Abstract
Clostridium difficile spores are considered the morphotype of infection, transmission and persistence of C. difficile infections. There is a lack of information on the composition of the outermost exosporium layer of C. difficile spores. Using recently developed exosporium removal methods combined with MS/MS, we have established a gel-free approach to analyze the proteome of the exosporium of C. difficile spores of strain 630. A total of 184 proteins were found in the exosporium layer of C. difficile spores. We identified 7 characterized spore coat and/or exosporium proteins; 6 proteins likely to be involved in spore resistance; 6 proteins possibly involved in pathogenicity; 13 uncharacterized proteins; and 146 cytosolic proteins that might have been encased into the exosporium during assembly, similarly as reported for Bacillus anthracis and Bacillus cereus spores. We demonstrate through Flag-fusions that CotA and CotB are mainly located in the spore coat, while the exosporium collagen-like glycoproteins (i.e. BclA1, BclA2 and BclA3), the exosporium morphogenetic proteins CdeC and CdeM, and the uncharacterized exosporium proteins CdeA and CdeB are mainly located in the exosporium layer of C. difficile 630 spores. This study offers novel candidates of C. difficile exosporium proteins as suitable targets for detection, removal and spore-based therapies.
Keywords: Clostridium difficile, Spores, Exosporium, spore surface proteins
1. Introduction
Clostridium difficile is a Gram positive, anaerobic, spore-forming nosocomial pathogen that has established itself as a major nosocomial pathogen1. The severity of Clostridium difficile infections (CDI) can vary from mild diarrhea to life threatening toxic megacolon, with a mortality rate of ~ 5%2. Clinical symptoms are caused by main virulence factors, the enterotoxin TcdA and the cytotoxin TcdB3. A striking feature of CDI is the high rates with which those patients that have cleared from the first CDI episode suffer recurrence of CDI and can reach up to 20 to 30% of CDI patients4, which is attributed primarily to the persistence of the dormant spore morphotype5. Indeed, C. difficile strains that are unable to form spores, are unable to persist and disseminate in a mouse model, indicating that spores are essential for persistence and dissemination of C. difficile5.
Bacterial spores have unique structural features that enable them to survive for extended periods of time in adverse environments6. From a spore-host interaction perspective, the outermost layer gains particular importance as it has the sites that will interact with the host7, 8. Identification of the protein composition of this outermost layer will uncover novel targets for the development of detection and/or spore removal strategies. The outermost layer of several bacterial species (i.e. members of the Bacillus cereus group and C. difficile) is the exosporium layer7, 9, which in B. anthracis and B. cereus is a loosely-fitted layer composed by ~ 15 proteins and/or enzymes7, and plays a relevant role during infection8, 10. In contrast, several C. difficile strains produce spores with an exosporium layer that has organized hair-like extensions and is tightly attached to the spore coat, while other strains such as 630 have a hairy-less electron-dense exosporium-like layer that is also attached to the spore coat9, 11, supporting the notion that significant ultrastructural differences exist with the Bacillus exosporium layer11. Regardless of the differences in exosporium ultrastructure, both types of layers (i.e. C. difficile and Bacillus exosporium layers) share common properties: i) both layers contribute to the spore surface hydrophobicity; and ii) both have conserved markers (i.e. collagen-like exosporium glycoproteins) responsible for the hair-like nap of the exosporium layer of members of the B. cereus group, which in C. difficile, are encoded by bclA1, bclA2 and bclA3 genes that become expressed in the mother cell compartment uniquely during sporulation11–13. The correct assembly of the C. difficile exosporium layer depends on the exosporium morphogenetic protein, CdeC9 indicating that the C. difficile exosporium layer has a unique assembly mechanism. Recent progress has been made in the identification of putative phenotypes for the exosporium layer of C. difficile spores, such as for adherence of C. difficile spores to epithelium cells in vitro14 and colonization in a mouse model15, possible disruption of the phagosome’s membrane during phagocytosis16, and for adherence to stainless steel17. The exosporium layer of C. difficile spores also seems to have some role in spore dormancy18. However, the proteins that are involved in these phenotypes remain elusive.
To date, only two studies have addressed the composition of C. difficile 630 spores19, 20; Lawley et al, (2009) used nano-liquid chromatography-tandem mass spectrometry (nano-LC-MS/-MS) to determine spore proteins previously fractionated on a NuPAGE gel, identifying ~ 336 proteins associated to C. difficile 630 spores19. However, their precise location in the different spore layers was unclear19. A recent study by Abhyankar et al. (2013) also used LC-MS-MS to identify proteins present in the spore coat fraction of C. difficile 630 spores. Among the 54 protein species identified in C. difficile spore coat fractions, none of the three C. difficile exosporium collagen-like glycoproteins (i.e. BclA1, BclA2 and BclA3), and the exosporium morphogenetic protein, CdeC, were found20. Both of the aforementioned studies used proteinase K during the purification of C. difficile spores19, 20, which we recently showed to efficiently remove the exosporium layer of C. difficile spores, as well as the exosporium BclA1 and CdeC proteins9, 12, 18; explaining, at least in part, the absence of these and other known exosporium proteins in the proteomic analysis of proteinase K-treated C. difficile 630 spore coat20. Therefore, in an effort to contribute with tools to study the exosporium layer of C. difficile spores, we recently published18 two different strategies based on sonication treatment and trypsin digestion. As demonstrated by transmission electron microscopy and spore surface hydrophobicity assay, both strategies remove most of the exosporium-like layer of C. difficile 630 spores, and apparently very little, if any, external spore coat material18. In the present work, in an effort to contribute towards the identification of proteins of the exosporium-like layer of spores of C. difficile strain 630, we extracted the exosporium layer by sonication and trypsin digestion and analyzed them by a highly sensitive “gel-free” ultra-high performance liquid chromatography (UPLC)-tandem MS/MS approach21. We have identified two exosporium proteins (i.e. BclA1 and CdeC), four previously described spore coat proteins (i.e. CotA, CotB, CotD, CotE) as well as several uncharacterized exosporium proteins including some with putative protective roles. Importantly, by creating translational fusions of a subset of these proteins with the FLAG epitope, we have demonstrated by western blot analysis of untreated and proteinase K-treated spores expressing Flag-fusions that, they localize to the exosporium-like layer of C. difficile 630 spores. These results provide the first proteomic study that focuses on the outermost exosporium-like layer of C. difficile 630 spores and when coupled with previous proteomic work on spores of strain 630, it provides comprehensive information of the proteins that are present in the outer layers of C. difficile 630 spores.
2. Methods
Spore preparation and purification.
For C. difficile strain 630 (tcdA+, tcdB+, tcdC+, ctdA−, and ctdB−)22 two independent biological spore crops were prepared as previously described on BHI agar plates14. Sporulating cultures were routinely washed with ice cold sterile distilled water, and free spores were separated using 50% HistoDenz and washed five times to eliminate traces of HistoDenz (Sigma). Spore suspensions were further washed 10 times with 150 mM NaCl in milliQ-grade water to remove cytosolic proteins from the exosporium layer. Suspensions were > 99% free of vegetative cells, sporulating cells and cell debris. To remove loosely bound proteins and intracellular contaminants, spores were washed 5 times with 1 M NaCl. Purified spores were quantified with a Neubauer chamber prior to storage at −80°C until use. This storage condition does not affect spore viability and ultrastructural properties (unpublished data).
Trypsin digestion of the exosporium-like layer of C. difficile 630 spores.
To uniquely remove exosporium proteins, two biological replicates of C. difficile 630 spores were treated with trypsin that has been previously demonstrated to remove exosporium material18. Briefly, C. difficile 630 spores (2 × 109 spores) were resuspended in 50 mM NH4HCO3 (pH 7.8)-0.4% Protease Max (Promega) with 10 mM DTT and incubated 30 min at 56°C. Protease Max is a surfactant that enhances trypsin digestion in solution. The reducing reaction was followed by an alkylation treatment with 55 mM iodoacetamide (IAA) in 100 mM NH4HCO3 for 45 min at room temperature in the dark. Samples were immediately digested for 18 h at 37°C with 1 μg of trypsin (Trypsin Sequencing Grade Promega, Madison, WI, USA) using a 1:2 w/w protease to protein ratio (Fig. S1 and Fig. 1). Spores were collected by centrifugation and the tryptic digests were desalted using PepClean C18 spin columns (30 μg capacity, Pierce Biotechnology, IL, USA). Purified peptides were quantified at 205 nm with a Nanodrop ND1000 spectrophotometer and stored at −80°C until analyzed.
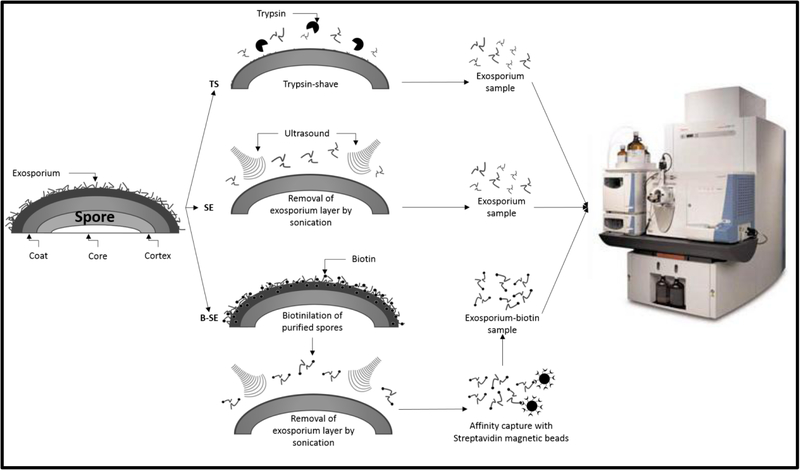
Figure 1. Gel-free strategy to remove and identify exosporium-like layer proteins of C. difficile 630 spores.
Experimental gel-free strategy to determine the proteome of the exosporium-like layer of C. difficile 630 spores. i) C. difficile spores were trypsin-digested (TS) for 18 h at room temperature and peptides analyzed by MS/MS as described in Methods section: ii) The exosporium-like layer of C. difficile 630 spores was removed by sonication, digested with trypsin and peptides analyzed by MS/MS; iii) C. difficile 630 spore surface proteins were biotin-labeled prior to sonication, and biotin-labeled exosporium extracts were captured by Streptavidin-magnetic beads and digested with trypsin and peptides were identified by MS/MS. All experiments were done with two biological replicates.
Extraction of exosporium fragments by sonication.
Sonication was previously shown to remove the exosporium layer leaving the spore coats intact, thus exosporium fragments of two biological replicates were extracted by sonication14, 18. Briefly, C. difficile 630 spores were resuspended in 50 mM Dulbecco’s Phosphate-Buffered Saline (DPBS)-0.5 mM EDTA buffer (pH 7.5), and sonicated with a Vibra-Cell VCX130 (Sonics & Materials, NewTown, CT, USA) in ice-cold conditions for 30 1-min bursts, separated by 3 min of cooling on ice. Sonicated spores were removed from the exosporium material by centrifugation, and the supernatant containing the exosporium fragments was filtered with syringe filters with 0.2 μM pores (Milipore, U.S.A.) to remove any remaining spores (Fig S1 and Fig. 1). Samples were concentrated by vacuum-centrifugation and resuspended in 50 mM NH4HCO3 with 0.2% Protease Max (Promega) and trypsin-digested as described above. The tryptic digests were desalted in C18 spin columns as described above and stored at −80°C until analyzed.
As an alternative strategy, the exosporium layer was biotinylated prior to sonication (Fig. 1). Briefly, two biological replicates of C. difficile spores (~ 2 × 109 spores) were biotinylated with 1 mg of biotinamidohexanoic acid N-hydroxysuccinimide ester (Sigma-Aldrich, U.S.A.) in 0.1 M sodium bicarbonate (pH 8.2) for 45 min at room temperature. Biotin-labeled spores were washed 10 times with sterile phosphate buffered saline (pH 7.4) with glycine (10 mg/ml) to inactivate and remove any excess of unbound biotin succinimide ester and three times with PBS to remove excess glycine. Biotin-labeled exosporium fragments were removed by sonication as described above and affinity captured with magnetic streptavidin-coated C1 Dynabeads (Invitrogen) as described by the manufacturer. Dynabeads containing exosporium fragments were resuspended in 50 mM NH4HCO3 with 0.2% Protease Max (Promega) and digested with trypsin as described above. Tryptic digests were desalted in a C18 spin column (Pierce), and stored at −80°C until use.
UPLC MS/MS analysis.
UPLC-MS/MS data were acquired with a LTQ-FT Ultra mass spectrometer with an IonMax ion source (Thermo, West Palm Beach, FL, USA) coupled to a nanoAcquity Ultra performance LC system (Waters, Milford, MA, USA) equipped with a Michrom Peptide CapTrap column (Michrom) and a C18 column (Zorbax 300SB-C18, 250 by 0.3 mm, 5-μm; Agilent). Prior to analysis, the digests were lyophilized using speedvac centrifuge then and resuspended to a final concentration of approximately 500 ng/μl in mass spectrometry (MS) loading solution (95% H2O, 5% [vol/vol] acetonitrile [ACN], 0,1% [vol/vol] formic acid). A binary gradient system was used consisting of solvent A (0.1 % aqueous formic acid) and solvent B (ACN containing 0.1% [vol/vol] formic acid). Two μl of C18 spin column-purified peptides was trapped and washed with 3% solvent B at a flow rate of 5μl/min for 3min. Trapped peptides were then eluted into an analytical column using a linear gradient from 3% B to 30% B at a flow rate of 4 μL/min over 35 minutes. The column was maintained at 37°C during the run. The mass spectrometer was operated in a data-dependent mode. A full FT-MS scan (m/z 350 to 2,000) was alternated with collision-induced dissociation (CID) MS/MS scans of the 5 most abundant doubly or triply charged precursor ions. As the survey scan was acquired in the ion cyclotron resonance (ICR) cell, the CID experiments were performed in the linear ion trap where precursor ions were isolated and subjected to CID in parallel with the completion of the full FT-MS scan. CID was performed with helium gas at a normalized collision energy of 35 % and activation time of 30 ms. Automated gain control (AGC) was used to accumulate sufficient precursor ions (target value, 5 × 104/micro scan; maximum fill time 0.2 s). Dynamic exclusion was used with a repeat count of 1 and exclusion duration of 60 s. Data acquisition was controlled by Xcalibur (version 2.0.5) software (Thermo Scientific).
Database search.
Thermo Scientific raw data files were processed with Proteome Discoverer version 1.2 using default parameters. A Mascot (version 2.2.04) search against a Clostridium difficile 630 database (obtained from UniProt; 3,982 sequences; 1,222,570 residues) was launched from Proteome Discoverer with the following parameters. The digestion enzyme was set to Trypsin/P and three missed cleavage sites were allowed as previously described19. The precursor ion mass tolerance was set to 5 ppm, while fragment ion tolerance of 0.8 Da was used. Dynamic modifications included: Deamidation (+0.984 Da) for asparagine and glutamine, oxidation (+15.9994 Da) for methionine, and carbamidomethyl (+57.0214 Da) for cysteine. For biotinylated exosporium extracts, an LC-biotin (+454.5) was also included to lysine and the N-terminus. Data from two experimental replicates were combined using Scaffold software (Proteome Software, Portland, OR) and applying the MudPIT (multidimensional protein identification technology) option, and the identified proteins from each sample were summarized with Scaffold 3 software. The inclusion of a protein in the final data set required that one or more unique peptides, with a score higher than 20, for that protein could be identified in at least one experimental condition. Only peptides that were identified with a confidence greater than 90% were utilized to calculate the overall probability of protein identification as specified by the Peptide Prophet algorithm23, 24.
Functional classification of identified proteins.
Where a direct homologue between C. difficile and other Clostridium species existed, functional annotations were drawn from the NCBI website (www.pubmed.com) or the BioCyc Database Collection (http://biocyc.org). Direct homologues were defined as proteins with an identity higher than 60%. The presence and direct annotation of direct homologues for C. difficile proteins identified in this study are detailed in Table S1 in the supplemental material.
Construction of plasmids.
To label exosporium proteins with the FLAG epitope at the C-terminal of selected genes, we used previously described plasmid pDP345 containing cdeC630-FLAG fusion as a template9. Briefly, to create a cdeCR20291-FLAG fusion, an NdeI-SalI fragment containing the promoter region and the entire cdeC630 with the exclusion of its stop codon, was PCR amplified from genomic DNA of C. difficile strain R20291 with Phusion High Fidelity DNA Polymerase (Thermo Scientific) using primer pairs P113-P166 (Table S1). The PCR fragment was inserted into NdeI and SalI sites of pDP345, giving plasmid pDP375 (Table S2). The resulting plasmid was sequenced to confirm that no mutations were generated during PCR or cloning. Plasmid pDP375 was transformed into E. coli HB101(pRK24) and subsequently conjugated into strain C. difficile 630ΔermB as previously described25. Sporulating cultures were obtained as described above and analyzed by Western blotting and immunofluorescence.
To construct FLAG fusions of the cotA (CD1613), cotB (CD1511), bclA1 (CD0332), bclA2 (CD3230), bclA3 (CD3349), cdeM (CD1581), cdeA (CD2375) and cdeB (CD2752) genes, the promoter region and the entire ORF with the exclusion of the respective stop codons was PCR amplified from genomic DNA of C. difficile strain 630 with Phusion High Fidelity DNA Polymerase (Thermo Scientific) using primer pairs described in Table S1. To facilitate cloning we used a KpnI site upstream of NdeI. KpnI-SalI fragments were introduced into KpnI and SalI sites of pDP345 giving plasmids described in Table S2. All plasmids were sequenced to confirm correct insertion, and were subsequently introduced into E. coli HB101(pRK24) and subsequently conjugated into strain C. difficile 630ΔermB. Sporulating cultures were obtained as described above and analyzed by Western blotting and immunofluorescence.
Sarkosyl-proteinase K treatment of C. difficile 630 spores.
To remove the exosporium layer as previously described18, spores (5 × 108 spores) were incubated at 37°C at 200 rpm with 30 μl of 1% sarkosyl, 0.3 mg/ml proteinase K, 25 mM phosphate buffer (pH 7.4) (PK) for 2 h. Treated spores were washed 10 times with sterile distilled water prior to storage at – 80°C until use. To confirm that the exosporium layer was removed, hydrophobicity of PK-treated spores was measured as previously described and decreased from 60–70% to 2%, meaning that only 2% of the spores were in the organic phase18.
Western blotting.
Samples (10μl) of SDS-PAGE loading buffer containing the spore coat and exosporium extracts from 4 × 107 untreated and only the spore coat extracts from 4 × 107 SPK treated spores of C. difficile 630 carrying plasmids pDP365 and pDP366 were electrophoresed on SDS-PAGE gels (12% acrylamide). Proteins were transferred to a nitrocellulose membrane (Bio-Rad) and blocked for 1 h at room temperature with 2% bovine serum albumin (BSA)–Tris-buffered saline (TBS) (pH 7.4). Western blots were probed with a 1:1,000 dilution of anti-FLAG overnight at 4°C and then with a 1:5,000 dilution of anti-mouse horseradish peroxidase (HRP) conjugate (Promega) for 1 h at room temperature in PBS–1% BSA–0.1% Tween 20. HRP activity was detected with a chemoluminescence detection system (Fotodyne Imaging system) by using PicoMax sensitive chemiluminescent detection system HRP substrate (RockLand Immunochemicals). Each Western blot also included 2μl of a PageRuler Plus prestained protein ladder (Fermentas). Each Western blot was repeated at least 3 times with independent spore extracts, and analyzed by densitometry to quantify the relative amounts of protein by ImageJ as previously described9.
3. Results and Discussion
Sonication and trypsin-digestion uniquely remove exosporium material of C. difficile 630 spores.
The precise composition of the outermost layer of C. difficile spores remains unknown. Previously, Lawley et al. (2009) provided the first proteomic study of C. difficile 630 spores19, while Abhyankar et al. (2013) defined the spore coat proteome of C. difficile 630 spores20. However, spores used in those studies were purified in the presence of proteinase K, which has been recently shown to remove the exosporium-like layer of C. difficile 630 spores18. Therefore, to pursue the identification of proteins of the outermost exosporium-like layer of C. difficile 630 spores, we used spores purified in the absence of proteases. To remove the electron-dense exosporium-like layer of C. difficile 630 spores (Fig. S1), we used two previously reported experimental approaches18. As expected, sonication and 18 h trypsin-digestion treatments removed most of the electron-dense exosporium-like layer of C. difficile 630 spores without apparent effect on the underlying layers (Fig. S1) as we have previously described18. However, we cannot rule out that some coat material could have been degraded without changing the ultrastructural appearance of the spore coat.
In addition to transmission electron micrographs, spore hydrophobicity was used as an additional marker for exosporium removal as we have previously described18. Untreated C. difficile 630 spores have a notional surface hydrophobicity value of 62%, meaning that nearly 62% of the spores are in the organic phase. As expected, 30 pulses of 1 min of sonication reduced the hydrophobicity of C. difficile 630 spores to 11%. Similarly, a trypsin-digestion treatment of 18 h at 37°C reduced the hydrophobicity to 9%. These results are consistent with most of the exosporium layer being removed by sonication and trypsin treatments as previously described18. The integrity of the spore coat of sonicated and trypsin-digested spores was further confirmed by lysozyme digestion. Phase contrast microscopy analysis of ~ 1,000 spores demonstrated that > 99% of either sonicated or trypsin-digested spores remained phase bright after lysozyme treatment. These results indicate that the spore coat of sonicated and trypsin-treated spores retained its permeability properties, but does not rule out that some spore coat material might have been removed. Collectively, these results suggest that these treatments remove sufficient exosporium material without affecting the spore coat properties.
Gel-free experimental approach to determine the proteome of the exosporium-like layer of C. difficile 630 spores.
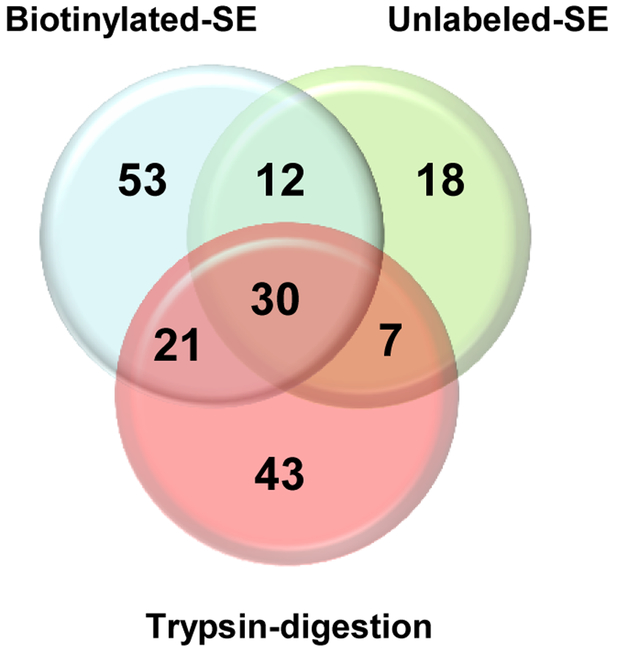
The aforementioned treatments were used to remove the exosporium-like layer for MS/MS analysis in an attempt to define the exosporium proteome of C. difficile 630 spores (Fig. 1). First, we used a trypsin digestion treatment to remove the exosporium-like layer of C. difficile 630 spores and detected a total of 101 protein species (Fig. 1 and Fig. 2). By reasoning that trypsin might encounter steric hindrance when digesting the intact exosporium-like layer, we also used an alternative approach where the exosporium-like layer was extracted by sonication and subsequently subjected to trypsin digestion (Fig. 1 and Fig. 2). This approach allowed the assignment of 67 polypeptides with high confidence, of which 30 polypeptides were not identified by trypsin-digestion of C. difficile 630 spores (Fig. 1 and Fig. 2). Biotinylation of bacterial surfaces prior to protein extraction has been used to identify the surface proteome in several species26. Consequently, we also included this alternative approach in our effort to identify the exosporium proteome. C. difficile 630 spores were biotinylated prior to exosporium extraction by sonication and sonicated-biotinylated-exosporium extracts were affinity captured and trypsin digested prior to MS/MS analysis (Fig. 1). This approach identified a total of 116 polypeptide species (Fig. 2) of which 53 were unique to this approach (Fig. 2). Combining these results, the exosporium of C. difficile 630 spores seems to harbor at least 184 polypeptide species (Table S1) of which 61 had not been found previously in proteomic studies with C. difficile 630 spores19, 20. Most of these proteins seem to be vegetative cell contaminants that became entrapped during exosporium assembly. The difference in the spectra of proteins detected by each treatment might rely in that some proteins were lost while others became accessible to trypsin during sonication providing a complementary protein spectrum to that of trypsin digestion of the entire spore.
Figure 2. Venn diagram of conserved superfamily domains in the identified spore exosporium proteins from C. difficile 630 spores.
A total of 183 superfamily domains were identified in the proteins extracted from the exosporium-like layer of C. difficile 630 spores using three gel-free experimental strategies: Boitinylated sonicated exosporium (Biotinylated-SE); Unlabeled sonicated exosporium (Unlabeled-SE); and trypsin digested spores (Trypsin-Digestion). Numbers corresponds to the number of superfamily domains identified.
Functional classification of exosporium proteins.
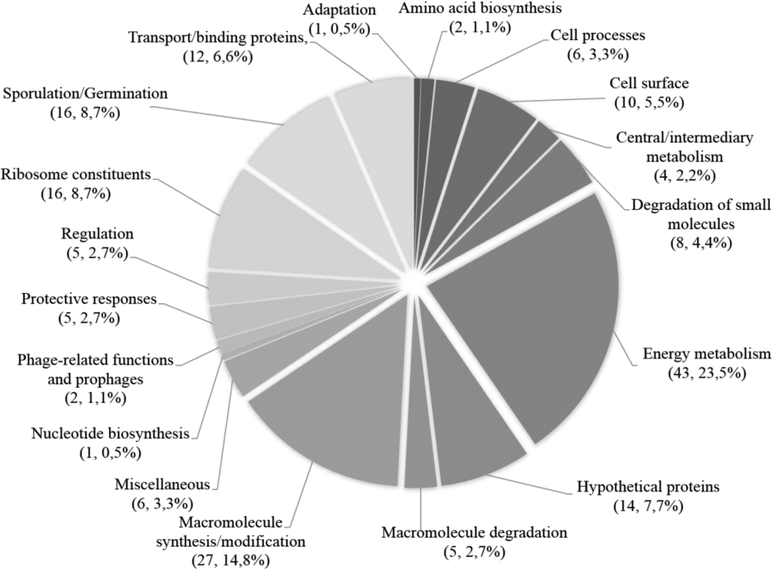
To analyze the exosporium-associated peptides, these were classified according to the functional scheme used for the C. difficile 630 genome27 (Fig. 3). Surprisingly, the most common functional classes found in the exosporium-like layer of C. difficile 630 spores were proteins involved in energy metabolism, with a total of 43 proteins (23.5 %) (Fig. 3); most likely these proteins were trapped during the assembly of the exosporium-like layer as has been similarly observed for the exosporium layer of spores of the B. cereus group28, 29, and easily detected by the highly sensitive UPLC MS/MS approach. The identified proteins found in the exosporium were divided into various categories based on their demonstrated and/or predicted functions: i) previously characterized; ii) putative roles in protective response; iii) possibly involved in pathogenesis; iv) uncharacterized proteins; v) other spore proteins.
Fig. 3. Representation of the distribution and abundance by functional classes of the exosporium proteins of C. difficile spores.
Shown is the percentage of each functional class in the exosporium of C. difficile 630 spores (Functional classes are described in reference27). The elevated number of energy metabolism and cytosolic proteins identified in this spore layer are most likely vegetative cell contaminants entrapped during the assembly of the exosporium layer, and therefore, it is likely unlikely that these proteins are genuine exosporium proteins.
1. Characterized exosporium proteins.
Recent work has characterized several C. difficile exosporium proteins9, 12, 30, 31. Polypeptides of the exosporium morphogenetic cysteine rich protein, CdeC9 (Table 1), were 10-fold more abundant than that of other species in the extracts from the trypsin digestion treatment. The high abundance of cysteine residues (9%) suggests that this protein might contribute to the crosslinking of the exosporium layer of C. difficile spores. CdeC is unique to C. difficile and thus may be used as a biomarker for C. difficile spores. Another spore morphogenetic factor, required for correct spore assembly, CotA, was also present in the exosporium extracts (Table 1 and S1). Polypeptides of the exosporium protein, CotB, which plays a slight role in spore resistance and the manganese catalase (CotD)31 were also found in the exosporium proteome of C. difficile 630 spores (Table 1 and Table S1).
TABLE 1.
Characterized exosporium proteins of Clostridium difficile 630 spores.
| Proteina (Gene identification) | N° of unique peptidesb | Percentage coveragec | MWd/pI | Proteome | Transcriptome | ||
|---|---|---|---|---|---|---|---|
| Sporee | Spore coat/exof | in vitrog | in vivoh | ||||
| Exosporium proteins | |||||||
| BclA1 (CD0332; bclA1) | 5 | 6.6 | 68.0/5.3 | ✓ | ✓ | σK | ✓ |
| CdeC (CD1067. cdeC) | 19 | 41 | 44.7/4.8 | ✓ | ✓ | σK | ✓ |
| Peroxiredoxin chitinase (CD1433, cotE) | 14 | 23 | 81.1/5.3 | ✓ | ✓ | σK | ✓ |
| CotB (CD1511, cotB) | 4 | 16 | 34.9/4.8 | ✓ | ✓ | σE | ✓ |
| Putative uncharacterized protein (CD1581, cdeM) | 10 | 51 | 19.1/5.3 | ✓ | ✓ | σK | ✓ |
| CotA (CD1613, cotA) | 4 | 14 | 34.0/4.6 | ✓ | ✓ | σK | ✓ |
| CotD (CD2401, cotD) | 1 | 9.5 | 21.4/4,8 | ✓ | ✓ | σK | ✓ |
Proteins identified by either of the three approaches.
Peptides certain by integrating the three approaches.
Represents the percentage of coverage by integrating the peptide sequences of the three approaches.
MW, molecular weight.
Protein present in the spore proteome by Lawley et al. (2009).
Protein present in the sproe coat proteome by Abhyanakr et al. (2013).
RNA polymerase sigma factors that governs the expression as described Fimlaid et al. (2013) and Saujet et al. (2013).
Expression in pathogenesis in vivo as described Janoir et al. (2013).
Three orthologues of exosporium collagen-like BclA glycoprotein of the B. cereus group, are encoded in the 630 genome (i.e. BclA1, BclA2 and BclA3). Despite the fact that all three orthologs are expressed during sporulation12, 13, 32 and localized to the surface of C. difficile 630 spores12, we could only detect peptides derived from BclA1, but not of BclA2 and BclA3 (Table 1 and Table S1). Analysis of the amino acid sequence of BclA2 and BclA3 shows the presence of 6 and 9 trypsin cleavage sites, respectively, suggesting that the BclA2 and BclA3 cleavage sites might not be accessible to trypsin. In previous work using in-gel trypsin digestion of electrophoresed spore coat and exosporium fractions, followed by UPLC MS/MS, we were not able to detect BclA2 and BclA3 derived peptides, yet detected significant peptides derived from the trypsin-digestion of BclA112. It was also surprising that the exosporium proteins CotF and CotG as well as the coat and exosporium protein CotCB31 were not detected in either of the three extraction methods (Table 1 and Table S1). These three proteins also have predicted trypsin-cleavage sites, suggesting that they might not be accessible to trypsin digestion. Alternatively, is also likely that these proteins might not be abundant enough to be detected using any of the approaches employed in this study.
Polypeptides of the uncharacterized protein CD1581, recently demonstrated to be highly expressed during in vivo sporulation, and which plays a role in persistence of C. difficile in a mouse model33, were present in the exosporium proteome (Table 1); however, whether it is uniquely localized in the exosporium layer and plays a role in the exosporium assembly or acts as an spore adhesin remains unclear. Polypeptides of C. difficile CotE, which has in vitro peroxiredoxin reductase and chitinase activity, should not be confused with the B. subtilis spore coat morphogenetic factor CotE, and were also detected in the exosporium proteome (Table 1)30, 31. Previous work localized C. difficile CotE to both the spore coat and the exosporium-like layer of C. difficile 630 spores31, suggesting that it might play a role during the assembly of both layers, albeit this role seems not noticeable since no ultrastructural defects were observed in spores of a cotE mutant31.
2. Exosporium proteins with roles in protective response.
In anaerobic organisms, such as C. difficile, defense against oxidative stress is of prime importance. Given the fact that C. difficile spores are metabolically dormant and possess similar resistance properties as those of B. subtilis that prevent small molecules from entering the spore core9, 31, they are naturally resistant to anaerobic conditions and germinate well in the presence of oxygen34. However, during the release of the nascent cell during spore outgrowth, proteins able to remove reactive oxygen species present in the exosporium of C. difficile spores might be of importance to the survival of the nascent cells. This may be the case of the C. difficile 630 exosporium-like layer, as three non-haem iron ruberythrin proteins (i.e. CD1474, CD1524 and CD2845) likely to be involved in protection against oxidative stress in anaerobic bacteria35 were identified in the putative exosporium proteome (Table 2). Interestingly, CD2845 has not been previously identified in the spore proteome19 nor in the spore coat proteome20 of C. difficile 630 spores, and has been demonstrated to be significantly upregulated during in vivo sporulation in a CDI mouse model33. Further validation of their location in the exosporium layer is needed.
TABLE 2.
Uncharacterized exosporium proteins of Clostridium difficile 630 sporesa.
| Proteina (Gene identification) | N° of unique peptidesb | Percentage coveragec | MWd/pI | Proteome | Transcriptome | ||
|---|---|---|---|---|---|---|---|
| Sporee | Spore coat/exof | in vitrog | in vivoh | ||||
| Proteins likely to play a role in spore resistance | |||||||
| Putative ferredoxin/flavodoxin oxidoreductase, alpha subunit (CD0116) | 4 | 19 | 39.1/5.4 | - | ✓ | - | ✓ |
| Putative ferredoxin/flavodoxin oxidoreductase, beta subunit (CD0117) | 5 | 36 | 26.7/6.9 | - | ✓ | - | ✓ |
| Putative ferredoxin/flavodoxin oxidoreductase, gamma subunit (CD0118) | 2 | 20 | 19.8/5.1 | - | - | - | ✓ |
| Putative rubrerythrin protein (CD1474) | 4 | 27 | 20.0/5.5 | ✓ | - | - | - |
| Putative rubrerythrin protein (CD1524) | 4 | 27 | 20.0/5.6 | ✓ | ✓ | - | - |
| Rubrerythrin (CD2845, rbr) | 2 | 15 | 22.4/6.5 | ✓ | ✓ | σG | ✓ |
| Proteins possibility involved in pathogenicity | |||||||
| Elongation factor Tu (CD0071, tuf1) | 12 | 43 | 44.0/5,1 | ✓ | ✓ | - | - |
| Chaperonin 60 kDa (CD0194, groL) | 1 | 2.2 | 57.7/4.8 | ✓ | ✓ | - | - |
| Chaperone protein (CD2461, dnaK) | 3 | 7.8 | 66.4/4.8 | ✓ | - | - | ✓ |
| Enolase (CD3170, eno) | 3 | 11 | 46.1/4.7 | ✓ | ✓ | - | ✓ |
| Glyceraldehyde-3-phosphate dehydrogenase (CD3174, gapA) | 6 | 24 | 36.0/6.0 | ✓ | ✓ | - | - |
| ATP-dependent Clp protease proteolytic subunit 1 (CD3305, clpP) | 1 | 3.6 | 21.3/5.3 | ✓ | ✓ | - | - |
| Other exosporium proteins | |||||||
| Putative uncharacterized protein (CD0311) | 1 | 7.1 | 31.4/4.6 | - | - | σE | ✓ |
| Putative cation efflux protein (CD0902) | 1 | 3.5 | 32.4/7.0 | - | - | σK | ✓ |
| Putative ATP-dependent protease (CD0564) | 3 | 5.2 | 85.4/5.2 | - | - | σK | ✓ |
| Putative ferrous ion transport protein (CD1517, feoB) | 3 | 6.4 | 78.2/6.6 | - | - | σK | - |
| Putative ferrous ion transport protein (CD1518, feoA) | 1 | 12 | 8.30/10 | - | - | σK | ✓ |
| Putative uncharacterized protein (CD1880) | 1 | 23 | 12.8/5.7 | - | - | σG | - |
| Putative uncharacterized protein (CD2245A) | 1 | 10 | 7.30/9.7 | ✓ | - | σG | - |
| Putative uncharacterized protein (CD2375, cdeA) | 2 | 37 | 11.3/5.6 | ✓ | - | σK | ✓ |
| Putative uncharacterized protein (CD2434) | 1 | 9.5 | 22.2/4.9 | ✓ | - | - | ✓ |
| Putative uncharacterized protein (CD2752, cdeB) | 2 | 9.1 | 25.5/5.2 | - | - | - | ✓ |
| Putative uncharacterized protein (CD3522) | 4 | 12 | 50.4/4.5 | ✓ | ✓ | σE | ✓ |
| Putative uncharacterized protein (CD3613) | 1 | 8.7 | 50.4/4.5 | ✓ | ✓ | σK | - |
| Putative peptidase, M1 family (CD3652) | 3 | 9.6 | 54.6/5.7 | ✓ | ✓ | σE | ✓ |
Proteins identified by either of the three approaches.
Peptides certain by integrating the three approaches.
Represents the percentage of coverage by integrating the peptide sequences of the three approaches.
MW, molecular weight.
Protein present in the spore proteome by Lawley et al. (2009).
Protein present in the sproe coat proteome by Abhyanakr et al. (2013).
RNA polymerase that governs the expression as described Fimlaid et al. (2013) and Saujet et al. (2013).
Expression in pathogenesis in vivo as described Janoir et al. (2013).
Polypeptides for all three subunits of a ferredoxin/flavodoxin oxidoreductase homologue (i.e. alpha, beta and gamma subunits) were found in the exosporium extracts (Table 2). Ferrodoxin is an electron acceptor for numerous low-potential oxidation-reduction reactions, including pyruvate:ferrodoxin oxidoreductase36, 37. Expression of all three proteins is upregulated during in vivo sporulation in a CDI mouse model33, suggesting that these proteins might play some role in the resistance to oxidative stress in the gut38, 39.
These rubrerythrins and oxidoreductases were previously reported in the proteomic study of the spore coat of C. difficile 630 spores treated with proteases during purification20 which coupled with the presence of their peptides in the exosporium-like layer, suggests that they are present in both layers of C. difficile 630 spores.
3. Exosporium proteins possibly involved in pathogenesis.
Some interesting proteins identified in the exosporium proteome of C. difficile 630 spores (Table 2 and S1) include those with high immunogenicity and present on the surface of B. anthracis spores28, 29. Two such proteins are the cytosolic Elongation Factor Tu (EF-Tu) and α-enolase (Table 2 and S1), which perhaps during the assembly of the exosporium-like layer of C. difficile 630 spores, become encased in the exosporium layers during its assembly. The surface location of these proteins on the exosporium layer of B. anthracis spores28 seems to be important during B. anthracis infection, acting as plasmin(ogen)-specific spore receptors that aid in the evasion B. anthracis spores from C3b-dependent innate immunity40. Peptides of the chaperonin protein, DnaK and glyceraldehyde-3-phosphate dehydrogenase (GADPH) were also identified in the spore exosporium proteome (Table 2), and have also been implicated in pathogenesis in other bacterial species28, 41. One last protein found in the exosporium proteome is the ATP-dependent Clp protease proteolytic subunit 1 (Table 2), which in several other bacteria has been shown to be required for survival to environmental stress28. Collectively, these results have identified several proteins located in the exosporium-like layer of C. difficile 630 spores and validation would be required to determine their predicted roles in pathogenesis.
4. Uncharacterized exosporium proteins.
Peptides from a number of uncharacterized proteins whose expression were shown to be under the control of sporulation specific sigma factors13, 32, and significantly upregulated during in vivo sporulation in a CDI mouse model33, where identified in the exosporium extracts of C. difficile 630 spores (Table 2). These candidates for exosporium proteins include the products of CD0311, CD0654, CD2375, CD2434, CD2752, CD3522 and CD3652 genes (Table 2). In addition, polypeptides of the products of CD1880, CD2245A and CD3613 genes which are under the control of sporulation specific sigma factors (Table 2) were also present in the exosporium proteome. Their roles in C. difficile spore biology remain unclear. Polypeptides of protein CD3613 (Table 2), that belongs to the Stay green family of proteins involved in chlorophyll degradation, and of the putative protease CD3652 that belongs to the family of thermolysin-like peptidases that includes several zinc-dependent metallopeptidases, were detected during this study. Notably, both of these proteins are highly expressed during in vivo sporulation in a mouse model33 and might play some role during CDI infection.
5. Cytosolic proteins.
Despite our effort to thoroughly wash C. difficile 630 spores to remove cytosolic contaminants with treatments that would not affect the exosporium integrity (i.e. distilled water and NaCl), we identified a number of cytosolic proteins such as ribosomal proteins, proteins involved in amino acid biosynthesis, cellular processes, central and intermediary metabolism, energy metabolism and transport of proteins were also detected in the exosporium proteome (Fig. 3 and Table S1). Indeed, we and others20, 42 believe that during the assembly of the exosporium, these proteins might be trapped during the polymerization of protein complexes, and represents the likely composition of the exosporium-like layer of C. difficile 630 spores found in the infective environment. The presence of cytosolic-proteins in the exosporium of B. anthracis spores also remain attached after extensive washes20, 28, 42. Thus, unlike the spore coat layer, which is mainly free of cytosolic proteins20, the exosporium layer, seems to have many cytosolic proteins that may be within the thick electron-dense layer removed by sonication or a trypsin digestion of the spore.
Validation of exosporium localization of selected proteins.
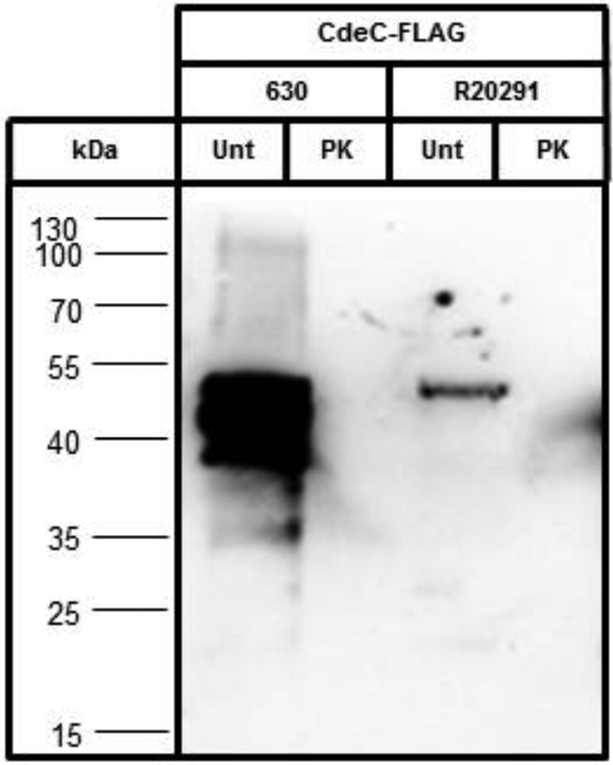
To validate our results, we used spores expressing Flag-tagged proteins identified in the exosporium proteome of C. difficile 630 spores, and selectively removed the exosporium layer using an exosporium-removal treatment based on proteinase K as previously described18. Nine proteins were selected based on the following criteria: i) several spore proteins whose localization to the spore coat and/or exosporium have not been defined (i.e, CotA, CotB, BclA2 and BclA3); ii) a putative novel exosporium morphogenetic protein (i.e. CdeM) (Paredes-Sabja unpublished work); iii) and two uncharacterized proteins (i.e. CdeA and CdeB). To demonstrate that this method removes exosporium proteins, we selected a C. difficile exosporium marker, CdeC, which is essential for the assembly of the exosporium layer of C. difficile R20291 spores9. Immunofluorescence analysis of untreated spores of strain 630 carrying CdeC630-FLAG fusion reveals that, as expected9, CdeC630 is accessible to anti-FLAG antibodies, similarly as in R20291 spores (Fig. S2A). Removal of the exosporium layer by proteinase K-treatment abolished the immunofluorescence signal of CdeC-FLAG fusion in both 630 and R20291 spores (Fig. S2A). However, since antibodies do not penetrate the spore coat located below the exosporium layer, possible spore coat-localized CdeC cannot be ruled out. Therefore, to confirm that CdeC is exosporium-located, we used SDS-PAGE loading buffer to remove the spore coat and exosporium fractions of untreated spores or only the spore coat fraction of proteinase K-treated (removes exosporium) spores from spores of strains 630(CdeC630-FLAG) and R20291(CdeCR20291-FLAG), and analyzed their removed fractions by western blotting. As expected, CdeC-FLAG fusion was detected in the spore coat and exosporium fractions of untreated spores of strains 630(CdeC630-FLAG) and R20291(CdeCR20291-FLAG), albeit signal was evidently more abundant in 630 spore coat and exosporium fractions (Fig. 4). No immunoreactive signal was observed by Western blot analysis in the spore coat fraction from proteinase K-treated spores of stains 630(CdeC630-FLAG) and R20291(CdeCR20291-FLAG), indicating that CdeC is located in the exosporium layer. The extent of exosporium removal can be confirmed by measuring spore hydrophobicity which drops to nearly 2% upon complete exosporium removal18. The hydrophobicity of 630(CdeC630-Flag) and R20291(CdeCR20291-FLAG) dropped from nearly 58% to 2% upon proteinase K-treatment (data not shown), indicating complete exosporium removal. However, we cannot rule out the possibility that CdeC might be located between the basal layer of the exosporium and the spore coat surface, as some proteins from the latter layer, above the protease impermeable layer of the spore coat, might also be removed by proteinase K treatment. Collectively, these results indicate that this method is, at most, adequate to determine the spore coat and/or exosporium localization of selected proteins present in the exosporium proteome.
Fig. 4. The exosporium morphogenetic protein, CdeC, localized uniquely to the exosporium layer of C. difficile 630 and R20291 spores.
Spore coat and exosporium extracts of untreated (Unt) and spore coat extracts of proteinase K-treated (PK) spores (4×107) were fractionated on SDS-PAGE, transferred to nitrocellulose membrane and analyzed by western blot using anti-FLAG specific antibody as described in Material and Methods. Results are representative of three independent experiments.
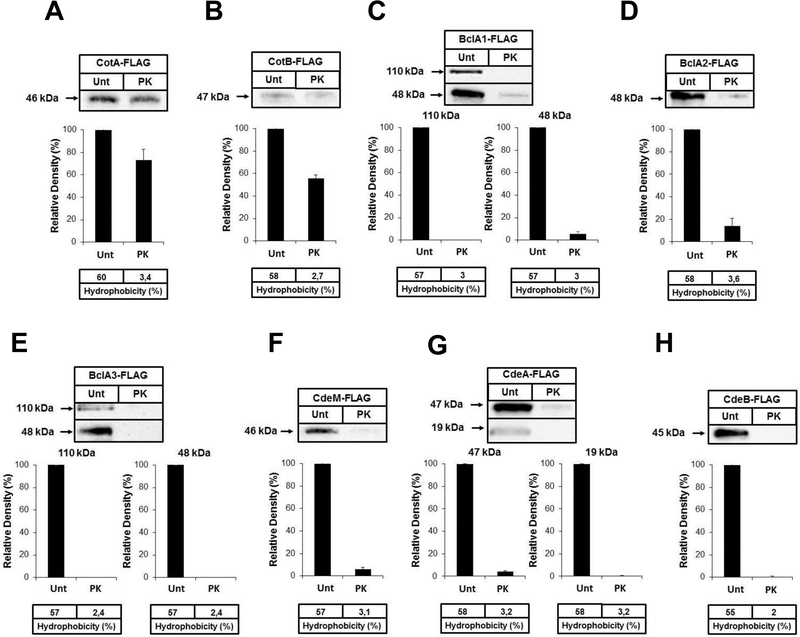
CotA and CotB are suggested to be spore coat proteins20, 31, although this has not been fully demonstrated11. To evaluate the relative spore coat/exosporium abundance of CotA and CotB, untreated and proteinase K-treated spores of strains 630(CotA-FLAG) and 630(CotB-FLAG) were analyzed by immunofluorescence and Western blot analysis. Both, CotA and CotB were accessible to anti-FLAG antibodies and signal was completely abolished by proteinase K-treatment (Fig. S3A and B). Western blot analysis demonstrate that upon removal of exosporium layer, levels of CotA and CotB in the spore coat fractions decrease by 30 and 45% relative to spore coat-exosporium fractions (untreated spores) (Fig. 5A and B). These results indicate that at least some CotA and CotB are surface exposed and accessible to antibodies, while most (65–70%) of CotA and CotB is inaccessible to anti-FLAG antibodies, proteinase K insensitive and located in the spore coat.
Fig. 5. Effect of removal of the exosporium layer on the relative abundance of selected outer surface spore proteins.
The spore coat and exosporium from untreated spores (Unt) and spore coat from proteinase K-treated (PK) spores of strains: A) 630(CotA-FLAG); B) 630(CotB-FLAG), C) 630(BclA1-FLAG); D) 630(BclA2-FLAG); E) 630(BclA3-FLAG); F) 630(CdeM-FLAG); G) 630(CdeA-FLAG); H) 630(CdeA-FLAG) were extracted with SDS-PAGE loading buffer, electrophoresed analyzed by Western blot analysis as described in Material and Methods. All experiments were done three independent times.
We have recently demonstrated that the C. difficile collagen-like BclA1 glycoprotein is located in the exosporium-like layer of C. difficile 630 spores12. However, whether BclA2 and BclA3 are also located in the exosporium, remains unclear. Immunofluorescence analysis of untreated and proteinase K-treated spores of strains 630(BclA1-FLAG), 630 (BclA2-FLAG) and 630(BclA3-FLAG) demonstrate that all three BclA are present in the spore surface and are absent in proteinase K-treated spores (Fig. S3C, D and E). To evaluate if any of the BclA proteins are also present in the spore coat, fractions of untreated and treated spores were analyzed by Western blotting. We found that all three BclA proteins are present as a 48-kDa immunoreactive species, which is smaller than their predicted molecular masses (i.e. predicted molecular mass of BclA1, BclA2 and BclA3 are 68-, 49- and 58-kDa, respectively), suggesting posttranslational cleavage and glycosylations43. Notably, BclA1 and BclA3 form stable high molecular mass complex of nearly 110–120-kDa (Fig. 5C, D and E) suggesting that similarly as with B. anthracis BclA44, C. difficile BclA1 and BclA3 might be forming stable dimers and/or trimers. Removal of the exosporium layer demonstrated that the majority (> 95 %) of BclA1 and BclA3 specific 44- and 110–120-kDa species are present in the exosporium fraction (Fig. 5C and E). Nearly 85 % of BclA2 was found to be present in the exosporium layer. Overall, these results indicate that BclA1, BclA2 and BclA3 are mostly exosporium proteins, where at least BclA1 and BclA3 form stable high molecular mass complex.
A recent study demonstrated that CD1581 is the most upregulated gene during sporulation of C. difficile strain 630 in vivo33. Genetic studies have demonstrated that CD1581 encodes an essential C. difficile exosporium morphogenetic protein termed CdeM (Paredes-Sabja unpublished work) that is found in the exosporium layer (Table 1); however, it is unclear if it is mainly present in the exosporium layer. Immunofluorescence of untreated 630(CdeM-FLAG) spores demonstrated that CdeM is located in the spore surface and accessible to antibodies (Fig. S3F). No immunofluorescence signal was detectable upon treatment with proteinase K (Fig. S3F), suggesting that CdeM is located in the exosporium-like layer. Western blot analysis demonstrated that CdeM is present as a 46-kDa immunoreactive species, suggesting that the 19.1 kDa spore morphogenetic protein is forming stable complexes with other spore proteins. Removal of the exosporium-like layer decreased the presence of CdeM by nearly 95% (Fig. 5F) indicating that CdeM is mostly localized in the exosporium-like layer of C. difficile 630 spores.
Two additional uncharacterized proteins identified in the exosporium proteome were selected to confirm their localization into the exosporium layer: i) a 11.3-kDa uncharacterized protein, encoded by CD2375 and named CdeA, which is present in the spore proteome19 and its expression decreases by 1.4-fold in a sigK mutant background13; ii) a 25.5-kDa uncharacterized protein, encoded by CD2752 and named CdeB, that has not been detected in previous proteome studies19, 20, but that its expression increases 3.3-fold during in vivo sporulation33. Immunofluroescence analysis of 630(CdeA-FLAG) and 630(CdeB-FLAG) spores demonstrates that both CdeA and CdeB are spore surface proteins removed by proteinase K (Fig. S3G and H), suggesting that they are located in the exosporiun layer. Analysis of spore coat and exosporium fractions of untreated 630(CdeA-FLAG) and 630(CdeB-FLAG) spores by Western blot shows that CdeA-FLAG is present as 19- and 47-kDa species, while CdeB-FLAG is forming a stable 45-kDa molecular complex (Fig. 5G and H). Removal of the exosporium layer with proteinase K demonstrated that 99% and 90% of CdeA’s 19-kDa and 47-kDa species, respectively, are present in the exosporium-like layer (Fig. 5G). Similarly, nearly 99% of CdeB was found to be located in the exosporium-like layer (Fig. 5H). These results indicate that CdeA and CdeB are exosporium proteins. Further validation of the remaining uncharacterized proteins (Table 1 and 2) is required to determine the precise composition of the exosporium-like layer of C. difficile 630 spores.
Concluding remarks.
In conclusion, this study offers a strategy to identify proteins located in the exosporium-like layer of C. difficile 630 spores and complements previous proteomic studies of C. difficile 630 spores19, 20, since it defines the proteome of the outermost exosporium-like layer of C. difficile 630 spores. Many cytosolic proteins were detected, similarly as for B. anthracis studies28, indicating that the exosporium layer of C. difficile spores is a complex layer that may entrap many cytosolic proteins during its assembly; or alternatively, that these contaminants are derived from vegetative contaminants that could not be removed by the procedures to remove loosely-bound proteins. Several proteins involved in protection against environmental stress as well as putative virulence factors that might interact with complement or plasminogen were identified which may control the fate of C. difficile spores in the host. Importantly, a list of uncharacterized putative exosporium proteins is provided. We demonstrate that previously characterized spore coat proteins (i.e, CotA and CotB) are also present in the exosporium layer, while the collagen-like glycoproteins (i.e. BclA1, BclA2 and BclA3) as well as a novel spore morphogenetic protein (i.e. CdeM) and two uncharacterized signature proteins (i.e. CdeA and CdeB) are exosporium proteins. Further on-going work to define the location and function of the remaining uncharacterized exosporium proteins (Table 1 and 2) will allow us to establish the composition of the exosporium layer of C. difficile spores. This work provides new protein targets for the diagnosis and/or therapeutics that may contribute to combat C. difficile infections.
Supplementary Material
Acknowledgements
This work was supported by grants from Fondo Nacional de Ciencia y Tecnología de Chile (FONDECYT Grant 1110569), from the Research Office of Universidad Andres Bello (DI-275–13/R 2013) and by a grant from Fondo de Fomento al Desarrollo Científico y Tecnológico (FONDEF) CA13I10077 (to D.P.-S); and by grants from the N. L. Tartar Foundation of Oregon State University, Agricultural Research Foundation of Oregon State University, Department of Defense Multi-disciplinary University Research Initiative (MURI) award through the U.S. Army Research Laboratory and the U. S. Army Research Office under contract number W911NF-09-1-0286 (to MRS). OSU’s mass spectrometry facility is supported in part by grant from NIH/NIEHS (P30 ES000210).
References
- 1.Rupnik M; Wilcox MH; Gerding DN, Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 2009, 7, (7), 526–36. [DOI] [PubMed] [Google Scholar]
- 2.Karas JA; Enoch DA; Aliyu SH, A review of mortality due to Clostridium difficile infection. J Infect 2010, 61, (1), 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Sarker MR; Paredes-Sabja D, Molecular basis of early stages of Clostridium difficile infection: germination and colonization. Future Microbiol 2012, 7, (8), 933. [DOI] [PubMed] [Google Scholar]
- 4.O’Donoghue C; Kyne L, Update on Clostridium difficile infection. Curr Opin Gastroenterol 2011, 27, (1), 38–47. [DOI] [PubMed] [Google Scholar]
- 5.Deakin LJ; Clare S; Fagan RP; Dawson LF; Pickard DJ; West MR; Wren BW; Fairweather NF; Dougan G; Lawley TD, Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 2012, 80, (8), 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setlow P, I will survive: DNA protection in bacterial spores. Trends. Microbiol 2007, 15, (4), 172–80. [DOI] [PubMed] [Google Scholar]
- 7.Henriques AO; Moran CP Jr., Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol 2007, 61, 555–88. [DOI] [PubMed] [Google Scholar]
- 8.Xue Q; Gu C; Rivera J; Hook M; Chen X; Pozzi A; Xu Y, Entry of Bacillus anthracis spores into epithelial cells is mediated by the spore surface protein BclA, integrin alpha2beta1 and complement component C1q. Cell Microbiol 2011, 13, (4), 620–34. [DOI] [PubMed] [Google Scholar]
- 9.Barra-Carrasco J; Olguin-Araneda V; Plaza-Garrido A; Miranda-Cardenas C; Cofre-Araneda G; Pizarro-Guajardo M; Sarker MR; Paredes-Sabja D, The Clostridium difficile exosporium cysteine (CdeC)-rich protein is required for exosporium morphogenesis and coat assembly. J Bacteriol 2013, 195, (17), 3863–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliva C; Turnbough CL Jr.; Kearney JF, CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proc Natl Acad Sci U S A 2009, 106, (33), 13957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paredes-Sabja D; Shen A; Sorg JA, Clostridium difficile spore biology: sporulation, germination and spore structural proteins. Trends Microbiol 2014, 22, (7), 406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizarro-Guajardo M; Olguin-Araneda V; Barra-Carrasco J; Brito-Silva C; Sarker MR; Paredes-Sabja D, Characterization of the collagen-like exosporium protein, BclA1, of Clostridium difficile spores. Anaerobe 2014, 25, 18–30. [DOI] [PubMed] [Google Scholar]
- 13.Fimlaid KA; Bond JP; Schutz KC; Putnam EE; Leung JM; Lawley TD; Shen A, Global Analysis of the Sporulation Pathway of Clostridium difficile. PLoS Genet 2013, 9, (8), e1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paredes-Sabja D; Sarker MR, Adherence of Clostridium difficile spores to Caco-2 cells in culture. J Med Microbiol 2012, 61, (9), 1208–1218. [DOI] [PubMed] [Google Scholar]
- 15.Phetcharaburanin J; Hong HA; Colenutt C; Bianconi I; Sempere L; Permpoonpattana P; Smith K; Dembek M; Tan S; Brisson MC; Brisson AR; Fairweather NF; Cutting SM, The spore-associated protein BclA1 affects the susceptibility of animals to colonization and infection by Clostridium difficile. Mol Microbiol 2014. [DOI] [PubMed] [Google Scholar]
- 16.Paredes-Sabja D; Cofre-Araneda G; Brito-Silva C; Pizarro-Guajardo M; Sarker MR, Clostridium difficile spore-macrophage interactions: spore survival. PLoS One 2012, 7, (8), e43635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi LT; Phillips DS; Williams CF; Alyousef A; Baillie L, The contribution of the spore to the ability of Clostridium difficile to adhere to surfaces. Appl Environ Microbiol 2012, 78, (21), 7671–7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escobar-Cortes K; Barra-Carrasco J; Paredes-Sabja D, Proteases and sonication specifically remove the exosporium layer of spores of Clostridium difficile strain 630. J Microbiol Methods 2013, 93, (1), 25–31. [DOI] [PubMed] [Google Scholar]
- 19.Lawley TD; Croucher NJ; Yu L; Clare S; Sebaihia M; Goulding D; Pickard DJ; Parkhill J; Choudhary J; Dougan G, Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 2009, 191, (17), 5377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abhyankar W; Hossain AH; Djajasaputra A; Permpoonpattana P; Ter Beek AS; Dekker HL; Cutting S; Brul S; de Koning LJ; de Koster CG, In Pursuit of Protein Targets: Proteomic characterization of Bacterial Spore Outer Layers. J Proteome Res 2013, 12, (10), 4507–21. [DOI] [PubMed] [Google Scholar]
- 21.Gumustas M; Ozkan SA, A validated stability-indicating RP-LC method for the simultaneous determination of amlodipine and perindopril in tablet dosage form and their stress degradation behavior under ICH-recommended stress conditions. J AOAC Int 2013, 96, (4), 751–7. [DOI] [PubMed] [Google Scholar]
- 22.Wust J; Sullivan NM; Hardegger U; Wilkins TD, Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J Clin Microbiol 1982, 16, (6), 1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A; Nesvizhskii AI; Kolker E; Aebersold R, Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry 2002, 74, (20), 5383–92. [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii AI; Keller A; Kolker E; Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003, 75, (17), 4646–58. [DOI] [PubMed] [Google Scholar]
- 25.Bouillaut L; McBride SM; Sorg JA, Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol 2011, Chapter 9, Unit 9A 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNamara M; Tzeng SC; Maier C; Zhang L; Bermudez LE, Surface proteome of “Mycobacterium avium subsp. hominissuis” during the early stages of macrophage infection. Infection and immunity 2012, 80, (5), 1868–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebaihia M; Wren BW; Mullany P; Fairweather NF; Minton N; Stabler R; Thomson NR; Roberts AP; Cerdeno-Tarraga AM; Wang H; Holden MT; Wright A; Churcher C; Quail MA; Baker S; Bason N; Brooks K; Chillingworth T; Cronin A; Davis P; Dowd L; Fraser A; Feltwell T; Hance Z; Holroyd S; Jagels K; Moule S; Mungall K; Price C; Rabbinowitsch E; Sharp S; Simmonds M; Stevens K; Unwin L; Whithead S; Dupuy B; Dougan G; Barrell B; Parkhill J, The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet 2006, 38, (7), 779–86. [DOI] [PubMed] [Google Scholar]
- 28.Delvecchio VG; Connolly JP; Alefantis TG; Walz A; Quan MA; Patra G; Ashton JM; Whittington JT; Chafin RD; Liang X; Grewal P; Khan AS; Mujer CV, Proteomic profiling and identification of immunodominant spore antigens of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Appl Environ Microbiol 2006, 72, (9), 6355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X; Wang D; Ren J; Tong C; Feng E; Wang X; Zhu L; Wang H, Identification of the immunogenic spore and vegetative proteins of Bacillus anthracis vaccine strain A16R. PLoS One 2013, 8, (3), e57959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Permpoonpattana P; Tolls EH; Nadem R; Tan S; Brisson A; Cutting SM, Surface layers of Clostridium difficile endospores. J Bacteriol 2011, 193, (23), 6461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Permpoonpattana P; Phetcharaburanin J; Mikelsone A; Dembek M; Tan S; Brisson MC; La Ragione R; Brisson AR; Fairweather N; Hong HA; Cutting SM, Functional Characterization of Clostridium difficile Spore Coat Proteins. J Bacteriol 2013, 195, (7), 1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saujet L; Pereira FC; Serrano M; Soutourina O; Monot M; Shelyakin PV; Gelfand MS; Dupuy B; Henriques AO; Martin-Verstraete I, Genome-Wide Analysis of Cell Type-Specific Gene Transcription during Spore Formation in Clostridium difficile. PLoS Genet 2013, 9, (10), e1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janoir C; Deneve C; Bouttier S; Barbut F; Hoys S; Caleechum L; Chapeton-Montes D; Pereira F; Henriques A; Collignon A; Monot M; Dupuy B, Insights into the adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 2013, 81, (10), 3757–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorg JA; Sonenshein AL, Bile salts and glycine as co-germinants for Clostridium difficile spores. J Bacteriol 2008, 190, (7), 2505–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sztukowska M; Bugno M; Potempa J; Travis J; Kurtz DM Jr., Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol 2002, 44, (2), 479–88. [DOI] [PubMed] [Google Scholar]
- 36.Drake HL; Hu SI; Wood HG, Purification of five components from Clostridium thermoaceticum which catalyze synthesis of acetate from pyruvate and methyltetrahydrofolate. Properties of phosphotransacetylase. J Biol Chem 1981, 256, (21), 11137–44. [PubMed] [Google Scholar]
- 37.Thauer RK, CO(2)-reduction to formate by NADPH. The initial step in the total synthesis of acetate from CO(2) in Clostridium thermoaceticum. FEBS Lett 1972, 27, (1), 111–115. [DOI] [PubMed] [Google Scholar]
- 38.Riebe O; Fischer RJ; Wampler DA; Kurtz DM Jr.; Bahl H, Pathway for H2O2 and O2 detoxification in Clostridium acetobutylicum. Microbiology 2009, 155, (Pt 1), 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehmann Y; Meile L; Teuber M, Rubrerythrin from Clostridium perfringens: cloning of the gene, purification of the protein, and characterization of its superoxide dismutase function. J Bacteriol 1996, 178, (24), 7152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung MC; Tonry JH; Narayanan A; Manes NP; Mackie RS; Gutting B; Mukherjee DV; Popova TG; Kashanchi F; Bailey CL; Popov SG, Bacillus anthracis interacts with plasmin(ogen) to evade C3b-dependent innate immunity. PLoS One 2011, 6, (3), e18119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal S; Kulshreshtha P; Bambah Mukku D; Bhatnagar R, alpha-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim Biophys Acta 2008, 1784, (7–8), 986–94. [DOI] [PubMed] [Google Scholar]
- 42.Liu H; Bergman NH; Thomason B; Shallom S; Hazen A; Crossno J; Rasko DA; Ravel J; Read TD; Peterson SN; Yates J 3rd; Hanna PC, Formation and composition of the Bacillus anthracis endospore. J Bacteriol 2004, 186, (1), 164–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strong PC; Fulton KM; Aubry A; Foote S; Twine SM; Logan SM, Identification and characterization of glycoproteins on the spore surface of Clostridium difficile. J Bacteriol 2014, 196, (14), 2627–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu CQ; Nuttall SD; Tran H; Wilkins M; Streltsov VA; Alderton MR, Construction, crystal structure and application of a recombinant protein that lacks the collagen-like region of BclA from Bacillus anthracis spores. Biotechnol Bioeng 2008, 99, (4), 774–82. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.