Abstract
Widespread therapeutic and commercial interest in recombinant mucin technology has emerged due to the unique ability of mucin glycoproteins to hydrate, protect, and lubricate biological surfaces. However, recombinant production of the large, highly repetitive domains that are characteristic of mucins remains a challenge in biomanufacturing likely due, at least in part, to the inherent instability of DNA repeats in the cellular genome. To overcome this challenge, we exploit codon redundancy to encode desired mucin polypeptides with minimal nucleotide repetition. The codon-scrambling strategy was applied to generate synonymous genes, or “synDNAs,” for two mucins of commercial interest: lubricin and mucin 1. Stable, long-term recombinant production in suspension-adapted human 293-F cells was demonstrated for the synonymous lubricin complementary DNA (cDNA), which we refer to as SynLubricin. Under optimal conditions, a 293-F subpopulation produced recombinant SynLubricin at more than 200 mg/L of media and was stable throughout 2 months of continuous culture. Functionality tests confirmed that the recombinant lubricin could effectively inhibit cell adhesion and lubricate cartilage explants. Together, our work provides a viable workflow for cDNA design and stable mucin production in mammalian host production systems.
Keywords: bioprocess, custom gene synthesis, lubrication, lubricin, Muc1, mucin, PRG4, recombinant, synthetic biology, tribology
1 |. INTRODUCTION
Mucins are membrane-bound or secreted glycoproteins containing a variable number of tandem repeats that are defined by their densely clustered sites for O-glycosylation (Hang & Bertozzi, 2005). This extensive glycosylation gives rise to a bottlebrush molecular structure that confers mucins with remarkable physical properties (Kuo, Gandhi, Zia, & Paszek, 2018). Mucins at biological interfaces can co-ordinate with water molecules to form hydrated layers that protect delicate cellular or tissue structures, deter biofouling, and resist pathological cellular deposition (Hattrup & Gendler, 2008). For instance, transmembrane mucins such as mucin 1 (Muc1) and mucin 16 (Muc16) are densely grafted on the ocular surface, where they maintain hydration, resist abrasion, and provide a selective barrier to macromolecules (Gipson, Spurr-Michaud, Tisdale, & Menon, 2014; Mauris & Argüeso, 2012) Similarly, the secreted mucin-like glycoprotein called proteoglycan 4 (PRG4), or lubricin, can bind to cells and tissue interfaces, including the articular cartilage and ocular surfaces, enabling low friction lubrication and protection from pathological cellular deposition and biofouling (Rhee et al., 2005; Schmidt, Sullivan, & Knop, 2013).
Alterations in mucin expression and glycosylation are observed in various pathological conditions, ranging from cancer and inflammatory bowel disease to ocular disease (Dhanisha, Guruvayoorappan, Drishya, & Abeesh, 2018). Patients with genetic mutations that preclude functional lubricin synthesis demonstrate symptoms of Camptodactyly-arthropathy-coxa vara-pericarditis syndrome, including early-onset polyarthropathy as a result of pannus formation and impaired joint lubrication (Bahabri et al., 1998; Marcelino et al., 1999). Decreased synovial fluid lubricin concentrations have also been observed in patients with anterior cruciate ligament injury, osteoarthritis, and rheumatoid arthritis (Elsaid et al., 2008; Kosinska et al., 2015). As such, there has been significant interest in the development of recombinant lubricin and other mucins as injectable therapeutics for osteoarthritis and rheumatic diseases (Le Graverand-Gastineau, 2010) and as topical treatments for chronic dry eye and other conditions that require the application of exogenous lubricants (Schmidt et al., 2013).
Despite this commercial interest, recombinant production has proven challenging for Muc1, lubricin, and other mucins that contain a high number of tandem repeats. Although highly productive clones of Chinese hamster ovary (CHO) cells have been isolated for a truncated Muc1 with approximately 1/3 of its native tandem repeats, similar attempts to isolate clones for full-length recombinant Muc1 have failed (Backstrom et al., 2003). Likewise, stable clones for recombinant lubricin with the complete 59 native tandem repeats produced the glycoprotein at low levels (Jones et al., 2007), but a modified recombinant lubricin protein construct (LUB:1), which contained only 1/3 of the tandem repeats, was more amenable to large-scale production (Flannery et al., 2009). More recently, the production of full-length recombinant human lubricin expressed in suspension-adapted CHO cells has been reported and has demonstrated potential as an ocular lubricant for treating dry eye disease or hydrating contact lenses (Samsom et al., 2014). The precise details of how recombinant production was achieved for the full-length lubricin remain proprietary, and at the current time, no published strategy for large-scale lubricin production is available.
The exact biology that underlies the difficulty of producing mucins at high levels remains unclear. However, long repetitive DNA sequences, such as those common in the complementary DNAs (cDNAs) of mucin tandem repeats, are relatively unstable in the cellular genome (Pearson, Edamura, & Cleary, 2005). The fidelity of nearly all DNA processing steps can be compromised by slippage and other errors linked to repetitive sequences (López Castel, Cleary, & Pearson, 2010). Consequently, repeats can mutate by addition or loss of their unit nucleotide sequence up to 100000 times more frequently than point mutations in nonrepetitive regions (Oren et al., 2016). The variation in tandem repeat numbers for Muc1 and other mucins in humans and mammals provides an evolutionary argument that these genomic DNAs may be mutational hotspots (Gemayel, Vinces, Legendre, & Verstrepen, 2010). Recombination and truncation of exogenous Muc1 cDNAs in bacteria have also been reported, suggesting a high level of instability for these repetitive sequences in host microbial cells, as well (Backstrom et al., 2003).
Now that advances in custom gene synthesis (CGS) enable fast and cost-effective synthesis of long cDNAs (Kosuri & Church, 2014), a new possibility for improved genomic stability of mucins would be to exploit codon redundancy to find synonymous gene sequences that are less repetitive but encode the same desired polypeptide. Such codon optimization algorithms have now been developed and successfully applied for elastin-like proteins and some other repetitive protein domains (Tang & Chilkoti, 2016). Optimized synthetic cDNAs have yet to be designed, synthesized, and tested for biomanufacturing of large mucins of commercial interest.
To date, most biologics, including mucins, have been produced in CHO cells due to their fast growth, adaptability to suspension culture, and capacity for glycosylation and other important posttranslational modifications. However, CHO cells can generate glycan epitopes that are now suspected to elicit adverse immunological responses in humans (Butler & Spearman, 2014). Namely, the α1,3-galactosyltransferases of CHO and other nonprimate cells produce glycans with Galα1,3-Gal residues that can be immunogenic to humans, apes, and other old-world monkeys that have lost α1,3-galactosyltransferase activity (Bosques et al., 2010; Brooks, 2004). CHO cells can also generate Neu5Gc, a terminal sialic acid that is common in most mammalian cells but has been lost in humans and primates (Ghaderi, Zhang, Hurtado-Ziola, & Varki, 2012). These glycans are of particular concern for recombinant mucins, which can consist of 75% or more carbohydrate by mass and are often highly sialylated (Estrella, Whitelock, Packer, & Karlsson, 2010). Recombinant production of the glycoproteins in human cells would avoid the risk of Galα1,3-Gal and Neu5Gc residues; but, to our knowledge, no successful attempts at large-scale mucin production in a human cell host production system have been reported.
Thus, our objective was to determine whether cDNA optimization through codon scrambling could be an effective strategy to achieve stable recombinant production of mucins and mucin-like glycoproteins and whether this strategy would be viable in suspension-adapted human 293-F cells. Notably, the United States Food and Drug Administration has recently approved several biologics produced in 293-F cells, establishing the cell platform as a viable alternative to CHO and other nonhuman systems for manufacturing specialized therapeutics (Dumont, Euwart, Mei, Estes, & Kshirsagar, 2016). Here, the codon-scrambling approach is demonstrated for Muc1 and lubricin, and the production strategy is further developed to achieve stable production of a functional, full-length recombinant lubricin.
2 |. MATERIALS AND METHODS
2.1 |. Antibodies and reagents
The following antibodies were used: mouse anti-human Muc1 (555925; BD Biosciences, San Jose, CA), mouse anti-human lubricin (MABT401, MilliporeSigma, Burlington, MA), goat anti-mouse immunoglobulin G–horseradish peroxidase (IgG–HRP; sc-2005; Santa Cruz Biotechnology, Dallas, TX), chicken anti-mouse IgG-HRP (AP126P; MilliporeSigma), mouse anti-SUMO (4G11E9; GenScript, Piscataway, NJ). Lectins used were biotinylated Peanut Agglutinin (PNA; B-1075; Vector Laboratories, Burlingame, CA). Biotinylated lectins were detected using ExtrAvidin-Peroxidase (E2886; MilliporeSigma). To induce transactivator cell lines, doxycycline was used (sc-204734; Santa Cruz Biotechnology). For neomycin selection, G418 was used (10131035; Thermo Fisher Scientific, Waltham, MA). Valproic acid (VPA) was used as a histone deacetylase inhibitor (P4543–100 G; MilliporeSigma).
2.2 |. Constructs
A tetracycline-inducible, transposon-based PiggyBac expression vector with an integrated, coexpressed reverse tetracycline transactivator gene (pPB tet rtTA NeoR) was used for stable line generation (kindly provided by Dr. Valerie Weaver, University of California San Francisco). The pPB tet rtTA NeoR plasmid was modified by the insertion of the internal ribosome entry site (IRES) of the encephalomyocarditis virus followed by the fluorescent protein copGFP into the NotI and XbaI sites of the plasmid (pPB tet IRES copGFP rtTA NeoR). Synthetic cDNA for a lubricin analog with 59 perfect repeats of KEPAPTTP, native N-and C-terminal domains, and an N-terminal SumoStar tag (LifeSensors, Malvern, PA) was generated through CGS (General Biosystems, Morrisville, NC) and cloned into the multiple cloning sites of pPB tet IRES copGFP rtTA NeoR using BamHI and EcoRI restriction sites. Similarly, cDNA for a soluble, codon-scrambled Muc1 having 42-perfect repeats of PDTRPAPGSTAPPAHGVTSA and a native human Muc1 N-terminus with SumoStar tag was generated by CGS in the pcDNA3 plasmid. For the construction of a mCherry2 IRES2 copGFP expression plasmid, a mCherry2 cDNA was isolated by EcoRI and NotI digestion of pmCherry2 N1 and cloned into the EcoRI and NotI digested pPB tet IRES copGFP rtTA NeoR vector to create pPB tet mCherry2 IRES copGFP rtTA NeoR.
2.3 |. Cell lines and culture
FreeStyle 293-F cells were obtained from Thermo Fisher Scientific. Cells were cultured and maintained according to the manufacturer’s guidelines in 100-mL Wheaton Celstir glass spinner flasks (DWK Life Sciences, Millville, NJ). Cells were maintained between 0.5 × 106 and 3 × 106 cells/mL at 120 rpm, 37°C, and 8% CO2 in FreeStyle 293 Expression Medium (Thermo Fisher Scientific). 293-F transfections were performed using polyethylenimine (PEI) as previously reported (Durocher, Perret, & Kamen, 2002). Stable cell lines were created by cotransfection of the pPB tet IRES copGFP rtTA NeoR plasmids described above with a hyperactive transposase plasmid (Shurer et al., 2018) and subsequently selected with 750 μg/mL of G418 for 2 weeks. Human embryonic kidney cells transformed with the SV40 large T antigen (293-T; American Type Culture Collection, Manassas, VA) were maintained in high-glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. 293-T cells were transfected through a standard calcium phosphate transfection protocol. Cell proliferation was quantified by cell counting on a hemocytometer with trypan blue exclusion.
2.4 |. Cell sorting and SynLubricin production
293-F cells with stable incorporation of SynLubricin IRES copGFP were expanded and induced at 1 × 106 cells/mL with 1 μg/mL doxycycline for 24 hr. The top 5% of copGFP-expressing cells were collected through fluorescence activated cell sorting (FACS) on a FACSAria Fusion (BD Biosciences). Cells were subsequently expanded in the absence of doxycycline to 1 × 106 cells/mL. Cells were induced with 1 μg/mL doxycycline for 24 hr and sorted a second time, collecting the top 10% of copGFP-expressing cells. For SynLubricin production, cells were transferred to a 1 L ProCulture® glass spinner flask (Corning, Corning, NY) and induced at 2 × 106 cells/mL with 1 μg/mL doxycycline and 3.5 mM VPA. Smaller scale production of lubricin was also conducted in 100-mL Wheaton Celstir glass spinner flasks for measurement of lubricin production rates and glucose consumption rates in the presence or absence of VPA. Glucose levels were recorded with a GlucCell® glucose monitoring system (CESCO BioProducts, Atlanta, GA). To test transgene stability in the 293-F genomes, PCR amplification was performed with Q5 Hot start high-fidelity DNA polymerase (New England Biolabs Inc., Ipswich, MA) using genomic DNA as the template for the following primers: the following primers: 5’-CAGGACCTGTCTAGCTGTGCC-3’ (FWD) and 5-AGGACAGTTGTACCACACTTTGCTC-3’ (REV). Genomic DNA was extracted from stable 293-F cell lines with the GeneJET genomic DNA purification kit (Thermo Fisher Scientific). A total of 80 ng of genomic DNA was used for PCR amplification.
2.5 |. Immuno- and lectin blot analysis
Proteins in culture supernatants or purified samples were separated on NuPAGE 3–8% Tris-acetate gels (Thermo Fisher Scientific) and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 3% bovine serum albumin (BSA) in tris-buffered saline + 0.1% Tween 20 (TBST) for 2 hr. Primary antibodies were diluted 1:1000 and lectins were diluted to 1 μg/mL in 3% BSA TBST and incubated on membranes overnight at 4°C. Secondary antibodies or ExtrAvidin were diluted 1:2000 in 3% BSA TBST and incubated for 2 hr at room temperature. Blots were developed in Clarity ECL (Bio-Rad, Hercules, CA) substrate and imaged on a ChemiDoc (Bio-Rad) documentation system. Fiji was used for image processing (Schindelin et al., 2012).
2.6 |. Enzyme-linked immunosorbent assay
A custom sandwich enzyme-linked immunosorbent assay (ELISA) was used to assess the concentration of SynLubricin. A 96-well Costar® plate (Corning) was incubated overnight at 4°C with 10 μg/mL PNA in 50 mM sodium bicarbonate buffer, pH 9.5. Plates were blocked with 3% BSA in phosphate-buffered saline (PBS) for 1 hr at room temperature. Serial dilutions of fast protein liquid chromatography (FPLC)-purified bovine synovial fluid lubricin were used as standards. Samples were loaded at 1:200 dilution in Dulbecco’s PBS for 1 hr at room temperature, followed by three washes in PBS + 0.1% Tween 20 (PBST). The primary antibody used (MABT401) binds to the native PRG4 tandem repeats of human and bovine lubricin, which have approximately 90% sequence similarity to the repeats of SynLubricin. Primary antibody and secondary antibody (AP126P) were diluted 1:5000 and 1:2000, respectively, and each incubated for 1 hr at room temperature, with three washes with PBST in between antibody incubations and following the secondary antibody incubation. The ELISA was developed at room temperature with 1-Step Ultra TMB (Thermo Fisher Scientific) for 9–12 min or until a royal blue color appeared, at which point the reaction was stopped with 2 N H2SO4. Absorbance was measured at 450 nm with 540 nm background subtraction on a Tecan Spark® microplate reader, and concentrations were calculated using Magellan software with a four parameter Marquardt fit (Tecan Life Sciences, Männedorf, Switzerland).
2.7 |. Purification of recombinant SynMuc1
293-F cells were transiently transfected using the PEI protocol as previously reported (Durocher, Perret, & Kamen, 2002). After 24 hr, the media supernatant was collected. The media supernatant was diluted 1:4 in 20 mM sodium phosphate, 0.5 M NaCl, pH 7.4 and incubated with 100 μL Ni Sepharose® excel resin (17371201; GE Healthcare, Chicago, IL) overnight at 4°C. Sample flow through was collected using a gravity column (29922; Thermo Fisher Scientific). The resin was washed with 5 mL 20 mM sodium phosphate, 0.5 M NaCl, 5 mM imidazole, pH 7.4. SynMuc1 was eluted with 5 mL of 20 mM sodium phosphate, 0.5 M NaCl, 500 mM imidazole, pH 7.4. SynMuc1 was desalted into PBS using a Zeba Spin Desalting Column (87766; Thermo Fisher Scientific).
2.8 |. Purification of recombinant SynLubricin
SynLubricin was purified from SynLubricin IRES copGFP positive 293-F cell culture supernatant by FPLC with Q Sepharose® resin (GE Healthcare). The supernatant was diluted 1:10 with 50 mM Tris-HCl buffer, pH 7.5, and loaded onto the column. The column was washed with 50 mM Tris-HCl, 500 mM NaCl, pH 7.5. Purified SynLubricin was collected by eluting with 50 mM Tris-HCl, 1 M NaCl, pH 7.5. The purified SynLubricin was dialyzed into PBS using a Tube-O-Dialyzer (G-Biosciences, St. Louis, MO) overnight at 4°C. The final purified product was obtained by concentrating with a SpeedVac Vacuum Concentrator on the low setting (Thermo Fisher Scientific).
2.9 |. Tribology
The performance of SynLubricin as a boundary lubricant was assessed using a custom linear reciprocating tribometer as previously described (Gleghorn & Bonassar, 2008). Briefly, cylindrical cartilage explants (6 mm diameter × 2 mm thickness) were harvested from the femoral condyles of neonatal bovine stifles. Endogenous cartilage-bound lubricin was extracted using a 30 min incubation in 1.5 M NaCl, followed by a 1 hr equilibration step in PBS. Explants were incubated in either PBS, PBS + SynLubricin, or bovine synovial fluid for 15–20 min before loading onto a tribometer in a 1 mL bath of the respective fluid. Explants were compressed to approximately 30% strain against a glass counter-face and permitted to depressurize over the course of 1 hr. After reaching an equilibrium normal load, the counter-face was linearly reciprocated at a speed of 0.3 mm/s for three cycles. Simultaneously, a biaxial load cell recorded the normal and shear loads. For both the forward and reverse directions and at each speed, the friction coefficient was calculated as the mean shear force while sliding divided by the equilibrium normal load.
2.10 |. Statistical analysis
Statistical significance was determined by one-way analysis of variance (ANOVA) or Student’s t test (two-tailed) as appropriate using Prism (GraphPad, La Jolla, CA). For the lubrication data, a one-way ANOVA with Tukey’s post hoc tests was performed to compare mean friction coefficients across all lubricants. All graphs were generated in Prism.
3 |. RESULTS AND DISCUSSION
3.1 |. Design and synthesis of cDNA for synonymous lubricin
As an approach for recombinant mucin production, we applied a codon scrambling and optimization strategy to design synthetic mucin cDNAs within minimal codon repetition (Figure 1a). A global codon optimization algorithm was applied to find the least repetitive gene sequence that encoded the desired-mucin tandem repeats (Tang & Chilkoti, 2016). To tailor the sequences for production in a human host system, such as 293-F, a subsequent optimization was conducted to replace any codons with less than 10% usage frequency in humans (Figure 1a). On the basis of previous findings with elastin-like proteins, we envisioned that the optimized mucin cDNAs could be synthesized through rapid- and low-cost services for CGS (Kosuri & Church, 2014; Tang & Chilkoti, 2016).
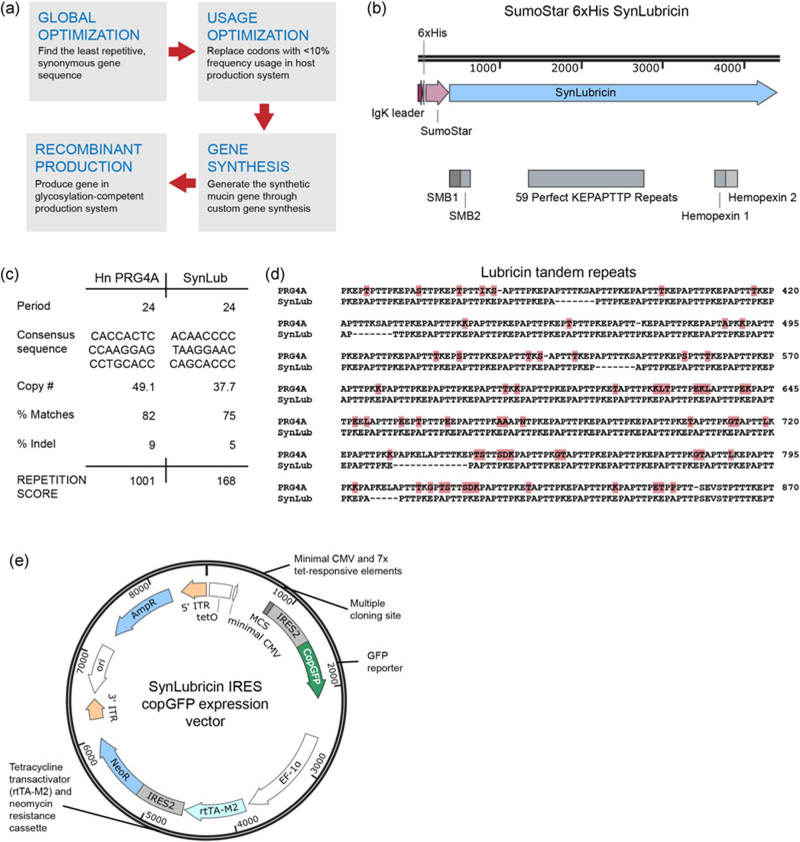
FIGURE 1.
Design and synthesis of synonymous lubricin (SynLubricin). (a) Overview of the design and production strategy for synthetic, codon-scrambled mucins. DNA sequences for the desired protein product were optimized through a global optimization to minimize repetitive DNA sequences by codon scrambling, followed by a second optimization that reassigned codons with infrequent usage in the host cell system. (b) SynLubricin was constructed of 59 perfect repeats of KEPAPTTP flanked by the native human N- and C-termini of PRG4. An IgK signal sequence, 6× histidine tag, and SumoStar tag were fused to SynLubricin for secretion and purification. SynLubricin also retains the two somatomedin B domains (SMB 1 and 2) and the two hemopexin domains of the native protein. (c) Calculated repetition score for the nucleotides encoding the tandem repeats of human PRG4 isoform A (PRG4A) and SynLubricin. (d) Alignment of the amino acid sequence of human PRG4 and SynLubricin. (e) Vector map illustrating the tetracycline-inducible promoter, multiple cloning site (MCS) for the cDNA of interest, bicistronic GFP reporter (IRES2 CopGFP), and the second expression cassette for the rtTA-M2 tetracycline transactivator and neomycin-resistance gene. cDNA: complementary DNA; IRES: internal ribosome entry site [Color figure can be viewed atwileyonlinelibrary.com]
We first tested the strategy for human lubricin, which has approximately 59 tandem repeats with a consensus sequence of KXPXPTTX, with KEPAPTTP being the most frequent repeat. For our synthetic lubricin, we optimized the codons for 59 perfect repeats of the KEPAPTTP consensus sequence (Figure 1b). The protein sequence for the perfect repeats had approximately 73% similarity to the native repeats of human PRG4 isoform A (Figure 1c). The synthetic tandem repeats were flanked by additional sequences encoding the native N- and C- termini of human PRG4. These sequences included the native somatomedin and hemopexin domains of lubricin. We also included an IgK leader sequence, 6× histidine tag, and N-terminal SumoStar tag to aid in protein secretion and purification (Figure 1b). We named the new semisynthetic gene “synonymous lubricin” or “SynLubricin.”
The nucleotides encoding SynLubricin were significantly less repetitive than native PRG4. We analyzed the nucleotide sequences with an alignment algorithm that detects tandem repeats and scores their degree of repetitiveness based on how frequently they repeat and how closely the identified consensus matches the nucleotides of the queried sequence (Benson, 1999). The detected repeats were aligned with the queried sequence through a Smith–Waterman style local alignment, and the overall repetitiveness was scored by assigning +2 for each nucleotide match and −7 for each mismatch or indel (Benson, 1999). Thus, a higher score was indicative of more nucleotide repetition. The tandem repeats of SynLubricin had a modest score of 168, whereas the native PRG4 repeats had a much higher repetition score of 1001.
We also aligned the amino acids of the SynLubricin tandem repeats to the tandem repeats of human PRG4 isoform A (Figure 1d). We noted that the perfect repeats of SynLubricin and the native repeats of human PRG4A have similar compositions of alanine, glutamic acid, lysine, and threonine, whereas proline content is slightly higher in the SynLubricin repeats (37% vs. 30.5%; Table S1). We also noted that the native repeats contain small amounts of asparagine (0.2%), aspartic acid (0.4%), glycine (0.8%), isoleucine(0.2%), leucine (1.4%), and serine (2.6%), which are not contained in SynLubricin (Table S1).
The low-repetition of nucleotides in the SynLubricin gene enabled synthesis of the desired cDNA through standard CGS services. We also commissioned the generation of a cDNA for the native human lubricin/PRG4 sequence through a specialized service for highly repetitive genes. However, our attempts to subsequently clone the native PRG4 cDNA sequence into a mammalian expression vector and recombinantly express the product in mammalian cells failed. Consequently, we discontinued further efforts at recombinant production of lubricin with the full-length, native cDNA.
Efforts to produce SynLubricin in transiently transfected mammalian cells were successful. The SynLubricin cDNA was fused to a bicistronic copGFP reporter and transiently transfected into adherent human embryonic kidney 293-T cells. The protein product of the SynLubricin gene was highly glycosylated and exhibited the antiadhesive properties that we expected. Transfected cells maintained large gaps between cells in the monolayer, particularly at locations where visible copGFP fluorescence reported high expression levels of the bicistronic messenger RNA (Figure S1A). We noted that these observations were consistent with the known antiadhesive functionality of native lubricin (Rhee et al., 2005). In contrast, mock transfected cells grew to a highly confluent monolayer in culture (Figure S1A). A Western blot of the media supernatant from the SynLubricin-transfected cultures revealed a high molecular weight protein of approximately 460 kDa, which was similar in size to the native lubricin that we detected in equine synovial fluid (Figure S1B). The expected molecular weight of the peptide backbone of SynLubricin was 145 kDa, indicating that SynLubricin was extensively glycosylated.
We next considered strategies for stable production of synthetic mucins in 293-F suspension cultures. On the basis of our previous findings that mucin cDNAs could be efficiently integrated into host cell genomes through transposition, we created a nonviral transposon vector for “all-in-one” inducible expression of mucins (Shurer et al., 2018). The vector contained a tetracycline-responsive promoter for inducible expression of the desired gene and a bicistronic copGFP reporter. The vector also contained a second cassette under control of an EF1α promoter for expression of the rtTA-M2 tetracycline transactivator and a bicistronic neomycin-resistance gene for selection (Figure 1e). To test the performance of the expression system, we cloned mCherry2 into the vector and transfected 293-F cells with cationic PEI condensates following standard protocols (Boussif et al., 1995; de los Milagros Bassani Molinas, Beer, Hesse, Wirth, & Wagner, 2014; Sonawane, Szoka Jr. & Verkman, 2003). Stable cell populations were isolated after 2 weeks of selection, and mCherry2 production was validated by flow cytometry. On the basis of the flow cytometric analysis, we found that stable cells produced high levels of mCherry2 and that the fluorescence readout of the copGFP reporter was generally a good indicator of recombinant protein production (Figure S2).
3.2 |. Design and synthesis of cDNA for synonymous Muc1
We tested whether our strategy for construction of mucin-type cDNAs was generalizable and could be applied to other mucins. We chose the mucin Muc1, which is important in the hydration and protection of the cornea and other epithelial surfaces (Mantelli & Argüeso, 2008). We noted that the native tandem repeats of Muc1 are polymorphic, with 42-perfect repeats being most frequent in humans (Nath & Mukherjee, 2014). We applied the codon optimization strategy to design a cDNA for 42-perfect Muc1 repeats, PDTRPAPGSTAPPAHGVTSA. The optimized sequence was fused to the codons for the native N-terminus of human Muc1. We also added the IgK leader sequence, 6× histidine tag, and SumoStar tag, similarly to SynLubricin (Figure S3A). We calculated a very high repetition score of 4,997 for the nucleotide coding sequence of the native human Muc1 tandem repeats. The repetition score was reduced to 220 in our synthetic cDNA, which we referred to as SynMuc1 (Figure S3B).
The optimized coding sequence for SynMuc1 was synthesized through standard CGS services, whereas our request to synthesize the extremely repetitious sequence of the native Muc1 cDNA was rejected by multiple commercial service providers. The custom synthesized SynMuc1 cDNA was transfected into 293-F cells. The recombinant protein was purified from the media supernatant via immobilized metal affinity chromatography (IMAC) and detected by Western blot with an antibody against the native human tandem repeats (Figure S3C). The recombinant mucin was extensively O-glycosylated, as indicated by the strong signal when probed with PNA, a lectin that is specific for a core-1, mucin-type disaccharide (Figure S3D).
During purification, we noticed that a significant percentage of the mucin failed to bind to the IMAC resin and was detected in the flow through (Figure S3C,D). Western blot analysis confirmed the presence of the 6× histidine SumoStar purification tag on the recombinant protein in the flow through and eluted fractions, suggesting that the N-terminus and purification tag were present but inaccessible to the immobilized IMAC cations as would be the case, for example, if the tag was buried in the random coil of the mucin biopolymer (Figure S3E). Because our goal was to demonstrate the production of the recombinant SynMuc1 and not to optimize its purification, alternative chromatography approaches were not explored.
3.3 |. Stable host production of recombinant SynLubricin
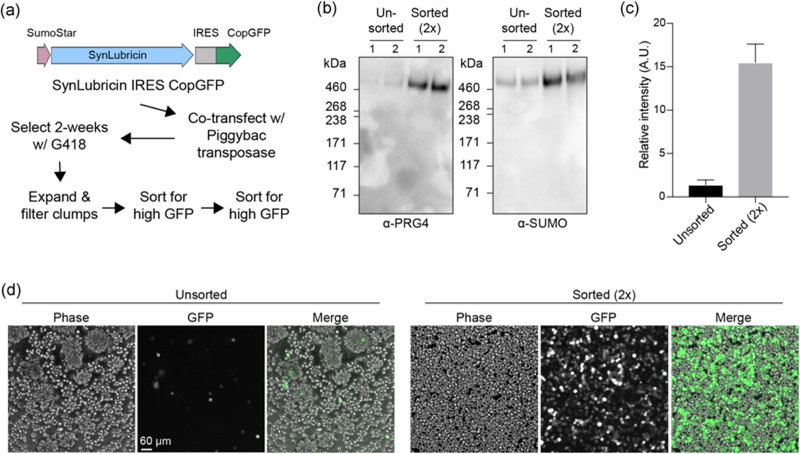
We attempted to generate a stable 293-F cell line for production of SynLubricin using the transposase system for genome editing (Figure 1e and Figure 2a). We found that after selection with G418, comparatively few cells exhibited high copGFP reporter levels following doxycycline induction (Figure 2b). To overcome the issue, we applied a two-round sorting strategy using the copGFP reporter to isolate a subpopulation of cells that expressed SynLubricin at high levels. Stable cells were expanded and sorted for the top 5% copGFP expressers, which were then expanded and sorted a second time for the top 10% expressers. We found that the sorting strategy improved SynLubricin production 15-fold and did not impact the molecular weight of the glycosylated protein product (Figure 2b,c). The sorted cell populations displayed noticeably higher levels of the copGFP reporter after induction with doxycycline, indicating successful isolation of a polyclonal population with higher gene expression levels (Figure 2d).
FIGURE 2.
Sorting strategy to isolate stable polyclonal cell populations that produce high levels of SynLubricin. (a) Strategy for the isolation of stable cell populations expressing high levels of SynLubricin. (b) Western blots of 293-F media supernatant showing relative SynLubricin production in unsorted and twice-sorted (2×) cell populations; 1 and 2 indicate samples from two independent experiments; probed with anti-PRG4 (MABT401) and SUMO antibodies. (c) Quantification of the relative intensity of signal from anti-PRG4 western blots in b. (d) Phase-contrast and fluorescence micrographs of unsorted and twice-sorted 293-F cells expressing SynLubricin [Color figure can be viewed at wileyonlinelibrary.com]
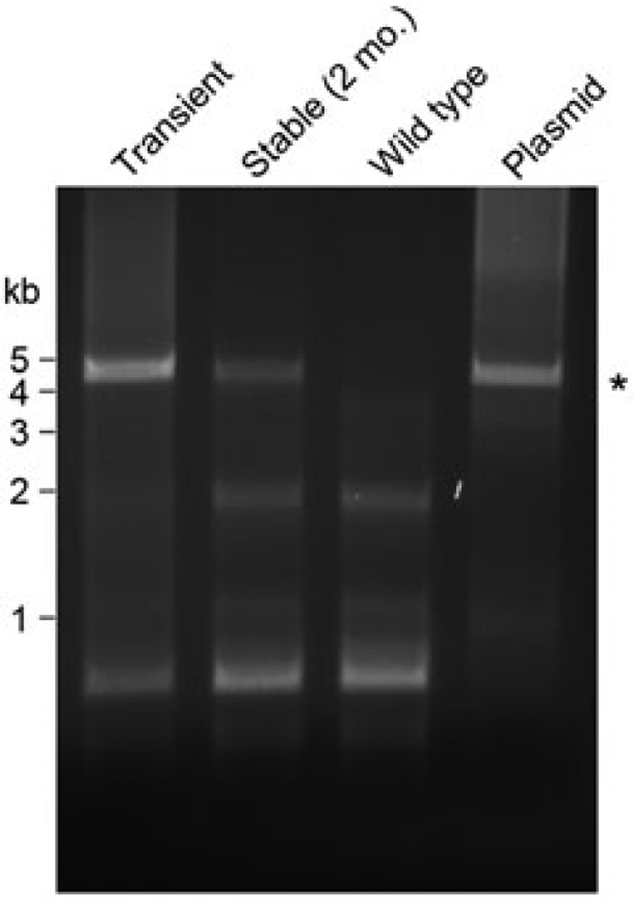
To confirm the cDNA stability of the integrated SynLubricin gene in our stable 293-F cells, genomic DNA was extracted from modified 293-F cells after 2 months of continuous culture. The SynLubricin cDNA was then amplified by polymerase chain reaction (PCR) using primers that were specific to SynLubricin (Figure 3). The amplified gene was approximately 4 kb in length, as expected, and indistinguishable in size from similarly amplified genes obtained using the original SynLubricin plasmid as the template or DNA extracted from transiently transfected cells (Figure 3). Even after culture for 2 months, the polyclonal cell population exhibited no apparent gene amplification or deletion, indicating a satisfactory level of genomic stability (Figure 3).
FIGURE 3.
Integrated SynLubricin cDNA is stable in the cellular genome. PCR amplification of SynLubricin coding region in genomic DNA extracts of wild-type and stably integrated 293-F cells cultured continuously for 2 months. As positive controls, PCR amplification of SynLubricin plasmid (Plasmid) and DNA extract from SynLubricin transiently transfected 293-F cells (Transient) are shown. The expected size of full-length SynLubricin is indicated by the star. cDNA: complementary DNA; PCR: polymerase chain reaction
3.4 |. Optimization of SynLubricin production
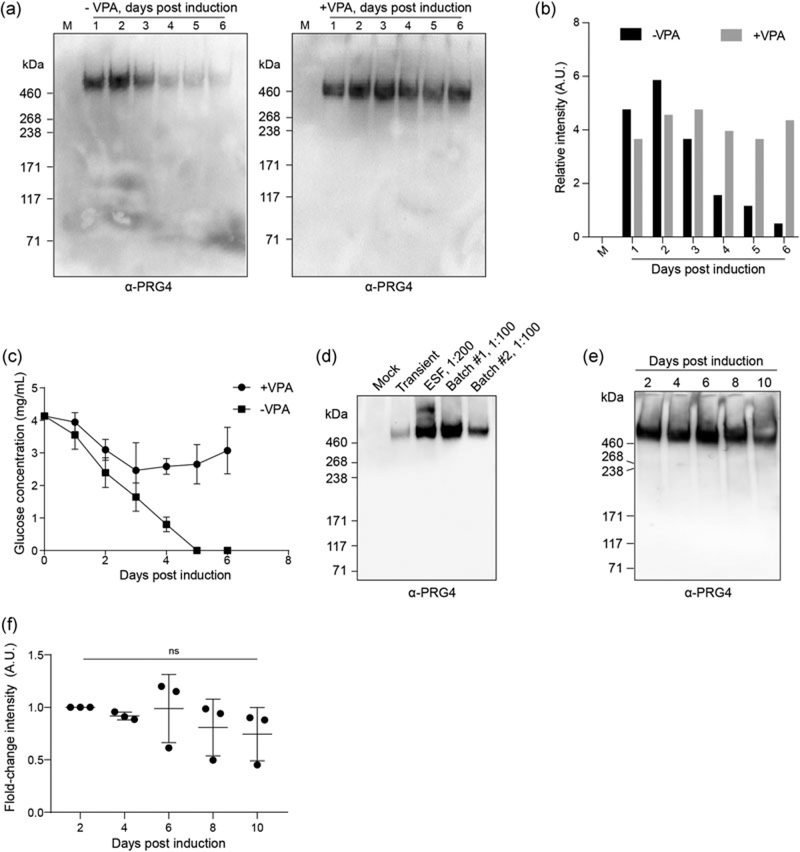
We considered whether SynLubricin productivity could be improved through the addition of the histone deacetylase inhibitor, VPA, which has previously been shown to drastically increase production of some recombinant proteins in 293-F cells (Backliwal et al., 2008). Our sorted cell population was induced with doxycycline in the presence or absence of 3.5 mM VPA, and media supernatants were sampled each subsequent day from batch cultures. The molecular weights of the protein products were similar, suggesting that VPA did not appreciably affect the total extent of glycosylation of the protein product (Figure 4a). Interestingly, the recombinant protein levels peaked at approximately 2–3 days postinduction in cultures without VPA and declined rapidly thereafter (Figure 4b). In VPA-treated cultures, SynLubricin levels in the media did not decline as significantly over time. We ruled out protein degradation as a likely explanation for the decline of recombinant protein in cultures without VPA because we saw no prominent degradation products for lubricin on Western blots (Figure 4a). We instead considered the possibility that the 293-F culture might consume the recombinant protein in conditions of reduced nutrient availability. Consistent with this possibility, we observed that the decline in recombinant protein levels coincided with the depletion of glucose in the cultures without VPA (Figure 4c). Glycolytic activity largely ceased in VPA-treated cultures after 3 days, as indicated by a sharp decline in glucose consumption (Figure 4c). Thus, VPA may prevent the loss of recombinant protein in batch cultures through slowing 293-F cellular metabolism.
FIGURE 4.
Optimization of SynLubricin production. (a) Western blots showing relative production of SynLubricin over time in control media (M) and media superntant from sorted 293-F cells induced with 1 μg/mL doxycycline for the indicated number of days in the absence or presence of the histone deacetylase inhibitor valproic acid (VPA; 3.5 mM). (b) Quantification of the relative intensity of signal for the blots shown in a. (c) Time course for glucose concentration in the media of sorted 293-F cells induced at Day 0 with 1 μg/mL doxycycline with or without 3.5 mM VPA. Mean and SD are shown, n = 3. (d) Western blot showing lubricin in the media harvested from nonproducing control cells (Mock), cells transiently transfected with SynLubricin cDNA (Transient), and two successive 1 L batch cultures of sorted 293-F cells induced for 3 days with 1 μg/mL doxycycline and 3.5 mM VPA (Batch #1 and Batch #2); equine synovial fluid (ESF) was loaded as a control. (e) Representative Western blot of SynLubricin produced from stably expressing 293-F cells collected at indicated time points after 1 μg/mL doxycycline induction on Day 0 with media replacement every other day. (f) Quantification of Western blot shown in (e) along with two additional replicates, n = 3. cDNA: complementary DNA; ns: not significant; SD: standard deviation
We next scaled up production to 1 L bioreactors operated in batch mode and conducted two independent production runs with VPA added. Each production run yielded a plentiful recombinant protein that was comparable in molecular weight to both recombinant protein isolated from transiently transfected cultures and native lubricin detected in equine synovial fluid (Figure 4d). An ELISA using purified bovine lubricin as a standard reported approximately 200 mg/L of SynLubricin in the batch runs with our stable 293-F lines. Less than 50% of the stable cell population showed strong expression of the copGFP reporter in the batch bioreactors, suggesting that increases in productivity could likely be achieved with clonal expansion of the production cell line (Figure 2d). We noted that an important limitation of our ELISA-based quantification was the use of a bovine standard, which may over- or under-estimate SynLubricin levels.
Finally, we tested whether stable protein production could be achieved with periodic media changes to avoid nutrient depletion. Conditioned media was harvested from doxycycline-induced cultures that were maintained for 10 consecutive days in the absence of VPA. Media in the batch cultures was exchanged every 48 hrs to replenish nutrients and remove metabolic waste products. Viable cell concentration was also reduced to 1 × 106 cells/mL every 48 hrs. SynLubricin production levels were stable over the 10 days of culture, and the SynLubricin molecular weight was constant, indicating that glycosylation levels were also stable (Figure 4e). While there potentially appeared to be a slight decrease in SynLubricin production with time, there was no significant difference in protein yield (Figure 4f).
3.5 |. SynLubricin is a functional biolubricant
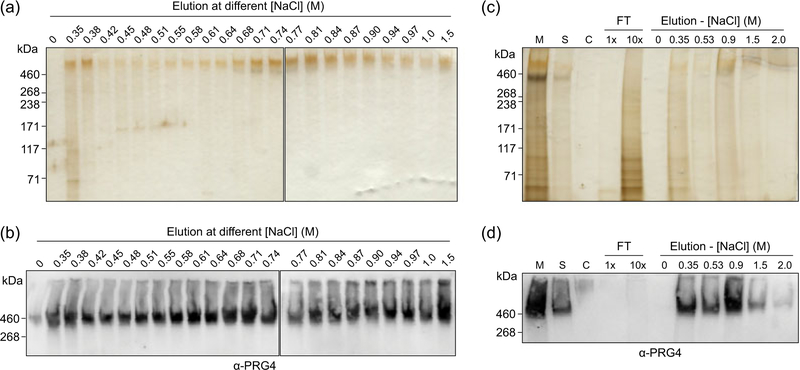
Recombinant SynLubricin was effectively purified with anion-exchange chromatography following our previously reported strategy for isolation of native lubricin from equine synovial fluid, with slight modification from using DEAE–Sepharose® to using Q Sepharose® (Reesink et al., 2016). We also attempted immobilized metal affinity chromatography (IMAC) for purification, but the recombinant SynLubricin had poor affinity to IMAC resins (Figure S4). We reasoned that the N-terminal histidine tag could be buried in the large, random coil of the SynLubricin tandem repeats and abandoned the IMAC approach. We found SynLubricin bound to the anion-exchange resin strongly and eluted continuously over high-salt concentrations ranging from approximately 350 mM to 1.5 M (Figure 5a,b). The continuous elution of SynLubricin was likely explained by a varying frequency of anionic sialic acids in the O-glycans of the recombinant SynLubricin (Estrella et al., 2010). We found that a stringent wash step of approximately 500 mM NaCl could remove most protein contaminants detectable by silver stain, although some SynLubricin was inevitably lost to this high-salt wash (Figure 5c,d).
FIGURE 5.
Purification of SynLubricin by anion-exchange chromatography. (a) Silver stain and (b) Western blot showing that SynLubricin elutes continuously from Q Sepharose® resin over a broad range of NaCl concentrations, as indicated. (c) Silver stain and (d) Western blot showing harvested SynLubricin media supernatant (M), 10-fold diluted SynLubricin media supernatant (S), wild-type 293-F conditioned media(C), flow through (FT-1×), 10-fold concentrated flow through (FT-10×), and eluted fractions at the indicated salt concentration [Color figure can be viewed at wileyonlinelibrary.com]
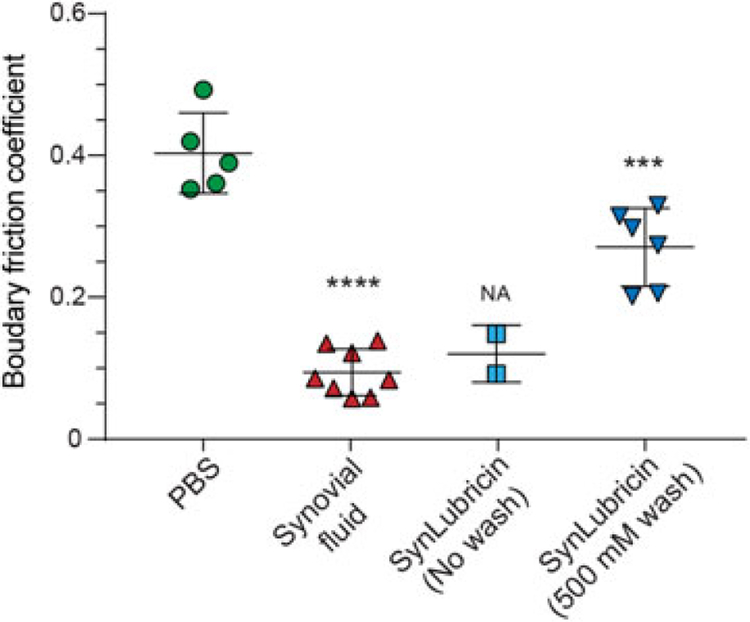
To ensure functionality of recombinant SynLubricin, we tested its ability to lubricate cartilage and reduce friction using a custom linear reciprocating tribometer (Jones et al., 2007). Recombinant SynLubricin was purified via anion-exchange chromatography using the stringent 500 mM NaCl wash step to eliminate most protein contaminants (Figure 5d). Following purification, SynLubricin was dialyzed in saline and diluted to physiological concentrations. Lubrication was tested on bovine articular cartilage explants where the native lubricin boundary layer had been extracted. Compared with saline control, we found that SynLubricin-containing solutions, as well as control synovial fluid, significantly reduced the boundary friction of cartilage explants (Figure 6; p < 0.001 and 0.0001, respectively).
FIGURE 6.
Lubrication of cartilage explants shows the functionality of SynLubricin. Friction coefficients of NaCl-extracted cartilage explants bathed in saline (PBS), bovine synovial fluid or SynLubricin eluted either without washing or with a stringent 500 mM NaCl wash. Mean and SD is shown with independent measurements indicated. ***p < 0.001, ****p < 0.0001. NA: statistical testing is not applicable due to sample size; PBS: phosphate-buffered saline; SD: standard deviation [Color figure can be viewed at wileyonlinelibrary.com]
We also tested a small quantity of a second SynLubricin sample that was purified without the stringent wash of the anion-exchange column with 500 mM NaCl. Notably, cartilage friction coefficients were markedly lower for this SynLubricin preparation than the more stringently washed SynLubricin preparations (Figure 6). It should be noted that a low sample volume for the unwashed SynLubricin preparation precluded obtaining enough independent measurements for meaningful statistical comparisons (Figure 6). Nevertheless, further optimization of purification conditions may yield recombinant lubricin fractions with improved performance in biolubrication. For instance, future studies should investigate whether the less negatively charged lubricin fractions that elute at lower salt concentrations (350–500 mM NaCl) are important for cartilage biolubrication, either by acting independently or in synergy with more negatively charged lubricin fractions. Alternatively, studies should consider whether any “contaminants” that are eliminated with the 500 mM NaCl wash might act synergistically with lubricin in cartilage lubrication.
4 |. CONCLUSIONS
Taken together, our studies provide a potential roadmap to larger-scale mucin biomanufacturing. Success in the design and synthesis of new semisynthetic genes for both Muc1 and lubricin, combined with our success in isolating stable, lubricin-expressing cell populations, suggests that this approach may be broadly applicable for recombinant mucins with long repetitive domains. Also of note is the successful demonstration of recombinant production in a human cell system that avoids the risk of immunogenic Galα1,3-Gal and Neu5Gc epitopes. We find that the recombinant product of our SynLubricin gene is functional in its ability to resist cellular adhesion (Figure S1A) and lubricate biological surfaces, such as cartilage (Figure 6). Thus, SynLubricin may have the potential for diverse applications ranging from injectables for osteoarthritis to topical treatments for chronic dry eye. Moreover, given the speed and low cost of CGS, the approach presented in this study could, in principle, be applied to rapidly prototype designer mucins with new or modified functional domains.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Weaver and J. Lakins for the original transposon plasmids that were further modified in this study. This investigation was supported by the National Institute of General Medicine Sciences Ruth L. Kirschstein National Research Service Award 2T32GM008267 (C. R. S.), Knight Family Foundation Graduate Research Fellowship in Nanoscience and Technology (C. R. S.), Samuel C. Fleming Family Graduate Fellowship (C. R. S. and Z. C.), National Science Foundation Graduate Research Fellowship (E.F., DGE-1650441), National Institute of Health T35 training grant OD010941 (M. A. S.), National Institute of Arthritis and Musculoskeletal and Skin Diseases/National Institute of Health K08AR068469(H. L. R.), National Institute of Health New Innovator DP2 GM229133 (M. J. P.), and National Cancer Institute U54 CA210184(M. J. P.). Flow cytometry was carried out at the Cornell University Biotechnology Resource Center.
Funding information
National Institute of Arthritis and Musculoskeletal and Skin Diseases, Grant/Award Number: K08AR068469; National Institutes of Health Director’s Program, Grant/Award Number: DP2 GM229133; National Cancer Institute, Grant/Award Number: U54 CA210184; National Institutes of Health, Grant/Award Number: T35 OD010941; National Institutes of Health National Research Service Award, Grant/Award Number: 2T32GM008267; National Science Foundation Graduate Research Fellowship, Grant/Award Number: DGE-1650441
Footnotes
CONFLICT OF INTERESTS
The authors have a pending patent related to cDNA sequences described in this work.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Backliwal G, Hildinger M, Kuettel I, Delegrange F, Hacker DL, & Wurm FM (2008). Valproic acid: A viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnology and Bioengineering, 101(1), 182–189. 10.1002/bit.21882 [DOI] [PubMed] [Google Scholar]
- Bäckström M, Link T, Olson FJ, Karlsson H, Graham R, Picco G, … Hansson GC (2003). Recombinant MUC1 mucin with a breast cancer-like O-glycosylation produced in large amounts in Chinese-hamster ovary cells. Biochemical Journal, 376(Pt 3), 677–686. 10.1042/bj20031130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahabri SA, Suwairi WM, Laxer RM, Polinkovsky A, Dalaan AA, & Warman ML (1998). The camptodactyly-arthropathy-coxa vara-pericarditis syndrome: Clinical features and genetic mapping to human chromosome 1. Arthritis & Rheumatism, 41(4), 730–735. [DOI] [PubMed] [Google Scholar]
- Benson G (1999). Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Research, 27(2), 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosques CJ, Collins BE, Meador JW, Sarvaiya H, Murphy JL, DelloRusso G, … Venkataraman G (2010). Chinese hamster ovary cells can produce galactose-α−1,3-galactose antigens on proteins. Nature Biotechnology, 28(11), 1153–1156. 10.1038/nbt1110-1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, & Behr JP (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America, 92(16), 7297–7301. 10.1073/pnas.92.16.7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA (2004). Appropriate glycosylation of recombinant proteins for human use. Molecular Biotechnology, 28(3), 241–255. 10.1385/MB:28:3:241 [DOI] [PubMed] [Google Scholar]
- Butler M, & Spearman M (2014). The choice of mammalian cell host and possibilities for glycosylation engineering. Current Opinion in Biotechnology, 30, 107–112. 10.1016/j.copbio.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Dhanisha SS, Guruvayoorappan C, Drishya S, & Abeesh P (2018). Mucins: Structural diversity, biosynthesis, its role in pathogenesis and as possible therapeutic targets. Critical Reviews in Oncology/Hematology, 122, 98–122. 10.1016/j.critrevonc.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Dumont J, Euwart D, Mei B, Estes S, & Kshirsagar R (2016). Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Critical Reviews in Biotechnology, 36(6), 1110–1122. 10.3109/07388551.2015.1084266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher Y, Perret S, & Kamen A (2002). High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Research, 30(2), E9–E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaid KA, Fleming BC, Oksendahl HL, Machan JT, Fadale PD, Hulstyn MJ, … Jay GD (2008). Decreased lubricin concentrations and markers of joint inflammation in the synovial fluid of patients with anterior cruciate ligament injury. Arthritis & Rheumatism, 58(6), 1707–1715. 10.1002/art.23495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella RP, Whitelock JM, Packer NH, & Karlsson NG (2010). The glycosylation of human synovial lubricin: Implications for its role in inflammation. Biochemical Journal, 429(2), 359–367. 10.1042/bj20100360 [DOI] [PubMed] [Google Scholar]
- Flannery CR, Zollner R, Corcoran C, Jones AR, Root A, Rivera-Bermúdez MA, … Glasson SS (2009). Prevention of cartilage degeneration in a rat model of osteoarthritis by intraarticular treatment with recombinant lubricin. Arthtitis and Rheumatism, 60(3), 840–847. 10.1002/art.24304 [DOI] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, & Verstrepen KJ (2010). Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual Review of Genetics, 44(1), 445–477. 10.1146/annurev-genet-072610-155046 [DOI] [PubMed] [Google Scholar]
- Ghaderi D, Zhang M, Hurtado-Ziola N, & Varki A (2012). Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnology & Genetic Engineering Reviews, 28, 147–175. 10.5661/bger-28-147 [DOI] [PubMed] [Google Scholar]
- Gipson IK, Spurr-Michaud S, Tisdale A, & Menon BB (2014). Comparison of the transmembrane Mucins MUC1 and MUC16 in epithelial barrier function. PLOS One, 9(6), e100393 10.1371/journal.pone.0100393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleghorn JP, & Bonassar LJ (2008). Lubrication mode analysis of articular cartilage using Stribeck surfaces. Journal of Biomechanics, 41(9), 1910–1918. 10.1016/j.jbiomech.2008.03.043 [DOI] [PubMed] [Google Scholar]
- Hang HC, & Bertozzi CR (2005). The chemistry and biology of mucin-type O-linked glycosylation. Bioorganic & Medicinal Chemistry, 13(17), 5021–5034. 10.1016/j.bmc.2005.04.085 [DOI] [PubMed] [Google Scholar]
- Hattrup CL, & Gendler SJ (2008). Structure and function of the cell surface (tethered) mucins. Annual Review of Physiology, 70(1), 431–457. 10.1146/annurev.physiol.70.113006.100659 [DOI] [PubMed] [Google Scholar]
- Jones ARC, Gleghorn JP, Hughes CE, Fitz LJ, Zollner R, Wainwright SD, … Flannery CR (2007). Binding and localization of recombinant lubricin to articular cartilage surfaces. Journal of Orthopaedic Research, 25(3), 283–292. 10.1002/jor.20325 [DOI] [PubMed] [Google Scholar]
- Kosinska MK, Ludwig TE, Liebisch G, Zhang R, Siebert H-C, Wilhelm J, … Steinmeyer J (2015). Articular joint lubricants during osteoarthritis and rheumatoid arthritis display altered levels and molecular species. PLOS One, 10(5), e0125192 10.1371/journal.pone.0125192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, & Church GM (2014). Large-scale de novo DNA synthesis: Technologies and applications. Nature Methods, 11, 499–507. 10.1038/nmeth.2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC-H, Gandhi JG, Zia RN, & Paszek MJ (2018). Physical biology of the cancer cell glycocalyx. Nature Physics, 14(7), 658–669. 10.1038/s41567-018-0186-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Graverand-Gastineau MP (2010). Disease modifying osteoarthritis drugs: Facing development challenges and choosing molecular targets. Current Drug Targets, 11(5), 528–535. 10.2174/138945010791011893 [DOI] [PubMed] [Google Scholar]
- López Castel A, Cleary JD, & Pearson CE (2010). Repeat instability as the basis for human diseases and as a potential target for therapy. Nature Reviews Molecular Cell Biology, 11, 165–170. 10.1038/nrm2854 [DOI] [PubMed] [Google Scholar]
- de los Milagros Bassani Molinas M, Beer C, Hesse F, Wirth M, & Wagner R (2014). Optimizing the transient transfection process of HEK-293 suspension cells for protein production by nucleotide ratio monitoring. Cytotechnology, 66(3), 493–514. 10.1007/s10616-013-9601-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantelli F, & Argüeso P (2008). Functions of ocular surface mucins in health and disease. Current Opinion in Allergy and Clinical Immunology, 8(5), 477–483. 10.1097/ACI.0b013e32830e6b04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino J, Carpten JD, Suwairi WM, Gutierrez OM, Schwartz S, Robbins C, … Warman ML (1999). CACP, encoding a secreted proteoglycan, is mutated in camptodactyly-arthropathy-coxa vara-pericarditis syndrome. Nature Genetics, 23, 319–322. 10.1038/15496 [DOI] [PubMed] [Google Scholar]
- Mauris J, & Argüeso P (2012). Mucins and Galectin-3 in ocular surface health and disease, Galectins and Disease Implications for Targeted Therapeutics (1115, 409–414). American Chemical Society. 10.1021/bk-2012-1115.ch025 [DOI] [Google Scholar]
- Nath S, & Mukherjee P (2014). Muc1: A multifaceted oncoprotein with a key role in cancer progression. Trends in Molecular Medicine, 20(6), 332–342. 10.1016/j.molmed.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M, Barela Hudgell MA, D’Allura B, Agronin J, Gross A, Podini D, & Smith LC (2016). Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genomics, 17, 900 10.1186/s12864-016-3241-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Edamura KN, & Cleary JD (2005). Repeat instability: Mechanisms of dynamic mutations. Nature Reviews Genetics, 6, 729–742. 10.1038/nrg1689 [DOI] [PubMed] [Google Scholar]
- Reesink HL, Bonnevie ED, Liu S, Shurer CR, Hollander MJ, Bonassar LJ, & Nixon AJ (2016). Galectin-3 binds to lubricin and reinforces the lubricating boundary layer of articular cartilage. Scientific Reports, 6, 25463 10.1038/srep25463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, … Carpten JD (2005). The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. Journal of Clinical Investigation, 115(3), 622–631. 10.1172/jci22263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsom ML, Morrison S, Masala N, Sullivan BD, Sullivan DA, Sheardown H, & Schmidt TA (2014). Characterization of full-length recombinant human Proteoglycan 4 as an ocular surface boundary lubricant. Experimental Eye Research, 127, 14–19. 10.1016/j.exer.2014.06.015 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, … Cardona A (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods, 9, 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TA, Sullivan DA, Knop E, Richards SM, Knop N, Liu S, … Sullivan BD (2013). Transcription, translation, and function of lubricin, a boundary lubricant, at the ocular surface. JAMA Ophthalmology, 131(6), 766–776. 10.1001/jamaophthalmol.2013.2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurer CR, Colville MJ, Gupta VK, Head SE, Kai F, Lakins JN, & Paszek MJ (2018). Genetically encoded toolbox for glycocalyx engineering: Tunable control of cell adhesion, survival, and cancer cell behaviors. ACS Biomaterials Science & Engineering, 4(2), 388–399. 10.1021/acsbiomaterials.7b00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane ND, Szoka FC Jr., & Verkman AS (2003). Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. Journal of Biological Chemistry, 278(45), 44826–44831. 10.1074/jbc.M308643200 [DOI] [PubMed] [Google Scholar]
- Tang NC, & Chilkoti A (2016). Combinatorial codon scrambling enables scalable gene synthesis and amplification of repetitive proteins. Nature Materials, 15(4), 419–424. 10.1038/nmat4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.