the interview or from the MHQ) and were excluded from all analyses (full details in eMethods 1 in the Supplement).
We selected 3 primary phenotypes for GWASs (eFigure 1 in the Supplement): (1) any psychotic experience defined as a positive response to any of the 4 symptom questions (UK Biobank field IDs: 20463, 20468, 20471, and 20474); (2) a distressing psychotic experience, defined as any psychotic experience that was rated as “a bit,” “quite,” or “very” distressing (UK Biobank field ID: 20462); and (3) multiple occurrences of psychotic experiences, defined as any psychotic experience that occurred on more than 1 occasion (UK Biobank field IDs: 20465, 20470, 20473, and 20476). As a comparator group, we included individuals who provided a negative response to all 4 psychotic experience symptom questions. In addition, we investigated each individual psychotic experience symptom for association with polygenic risk scores (PRSs).
Genetic Data
Genetic data for the study participants were provided by UK Biobank and the imputation and quality control procedures are fully described elsewhere.16 The data release contained 488 377 participants assayed on either the UK Biobank Axiom or the UK BiLEVE Axiom purpose-built arrays at the Affymetrix Research Services Laboratory. Standard quality control procedures were applied prior to imputation using Haplotype Reference Consortium17 and UK10K haplotype reference18 panels. We applied additional quality control filters to select high-quality SNPs,19 minor allele frequency greater than 0.01, imputation score greater than 0.8, missingness less than 0.05, Hardy-Weinberg equilibrium P value greater than 1 × 10−6 and removed SNPs imputed by the UK10K haplotype reference18 data set in accordance with guidance from the UK Biobank (http://www.ukbiobank.ac.uk/2017/07/important-note-about-imputed-genetics-data/). One member from each related pair with a kinship coefficient greater than 0.15 was excluded from analyses, preferentially retaining individuals who had experienced a psychotic experience, and were otherwise removed at random.
Analyses were restricted to individuals with a self-reported British and Irish ethnicity (UK Biobank field ID: 21000) and principal components supplied by UK Biobank16 (UK Biobank field ID: 22009) were used to make additional exclusions and control for population structure (described in eMethods 2 and eFigures 2 and 3 in the Supplement).
GWAS Analysis
To identify genetic risk variants for psychotic experiences, association analysis was performed in SNPTEST, version 2.5.420 using bgen, version 1.2 imputed dosage data.21 More than 7.5 million SNPs were included in each GWAS. An additive logistic regression model was used including as covariates the genotyping array, the top 5 principal components (as recommended for most GWAS approaches22), and any additional principal components from the first 20 that were nominally associated (P < .05) with the GWAS phenotype in a logistic regression. To obtain relatively independent index SNPs, linkage disequilibrium (LD) clumping was performed in PLINK23 (r2 < 0.1; P < 1 × 10−4; window size, <3 MB) for each GWAS using a reference panel of 1000 randomly selected individuals with confirmed European ancestry in the UK Biobank. Functional annotation was conducted using FUMA24 and LDSC25 was used to calculate the LD score intercept and heritability on the observed scale using the summary statistics from each psychotic experience GWAS.
Validation Analyses in the Avon Longitudinal Study of Parents and Children Cohort
To assess the reproducibility of the psychotic experience GWAS, we used the summary statistics from the GWAS of any psychotic experience to target psychotic experiences in the Avon Longitudinal Study of Parents and Children (ALSPAC) cohort,26,27 which have been previously described.11,28 The psychotic experience PRS was generated for each ALSPAC participant using the widely used method29 and logistic regression was used to test for an association between PRS and psychotic experiences reported at 12 and 18 years of age (eMethods 3 in the Supplement).
Genetic Correlations
LDSC25,30 was used to calculate the genetic correlation between the GWAS of any psychotic experience and other psychiatric and personality traits. External GWAS data sets (that did not include data from the UK Biobank where possible) used to generate the correlations included schizophrenia,31 bipolar disorder,32 major depressive disorder,33 attention-deficit/hyperactivity disorder (ADHD),34 autism spectrum disorder,35 neuroticism,36 and intelligence.37 A Bonferroni correction was applied to control for multiple testing.
Polygenic Risk Scores
PRSs were generated to investigate additional psychotic experience phenotypes that were not sufficiently powered for genetic correlation analyses. We selected the same summary statistics used for genetic correlations to create the risk scores for UK Biobank participants using the method described by the Psychiatric Genomics Consortium29 and detailed in eMethods 4 in the Supplement. The intelligence GWAS summary statistics used in this study specifically excluded participants from the UK Biobank (74 214 individuals remaining). None of the other training sets included the UK Biobank as a contributing sample, although we were not able to test for duplicates at the genotype level and so cannot rule duplicates out. Given this potential for sample overlap with the training sets, PRS results should be treated with a degree of caution and are used primarily to support and extend findings from genetic correlation analysis (which allows for overlapping samples). The primary analysis used standardized scores generated from SNPs with a discovery sample P value threshold of P ≤ .05, but associations at 10 other P value thresholds were also tested. A logistic regression model was used to test the association of each PRS with various psychotic experience phenotypes, covarying for the first 5 principal components and genotyping array.
Copy Number Variation
Copy number variation (CNV) calling for the UK Biobank has been described in detail elsewhere38 and is detailed in eMethods 5 in the Supplement. We compared carrier status of rare CNVs previously associated with schizophrenia39 and neurodevelopmental disorders more widely40 with the 3 primary psychotic experience phenotypes used for GWASs. Association analyses were carried out using logistic regression and included age, sex, and genotyping array as covariates.
Results
A total of 7803 (5.0%; 60.0% women and 40.0% men; mean [SD] age, 62.7 [7.7] years) individuals from the UK Biobank who completed the MHQ reported at least 1 psychotic experience, 3012 individuals rated the psychotic experience as distressing, and a total of 4388 individuals reported multiple occurrences of at least 1 psychotic experience. A total of 147 461 individuals (56.0% women and 44.0% men; mean [SD] age, 64.1 [7.6] years) who reported no psychotic experiences constituted the comparison group for our association analyses. The mean (SD) age of the first psychotic experience was 31.6 (17.6) years (eFigure 4 in the Supplement), with 2341 of 6654 (35.2%) first occurring before the age of 20 years or for as long as the participant could remember, 2137 (32.1%) between the ages of 20 and 39 years, and 2176 (32.7%) between the ages of 40 and 76 years. We excluded 198 individuals who had a diagnosis of schizophrenia, 818 with bipolar disorder, and 346 with other psychotic disorders from all genetic analyses (detailed in eMethods 1 in the Supplement).
Genome-Wide Association Studies
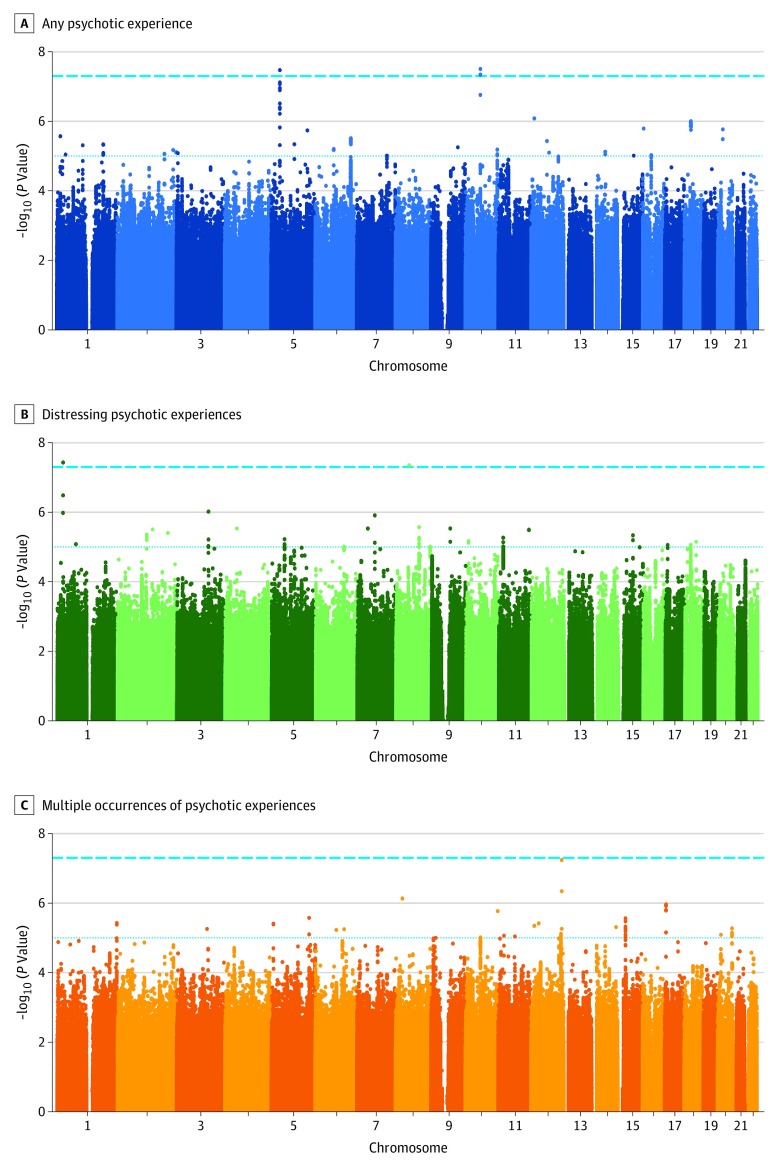
The GWAS of any psychotic experience in 6123 cases and 121 843 controls (after quality control; exclusions detailed in eMethods 6 in the Supplement) identified 2 variants that were associated at the genome-wide significance level of P < 5 × 10−8 (Figure 1, Table 1; λGC = 1.05, LD score regression intercept = 1.00): rs10994278, an intronic variant within Ankyrin-3 (ANK3 [GenBank NM_020987]) (odds ratio [OR], 1.16; 95% CI, 1.10-1.23; P = 3.06 × 10−8), and intergenic variant rs549656827 (OR, 0.61; 95% CI, 0.50-0.73; P = 3.30 × 10−8).
Figure 1. Manhattan Plot for Genome-Wide Association Study Analyses of Any Psychotic Experience, Distressing Psychotic Experiences, and Multiple Occurrences of Psychotic Experiences.
Dashed lines represent the genome-wide significance level of P < 5 × 10−8, and dotted lines represent P < 1 × 10−5.
Table 1. Genome-Wide Significant Associationsa.
| GWAS Phenotype | Single-Nucleotide Polymorphism | Chromosome | Base Position | A1 | OR (95% CI) | P Value | Position | Nearest Gene |
|---|---|---|---|---|---|---|---|---|
| Any PE | rs10994278 | 10 | 62009219 | T | 1.16 (1.10-1.23) | 3.06 × 10−8 | Intronic | ANK3 |
| Any PE | rs549656827 | 5 | 35349428 | G | 0.61 (0.50-0.73) | 3.30 × 10−8 | Intergenic | PRLR |
| Distressing PE | rs75459873 | 1 | 24256312 | G | 0.66 (0.56-0.78) | 3.78 × 10−8 | Intronic | CNR2 |
| Distressing PE | rs3849810 | 8 | 53009606 | A | 1.22 (1.13-1.31) | 4.55 × 10−8 | Intergenic | ST18 |
Abbreviations: A1, risk allele; GWAS, genome-wide association study; OR, odds ratio; PE, psychotic experience.
Genome-wide significant associations (P < 5 × 10−8) from GWAS analyses.
A second GWAS restricting the cases to 2143 individuals with distressing psychotic experiences identified 2 genome-wide significance variants (Table 1; λGC = 1.03, LD score intercept = 1.01): rs75459873, intronic to cannabinoid receptor 2 (CNR2 [GenBank NM_001841]) (OR, 0.66; 95% CI, 0.56-0.78; P = 3.78 × 10−8), and intergenic variant rs3849810 (OR, 1.22; 95% CI, 1.13-1.31; P = 4.55 × 10−8).
The third GWAS restricting the cases to 3337 individuals who reported multiple occurrences of psychotic experiences did not identify any associated variants at genome-wide significance (λGC = 1.03, LD score intercept = 1.00). QQ plots for each GWAS are displayed in eFigure 5 in the Supplement and LocusZoom plots for each genome-wide significant locus are provided in eFigure 6 in the Supplement.
Single-nucleotide polymorphism–based heritability estimates calculated by LDSC25 for the GWAS of any psychotic experience was h2 = 1.71% (95% CI, 1.02%-2.40%). The other GWAS analyses did not requirements (case n >5000 and z score >441) for heritability or genetic correlation analyses with LDSC. None of the associated regions from the GWAS for any psychotic experience and distressing psychotic experience showed evidence of colocalization42 with schizophrenia,31 bipolar disorder,32 or major depressive disorder.33
Validation Analyses in ALSPAC
There was evidence of an association between the PRS calculated using the GWAS of any psychotic experience from the UK Biobank sample at the P value threshold of ≤.5 and definite psychotic experiences between 12 and 18 years of age in ALSPAC (OR, 1.13; 95% CI, 1.02-1.25; R2 = 0.002; P = .02). This finding was consistent for thresholds above P < .05 and when using measures from age 18 years only (eFigure 7 and eTable 1 in the Supplement). However, the psychotic experiences PRS was also associated with the presence of major depressive disorder at 18 years of age (OR, 1.19; 95% CI, 1.05-1.35; R2 = 0.004; P = .01).
Link Between Association at CNR2 and Cannabis Use
We investigated, but found no evidence for, a mediating or moderating association of cannabis use with the association between distressing psychotic experiences and rs75459873 at the cannabinoid receptor gene CNR2. Cannabis use itself (UK Biobank field ID: 20453) was significantly associated with distressing psychotic experiences (OR, 1.36; 95% CI, 1.32-1.40; P = 9.16 × 10−88), but rs75459873 was not associated with cannabis use (OR, 0.99; 95% CI, 0.95-1.04; P = .70). Furthermore, the association between rs75459873 and distressing psychotic experiences was unchanged in a model including cannabis use as a covariate (OR, 0.62; 95% CI, 0.52-0.75) or as an interaction term (OR, 0.59; 95% CI, 0.48-0.73; P = .31 for interaction).
Genetic Correlations
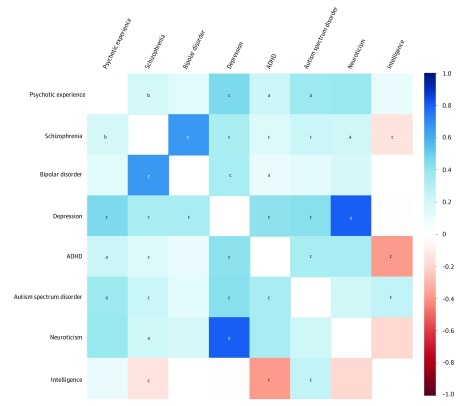
Significant genetic correlations (rg) were observed between any psychotic experience and major depressive disorder (rg = 0.46; P = 4.64 × 10−11), autism spectrum disorder (rg = 0.39; P = 1.68 × 10−4), ADHD (rg = 0.24; P = 4.61 × 10−3), and schizophrenia (rg = 0.21; P = 7.29 × 10−5). Figure 2 displays these genetic correlations and eTable 2 in the Supplement details the full results. The other psychotic experience GWASs did not meet requirements41 for genetic correlation analysis.
Figure 2. Genetic Correlation Analysis.
Color corresponds to the strength of the correlation, and the letters (a, b, and c) correspond to the statistical significance of the correlation. Positive correlations are shown in blue and negative correlations in red. ADHD indicates attention deficit/hyperactivity disorder.
aP < 6.25 × 10−3.
bP < 1.00 × 10−4.
cP < 1.00 × 10−5.
PRS Analysis
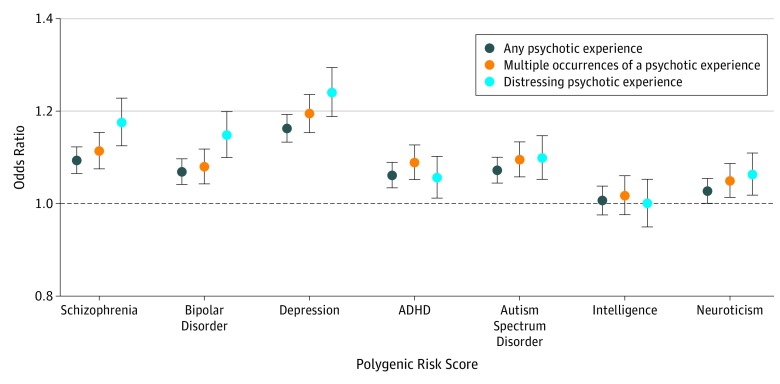
We found evidence of a weak association between any psychotic experience and genetic liability, indicated by PRS at a P < .05 threshold for SNP inclusion, for schizophrenia (OR, 1.09; 95% CI, 1.06-1.12; adjusted R2 = 0.001; P = 2.96 × 10−11), major depressive disorder (OR, 1.16; 95% CI, 1.13-1.19; R2 = 0.003; P = 1.48 × 10−30), bipolar disorder (OR, 1.07; 95% CI, 1.04-1.10; R2 = 0.001; P = 5.11 × 10−7), ADHD (OR, 1.06; 95% CI, 1.03-1.09; R2 = 0.0005; P = 5.73 × 10−6), and autism spectrum disorder (OR, 1.07; 95% CI, 1.04-1.10; R2 = 0.001; P = 1.34 × 10−5). These associations were stronger for distressing psychotic experiences (Figure 3; eTable 2 in the Supplement) and consistent across most P value thresholds (eFigures 8 and 9 in the Supplement). We also considered individual psychotic symptoms and found that PRSs for schizophrenia, bipolar disorder, depression, and ADHD were more strongly associated with delusions of persecution than with the other psychotic symptoms (eTable 3 and eFigures 10-12 in the Supplement). The association with psychotic experience phenotypes for bipolar disorder PRS and major depressive disorder PRS remained significant in analyses controlling for schizophrenia PRS (eTable 4 in the Supplement). The association with major depressive disorder PRS remained significant when individuals with a diagnosis of depression were removed (eTable 5 in the Supplement). Given the potential for sample overlap with the training sets, PRS findings should be treated with a degree of caution.
Figure 3. Polygenic Risk Score (PRS) Analysis.
The x-axis refers to the PRS tested (schizophrenia, bipolar disorder, major depressive disorder, attention deficit/hyperactivity disorder [ADHD], autism spectrum disorder, neuroticism, and intelligence), and the y-axis represents the odds ratio (OR). Points display the OR and 95% CIs (error bars) for each PRS (P < .05 single-nucleotide polymorphism [SNP] inclusion threshold) regressed against each psychotic experience phenotype. Plots displaying multiple P value SNP inclusion thresholds are shown in eFigures 8 and 9 in the Supplement.
Copy Number Variation
Individuals reporting distressing psychotic experiences in particular had an increased burden of CNVs previously associated with schizophrenia (OR, 2.04; 95% CI, 1.39-2.98; P = 2.49 × 10−4) and neurodevelopmental disorders (OR, 1.75; 95% CI, 1.24-2.48; P = 1.41 × 10−3). There was evidence of an increased burden of these CNVs in individuals reporting any psychotic experience, but not multiple occurrences of psychotic experiences (Table 2).
Table 2. Association of Psychotic Experience Phenotypes With CNVsa.
| Phenotype | Rate, No./Total No. (%) | OR (95% CI) | P Value | |
|---|---|---|---|---|
| Case | Control | |||
| Schizophrenia CNVs | ||||
| Any PE | 59/5829 (1.0) | 572/115807 (0.5) | 1.54 (1.18-2.01) | 1.60 × 10−3 |
| Distressing PE | 28/2046 (1.4) | 572/115807 (0.5) | 2.04 (1.39-2.98) | 2.49 × 10−4 |
| Multiple occurrence of PE | 28/3177 (0.9) | 572/115807 (0.5) | 1.34 (0.91-1.94) | .14 |
| Neurodevelopmental disorder CNVs | ||||
| Any PE | 83/5829 (1.4) | 1058/115807 (0.9) | 1.54 (1.23-1.92) | 1.89 × 10−4 |
| Distressing PE | 34/2046 (1.7) | 1058/115807 (0.9) | 1.75 (1.24-2.48) | 1.41 × 10−3 |
| Multiple occurrence of PEs | 38/3177 (1.2) | 1058/115807 (0.9) | 1.30 (0.92-1.78) | .14 |
Abbreviations: CNV, copy number variant; OR, odds ratio; PE, psychotic experience.
All schizophrenia-associated CNVs are also included in neurodevelopmental disorders.
Discussion
We conducted the largest genetic-association study of psychotic experiences, to our knowledge, using the population-based UK Biobank sample and found evidence of a shared genetic liability between psychotic experiences and several psychiatric disorders, which included, but was not specific to, schizophrenia. Genetic correlation analysis identified significant genetic correlations between psychotic experiences and major depressive disorder (rg = 0.46), autism spectrum disorder (rg = 0.39), ADHD (rg = 0.24), and schizophrenia (rg = 0.21). Polygenic risk score analyses identified associations between psychotic experiences and genetic liability for schizophrenia, major depressive disorder, bipolar disorder, ADHD, and autism spectrum disorder, and we found particular enrichment of these PRS scores in distressing psychotic experiences and for delusions of persecution. However, given the possibility of sample overlap between the UK Biobank and the training sets, the PRS findings should be treated with caution. The mechanisms underlying the high genetic correlation between depression and psychotic experiences cannot be discerned from our analysis, but one possibility is that the psychotic experiences have arisen in the context of mood-related changes, consistent with the strong associations between psychotic experiences and depressive symptoms observed in population-based studies.43 Nonetheless, when individuals with a lifetime history of depression were excluded from PRS analyses, the association remained, indicating that the findings are not wholly attributable to depression.
We found an increased burden of CNVs previously associated with schizophrenia and neurodevelopmental disorders more widely in individuals with any psychotic symptoms and distressing psychotic symptoms, although the association was stronger for those with distressing psychotic experiences. All schizophrenia-associated CNVs are also associated with neurodevelopmental disorders such as intellectual disability and autism spectrum disorder; in fact, penetrance is higher in individuals with these disorders.39 Furthermore, CNVs in the UK Biobank have been associated with a range of outcomes, including cognitive performance38,44 and depression,45 adding strength to our findings of a lack of specificity for genetic risk of psychotic experiences.
Several studies have demonstrated that psychopathologic conditions in the population are best described by a bifactor model with a common latent trait as well as specific traits, and that psychotic experiences index the more severe end of the common or shared trait.46,47 Our findings of nonspecificity of genetic risk for psychotic experiences with risk for other disorders are consistent with those of previous studies.7,8,11,12 Nonetheless, despite lacking specificity, our results suggest that incorporating questions about distress to self-reported assessments of psychotic experiences may allow a more valid identification of experiences that index liability of schizophrenia and major mental health disorders.
In the largest GWAS of psychotic experiences to date, we identified 4 genome-wide significant loci. However, consistent with other studies,7,11 the heritability estimate (1.71%) was low and, given that the variance explained in our PRS analysis was also low, the findings suggest that understanding the genetics of psychotic experiences is unlikely to have an important effect on understanding the genetics of schizophrenia specifically.
The primary GWAS findings are related to intronic variants in ANK3 and CNR2. The GWAS of any psychotic experience identified 2 significant loci, the most significant of which was indexed by rs10994278, an intronic variant to ANK3 (OR, 1.16; 95% CI, 1.10-1.23; P = 3.06 × 10−8). The ANK3 gene encodes ankyrin-G, a protein that has been shown to regulate the assembly of voltage-gated sodium channels and is essential for normal synaptic function.48 ANK3 is one of strongest and most replicated genes for bipolar disorder,32 and variants within ANK3 have also been associated in the Psychiatric Genomics Consortium cross-disorder GWAS,49 and in a rare variant analysis of autism spectrum disorder.50
The GWAS of distressing psychotic experiences also identified 2 significant loci, the most significant of which was indexed by rs75459873, an intronic variant to CNR2 (OR, 0.66; 95% CI, 0.56-0.78; P = 3.78 × 10−8). CNR2 encodes for CB2, 1 of 2 well-characterized cannabinoid receptors (CB1 being the other). Several lines of evidence have implicated the endocannabinoid system in psychiatric disorders, including schizophrenia51,52 and depression.53 The main psychoactive agent of cannabis, Δ9-tetrahydrocannabinol, can cause acute psychotic symptoms and cognitive impairment.54 Given that cannabis use is strongly associated with psychotic experiences, we tested, but found no evidence for, a mediating or moderating effect of cannabis use on the association of rs75459873 and distressing psychotic experiences. However, while no evidence was found in this study, a mediating effect of cannabis use cannot be ruled out given the relatively low power of such analyses and the potential measurement error in cannabis use assessed via lifetime self-report. Independent replication of genetic loci identified in this study will be required to further understand their role in subclinical psychotic experiences.
Strengths and Limitations
Strengths of this study include the large sample size (approximately 10 times that of previous studies), the use of an adult cohort, and the use of multiple psychotic experience phenotypes, which increase confidence in our findings. This study also has some limitations. Although we used all available information to identify and remove individuals with a psychotic disorder, it remains possible that some individuals were not identified and remained in the analysis. A further limitation of this study relates to the retrospective measurement of lifetime psychotic experiences by self-report from an online questionnaire, as this increases the likelihood of measurement error. A further limitation is the evidence of a “healthy volunteer” selection bias for the participants recruited to the UK Biobank and the sample cannot be therefore considered representative of the general population.55 We also found that the participants who completed the MHQ had significantly higher intelligence and lower schizophrenia, depression, and neuroticism PRS compared with UK Biobank participants who did not complete the MHQ (eTable 6 in the Supplement). Last, we were not able to entirely deduplicate the UK Biobank individuals from all the external data sets used for the PRS analysis; thus, these findings should be treated with caution.
Conclusions
In the largest GWAS of psychotic experiences from the population-based UK Biobank sample, we found support for a shared genetic liability between psychotic experiences and several psychiatric disorders including schizophrenia, major depressive disorder, bipolar disorder, and neurodevelopmental disorders, indicating that psychotic experiences are not specifically associated with schizophrenia, but rather with a general risk for mental health disorders.
eAppendix. Unusual and Psychotic Experience Questions in UK Biobank
eMethods 1. Defining Schizophrenia, Bipolar Disorder and Psychotic Disorder
eMethods 2. Defining European Genetic Ancestry
eMethods 3. Replication in ALSPAC Cohort
eMethods 4. Calculation of Polygenic Risk Scores in UK Biobank
eMethods 5. CNV Calling
eMethods 6. Individuals Excluded
eFigure 1. Psychotic Experience Phenotypes
eFigure 2. Principal Component Analysis
eFigure 3. Principal Component Analysis Post-Exclusions
eFigure 4. Age of Onset of Psychotic Experiences
eFigure 5. QQ Plots of Psychotic Experience GWAS
eFigure 6. LocusZoom Plots of Genome-Wide Significant Loci
eFigure 7. PRS Analysis of Psychotic Experience Symptoms in ALSPAC
eFigure 8. PRS Analysis of Psychotic Experience Phenotypes
eFigure 9. PRS Analysis of Psychotic Experience Phenotypes
eFigure 10. PRS Analysis of Psychotic Experience Symptoms
eFigure 11. PRS Analysis of Psychotic Experience Symptoms
eFigure 12. PRS Analysis of Psychotic Experience Symptoms
eTable 1. Validation of Psychotic Experience GWAS in ALSPAC Cohort
eTable 2. Genetic Correlations
eTable 3. Polygenic Risk Score Analysis
eTable 4. Polygenic Risk Score Analysis—Covarying for Schizophrenia PRS
eTable 5. Depression Polygenic Risk Score Analysis—Removing Individuals With Depression
eTable 6. Association of Neuropsychiatric Polygenic Risk Scores With Completion of the Mental Health Questionnaire
References
- 1.McGrath JJ, Saha S, Al-Hamzawi A, et al. Psychotic experiences in the general population: a cross-national analysis based on 31,261 respondents from 18 countries. JAMA Psychiatry. 2015;72(7):697-705. doi: 10.1001/jamapsychiatry.2015.0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Os J, Linscott RJ, Myin-Germeys I, Delespaul P, Krabbendam L. A systematic review and meta-analysis of the psychosis continuum: evidence for a psychosis proneness-persistence-impairment model of psychotic disorder. Psychol Med. 2009;39(2):179-195. doi: 10.1017/S0033291708003814 [DOI] [PubMed] [Google Scholar]
- 3.Zavos HM, Freeman D, Haworth CM, et al. Consistent etiology of severe, frequent psychotic experiences and milder, less frequent manifestations: a twin study of specific psychotic experiences in adolescence. JAMA Psychiatry. 2014;71(9):1049-1057. doi: 10.1001/jamapsychiatry.2014.994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronald A. Recent quantitative genetic research on psychotic experiences: new approaches to old questions. Curr Opin Behav Sci. 2015;2:81-88. doi: 10.1016/j.cobeha.2014.10.001 [DOI] [Google Scholar]
- 5.Ronald A, Pain O. A systematic review of genome-wide research on psychotic experiences and negative symptom traits: new revelations and implications for psychiatry. Hum Mol Genet. 2018;27(R2):R136-R152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sieradzka D, Power RA, Freeman D, Cardno AG, Dudbridge F, Ronald A. Heritability of individual psychotic experiences captured by common genetic variants in a community sample of adolescents. Behav Genet. 2015;45(5):493-502. doi: 10.1007/s10519-015-9727-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pain O, Dudbridge F, Cardno AG, et al. Genome-wide analysis of adolescent psychotic-like experiences shows genetic overlap with psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. 2018;177(4):416-425. doi: 10.1002/ajmg.b.32630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zammit S, Hamshere M, Dwyer S, et al. A population-based study of genetic variation and psychotic experiences in adolescents. Schizophr Bull. 2014;40(6):1254-1262. doi: 10.1093/schbul/sbt146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega-Alonso A, Ekelund J, Sarin AP, et al. Genome-wide association study of psychosis proneness in the Finnish population. Schizophr Bull. 2017;43(6):1304-1314. doi: 10.1093/schbul/sbx006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher HL, Caspi A, Poulton R, et al. Specificity of childhood psychotic symptoms for predicting schizophrenia by 38 years of age: a birth cohort study. Psychol Med. 2013;43(10):2077-2086. doi: 10.1017/S0033291712003091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones HJ, Stergiakouli E, Tansey KE, et al. Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry. 2016;73(3):221-228. doi: 10.1001/jamapsychiatry.2015.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sieradzka D, Power RA, Freeman D, et al. Are genetic risk factors for psychosis also associated with dimension-specific psychotic experiences in adolescence? PLoS One. 2014;9(4):e94398. doi: 10.1371/journal.pone.0094398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones HJ, Heron J, Hammerton G, et al. ; 23 and Me Research Team . Investigating the genetic architecture of general and specific psychopathology in adolescence. Transl Psychiatry. 2018;8(1):145. doi: 10.1038/s41398-018-0204-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGrath JJ, Saha S, Al-Hamzawi AO, et al. Age of onset and lifetime projected risk of psychotic experiences: cross-national data from the World Mental Health Survey. Schizophr Bull. 2016;42(4):933-941. doi: 10.1093/schbul/sbw011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. doi: 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bycroft C, Freeman C, Petkova D, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint. Posted online July 20, 2017. bioRxiv 166298. doi: 10.1101/166298 [DOI]
- 17.McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279-1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Howie B, McCarthy S, et al. ; UK10K Consortium . Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DJ, Escott-Price V, Davies G, et al. Genome-wide analysis of over 106 000 individuals identifies 9 neuroticism-associated loci. Mol Psychiatry. 2016;21(11):1644. doi: 10.1038/mp.2016.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906-913. doi: 10.1038/ng2088 [DOI] [PubMed] [Google Scholar]
- 21.Band G, Marchini J. BGEN: a binary file format for imputed genotype and haplotype data. Preprint. Posted online May 2, 2018. bioRxiv 308296. doi: 10.1101/308296 [DOI]
- 22.Tucker G, Price AL, Berger B. Improving the power of GWAS and avoiding confounding from population stratification with PC-Select. Genetics. 2014;197(3):1045-1049. doi: 10.1534/genetics.114.164285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyd A, Golding J, Macleod J, et al. Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42(1):111-127. doi: 10.1093/ije/dys064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser A, Macdonald-Wallis C, Tilling K, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC Mothers Cohort. Int J Epidemiol. 2013;42(1):97-110. doi: 10.1093/ije/dys066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zammit S, Kounali D, Cannon M, et al. Psychotic experiences and psychotic disorders at age 18 in relation to psychotic experiences at age 12 in a longitudinal population-based cohort study. Am J Psychiatry. 2013;170(7):742-750. doi: 10.1176/appi.ajp.2013.12060768 [DOI] [PubMed] [Google Scholar]
- 29.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421-427. doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan B, Finucane HK, Anttila V, et al. ; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 . An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236-1241. doi: 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium; CRESTAR Consortium . Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381-389. doi: 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahl EA, Breen G, Forstner AJ, et al. ; eQTLGen Consortium; BIOS Consortium; Bipolar Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51(5):793-803. doi: 10.1038/s41588-019-0397-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wray NR, Ripke S, Mattheisen M, et al. ; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50(5):668-681. doi: 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demontis D, Walters RK, Martin J, et al. ; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team . Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63-75. doi: 10.1038/s41588-018-0269-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grove J, Ripke S, Als TD, et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team . Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51(3):431-444. doi: 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Moor MH, van den Berg SM, Verweij KJ, et al. ; Genetics of Personality Consortium . Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with major depressive disorder. JAMA Psychiatry. 2015;72(7):642-650. doi: 10.1001/jamapsychiatry.2015.0554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savage JE, Jansen PR, Stringer S, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50(7):912-919. doi: 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kendall KM, Rees E, Escott-Price V, et al. Cognitive performance among carriers of pathogenic copy number variants: analysis of 152,000 UK Biobank subjects. Biol Psychiatry. 2017;82(2):103-110. doi: 10.1016/j.biopsych.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 39.Rees E, Kendall K, Pardiñas AF, et al. Analysis of intellectual disability copy number variants for association with schizophrenia. JAMA Psychiatry. 2016;73(9):963-969. doi: 10.1001/jamapsychiatry.2016.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46(10):1063-1071. doi: 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium . LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272-279. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickrell JK, Berisa T, Liu JZ, Ségurel L, Tung JY, Hinds DA. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet. 2016;48(7):709-717. doi: 10.1038/ng.3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Rossum I, Dominguez MD, Lieb R, Wittchen HU, van Os J. Affective dysregulation and reality distortion: a 10-year prospective study of their association and clinical relevance. Schizophr Bull. 2011;37(3):561-571. doi: 10.1093/schbul/sbp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kendall KM, Bracher-Smith M, Fitzpatrick H, et al. Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: analysis of the UK Biobank. Br J Psychiatry. 2019;214(5):297-304. doi: 10.1192/bjp.2018.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kendall KM, Rees E, Bracher-Smith M, et al. Association of rare copy number variants with risk of depression. JAMA Psychiatry. 2019;76(8):818-825. doi: 10.1001/jamapsychiatry.2019.0566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lahey BB, Krueger RF, Rathouz PJ, Waldman ID, Zald DH. A hierarchical causal taxonomy of psychopathology across the life span. Psychol Bull. 2017;143(2):142-186. doi: 10.1037/bul0000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stochl J, Khandaker GM, Lewis G, et al. Mood, anxiety and psychotic phenomena measure a common psychopathological factor. Psychol Med. 2015;45(7):1483-1493. doi: 10.1017/S003329171400261X [DOI] [PubMed] [Google Scholar]
- 48.Leussis MP, Madison JM, Petryshen TL. Ankyrin 3: genetic association with bipolar disorder and relevance to disease pathophysiology. Biol Mood Anxiety Disord. 2012;2:18. doi: 10.1186/2045-5380-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381(9875):1371-1379. doi: 10.1016/S0140-6736(12)62129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bi C, Wu J, Jiang T, et al. Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum Mutat. 2012;33(12):1635-1638. doi: 10.1002/humu.22174 [DOI] [PubMed] [Google Scholar]
- 51.Lu HC, Mackie K. An introduction to the endogenous cannabinoid system. Biol Psychiatry. 2016;79(7):516-525. doi: 10.1016/j.biopsych.2015.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore TH, Zammit S, Lingford-Hughes A, et al. Cannabis use and risk of psychotic or affective mental health outcomes: a systematic review. Lancet. 2007;370(9584):319-328. doi: 10.1016/S0140-6736(07)61162-3 [DOI] [PubMed] [Google Scholar]
- 53.Huang WJ, Chen WW, Zhang X. Endocannabinoid system: role in depression, reward and pain control. Mol Med Rep. 2016;14(4):2899-2903. doi: 10.3892/mmr.2016.5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manseau MW, Goff DC. Cannabinoids and schizophrenia: risks and therapeutic potential. Neurotherapeutics. 2015;12(4):816-824. doi: 10.1007/s13311-015-0382-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fry A, Littlejohns TJ, Sudlow C, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186(9):1026-1034. doi: 10.1093/aje/kwx246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Unusual and Psychotic Experience Questions in UK Biobank
eMethods 1. Defining Schizophrenia, Bipolar Disorder and Psychotic Disorder
eMethods 2. Defining European Genetic Ancestry
eMethods 3. Replication in ALSPAC Cohort
eMethods 4. Calculation of Polygenic Risk Scores in UK Biobank
eMethods 5. CNV Calling
eMethods 6. Individuals Excluded
eFigure 1. Psychotic Experience Phenotypes
eFigure 2. Principal Component Analysis
eFigure 3. Principal Component Analysis Post-Exclusions
eFigure 4. Age of Onset of Psychotic Experiences
eFigure 5. QQ Plots of Psychotic Experience GWAS
eFigure 6. LocusZoom Plots of Genome-Wide Significant Loci
eFigure 7. PRS Analysis of Psychotic Experience Symptoms in ALSPAC
eFigure 8. PRS Analysis of Psychotic Experience Phenotypes
eFigure 9. PRS Analysis of Psychotic Experience Phenotypes
eFigure 10. PRS Analysis of Psychotic Experience Symptoms
eFigure 11. PRS Analysis of Psychotic Experience Symptoms
eFigure 12. PRS Analysis of Psychotic Experience Symptoms
eTable 1. Validation of Psychotic Experience GWAS in ALSPAC Cohort
eTable 2. Genetic Correlations
eTable 3. Polygenic Risk Score Analysis
eTable 4. Polygenic Risk Score Analysis—Covarying for Schizophrenia PRS
eTable 5. Depression Polygenic Risk Score Analysis—Removing Individuals With Depression
eTable 6. Association of Neuropsychiatric Polygenic Risk Scores With Completion of the Mental Health Questionnaire