Abstract
Protein N-glycosylation stands out for its intrinsic and functionally related heterogeneity. Despite its biomedical interest, Glycoprofile analysis still remains a major scientific challenge. Here, we present an NMR-based strategy to delineate the N-glycan composition in intact glycoproteins and under physiological conditions. The employed methodology allowed dissecting the glycan pattern of the IgE high-affinity receptor (FcεRIα) expressed in human HEK 293 cells, identifying the presence and relative abundance of specific glycan epitopes. Chemical shifts and differences in the signal line-broadening between the native and the unfolded states were integrated to build a structural model of FcεRIα that was able to identify intramolecular interactions between high-mannose N-glycans and the protein surface. In turn, complex type N-glycans reflect a large solvent accessibility, suggesting a functional role as interaction sites for receptors. The interaction between intact FcεRIα and the lectin hGal3, also studied here, confirms this hypothesis and opens new avenues for the detection of specific N-glycan epitopes and for the studies of glycoprotein–receptor interactions mediated by N-glycans.
Short abstract
An NMR-based Glycoprofile analysis of an intact glycoprotein allows the semiquantitative characterization of the protein decoration by N-glycans, unraveling an extra layer of functional complexity.
Introduction
N-glycosylation is the most abundant post-translational modification in proteins.1 Since N-glycosylation is not template-driven, the hallmark is heterogeneity, which constitutes an extra layer of complexity, directly bound to its biological function. In fact, the N-glycan’s ensemble is directly responsible for modulating molecular recognition events in cell signaling, tissue differentiation, host–pathogen recognition, infections, immune response, and cancer, and it also contributes to the proper folding of the protein.2,3 Alteration of the glycoform pool composition has deleterious effects. For instance, tumor cells display a wide range of glycosylation alterations (i.e., sialylation, fucosylation, and glycan branching) when compared to the nonaltered counterparts.4,5
Given their abundance on the cell surface and vast structural diversity, carbohydrates have been used as epitopes to stage and subtype cell lines.6−8 For example, cell transitions from pluripotency to differentiated progenitors result in elevated α2–6 linked sialic acid on the cellular surface,9 while enzymatic removal of sialic acid triggers differentiation along the ectoderm lineage.10 However, these studies only provide information at the cellular level, and a more detailed characterization of the glycoform ensemble at atomic resolution is desirable. Moreover, there is a wide spectrum of therapeutic opportunity that emerges from a detailed knowledge of the glycoprotein profile.11−14
Glycoprofile analysis remains technically difficult due to the tremendous range of possible monosaccharide combinations and the different ways they might be linked.15 The most common strategy involves a combination of mass spectrometry (MS) and enzymatic digestion or degradation, liquid chromatography, enrichment, and affinity separation.16 In this context, recent methods based on the spectroscopic discrimination of fragments by mass spectrometry17 provide insight on the monosaccharide configuration (α/β) along with the attachment of the glycosidic linkages between the monosaccharide constituents, expanding other methods based on enzymatic digestion and lectin-based analysis. However, glycan release from the glycoprotein hampers the elucidation of the structural–functional role that glycans may play on the protein and vice versa. Ideally, glycoprotein analysis should involve simpler procedures that only minimally alter the test samples.
NMR spectroscopy has contributed to glycoprotein structural characterization, especially focusing on the anomeric region, best resolved in 2D experiments.18−20 Because signal intensities from 2D experiments are impacted by molecular tumbling, the quantification of the N-glycan structure in glycoproteins is best achieved under denaturing conditions,21 which minimize glycan–protein interactions while preserving the glycan composition. However, non-native conditions do not allow glycoprotein structural nor functional analyses. Here, we present an NMR-based strategy to structurally characterize the intact protein glycan content and dynamics. The analysis carried out under physiological conditions was further complemented with the comparison under denaturing conditions, providing leads about glycan presentation and dynamics, information used to build an integrated N-glycosylation model. As a case study we have chosen the soluble domain of the human high-affinity Fc receptor for IgE (FcεRI),22−25 a protein of 20 kDa that contains seven N-linked glycosylation sites (N18, N39, N47, N71, N132, N137, and N163), but the proposed strategy can be potentially applied to other glycoproteins, always within the intrinsic limitations of NMR spectroscopy which include signal overlap of the glycan moieties and the molecular size limit for the glycoprotein, among others. The method entails the determination of the precise glycan structure, including the glycosidic linkages, and a semiquantitative characterization of the protein’s intrinsic glycan heterogeneity, as previously demonstrated.21 The experimental data set is integrated with the aid of computational analysis, and we present a valid model for the glycoprotein in solution. As proof of concept for application and the structural model, we have also studied the glycan-mediated FcεRI interaction with a lectin of biomedical interest, human galectin-3.26,27
Results
NMR-Based Glycoprofile Characterization of FcεRIα
To characterize the Glycoprofile of the soluble portion of the FcεRI glycoprotein (FcεRIα) in physiological conditions by NMR spectroscopy, the uniformly 13C,15N double-labeled protein was produced in human HEK 293 cells as previously described.28 HEK 293 cells are challenging because they incorporate many more glycoforms than other cell lines like CHO DG44, which lead to almost exclusively sialylated biantennary N-glycans.29 The protocol ensured large amounts (360 μg of prot/20 mL of culture) of glycoprotein, uniformly labeled in both the peptide and glycan chains. The obtained glycoprotein (350 μL at 60 μM concentration) after proper purification was suitable for its investigation by NMR spectroscopy (Figure S1). Glycoproteins exhibit slower molecular tumbling because of their bulky glycan moieties and often because of their multidomain and/or oligomeric structures. Therefore, and except for counted cases limited to glycoproteins with homogeneous tailored glycosylation,19,30 the NMR characterization of the peptidic portion is precluded. Instead, the inherent flexibility of glycans allows for their structural characterization by standard NMR experiments.
A canonical NMR-based approach31 was employed to characterize the Glycoprofile of FcεRIα under denaturing conditions. The anomeric region of the 1H,13C-HSQC spectrum (Figure 1d) revealed differentiated cross-peaks that allowed determining the precise structure of the glycans along with the nature of the glycosidic linkages between monosaccharides. To that end, the combined analysis of 1H,13C-HSQC-TOCSY and 1H,13C-HSQC-NOESY was contrasted with the existing literature.21,32−34 To avoid miss-assignments arising from secondary chemical shifts due to interactions with the protein, a parallel assignment was carried out under denaturing conditions (Figure 1e). Fittingly, the anomeric region showed no significant changes in chemical shifts between the folded and denatured samples (see below).
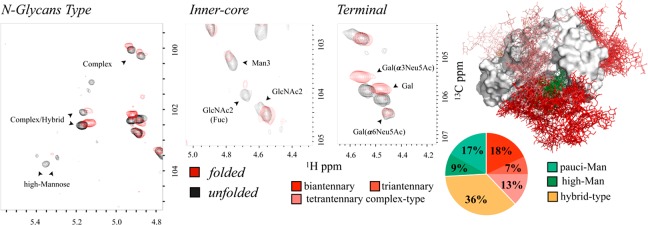
Figure 1.
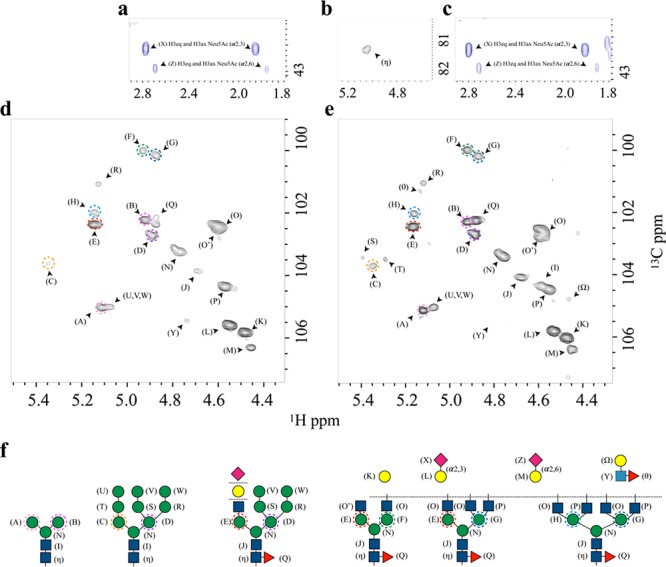
Glycan content. FcεRIα glycosylation patterns as deduced by direct analysis of the [1H,13C]-HSQC NMR spectra. Expansion of the region containing the axial and equatorial H3 protons of the Neu5Ac residues in the folded (a) and the unfolded (c) state. (b) Expansion of the spectral region showing the anomeric (C1–H1) cross-peaks of the Asn-linked GlcNAc 1, which appears only under denaturing conditions. (d, e) Expansion of the spectral region showing the anomeric (C1–H1) cross-peaks of the linked saccharides in native and denaturing conditions, respectively. (f) Schematic representation of the different N-glycans identified. Each monosaccharide with a unique 1H,13C resonance is labeled with a different letter. The Man residues in the antennae (A–H) are further identified through different colored dashed circles. These residues differ in terms of their chemical linkages and, consequently, in their NMR chemical shifts.
The analysis shows that the N-glycans on the protein contain sialic acid (Neu5Ac), galactose (Gal), N-acetylglucosamine (GlcNAc), mannose (Man), and fucose (Fuc) residues (Figure 1). The detailed resonance assignment is given in the Supporting Information (Table S1). To validate the anomeric protons assignment, we used trimming enzymes as shown in Figure S2, which was crucial in the assignment of partially overlapping peaks. The deduced structures include the presence of pauci-mannose, high-mannose, hybrid, and bi-, tri-, and tetra-antennary complex type N-glycans with different degrees of fucosylation and sialylation.
Protein’s Glycan Heterogeneity in Denaturing Conditions
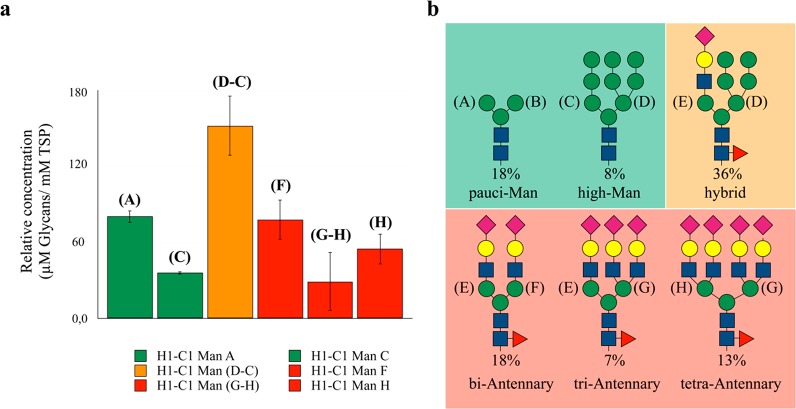
A significant degree of signal overlap underlines the structural similarity among the N-glycans. Still, branched Man residues, which are unique in the different N-glycan types, generate unequivocal resonances in the 1H,13C-HSQC spectrum (Figure 1d,e). Information from the peaks volumes in HSQC spectra is complex since it may be modulated by scalar couplings and for the signal’s dependence on the polarization transfer scheme. However, C–H cross-peaks corresponding to the anomeric groups of different Man residues (A–H) are much more comparable since they all share similar J-couplings and relaxation properties, and they were used to define the glycan heterogeneity of FcεRIα (A–H in Figure 1). Peak integration compared to the set of signals or to a reference standard provides relative or absolute quantification: 110 μM oligo-mannose (26%, ∼1.8 mol glyc/mol prot), 150 μM hybrid (36%, ∼2.5 mol glyc/mol prot), and 160 μM complex type (38%, ∼2.7 mol glyc/mol prot). Among the complex type, biantennary was detected at the highest amount (80 μM, 18% of the total), whereas tri- and tetra-antennary were 30 μM (7%) and 50 μM (13%), respectively. Among oligo-mannose N-glycans, 76 μM (18%) are pauci-mannose and 34 μM (8%) are high-mannose types, as shown in Figure 2. Thus, since the absolute quantification adds little value to the characterization, it was no longer considered.
Figure 2.
Quantitative Glycoprofile analysis. (a) Relative concentration of the different glycoforms with respect to the protein concentration (60 μM) as estimated by the integration of the HSQC signals of the branched Man residues compared to internal reference TSP (trimethylsilylpropanoic acid 1 mM). Data analysis has been done over three different samples, and error bars are calculated by standard deviation analysis. The quantification of the hybrid and triantennary glycans was carried out by subtraction of the integrals arising from C to those of D, and H to G, respectively. The color code bar corresponds to the N-glycans topology as represented in part b.
Composition of the Specific Glycan Epitopes
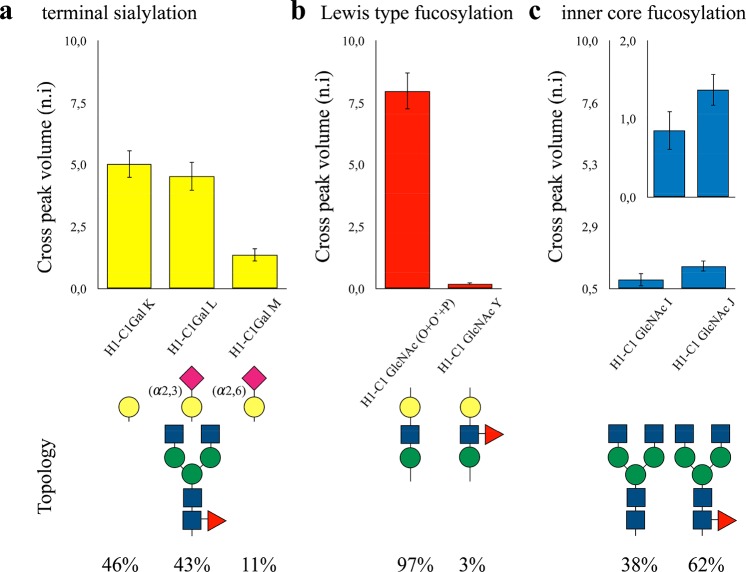
Glycoprofile analysis of intact glycoproteins by NMR provides a unique tool to identify specific glycan epitopes. Neu5Ac residues are unequivocally identified by the high field resonances of the axial and equatorial H3 protons of the sugar ring. Fittingly, the chemical shifts for these protons are sensitive enough to discriminate between the α2,3- and α2,6-linked Neu5Ac residues (Figure 1c).35 Moreover, the Gal residues show discriminant chemical shifts between the terminal position and the α2,3- or α2,6-substituted by sialic acid residues (K–M, Figure 1d,e). Thus, the relative abundance of these glycan epitopes is quantified by comparing the anomeric H–C cross-peak volumes of Gal residues K–M. FcεRIα presents an almost equal distribution of terminal and α2,3-linked Gal moieties (46% and 43%, respectively), while α2,6-linked Neu5Ac are significantly less populated (11%) (Figure 3a). The chemical shifts of the anomeric protons of the GlcNAc residues are fairly sensitive to Fuc substitution in the antennae and in the core. Thus, a similar analysis also allowed quantifying the relative abundance of fucosylated N-glycans as well as their substitution position (i.e., branch modification or inner-core). Specifically, only 3% of N-glycans in FcεRIα contained the Lewis type antigens in the antennae (Figure 3b) as determined by a comparison of the anomeric signal of (Y) with respect to the equivalent in O′, O, and P. On the other hand, 62% presented inner-core fucosylation (Figure 3c), as determined by comparing the J and I spin systems. Lewis antigens (Lea, Leb, Ley, Lex) can be identified due to the characteristic downfield chemical shift of H5 of the Fuc residue, above 4.6 ppm.36 In here, the direct comparison of the HSQC of the free Lewis X with the HSQC of the FcεRIα showed a perfect match of the cross-peaks, suggesting that the Lewis antigen in the antennae is LeX. Again, only equivalent residues were compared, sharing the same position along the glycan chain as well as the chemical nature, to avoid intensity differences due to the different mobility (different rotational motion effective correlation times) or C–H coupling.
Figure 3.
Specific glycan epitope analysis. The different biorelevant N-glycans and epitopes contained in FcεRIα, together with their relevant abundance. (a) Estimation of the specific terminal sialylation obtained by comparing the cross-peak intensities of the galactose signals measured for the terminal Gal, Neu5Ac(α2–3)Gal, and Neu5Ac(α2–6)Gal moieties.35 (b) Estimation of the degree of fucosylation in the antennae as well as its relative abundance, calculated comparing (O′ + O + P) with (Y). (c) Estimation of the degree of inner-core fucosylation, calculated comparing (I) and (J). The lowest signal intensities observed for the GlcNAc2 residues are related to their different rotational motion effective correlation times with respect to the terminal residues, which are fairly more mobile. The inset plot in a different scale clarifies the relative abundance of this glycan modification.
N-Glycan Presentation and Dynamics at the Glycoprotein Surface
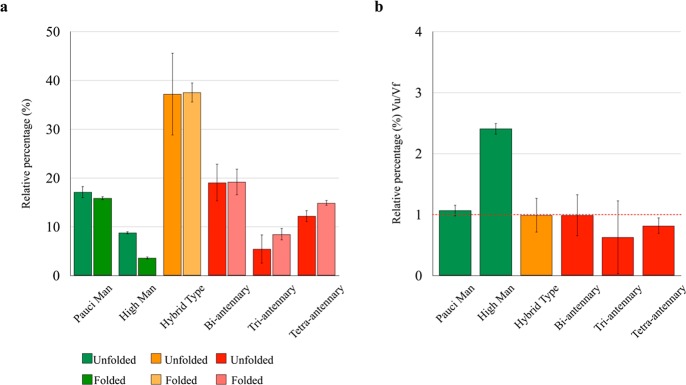
The high sensitivity of the NMR chemical shift to the environment37,38 was now employed to investigate how the protein scaffold presents the linked N-glycans. In fact, this feature remains an open question for the full characterization of the structure–function relationship in glycoproteins. The close chemical shift coincidence between most of the N-glycans in FcεRIα between the folded and the unfolded states (Figure 1d,e) and with the reported values for the free saccharides is consistent with a solvent-exposed state of the N-glycans, which merely undergo transient intramolecular interactions with the protein chain. However, the semiquantitative analysis carried out in the unfolded state may significantly differ from that obtained under native conditions (folded state) if the N-glycan establishes stable interactions with the protein moiety that will increase its tumbling time, reducing its intensity and yielding an apparent composition.18 This is the case for the high-mannose N-glycans, whose composition becomes 3.3% under native conditions, as compared to 8.3% in denaturing solvent. On the other hand, pauci-mannose and complex N-glycans are largely insensitive to solvent composition (Figure 4), indicating that oligo-mannose N-glycans are more prone to participate in glycan–protein intramolecular interactions than the hybrid or complex type N-glycans.
Figure 4.
Comparison of the N-glycan manifold in FcεRIα under denaturing and native-preserving conditions. (a) Relative percentage of the different types of N-glycans in FcεRIα, as determined in denatured and native conditions. Under denaturing conditions, the percentage always reflects the N-glycan composition while the apparent value obtained in native-preserving solvent is lowered when the N-glycan is interacting with the protein moiety. (b) Ratio of the population in the native and denatured states for the different N-glycan types, where the high mannoses show a statistically significant deviation from unity. Error bars are obtained from independent triplicate measurements.
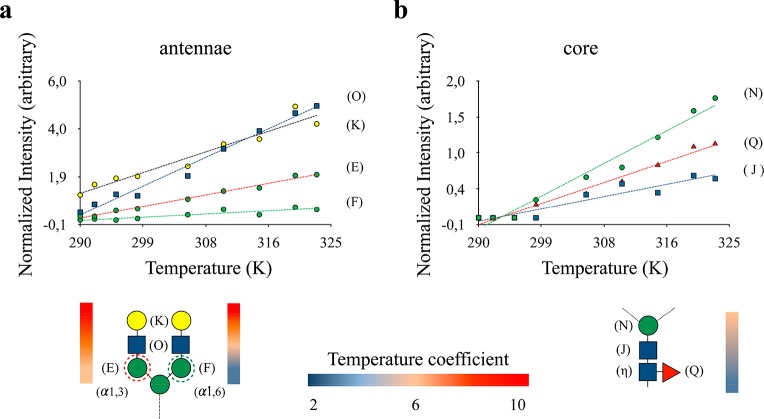
In the absence of chemical–conformational exchange, relaxation in the ns–ps regime dictates the observed line-broadening of the cross-peak intensities in the spectrum, providing information about the segmental dynamics of the N-glycans, with intense (weak) resonances indicating the existence of fast (limited) rotational diffusion. A CPMG experiment39 measured at two distant values of νCPMG demonstrates that there is no significant exchange contribution to the line-broadening of the N-glycan moieties in FcεRIα. Under these circumstances, temperature coefficients can also provide useful information regarding N-glycan dynamic behavior and their relative interactions with the solvent. Figure 5 shows the normalized intensities of the 1H–13C cross-peak intensities and their temperature dependence, using TSP as an internal reference. As expected, the core saccharides always show extremely weak signals as compared to the terminal Neu5Ac or Gal residues. In fact, the N-glycans with terminal Gal residues present a direct correlation between the position of the sugar along the glycan chain and its temperature susceptibility, with the terminal sugars being significantly more sensitive to temperature changes than those closer to the protein.
Figure 5.
Analysis of N-glycans dynamics. The plots represent the signal intensities measured for the anomeric C1–H1 cross-peaks of each monosaccharide for a biantennary N-glycan, recorded as a function of temperature. (a) The antennae monosaccharides show intense resonances, which increase with temperature in a linear way. The differences among the sugar positions, at either the 1,3 or 1,6 arms, are also highlighted. (b) The inner-core sugars (Fuc, GlcNAc2, and Man3) show extremely weak signals.
Remarkably, the combined line broadening and temperature coefficient analysis allowed discriminating between the 1,3 and 1,6 arms. Indeed, because of the higher flexibility provided by the additional glycosidic torsion angle, the Man residues in the 1,6 antennae are much less sensitive to temperature, reflecting their intrinsic higher mobility. This is functionally relevant, and for a glycoengineered variant of the IgG-Fc antibody, whose single N-glycan was uniformly remodeled with 13C-Gal and Neu5Ac, it has been demonstrated that the degree of flexibility in the 1,6 arm allows for transient glycan–protein contacts, which are otherwise unfavorable in the 1,3 arm.40,41 Finally, the subtle dynamic differences observed between the antennae are also based on structural details, since they are not observed in the unfolded state of FcεRIα (Figure S3).
Integrative Structural Model of the FcεRIα Glycoprotein
The experimental and computational results were integrated to generate a solution model of FcεRIα. Any proposed model should consider the intrinsic glycan heterogeneity as well as the observed glycan dynamic behavior. Additionally, it is required for the N-glycan moieties to link the corresponding asparagine (Asn) residues in the protein. It has been demonstrated that the less processed high-mannose glycans are regularly attached to Asn residues that are highly inaccessible to the action of the enzymes, while the complex type glycans are usually solvent-exposed.42,43
The direct comparison of the magnitude of the NMR signal intensities between native and unfolded states suggests that this is also true for the FcεRIα protein. In fact, NMR signals belonging to the high-mannose glycans experience the most pronounced intensity gain and chemical shift perturbation as result of protein unfolding (Figure 4b and Figure S3). Thus, we suggest that the high-mannose N-glycans are mainly linked to the less solvent-exposed Asn residues of the FcεRIα glycoprotein. The analysis of the solvent accessible surface area (SASA) of FcεRIα revealed that Asn132 is the most protected residue. Therefore, a high-mannose N-glycan was built at this locus. On the contrary, Asn71, 137, and 163 are highly solvent-exposed and are the perfect targets to allocate the tetra-antennary N-glycans, with different degrees of sialylation. Finally, Asn47, 39, and 18 show intermediate SASA. These positions were decorated with hybrid and biantennary sialylated and nonsialylated N-glycans, respectively (Figure S4).
To confirm these results, we produced the A132N mutant of the FcεRIα. The analysis of the glycan content for this mutant demonstrates a significative reduction in the population of high-mannose species (Figure S5). To adequately explore the conformational space accessible to the N-glycans we performed a 1 μs MD simulation on the above-mentioned glycoprotein configuration. In the 10-conformer ensemble extracted from the MD simulation, the majority of the N-glycans protrude outside the protein (Figure 6). According to this model, a structural role can be attributed to the glycans attached to Asn39 and Asn132. Indeed, the simulation suggests that the glycan at Asn39 (Figure 6, light blue) shows multiple orientations that bridge the two immunoglobulin-like domains of the protein through intramolecular interactions with the sugar residues at the 1,6 and 1,3 arms. In turn, the high-mannose glycan at Asn132 is buried within the two protein’s domains and could contribute to keep the structural integrity of the glycoprotein (Figure 6, green). In general, all N-glycans besides the confined high-mannose glycan show a significantly dynamic behavior, being prevalently solvent-exposed, providing putative interaction sites for receptors, including lectins.44
Figure 6.
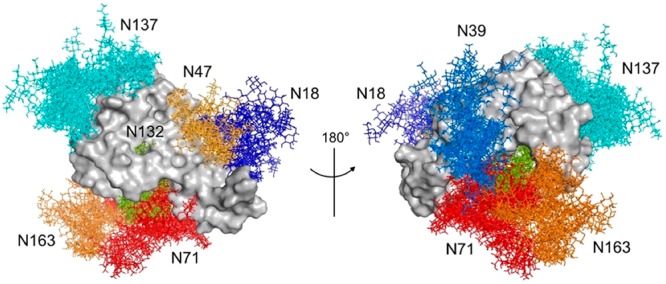
Proposed model of the FcεRIα glycoprotein. Front and back view for an ensemble of 10 conformers extracted from the 1 μs MD simulation. Structures were superimposed on the peptide backbone atoms. The N-glycans at each N-glycosylation site are colored as follows: biantennary N-glycan at Asn18 (dark-blue); biantennary N-glycan at Asn39 (light-blue); hybrid N-glycan at Asn47 (light-orange); tetra-antennary N-glycan at Asn71 (red); high-mannose N-glycan at Asn132 (green); tetra-antennary N-glycan at Asn137 (cyan); tetra-antennary N-glycan at Asn163 (orange).
Glycoprotein–Protein (Lectin) Interactions Mediated by Glycans
To further validate the structural model and the presentation of the glycans, we have experimentally investigated the interaction between FcεRIα and the carbohydrate recognition domain (CRD) of a lectin of biomedical interest, the human galectin-3 (hGal3). hGal3 recognizes 3′-sialyl N-acetyllactosamine (3′SLN) and N-acetyllactosamine (LN) but does not bind 6′-sialyl N-acetyllactosamine.45 Therefore, the lectin should show selectivity for the different glycan epitopes present in FcεRIα. 1H–15N TROSY spectra of isotopically labeled hGal3 were measured in the absence and in the presence of unlabeled FcεRIα, and the changes in the signal line width were monitored. As shown in Figure 7, hGal3 exhibited significant reductions of varying magnitudes in peak intensity. Interestingly, intensity attenuation was far more pronounced in the peaks originating from residues belonging to the S2, S3, S4, S5, and S6 strands of the β-sheet containing the ligand binding site.46 These data allowed the generation of a docking model for the interaction between FcεRIα and hGal3 (Figure S6).
Figure 7.
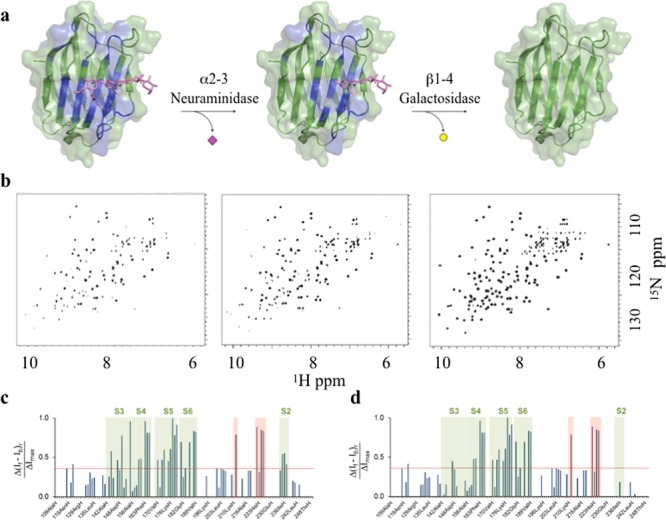
Lectin binding experiments with intact FcεRIα. The recognition of specific exposed epitopes in FcεRIα by hGal3 was monitored by NMR signals intensity analysis of 15N-hGal3 in 1H–15N TROSY NMR experiments. (a) The model for the lectin–glycoprotein interaction was built by manually docking the N-glycan at the Asn137 of the glycoprotein obtained from the MD simulation approach (see materials and methods) to the crystal structure of the human galectin-3 bound to N-acetyllactosamine (PDB code: 1A3K). In this cartoon, the glycoprotein peptide portion and the other N-glycans have been removed for the sake of clarity (see Figure S6 for the complete picture). The hGal3 residues affected by glycoprotein binding are mapped in blue. (b) 1H–15N TROSY NMR spectra of hGal3 in the presence of the complete FcεRIα glycoprotein (left), after enzymatic hydrolysis of the terminal α2,3-linked Neu5NAc moieties, and after subsequent hydrolysis of the β1,4-linked Gal residues. (c) Plot bars of the intensity differences per residue measured between apo hGal3 and FcεRIα-bound hGal3. (d) Plot bars of the intensity differences per residue measured between apo hGal3 and FcεRIα-bound hGal3 after treatment with α2–3 neuraminidase S. Specific residues show meaningful intensity differences and belong to the S2–S6 strands (green boxes). Additional residues located far away from the canonical carbohydrate binding site are also perturbed (red box), an effect previously observed.46
To demonstrate ligand specificity, the terminal sialyl residues of the Neu5NAc(α2–3)Gal moieties were selectively trimmed after treating FcεRIα with α2–3 neuraminidase S, which is highly specific for these entities while leaving the Neu5NAc(α2–6)Gal fragments unperturbed. The TROSY spectrum of hGal3 recorded in the presence of this variant displayed a clear recovery of the observed intensities of those cross-peaks belonging to amino acids in the S2–S3 region. Fittingly, this is the specific lectin region that provides interaction to Neu5NAc moieties.47 In contrast, the signals arising from the amino acids at S4–S6 strands remained attenuated. In a further modification of the same sample, the external β-galactose epitopes were removed by treating the receptor with Escherichia coli β-galactosidase, which is an exo-galactosidase with preference for terminal β-Gal moieties. The new TROSY spectrum now fully coincides with the canonical spectrum for apo hGal3, demonstrating that the FcεRIα–hGal3 interaction is exclusively on cargo of the exposed specific N-glycans, in particular those exposing terminal 3′SLN and LN moieties.
Concluding Remarks
We demonstrated that the use of a relatively inexpensive and standard isotopically labeled glycoprotein produced in human cells lines allows for the structural and functional studies of the full glycoprotein, keeping its intrinsic complexity, under physiological conditions. The detailed analysis of the intrinsic glycan heterogeneity has been solved by combining a suite of standard NMR experiments. Since NMR is also able to clinch the dynamic behavior of glycan chains in solution,48 the relative dynamic features of the different glycans and epitopes have also been explored. Moreover, an educated guess for the site-specific glycan assignment may be extracted by determining the accessibility of the glycosylation site to the enzymatic processing machinery,42,43 combined with the analysis of NMR signals perturbations as a function of the folded state of the protein.
Among the limitations of the method, the requirement for the protein to be isotopically labeled (15N and 13C) limits its breath. However, we and others28 demonstrated that this strategy is suitable for very different systems, at a reasonable economic cost. Size of the glycoprotein is also limited by the maximum correlation time that can be studied by NMR, and the complexity in the glycan moiety may also compromise the study due to signal overlap in the spectrum. The incorporation of TROSY techniques and multidimensional spectroscopy may alleviate, in part, these shortcomings. Another significant limitation of the method is that it does not directly provide the site-specific occupancy of the N-glycans at the protein. The recently presented strategy for site-specific analysis of glycoproteins49 represents an interesting complement to the methodology presented herein.
As future perspectives we propose that the herein presented methodologies could be used for the unambiguous detection of specific glycan signatures found on tumor cells, which are considered as a novel type of immune checkpoint,7 and to detect the effects of therapeutic interventions that relate to the tumor glyco-code.
Safety Statement
No unexpected or unusually high safety hazards were encountered in this research.
Acknowledgments
We thank Agencia Estatal de Investigación (Spain) for Grants CTQ2015-64597-C2-1-P and CTQ2015-68756-R, for an FPI fellowship to M.P.L., and for the Severo Ochoa Excellence Accreditation (SEV-2016-0644). J.J.-B. also thanks the European Research Council (RECGLYCANMR, Advanced Grant 788143).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.9b00540.
Additional experimental data including chemical shift assignments and the outcome of enzymatic reactions and additional figures for the structural model of the glycoprotein (PDF)
Author Contributions
L.U., O.M., and J.J.-B. designed the research and wrote the paper. L.U., M.P.L., and A.L. performed the experiments, and L.U., P.U., M.P.L., J.M.F.-P., and A.A. analyzed the data.
The authors declare no competing financial interest.
Supplementary Material
References
- Varki A. e. a.Essentials of Glycobiology; 3rd ed.; Cold Spring Harbor Laboratory Press, 2017. [PubMed] [Google Scholar]
- Helenius A.; Aebi M. Intracellular functions of N-linked glycans. Science (Washington, DC, U. S.) 2001, 291 (5512), 2364–2369. 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K.; Marth J. D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126 (5), 855–867. 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Moginger U.; Grunewald S.; Hennig R.; Kuo C. W.; Schirmeister F.; Voth H.; Rapp E.; Khoo K. H.; Seeberger P. H.; Simon J. C.; et al. Alterations of the Human Skin N- and O-Glycome in Basal Cell Carcinoma and Squamous Cell Carcinoma. Front. Oncol. 2018, 8, 70. 10.3389/fonc.2018.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho S. S.; Reis C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15 (9), 540–555. 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Gooi H. C.; Feizi T.; Kapadia A.; Knowles B. B.; Solter D.; Evans M. J. Stage-specific embryonic antigen involves alpha 1 goes to 3 fucosylated type 2 blood group chains. Nature 1981, 292 (5819), 156–158. 10.1038/292156a0. [DOI] [PubMed] [Google Scholar]
- Rodríguez E.; Schetters S. T. T.; van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018, 18 (3), 204–211. 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- Solter D.; Knowles B. B. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc. Natl. Acad. Sci. U. S. A. 1978, 75 (11), 5565–5569. 10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasehira K.; Tateno H.; Onuma Y.; Ito Y.; Asashima M.; Hirabayashi J. Structural and quantitative evidence for dynamic glycome shift on production of induced pluripotent stem cells. Mol. Cell. Proteomics 2012, 11 (12), 1913–1923. 10.1074/mcp.M112.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisson-Silva F.; de Carvalho Rodrigues D.; Vairo L.; Asensi K. D.; Vasconcelos-Dos-Santos A.; Mantuano N. R.; Dias W. B.; Rondinelli E.; Goldenberg R. C.; Urmenyi T. P.; et al. Evidences for the involvement of cell surface glycans in stem cell pluripotency and differentiation. Glycobiology 2014, 24 (5), 458–468. 10.1093/glycob/cwu012. [DOI] [PubMed] [Google Scholar]
- Dalziel M.; Crispin M.; Scanlan C. N.; Zitzmann N.; Dwek R. A. Emerging principles for the therapeutic exploitation of glycosylation. Science (Washington, DC, U. S.) 2014, 343 (6166), 1235681. 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- Jennewein M. F.; Alter G. The Immunoregulatory Roles of Antibody Glycosylation. Trends Immunol. 2017, 38 (5), 358–372. 10.1016/j.it.2017.02.004. [DOI] [PubMed] [Google Scholar]
- Li J.; Chen M.; Liu Z.; Zhang L.; Felding B. H.; Moremen K. W.; Lauvau G.; Abadier M.; Ley K.; Wu P. A Single-Step Chemoenzymatic Reaction for the Construction of Antibody-Cell Conjugates. ACS Cent. Sci. 2018, 4 (12), 1633–1641. 10.1021/acscentsci.8b00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeem T.; Khan M. A.; Ijaz B.; Ahmed N.; Rahman Z. U.; Latif M. S.; Ali Q.; Rana M. A. Glycosylation of Recombinant Anticancer Therapeutics in Different Expression Systems with Emerging Technologies. Cancer Res. 2018, 78 (11), 2787–2798. 10.1158/0008-5472.CAN-18-0032. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Luo S.; Zhang B. Glycan analysis of therapeutic glycoproteins. MAbs 2016, 8 (2), 205–215. 10.1080/19420862.2015.1117719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarich D.; Jensen P. H.; Altmann F.; Packer N. H. Determination of site-specific glycan heterogeneity on glycoproteins. Nat. Protoc. 2012, 7 (7), 1285–1298. 10.1038/nprot.2012.062. [DOI] [PubMed] [Google Scholar]
- Schindler B.; Barnes L.; Renois G.; Gray C.; Chambert S.; Fort S.; Flitsch S.; Loison C.; Allouche A. R.; Compagnon I. Anomeric memory of the glycosidic bond upon fragmentation and its consequences for carbohydrate sequencing. Nat. Commun. 2017, 8 (1), 973. 10.1038/s41467-017-01179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert M.; Walczak M. J.; Aebi M.; Wider G. Posttranslational modifications of intact proteins detected by NMR spectroscopy: application to glycosylation. Angew. Chem., Int. Ed. 2015, 54 (24), 7096–7100. 10.1002/anie.201502093. [DOI] [PubMed] [Google Scholar]
- Slynko V.; Schubert M.; Numao S.; Kowarik M.; Aebi M.; Allain F. H. NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation. J. Am. Chem. Soc. 2009, 131 (3), 1274–1281. 10.1021/ja808682v. [DOI] [PubMed] [Google Scholar]
- Yanaka S.; Yagi H.; Yogo R.; Yagi-Utsumi M.; Kato K. Stable isotope labeling approaches for NMR characterization of glycoproteins using eukaryotic expression systems. J. Biomol. NMR 2018, 71 (3), 193–202. 10.1007/s10858-018-0169-2. [DOI] [PubMed] [Google Scholar]
- Peng J.; Patil S. M.; Keire D. A.; Chen K. Chemical Structure and Composition of Major Glycans Covalently Linked to Therapeutic Monoclonal Antibodies by Middle-Down Nuclear Magnetic Resonance. Anal. Chem. 2018, 90 (18), 11016–11024. 10.1021/acs.analchem.8b02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U.; Ra C. S.; Kinet J. P. Characterization of truncated alpha chain products from human, rat, and mouse high affinity receptor for immunoglobulin E. J. Biol. Chem. 1991, 266 (4), 2639–2646. [PubMed] [Google Scholar]
- Garman S. C.; Kinet J. P.; Jardetzky T. S. Crystal structure of the human high-affinity IgE receptor. Cell 1998, 95 (7), 951–961. 10.1016/S0092-8674(00)81719-5. [DOI] [PubMed] [Google Scholar]
- Garman S. C.; Sechi S.; Kinet J. P.; Jardetzky T. S. The analysis of the human high affinity IgE receptor Fc epsilon Ri alpha from multiple crystal forms. J. Mol. Biol. 2001, 311 (5), 1049–1062. 10.1006/jmbi.2001.4929. [DOI] [PubMed] [Google Scholar]
- Garman S. C.; Wurzburg B. A.; Tarchevskaya S. S.; Kinet J. P.; Jardetzky T. S. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature 2000, 406 (6793), 259–266. 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- Chou F. C.; Chen H. Y.; Kuo C. C.; Sytwu H. K. Role of Galectins in Tumors and in Clinical Immunotherapy. Int. J. Mol. Sci. 2018, 19 (2), E430. 10.3390/ijms19020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacchitano S.; Lavra L.; Morgante A.; Ulivieri A.; Magi F.; De Francesco G. P.; Bellotti C.; Salehi L. B.; Ricci A. Galectin-3: One Molecule for an Alphabet of Diseases, from A to Z.. Int. J. Mol. Sci. 2018, 19 (2), E379. 10.3390/ijms19020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri L.; Luchinat E.; Banci L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nat. Protoc. 2016, 11 (6), 1101–1111. 10.1038/nprot.2016.061. [DOI] [PubMed] [Google Scholar]
- Shibata-Koyama M.; Iida S.; Okazaki A.; Mori K.; Kitajima-Miyama K.; Saitou S.; Kakita S.; Kanda Y.; Shitara K.; Kato K.; et al. The N-linked oligosaccharide at Fc gamma RIIIa Asn-45: an inhibitory element for high Fc gamma RIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology 2008, 19 (2), 126–134. 10.1093/glycob/cwn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y.; Nishimura M.; Nagano M.; Yagi H.; Sasakawa H.; Uchida K.; Shitara K.; Kato K. Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim. Biophys. Acta, Gen. Subj. 2006, 1760 (4), 693–700. 10.1016/j.bbagen.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Weller C. T.; Lustbader J.; Seshadri K.; Brown J. M.; Chadwick C. A.; Kolthoff C. E.; Ramnarain S.; Pollak S.; Canfield R.; Homans S. W. Structural and conformational analysis of glycan moieties in situ on isotopically 13C, 15N-enriched recombinant human chorionic gonadotropin. Biochemistry 1996, 35 (27), 8815–8823. 10.1021/bi960432f. [DOI] [PubMed] [Google Scholar]
- Canales A.; Boos I.; Perkams L.; Karst L.; Luber T.; Karagiannis T.; Dominguez G.; Canada F. J.; Perez-Castells J.; Haussinger D.; et al. Breaking the Limits in Analyzing Carbohydrate Recognition by NMR Spectroscopy: Resolving Branch-Selective Interaction of a Tetra-Antennary N-Glycan with Lectins. Angew. Chem., Int. Ed. 2017, 56 (47), 14987–14991. 10.1002/anie.201709130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Toro B.; Peng W.; Thompson A. J.; Dominguez G.; Canada F. J.; Perez-Castells J.; Paulson J. C.; Jimenez-Barbero J.; Canales A. Avenues to Characterize the Interactions of Extended N-Glycans with Proteins by NMR Spectroscopy: The Influenza Hemagglutinin Case. Angew. Chem., Int. Ed. 2018, 57 (46), 15051–15055. 10.1002/anie.201807162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno A.; Reichardt N. C.; Canada F. J.; Perkams L.; Unverzagt C.; Jimenez-Barbero J.; Arda A. NMR and Molecular Recognition of N-Glycans: Remote Modifications of the Saccharide Chain Modulate Binding Features. ACS Chem. Biol. 2017, 12 (4), 1104–1112. 10.1021/acschembio.6b01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klukowski P.; Schubert M. Chemical shift-based identification of monosaccharide spin-systems with NMR spectroscopy to complement untargeted glycomics. Bioinformatics 2019, 35 (2), 293–300. 10.1093/bioinformatics/bty465. [DOI] [PubMed] [Google Scholar]
- Aeschbacher T.; Zierke M.; Smiesko M.; Collot M.; Mallet J. M.; Ernst B.; Allain F. H.; Schubert M. A Secondary Structural Element in a Wide Range of Fucosylated Glycoepitopes. Chem. - Eur. J. 2017, 23 (48), 11598–11610. 10.1002/chem.201701866. [DOI] [PubMed] [Google Scholar]
- Kato K.; Yamaguchi Y.; Arata Y. Stable-isotope-assisted NMR approaches to glycoproteins using immunoglobulin G as a model system. Prog. Nucl. Magn. Reson. Spectrosc. 2010, 56 (4), 346–359. 10.1016/j.pnmrs.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Matsumiya S.; Yamaguchi Y.; Saito J.; Nagano M.; Sasakawa H.; Otaki S.; Satoh M.; Shitara K.; Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J. Mol. Biol. 2007, 368 (3), 767–779. 10.1016/j.jmb.2007.02.034. [DOI] [PubMed] [Google Scholar]
- Millet O.; Bernado P.; Garcia J.; Rizo J.; Pons M. NMR measurement of the off rate from the first calcium-binding site of the synaptotagmin I C2A domain. FEBS Lett. 2002, 516 (1–3), 93–96. 10.1016/S0014-5793(02)02508-5. [DOI] [PubMed] [Google Scholar]
- Barb A. W.; Meng L.; Gao Z.; Johnson R. W.; Moremen K. W.; Prestegard J. H. NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 2012, 51 (22), 4618–4626. 10.1021/bi300319q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barb A. W.; Prestegard J. H. NMR analysis demonstrates immunoglobulin G N-glycans are accessible and dynamic. Nat. Chem. Biol. 2011, 7 (3), 147–153. 10.1038/nchembio.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang I.; Lin C. W.; Grant O. C.; Fleurkens S.; Villiger T. K.; Soos M.; Morbidelli M.; Woods R. J.; Gauss R.; Aebi M. Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi. Glycobiology 2015, 25 (12), 1335–1349. 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri K.; Klein J. A.; White M. R.; Grant O. C.; Leymarie N.; Woods R. J.; Hartshorn K. L.; Zaia J. Integrated Omics and Computational Glycobiology Reveal Structural Basis for Influenza A Virus Glycan Microheterogeneity and Host Interactions. Mol. Cell. Proteomics 2016, 15 (6), 1895–1912. 10.1074/mcp.M116.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfert M. A.; Boons G. J. Adaptive immune activation: glycosylation does matter. Nat. Chem. Biol. 2013, 9 (12), 776–784. 10.1038/nchembio.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell S. R.; Arthur C. M.; Mehta P.; Slanina K. A.; Blixt O.; Leffler H.; Smith D. F.; Cummings R. D. Galectin-1, −2, and −3 exhibit differential recognition of sialylated glycans and blood group antigens. J. Biol. Chem. 2008, 283 (15), 10109–10123. 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl C.; Engstrom O.; Delaine T.; Hakansson M.; Genheden S.; Modig K.; Leffler H.; Ryde U.; Nilsson U. J.; Akke M. Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J. Am. Chem. Soc. 2010, 132 (41), 14577–14589. 10.1021/ja105852y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P. M.; Bum-Erdene K.; Yu X.; Blanchard H. Galectin-3 interactions with glycosphingolipids. J. Mol. Biol. 2014, 426 (7), 1439–1451. 10.1016/j.jmb.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Subedi G. P.; Falconer D. J.; Barb A. W. Carbohydrate-Polypeptide Contacts in the Antibody Receptor CD16A Identified through Solution NMR Spectroscopy. Biochemistry 2017, 56 (25), 3174–3177. 10.1021/acs.biochem.7b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L.; Diedrich J. K.; Ma Y.; Wang N.; Pauthner M.; Park S. R.; Delahunty C. M.; McLellan J. S.; Burton D. R.; Yates J. R.; et al. Global site-specific analysis of glycoprotein N-glycan processing. Nat. Protoc. 2018, 13 (6), 1196–1212. 10.1038/nprot.2018.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.