Abstract
Super-resolution microscopy, or nanoscopy, revolutionized the field of cell biology, enabling researchers to visualize cellular structures with nanometric resolution, single-molecule sensitivity, and in multiple colors. However, the impact of these techniques goes beyond biology as the fields of nanotechnology and nanomedicine can greatly benefit from them, as well. Nanoscopy can visualize nanostructures in vitro and in cells and can contribute to the characterization of their structures and nano–bio interactions. In this Perspective, we discuss the potential of super-resolution imaging for nanomedicine research, its technical challenges, and the future developments we envision for this technology.
In the past few decades, the advent of super-resolution optical microscopy, or nanoscopy, overcame the diffraction limit of light and extended the realm of fluorescence microscopy to the nanoscale.1 This advance had significant impact on cell biology, enabling researchers to unveil the structural details of subdiffraction cellular architectures. Although the initial application of nanoscopy was imaging cellular structures,2 its potential goes beyond biology. In the past 5 years, the use of fluorescence nanoscopy has been extended to include nanotechnology and material science, as well.3 In this Perspective, we reflect on the potential of super-resolution microscopy to contribute to the field of nanomedicine with a focus on its ability to shine new light on the properties and behavior of nanomaterials in vitro and in cells. We discuss the main technical challenges and abilities of the different methods (see Figure 1), providing a guide to nanotechnologists approaching these new and exciting techniques. Finally, we envision the role of nanoscopy in promoting a more rational design of nanomaterials for medicine.
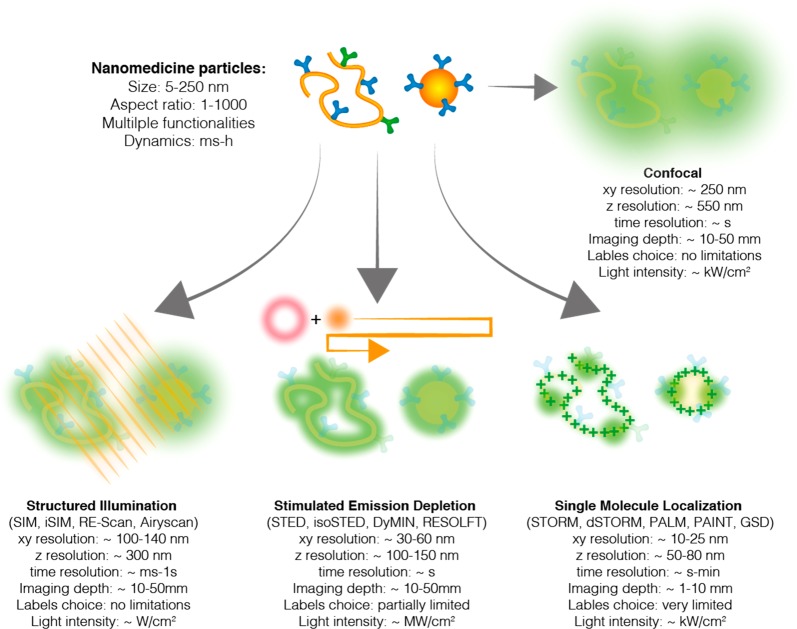
Figure 1.
Super-resolution microscopy. Schematic representation of super-resolution methods and their performances. Three main families can be identified: (i) structured illumination microscopy (SIM) methods and their point scanning variations where the sample is irradiated with patterned illumination and the resolution is enhanced through mathematical reconstruction; (ii) stimulated emission depletion (STED) where a de-excitation doughnut is scanned around the excitation beam, resulting in the confinement of the excitation and subsequent enhancement of resolution; and (iii) single-molecule localization microscopy (SMLM) where individual fluorophores are sequentially localized and the image reconstructed in a pointillistic fashion. Many SMLM variants are available, depending on the mechanisms of single-molecule control: stochastic optical reconstruction microscopy (STORM), photoactivated localization microscopy (PALM), ground-state depletion (GSD), and point accumulation for imaging nanotopography (PAINT). Notably, it is important to compare the techniques’ performances with the properties of the material under study (top left).
Why Super-resolution for Nanomedicine?
The field of nanomedicine is currently in a critical moment. Despite numerous reports in the past decade describing novel nanomaterials with therapeutic potential, clinical translation is still unsatisfactory and only a few drug and gene carriers are FDA- and EMA-approved.4 Several recent reviews have discussed what must be improved upon in the current approach to design the next generation of effective therapeutic nanomaterials.5−9 In this framework, there is consensus that one of the limiting factors is our lack of basic knowledge on nanocarrier behavior in the biological environment, that is, the nanomedicine black box (see Figure 2). Understanding key phenomena such as protein corona formation, immune escape, extravasation, and targeting is critical for the rational design of effective materials. Novel spectroscopy and microscopy techniques that can reveal the behavior of nanomaterials in complex cellular and tissue environments are of outmost importance, and super-resolution imaging can play an important role due to its appealing features.
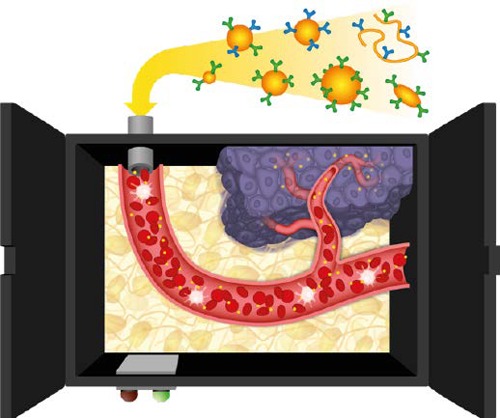
Figure 2.
Opening the nanomedicine black box. Pictorial representation of the journey of a nanoparticle from the injection site to the target tissue (cancer). Several barriers have to be overcome in blood (protein corona, immune system), tissues (extravasation, matrix diffusion), and cells (membrane targeting, cell uptake, endosomal escape, and cell trafficking). Super-resolution imaging can shed light on the mechanisms of these phenomena, contributing to opening the black box of nanomedicine.
First, super-resolution microscopy enables nanometric resolution whereby researchers can image nanostructured materials down to tens of nanometers in vitro and in cells. This functionality paves the way for determining nanoparticles’ sizes and morphologies accurately, both before and after cell administration. Second, nanoscopy retains a key feature of conventional fluorescence imaging, the multicolor ability, which is of paramount importance for imaging interactions between two or more molecular partners and is one of the reasons for the success of nanoscopy in biology. Being able to label the materials of interest in one or more colors and biomolecular partners in different colors enables the study of key binding events such as protein corona formation, immune recognition, and targeting. Finally, the molecular specificity of nanoscopy labeling enables researchers not only to track single nanoparticles but also to follow a specific molecular species in space and time, including tracking payload molecules’ loading and release and identifying changes in the nanoparticles’ molecular structures and compositions.
This information, which is typically inaccessible or accessible only with indirect methods, is critical for the design of new nanoparticles and elucidating their function, contributing to opening the “nanomedicine black box” and providing a guide to understand the biological barriers to the therapeutic target.
Nanomedicine in Multicolor
Researchers currently study the structure of nanoparticles with a combination of ensemble techniques (dynamic light scattering,10 DLS; small-angle X-ray scattering11) and imaging (atomic force microscopy,12 electron microscopy13). Super-resolution microscopy can complement these methods, providing molecule-specific information at the single-nanoparticle level. Due to fluorescence labeling, a precise molecular component can be tracked. As a relevant example, a recent report demonstrated the ability of stochastic optical reconstruction microscopy (STORM) to measure the loss of antibodies due to interactions with serum proteins.14 Moreover, multiple components can be visualized at the same time, paving the way for the intraparticle mapping of features and functionalities. Meijer and co-workers used this capability to image multiple monomer types inside a supramolecular polymer, highlighting their positions and their different dynamics inside the fibers.15 Notably, super-resolution microscopy routinely enables two- or three-color imaging and can be expanded to a theoretically unlimited number of channels in the case of DNA point accumulation for imaging nanotopography (DNA-PAINT),16 enabling the full characterization of complex and multifunctional drug and gene carriers.
Dynamic Information from Static Pictures
The multicolor ability of nanoscopy can also be exploited for an unexpected use: studying the dynamic behavior of self-assembled materials. Supramolecular drug carriers are endowed with novel properties such as adaptivity and responsivity to external stimuli because they continuously rearrange their conformations through molecule exchange or diffusion. However, most super-resolution methods have limited temporal resolution (see Figure 1) and appear unsuitable for live imaging of materials. Labeling two batches of the same material with two separate colors and mixing them after equilibration enables researchers to follow the dynamic exchange of molecules between the species. In this case, the temporal information is encrypted in the color: It is possible to track a “red” molecule jumping into a “green” structure and, thus, follow monomer exchange in time and space. This strategy has been used to probe the dynamics of supramolecular fibers17−19 and in polymer assemblies20in vitro at the single-particle and molecular levels by two-color STORM. Tracking the exchange of monomers between structures reveals the underlying kinetics and mechanisms. The use of smart labeling strategies will enable researchers to probe the dynamics of supramolecular drug carriers such as micelles, nanoparticles, and liposomes while in biological fluids and cells.21
A Quantitative Look: Toward Molecular Counting
A typical strategy for obtaining functional materials is to conjugate moieties such as targeting ligands, contrast agents, and bioactive compounds covalently or noncovalently on the particle surface. However, after synthesis, it is not straightforward to get quantitative estimates of the numbers and the activities of the active functionalities on a single-particle basis. Estimates can be achieved using single-molecule-based techniques such as STORM, photoactivated localization microscopy (PALM), and DNA-PAINT, as they are endowed with single-molecule sensitivity and enable a reliable quantification.22−27 In a recent example, van Oijen and co-workers quantified the amount of antibodies present on liposome surfaces and their stoichiometry using single-molecule imaging.28 Moreover, a smart labeling strategy can be used to quantify the fraction of antibody that is correctly oriented for binding using DNA-PAINT.29 The extension of these methods to other types of materials and functionalizations is a logical next step to understanding the behavior of targeted materials quantitatively.
More than Structure: Toward Functional Imaging
The environmental sensitivity of fluorescent dyes opens new avenues in the functional imaging of nanostructures, as properties such as hydrophobicity and charge are important drivers of nanoparticles’ behavior in cells and in vivo. For this purpose, sensing fluorophores can be used in combination with single-molecule localization microscopy (SMLM) imaging. With a small change in the optical setup, it is possible to record both the position of an emitter and its spectrum; because many dyes change their emission spectra depending on the environment, this method can be used for functional imaging.30 The pioneering work of Lee and Xu demonstrated the use of spectrally resolved PAINT (sPAINT) to image the hydrophobicity of liposomes with different compositions31 and liquid nanodemixing at the interface, respectively.32 Moreover, the chemical reactivity of nanoparticles can be measured using turn-on probes and SMLM. In these methods, such as nanometer accuracy by stochastic catalytic reactions microscopy (NASCA),33 individual chemical reactions are probed in space and time, enabling researchers to map the surface reactivity of catalytic materials. Altogether, these pioneering results show that the combination of smart probes and super-resolution imaging enables researchers to go beyond simple structure visualization, offering wider functional characterization.
Imaging Materials’ Nano–Bio Interactions
A critical step for the design of effective nanomedicines is understanding and controlling the interactions of synthetic materials with biological molecules and machinery. To date, fluorescence, due to its multicolor ability, has been the primary technique for this purpose. Nanoscopy will extend this potential to nanoscale spatial resolution, unveiling nano–bio interactions at the molecular level. The colocalization of nanoparticles’ fluorescent signal with the signal of a specific cellular structure is the most used method to probe the localization and interactions of synthetic objects with a specific cellular compartment,34 for example, with endosomal vesicles to understand the pathway of internalization of nanoparticles. However, the limited resolution of conventional microscopy often makes colocalization unclear and ambiguously interpreted. Super-resolution colocalization assays will offer dramatic improvements. The next generation of nanoscale colocalization will change from correlation analysis (e.g., Pearson coefficient) to evaluations of the spatial proximity of two objects as a parameter of interaction.35−37 Recent work by Caruso, Cavalieri, and co-workers demonstrated the potential of this approach, showing how nanoscale colocalization can provide new information on endosome escape.38
Nanoscopy will also enable the direct visualization of molecular interactions. A relevant case is represented by the study of protein corona formation, where the binding between serum proteins and nanoparticles can be investigated at the single-protein level. Two recent reports demonstrated the possibility of using STORM to image corona formation with nanometric resolution and protein specificity.14,39 Dynamic light scattering and proteomics are high-throughput methods to study corona in vitro: super-resolution will enable researchers to “zoom in” at the molecular level and to follow the corona in real time inside cells. The extension of these methods to other relevant biointeractions, both in vitro or in cells, will extend our knowledge of the nanoparticles’ interactions.
Not Only Where, But Also How
Thus far, we have described the potential of super-resolution imaging to contribute to understanding cellular localization (“where”) and interactions (“with whom”) of nanoparticles. The nanometric resolution of super-resolution microscopy opens the further possibility of understanding the state of the nanostructure materials during their cellular journey (“how”). After injection into the biological environment, interactions with biomolecules and cellular structures can modify nanoparticles’ sizes, shapes, and compositions. As an example, structure illumination microscopy (SIM) has been used to monitor particle deformation during internalization.40 Interestingly, different cell lines exert different mechanical forces, inducing a proportional polymer capsule’s shrinkage. Moreover, the stability of supramolecular aggregates in serum and in cells can be followed by STORM, revealing in what state (assembled or disassembled) the carriers reach different locations.27 This information is critical because unstable carriers will disassemble too quickly and be unable to perform their functions, whereas carriers that are too stable will result in bioaccumulation, toxicity, and poor drug release.
Super-resolution for Nanomedicine Roadmap: Faster, Deeper, and Quantitative
Despite noteworthy accomplishments, super-resolution imaging is still a young technique, and future technical improvements are envisioned, several of which will be relevant for the field of nanomedicine. In this development, the increase of temporal resolution plays a pivotal role. The relatively slow nature of nanoscopy limits the imaging of fast events and/or makes the technique low-throughput. New, parallelized experimental setups,41 machine-learning analysis,42 and new fluorophores with improved photophysics43,44 are increasing speed, making fast-occurring phenomena accessible. Notably, minimal emission fluxes (MINFLUX) broke new records in speed and resolution, enabling researchers to track molecular movements with 2 nm spatial and microsecond temporal resolution.45 Recent improvements have overcome the limited permeation of nanoscopy into thick samples such as tissues and organs. The combination of clearing methods with light-sheet illumination schemes46,47 and adaptive optics48 holds great promise to image nanoparticles in thick tissues, organs, and organisms. The combination of these technologies will open the way for the use of nanoscopy in ex vivo samples, such as sections and whole organs of model animals treated with nanomedicine, and potentially also in vivo. However, so far, there are few examples of in vivo nanoscopy, including stimulated emission depletion (STED) imaging in rodents’ brains49 and adaptive optics-aided lattice light-sheet imaging on a transparent model organism.50 The use of intravital nanoscopy for the imaging of nanocarriers and nanodevices holds great promise for understanding the interactions of synthetic materials in vivo. Lastly, the development of methods for the correlative imaging of nanoparticles in vitro and in cells could represent a way to overcome the limitation of the individual techniques. In correlative imaging, the sample is imaged with two different modalities and the two images are overlaid to obtain different information from both methods and get the “best of both worlds”.51 However, this technique comes at the cost of cumbersome and technically difficult methods, and therefore, applications in nanomedicine remain limited.52,53 Combining the atomic resolution of electron microscopy, the structural information on X-ray imaging and spectroscopy, and the specific labeling and molecular imaging of nanoscopy could represent the ultimate tool to characterize therapeutic nanoparticle structures and interactions, solving open questions in nanomedicine that are currently under debate due to the lack of suitable methods of investigation.
Acknowledgments
The authors thank the entire Nanoscopy for Nanomedicine group for enlightening discussions and their contributions to shaping the ideas in this Perspective. The authors thank the Spanish Ministry of Economy, Industry and Competitiveness through Project SAF2016-75241-R, the Generalitat de Catalunya through the Centres de Recerca de Catalunya (CERCA) programme, the EuroNanoMed II platform through the NanoVax project, the Obra Social La Caixa foundation and the European Research Council (ERC- StG-757397).
The authors declare no competing financial interest.
References
- Schermelleh L.; Ferrand A.; Huser T.; Eggeling C.; Sauer M.; Biehlmaier O.; Drummen G. P. C. Super-Resolution Microscopy Demystified. Nat. Cell Biol. 2019, 21, 72–84. 10.1038/s41556-018-0251-8. [DOI] [PubMed] [Google Scholar]
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence Nanoscopy in Cell Biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Pujals S.; Feiner-Gracia N.; Delcanale P.; Voets I.; Albertazzi L. Super-Resolution Microscopy as a Powerful Tool To Study Complex Synthetic Materials. Nat. Rev. Chem. 2019, 3, 68. 10.1038/s41570-018-0070-2. [DOI] [Google Scholar]
- Anselmo A. C.; Mitragotri S. Nanoparticles in the Clinic. Bioeng. Transl. Med. 2016, 1, 10–29. 10.1002/btm2.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. W. Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater. 2016, 1, 16014. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- Ledford H. Bankruptcy Filing Worries Developers of Nanoparticle Cancer Drugs. Nature 2016, 533, 304–305. 10.1038/533304a. [DOI] [PubMed] [Google Scholar]
- Lammers T.; Kiessling F.; Ashford M.; Hennink W.; Crommelin D.; Storm G. Cancer Nanomedicine: Is Targeting Our Target?. Nat. Rev. Mater. 2016, 1, 16069. 10.1038/natrevmats.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrice M. Does Nanomedicine Have a Delivery Problem?. ACS Cent. Sci. 2016, 2, 434–437. 10.1021/acscentsci.6b00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. C. W. Nanomedicine 2.0. Acc. Chem. Res. 2017, 50, 627–632. 10.1021/acs.accounts.6b00629. [DOI] [PubMed] [Google Scholar]
- Ramos A. P.Dynamic Light Scattering Applied to Nanoparticle Characterization. In Nanocharacterization Techniques; Da Róz A. L., Ferreira M., de Lima Leite F., Oliveira O. N., Eds.; Elsevier: Amsterdam, 2017; pp 99–110. [Google Scholar]
- Li T.; Senesi A. J.; Lee B. Small Angle X-Ray Scattering for Nanoparticle Research. Chem. Rev. 2016, 116, 11128–11180. 10.1021/acs.chemrev.5b00690. [DOI] [PubMed] [Google Scholar]
- Rao A.; Schoenenberger M.; Gnecco E.; Glatzel T.; Meyer E.; Brändlin D.; Scandella L. Characterization of Nanoparticles Using Atomic Force Microscopy. J. Phys.: Conf. Ser. 2007, 61, 971–976. 10.1088/1742-6596/61/1/192. [DOI] [Google Scholar]
- Malatesta M. Transmission Electron Microscopy for Nanomedicine: Novel Applications for Long-Established Techniques. Eur. J. Histochem. 2016, 60, 2751. 10.4081/ejh.2016.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner-Gracia N.; Beck M.; Pujals S.; Tosi S.; Mandal T.; Buske C.; Linden M.; Albertazzi L. Super-Resolution Microscopy Unveils Dynamic Heterogeneities in Nanoparticle Protein Corona. Small 2017, 13, 1701631. 10.1002/smll.201701631. [DOI] [PubMed] [Google Scholar]
- Baker M. B.; Gosens R. P. J.; Albertazzi L.; Matsumoto N. M.; Palmans A. R. A.; Meijer E. W. Exposing Differences in Monomer Exchange Rates of Multicomponent Supramolecular Polymers in Water. ChemBioChem 2016, 17, 207–213. 10.1002/cbic.201500606. [DOI] [PubMed] [Google Scholar]
- Jungmann R.; Avendaño M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. Multiplexed 3D Cellular Super-Resolution Imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11, 313–318. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertazzi L.; Martinez-Veracoechea F. J.; Leenders C. M. A.; Voets I. K.; Frenkel D.; Meijer E. W. Spatiotemporal Control and Superselectivity in Supramolecular Polymers Using Multivalency. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 12203–12208. 10.1073/pnas.1303109110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujals S.; Tao K.; Terradellas A.; Gazit E.; Albertazzi L. Studying Structure and Dynamics of Self-Assembled Peptide Nanostructures Using Fluorescence and Super Resolution Microscopy. Chem. Commun. 2017, 53, 7294–7297. 10.1039/C7CC02176C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox H.; Georgiades P.; Xu H.; Waigh T. A.; Lu J. R. Self-Assembly of Mesoscopic Peptide Surfactant Fibrils Investigated by STORM Super-Resolution Fluorescence Microscopy. Biomacromolecules 2017, 18, 3481–3491. 10.1021/acs.biomac.7b00465. [DOI] [PubMed] [Google Scholar]
- Duro-Castano A.; Nebot V. J.; Niño-Pariente A.; Armiñán A.; Arroyo-Crespo J. J.; Paul A.; Feiner-Gracia N.; Albertazzi L.; Vicent M. J. Capturing “Extraordinary” Soft-Assembled Charge-Like Polypeptides as a Strategy for Nanocarrier Design. Adv. Mater. 2017, 29, 1702888. 10.1002/adma.201702888. [DOI] [PubMed] [Google Scholar]
- Raulf A.; Spahn C. K.; Zessin P. J. M.; Finan K.; Bernhardt S.; Heckel A.; Heilemann M. Click Chemistry Facilitates Direct Labelling and Super-Resolution Imaging of Nucleic Acids and Proteins. RSC Adv. 2014, 4, 30462–30466. 10.1039/C4RA01027B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis C.; Fricke F.; Hummer G.; Heilemann M. Molecule Counts in Localization Microscopy with Organic Fluorophores. ChemPhysChem 2017, 18, 942–948. 10.1002/cphc.201601425. [DOI] [PubMed] [Google Scholar]
- Nino D.; Djayakarsana D.; Milstein J. N. Nanoscopic Stoichiometry and Single-Molecule Counting. Small Methods 2019, 1900082. 10.1002/smtd.201900082. [DOI] [Google Scholar]
- Fricke F.; Beaudouin J.; Eils R.; Heilemann M. One, Two or Three? Probing the Stoichiometry of Membrane Proteins by Single-Molecule Localization Microscopy. Sci. Rep. 2015, 5, 14072. 10.1038/srep14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschout H.; Shivanandan A.; Annibale P.; Scarselli M.; Radenovic A. Progress in Quantitative Single-Molecule Localization Microscopy. Histochem. Cell Biol. 2014, 142, 5–17. 10.1007/s00418-014-1217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post R. A. J.; van der Zwaag D.; Bet G.; Wijnands S. P. W.; Albertazzi L.; Meijer E. W.; van der Hofstad R. W. A Stochastic View on Surface Inhomogeneity of Nanoparticles. Nat. Commun. 2019, 10, 1663. 10.1038/s41467-019-09595-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner-Gracia N.; Olea R. A.; Fitzner R.; El Boujnouni N.; van Asbeck A. H.; Brock R.; Albertazzi L. Super-Resolution Imaging of Structure, Molecular Composition, and Stability of Single Oligonucleotide Polyplexes. Nano Lett. 2019, 19, 2784–2792. 10.1021/acs.nanolett.8b04407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore L.; Spenkelink L. M.; Ranson M.; van Oijen A. M.; Vine K. L. Quantification of Ligand Density and Stoichiometry on the Surface of Liposomes Using Single-Molecule Fluorescence Imaging. J. Controlled Release 2018, 278, 80–86. 10.1016/j.jconrel.2018.03.022. [DOI] [PubMed] [Google Scholar]
- Delcanale P.; Miret-Ontiveros B.; Arista-Romero M.; Pujals S.; Albertazzi L. Nanoscale Mapping Functional Sites on Nanoparticles by Points Accumulation for Imaging in Nanoscale Topography (PAINT). ACS Nano 2018, 12, 7629–7637. 10.1021/acsnano.7b09063. [DOI] [PubMed] [Google Scholar]
- Yan R.; Moon S.; Kenny S. J.; Xu K. Spectrally Resolved and Functional Super-Resolution Microscopy via Ultrahigh-Throughput Single-Molecule Spectroscopy. Acc. Chem. Res. 2018, 51, 697–705. 10.1021/acs.accounts.7b00545. [DOI] [PubMed] [Google Scholar]
- Bongiovanni M. N.; Godet J.; Horrocks M. H.; Tosatto L.; Carr A. R.; Wirthensohn D. C.; Ranasinghe R. T.; Lee J.-E.; Ponjavic A.; Fritz J. V.; Dobson C. M.; Klenerman D.; Lee S. F. Multi-Dimensional Super-Resolution Imaging Enables Surface Hydrophobicity Mapping. Nat. Commun. 2016, 7, 13544. 10.1038/ncomms13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L.; Wojcik M.; Kenny S. J.; Yan R.; Moon S.; Li W.; Xu K. Optical Characterization of Surface Adlayers and Their Compositional Demixing at the Nanoscale. Nat. Commun. 2018, 9, 1435. 10.1038/s41467-018-03820-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeffaers M. B. J.; De Cremer G.; Libeert J.; Ameloot R.; Dedecker P.; Bons A.-J.; Bückins M.; Martens J. A.; Sels B. F.; De Vos D. E.; Hofkens J. Super-Resolution Reactivity Mapping of Nanostructured Catalyst Particles. Angew. Chem., Int. Ed. 2009, 48, 9285–9289. 10.1002/anie.200904944. [DOI] [PubMed] [Google Scholar]
- Vercauteren D.; Deschout H.; Remaut K.; Engbersen J. F. J.; Jones A. T.; Demeester J.; De Smedt S. C.; Braeckmans K. Dynamic Colocalization Microscopy To Characterize Intracellular Trafficking of Nanomedicines. ACS Nano 2011, 5, 7874–7884. 10.1021/nn2020858. [DOI] [PubMed] [Google Scholar]
- Pageon S. V.; Nicovich P. R.; Mollazade M.; Tabarin T.; Gaus K. Clus-DoC: A Combined Cluster Detection and Colocalization Analysis for Single-Molecule Localization Microscopy Data. Mol. Biol. Cell 2016, 27, 3627–3636. 10.1091/mbc.e16-07-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg W.; Leutenegger M.; Lasser T.; Hofkens J.; Dedecker P. Diffraction-Unlimited Imaging: From Pretty Pictures to Hard Numbers. Cell Tissue Res. 2015, 360, 151–178. 10.1007/s00441-014-2109-0. [DOI] [PubMed] [Google Scholar]
- Malkusch S.; Endesfelder U.; Mondry J.; Gelléri M.; Verveer P. J.; Heilemann M. Coordinate-Based Colocalization Analysis of Single-Molecule Localization Microscopy Data. Histochem. Cell Biol. 2012, 137, 1–10. 10.1007/s00418-011-0880-5. [DOI] [PubMed] [Google Scholar]
- Wojnilowicz M.; Glab A.; Bertucci A.; Caruso F.; Cavalieri F. Super-Resolution Imaging of Proton Sponge-Triggered Rupture of Endosomes and Cytosolic Release of SiRNA. ACS Nano 2019, 13, 187–202. 10.1021/acsnano.8b05151. [DOI] [PubMed] [Google Scholar]
- Clemments A. M.; Botella P.; Landry C. C. Spatial Mapping of Protein Adsorption on Mesoporous Silica Nanoparticles by Stochastic Optical Reconstruction Microscopy. J. Am. Chem. Soc. 2017, 139, 3978–3981. 10.1021/jacs.7b01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Cui J.; Sun H.; Müllner M.; Yan Y.; Noi K. F.; Ping Y.; Caruso F. Analysing Intracellular Deformation of Polymer Capsules Using Structured Illumination Microscopy. Nanoscale 2016, 8, 11924–11931. 10.1039/C6NR02151D. [DOI] [PubMed] [Google Scholar]
- Bingen P.; Reuss M.; Engelhardt J.; Hell S. W. Parallelized STED Fluorescence Nanoscopy. Opt. Express 2011, 19, 23716–23726. 10.1364/OE.19.023716. [DOI] [PubMed] [Google Scholar]
- Ouyang W.; Aristov A.; Lelek M.; Hao X.; Zimmer C. Deep Learning Massively Accelerates Super-Resolution Localization Microscopy. Nat. Biotechnol. 2018, 36, 460–468. 10.1038/nbt.4106. [DOI] [PubMed] [Google Scholar]
- Uno S.-N.; Kamiya M.; Yoshihara T.; Sugawara K.; Okabe K.; Tarhan M. C.; Fujita H.; Funatsu T.; Okada Y.; Tobita S.; Urano Y. A Spontaneously Blinking Fluorophore Based on Intramolecular Spirocyclization for Live-Cell Super-Resolution Imaging. Nat. Chem. 2014, 6, 681–689. 10.1038/nchem.2002. [DOI] [PubMed] [Google Scholar]
- Grimm J. B.; English B. P.; Chen J.; Slaughter J. P.; Zhang Z.; Revyakin A.; Patel R.; Macklin J. J.; Normanno D.; Singer R. H.; Lionnet T.; Lavis L. D. A General Method to Improve Fluorophores for Live-Cell and Single-Molecule Microscopy. Nat. Methods 2015, 12, 244–250. 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers Y.; Ta H.; Gwosch K. C.; Balzarotti F.; Hell S. W. MINFLUX Monitors Rapid Molecular Jumps with Superior Spatiotemporal Resolution. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 6117–6122. 10.1073/pnas.1801672115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson A.-K.; Petrov P. N.; Lee M. Y.; Shechtman Y.; Moerner W. E. 3D Single-Molecule Super-Resolution Microscopy with a Tilted Light Sheet. Nat. Commun. 2018, 9, 123. 10.1038/s41467-017-02563-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant W. R.; Shao L.; Grimm J. B.; Brown T. A.; Milkie D. E.; Avants B. B.; Lavis L. D.; Betzig E. High Density Three-Dimensional Localization Microscopy across Large Volumes. Nat. Methods 2016, 13, 359–365. 10.1038/nmeth.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth M.; Andrade D.; Burke D.; Patton B.; Zurauskas M. Aberrations and Adaptive Optics in Super-Resolution Microscopy. Microscopy 2015, 64, 251–261. 10.1093/jmicro/dfv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter Veer M. J. T.; Pfeiffer T.; Nägerl U. V. Two-Photon STED Microscopy for Nanoscale Imaging of Neural Morphology In Vivo. Methods Mol. Biol. 2017, 1663, 45–64. 10.1007/978-1-4939-7265-4_5. [DOI] [PubMed] [Google Scholar]
- Liu T.-L.; Upadhyayula S.; Milkie D. E.; Singh V.; Wang K.; Swinburne I. A.; Mosaliganti K. R.; Collins Z. M.; Hiscock T. W.; Shea J.; Kohrman A. Q.; Medwig T. N.; Dambournet D.; Forster R.; Cunniff B.; Ruan Y.; Yashiro H.; Scholpp S.; Meyerowitz E. M.; Hockemeyer D.; et al. Observing the Cell in Its Native State: Imaging Subcellular Dynamics in Multicellular Organisms. Science 2018, 360, eaaq1392 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T.; Bhamidimarri S. P.; Brending N.; Colin-York H.; Collinson L.; De Jonge N.; de Pablo P. J.; Debroye E.; Eggeling C.; Franck C.; Fritzsche M.; Gerritsen H.; Giepmans B. N. G.; Grunewald K.; Hofkens J.; Hoogenboom J. P.; Janssen K. P. F.; Kaufmann R.; Klumperman J.; Kurniawan N.; et al. The 2018 Correlative Microscopy Techniques Roadmap. J. Phys. D: Appl. Phys. 2018, 51, 443001. 10.1088/1361-6463/aad055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondia P.; Jurado R.; Casado S.; Domínguez-Vera J. M.; Gálvez N.; Flors C. Hybrid Nanoscopy of Hybrid Nanomaterials. Small 2017, 13, 1603784. 10.1002/smll.201603784. [DOI] [PubMed] [Google Scholar]
- Hua X.; Szymanski C.; Wang Z.; Zhou Y.; Ma X.; Yu J.; Evans J.; Orr G.; Liu S.; Zhu Z.; Yu X. Y. Chemical Imaging of Molecular Changes in a Hydrated Single Cell by Dynamic Secondary Ion Mass Spectrometry and Super-Resolution Microscopy. Integr. Biol. Quant. Biosci. Nano Macro 2016, 8, 635–644. 10.1039/c5ib00308c. [DOI] [PubMed] [Google Scholar]