Abstract
Background
Microbial shifts have been associated with disease activity in Crohn’s disease [CD], but findings on specific taxa are inconsistent. This may be due to differences in applied methods and cross-sectional study designs. We prospectively examined the faecal microbiota in adult CD patients with changing or stable disease course over time.
Methods
Faeces were collected at two time-points from 15 healthy control individuals [HCs], 35 CD patients who were in remission and who maintained remission [RRs], and 22 CD patients during remission and also during subsequent exacerbation [RAs]. The microbial composition was assessed by 16S rRNA [V4] gene sequencing.
Results
Compared with HCs, patients with CD had a lower microbial richness [p = 0.0002] and diversity [p = 0.005]. Moreover, the microbial community structure of a subset of patients, clustered apart from HCs, was characterized by low microbial diversity and Faecalibacterium abundance. Patients within this cluster did not differ with respect to long-term disease course compared with patients with a ‘healthy-appearing’ microbiota.
Over time, microbial richness and diversity did not change in RR versus RA patients. Although the microbial community structure of both RR and RA patients was less stable over time compared with that of HCs, no differences were observed between the patient groups [p = 0.17]; nor was the stability impacted by Montreal classification, medication use, or surgery.
Conclusion
The altered microbiota composition and stability in CD was neither associated with disease activity nor long-term disease course, questioning its involvement in the development of an exacerbation. The aberrant microbiota composition in a subset of CD patients warrants further exploration of a more microbiota-driven etiology in this group.
Keywords: Crohn’s disease, microbiota, disease course, 16S rRNA gene, microbiota dynamics
1. Introduction
Crohn’s disease [CD] is a chronic gastrointestinal inflammatory disease of which the incidence is increasing worldwide.1 It is a relapsing disease characterized by periods of active inflammation with symptoms of abdominal pain and [bloody] diarrhea, alternating with periods of remission. The disease course varies between patients and has a poor predictability,2 hindering clinical decision-making. Crohn’s disease has a significant impact on the patient’s quality of life and health-related costs, especially during active disease.3,4 Further insights into factors contributing to disease activity may provide leads for preventive strategies and thus improve disease outcome.
Although the exact cause is unclear, the generally accepted hypothesis is that CD results from an aberrant immune response against commensal bacteria in genetically susceptible hosts. Previous studies have reported microbiota perturbations, characterized by a decreased diversity and changes in the abundance of specific taxa [e.g. reduction in Faecalibacterium prausnitzii and increase in Enterobacteriaceae] in patients with CD compared with healthy individuals.5–10 Moreover, several studies have reported microbial shifts in relation to disease activity. When compared with patients with inactive CD, the microbiota of CD patients during an exacerbation is characterized by increased members of Enterobacteriaceae11,12 and Bacteroides spp.,11,13 and a reduction of F. prausnitzii14–16 and Clostridium coccoides group,14,17 although these associations vary between studies. These inconsistencies may in part be due to differences in assessing disease activity, in applied molecular methods, and in study populations, but also to potential confounding factors such as medication use. Many studies are based on a cross-sectional design, comparing patients with active CD with those with inactive CD. Considering the inter-individual variation in microbiota composition and the heterogeneous nature of CD, longitudinal studies are particularly relevant.
To our knowledge, nine studies have investigated the microbiota in adult patients with CD in relation to changing disease course over time.8,13,17–23 Four focused on the predictive value of the microbiota regarding treatment response13,20 or post-surgery recurrence.18,19 The other five focused on remission patients subsequently developing an exacerbation.8,17,21–23 With the exception of two studies,8,23 these studies included only a few subjects, and only three studies comprehensively assessed the microbiota with next-generation sequencing techniques.8,22,23 Although the other techniques used in previous studies yield valuable information, they do not provide the same resolution as next-generation sequencing. Moreover, the extent to which microbiota composition and stability are related to long-term disease course is largely unknown.
Within this study, we therefore aimed [i] to compare the faecal microbiota stability of patients with CD with that of healthy individuals, [ii] to compare the stability of the faecal microbiota of patients with CD with either changing or stable disease activity over time, and [iii] to explore the association between either microbiota composition or stability and long-term disease course, by means of next-generation sequencing.
2. Material and Methods
2.1. Study population
A total of 57 patients with CD and 15 healthy subjects were included in this study.24 The CD patients participated in a prospective follow-up study25 of the deeply phenotyped IBDSL cohort. Clinical data, blood, and faeces were collected at each outpatient visit and during an exacerbation throughout follow-up. As the current standard endoscopy is too invasive for disease monitoring over time, and clinical indices do not correlate well with mucosal inflammation,26 disease activity was defined in terms of the combination of faecal calprotectin [FC], serum CRP, and the Harvey–Bradshaw Index [HBI]: i.e. FC > 250 μg/g or FC > 100 μg/g with at least a 5-fold increase from baseline. Remission was defined by FC < 100 μg/g and CRP < 5 mg/L or FC < 100 μg/g, CRP < 10 mg/L, and HBI ≤ 4. Patients in remission at baseline were eligible for further analyses. Healthy control subjects [HCs], all without any GI disease, GI symptoms, or comorbidities, were recruited from among the controls who participated in the Maastricht IBS cohort as a reference group.27
Faecal samples were collected from all CD and HC subjects at two time-points. The CD group comprised 22 patients with baseline sampling at time of remission and subsequent sampling during an exacerbation [i.e. RA group], and 35 patients with two subsequent samples while maintaining remission [i.e. RR group, without any flares in between subsequent samples]. Complete defecations were collected at home, kept at room temperature and brought to the hospital within 12 h after defecation. Part of the faecal sample of the CD patients was sent to the laboratory of Clinical Chemistry for routine analysis of FC. The remaining part was aliquoted and frozen at –80°C for microbiota analysis. We have previously shown that this sample collection procedure does not significantly alter the microbiota composition when compared with immediate freezing of samples upon defecation at –80°C.28 Blood was collected for routine analysis of CRP.
The standardized computer registration of the IBDSL and Maastricht IBS cohort [for HCs] was used to retrieve demographics, data on disease phenotype by the Montreal classification, surgery (including [hemi]colectomy and ileocecal resection), medication use, and clinical activity scores [HBI].
All study subjects gave written informed consent prior to participation. Both studies have been approved by the Medical Ethics Committee of Maastricht University Medical Center+ and have been registered in the US National Library of Medicine [http://www.clinicaltrials.gov: NCT02130349 and NCT00775060, respectively].
2.2. Microbiota analysis and statistics
The faecal microbiota composition was assessed by Ilumina Miseq sequencing of the V4-region of the 16S rRNA gene. A detailed description of metagenomic DNA isolation, sequencing, and quality control is provided in the supplemental information.
Statistical analysis were performed in R Studio 1.0.143 [R 3.4.1] using vegan, Rhea, stats, igraph, ggraph, GUniFrac, and DirichletMultinomial packages. Alpha diversity estimates [observed species, Chao1, and Shannon index] were computed using Rhea standard script and settings.29 Those indices were computed, per individual, and the average differences were subsequently compared between the three study groups [RRs, RAs, and HCs]. Significance was tested using the Kruskal–Wallis test and the Mann–Whitney U test for post-hoc analysis.
Bray–Curtis and [un]weighted UniFrac dissimilarities within subjects were used to investigate both the changes in the microbiota community structure between subjects at baseline and within subjects over time. Enterotype analysis was performed at baseline using the Dirichlet multinomial mixture method as described previously.30 Principal Coordinate Analysis [PCoA] was used as an unconstrained ordination technique. To investigate whether the microbiota was more stable in HCs than in patients with CD, the Kruskal–Wallis and Mann–Whitney U tests were used to check for statistically significant differences between the groups with respect to the variation in relative abundance of bacterial genera as well as to the within-subject beta-diversity [distance between first and second sample]. A linear model was used to test whether the time-span between collection of the first and second samples was affecting the within-subject weighted UniFrac distance, as well as to investigate the correlation between the within-subject weighted UniFrac and the variation over time of the faecal Calprotectin and plasma CPR levels.
To examine the variation in microbial community structure, we first performed a PERMANOVA using Montreal classification factors [age at diagnosis, disease localization, and disease behaviour],31 medications used, number of liquid stools per day, surgery, and smoking habits as explanatory variables for the microbial community structure. We then performed a distance-based redundancy analysis [dbRDA]32 to test whether CD patients clustered according to the disease activity or the medications used, with and without removing the effect of age, gender, and Bacteroides/Prevotella ratio.
Finally, we examined whether the history of disease activity and/or disease activity in the years following sample collection were associated with microbial community structure at baseline. For this purpose, the disease course of each individual patient was reviewed from the year before until 5 years after inclusion. Each yearly quarter was assessed for disease activity, defined by [i] active disease on endoscopy or imaging, [ii] hospitalization due to an exacerbation, [iii] surgery for active IBD, or [iv] treatment adjustment for increased symptoms. The number of active quarters before inclusion was used as a marker for ‘disease course before sample collection’ and the number of active quarters after inclusion was used as a marker for ‘disease course after baseline sample collection’.
3. Results
3.1. Study population
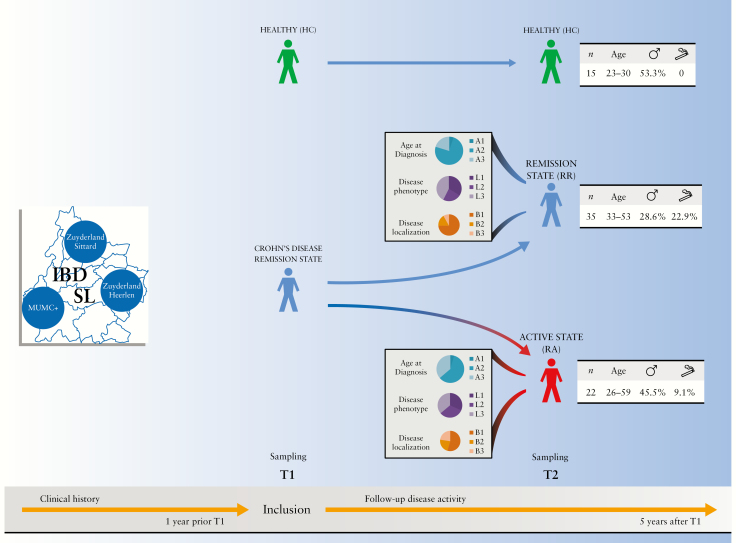
A total of 144 faecal samples of 57 patients with CD [35 RRs, 22 RAs] and 15 HCs were available for analysis [Figure 1 and Supplementary Table 1]. The median time between baseline and follow-up samples was 14 [IQR 11–21], 20 [8–36], and 13 [12–16] weeks for RR patients, RA patients, and HCs, respectively. No substantial differences were found in overall medication use between the different CD patient populations, or within each patient group over time [Supplementary Table 2].
Figure 1.
Graphic schematization of the study design and demographics of the study population. Age legend: A1 [≤16 years]; A2 [17–40 years]; A3 [>40 years]. Phenotype legend: B1 [non-structuring/non-penetrating]; B2 [structuring]; B3 [penetrating]. Disease localization legend: L1 [ileal]; L2 [colonic]; L3 [ileocolonic].
3.2. Baseline microbial richness, diversity, and community structure
At baseline, CD patients had a significantly lower faecal microbial richness and diversity when compared with HCs as indicated by the number of observed species (median [IQR]: 170 [97–233] and 209 [135–251], respectively; p = 0.0002), Chao1 index (173 [107–236] and 209 [135–251], respectively; p = 0.0006), and Shannon index (3.5 [1.8–4.1] and 3.8 [2.7–4.4], respectively; p = 0.005) [Supplementary Figure 1].
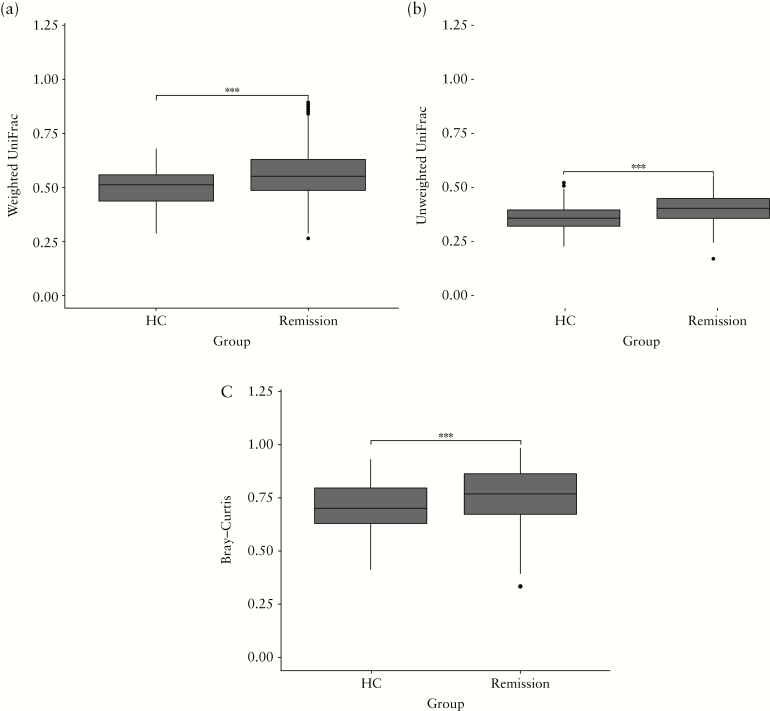
Differences in the faecal microbial community structure between samples at baseline were assessed using the Bray–Curtis and [un]weighted UniFrac. Microbial community structure was more heterogeneous among patients with CD, as indicated by the significantly higher distances in CD when compared with HCs for weighted UniFrac as well as for the other beta-diversity indices [Figure 2].
Figure 2.
Within-group dissimilarity in the microbial community structure based upon [a] weighted UniFrac, [b] unweighted UniFrac and [c] Bray–Curtis for healthy controls [HCs] and Crohn’s disease patients [remission] at baseline [T1]. All three beta-diversity indices indicate that the microbial community structure is significantly more heterogenous between CD patients than between HCs. Significance was tested using the Wilcoxson Signed-Ranks Test; *** indicates p < 0.001.
Enterotype analysis revealed the presence of three enterotypes driven by high abundances of Bacteroides spp. [E1] and Prevotella spp. [E2] and a low abundance of Faecalibacterium spp. [E3], but none of the enterotypes was significantly associated with one of the subject groups.
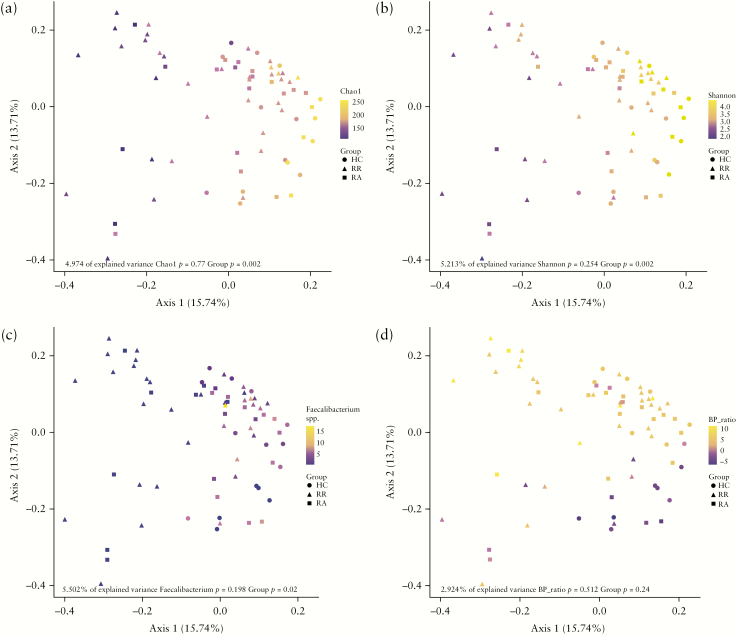
Next, we performed a PCoA based on the weighted UniFrac distance of the faecal microbiota of CD patients and HCs at baseline, aiming to visualize differences between sample groups and to identify factors driving the separation of samples. The results highlighted a subgroup of CD patients who clustered apart from HCs along the first principal coordinate. This subgroup was characterized by a lower relative abundance of Faecalibacterium spp., as well as a lower microbial richness and diversity [Figure 3a–c]. In addition, the PCoA showed that the ratio of the relative abundance between Bacteroides spp. and Prevotella spp. seemed to drive the separation along the second principal coordinate [Figure 3d].
Figure 3.
PCoA based on weighted UniFrac distance metric of baseline [T1] faecal microbial community structure in healthy controls and Crohn’s disease patients. Samples are coloured based on [a] Chao1 index, [b] Shannon index, [c] relative abundance of Faecalibacterium spp. and [d] log2 ratio of the relative abundance of Bacteroides spp. and Prevotella spp. Alpha diversity and abundance of Faecalibacterium spp. drive separation along the first principle coordinate, whereas the Bacteroides to Prevotella ratio drives separation along the second coordinate.
3.3. Baseline microbial community structure association with preceding and subsequent disease course
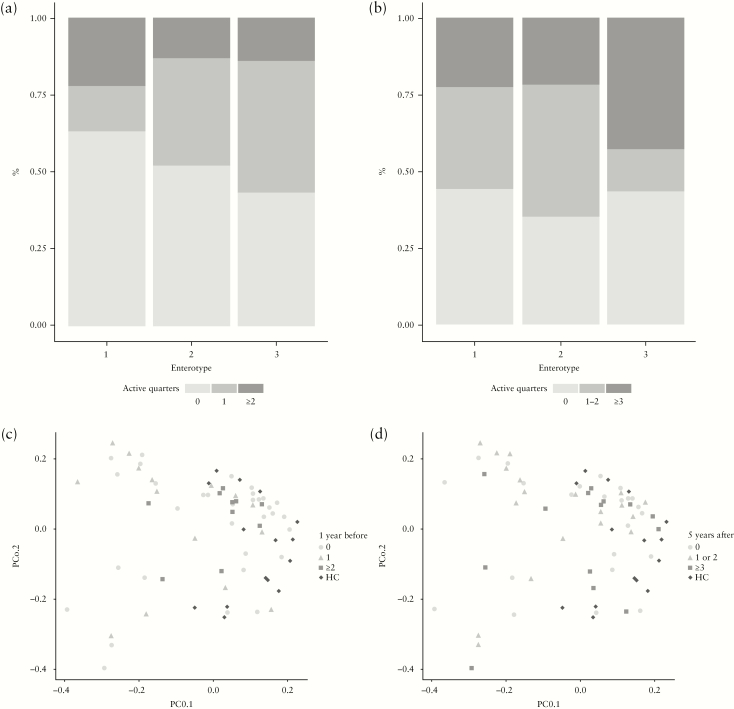
Finally, we examined whether disease activity in the year preceding the baseline sampling was predictive of the microbial community structure, or whether the microbial community structure was predictive of the disease activity up to 5 years after sampling. Although Enterotype 1 comprised slightly more patients without active quarters within the year prior to baseline when compared with the other enterotypes, the distribution of the number of active quarters in the year prior to sampling was not statistically significantly different between the three enterotypes, as examined by a generalized linear model using a logistic regression [Figure 4a, Supplementary Table 3]. Similarly, PCoA and PERMANOVA based on weighted UniFrac distance of the overall community structure did not show any separation according to the clinical history [Figure 4c, Supplementary Table 5].
Figure 4.
The number of active disease quarters is not associated with the microbial community structure: [a] bar plot representing the proportion of CD patients among each enterotype with 0, 1, or ≥2 active quarters within the year prior to inclusion; [b] bar plot depicting the proportion of CD patients among each of the enterotypes with 0, 1–2, or ≥3 active quarters in the 5 years after inclusion; [c+d] PCoA based on weighted UniFrac distance metric of baseline faecal microbiota samples of healthy controls and Crohn’s disease patients. Samples are coloured based on the number of active quarters during [c] 1 year before inclusion, and [d] 5 years after inclusion.
Also, the disease activity in the 5 years following baseline sampling showed no association with baseline enterotype [Figure 4b] [Supplementary Table 4] or with the microbial community structure based upon the weighted Unifrac [Figure 4d, Supplementary Tables 6]. This indicates that the overall microbial community structure appeared not to be predictive of future disease course.
3.4. Temporal dynamics of the microbial richness, diversity, and community structure
Changes in alpha and beta diversity indices within study subjects over time were compared between RR and RA patients and HCs. Although the microbial richness and diversity of CD patients was lower than those of HCs at baseline [Supplementary Figure 1], the temporal dynamics of these parameters did not differ significantly between HCs and CD patients who either maintained remission [RRs] or developed an exacerbation during follow-up [RAs] [Figure 5].
Figure 5.
Changes in alpha-diversity indices in samples collected at first and second time-point [T1–T2] from healthy controls [HCs], CD patients staying in remission [RR], and CD patients in remission followed by an exacerbation [RA]. Panels depict: [a] observed species, [b] Chao1 index, and [c] Shannon index between. Changes in alpha-diversity were not significantly different between HC, RR, and RA groups. Significance was tested using Wilcoxson Signed-Ranks Test.
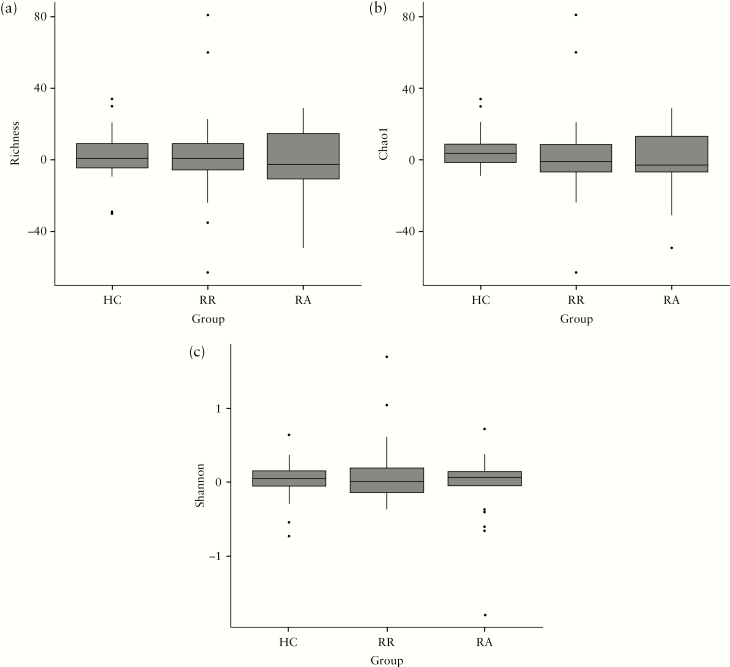
We next examined the fluctuation over time of the individual bacterial genera in association with the disease groups. Although some bacterial genera seemed to increase over time in the RA group and decrease in the RR group [Supplementary Figure 2], the Kruskall–Wallis test showed that, after false-discovery rate adjustment for multiple comparisons, this trend was not significant [Supplementary Table 7].
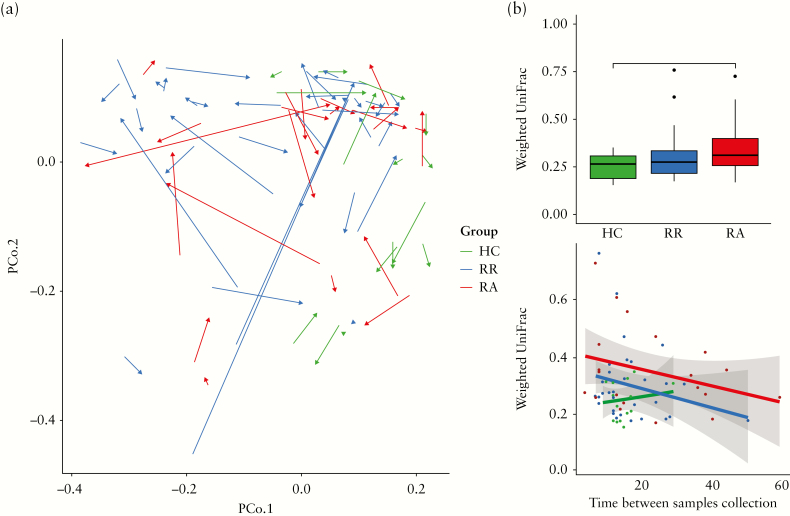
We then investigated the temporal dynamics in the microbial community structure as indicated by the within-subject beta-diversity. First, a PCoA was performed based on the weighted UniFrac distance of faecal samples from HCs and CD patients at baseline and the second time-point [Figure 6a]. As for the baseline data, no discrete separation between RR and RA samples could be observed. However, the temporal stability in microbial community structure between two subsequent samples, as indicated by the weighted Unifrac distance, was significantly higher in HCs than in the RA patients [Figure 6b]. When performing similar analyses based upon the unweighted UniFrac, the microbial community structure of HCs appeared to be more stable than that of both RR and RA patients [Supplementary Figure 3]. We next examined whether subjects switched enterotypes over time. None of the HCs changed enterotype during the sampling period, whereas 9 CD patients switched from one enterotype to another, again indicating a lower stability in [some] CD patients as compared with HCs. The proportion of patients who changed enterotype [6/35 for RRs and 3/22 for RAs] did not differ, however, between the two disease groups.
Figure 6.
[a] PcoA based on within-subject weighted Unifrac distance of faecal microbiota at baseline [T1] and second sampling time-point [T2]. Samples of healthy controls [HCs] are indicated in green, whereas samples of CD patients that remain in remission [RR] are indicated in blue and patients that develop an exacerbation [RA] in red. The arrows connect two samples from the same individual. The direction goes from T1 to T2. [b] Healthy controls show a statistically significantly smaller within-subject UniFrac distance between the two subsequent time-points, when compared with patients that develop an exacerbation [RA], whereas no difference is observed when compared with patients that remain in remission. Significance was tested using the Wilcoxson Signed-Ranks Test; * indicates p < 0.05. [c] There is no association between the within-subject distance and the actual time [in weeks] between subsequent sampling time-points as assessed by a linear model.
We then investigated whether the temporal stability of the microbiota composition of CD patients was related to the variation over time of Calprotectin or CRP. As expected, the RA group showed higher levels of faecal Calprotectin over time than the RR group. Nonetheless, our results proved that the variation over time of neither Calprotectin nor CRP affected the temporal stability of the microbiota composition [Supplementary Figure 4].
To examine the possibility that the temporal stability was confounded by variation in the time period [in weeks] between the collection of the two subsequent samples, we constructed a linear model between time and the weighted UniFrac distance per study group [Figure 6c]. These analyses did not reveal any evidence that the difference in collection time acted as a confounding factor [p-values HCs p = 0.64; RRs p = 0.16; RAs p = 0.16].
We subsequently examined whether the stability of the gut microbiota might also differ for CD patients according to disease localizations [ileal, colonic, or ileocolonic CD] or abdominal surgery. We found that only the subgroup of colonic CD patients had a gut microbiota composition that was statistically significantly less stable when compared with that of the HCs [Supplementary Figure 5a]; abdominal surgery did not affect microbiota stability [Supplementary Figure 5b].
To better understand the covariates that drive the microbial variation between samples, we next used distance-based Redundancy-Analysis [dbRDA] as an additional ordination method. The results showed that, when looking at the CD patients only, disease activity only created a minor shift in the spatial distribution of the data, which was insufficient to create separate clusters [Supplementary Figures 6 and 7].
Using permutational multivariate analyses of variance [PERMANOVA], we confirmed that our study results were not confounded by Montreal classification factors [age at diagnosis, disease localization, or disease phenotype], medication use [mesalazines, thiopurines, biologics, corticosteroids, or proton pump inhibitors] prior to or during the study period, age, gender, number of liquid stools/day, surgery, or original sequencing depth. Only disease phenotype was statistically significantly associated with the microbial community structure of CD patients [Supplementary Table 8]. This association was mainly related to a significantly different microbial community structure in CD patients with a penetrating [B3] phenotype when compared with the non-structuring/non-penetrating [B1] phenotype [Supplementary Table 9].
Altogether, our findings suggested that the microbiota community structure only marginally differed between RA and RR patients.
4. Discussion
To the best of our knowledge, together with the work from Pascal et al.,8 and Halfvarson et al.,23 this is one of the largest longitudinal studies that has comprehensively investigated the stability of the faecal microbiota of adult CD patients during their disease course. First, CD patients showed a lower microbial richness and diversity when compared with HCs. Second, a subset of CD patients were clustered separately from HCs and were characterized by a low microbial diversity and a relatively low abundance of Faecalibacterium spp. Third, the temporal stability of the microbial community structure was lower in CD patients when compared with HCs, but the microbial stability was not affected by changes in disease activity. And finally, the overall microbial community structure was not associated with disease history or subsequent disease course.
By collecting multiple samples of HCs and CD patients both with and without a changing disease activity over time, we were able to assess the microbial stability and to investigate the microbial changes during remission and active disease, thereby limiting potential confounding associated with cross-sectional studies. The present study confirmed previous observations that the faecal microbiota of CD patients is less diverse compared with that of HCs.33 This lower stability in CD was for some indices [weighted Unifrac] only statistically significant for the RA patients, whereas for other indices of microbial stability [unweighted Unifrac], both RR and RA patients showed a lower temporal stability when compared with HCs. However, for none of the indices of microbial stability did we find a significant difference between the RA and RR patient groups. Altogether, these results showed that HCs had a stronger temporal stability of the microbial community structure when compared with CD patients, regardless of whether these patients maintained remission or developed an exacerbation. Our results thus can be used to confirm that CD patients have a less diverse microbiota with larger intra-individual variations. Moreover, we observed that it was especially patients with colonic disease who had lower temporal microbiota stability.
Although the absence of an altered microbial composition and stability in patients developing active disease as compared with patients maintaining remission is in agreement with previous longitudinal studies,8,23 it contrasts with several previous cross-sectional studies.11,16 As indicated by the study of Halfvarson et al.,23 changes in medication use are more strongly linked to the dynamics in the microbial community structure than changes in disease activity. This might explain why previous cross-sectional studies, with large variations in medication use between subjects, have reported stronger associations between disease activity and microbiota composition. This suggests the need for longitudinal studies with repeated sampling to rule out confounding, but also calls into question the involvement of the overall microbiota in the development of exacerbations. Nevertheless, small shifts in [a combination of] specific taxa may be present in CD patients with changing disease activity over time. It is also plausible that patient-specific changes are present, but due to our focus on the overall microbiota composition, these changes remained undetected. Therefore, further analyses focusing on [small] changes in individual taxa are warranted in large groups of patients with longitudinal follow-up, taking into account disease phenotype, medication use, surgery, and dietary habits. The latter factor was not included in the present study because dietary information was not available, but it could be a potential reason for the lack of consistent changes in the microbiota community structure among CD patients.
We found a subgroup of CD patients for whom the microbiota composition, characterized by a low microbial richness and diversity and a low relative abundance of Faecalibacterium spp., deviated from the microbiota of HCs. Subgroups of CD patients clustered apart from other CD patients and HCs have also been demonstrated in previous studies.6,23,34,35 Consistent with the recent study of Halfvarson et al.,23 we found this subgroup of CD patients to be characterized by lower F. prausnitzii abundance and low microbial richness, but in our study this subcluster was not restricted to patients with ileal CD. The existence of a subgroup of CD patients with a deviating microbiota composition might [in part] be explained by disease-related factors. We could, however, not find clear differences in age at onset, disease localization, disease behavior, disease activity before and after inclusion in the study, number of liquid stools per day, surgery, or medication use when studying the CD patients with a more deviant microbiota profile versus those with a more ‘healthy’ microbiota profile. It should, however, be noted that numbers were relatively small. Larger studies are needed to further characterize the subgroup of CD patients that does not cluster with HCs, and to investigate whether this altered microbiota might be related to a more microbiota-driven disease etiology or certain host or environmental factors. Incorporation of host genetics, metabolomics, and/or transcriptomics data in future large-scale studies could potentially explain the reason for this subgroup of CD patients. Using PERMANOVA, we did rule out the possibility that our study results were confounded by Montreal classification factors, medication use, age, gender, number of liquid stools/day, surgery, or original sequencing depth. We did find, however, a different microbiota structure in patients with penetrating disease. So far, the limited number of studies available has not been able to provide clear evidence for increased numbers of bacteria within fistula tracts,36,37 but it would be interesting to investigate whether such perturbations may impact the physical and/or immunological intestinal barrier, and thereby fistula formation.
A potential limitation of our study was the collection and transport of faecal samples at room temperature. However, we already proved previously that the sample processing as applied in the present study neither significantly altered the microbial diversity and community structure, nor led to the overgrowth of specific bacterial groups when compared with immediate freezing of samples upon defecation.28 On the other hand, our study had several strengths—in particular, the longitudinal study design. Although cross-sectional microbiota studies are restricted by the large inter-individual variation of the microbiota, most microbiota studies on disease activity in adult CD patients are based on a cross-sectional design. Longitudinal studies are able to circumvent this limitation. Furthermore, only small numbers of patients in each group [RR and RA] had a change in medication use between the consecutive samples, further limiting potential bias due to confounding. Moreover, repeating our analyses without these patients did not impact our findings [data not shown].
Another strength of our study was the use of a composite score, including both clinical and inflammation markers, to determine disease activity. Repeated endoscopy, which is the golden standard for disease activity assessment, is too invasive in a longitudinal patient cohort. Most previous studies, including the longitudinal study of Pascal and colleagues,8,38,39 have used clinical activity indices to assess disease activity,11,13,17,20,21 and these have been shown to correlate only moderately with mucosal inflammation.26 A combination of inflammatory and clinical markers, as used in the present study, provides a more reliable and accepted surrogate for mucosal inflammation.26 Finally, due to the deeply phenotyped patients included, we were able to explore the association of the microbiota composition and stability with long-term disease course.
In conclusion, our prospective longitudinal study showed that the altered microbiota composition and stability in CD was not associated with disease activity or with long-term disease course. This result calls into question the involvement of the overall microbiota structure in the development of exacerbations. The aberrant microbiota composition in a subset of CD patients warrants further exploration of a more microbiota-driven etiology in this group.
Funding
This study was financially supported by a grant from the Academic Fund of Maastricht UMC+.
Conflict of Interest
The contributing authors have no conflicts of interest regarding this manuscript.
Author Contributions
DMAEJ, MJP, and AGLB conceived and coordinated the follow-up of the IBD SL study. DIT performed the lab analyses. GG, DIT, and DSJW performed the bioinformatics and data analyses of the study, guided by JP and DMAJ. JP, DMAEJ, DIT, and GG interpreted the data. GG, JP, DMAEJ, and DIT wrote the manuscript. PHMS, AAMM, MJP, AGLB, DMAEJ, JP, DSJW, GG, and DIT participated in discussions and revisions. All authors read and approved the final version of the manuscript.
Supplementary Material
References
- 1. Molodecky NA, Soon IS, Rabi DM, et al. . Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 2. Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 2012;380:1590–605. [DOI] [PubMed] [Google Scholar]
- 3. Burisch J, Jess T, Martinato M, Lakatos PL; ECCO -EpiCom The burden of inflammatory bowel disease in Europe. J Crohns Colitis 2013;7:322–37. [DOI] [PubMed] [Google Scholar]
- 4. Romberg-Camps MJ, Bol Y, Dagnelie PC, et al. . Fatigue and health-related quality of life in inflammatory bowel disease: results from a population-based study in the Netherlands: the IBD-South Limburg cohort. Inflamm Bowel Dis 2010;16:2137–47. [DOI] [PubMed] [Google Scholar]
- 5. Fujimoto T, Imaeda H, Takahashi K, et al. . Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol 2013;28:613–9. [DOI] [PubMed] [Google Scholar]
- 6. Gevers D, Kugathasan S, Denson LA, et al. . The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morgan XC, Tickle TL, Sokol H, et al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pascal V, Pozuelo M, Borruel N, et al. . A microbial signature for Crohn’s disease. Gut 2017;66:813–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker AW, Sanderson JD, Churcher C, et al. . High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol 2011;11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Willing BP, Dicksved J, Halfvarson J, et al. . A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010;139:1844–54.e1. [DOI] [PubMed] [Google Scholar]
- 11. Kolho KL, Korpela K, Jaakkola T, et al. . Fecal microbiota in pediatric inflammatory bowel disease and its relation to inflammation. Am J Gastroenterol 2015;110:921–30. [DOI] [PubMed] [Google Scholar]
- 12. Papa E, Docktor M, Smillie C, et al. . Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 2012;7:e39242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andoh A, Kobayashi T, Kuzuoka H, et al. . Characterization of gut microbiota profiles by disease activity in patients with Crohn’s disease using data mining analysis of terminal restriction fragment length polymorphisms. Biomed Rep 2014;2:370–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sokol H, Seksik P, Furet JP, et al. . Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 2009;15:1183–9. [DOI] [PubMed] [Google Scholar]
- 15. Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis 2008;14:147–61. [DOI] [PubMed] [Google Scholar]
- 16. Wang W, Chen L, Zhou R, et al. . Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 2014;52:398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seksik P, Rigottier-Gois L, Gramet G, et al. . Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 2003;52:237–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Cruz P, Kang S, Wagner J, et al. . Association between specific mucosa-associated microbiota in Crohn’s disease at the time of resection and subsequent disease recurrence: a pilot study. J Gastroenterol Hepatol 2015;30:268–78. [DOI] [PubMed] [Google Scholar]
- 19. Dey N, Soergel DA, Repo S, Brenner SE. Association of gut microbiota with post-operative clinical course in Crohn’s disease. BMC Gastroenterol 2013;13:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajca S, Grondin V, Louis E, et al. . Alterations in the intestinal microbiome [dysbiosis] as a predictor of relapse after infliximab withdrawal in Crohnʼs disease. Inflamm Bowel Dis 2014:20:978–86. [DOI] [PubMed] [Google Scholar]
- 21. Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol 2006;44:3980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wills ES, Jonkers DM, Savelkoul PH, Masclee AA, Pierik MJ, Penders J. Fecal microbial composition of ulcerative colitis and Crohn’s disease patients in remission and subsequent exacerbation. PLoS One 2014;9:e90981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Halfvarson J, Brislawn CJ, Lamendella R, et al. . Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van den Heuvel TR, Jonkers DM, Jeuring SF, et al. . Cohort profile: the inflammatory bowel disease south Limburg cohort [IBDSL]. Int J Epidemiol 2017;46:e7. [DOI] [PubMed] [Google Scholar]
- 25. Bodelier AG, Pierik MJ, van den Heuvel T, et al. . Pancreatitis-associated protein has no additional value as a marker of disease activity in a real-life cohort of IBD patients. Eur J Gastroenterol Hepatol 2014;26:902–9. [DOI] [PubMed] [Google Scholar]
- 26. Falvey JD, Hoskin T, Meijer B, et al. . Disease activity assessment in IBD: clinical indices and biomarkers fail to predict endoscopic remission. Inflamm Bowel Dis 2015;21:824–31. [DOI] [PubMed] [Google Scholar]
- 27. Mujagic Z, Ludidi S, Keszthelyi D, et al. . Small intestinal permeability is increased in diarrhoea predominant IBS, while alterations in gastroduodenal permeability in all IBS subtypes are largely attributable to confounders. Aliment Pharmacol Ther 2014;40:288–97. [DOI] [PubMed] [Google Scholar]
- 28. Tedjo DI, Jonkers DM, Savelkoul PH, et al. . The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One 2015;10:e0126685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lagkouvardos I, Fischer S, Kumar N, Clavel T. Rhea: a transparent and modular R pipeline for microbial profiling based on 16S rRNA gene amplicons. PeerJ 2017;5:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan M. DirichletMultinomial: Dirichlet–Multinomial Mixture Model Machine Learning for Microbiome Data. https://rdrr.io/bioc/DirichletMultinomial/ Accessed June 2018. [Google Scholar]
- 31. Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006;55:749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Legendre P, Anderson MJ. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 1999;69:1–24. [Google Scholar]
- 33. Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012;9:599–608. [DOI] [PubMed] [Google Scholar]
- 34. Frank DN, St. Amand AL, Feldman RA, et al. . Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis JD, Chen EZ, Baldassano RN, et al. . Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015;18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sugrue J, Nordenstam J, Abcarian H, et al. . Pathogenesis and persistence of cryptoglandular anal fistula: a systematic review. Tech Coloproctol 2017;21:425–32. [DOI] [PubMed] [Google Scholar]
- 37. Siegmund B, Feakins RM, Barmias G, et al. . Results of the fifth scientific workshop of the ECCO [II]: pathophysiology of perianal fistulizing disease. J Crohns Colitis 2016;10:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. af Björkesten CG, Nieminen U, Turunen U, Arkkila P, Sipponen T, Färkkilä M. Surrogate markers and clinical indices, alone or combined, as indicators for endoscopic remission in anti-TNF–treated luminal Crohn’s disease. Scand J Gastroenterol 2012;47:528–37. [DOI] [PubMed] [Google Scholar]
- 39. D’Haens G, Ferrante M, Vermeire S, et al. . Fecal calprotectin is a surrogate marker for endoscopic lesions in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:2218–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.