Abstract
Scope: Intermittent fasting (IF) has been extensively reported to promote improved energy homeostasis and metabolic switching. While IF may be a plausible strategy to ameliorate the epidemiological burden of disease in many societies, our understanding of the underlying molecular mechanisms behind such effects is still lacking. The present study has sought to investigate the relationship between IF and changes in gene expression. We focused on the liver, which is highly sensitive to metabolic changes due to energy status. Mice were randomly assigned to ad libitum feeding or IF for 16 hours per day or for 24 hours on alternate days for 3 months, after which genome-wide transcriptome analysis of the liver was performed using RNA sequencing. Our findings revealed that IF caused robust transcriptomic changes in the liver that led to a complex array of metabolic changes. We also observed that the IF regimen produced distinct profiles of transcriptomic changes, highlighting the significance of temporally different periods of energy restriction. Our results suggest that IF can regulate metabolism via transcriptomic mechanisms and provide insight into how genetic interactions within the liver might lead to the numerous metabolic benefits of IF.
Keywords: transcriptome, metabolism, liver, intermittent fasting, RNA sequencing
Introduction
There is a great need to address the increased global burden of noncommunicable diseases, such as obesity, type II diabetes mellitus, and cardiovascular and neurodegenerative diseases.1 There is a consensus that excessive dietary energy intake increases the risk of development of such diseases.2 It has been observed in rodent studies that chronic ad libitum (AL) feeding conditions also results in the development of a metabolic syndrome-like phenotype.3 Moreover, energy restriction through continuously limiting caloric intake (ie, caloric restriction [CR]) results in an increased life span in many animal models and the amelioration of several age-associated diseases.4-6 It has been reported that during CR, metabolic switching from glucose utilization to preferential ketone metabolism alters both glucose and ketone homeostasis, which in turn promotes metabolic health and reduces metabolic syndrome development.7 Intermittent fasting (IF) is another variant of energy restriction that has gained public and scientific interest. Compared to CR, IF involves restricting daily food intake to within defined periods which has been found to be a more sustainable long-term eating pattern.8 Intermittent fasting has also been found to induce longevity in many animal models and has extensive systemic effects that may trigger biological pathways beneficial for metabolic health. For example, IF has been reported to influence the circadian clock, intestinal microbiota niche, and to metabolically regulate insulin sensitivity, lipid metabolism, and inflammatory responses which likely contribute to the development of resistance to cardiovascular, neurodegenerative, and metabolic diseases.9-11
Despite many reported health benefits from IF, studies have also shown that IF may not be sufficiently robust to ameliorate many age-related degenerative pathways, and this lack of translation in slowing down systemic aging has led to poor understanding of IF-induced benefits still.12 Besides that, the transcriptome-associated changes mediated by IF in the metabolic regulation arm is relatively not well-understood. Therefore, we designed an experimental study to investigate the transcriptomic changes resulting from 2 common IF regimens, time-restricted fasting for 16 hours (IF16) or for 24 hours on alternate days (ie, every other day [EOD]), in mice for a period of 3 months. The liver, which is highly sensitive to metabolic changes due to energy status, was selected for this study. We identified that both IF16 and EOD regimens differentially induce changes in gene expression profiles and signaling pathways to produce distinct metabolic profiles. Our data provide novel insights to the varied transcriptomic changes that can occur as a result of IF.
Material and Methods
Animals and IF Regimen
C57/BL6NTac male mice (InVivos Pte Ltd, Singapore) were raised to 3-month-old with AL food using standard Teklad Global 18% protein rodent diet (Envigo, Huntingdon, UK) and water. Mice were then randomly assigned and subjected to AL feeding, or IF for 16 hours per day (IF16) or for 24 hours on alternate days (ie, EOD) for 3 months. For the IF16 regimen, food was provided at 7 AM and then removed at 3 PM for 16 hours daily. For the EOD regimen, food was provided at 7 AM and removed at 7 AM the next day for a further 24 hours. All mice had AL access to water, with AL mice having free access to both food and water. During the entire experiment, the mice were housed in animal rooms at 20°C to 22°C with 30% to 40% relative humidity under a 12-hour light/dark cycle. Blood glucose and ketones were measured using a FreeStyle Optimum Meter and corresponding test strips (Abbott Laboratories, UK) at baseline and monthly via the tail bleed method. Both tests were performed at 7 am. Body weight was measured weekly. Monthly food/energy consumption was recorded (weight of food consumed × kcal/g of food). Data from these additional tests are not provided in this article. All animal procedures were approved by the National University of Singapore Animal Care and Use Committee and performed according to the guidelines set forth by the National Advisory Committee for Laboratory Animal Research, Singapore. All sections of the article were performed in accordance with Animal Research: Reporting in Vivo Experiments guidelines. The entire experimental workflow can be visualized in the Supplemental Information Section (Figure S1).
Liver Tissue Collection
After the 3-month AL/IF protocol, animals were anesthetized and euthanized. Every other day mice were euthanized on a food-deprivation day. All mice were euthanized between 7 am and noon. The left lateral liver lobe was harvested, immediately flash frozen, and stored at −80°C.
Total RNA Extraction and Quality Control Validation
RNA from the liver was isolated using EZ-10 DNAaway RNA Mini-Preps Kit (Bio Basic, Markham, Canada) according to the manufacturer’s protocol. Briefly, frozen liver samples were homogenized and lysed in the provided lysis buffer. Prevention of contamination by genomic DNA was achieved using the provided gDNA eliminator column. RNA purity was determined using Nanodrop ND-1000 (Thermo Fisher Scientific, Waltham, USA), whereas the RNA integrity was assessed via agarose gel electrophoresis and the Agilent 2100 Bioanalyzer (Agilent). Enrichment of samples with high-quality RNA should demonstrate an OD260/OD280 ratio of 1.9 to 2.0 from Nanodrop readings, 2 distinct bands indicating 28S and 18S following agarose gel electrophoresis, and ≥6.8 RNA integrity number with a smooth baseline using the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, USA).
cDNA Library Construction and Sequencing
Following the isolation of high-quality and pure total RNA samples from liver tissue, complementary DNA (cDNA) library construction was performed using the NEBNext UltraTM RNA library preparation kit as per the manufacturer’s protocol (New England BioLabs, Ipswich, USA). Messenger RNA was first purified by addition of poly-T-oligo-attached magnetic beads, before subjecting them to random fragmentation by a fragmentation buffer. The initial strand of the cDNA is first synthesized through the utilization of a random hexamer primer and RNase H (M-MuLV reverse transcriptase). The second strand of cDNA is then synthesized using DNA polymerase I and RNase H, and the resulting double stranded cDNA is purified using AMPure XP beads. The overhangs of these purified double-stranded cDNA were then processed by exonuclease and polymerase to become blunt ends, before subjecting the 3′ ends of the DNA fragments to adenylation and subsequent ligation of NEBNext hairpin loop structure adaptor on both ends for hybridization. For optimal isolation of cDNA fragments of approximately 150 to 200 base pairs in length, DNA fragments were purified using the AMPure XP system (Beckman Coulter, Brea, USA), before utilizing both PCR amplification and purification by AMPure XP beads to obtain the completed library DNA fragments. The resultant libraries were then sequenced using HiSeqTM 2500 Illumina platforms to obtain 12 gB of raw data per sample (Illumina, San Diego, USA).
Transcriptome Reads Mapping to the Reference Genome
The reference genome for Mus musculus (mm10) and gene model annotation files were downloaded from the National Center for Biotechnology Information genome website browser. Indexes of the reference genome were constructed before the paired-end and clean reads were then mapped to the reference genome using STAR (version 2.5)13 for the liver transcriptome.
Quantification of Gene Expression Levels
HTSeq (version 0.6.1)14 was used to compute the reads mapped onto each gene. Then, the Reads Per Kilobase of exon model per Million mapped reads (FPKM) of each gene was then computed using the length of the gene and reads count mapped to that particular gene. The FPKM value was then used for estimation of gene expression levels. A total of 35 275 unique RNA transcripts each were quantified in total for liver data sets.
Differential Expression Analysis
Differential gene expression analysis between 5 biological replicates per condition was performed using the DESeq2R Package (v2_1.6.3). A negative binomial distribution was used to model a statistical analysis for differential gene expression as achieved in DESeq2. The resultant P values were then adjusted using Benjamini and Hochberg’s test to control false discovery rate. Genes with adjusted P < .05 were assigned as being differentially expressed.15 Principal component plots, Venn diagrams, and volcano plots were prepared using the ggplot2R package (version 3.0.0).16
Correlations and Clustering
To allow logarithm adjustment, genes showing FPKM values of zero were assigned a corresponding value of 0.01. Correlation was then determined using the “cor.test” function in R with options set alternative = “greater” and method = “Spearman”. All experiments were performed using at least 5 biological replicates per condition, and Pearson correlation coefficients obtained of at least 0.9 demonstrated high coverage and reproducibility (Figure S2). To then establish relationships between samples, clustering was performed using the FPKM expression level by utilizing the hierarchical clustering distance method with the aid of heatmap, self-organization mapping and k-means via silhouette coefficient in order to adapt optimal classification with R’s default parameters imbued. Heatmap illustrated in this article was prepared using pheatmap R package (version 1.0.10).17
Enrichment Analysis of Differentially Expressed Genes
To understand the gene ontology (GO) as well as the pathway association for differentially expressed genes, gprofile R package (version 0.6.7)18 and clusterProfiler R package (version 3.8)19 were utilized. After correction of gene length bias, GO terms with adjusted P value <.05 were assigned as being significantly enriched among the pool of differentially expressed genes. Diagrams demonstrating enrichment analysis was plotted using ggplot2R package (version 3.0.0) and Sigmaplot (version 1.3).
Results
Intermittent Fasting Induces Differential Transcriptomic Profiling in the Liver
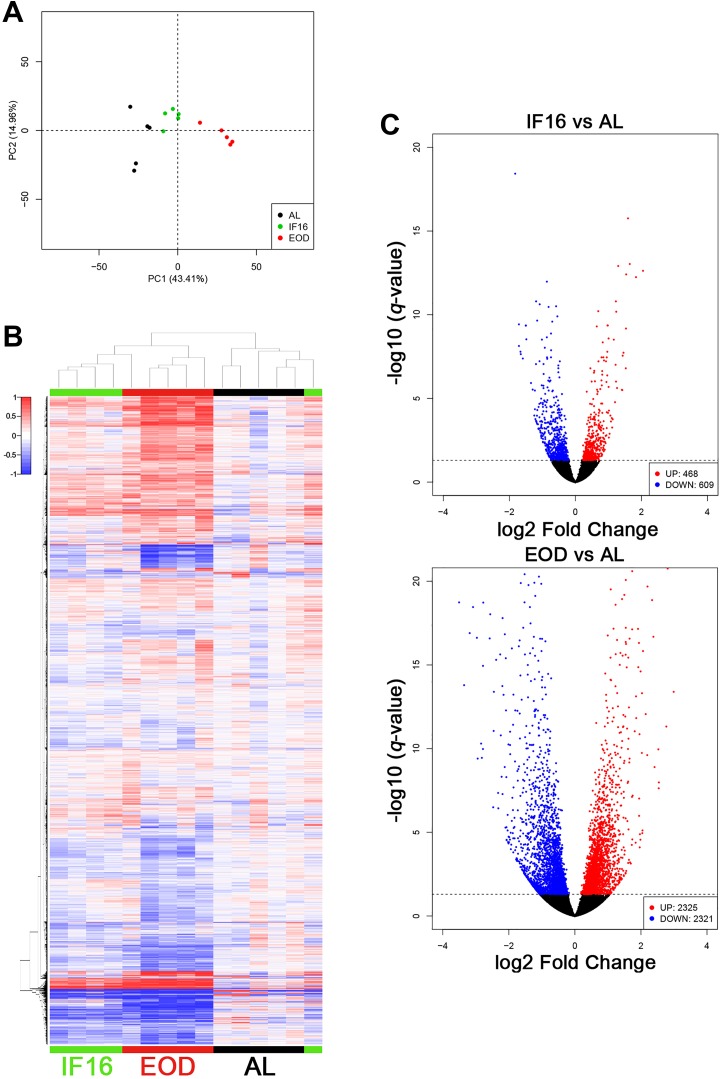
Our initial goal was to characterize the effects of 2 IF regimens on the liver, an organ that is highly responsive to energy restriction and, also a central metabolic adaptor in response to energy status.20,21 Principal component analysis (PCA) showed that biological replicates have high similarity to each other within AL, IF16, and EOD groups through occupation of unique cluster regions (Figure 1A). However, unique cluster regions representing each condition showed minimal overlap, suggesting that transcriptome patterns of IF16 and EOD largely differ from that of AL. Notably, the 2 different IF regimens seems to distinctly modulate transcript expression changes.
Figure 1.
Intermittent fasting regimens induce differential gene expression in the liver. (A) Principal component (PCA) analysis discriminates the variance in a data set in terms of principal components. The two most significant principal components (PC1 and PC2) are displayed on the x- and y-axes, respectively. Principal component analysis discriminated AL, IF16, and EOD into three unique cluster regions. (B) Unsupervised hierarchical clustering segregated AL, IF16 and EOD into three distinct cluster regions shown using a heatmap (black, green and red, respectively). Intermittent fasting16 transcriptomic pattern appears to be more similar to EOD than to AL. Notably, one IF16 sample appeared to be clustered with AL replicates. Despite this observation, this IF16 anomaly was being clustered as green, which indicates higher similarity to its respective replicates than to AL. Red indicates high expression of genes whereas blue indicates low expression of genes. (C) Volcano plot of statistical significance (−log10 q-value) against enrichment (log2-fold change) of differentially expressed genes in IF16 and EOD against AL. Number of upregulated genes are expressed in red, whereas those that are downregulated are expressed in blue. Insignificant differentially expressed genes are expressed in black. IF indicates intermittent fasting. AL indicates ad libitum; EOD, every other day; IF, intermittent fasting.
Unsupervised hierarchical clustering of global RNA transcripts sequenced revealed distinct segregation of AL, IF16, and EOD groups under 3 separate clusters (Figure 1B). This demonstrated that both IF16 and EOD induces differential expression of transcripts as compared to AL. Moreover, the IF16 transcriptomic pattern appears to be more similar to EOD than to AL. Despite this observation, IF16 and EOD groups were discriminated after clustering, which reinforced the finding that the patterns of gene expression were still largely distinct between IF16 and EOD, consistent with the PCA analysis (Figure 1A and B).
Next, we quantified the differentially expressed transcripts between individual IF regimens and AL. We observed a total of 1077 significantly differentially expressed transcripts (468 up- and 609 downregulated) between IF16 and AL, and 4646 significantly differentially expressed transcripts (2325 up- and 2321 downregulated) between EOD and AL. Notably, the number of differentially expressed transcripts for EOD was higher than for IF16 when compared to AL (Figure 1C).
Intermittent Fasting Results in Distinct Gene Ontologies Enrichment Profiles in the Liver
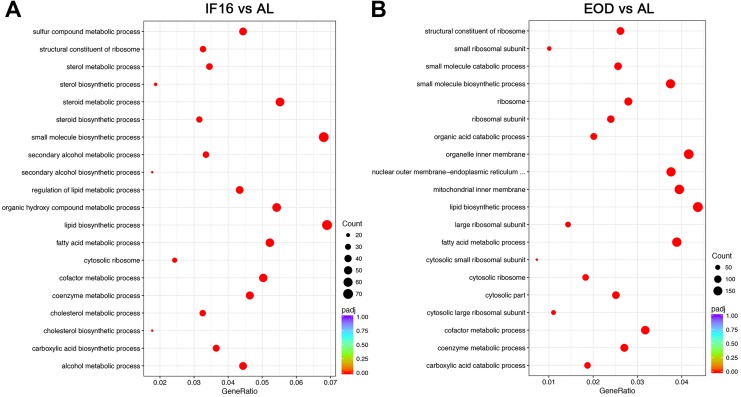
We next annotated these differentially expressed transcripts that are modulated as a result of the 2 IF regimens. To achieve this, differential enrichment analysis was performed using clusterProfiler. The results showed that the top 20 significantly enriched gene ontologies for IF16 as compared to AL belonged to a myriad of metabolic processes, such as sulfur compound (GO:0006790), sterol (GO:0016125), alcohol (GO:1902652), lipid (GO:0008610 and GO:0006631), and cholesterol modulation (GO:0008203 and GO:0006695) in the liver (Figure 2A). This analysis also revealed that IF16 induces a vast number of other key processes in the liver as compared to AL, such as regulation of carbohydrate metabolism (GO:0006109) and circadian rhythm (GO:0007623; Table S1).
Figure 2.
Transcriptomic gene ontologies (GO) analysis of liver following IF. (A) Intermittent fasting 16 induces modulation of a myriad of metabolic processes such as sulfur compound, sterol, alcohol, lipid and cholesterol in the liver. (B) Every other day induces changes in key metabolic processes such as fatty acid metabolism, a plethora of cellular changes involving both mitochondria and endoplasmic reticulum, as well as major modulation of ribosomal involvement. Top 20 significantly enriched gene ontologies are reflected in this illustration. Gene ontology terms are listed on the left whereas GeneRatio is calculated and reflected on the x-axis. The size of the dots represents gene counts, and red symbolizes highly significant adjusted P value whereas blue symbolizes nonsignificant P adj value. Nonsignificant GO terms are not displayed. IF indicates intermittent fasting; GO, gene ontology.
However, the top 20 significantly enriched gene ontologies for EOD as compared to AL belong to key metabolic processes such as fatty acid metabolism (GO:0008610 and GO:0006631), a plethora of cellular changes involving both mitochondria (GO:0005743) and endoplasmic reticulum (GO:0042175), and major modulation of ribosomal involvement (GO:0005840 and GO:0003735; Figure 2B). Furthermore, EOD also induced other key processes in the liver as compared to AL, such as nucleotide metabolism (GO:0019362 and GO:0006163), autophagy (GO:0006914), carbohydrate (GO:0033500), and protein metabolism (GO:1903052 and GO:0042177) as well as circadian rhythm (GO:0007623; Table S2).
Intermittent Fasting Results in Distinct Pathway Enrichment Profiles in the Liver
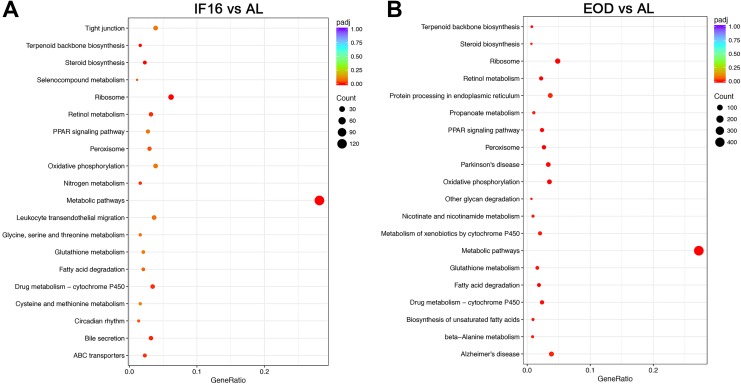
Our next goal was to decipher the roles of the differentially expressed transcripts modulated by IF16 and EOD in various pathways. For this we used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways statistical enrichment tool from clusterProfiler. Notably, a high number of IF16-induced differentially expressed genes showed a significant functional bias toward metabolic pathways (mmu01100) in the liver. A deeper exploration of metabolic pathways revealed that IF16 modulated transcriptomic changes in steroid biosynthesis (mmu00100), terpenoid backbone biosynthesis (mmu00900), retinol (mmu00830), drug (mmu00982), and nitrogen (mmu00910) metabolism. Intermittent Fasting 16 was also found to influence the peroxisome (mmu04146) and peroxisome proliferator activator receptor (PPAR) signaling pathways (mmu03320) in the liver, as compared to AL. In addition, consistent with gene ontologies data, IF16 influenced the circadian rhythm axis (mmu04710) (Figure 3A and Table S3).
Figure 3.
Transcriptomic pathways enrichment analysis of liver following intermittent fasting. (A) and (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis revealed that both IF16 and EOD regimens share modulation of similar pathways. Notably, IF16 and EOD are able to most significantly modulate changes in metabolic pathways. IF indicates intermittent fasting; EOD, every other day.
Similar to IF16, KEGG analysis revealed that EOD-induced differentially expressed genes were significantly involved in metabolic pathways (mmu01100) in the liver. However, it was evident that EOD induced a higher number of gene changes than IF16. Many of the pathway enrichments observed in IF16 were also seen during EOD, but the quantity of differentially expressed transcripts was consistently higher in EOD. We also noted that EOD modulates a wider spectrum of pathway changes as compared to IF16 that included similar pathways as well as other areas of metabolic changes such as fatty acid degradation (mmu00071) and biosynthesis (mmu00061), glutathione (mmu00480) and beta-alanine (mmu00410) metabolism. In brief, it appears that EOD can influence more metabolic cascades than IF16 in the liver. In addition, EOD is able to induce changes in the circadian rhythm (mmu04710) and p53 signaling pathway (mmu04115) axes (Figure 3B and Table S4).
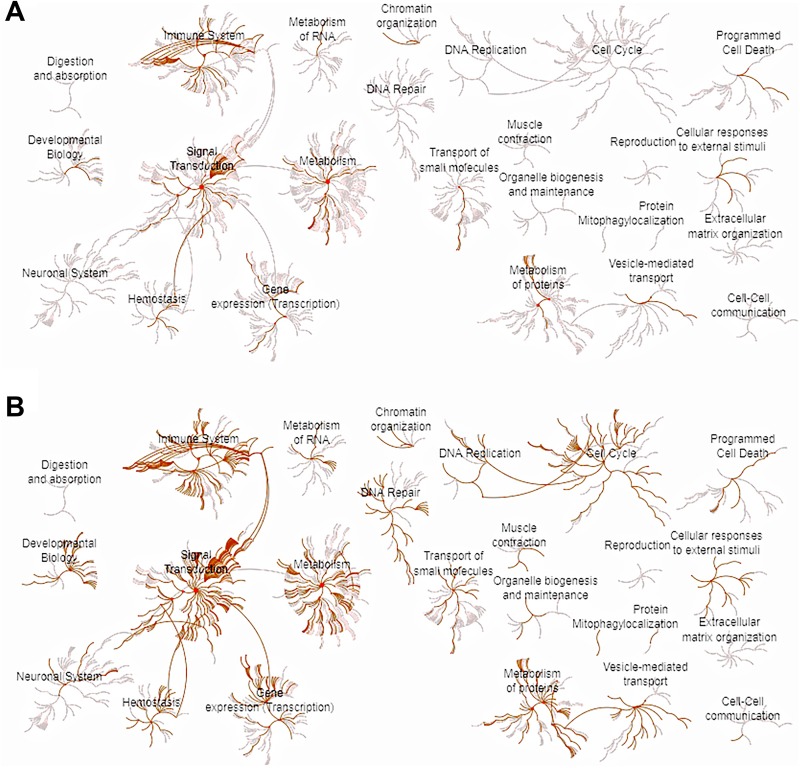
Considering the plethora of transcriptomic changes associated with significant biological pathways as a result of these 2 IF regimens, we investigated the coverage distribution of the interaction between pathway enrichment and the transcriptomic changes. Reactome analysis results indicated that IF16 induces the immune system, and signal transduction and metabolism, as compared to AL. On the other hand, pathway coverage of EOD was strikingly different from that of IF16 with better representation of pathways in signal transduction, immune system, cell cycle, and development, in addition to enrichment of metabolic pathways as compared to AL (Figure 4A and B, and Table S5-6).
Figure 4.
Biological pathway analysis using reactome in the liver. (A) Biological pathway enrichment for differentially expressed genes for the IF16 regimen as compared to AL falls under the categories of immune system, signal transduction and metabolism. (B) Biological pathway enrichment for differentially expressed genes for the EOD regimen as compared to AL falls under the categories of immune system, signal transduction and metabolism, metabolism of proteins, cell cycle, and developmental biology. Every other day appears to induce more robust pathway enrichment as compared to IF16. AL indicates ad libitum; EOD, every other day; IF, intermittent fasting.
Both IF16 and EOD Transcriptomic Changes Induce Common and Unique Metabolic Changes in the Liver
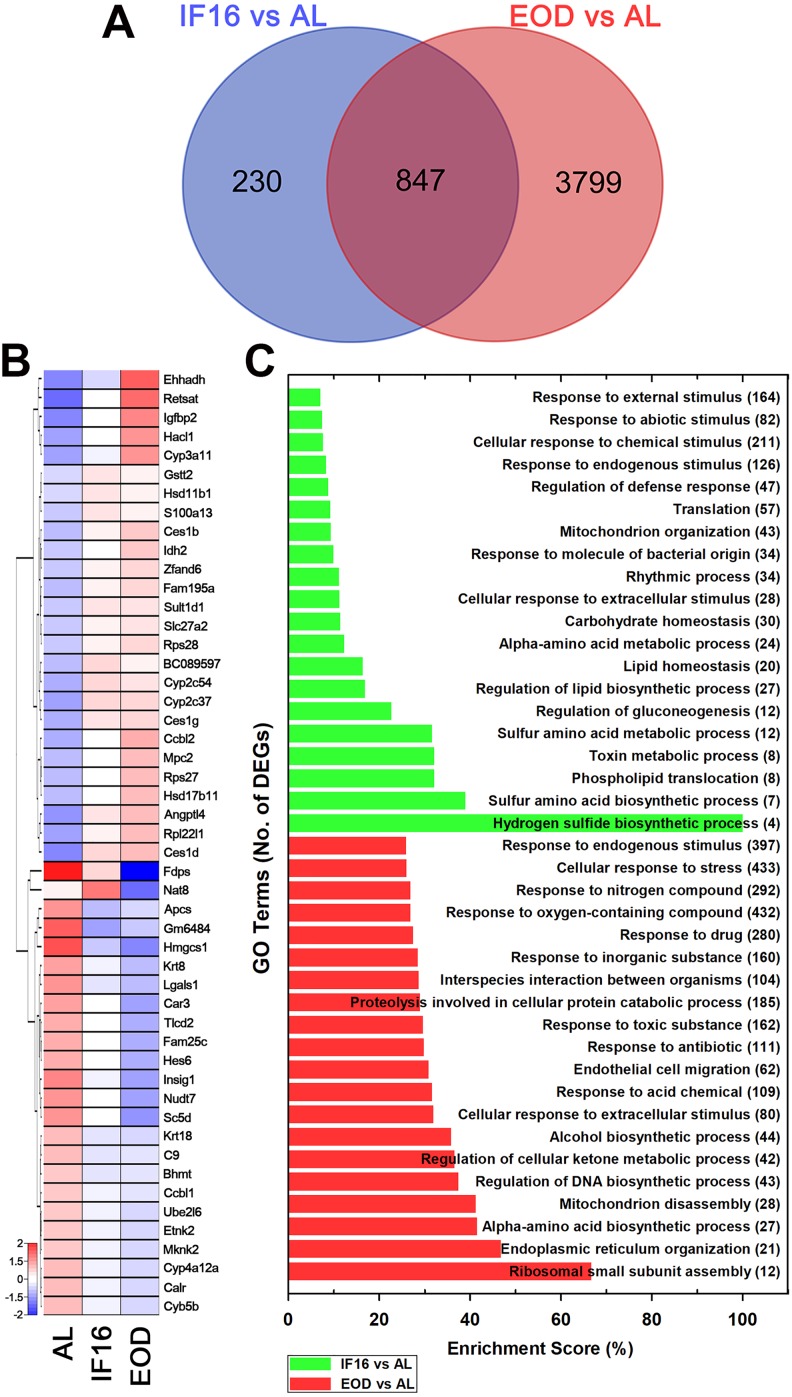
Since both IF regimens were able to modulate key metabolic changes in the liver, we next investigated the common and unique metabolic pathway changes that occurred. We observed a total of 847 differentially expressed genes that were common to IF16 and EOD as compared to AL. Two hundred thirty genes were differentially expressed uniquely in IF16 as compared to AL, whereas 3799 genes were differentially expressed uniquely in EOD as compared to AL (Figure 5A). Subsequently, we analyzed the top 50 commonly expressed genes exhibited by both IF16 and EOD as compared to AL by ranking FPKM of each gene in a descending manner. Gene ontology analysis of these transcripts using gProfiler showed that many of these common genes fall under the biological function categories of lipid (GO: 0006629) or sulfur (GO: 0006790) compound metabolic processes, oxidation-reduction (GO: 0055114), and cofactor metabolic processes (GO: 0051186; Figure 5B and Table S7). Other metabolic pathways associated with these common genes include linoleic acid (mmu00591) and retinol metabolism (mmu00830), peroxisome (mmu04146) and PPAR signaling (mmu03320), steroid hormone biosynthesis (mmu00140), and chemical carcinogenesis pathways (mmu05240; Table S8).
Figure 5.
Both IF16 and EOD transcriptomic changes induce common and unique metabolic changes in the liver. (A) Venn diagram illustrates the number of differentially expressed genes that are common and distinct in each type of IF regimen when compared to AL. (B) Unsupervised hierarchical clustering of the top 50 common genes that were differentially expressed in both IF16 and EOD against AL using a heatmap. Both IF16 and EOD expression patterns showed distinct segregation to AL and between themselves. Red indicates high expression of genes whereas blue indicates low expression of genes. Top 50 common gene identity is illustrated in each row on the right. (C) Gene ontologies of differentially expressed genes of IF16 and EOD compared to AL plotted against enrichment score (represented as percentage). Number of differentially expressed genes belonging to a single gene ontology term is shown in brackets beside the term. Differentially expressed genes of IF16 relative to AL are shown in green, and the gene ontology with the highest enrichment score belonged to hydrogen sulfide biosynthetic process. On the other hand, differentially expressed genes of EOD against AL are shown in red, and the gene ontology with the highest enrichment score belonged to ribosomal small subunit assembly. AL indicates ad libitum; EOD, every other day; IF, intermittent fasting.
Based on this observation, we analyzed differential effects of the 2 IF regimens independently as compared to AL using gProfiler software. The highest cluster enrichment that was differentially expressed during IF16 when compared to AL belonged to the hydrogen sulfide biosynthetic process (GO:0070814). In addition, IF16 appeared to induce differentiation of other genes belonging to regulation of immunity (GO:0031347), translation (GO:006412), mitochondrion organization (GO:0007005), rhythmic process (GO:0048511), phospholipid translocation (GO:0045332), and various metabolic processes related to carbohydrate (GO:0033500), amino acid (GO: 0000096), lipid (GO: 0055088), and toxin (GO: 0009404) homeostasis (Figure 5C and Table S9). On the other hand, the highest enrichment of genes that were differentially expressed during EOD when compared to AL in the liver belonged to ribosomal small subunit assembly (GO:0000028). Moreover, EOD was able to induce distinct differentiation of other genes as compared to AL with profiles belonging to endothelial cell migration (GO:0043542), mitochondrial disassembly (GO:0061726), endoplasmic reticulum organization (GO:0007029), and metabolic processes related to alcohol (GO:0046165), DNA (GO:2000278), α amino acid (GO:1901607), protein (GO:1903050), and ketones (GO:0010565; Figure 5C and Table S10). Altogether, these results supported our hypothesis that the temporal period of energy restriction can differentially regulate transcriptomic and pathway changes and could therefore eventually be translated to distinct cellular changes being achieved through adoption of different IF regimens.
Discussion
Intermittent fasting has gained increasing popularity in recent years as a plausible intervention for metabolic syndrome.19-21 However, there is limited mechanistic information as to how fasting might lead to such beneficial alterations in metabolism. The findings of our present study reveal that IF triggers robust and complex changes in gene expression in the liver and that different fasting regimens can have profoundly different profiles of effect.
The liver is a key metabolic organ that strictly controls different facets of carbohydrate, fat, and protein metabolism. During IF, the liver is highly responsive to energy deficiency and triggers a plethora of cellular responses to achieve energy homeostasis. Transcriptomic data sets of liver subjected to IF16 or EOD reveal robust changes in gene expression when compared to AL, indicating that IF-induced metabolic changes are regulated via a transcriptomic axis. Interestingly, the duration of fasting distinctly impacts the transcriptome. Our data set shows that the gene signatures exhibited by IF16 and EOD remained highly varied according to the PCA and distinct clustering of experimental groups using heatmap analysis. Despite this observation, the anomaly in IF16 gene expression patterns remain similar to the other IF16 replicates, demonstrating a higher degree of similarity in gene expression within IF16 replicates than in AL. Moreover, volcano map analysis provided a clearer depiction that the differential gene expression patterns of IF16 and EOD are highly disparate. Even though common genes that are differentially expressed by both IF regimen when compared to AL are present, it can be further observed that a large number of differentially expressed genes are unique to either the IF16 or the EOD regimen. Moreover, relative to AL, EOD exhibited a higher number of differentially expressed genes than IF16. Interestingly, our data set has revealed common robust transcriptomic differences induced by both IF16 and EOD that have been reported by other studies. For instance, common genes such as Ehhadh and Angptl4 were both upregulated following IF16 and EOD and seemed to regulate sulfur compound and lipid metabolic processes, respectively. During fasting, Ehhadh has been reported to be involved in the mitochondrial and peroxisomal β-oxidation fatty acid pathway that is highly dependent on PPARα signaling in the liver.22,23 This hepatic sulfur compound metabolism induced by Ehhadh also seems to be implicated in lipid metabolic processes through a synergistic relationship with Angptl4, which catalyses the release of fatty acids from adipose tissue.24 While there have been previous reports that both protein products are implicated during fasting, our data set implies that upregulation of these 2 genes may represent cross talk between liver and adipose tissue in mediating a preferential utilization of fat to meet energy demands. Besides that, other common differentially expressed genes, such as Fsn, Cry1, and Ppar were also modulated as a result of both IF regimen which have been involved in lipid homeostasis during time-restricted feeding.25-27 Our study has provided us with information that was previously reported by other groups, yet the adoption of different IF regimens allow us to compare the similarities and differences in the transcriptomic signatures in the liver.
In liver tissue from mice subjected to the IF16 regimen, gene ontologies that possessed the highest enrichment scores belonged to the hydrogen sulfide biosynthetic process. Hydrogen sulfide is an important gaseous modulator reported to be a key factor in protecting against various liver diseases.28,29 Moreover, IF16 also seemed to induce expression of genes that modulate metabolic processes such as carbohydrate and lipid homeostasis. In contrast, during EOD, gene ontologies that possessed the highest enrichment scores belonged to ribosomal small subunit assembly, suggesting that translational processes and protein synthesis are being significantly modulated in the liver. These findings are consistent with the concomitant upregulation of many key genes in α-amino acid biosynthetic and cellular protein catabolic processes. Interestingly, many genes that were modulated by the EOD regimen have not yet been reported. Furthermore, EOD appears to induce greater disparity in gene expression changes than IF16.
Our study has provided a detailed analysis of the transcriptomic changes which occur in the liver as a result of 2 IF regimens, allowing a better understanding of the temporal regulation of genes to bring about the IF-induced cellular changes previously reported.30 Thus, a novel aspect of our findings is that there appears to be strong evidence that the adoption of different IF regimens may produce common or distinct metabolic changes in an organism that may be translated into differential cellular effects. We also have gained a better understanding that different IF regimens may induce differential metabolic switching processes, which may result in varying degrees of metabolic adaptation. However, there is a need to consider confounding factors in our present study. For instance, the present study provided mice with food at 0700 hours for both IF16 and EOD groups, which represent 12 hours off synchronization from AL mice’s normal nocturnal circadian rhythm for feeding. The lack of synchronization in feeding and fasting chronobiology of mice in this study may thereby manifest as a confounding variable to this study. This in turn may also affect the chronobiology of the transcriptome, and nonconsistent adoption of time-restricted feeding at different time points may eventually lead to nontranslatable results.25,31 Despite this, our study has provided the impetus to consider in more detail the circumstances of the effects reported in many IF studies, as differences in IF regimens may be a confounding factor in reproducibility of results. Ultimately, the complexity of these IF-induced cellular changes in the liver may substantially contribute to the metabolic adaptations commonly observed during fasting.
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Research Data
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 1._Ng_et_al.,_2019-Table_S1_IF16_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 2._Ng_et_al.,_2019-Table_S2_EOD_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 3._Ng_et_al.,_2019-Table_S3_IF16_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 4._Ng_et_al.,_2019-Table_S4_EOD_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 5._Ng_et_al.,_2019-Table_S5_Reactome_IF16vsAL_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 6._Ng_et_al.,_2019-Table_S6_Reactome_EODvsAL_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 7._Ng_et_al.,_2019-Table_S7_Top_50_Common_Genes_Liver_gProfiler for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 8._Ng_et_al.,_2019-Table_S8_Top_50_Common_Genes_Liver_List for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 9._Ng_et_al.,_2019-Table_S9_Gene_Ontologies_IF16_against_AL_Liver. for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, 10._Ng_et_al.,_2019-Table_S10_Gene_Ontologies_EOD_against_AL_Liver. for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material
Supplemental Material, Supplemental_Information for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Acknowledgments
The authors thank the Novogene (Beijing, China) for kind assistance in data processing.
Authors’ Note: High-throughput sequencing data from this manuscript have been submitted to the NCBI Sequence Read Archive (SRA) under accession number GSE130127. Gavin Yong-Quan Ng, Sung-Wook Kang contributed equally.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Singapore National Medical Research Council Research Grant (NMRC-CBRG-0102/2016) supported this work.
ORCID iD: Thiruma V. Arumugam  https://orcid.org/0000-0002-3377-0939
https://orcid.org/0000-0002-3377-0939
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Salinas AM, Kones R. Barriers to global action plan for the prevention and control of noncommunicable diseases: proposal modifications to the voluntary targets. J Prev Med. 2018;03(01):1–5. [Google Scholar]
- 2. Uauy R, Díaz E. Consequences of food energy excess and positive energy balance. Public Health Nutr. 2005;8(7A):1077–1999. [DOI] [PubMed] [Google Scholar]
- 3. Martin B, Ji S, Maudsley S, Mattson MP. “Control” laboratory rodents are metabolically morbid: Why it matters. Proc Natl Acad Sci. 2010;107(14):6127–6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17(3):313–321. [DOI] [PubMed] [Google Scholar]
- 5. Manzanero S, Gelderblom M, Magnus T, Arumugam TV. Calorie restriction and stroke. Exp Transl Stroke Med. 2011;3(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fontana L, Meyer TE, Klein S, Holloszy JO. Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc Natl Acad Sci. 2004;101(17):6659–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mattson MP, Arumugam TV. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. 2018;27(6):1176–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mattson MP, Longo VD, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. 2017;39:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim J, Kang SW, Mallilankaraman K, et al. Transcriptome analysis reveals intermittent fasting-induced genetic changes in ischemic stroke. Hum Mol Genet. 2018;27(9):1497–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fann DY, Ng GYQ, Poh L, Arumugam TV. Positive effects of intermittent fasting in ischemic stroke. Exp Gerontol. 2017;89:93–102. [DOI] [PubMed] [Google Scholar]
- 11. Fann DY, Santro T, Manzanero S, et al. Intermittent fasting attenuates inflammasome activity in ischemic stroke. Exp Neurol. 2014;257:114–149. [DOI] [PubMed] [Google Scholar]
- 12. Xie K, Neff F, Markert A, Rozman J, et al. Every-other-day feeding extends lifespan but fails to delay many symptoms of aging in mice. Nat Commun. 2017;8(1):155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dobin A, Davis CA, Schlesinger F, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anders S, Pyl PT, Huber W. HTSeq: Analysing high-throughput sequencing data with Python. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wickham H. Elegant Graphics for Data Analysis. Vol. 35 New York, NY: Springer-Verlag Media; 2015. p 211. [Google Scholar]
- 17. Diao C, Xi Y, Xiao T. Identification and analysis of key genes in osteosarcoma using bioinformatics. Oncol Lett. 2018;15(3):2789–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reimand J, Arak T, Adler P, et al. g: Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016;44(W1):W83–W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu G, Wang L, Han Y, He Q. . clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fischer W. Gewebeersatz in der Urogynäkologie. Gynakol Prax. 2003;27(4):705–715. [Google Scholar]
- 21. Browning JD, Baxter J, Satapati S, Burgess SC. The effect of short-term fasting on liver and skeletal muscle lipid, glucose, and energy metabolism in healthy women and men. J Lipid Res. 2012;53(3):577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Izumida Y, Yahagi N, Takeuchi Y, et al. Glycogen shortage during fasting triggers liver-brain-adipose neurocircuitry to facilitate fat utilization. Nat Commun. 2013;4(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vendelbo MH, Møller AB, Christensen B, et al. Fasting increases human skeletal muscle net phenylalanine release and this is associated with decreased mTOR signaling. Plos One. 2014;9(7):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen AE, Tunstall RJ, Carey KA, et al. Actions of short-term fasting on human skeletal muscle myogenic and atrogenic gene expression. Ann Nutr Metab. 2006;50(5):476–481. [DOI] [PubMed] [Google Scholar]
- 25. Chaix A, Lin T, Le HD, et al. Time-restricted feeding prevents obesity and metabolic syndrome in mice. Cell Metab. 2019;29(2):303–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato S, Solanas G, Peixoto FO, et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. 2017;170(4):664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marosi K, Moehl K, Navas-enamorado I, et al. Metabolic and molecular framework for the enhancement of endurance by intermittent food deprivation. FASEB J. 2018;32(7):3844–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li C, Sadraie B, Steckhan N, et al. Effects of a one-week fasting therapy in patients with Type-2 diabetes mellitus and metabolic syndrome - a randomized controlled explorative study. Exp Clin Endocrinol Diabetes. 2017;125(9):618–624. [DOI] [PubMed] [Google Scholar]
- 29. Patterson RE, Sears DD. Metabolic effects of intermittent fasting. Annu Rev Nutr. 2017;37(1):371–393. [DOI] [PubMed] [Google Scholar]
- 30. Sokolovic M, Sokolovic A, Wehkamp D, et al. The transcriptomic signature of fasting murine liver. BMC Genomics. 2008;9(528):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106(50):21453–21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access page (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data abstract for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 1._Ng_et_al.,_2019-Table_S1_IF16_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 2._Ng_et_al.,_2019-Table_S2_EOD_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 3._Ng_et_al.,_2019-Table_S3_IF16_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 4._Ng_et_al.,_2019-Table_S4_EOD_vs_AL_clusterProfiler_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 5._Ng_et_al.,_2019-Table_S5_Reactome_IF16vsAL_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 6._Ng_et_al.,_2019-Table_S6_Reactome_EODvsAL_Liver for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 7._Ng_et_al.,_2019-Table_S7_Top_50_Common_Genes_Liver_gProfiler for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 8._Ng_et_al.,_2019-Table_S8_Top_50_Common_Genes_Liver_List for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 9._Ng_et_al.,_2019-Table_S9_Gene_Ontologies_IF16_against_AL_Liver. for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, 10._Ng_et_al.,_2019-Table_S10_Gene_Ontologies_EOD_against_AL_Liver. for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response
Supplemental Material, Supplemental_Information for Genome-Wide Transcriptome Analysis Reveals Intermittent Fasting-Induced Metabolic Rewiring in the Liver by Gavin Yong-Quan Ng, Sung-Wook Kang, Joonki Kim, Asfa Alli-Shaik, Sang-Ha Baik, Dong-Gyu Jo, M. Prakash Hande, Christopher G. Sobey, Jayantha Gunaratne, David Yang-Wei Fann and Thiruma V. Arumugam in Dose-Response