Abstract
Descriptive and translational investigations into the human gut microbiome (GM) are rapidly expanding; however, studies are largely restricted to industrialized populations in the USA and Europe. Little is known about microbial variability and its implications for health and disease in other parts of the world. Populations in Africa are particularly underrepresented. What limited research has been performed has focused on a few subject domains, including the impact of long-term lifestyle and dietary factors on GM ecology, its maturation during infancy, and the interrelationships between the microbiome, infectious disease, and undernutrition. Recently, international consortia have laid the groundwork for large-scale genomics and microbiome studies on the continent, with a particular interest in the epidemiologic transition to noncommunicable disease. Here, we survey the current landscape of GM scholarship in Africa and propose actionable recommendations to improve research capacity and output.
Background about Worldwide Gut Microbiome Research
With thousands of manuscripts published yearly on the topic, investigations into the human gut microbiome (GM) (see Glossary) have reached an inflection point. Research is beginning to leverage an expanding foundational knowledge base to inform novel interventions for disease treatment and prevention [1]. As the field transitions to translational applications, it is important to evaluate the generalizability of these findings. To date, most microbiome and genomics research has focused on human populations in Europe and the USA (often termed ‘western’); more recently, studies are emerging from a few additional countries with similar profiles of lifestyle and industrialization, including Israel, Japan, and China [2–4]. In these studies, lifestyle, diet, and environmental exposures have been shown to strongly affect GM composition. Little is known about the GM in nonwestern and other groups historically understudied in institutional research. Populations in South America, Africa, and regions in Asia, such as India, have been scarcely studied despite genetic, ethnic, sociocultural, lifestyle, and dietary diversity. As a result, we are left with significant biases and an incomplete understanding of microbiome variability around the world.
Over the past century, Africa has undergone a rapid period of industrialization, globalization, and economic expansion. Concomitantly, disease epidemiology, urban settlement, and population demographics have changed profoundly [5–7]. The adoption of western lifestyle and dietary practices has precipitated a rising burden of noncommunicable diseases (NCDs), such as obesity, cancer, cardiovascular disease, and type 2 diabetes. That said, the pace of transition has been heterogeneous, and most populations lie on a continuum of westernization, varying by country and along the urban–rural divide [5,6]. Undernutrition, poor sanitation, food insecurity, and communicable diseases remain formidable causes of morbidity and mortality.
Several genomic and bioinformatic initiatives have formed to better understand Africa’s unique and multiform evolutionary and epidemiologic trajectories, beginning with the HapMap Project in 2002 [8]. Today, Human Heredity and Health and Africa (H3Africa) and other consortia are harnessing genomic technologies toward improving health outcomes [9]. Building on the success of these organizations, the GM is now emerging as a subject of substantial scientific inquiry in Africa. Here, we review the landscape of GM scholarship in Africa to date, emphasizing contributions to the field and outstanding knowledge gaps. Additionally, we propose actionable recommendations to overcome limitations that have slowed advances in GM research.
Insights from Agriculturalists and Hunter-Gatherers: The Effects of Western Lifestyles and Diet
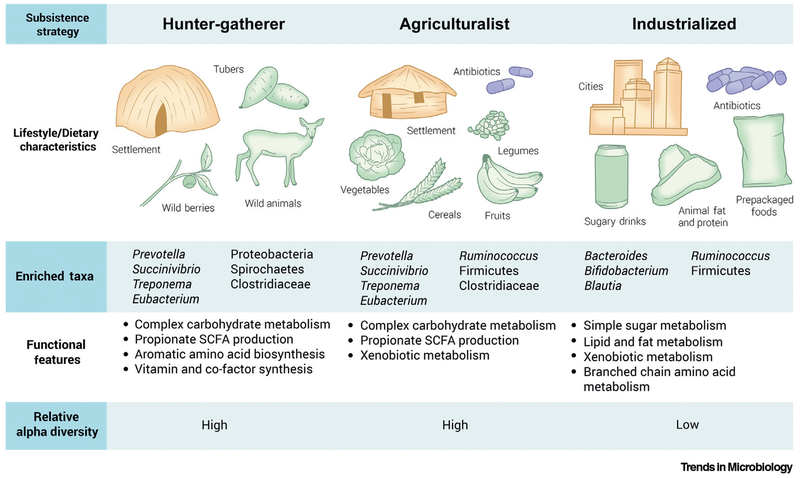
Today, several remote African agriculturalist and hunter-gatherer societies practice subsistence strategies that resemble those dating back to before the Neolithic era, more than 10 000 years ago. Juxtaposing these traditional African groups with western populations has informed how changes in dietary and environmental factors accompanying industrialization can profoundly impact the GM (Figure 1).
Figure 1. Gut Microbiome Features across Host Subsistence Strategies.
Lifestyle and dietary factors exert a profound effect on gut microbiome configuration. Hunter-gatherer populations, who rely on foraging and have little exposure to therapeutic agents, are highly enriched in Prevotella and other fiber-degrading taxa. Conversely, the dietary profile and sanitation infrastructure found in westernized societies is associated with Bacteroides dominance and a concomitant decrease in microbial diversity. Traditional agriculturalists occupy a putative intermediate state, sharing features of both western and hunter-gatherer communities. Abbreviation: SCFA, short-chain fatty acid.
It has been well established that long-term diet influences the taxonomic and functional signature of the microbiome. A consistent pattern is the inverse relationship between two members of the Bacteroidetes phylum, Prevotella and Bacteroides. Whereas Prevotella associates with a diet rich in plant-based foods found predominately in nonindustrialized populations, a higher intake of animal fats and proteins, characteristic of industrialized populations, correlates with increased relative abundance of Bacteroides [10–14]. Hunter-gatherer communities, such as the Hadza of Tanzania and the BaAka of the Central African Republic, consume diverse simple and complex carbohydrates in the form of wild tubers, roots, and berries. For the Hadzaand BaAka, a foraging lifestyle is associated with an enrichment in Prevotella, as well as Treponema, and Succinivibrio, all of which have fiber-degrading activity and may be involved in starch, hemicellulose, and xylan degradation [11,12,15–17]. Metagenomic analyses in the Hadza further reveal a high representation of genes related to broad-spectrum carbohydrate and amino acid utilization [15,18]. Notably, increased abundance of fibrinolytic lineages may be characteristic of an ancestral GM state, as observed by previous studies of coprolite samples from ancient civilizations [19]. Taxonomic and dietary parallels make it tempting to use extant hunter-gatherer populations as a direct proxy for the gut ecosystem of our ancestors. However, it should be noted that we are limited in our ability to confidently assess the appropriateness of this proxy without direct characterization of past GM composition. Furthermore, modern traditional societies have likely been exposed to classically western microbes and xenobiotics through contact with researchers.
Agriculturalist societies, marked by subsistence farming and animal domestication, share microbial features with hunter-gatherer communities. Populations from rural Burkina Faso and the Bantu community in the Central African Republic follow a vegetarian diet comprised of grains, vegetables, and legumes. Accordingly, taxa capable of metabolizing plant polysaccharides and fibers, including Prevotella and Treponema, dominate the gut, though at lower abundances than those in hunter-gatherers [10,12]. While the GM profiles of agriculturalist individuals more closely resemble the GM of hunter-gatherers, some features are shared with western populations – such as bacterial taxa and gene pathways associated with digestible sugars [12,16]. Robust evidence links the typical western diet – high in simple sugars and animal fats and proteins – to shifts in GM ecology. Bacteroides spp. emerge as the most prominent bacteria in western populations, with lower levels of fibrinolytic species, such as Prevotella and Treponema [10,12,13,17] (Box 1). This is consistent with increased access to refined sugars and animal fats and proteins. Additionally, an enrichment in genes involved in amino acid and bile salt metabolism has been described in individuals living in the USA who consume a typical western diet [20].
Box 1. The Influence of Short-chain Fatty Acids on Chronic Inflammatory Diseases.
The higher intake of plant-derived carbohydrates by traditional populations compared with industrialized populations correlates positively with the synthesis of short-chain fatty acids (SCFAs) [20]. SCFAs are metabolites derived from the fermentation of soluble fibers and serve as an energy source for gut epithelial cells [21]. Low SCFA levels from insufficient dietary fiber consumption may predispose individuals to chronic inflammatory diseases. In a diet-switch study, African-Americans who consumed a low-fat, high-fiber South African-style diet exhibited increased GM saccharolytic fermentation and SCFA production as well as reduced markers of inflammation [22]. SCFAs are thought to protect against colorectal cancer and may partially explain the lower incidence of the disease in South Africa compared with the USA, although it should be noted that colorectal cancer screening strategies differ substantially in these two countries and diagnosis bias may contribute to the observed increased incidence of colorectal cancer in the USA [23].
In addition to diet, changes in sanitation and hygiene practices have placed new selective pressures on the gut microbiota of industrialized populations. Microbiomes of western and even some agriculturalist communities are enriched in xenobiotic-processing genes, potentially due to therapeutic medication and food additive exposure [12,16]. The disruptive effects of antibiotics on the GM are well-established [24,25]. In studies of children in Burkina Faso and Nigeria, oral administration of azithromycin significantly decreased GM alpha-diversity and altered taxonomic composition [26,27]. Antibiotic exposure via food and medicine may increase the reservoir of antibiotic-resistance genes (ARGs) in the GM, collectively known as the resistome. However, ARGs exist not only in the metagenomes of industrialized populations, but also in hunter-gatherer and other communities with little exposure to pharmacological agents [15]. Their resistome contains genes resembling those that naturally occur in soil, suggesting an alternative, environmentally-based model of ARG acquisition. Microbial transmission secondary to interactions with individuals who have been exposed to antibiotics may also contribute to the resistome of hunter-gatherers.
At the community level, microbial diversity differs between populations with distinct lifestyle practices. Hunter-gatherer and agriculturalist cohorts have consistently higher GM alpha-diversity than western populations, contributing to the hypothesis that GM diversity declines with industrialization [10,12–14,16,20]. This trend is attributed to differences in dietary habits and sanitation standards. Colonization with parasites, which have been largely eradicated in western countries, may also promote GM diversity [28]. Interestingly, alpha-diversity tends to be inversely related to beta-diversity [10–13,16,20]. Traditional societies may have greater contact with their natural environment as well as with other village- and family-members and thus may have comparatively fewer barriers to microbial transmission and dispersal. Consequently, population-level GM composition tends to converge while local host diversity increases [14,29]. Collectively, descriptions of traditional African groups have revealed shifts in GM structure and function, driven primarily by diet and lifestyle, along the axis of westernization, (Box 2).
Box 2. Lifestyle and Dietary Factors Override Host Genetics in Determining GM Composition.
Mounting evidence indicates that lifestyle exerts a much stronger effect on microbiome composition than genetics and ethnicity [2]. This was initially reported by Yatsunenko et al. who analyzed hunter-gatherer, agriculturalist, and industrial populations from rural Venezuela, Malawi, and the USA [20]. Principal coordinate analysis showed a clear segregation of the populations based on lifestyle with little relationship to genetic identity. Reciprocally, individuals from a similar genetic background can diverge in GM composition if subjected to different environmental exposures. In a study of 2- to 8-year-old children in Burkina Faso of the same ethnicity, but living in rural, suburban, or urban settings, the microbial communities of urban subjects differed in accordance with the metabolic and dietary needs associated with industrialization [13].
Early Microbial Colonization and Maturation
Differences in GM composition can be observed well before adult life between traditional and industrialized populations. In the immediate postparturition time period, the microbial consortium undergoes a complex succession of growth and maturation [30,31]. It is influenced by exposures, including mode of delivery, the maternal gut and skin microbiome, and feeding practices. The process of GM maturation may carry important health implications. Aberrant or inadequate colonization has been associated with an increased susceptibility to immune- and inflammatory-related diseases as well as adverse metabolic outcomes that disproportionately burden industrialized countries [32]. Moreover, differences in GM composition between western and ‘non-western’ infants may be partially responsible for divergences in vaccine responsiveness (Box 3). Toward this end, investigations into the temporal and microbial features of the GM in African infants may help to elucidate risk factors related to these outcomes.
Box 3. Clinically-Relevant Phenotypes May Be Shaped by GM Ecology.
There is evidence that differences in infant GM composition have important clinical implications. For example, differences in the early microbiota can impact the efficacy of early and important clinical interventions, such as vaccines. Children in low-income countries in Africa generally have poor responsiveness to the oral rotavirus vaccines (RVV), with rates as low as 48–64% [33,34]. RVV response in Europe and other industrialized populations reaches as high as 98% [35,36]. Microbiome composition has been hypothesized to play a role in this discrepancy: in a study of 6-week old infants from Ghana and the Netherlands, overall taxonomic composition significantly differed between RVV responders and nonresponders [37]. Taken together, these data suggest that prospective clinical trials will be necessary to determine whether the GM can explain or drive variability in vaccine efficacy.
Delivery mode and setting are among the earliest external factors that shape the nascent GM [38–40]. Brazier et al. noted significant differences in taxonomic abundance in African neonates based on birth mode. Bacteroides and Collinsella were enriched with vaginal birth, while Sarcina and Klebsiella were associated with Cesarean section [41]. Sanitary conditions in the delivery environment can also influence the infant GM. A hospital-based study in Nigeria found that greater than 90% of newborns had GM colonization with principally facultative anaerobes on the first day of life [42]. European neonates appear to have a lower rate of first-day GM colonization , perhaps due to differences in hospital hygiene practices [43,44].
In the weeks and months after parturition, feeding practices largely mediate GM dynamics in developing infants [40,45]. The gut becomes increasingly diversified after birth and, after a period of dominance by facultative anaerobic or aerobic lineages, transitions to predominately strictly anaerobic bacteria in the first 6 to 9 months of life. Among the most prevalent genera is Bifidobacterium, which can be transferred in breast milk [46]. Bifidobacteria modulate education of the immune system and govern early gut community structure [47]. The relative proportions of high-abundance taxa differ between African and western guts. Malawian infants had higher levels of Bifidobacterium, Clostridium histolyticum, and the Bacteroides–Prevotella group than did Finnish infants at 6 months of age [48]. Additionally, Lactobacillus species, which are also transmitted from mother to infant, were more abundant and diverse in the Malawian cohort [49]. Other studies have identified functional signatures in Malawian and nonindustrialized neonates, namely an enrichment in host glycan metabolism and urease genes [20]. This is primarily explained by differences in breast milk oligosaccharide composition and the earlier introduction to complementary feeding (solid foods) in traditional groups [13]. Interestingly, extensive environmental contact nullifies the classical progression of GM maturation in hunter-gatherer infants [14]. In a study of Bassa infants, Bifidobacterium was largely absent, and GM stability and diversity profiles resembled those of adults. The authors speculated that GM assembly can be altered or shortened through dispersal of microbes amongst community members.
The weaning process marks the next major milestone in GM development. Around 1–2 years of age, the presence of Bacteroidetes and Firmicutes increases as the microbiome further diversifies [48,49]. The transition to a varied solid-food diet coincides with the divergence in microbial ecology based on subsistence strategy. De Filipo et al. described similar overall community structure and Bifidobacterium profiles in breast-fed children from Burkina Faso and Italy [10]. A distinct separation into geographical groupings occurred after weaning, underscoring the dominance of dietary specialization in shaping the young GM. Dietary influences persist through 18–36 months, when the GM stabilizes and approaches the adult state with respect to taxonomic composition and diversity [50,51]. Ultimately, patterns of GM maturation in African relative to non-African infants may be an important source of variation that drives differences in host phenotypes later in life.
The Gut Microbiome and Nutritional Status
Clearly, lifestyle and diet contribute substantially to GM development and composition. One of the major health issues faced in low- and middle-income settings is undernutrition. Chronic deficiencies in carbohydrate, fat, protein, and/or micronutrient intake are a leading cause of under-five mortality, primarily affecting low- and middle-income countries [52]. The World Health Organization (WHO) estimates that sub-Saharan Africa accounts for nearly 40% of cases globallyi.
Severe acute malnutrition (SAM) is the most extreme form of undernutrition. Just as SAM can drive significant alterations in GM, the GM can contribute to, or exacerbate, SAM by impeding the ability to extract dietary nutrients [53]. Furthermore, numerous reports link the GM as a causative agent of SAM. This was first revealed in a seminal transplantation study, wherein gnotobiotic mice receiving fecal samples from Malawian children with kwashiorkor, a SAM subtype, reproduced the weight-loss phenotype [54]. This supported the hypothesis that the GM is sufficient to induce substantial weight loss even when controlling for oral dietary intake. Microbial communities from malnourished hosts have lowered alpha-diversity levels and are taxonomically immature compared with healthy controls, reflecting a disruption in normal GM assembly. Malnutrition also impacts the viral component of the GM, otherwise known as the virome. Studying the same Malawian cohort, Reyes et al. showed that SAM significantly perturbs the normal program of virome development [55].
SAM is also associated with a dramatic loss of obligate anaerobes and oxygen-sensitive prokaryotes, such as Methanobacter, in conjunction with overgrowth of aerobic taxa and increased redox potential [56,57]. The enriched aerobic fraction includes numerous enteropathogenic organisms, most notably Enterobacteriaceae and Enterococcus. Overall, it is unclear whether this enrichment for Enterobacteriaceae and Enterococcus is sufficient to induce adverse clinical phenotypes. On the one hand, transplantation experiments implicate this pathogenic consortium in causing increased diarrhea, enteric inflammation, sepsis, and gut mucosal barrier permeability [58]. However, on the other hand, the presence of intestinal pathogens did not correlate with either enteropathy or death in a study of hospitalized malnourished Malawian children [59]. The leading hypothesis suggests that the loss of protective factors conferred by a predominantly anaerobic GM rather than pathogen overgrowth underlies SAM pathophysiology.
Stunting, the most common manifestation of undernutrition worldwide, also correlates with GM dysbiosis. In an investigation of stunted infants in the Central African Republic and Madagascar, the GM harbored an overrepresentation of taxa that normally reside in the oropharyngeal cavity [60]. The authors propose that bacterial translocation to lower GI regions – or ‘decompartmentalization’ – could contribute to linear growth deficits and enteropathy.
Given the complex interplay with the microbiome, effectively addressing malnutrition demands an intervention that resolves both nutritional deficiencies and alterations in GM composition. Ready-to-use therapeutic food (RUTF), a calorie-dense peanut mixture, is the current international standard for SAM therapyii. While short-term RUTF administration has dramatically reduced malnutrition-associated morality, high rates of relapse and comorbidities, from stunting to immune dysfunction, have limited its efficacy [61]. Disappointing findings have been partially attributed to interactions with the GM. In Malawian co-twins discordant for kwashiorkor, RUTF yielded temporary improvements in microbial composition, namely enrichments in Bifidobacteria and Lactobacillus, and amino acid and carbohydrate processing, but the effect rapidly diminishes after treatment cessation [54]. Additionally, it fails to repair aberrations in the proper assembly of the gut virome [55].
Other nutrient replacement strategies have caused similar concerns. Oral iron supplementation or fortification through micronutrient powders (MNPs) are effective for iron-deficiency anemia, but early high-dose formulations led to increased hospitalizations, deaths, and diarrhea [62,63]. A putative mechanism lies in the GM, given the increased colonic iron secondary to poor fractional absorption in the duodenum. Many gut bacteria rely on iron as a key, growth-limiting factor [64]. Thus, high iron levels in the colon favor colonization by, and virulence of, enteric Gram-negative pathogens while exerting little effect on Bifidobacteria, Lactobacillus, and other beneficial commensal microbes. Unsurprisingly, iron fortification and supplementation has been found to produce adverse shifts in GM ecology [65,66]. The guts of Kenyan infants receiving iron-fortified maize porridge exhibited acute gut inflammation, an increased and decreased prevalence of Enterobacteria and Bifidobacterium, respectively, as well as an enrichment in enteropathogens [65]. Results have been mixed, however, raising the possibility that environmental factors play a role in observed outcomes. In a trial of high-dose iron supplementation, South African children, who have comparatively better sanitation and hygiene standards, were spared dysbiosis and gut inflammation [39]. Another complicating factor is antibiotic coadministration. Compared with those without fortification, oral broad-spectrum antibiotic use in conjunction with iron-containing MNPs in Kenyan infants resulted in elevated pathogen carriage and decreased abundances of commensal Bifidobacterium, suggesting that iron modifies therapeutic efficacy [67]. As novel nutritional replacement strategies emerge (Box 4), potential solutions are compelled to navigate the complex interrelationship between malnutrition and the microbiome.
Box 4. ‘Rehabilitating’ Microbial Communities with GM-sensitive Nutritional Replacement Strategies.
The lack of efficacy of contemporary nutrient supplementation strategies has made way for efforts to explore alternative, GM-sensitive methods to treat malnutrition and associated pathologies [68–70]. For example, lipid-based nutrient supplementation comprises essential macro- and micronutrients in edible fats. Although their impact on undernourishment and linear growth are not clear, evidence indicates that lipid-based supplementation does not negatively affect GM composition or development [68,71]. There is also emerging interest in microbiome-based therapies. A metagenomic and culturomic study of malnourished and healthy children from Senegal and Niger identified a distinct microbial signature of 12 bacterial species in healthy samples that included Alistipes indistinctus, Anaerostipes caccae, and Bacillus licheniformis [72]. Prebiotics or probiotics designed with selected taxa could restore normal GM configuration and prevent long-term complications related to enteropathogen colonization [73,74]. That said, early findings from a controlled interventional trial in Malawi have been inconclusive [75]. While further work is needed to validate their efficacy and safety, targeted microbiome interventions may help to overcome the shortcomings of current approaches.
Microbial Dynamics in Infectious Disease
Despite enormous strides in public health infrastructure, communicable diseases – mainly malaria, tuberculosis, HIV/AIDS, and respiratory and diarrheal pathologies – remain the foremost cause of mortality in Africaiii. Disease risk is, in part, governed by the GM, which modulates the mucosal immune system and is thought to confer resistance to pathogen colonization [76]. The high infectious disease burden makes Africa an appropriate setting to better understand these dynamics.
HIV/AIDS, in particular, is a major public health concern, with Africa accounting for more than 70% of the global burden of infection [77]. The relationship of HIV infection to the GM, namely dysregulation of the host immune–epithelial interface, is a subject of active inquiry. The virus leads to disruption of key mucosal lymphoid compartments, including T helper (Th)17 and dendritic cells [78]. Disrupting the intestinal barrier can permit microbial translocation into the bloodstream and lead to chronic systemic inflammation and GM dysbiosis. Low CD4 counts in HIV+ patients in Uganda correlated with an expansion in adenovirus populations in the GM [79]. Furthermore, bacterial communities in HIV+ compared with HIV− individuals exhibited reduced alpha-diversity and increases in Enterobacteraceiae, which may contribute to the enteropathy, diarrhea, and related inflammatory comorbidities associated with disease progression. Many millions of HIV+ patients receive long-term antiretroviral therapy (ART). Although effective, ART carries significant side-effects, and is known to have a potent in vitro antibacterial effect [80]. How these lifesaving medications interact with the GM is poorly understood and is in need of urgent research [81].
Moderate-to-severe diarrhea (MSD) is another major public health problem, particularly amongst children under the age of five [82]. This enteritis has been linked to large-scale GM alterations, from loss of alpha-diversity to reduced microbial community stability [83,84]. Additionally, there is an inverse relationship in the proportions of obligate anaerobes and facultative anaerobes, many of which cause diarrhea. In a report on children from countries in West Africa, East Africa, and South Asia, Pop et al. identified an enrichment of not only known enteropathogens in MSD relative to controls, such as Shigella and Campylobacter jejuni, but also several novel pathogenic lineages, including Streptococcus and Granulicatella species [84].
Much like bacterial and viral components, parasites – including protozoa and helminths – have complex transkingdom interactions within the GM. On one hand, they exert pathogenic effects that are partially mediated through the gut [85,86]. Colonization with Giardia duodenalis, for example, significantly alters microbial community structure and supports the expansion of facultative anaerobic and potentially pathogenic taxa [85]. Meanwhile, GM composition may influence malaria disease progression and outcome, such as the predisposition to infection with Plasmodium falciparum [87]. Specifically, enrichments in Lactobacillus, Bifidobacterium, and Streptococcus were associated with reduced P. falciparum infection in a Malian cohort. Only recently have resident eukaryotic microbes been appreciated for their potentially beneficial role in GM ecology. The presence of Entamoeba is associated with increased alpha-diversity and favorable shifts in taxonomic composition in studies of various rural African populations [28]. Nonwestern groups also exhibit a high prevalence of Blastocystis, but its clinical significance and relationship to the gut ecosystem are unresolved [88]. According to the ‘old friends’ hypothesis, parasites belonged to an ancient GM consortium, exerting important immunoregulatory functions [89]. The loss of persistent colonization connected with modern lifestyle and sanitation practices may have precipitated decreased bacterial diversity and the rising prevalence of allergic and other inflammatory disorders in industrialized populations.
Concluding Remarks
Research in African populations has deepened our understanding of GM ecology by leveraging the unique make-up of sociocultural, demographic, and epidemiologic features that cannot be captured in other geographies. Rural African hunter-gatherers and agriculturalists have provided a window into the relative influence of subsistence strategy and industrialization. These factors begin to shape GM composition as early as infancy, leading to differences in microbial development and maturation. Moreover, we have come to appreciate the complex interplay between the GM and infectious disease and malnutrition, both of which are pervasive public health issues on the continent.
Despite major progress, there are still gaps in our knowledge of GM characteristics in African populations (see Outstanding Questions). The current research landscape has not kept pace with the contemporary scientific and public health needs on the continent. Comparative studies have focused on the ‘extremes’ of lifestyle characteristics, most commonly rural traditional and industrialized subsistence modes in African and western countries, respectively. The relative dearth of studies invites the tendency to over-apply the findings from one subgroup, such as hunter-gatherers and agriculturalists, which actually constitute a small proportion of the population, to the entire African continent. This also comes at the expense of understanding populations that have transitioned or are transitioning to western lifestyles. Relatedly, little attention has been given to the epidemiological shift from communicable, maternal, neonatal, and nutritional pathologies to NCDs [5,6]. The WHO estimates that NCDs will become the leading causes of morbidity and mortality in Africa by 2050. Yet, we found little scholarship – and no large-scale studies – that examine the relationship between the GM and most prevalent NCDs, namely cardiovascular and respiratory disease, cancer, and type II diabetes [90].
Outstanding Questions.
How does the GM vary with and influence noncommunicable disease in African populations?
Do HIV medications influence the GM and infectious risk? And if so, how?
How does the GM change in populations in the midst of the nutritional and epidemiological shifts associated with westernization?
To what extent can the GM explain differential vaccine responsiveness between ‘western’ and African populations?
What are the most effective strategies for managing malnutrition-associated pathologies while minimizing harm to and/or restoring healthy microbial ecology?
What is the mechanistic relationship between the GM, host immunity, and parasites and other ‘old friends’ that may have been depleted by ‘westernization?’
How can metagenomic approaches be harnessed to capture yet unexplored GM variability?
How can the international scientific community best support African institutions and researchers to perform large-scale microbiome studies?
Limitations in the current microbiome literature on Africans are compounded by constraints in sequencing methodology. To date, most investigations have used 16S ribosomal RNAgene sequencing. While cost-effective and valuable for quantitative descriptions of microbial composition, this approach is constrained to bacterial and archaeal classification and is fraught with issues of amplification bias [91]. Shotgun metagenomic sequencing affords more comprehensive taxonomic characterization as well as enhanced prediction of gene and pathway content. However, the associated technical and computational costs have prevented its widespread adoption on a global scale. Consequently, our present understanding likely dramatically underestimates GM diversity in African populations, restricting our ability to understand and act on key scientific and medical issues.
We urgently need greater investment in GM research in Africa, but proposed solutions must be prepared to reckon with systemic and multifaceted barriers, not the least of which include: resource and financial constraints; challenges related to funding and government regulation; and limited participation of local scientists. These and other obstacles are the targets of several international organizations, most notably H3Africa, that seek to improve the ecosystem for modern health-related research [9]. Funded by the US National Institutes of Health, the Africa Academy of Sciences, and the Wellcome Trust, H3Africa supports large-scale genomic and epidemiological studies of specific relevance to Africa. Additionally, it advances capacity-building measures focused on developing local scientific infrastructure and educational programming. The strategies espoused by H3Africa and related efforts are being directly and indirectly harnessed to augment GM scholarship. In this setting, we offer the following recommendations on areas of high priority.
Build Diverse Reference Databases
Accurate taxonomic and functional characterization of GM samples relies on comprehensive reference databases. Compared with traditional culture-based methods, culture-independent metagenomic approaches – such as the assembly of genomes from metagenomes – have dramatically expanded the catalog of identifiable organisms. Reference databases derived from these methods are biased toward well-profiled western populations. As a result, a large fraction of the microbial composition in understudied groups has been unexplored. Recently, Pasolli et al. reconstructed 154 723 microbial genomes and 4930 species-level genome bins from metagenomes of various ages, lifestyles, and geographies, including Madagascar and Ethiopia [92]. Acomparable meta-analysis was performed by Nayfach et al. who recovered more than 60 000 genomes corresponding to 2058 novel species [93]. These newly described genomes particularly avail the taxonomic classification of non-western populations, dramatically improving the mapability of metagenomic reads. Continuing to incorporate diverse cohorts in large-scale metagenomic assembly efforts will enable greater insights into microbial variability around the world.
Invest in Bioinformatics and Biotechnology Capacity
Authors of GM studies from Europe and the USA vastly outnumber those from African institutions. While the cause of this disparity is unknown, we suspect that the underrepresentation of local scientists reflects the limited opportunities for specialized training [94]. GM researchers can tremendously benefit from H3Africa’s commitment to education, which includes fostering competencies in bioinformatics, genomics, statistical analysis, and high-throughput technologies. Furthermore, organizations like H3ABioNet, a subsidiary of H3Africa, are poised to further research capacity by formalizing a pan-African bioinformatics network comprising conferences, workshops, and forums [95].
Forge International Partnerships
Given limitations in obtaining strong, ongoing domestic funding and/or technical resources, longitudinal collaborations with international research institutions can fill resource gaps in African countries. Indeed, H3Africa identifies links between African and non-African organizations as sustainable channels for financial support, equipment, and training. Importantly, relationships must be premised on the meaningful participation and contribution of both parties so as to minimize ‘parachute research’, that is, the practice of western institutions mining scientific data from low- and middle-income countries without consulting or involving academics from those institutions. A precedent set by H3Africa, this dynamic can be combated with agreed upon ethical guidelines for empowering local researchers, data and sample management, and community engagement.
Leverage Advances in Sequencing Technologies
Developments in next-generation sequencing approaches have been accompanied by innovative products that circumvent the requirement for extensive computational or laboratory infrastructure. For example, the MinlON, engineered by Oxford Nanopore Technologies, affords lower-cost, portable, real-time generation of high-quality GM sequencing data [96]. While such technologies have only recently been commercialized, they have and will continue to expand the penetration of modern genomics tools in low-resource settings.
Beyond Africa, we believe that these recommendations can be generalized to analogous microbiome efforts in other geographic contexts. It will be crucial to integrate ethical research principles and multilateral partnerships in the Global Microbiome Conservancy, for example, which aims to comprehensively catalog global gut microbial diversity with an emphasis on traditional and indigenous communities. [97] Additionally, our proposals can potentially inform emerging national-level associations, such as the Indian Human Microbiome Initiative, in providing a framework for large-scale microbiome studies in underrepresented settingsiv.
As our knowledge of GM ecology rapidly evolves, so too must the inclusion of diverse populations be prioritized. Africa contains some of the richest genetic diversity with unique sociocultural, epidemiologic, and biological characteristics. Future studies promise to elucidate novel host–microbiome interactions that influence human health and disease. Scientific imperatives are coupled with critical matters of justice. For as long as GM scholarship stays concentrated in predominantly western, industrialized countries, disparities in characterizing and serving historically understudied populations will persist. Still in its relative infancy, the global GM community has the opportunity to codify practices that value the full ensemble of microbial variability around the world. We envision a future where the GM in Africa not only receives its due attention, but also where such knowledge is produced by, and for, those it affects most.
Highlights.
Most human GM research has concentrated on westernized societies. Whether these findings can be generalized to African populations is poorly understood.
Comparative studies between western individuals and hunter-gatherer and agriculturalist communities in Africa have revealed distinct taxonomic signatures based on lifestyle and dietary factors.
Microbiome alterations are associated with and may underlie malnutrition-associated pathologies. Contemporary nutrient replacement strategies have failed to durably correct these changes, limiting their therapeutic efficacy.
Infectious diseases have been linked to disruptions in microbial ecology. Colonization with parasites may be an important determinant of microbiome structure.
Representing diverse populations must be prioritized in future microbiome studies to capture the full ensemble of microbial diversity worldwide.
Acknowledgments
We thank the Dr Henry Lee and the Lane Medical Library and Knowledge Management Center at the Stanford University School of Medicine for helping develop our literature search strategy. Additionally, we are grateful to Tessa Andermann, Pagé Goddard, Amee Azad, and Yiran Liu for critical feedback on the manuscript. This work was supported by the South Africa National Research Foundation (Grant CPRR160421162721; to O.O. and S.H.), the Rosenkranz Prize (to A.S.B.), a Stanford Center for Innovation in Global Health seed award (to A.S.B.), a Fogarty Global Health Equity Scholar award (TW009338; to O.O.), the Center for Computational, Evolutionary and Human Genomics (to F.T.), and Stanford MedScholars program funding (to E.A. and R.B.).
Glossary
- Agriculturalist:
the primary subsistence strategy of the Neolithic age (~10 000 years ago). In contrast to hunter-gatherers, agriculturalists have stable settlements, and practice food-crop and animal domestication
- Alpha-diversity:
a measure of the number of different species, or richness, within a microbial community
- Beta-diversity:
a measure of the difference in microbial composition between samples
- Dysbiosis:
a breakdown in the balance between putative species of ‘protective’ vs ‘harmful’ intestinal microorganisms
- Gut microbiome (GM):
the genomes (genetic material [DNA or RNA]) of the ensemble of microorganisms – including eukaryotes, archaea, bacteria, and viruses, residing in the intestinal tract
- Hunter-gatherer:
a person living in a society whose primary subsistence strategy involves. Hunting and gathering was dominant amongst early modern humans prior to the advent of agriculture (>10 000 years ago)
- Metagenomics:
the study of a collection of genetic material (genomes) from a mixed ecological community of organisms
- Ready-to-use therapeutic food (RUTF):
a nutrient- and energy-dense peanut-based food indicated as a therapeutic food for severe malnutrition
- Severe acute malnutrition (SAM):
is defined by a weight-for-height below three standard deviations from the World Health Organization (WHO) Standards median, visible severe wasting, or the presence of bilateral pitting edema
- Stunting:
refers to a child with a length-for-age below two standard deviations from the WHO Standards median
- Westernization:
the process by which a society adopts the customs and practices typical of Western European and North American culture. ‘Western’ populations are generally characterized by a more sedentary lifestyle, decreased exposure to microbes and pathogens, and a diet rich in energy-dense foods and animal fat and protein
- 16S rRNA sequencing:
16S ribosomal RNA/DNA of prokaryotes is amplified via PCR and sequenced to identify the taxonomic classification of bacteria and archaea
Footnotes
References
- 1.Gilbert JA et al. (2018) Current understanding of the human microbiome. Nat. Med. 24, 392–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothschild D et al. (2018) Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 [DOI] [PubMed] [Google Scholar]
- 3.Qin J et al. (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 [DOI] [PubMed] [Google Scholar]
- 4.Hattori M and Taylor TD (2009) The human intestinal microbiome: a new frontier of human biology. DNA Res 16, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahams Z et al. (2011) Diet and mortality rates in sub-Saharan Africa: stages in the nutrition transition. BMC Public Health 11,801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forouzanfar MH et al. (2016) Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1659–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peer N (2015) The converging burdens of infectious and non-communicable diseases in rural-to-urban migrant sub-Saharan African populations: a focus on HIV/AIDS, tuberculosis and cardio-metabolic diseases. Trop. Dis. Travel Med. Vaccines 1, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The International HapMap Consortium et al. (2003) The International HapMap Project. Nature 426, 789–796 [DOI] [PubMed] [Google Scholar]
- 9.Mulder N et al. (2018) H3Africa: current perspectives. Pharmacogenom. Personal. Med 11, 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Filippo C et al. (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A 107, 14691–14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnorr SL et al. (2014) Gut microbiome of the Hadza hunter-gatherers. Nat. Commun 5, 3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez A et al. (2016) Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep 14, 2142–2153 [DOI] [PubMed] [Google Scholar]
- 13.De Filippo C et al. (2017) Diet, environments, and gut microbiota. a preliminary investigation in children living in rural and urban Burkina Faso and Italy. Front. Microbiol 8, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ayeni FA et al. (2018) Infant and adult gut microbiome and metabolome in rural Bassa and urban settlers from Nigeria. Cell Rep 23, 3056–3067 [DOI] [PubMed] [Google Scholar]
- 15.Rampelli S et al. (2015) Metagenome sequencing of the Hadza hunter-gatherer gut microbiota. Curr. Biol 25, 1682–1693 [DOI] [PubMed] [Google Scholar]
- 16.Hansen MEB et al. (2019) Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana. Genome Biol 20, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angelakis E et al. (2019) Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals. New Microbes New Infect 27, 14–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smits SA et al. (2017) Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tito RY et al. (2012) Insights from characterizing extinct human gut microbiomes. PLoS One 7, e51146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yatsunenko T et al. (2012) Human gut microbiome viewed across age and geography. Nature 486, 222–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh A et al. (2016) From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 [DOI] [PubMed] [Google Scholar]
- 22.O’Keefe SJD et al. (2015) Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun 6, 6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bénard F et al. (2018) Systematic review of colorectal cancer screening guidelines for average-risk adults: Summarizing the current global recommendations. World J. Gastroenterol 24, 124–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dethlefsen L and Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A 108, 4554–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dethlefsen L et al. (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6, e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doan T et al. (2017) Gut microbial diversity in antibiotic-naive children after systemic antibiotic exposure: a randomized controlled trial. Clin. Infect. Dis 64, 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldenburg CE et al. (2018) Effect of commonly used pediatric antibiotics on gut microbial diversity in preschool children in Burkina Faso: a randomized clinical trial. Open Forum Infect. Dis 5, ofy289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morton ER et al. (2015) Variation in rural African gut microbiota is strongly correlated with colonization by entamoeba and subsistence. PLoS Genet 11, e1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez I et al. (2015) The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Rep 11, 527–538 [DOI] [PubMed] [Google Scholar]
- 30.Robertson RC et al. (2019) The human microbiome and child growth – first 1000 days and beyond. Trends Microbiol 27, 131–147 [DOI] [PubMed] [Google Scholar]
- 31.Ojo-Okunola A et al. (2018) Human breast milk bacteriome in health and disease. Nutrients 10, 1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson CC and Ownby DR (2017) The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Translat Res 179, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armah GE et al. (2010) Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 376, 606–614 [DOI] [PubMed] [Google Scholar]
- 34.Madhi SA et al. (2010) Effect of human rotavirus vaccine on severe diarrhea in African infants. N. Engl. J. Med 362, 289–298 [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Palacios GM et al. (2006) Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Eng . J. Med 354, 11–22 [DOI] [PubMed] [Google Scholar]
- 36.Vesikari T et al. (2007) Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. Lancet 370, 1757–1763 [DOI] [PubMed] [Google Scholar]
- 37.Harris VC et al. (2017) Significant correlation between the infant gut microbiome and rotavirus vaccine response in rural Ghana. J. Infect. Dis 215, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferretti P et al. (2018) Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Celt Host Microbe 24,133–145.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill CJ et al. (2017) Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claassen-Weitz S et al. (2018) HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a South African birth cohort. Sci. Rep 8, 5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brazier L et al. (2017) Evolution in fecal bacterial/viral composition in infants of two central African countries (Gabon and Republic of the Congo) during their first month of life. PLoS One 12, e0185569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kigbu A et al. (2016) Intestinal bacterial colonization in the first 2 weeks of life of Nigerian neonates using standard culture methods. Front. Pediatr 4, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adlerberth I et al. (1991) Intestinal colonization with Enterobacteriaceae in Pakistani and Swedish hospital-delivered infants. Acta Paediatr. Scand 80, 602–610 [DOI] [PubMed] [Google Scholar]
- 44.Adlerberth I et al. (2006) Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr. Res 59, 96–101 [DOI] [PubMed] [Google Scholar]
- 45.O’Sullivan A et al. (2015) The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutrit. Metab. Insights 8s1 NMI.S29530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Underwood MA et al. (2015) Bifidobacterium longum subspecies infants: champion colonizer of the infant gut. Pediatr. Res 77, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz L et al. (2017) Bifidobacteria and their molecular communication with the immune system. Front. Microbiol 8, 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grześkowiak Ł. et al. (2012) Distinct gut microbiota in Southeastern African and Northern European infants. J. Pediatr. Gastroenterol. Nutrit 54, 812–816 [DOI] [PubMed] [Google Scholar]
- 49.Aakko J et al. (2015) Distinctive intestinal Lactobacillus communities in 6-month-old infants from rural Malawi and Southwestern Finland. J. Pediatr. Gastroenterol. Nutrt 61, 641–648 [DOI] [PubMed] [Google Scholar]
- 50.Bergstrom A et al. (2014) Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol 80, 2889–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koenig JE et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A 108, 4578–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Black RE et al. (2013) Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 382, 427–451 [DOI] [PubMed] [Google Scholar]
- 53.Kane AV et al. (2015) Childhood malnutrition and the intestinal microbiome. Pediatr. Res 77, 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith MI et al. (2013) Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339, 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reyes A et al. (2015) Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc. Natl. Acad. Sci. U. S. A 112,11941–11946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Million M et al. (2016) Increased gut redox and depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci. Rep 6, 26051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Ordiz MI et al. (2017) Environmental enteric dysfunction and the fecal microbiota in Malawian children. Am. J. Trop. Med. Hyg 96, 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kau AL et al. (2015) Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci. Trans . Med 7 276ra24–276ra24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Attia S et al. (2016) Mortality in children with complicated severe acute malnutrition is related to intestinal and systemic inflammation: an observational cohort study. Am. J. Clin. Nutr 104, 1441–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vonaesch P et al. (2018) Stunted childhood growth is associated with decompartmentalization of the gastrointestinal tract and overgrowth of oropharyngeal taxa. Proc. Natl. Acad. Sci. U. S. A 115, E8489–E8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Sullivan NP et al. (2018) Follow-up between 6 and 24 months after discharge from treatment for severe acute malnutrition in children aged 6-59 months: A systematic review. PLoS One 13, e0202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasricha S-R et al. (2013) Effect of daily iron supplementation on health in children aged 4–23 months: a systematic review and meta-analysis of randomised controlled trials. Lancet Global Health 1, e77–e86 [DOI] [PubMed] [Google Scholar]
- 63.Zlotkin S et al. (2013) Effect of iron fortification on malaria incidence in infants and young children in Ghana: a randomized trial. JAMA 310, 938. [DOI] [PubMed] [Google Scholar]
- 64.Andrews SC et al. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 65.Jaeggi T et al. (2015) Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 64, 731–742 [DOI] [PubMed] [Google Scholar]
- 66.Zimmermann MB et al. (2010) The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Côte d’Ivoire. Am. J. Clin. Nutr 92, 1406–1415 [DOI] [PubMed] [Google Scholar]
- 67.Paganini D et al. (2019) Iron-containing micronutrient powders modify the effect of oral antibiotics on the infant gut microbiome and increase post-antibiotic diarrhoea risk: a controlled study in Kenya. Gut 68, 645–653 [DOI] [PubMed] [Google Scholar]
- 68.Aakko J et al. (2017) Lipid-based nutrient supplements do not affect gut Bifidobacterium microbiota in Malawian infants: a randomized trial. J. Pediatr. Gastroenterol. Nutrit 64, 610–615 [DOI] [PubMed] [Google Scholar]
- 69.Ordiz MI et al. (2015) The effect of dietary resistant starch type 2 on the microbiota and markers of gut inflammation in rural Malawi children. Microbiome 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bisanz JE et al. (2015) Microbiota at multiple body sites during pregnancy in a rural Tanzanian population and effects of Moringa-supplemented probiotic yogurt. Appl. Environ. Microbiol 81, 4965–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheung YB et al. (2016) Gut microbiota in Malawian infants in a nutritional supplementation trial. Trop. Med. Int Health 21, 283–290 [DOI] [PubMed] [Google Scholar]
- 72.Tidjani Alou M et al. (2017) Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front. Microbiol 8, 899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allen SJ et al. (2010) Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst. Rev CD003048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Westerik N et al. (2018) Lactobacillus rhamnosus probiotic food as a tool for empowerment across the value chain in Africa. Front. Microbiol 9, 1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kerac M et al. (2009) Probiotics and prebiotics for severe acute malnutrition (PRONUT study): a double-blind efficacy randomised controlled trial in Malawi. Lancet 374, 136–144 [DOI] [PubMed] [Google Scholar]
- 76.Kim S et al. (2017) The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev 79,90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kharsany ABM and Karim QA (2016) HIV infection and AIDS in sub-Saharan Africa: current status, challenges and opportunities. Open AIDS J. 10, 34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zevin AS et al. (2016) Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr. Opin. HIV AIDS 11, 182–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monaco CL et al. (2016) Altered virome and bacterial quired immunodeficiency syndrome. Cell Host Microbe 19, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shilaih M et al. (2017) Antibacterial effects of antiretrovirals, potential implications for microbiome studies in HIV. Antiviral Ther 23, 91–94 [DOI] [PubMed] [Google Scholar]
- 81.Pinto-Cardoso S et al. (2018) Impact of antiretroviral drugs on the microbiome: unknown answers to important questions. Curr. Opin. HIV AIDS 13, 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Troeger C et al. (2018) Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis 18,1211–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schneider D et al. (2017) Gut bacterial communities of diarrheic patients with indications of Clostridioides difficile infection. Sci. Data 4, 170152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pop M et al. (2014) Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol 15, R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lebba V et al. (2016) Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte d’Ivoire. J. Infect. Develop. Countries 10, 1035. [DOI] [PubMed] [Google Scholar]
- 86.Ajibola O et al. (2019) Urogenital schistosomiasis is associated with signatures of microbiome dysbiosis in Nigerian adolescents. Sci. Rep 9, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yooseph S et al. (2015) Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genom 16, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beghini F et al. (2017) Large-scale comparative metagenomics of Blastocystis, a common member of the human gut microbiome. ISME J. 11,2848–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rook GAW (2005) Microbes, immunoregulation, and the gut. Gut 54, 317–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Allali I et al. (2018) Gut microbiome of Moroccan colorectal cancer patients. Med. Microbiol. Immunol 207, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kennedy K et al. (2014) Evaluating bias of Illumina-based bacterial 16S rRNA gene profiles. Appl. Environ. Microbiol 80, 5717–5722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pasolli E et al. (2019) Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nayfach S et al. (2019) New insights from uncultivated genomes of the global human gut microbiome. Nature 568, 505–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Karikari TK et al. (2015) Developing expertise in bioinformatics for biomedical research in Africa. Appl. Translat. Genom 6, 31–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulder NJ et al. (2016) H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa. Genome Res 26, 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tyler AD et al. (2018) Evaluation of Oxford Nanopore’s MinION sequencing device for microbial whole genome sequencing applications. Sci. Rep 8, 10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rabesandratana T (2018) Microbiome conservancy stores global fecal samples. Science 362, 510–511 [DOI] [PubMed] [Google Scholar]