Abstract
Chemical cross-linking mass spectrometry is rapidly emerging as a prominent technique to study protein structures. Structural information is obtained by covalently connecting peptides in close proximity by small reagents and identifying the resulting peptide pairs by mass spectrometry. However, substoichiometric reaction efficiencies render routine detection of cross-linked peptides problematic. Here, we present a new trifunctional cross-linking reagent, termed PhoX, which is decorated with a stable phosphonic acid handle. This makes the cross-linked peptides amenable to the well-established immobilized metal affinity chromatography (IMAC) enrichment. The handle allows for 300× enrichment efficiency and 97% specificity. We exemplify the approach on various model proteins and protein complexes, e.g., resulting in a structural model of the LRP1/RAP complex. Almost completely removing linear peptides allows PhoX, although noncleavable, to be applied to complex lysates. Focusing the database search to the 1400 most abundant proteins, we were able to identify 1156 cross-links in a single 3 h measurement.
Short abstract
A novel enrichable cross-linking reagent amenable to IMAC enrichment produces high purity in cross-linked peptides and leads to vastly simplified XL-MS workflows and increased analytical depth.
Introduction
Cross-linking mass spectrometry (XL-MS) is a powerful tool that uses chemical reagents to investigate the structure of proteins and the complexes they form.1−5 The employed reagents are typically small bifunctional chemicals that covalently connect amino acids in close proximity. Most commonly highly efficient NHS-chemistry is used to capture the side chains of lysines in a protein.6−12 A spacer separates the reactive groups, and as such, the cross-linking reagent acts as a distance constraint between the captured amino acids.13 After cross-linking the proteins in their native state, the protein sample is typically alkylated, reduced, and finally digested into peptides by a protease, often trypsin. The full mixture of linear and cross-linked peptides is subsequently subjected to liquid chromatography tandem mass spectrometry (LC-MS/MS) for identification. After detection, cross-linked peptides provide valuable distance information on protein tertiary structure in the form of intralinks (two peptides originating from the same protein) or on protein quaternary structure in the form of interlinks (two peptides originating from different proteins).14 From these measurements, we and others found that the sought-for cross-linked peptides are completely overwhelmed by the linear peptides in terms of both numbers and abundance. To illustrate, from available data we estimate that the cross-link reaction efficiency is only 1–5%, and typically, relatively few lysine pairs are in close enough proximity to be cross-linked.15,16 Attempts to alleviate this situation have been made by extensive prefractionation of the peptide products using several chromatographic techniques.10,17−20 These techniques use properties that are exuberated for cross-linked peptides (e.g., size or charge), but are not unique. This typically results in large amounts of samples of which each still contains a high background of linear peptides. Other attempts integrate an enrichment handle directly on the cross-linking reagent resulting in a trifunctional reagent to separate cross-linked peptides from linear peptides. Two distinct approaches are typically used; the first one uses, e.g., biotin as an enrichment handle on the spacer region.21−24 The second approach utilizes smaller functionalities on the spacer region like azides that allow performing bioorthogonal transformations after the cross-linking reaction. The cross-linked peptides are consequently functionalized by the enrichment handle using 1,3 dipolar cycloadditions (click-chemistry).25−27
The new strategy demonstrated here places a very small enrichable tag on the cross-linking reagent that possesses excellent enrichment properties in combination with the most efficient cross-linking chemistry. Inspired by the groundbreaking developments in phosphoproteomics, where immobilized metal affinity chromatography (IMAC) has reached levels of >95% enrichment specificity,28 we hypothesized that a phosphate group could be an ideal enrichment handle. Such a handle has the additional advantage that IMAC enrichment is nowadays a nearly routine protocol in most proteomics laboratories,29−31 and high levels of automation with liquid handling platforms can be achieved.32 The lability of the phosphate group during sample handling and mass spectrometric acquisition makes detection however problematic.33,34 We tackled this issue by altering the phosphate group to a phosphonic acid, replacing the labile P–O bond by a stable P–C bond. Such a phosphonic acid handle is still amenable to IMAC enrichment, and thus, PhoX enables a completely novel and highly efficient enrichment approach for the field of XL-MS. We demonstrate the versatility of PhoX on various proteins and protein complexes and even a full cell lysate (see the Supporting Information, Notes 1–4, for further details on sample preparation, enrichment, data acquisition, and data analysis). For interpreting the acquired RAW data we used widely accepted data analysis software like the Mascot peptide search engine35 and the XlinkX-cross-linked peptide search engine;14 the resulting identifications were false discovery rate controlled at 1% (i.e., for 100 identifications an estimated 1 can be a false positive).36 From our experiments we find a roughly 2/58/40 ratio for linear peptides/monolinks/cross-links, making detection of cross-linked peptides easy to perform with modern mass spectrometry equipment.
Synthesis
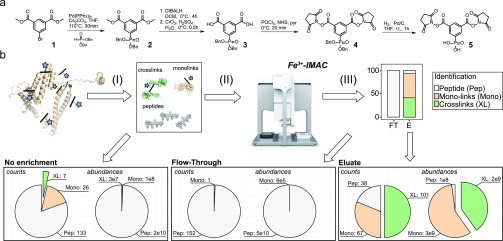
In the design of PhoX we attempted to minimize the spacer length and occupied chemical space and optimized the hydrophobicity of the reagent to ensure maximum reaction efficiency (see the Supporting Information, Note 5). We make use of an aromatic core structure as the vehicle for two NHS-esters and the enrichable phosphonic acid (compound 5; see scheme in Figure 1a). This results in a planar rigid spacer providing a fixed length of 5 Å, leading to a maximum distance constraint of about 20 Å when including the flexible lysines side chains. A major hurdle in the synthesis of such a reagent lies in the combination of NHS-ester and phosphonic acid functionalities. Carboxylic acids, common precursors for NHS-esters, as well as phosphonic acids, can be activated by carbodiimide reagents, leading to a mixture of undesired NHS-ester functionalities attached to the phosphonic acid moiety.37 To counter this, the phosphonic acid was protected by a dibenzyl-ester during the NHS-ester synthesis. This is also a common protection group strategy for the synthesis of peptides containing phosphorylated serine and its phosphonate mimic.38,39 Dibenzyl-esters can be selectively removed by hydrogenation with palladium on carbon (Pd/C), leaving the NHS-esters intact and thus solving the hurdle.
Figure 1.
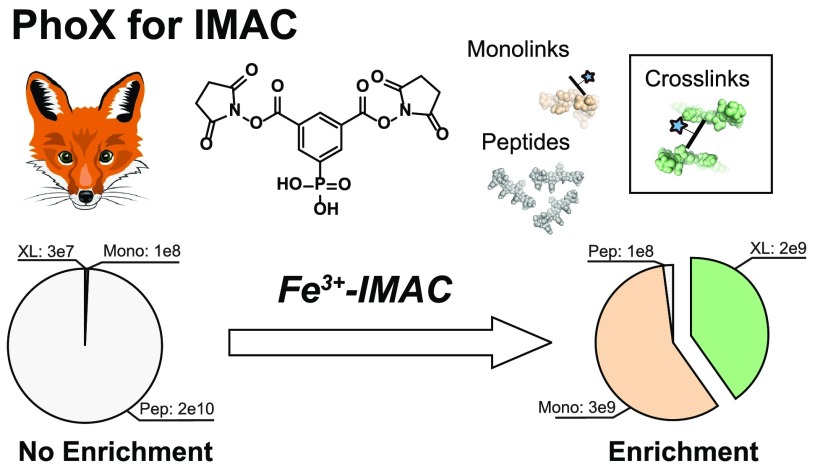
PhoX synthesis, workflow, and performance. (a) The synthesis of PhoX can be achieved in five steps starting from commercially available precursors. (b) After cross-linking the proteins in their native state, the proteins are denatured, reduced, alkylated, and digested to peptides (I). The mixture of peptides contains 3 different kinds of products: unmodified peptides (gray), monolinks (orange), and cross-links (green) (II); these colors are reused. Cross-linked and monolinked peptides are enriched using Fe-IMAC on a liquid sample handling platform providing high sample throughput (III; used data is from the BSA experiment presented in the Supporting Information, Note 8). Direct measurement of the cross-linked peptides produces low counts of cross-link identifications due to their extremely low abundances (left panel), while measurement following Fe-IMAC enrichment results in no cross-link identifications in the flow-through with similar abundance levels for the linear peptides as detected in the No enrichment (middle panel) and many in the eluate due to their enhanced abundance levels (right panel).
The final cross-linking reagent 5 was synthesized using the commercially available dimethyl 5-bromoisophthalate 1 as the starting compound, which acts as our spacer (see Figure 1a; see the Supporting Information, Note 12). Compound 1 was subjected to a palladium-catalyzed cross-coupling reaction to form a C–P bond in the coupling product 2. As deprotection of the methyl-ester of 2 under basic conditions yielded partly the free phosphonic acid, we reduced the methyl-ester by treatment with diisobutylaluminum hydride at 0 °C to the alcohols. The dialcohol was oxidized in quantitative yields to the corresponding dicarboxylic acid 3 with the Jones reagent. No byproducts were observed using this reaction, and only a crude workup was necessary. Activation of compound 3 with phosphoroxychlorid in pyridine at 0 °C and subsequent addition of N-hydroxysuccinimide yielded the NHS-ester functionalized reagent 4 within 20 min. As the final step, the dibenzyl phosphonic acid 4 was subjected to a hydrogenation reaction using Pd/C to yield the free phosphonic acid while keeping the NHS-esters intact. After filtering of the Pd/C, the cross-linking reagent 5 could be used without further purification. This strategy can potentially also be used for a cleavable version of PhoX; however, the integration of a cleavable moiety (such as a sulfoxide) will complicate the synthesis even further.
Performance on Model Proteins
Verification by tandem mass spectrometry showed that, in contrast to the P–O bond, the P–C bond is indeed very stable (see the Supporting Information, Note 6). To visualize the performance of our reagent, we cross-linked human hemoglobin and ran the cross-linked protein on SDS-PAGE. We tested a range between 0.5 and 5 mM for our cross-linking reagent and benchmarked it against the commonly used cross-linking reagent disuccinimidyl suberate (DSS). On the basis of the SDS-PAGE we estimated for both linkers that the optimal concentration is 1–2 mM, although DSS appears a little more reactive which can easily be compensated with a higher cross-linker concentration (see the Supporting Information, Note 7). For further performance tests we used BSA, a protein which has emerged as a standard for testing cross-linking performance.40−42 Cross-linking with 1 mM PhoX resulted in the best compromise between producing sufficient amounts of cross-links and preventing over-cross-linking as at this point not all monomeric molecules have disappeared (see the Supporting Information, Note 7). In-solution digesting the sample after cross-linking provided the input for our enrichment (see Figure 1b and the Supporting Information, Note 8). We retained part of the input as a control (i.e., nonenriched) prior to enrichment with Fe-IMAC and collected both the flow-through as well as the eluate. The control resulted in the identification of 7 cross-linked peptides, 26 monolinks (one NHS-ester of the reagent reacted with the peptide, and the other is hydrolyzed, providing no structural information), and 133 linear peptides. The abundance levels of the cross-linked peptides—constituting 0.14% of total abundance—are not even visible in the high background levels of linear peptides and as such will be difficult to analyze (Figure 1b; left panel). We retrieved from the flow-through no cross-linked peptides, 1 monolink, and 152 linear peptides. As expected, investigation of the abundances shows the same general trend as the control (Figure 1b; middle panel). The fact that the flow-through contained no cross-linked peptides indicates that the cross-linked peptides were successfully bound to the IMAC material. In sharp contrast, we retrieved from the eluate 101 cross-linked peptides, 67 monolinked peptides, and 38 linear peptides. Investigation of the abundance values showed a radically different picture from the control (Figure 1b; right panel). Even though there are more cross-linked peptides than monolinks, monolinks show a ∼1.3× higher abundance as it is easier to form a monolink than a cross-link, and both bind to the IMAC material. Calculation of enrichment efficiency and specificity from the detected abundance levels demonstrates a 300× enrichment of cross-linked peptides (comparison of the abundance levels of cross-linked peptides from the nonenriched sample divided by total abundances of identifications versus those from the enriched sample), and a 97% enrichment specificity (comparison of cross-linked peptides versus the linear/monolink peptides within the enriched sample).
To assess the performance of PhoX cross-linking in a protein environment of similar complexity as those typically encountered for purified protein complexes, we applied the cross-linker on the Pierce intact protein standard mix consisting of six proteins (Supporting Information, Note 9). As these proteins do not naturally interact, no interlinks are expected. After optimization, as before for BSA, the best PhoX cross-link reagent concentration was found to be 2 mM (see the Supporting Information, Note 7). From the resulting enriched peptide mixture, we detected after IMAC enrichment 134 cross-links. We found two interlinks between DNA polymerase I and thioredoxin as well as DNA polymerase I and protein G, which are most likely false positives. An additional 5 interlinks are reported in the table, which all derive from protein A and G and should actually be considered intralinks as the sample contains the fusion protein protein A/G. A similar analysis of the flow-through revealed no cross-linked peptides, demonstrating the effectiveness of the enrichment approach also in analyzing this more complex protein mixture.
Enrichment Efficiency
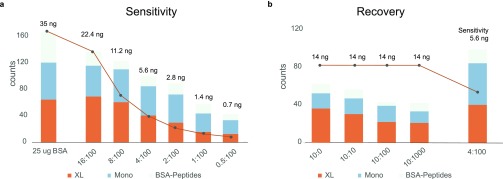
To further test the efficiency of our approach for enriching low-abundant cross-linked peptides, we next spiked PhoX-cross-linked BSA peptides into a background of an Escherichia coli tryptic digest in a broad dilution range. We chose the E. coli digest as background, as it contains relatively low levels of potentially coenriched phosphorylated peptides.43 Such a dilution range can be done in two ways, each representing a different outcome (Table 1; Figure 2). The first approach consists of mixing a decreasing amount of PhoX-cross-linked BSA peptides in a fixed background of E. coli tryptic peptides—we term this approach “sensitivity”. This mimics the concentration levels at which cross-links can still be recovered from a full lysate using our approach. For this, a concentration of cross-linked BSA peptides ranging from 16 μg down to 0.5 μg was spiked into a background of 100 μg of E. coli tryptic peptides. Given that we observed that only 0.14% of the total peptide weight was represented by cross-linked peptides, we estimate at its highest dilution point cross-linked peptides represent approximately 700 pg. These detectable picogram levels are within the range typically achieved by multiple reaction monitoring (MRM),44 although our measurements do not make use of a targeted approach. MS analysis of the enriched samples reveals that cross-linked peptides are reliably identified at approximately 10 ng levels with 56 identified cross-links (93% of undiluted cross-linked BSA—slightly higher compared to the amount detected for the recovery experiment which we attribute to experimental variation), although at the lowest level of 700 pg still two cross-links can be identified (Figure 2a).
Table 1. Setup and Results for the Spike-In Experiments.
| sensitivity |
recovery |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ratio | BSA (μg) | E. coli (μg) | final XLs (ng) | XL count | ratio | BSA (μg) | E. coli (μg) | final XLs (ng) | XL count |
| 16:100 | 16 | 100 | 22.4 | 69 | 0 | 10 | 0 | 14 | 36 |
| 8:100 | 8 | 100 | 11.2 | 60 | 10:10 | 10 | 10 | 14 | 30 |
| 4:100 | 4 | 100 | 5.6 | 40 | 10:100 | 10 | 100 | 14 | 22 |
| 2:100 | 2 | 100 | 2.8 | 29 | 10:1000 | 10 | 1000 | 14 | 21 |
| 1:100 | 1 | 100 | 1.4 | 15 | |||||
| 0.5:100 | 0.5 | 100 | 0.7 | 12 | |||||
Figure 2.
Testing PhoX peptide enrichment in a complex background. (a) Sensitivity—mixing cross-linked BSA peptides in decreasing amounts in a fixed background of E. coli peptides provides an estimate of the detection limit of the methodology. (b) Recovery method—mixing cross-linked BSA peptides in equal amounts in an increasing background of E. coli peptides allows for recovery of cross-linked BSA peptides upon enrichment. The estimated cross-link amounts for the given cross-linked BSA peptides are shown above the bars.
The second approach consists of mixing equal amounts of cross-linked BSA peptides in an increasing background of E. coli peptides—we term this approach “recovery”—for which we used 10 μg of cross-linked BSA. For this experiment the maximum loading amounts for enrichment were rate-limited by the 10:1000 mixture (i.e., the highest amount of E. coli) leading to far lower amounts than what could be loaded for the sensitivity experiment. Hence, the number of identifications is expected to be lower. From this experiment, however, an equal amount of cross-linked peptides can be recovered and measured, removing effects of low concentration prohibiting successful detection by LC-MS/MS. Here, we estimate that the cross-linked peptides represent approximately 14 ng. Approximately 30 cross-linked peptides at these concentration levels are recovered efficiently without much loss, indicating that the enrichment is not significantly affected by high background levels of linear peptides (Figure 2b).
Application to Full Human Lysate
Advanced mass spectrometry in combination with cross-linking has the potential to unbiasedly map protein–protein interaction networks in situ of the cell. Major hurdles associated with the low stoichiometry of the cross-linking reaction, the presence of two peptides during fragmentation, and the overwhelming complexity for database searches remain. Typically, in such an experiment, extensive fractionation has to be performed to reduce the complexity to successfully capture cross-linked peptides—an area where PhoX could provide an excellent alternative. We tested the enrichment power of PhoX on a natively lysed human substrate without fractionation. As PhoX is not (yet) an MS-cleavable cross-linker, database searches against a whole human proteome are still a nearly unsurmountable problem. To tackle this, we chose to search with a reduced FASTA file of one-third of the most abundant proteins detected in a normal shotgun proteomics run (resulting in a list of 1339 proteins) for the search in Proteome Discoverer with XlinkX integrated.7,14 The search time was 1 day, demonstrating that PhoX as a noncleavable cross-linker can be used for the data analysis of complex systems. From a single 180 min LC-MS/MS run on the IMAC eluate we could identify 1028 cross-links (from 2208 spectra). As enrichment of cross-links by Fe-IMAC also involves simultaneous enrichment of phosphorylated peptides, we envisioned that dephosphorylation of peptides prior to enrichment could further improve this result. Whereas phosphate-esters present on phosphorylated peptides are hydrolyzed upon phosphatase treatment, the carbon–phosphor bond present on the phosphonic acid group of the cross-linking reagent would not be affected. After enrichment of the CIP phosphatase treated cross-linked lysate, we could increase the amount of detected cross-links to 1156 (from 2374 spectra), an improvement of 11%. Further indication that the phosphatase treatment was successful came from the observed drop of 533 to 42 phosphorylated peptides (from 1120 and 64 PSMs, respectively) for phosphorylated peptides in the nontreated lysate versus treated lysate. Without phosphatase treatment, the enrichment purity was 91% (combined cross-links and monolink abundance; 8.3% of the total abundance consists of phosphorylated peptides). After phosphatase treatment, the enrichment purity increased to 99% (see the Supporting Information, Note 10).
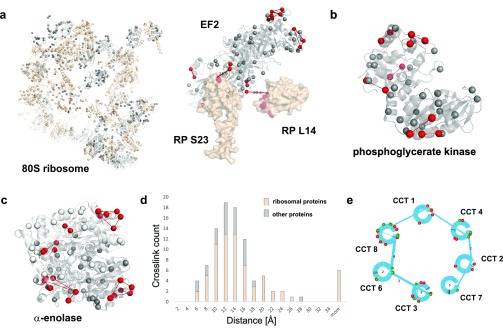
In this data set, 140 cross-links (identified by 299 spectra) are solely on the 80S ribosome, a 50% higher number than reported in previous reports.7,45 Of the 140 cross-links, 70 cross-links could be mapped on the cryo-EM structure of the 80S human ribosome (PDB, 4v6x; Figure 3a, left panel). The remaining 70 mapped to locations not present in the PDB file. Our identified inter-cross-links between elongation factor 2 and ribosomal proteins support the interaction site uncovered in the cryo-EM map of the 80S ribosome (PDB, 4v6x; Figure 3a, right panel). Of interest, though, is that these numbers were obtained with a single shot measurement of 180 min. For previous efforts, extensive fractionation was required to get similar numbers (e.g., for the XL-MS study on the nucleus, 86 measurements, totaling 172 h of measurement time, were needed to extract 87 cross-links on the ribosome45), clearly demonstrating the sensitivity and efficiency of PhoX. We validated cross-links by mapping on crystal structures of ribosome and other well-characterized proteins, including nucleolin, phosphoglycerate kinase (Figure 3b), stress-induced phosphoprotein 1, α-enolase (Figure 3c), heat shock protein 90-α, and Hsc-interacting protein, and found 93% of the cross-links within the distance constraint (Figure 3d). We detected 6 cross-links on the ribosome as overlength (see the Supporting Information, Table S2), which we attribute as arising from polysomes in line with observations from earlier work.7 Furthermore, we validated our data set on the TCP-1-ring complex (TRiC/CCT). Here, PhoX inter-cross-links match with the commonly known interaction network of the TRiC/CCT complex (Figure 3e).
Figure 3.
Application of PhoX to investigate human cell lysates. Cross-linked lysines are depicted as red spheres; non-cross-linked lysines are depicted as gray spheres. (a) Ribosome cryo-EM map (PDB, 4v6x). Proteins found cross-linked are shown in light brown; proteins without cross-links are shown in gray (RNA is not shown). Intra-cross-links mapped on the elongation factor 2 (part of PDB, 4v6x). Inter-cross-links of elongation factor to ribosomal proteins mapped on the ribosome (PDB, 6v6x). (b) Intra-cross-links of phosphoglycerate kinase mapped on its crystal structure (PDB, 3c39) (c) Intra-cross-links of α-enolase mapped on the dimeric crystal structure of human enolase 1 (PDB, 3b97). (d) Histogram of observed Lys–Lys distances in PhoX cross-links on the 80S ribosome (colored in light brown) and other proteins (nucleolin, PDB, 2kkr; nucleophosmin, PDB, 2llh; elongation factor 2, PDB, 4v6x; stress-induced-phosphoprotein 1, PDB, 1ewl; α-enolase, PDB, 3b97; phosphoglycerate kinase, PDB, 3c39). (e) Interaction network found for the TRiC/CCT complex. Proteins of the TRiC/CCT complex are shown as blue circles. Interlinks between the proteins are shown as blue lines; the thickness of the lines indicates the amount of interlinks identified. Green lines in the circles are the lysine positions; red balls indicate phosphorylation sites as extracted from uniprot, and green balls indicate lysines involved in interlinks.
Elucidation of LRP1-RAP Binding
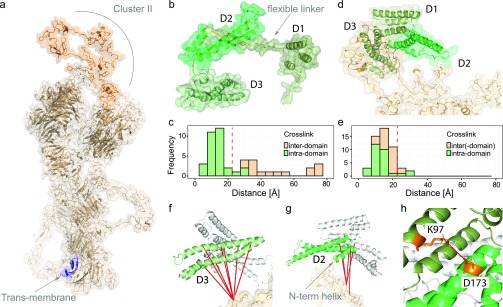
Its enrichable nature, small footprint, and short spacer make PhoX attractive for the elucidation of interfaces between tightly interacting proteins. We demonstrate this here by investigating the interaction of the lipoprotein receptor-related protein 1 (LRP1; Figure 4a) and its antagonist receptor associated protein (RAP; Figure 4b); further information on these proteins is provided in the Supporting Information, Note 11. It was biochemically shown that RAP, which is comprised of three domains (D1–3), has a high affinity for Cluster II.46 D3 tightly interacts via exposed lysines (K293 and K307) to acidic pockets exposed on the surface of LRP1 (D283 and D326). The D2 domain exhibits lower affinity binding, while the separate D1 domain showed no binding. Nevertheless, it is so far unknown how full-length RAP binds to LRP1, despite previous attempts.47 Here, we investigate the binding of full-length RAP to Cluster II of LRP1 with PhoX (see the Supporting Information, Note 11, for information on the integrative structural modeling). Using PhoX on recombinantly expressed versions of the proteins reconstituted in vitro, and we found 31 intralinks on RAP, 3 intralinks on LRP1, and 16 interlinks between the two proteins. Investigation of the intralinks within the individual domains of RAP revealed that the observed distance constraints satisfy the maximum length of PhoX, indicating that the experiment successfully yielded useful structural information. Investigation of the interdomain cross-links revealed a considerably different picture where almost all distances are outside the range (on average 37.3 ± 20.6 Å) indicating that RAP undergoes extensive structural rearrangement upon binding LRP1 (Figure 4c). Notably, on the basis of NMR data, the possibility of such rearrangement had already been predicted.48
Figure 4.
Application of PhoX to investigate the binding of full-length RAP to LRP1 Cluster II. (a) Predicted structural model for full-length LRP1, with Cluster II (PDB, 1n7d) and the transmembrane region highlighted. (b) Structure of RAP with the individual domains indicated (predicted from PDB, 2p03). (c) Measured Cα–Cα distances for intra- and interdomain cross-links on the structure of RAP (maximum distance constraint indicated with red, dashed line). (d) Multibody docking of the individual domains of RAP to LRP1 using HADDOCK. (e) Measured Cα–Cα distances after docking (maximum distance constraint indicated with red, dashed line). (f) Cross-links between the D3 domain of RAP and LRP1. (g) Cross-links between the D2 domain of RAP and LRP1. (h) Domain D1 (K97) in close proximity to domain D2 (D173).
To further investigate this major structural rearrangement, we performed multibody docking to LRP1 using HADDOCK.49 For this, the individual domains of RAP were separated from each other by removing the long and highly flexible linker regions. The resulting best scoring structural model, which retains the connectivity of the lysines and the acidic pockets, was selected for further analysis (Figure 4d). This model places K293 of RAP in the acidic pocket around D326 of LRP1 and K307 of RAP in the acidic pocket around D283 of LRP1. In this model, the domains D2 and D3 connect with LRP1, while the domain D1 exclusively interacts with the D2 and D3 domains. Inspection of the measured distances shows that over 90% of the cross-links in this model are within the set distance constraints (Figure 4e). Inspection of the cross-links between LRP1 and the D3 domain of RAP shows that cross-links are detected over the full length of this domain (Figure 4f). This can readily be explained by the strong interactions of the two lysines on D3 for which no cross-links were found, enforcing a close interaction of the full length of this domain to LRP1. In contrast, cross-links between LRP1 and the D2 domain of RAP are located on the short N-terminal helix of this domain lifting a large portion of this domain of the surface of LRP1 (Figure 4g). This suggests that this helix is interacting solely with LRP1, previously suggested on the basis of its high sequence conservation.46 The placement of the D1 domain suggests a role for this domain to lock the domain D2 into place upon LRP1 binding. Structurally a connection could potentially be made through a salt bridge between D173 and K97, a lysine which previously was shown to be shielded upon binding to LRP150 and for which we observe no cross-links.
Conclusion
Here, we presented a novel enrichable cross-linking reagent functionalized with a phosphonic acid group on the spacer region termed PhoX (full details on the synthesis are provided in the Supporting Information, Note 12). PhoX integrates a small phosphonic acid functionality on the spacer region that is amenable to large-scale and efficient IMAC enrichment as commonly used in phosphoproteomics. The linker however incorporates a stable C–P bond ensuring permanent decoration of the cross-linked peptides with the enrichment handle. IMAC not only is a unique approach for the enrichment of cross-linked peptides, but also allows for a best-case 300× enrichment efficiency and 97% enrichment specificity according to our data. The described workflows in combination with PhoX, we expect, will simplify XL-MS approaches on several fronts:
-
(I)
efficient cross-linking with a small, nonsterically hindered cross-linking reagent;
-
(II)
a condensed spacer length facilitating higher precision for modeling protein structures;
-
(III)
a simplified and fast workflow, which uses standardized phosphoproteomics techniques for the enrichment of cross-linked peptides (these techniques have been implemented in many proteomics laboratories, facilitating adoption of the reagent; competing molecules for the IMAC enrichment such as phosphopeptides and nucleic acids can easily be removed, while the much more stable phosphonic acid moiety for the enrichment of cross-linked peptides remains intact);
-
(IV)
Fe-IMAC for the enrichment of cross-linked peptides enabling high-throughput sampling in a 96-well plate format;
-
(V)
LC-MS measurement time can be decreased as one fraction is sufficient to achieve in-depth cross-link identification (in those cases where further fractionation is required, PhoX is highly compatible with orthogonal approaches like high-pH fractionation);
-
(VI)
removal of linear peptides enabling reliable identification of cross-linked peptides resulting in high-quality fragmentation spectra for cross-linked peptides.
As always, there remains room for improvement. For example, the negative charges on PhoX likely preclude it from entering intact cells through the cellular membrane. Even though this can be circumvented by protecting the phosphonic acid moiety and thus masking the negative charge, the used NHS-esters will be prone to quenching while passing through the membrane. Likely, next versions of PhoX will come with different reactive groups and incorporate stable isotopes and gas-phase cleavable moieties. However, a gas-phase cleavable version of PhoX is difficult to synthesize since, next to the protection group intensive combination of a phosphonic acid with a NHS-ester functionality, a cleavable moiety such as a sulfoxide has to be incorporated. Still, we envision already widespread adoption of XL-MS supported by this first version of PhoX, as it already tremendously facilitates in-depth studies into the dynamics of proteins, protein complexes, and interactomes, diminishing analysis time and increasing sensitivity by sizable factors.
Safety Statement
No unexpected or unusually high safety hazards were encountered.
Acknowledgments
We thank all members of the Heck group for their helpful contributions, Dr. Domenico Fasci for providing the cell lysate, and Prof. Dr. Sander Meier and Carmen van der Zwaan from Sanquin for the kind gift of the LRP1 Cluster II and RAP proteins. We acknowledge financial support by the large-scale proteomics facility Proteins@Work (Project 184.032.201) embedded in The Netherlands Proteomics Centre and supported by The Netherlands Organization for Scientific Research (NWO). Additional support came through the European Union Horizon 2020 program FET-OPEN project MSmed (Project 686547), the European Union Horizon 2020 program INFRAIA project Epic-XS (Project 823839), and a seed grant kindly provided by the Utrecht Institute for Pharmaceutical Sciences (UIPS).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00416.
Additional notes, data, and figures including workflows, simulations, cLogP values, structures, reagent stability test, cross-linker concentration optimization, and 1H and 31P NMR results (PDF)
Author Contributions
R.A.S. and A.J.R.H. conceived of the study. R.J.P. supported the synthesis and provided input for the design of the reagent. B.S. developed and synthesized the reagents and performed the experiments. B.S. and R.A.S. performed data analysis. All authors critically read and edited the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Leitner A.; Faini M.; Stengel F.; Aebersold R. Crosslinking and Mass Spectrometry: An Integrated Technology to Understand the Structure and Function of Molecular Machines. Trends Biochem. Sci. 2016, 41 (1), 20–32. 10.1016/j.tibs.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Holding A. N. XL-MS: Protein Cross-Linking Coupled with Mass Spectrometry. Methods 2015, 89, 54–63. 10.1016/j.ymeth.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Rappsilber J. The Beginning of a Beautiful Friendship: Cross-Linking/Mass Spectrometry and Modelling of Proteins and Multi-Protein Complexes. J. Struct. Biol. 2011, 173 (3), 530–540. 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkley E. D.; Cort J. R.; Adkins J. N. Cross-Linking and Mass Spectrometry Methodologies to Facilitate Structural Biology: Finding a Path through the Maze. J. Struct. Funct. Genomics 2013, 14 (3), 77–90. 10.1007/s10969-013-9160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A. Chemical Cross-Linking and Mass Spectrometry to Map Three-Dimensional Protein Structures and Protein–Protein Interactions. Mass Spectrom. Rev. 2006, 25 (4), 663–682. 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- Kao A.; Chiu C.; Vellucci D.; Yang Y.; Patel V. R.; Guan S.; Randall A.; Baldi P.; Rychnovsky S. D.; Huang L. Development of a Novel Cross-Linking Strategy for Fast and Accurate Identification of Cross-Linked Peptides of Protein Complexes. Mol. Cell. Proteomics 2011, 10 (1), M110.002212. 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Rijkers D. T. S.; Post H.; Heck A. J. R. Proteome-Wide Profiling of Protein Assemblies by Cross-Linking Mass Spectrometry. Nat. Methods 2015, 12 (12), 1179–1184. 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- Müller M. Q.; Dreiocker F.; Ihling C. H.; Schäfer M.; Sinz A. Cleavable Cross-Linker for Protein Structure Analysis: Reliable Identification of Cross-Linking Products by Tandem MS. Anal. Chem. 2010, 82 (16), 6958–6968. 10.1021/ac101241t. [DOI] [PubMed] [Google Scholar]
- Herzog F.; Kahraman A.; Boehringer D.; Mak R.; Bracher A.; Walzthoeni T.; Leitner A.; Beck M.; Hartl F.-U.; Ban N.; et al. Structural Probing of a Protein Phosphatase 2A Network by Chemical Cross-Linking and Mass Spectrometry. Science (Washington, DC, U. S.) 2012, 337 (6100), 1348–1352. 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- Leitner A.; Reischl R.; Walzthoeni T.; Herzog F.; Bohn S.; Förster F.; Aebersold R. Expanding the Chemical Cross-Linking Toolbox by the Use of Multiple Proteases and Enrichment by Size Exclusion Chromatography. Mol. Cell. Proteomics 2012, 11 (3), M111.014126. 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund R. D.; Wilkinson M. E.; Klykov O.; Barendregt A.; Pearce F. G.; Kieper S. N.; Maxwell H. W. R.; Capolupo A.; Heck A. J. R.; Krause K. L.; et al. Spacer Capture and Integration by a Type I-F Cas1–Cas2–3 CRISPR Adaptation Complex. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (26), E5122–E5128. 10.1073/pnas.1618421114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurtu F.; Zolog O.; Iacob B.; Silaghi-Dumitrescu R. Hemoglobin–Albumin Cross-Linking with Disuccinimidyl Suberate (DSS) and/or Glutaraldehyde for Blood Substitutes. Artif. Cells, Nanomed., Biotechnol. 2014, 42 (1), 13–17. 10.3109/21691401.2012.762652. [DOI] [PubMed] [Google Scholar]
- Green N. S.; Reisler E.; Houk K. N. Quantitative Evaluation of the Lengths of Homobifunctional Protein Cross-Linking Reagents Used as Molecular Rulers. Protein Sci. 2001, 10 (7), 1293–1304. 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klykov O.; Steigenberger B.; Pektaş S.; Fasci D.; Heck A. J.; Scheltema R. A. Efficient and Robust Proteome-Wide Approaches for Crosslinking Mass Spectrometry. Nat. Protoc. 2018, 13, 2964–2990. 10.1038/s41596-018-0074-x. [DOI] [PubMed] [Google Scholar]
- Chavez J. D.; Bruce J. E. Chemical Cross-Linking with Mass Spectrometry: A Tool for Systems Structural Biology. Curr. Opin. Chem. Biol. 2019, 48, 8–18. 10.1016/j.cbpa.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A.; Walzthoeni T.; Kahraman A.; Herzog F.; Rinner O.; Beck M.; Aebersold R. Probing Native Protein Structures by Chemical Cross-Linking, Mass Spectrometry, and Bioinformatics. Mol. Cell. Proteomics 2010, 9 (8), 1634–1649. 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinnefeld V.; Venne A. S.; Sickmann A.; Zahedi R. P. Enrichment of Cross-Linked Peptides Using Charge-Based Fractional Diagonal Chromatography (ChaFRADIC). J. Proteome Res. 2017, 16 (2), 459–469. 10.1021/acs.jproteome.6b00587. [DOI] [PubMed] [Google Scholar]
- Fritzsche R.; Ihling C. H.; Götze M.; Sinz A. Optimizing the Enrichment of Cross-Linked Products for Mass Spectrometric Protein Analysis. Rapid Commun. Mass Spectrom. 2012, 26 (6), 653–658. 10.1002/rcm.6150. [DOI] [PubMed] [Google Scholar]
- Schmidt R.; Sinz A. Improved Single-Step Enrichment Methods of Cross-Linked Products for Protein Structure Analysis and Protein Interaction Mapping. Anal. Bioanal. Chem. 2017, 409 (9), 2393–2400. 10.1007/s00216-017-0185-1. [DOI] [PubMed] [Google Scholar]
- Rampler E.; Stranzl T.; Orban-Nemeth Z.; Hollenstein D. M.; Hudecz O.; Schlögelhofer P.; Mechtler K. Comprehensive Cross-Linking Mass Spectrometry Reveals Parallel Orientation and Flexible Conformations of Plant HOP2–MND1. J. Proteome Res. 2015, 14 (12), 5048–5062. 10.1021/acs.jproteome.5b00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.; Bruce J. E. A New Cross-Linking Strategy: Protein Interaction Reporter (PIR) Technology for Protein-Protein Interaction Studies. Mol. BioSyst. 2010, 6 (6), 939–947. 10.1039/b920876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Tang X.; Munske G. R.; Tolic N.; Anderson G. A.; Bruce J. E. Identification of Protein-Protein Interactions and Topologies in Living Cells with Chemical Cross-Linking and Mass Spectrometry. Mol. Cell. Proteomics 2009, 8 (3), 409–420. 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; Mou L.; Lanman J.; Velu S.; Brouillette W. J.; Prevelige P. E. Synthesis of Biotin-Tagged Chemical Cross-Linkers and Their Applications for Mass Spectrometry. Rapid Commun. Mass Spectrom. 2009, 23 (11), 1719–1726. 10.1002/rcm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinz A.; Kalkhof S.; Ihling C. Mapping Protein Interfaces by a Trifunctional Cross-Linker Combined with MALDI-TOF and ESI-FTICR Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2005, 16 (12), 1921–1931. 10.1016/j.jasms.2005.07.020. [DOI] [PubMed] [Google Scholar]
- Kaake R. M.; Wang X.; Burke A.; Yu C.; Kandur W.; Yang Y.; Novtisky E. J.; Second T.; Duan J.; Kao A.; et al. A New in Vivo Cross-Linking Mass Spectrometry Platform to Define Protein–Protein Interactions in Living Cells. Mol. Cell. Proteomics 2014, 13 (12), 3533–3543. 10.1074/mcp.M114.042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.; Li Q.; Zhang M.-J.; Liu C.; Ma C.; Zhang P.; Ding Y.-H.; Fan S.-B.; Tao L.; Yang B.; et al. Trifunctional Cross-Linker for Mapping Protein-Protein Interaction Networks and Comparing Protein Conformational States. eLife 2016, 5, e12509. 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey M.; Dupré M.; Lopez-Neira I.; Duchateau M.; Chamot-Rooke J. EXL-MS: An Enhanced Cross-Linking Mass Spectrometry Workflow To Study Protein Complexes. Anal. Chem. 2018, 90 (18), 10707–10714. 10.1021/acs.analchem.8b00737. [DOI] [PubMed] [Google Scholar]
- Ruprecht B.; Koch H.; Medard G.; Mundt M.; Kuster B.; Lemeer S. Comprehensive and Reproducible Phosphopeptide Enrichment Using Iron Immobilized Metal Ion Affinity Chromatography (Fe-IMAC) Columns. Mol. Cell. Proteomics 2015, 14 (1), 205–215. 10.1074/mcp.M114.043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.; Ye M.; Dong J.; Corradini E.; Cristobal A.; Heck A. J. R.; Zou H.; Mohammed S. Robust Phosphoproteome Enrichment Using Monodisperse Microsphere–Based Immobilized Titanium (IV) Ion Affinity Chromatography. Nat. Protoc. 2013, 8 (3), 461–480. 10.1038/nprot.2013.010. [DOI] [PubMed] [Google Scholar]
- Ficarro S. B.; McCleland M. L.; Stukenberg P. T.; Burke D. J.; Ross M. M.; Shabanowitz J.; Hunt D. F.; White F. M. Phosphoproteome Analysis by Mass Spectrometry and Its Application to Saccharomyces Cerevisiae. Nat. Biotechnol. 2002, 20 (3), 301–305. 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- Villén J.; Gygi S. P. The SCX/IMAC Enrichment Approach for Global Phosphorylation Analysis by Mass Spectrometry. Nat. Protoc. 2008, 3 (10), 1630–1638. 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post H.; Penning R.; Fitzpatrick M. A.; Garrigues L. B.; Wu W.; MacGillavry H. D.; Hoogenraad C. C.; Heck A. J. R.; Altelaar A. F. M. Robust, Sensitive, and Automated Phosphopeptide Enrichment Optimized for Low Sample Amounts Applied to Primary Hippocampal Neurons. J. Proteome Res. 2017, 16 (2), 728–737. 10.1021/acs.jproteome.6b00753. [DOI] [PubMed] [Google Scholar]
- DeGnore J.; Qin J. Fragmentation of Phosphopeptides in an Ion Trap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 1998, 9 (11), 1175–1188. 10.1016/S1044-0305(98)00088-9. [DOI] [PubMed] [Google Scholar]
- Huang H.; Haar Petersen M.; Ibañez-Vea M.; Lassen P. S.; Larsen M. R.; Palmisano G. Simultaneous Enrichment of Cysteine-Containing Peptides and Phosphopeptides Using a Cysteine-Specific Phosphonate Adaptable Tag (CysPAT) in Combination with Titanium Dioxide (TiO2) Chromatography. Mol. Cell. Proteomics 2016, 15 (10), 3282–3296. 10.1074/mcp.M115.054551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J. C.; Creasy D. M.; Cottrell J. S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20 (18), 3551–3567. . [DOI] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-Decoy Search Strategy for Mass Spectrometry-Based Proteomics. Methods Mol. Biol. 2010, 604, 55–71. 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinglé C.; Kuntz L.; Tritsch D.; Grosdemange-Billiard C.; Rohmer M. Isoprenoid Biosynthesis via the Methylerythritol Phosphate Pathway: Structural Variations around Phosphonate Anchor and Spacer of Fosmidomycin, a Potent Inhibitor of Deoxyxylulose Phosphate Reductoisomerase. J. Org. Chem. 2010, 75 (10), 3203–3207. 10.1021/jo9024732. [DOI] [PubMed] [Google Scholar]
- Vorherr T.; Bannwarth W. Phospho-Serine and Phospho-Threonine Building Blocks for the Synthesis of Phosphorylated Peptides by the Fmoc Solid Phase Strategy. Bioorg. Med. Chem. Lett. 1995, 5 (22), 2661–2664. 10.1016/0960-894X(95)00475-9. [DOI] [Google Scholar]
- Kang J.; Chen H.-X.; Huang S.-Q.; Zhang Y.-L.; Chang R.; Li F.-Y.; Li Y.-M.; Chen Y.-X. Facile Synthesis of Fmoc-Protected Phosphonate PSer Mimetic and Its Application in Assembling a Substrate Peptide of 14–3-3 ζ. Tetrahedron Lett. 2017, 58 (26), 2551–2553. 10.1016/j.tetlet.2017.05.037. [DOI] [Google Scholar]
- Belsom A.; Schneider M.; Fischer L.; Brock O.; Rappsilber J. Serum Albumin Domain Structures in Human Blood Serum by Mass Spectrometry and Computational Biology. Mol. Cell. Proteomics 2016, 15 (3), 1105–1116. 10.1074/mcp.M115.048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer L.; Chen Z. A.; Rappsilber J. Quantitative Cross-Linking/Mass Spectrometry Using Isotope-Labelled Cross-Linkers. J. Proteomics 2013, 88, 120–128. 10.1016/j.jprot.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbowski L.; Mendes M. L.; Rappsilber J. Optimizing the Parameters Governing the Fragmentation of Cross-Linked Peptides in a Tribrid Mass Spectrometer. Anal. Chem. 2017, 89 (10), 5311–5318. 10.1021/acs.analchem.6b04935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B.; Gnad F.; Soufi B.; Kumar C.; Olsen J. V.; Mijakovic I.; Mann M. Phosphoproteome Analysis of E. Coli Reveals Evolutionary Conservation of Bacterial Ser/Thr/Tyr Phosphorylation. Mol. Cell. Proteomics 2008, 7 (2), 299–307. 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- Gillette M. A.; Carr S. A. Quantitative Analysis of Peptides and Proteins in Biomedicine by Targeted Mass Spectrometry. Nat. Methods 2013, 10 (1), 28–34. 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasci D.; van Ingen H.; Scheltema R. A.; Heck A. J. R. Histone Interaction Landscapes Visualized by Crosslinking Mass Spectrometry in Intact Cell Nuclei. Mol. Cell. Proteomics 2018, 17 (10), 2018–2033. 10.1074/mcp.RA118.000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.; Beglova N.; Blacklow S. C. Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors. Mol. Cell 2006, 22 (2), 277–283. 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- De Nardis C.; Lössl P.; van den Biggelaar M.; Madoori P. K.; Leloup N.; Mertens K.; Heck A. J. R.; Gros P. Recombinant Expression of the Full-Length Ectodomain of LDL Receptor-Related Protein 1 (LRP1) Unravels PH-Dependent Conformational Changes and the Stoichiometry of Binding with Receptor-Associated Protein (RAP). J. Biol. Chem. 2017, 292 (3), 912–924. 10.1074/jbc.M116.758862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.; Walsh J. D.; Migliorini M.; Yu P.; Cai T.; Schwieters C. D.; Krueger S.; Strickland D. K.; Wang Y.-X. The Structure of Receptor-Associated Protein (RAP). Protein Sci. 2007, 16 (8), 1628–1640. 10.1110/ps.072865407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zundert G. C. P.; Rodrigues J. P. G. L. M.; Trellet M.; Schmitz C.; Kastritis P. L.; Karaca E.; Melquiond A. S. J.; van Dijk M.; de Vries S. J.; Bonvin A. M. J. J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428 (4), 720–725. 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Bloem E.; Ebberink E. H. T. M.; van den Biggelaar M.; van der Zwaan C.; Mertens K.; Meijer A. B. A Novel Chemical Footprinting Approach Identifies Critical Lysine Residues Involved in the Binding of Receptor-Associated Protein to Cluster II of LDL Receptor-Related Protein. Biochem. J. 2015, 468 (1), 65–72. 10.1042/BJ20140977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.