Abstract
Drug addiction is a chronic relapsing brain disorder that is characterized by compulsive drug seeking and continued use despite negative outcomes. Current pharmacological therapies target neuronal receptors or transporters upon which drugs of abuse act initially, yet these treatments remain ineffective for most individuals and do not prevent disease relapse after abstinence. Drugs of abuse, in addition to their acute effects, cause persistent plasticity after repeated use, involving dysregulated gene expression in the brain’s reward regions, which are thought to mediate the persistent behavioral abnormalities that characterize addiction. Emerging evidence implicates epigenetic priming as a key mechanism that underlies the long-lasting alterations in neuronal gene regulation, which can remain latent until triggered by re-exposure to drug-associated stimuli or the drug itself. Thus, to effectively treat drug addiction, we must identify the precise epigenetic mechanisms that establish and preserve the drug-induced pathology of the brain reward circuitry.
TRANSCRIPTIONAL PRIMING AS A DISEASE MECHANISM IN DRUG ADDICTION
Drug addiction is a debilitating neuropsychiatric disorder with severe health, financial, and societal consequences (Leshner 1997). The urgent need for advanced mechanistic insight into this complex disorder is driven by sharp increases in drug use and ineffective conventional pharmacotherapeutics (Bose et al. 2018). Drug addiction is characterized by dysregulated learning involving drugs of abuse and their predictive cues, associated with long-lasting changes at the molecular, cellular, and circuit levels that lead to lifelong behavioral abnormalities and relapse, even after prolonged periods of abstinence (Kreek et al. 2005; Walker et al. 2015; Koob and Volkow 2016). Over the past decades, there have been tremendous steps forward in our understanding of the neurobiology of addiction, and there is clear evidence, both in humans and in animal models, that changes in gene expression and epigenetic regulation contribute importantly to drug-induced maladaptive neuronal plasticity (Robison and Nestler 2011; Walker et al. 2015). Epigenetic processes are known to play key roles in the synaptic plasticity that mediates learning and memory formation, involving a multitude of chromatin-modifying enzymes and regulatory proteins that control activity-dependent transcription (Gräff and Tsai 2013; Mews et al. 2017) (see Box 1: Epigenetics and Chromatin). Similarly, chromatin-based mechanisms establish and adapt neuronal ensembles, or “engrams,” across many brain regions to encode drug-associated information and promote continued drug use (Mews and Calipari 2017; Whitaker and Hope 2018). Notably, repeated drug exposure remodels neuronal cells in a way that changes their response to future stimuli, involving altered patterns of gene regulation that persist even when the original drug-induced stimulus has long faded (Walker et al. 2015). Permanent (or at least highly stable) changes in neuronal chromatin structure are assumed to underlie such latent dysregulation of transcription, which is referred to as gene “priming” and “desensitization” (Robison and Nestler 2011; Bastle and Maze 2019). In this review, we highlight recent research findings in support of this hypothesis and further discuss ongoing investigations that explore epigenetic and transcriptional priming as a key disease mechanism in drug addiction.
BOX 1. EPIGENETICS AND CHROMATIN.
Recent years have seen spectacular advances in the field of epigenetics, and additional aspects of epigenetic control keep emerging that extend its modern definition. These qualities transcend the mechanisms that hardwire phenotypes at the cellular level during development and further implicate them in the acute regulation of dynamic gene expression that must be adapted to the ever-changing cellular environment. It is through epigenetic mechanisms that the relatively fixed genetic blueprint can flexibly accommodate variability resulting from environmental stimuli, such as dietary signals, stress, and life experiences, among other influences. The primary focus of the field has been to illuminate the mechanistic layers that underlie environment–genome interaction at the level of chromatin. Chromatin describes the DNA–protein packaging complex that determines the accessibility of DNA in eukaryotic cells, making it the focal point of transcriptional gene regulation. The basic repeating unit of chromatin structure is the nucleosome: 146 base pairs of genomic DNAwrapped around a protein octamer, assembled from two molecules each of histones H2A, H2B, H3, and H4. The nucleosome constitutes a platform for complex chemical modifications—that is, “epigenetic marks”—that dynamically regulate chromatin architecture and gene transcription (Rivera and Ren 2013). Each histone protein can undergo numerous posttranslational modifications (e.g., acetylation, methylation, phosphorylation, ubiquitination, or SUMOylation) that not only alter the structure of the nucleosome but also change the interaction of DNA with the associated histones and other regulatory proteins, thus increasing or decreasing the likelihood of transcription at a given locus. The entirety of these epigenetic features has been denoted the epigenome, which defines neuronal identity and expresses the regulatory channels that operate at the interface of genome and environment (Day and Sweatt 2011). Although it is thought that a multitude of modifications in histones, DNA, and associated regulatory proteins, and their interactions, are involved in the acquisition and maintenance of the addicted phenotype, histone acetylation and methylation are currently the most extensively studied posttranslational modifications in the addiction field. The enzymes that “write” and “erase” these dynamic epigenetic modifications have emerged as key players in drug addiction, even gaining attention as potential therapeutic targets to prevent or reverse drug-induced changes in neuronal chromatin.
DRUGS OF ABUSE CORRUPT “BRAIN REWARD REGIONS”
A continuing focus of studies examining the molecular pathology of drug addiction has been the exploration of mechanisms that maintain changes in the brain’s reward circuitry over long periods of time. Neural circuits of reward integrate information across a wide range of sensory modalities and balance that information with ever-changing internal states to control and effect appropriate behaviors. However, in drug addiction, these systems become dysregulated in an activity-dependent manner. The enhanced activation of the dopaminergic system and related pathways by drugs of abuse leads to the remodeling of reward circuits, which involves stable changes in synaptic connectivity and strength that, in still incompletely understood ways, foster continued drug seeking and relapse. These long-lasting alterations in circuit dynamics raise a challenging question: How are changes at the cellular and circuit levels maintained when the primary drug-induced signaling and transcriptional events are exceptionally dynamic and short-lived? New evidence suggests that activity-dependent, transient changes in chromatin structure and gene expression create a platform for long-term epigenetic adaptations. These adaptations include posttranslational modifications of histone proteins and of the DNA itself, resulting in altered epigenetic signatures that demarcate genes with key functions in synaptic remodeling and that act to regulate genes on a timescale that extends far beyond the initial transient drug-associated signals. However, the interplay between short- and long-lived epigenetic effects is relatively unstudied, and better insight into how transient changes in chromatin structure lead to long-lasting epigenetic regulation of gene expression in the context of drug exposure remains a critical objective in the field.
DRUG ADDICTION IS LINKED TO CHANGES IN NEURONAL PLASTICITY
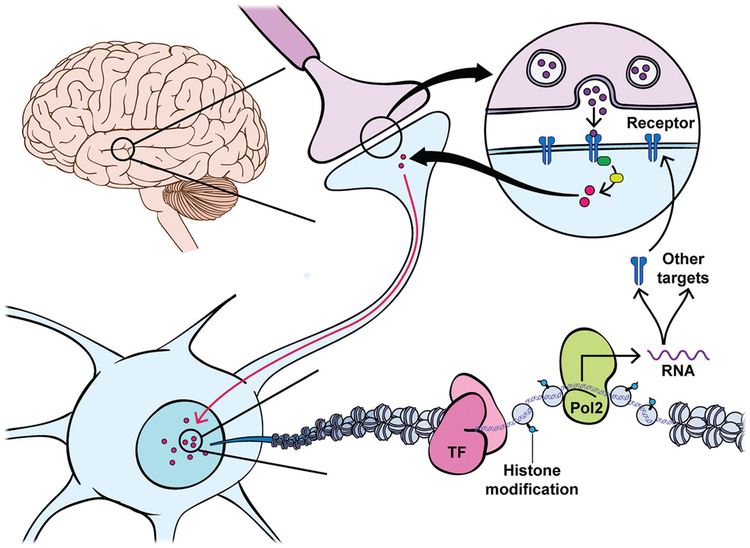
Although their initial mechanisms of action differ, all drugs of abuse increase dopaminergic transmission from VTA dopamine (DA) neurons into the striatum. In particular, the nucleus accumbens (NAc), part of the ventral striatum, is important for the integration of information in response to salient stimuli through coordination of the cellular response to DA released from the VTA and to glutamatergic inputs from several reward-associated cortical and thalamic regions. The NAc is primarily composed (>90%) of medium spiny neurons (MSNs), which are categorized by their expression of different DA receptors (Drds), either Drd1 (D1)- or Drd2 (D2)-containing cells (Surmeier et al. 2007). Drds are G protein–coupled receptors and differ in their activation (Gs for D1) or inhibition (Gi for D2) of cyclic AMP and ultimately protein kinase A (PKA), thus leading to the activation (D1) or inhibition (D2) of DA-sensitive cells within the NAc. Chronic drug exposure and activation of D1 neurons enhances the synaptic connectivity of glutamatergic inputs onto this neuronal population, which, coupled to elevated ratios of AMPA to NMDA receptors during prolonged withdrawal (Pierce et al. 1996; Reid and Berger 1996; Baker et al. 2003; Kourrich et al. 2007; Conrad et al. 2008; Kau et al. 2008), increases the responsiveness of the neurons to glutamatergic projections from the extended reward circuitry. In addition to affecting glutamate receptors, DA binding its receptors activates second messenger pathways that ultimately influence transcription through signaling cascades within the cell. This process is crucial for the integration of information in response to rewarding stimuli, including drugs of abuse, and leads to transcriptional changes within each cell type that influence subsequent exposures to drugs of abuse (Fig. 1; Robison and Nestler 2011). It is thought that such transcriptional changes within the NAc are important for the transition to different stages of addiction from use, to abuse, to substance use disorder and relapse (i.e., addiction). Therefore, a large body of work is devoted to understanding the transcriptional changes within the NAc at different stages of the “addiction cycle.”
Figure 1.
Gene expression and its spatiotemporal regulation are central to the development of drug addiction. Activity-dependent changes at the circuit level remodel chromatin within individual neurons to regulate gene expression involved in synaptic and neural plasticity. Circuit activity and neurotransmitter release trigger intracellular signaling cascades such as the PKA or MAP-kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathways that are activated by G protein–coupled receptors, and activated calcium pathways that occur upon receptor stimulation or neural activity, and transmit circuit activity information to the cell nucleus. In the nucleus, DNA is organized by wrapping around histone octamers to form nucleosomes. Only by temporarily unraveling compacted chromatin can the DNA of a specific gene be made accessible to the transcriptional machinery. This process involves the recruitment of chromatin remodeling and modifying enzymes that mediate acute and transient gene expression in response to upstream neural activity. Induction of immediate early genes (e.g., cFos, Egr1, NPas4, Arc) leads to the activation or inhibition of many other transcription factors and nuclear targets, including chromatin-regulatory proteins, that ultimately alter the composition or levels of membrane receptors and many other classes of proteins in neuronal signaling pathways, leading to changes in the excitability and structural connectivity of neurons in the reward circuitry.
Early studies investigating these transcriptional changes discovered that the transcription factor ΔFosB is induced in the NAc and other brain reward regions by chronic drug exposure and accumulates to high levels of expression because of the protein’s unusual stability (Robison and Nestler 2011). This distinct member of the AP1 family of transcription factors presented an attractive candidate mechanism for how drug-induced transient gene expression can be integrated to produce long-lasting changes in gene regulation. Notably, whereas ΔFosB protein levels gradually recover to normal during prolonged periods of withdrawal, several studies have shown that certain drug-induced changes in the epigenetic landscape persist for longer periods of time and remain stable even after prolonged withdrawal. Akin to epigenetic control of cellular differentiation, chromatin-based mechanisms could maintain drug-induced neuronal adaptations over extended periods of time that far exceed the turnover rate of any individual protein or synapse involved. Recent studies, including research from our own laboratory outlined below, have revealed that transcriptional priming occurs in NAc and other reward-related brain regions following repeated exposure to drugs of abuse and involves key genes that regulate AMPA and NMDA receptor levels and their subunit composition, among many other classes of synaptic and neural-signaling proteins, to produce plasticity at both the synaptic and circuit levels.
COCAINE INDUCES LONG-LASTING CHANGES IN GENE EXPRESSION
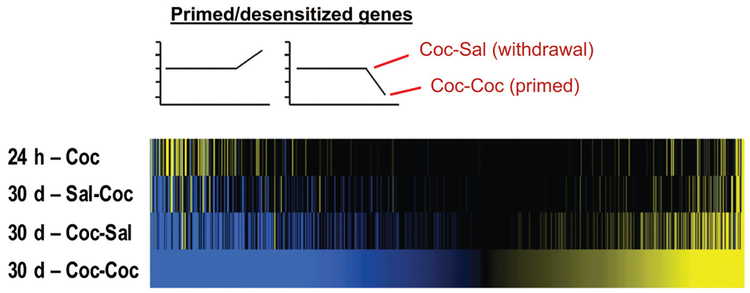
A recent study from our laboratory used RNA sequencing to identify transcriptome-wide changes within the NAc and five other brain reward regions, after short-and long-term withdrawal (1 and 30 d, respectively) from cocaine self-administration in an unbiased manner (Walker et al. 2018). To determine how the transcriptional profile was affected by re-exposure to drug-paired context or re-exposure to the context plus drug, animals were given an intraperitoneal injection of saline or cocaine and returned to their original self-administration chamber after long-term withdrawal. One important finding from this study was the identification of expression changes in large sets of genes within the NAc that occurred upon re-exposure to the drug-paired context alone or to drug plus context or both (Fig. 2). Interestingly, many of these genes were also affected by an animal’s past exposure to cocaine, but with significantly increased or decreased magnitudes of regulation seen at the prolonged withdrawal time point. These data reveal those transcripts that are either primed or desensitized within the NAc by past cocaine experience. Similar patterns of regulation were seen in the other brain regions studied, although with mostly distinct genes affected in each region. We next deduced predicted upstream transcriptional regulators of sets of genes that display similar patterns of expression across the different experimental conditions of the study. Among the highest ranked transcription factors are AP1—validating prior work with ΔFosB cited above—and cAMP response element-binding protein (CREB), also implicated previously in the actions of drugs of abuse (Robison and Nestler 2011). As well, this analysis suggested the prominent involvement of several additional transcription factors, such as E2F and nuclear receptor family proteins, which had not to date been implicated in addiction. The validity of these bioinformatics predictions was verified by a subsequent study, which indeed showed directly the essential role played by E2F3a, in particular, in the NAc in mediating the transcriptional and behavioral actions of cocaine (Cates et al. 2018). Importantly, each identified transcription factor appears to target largely different sets of transcripts under different experimental conditions (e.g., context re-exposure vs. drug plus context re-exposure) within a given brain region. This finding adds credence to the hypothesis that epigenetic mechanisms change the chromatin landscape to influence the inducibility of transcription mediated by key transcription factors within the NAc after cocaine self-administration (Fig. 3; Walker et al. 2018).
Figure 2.
Patterns of time-dependent gene regulation in NAc after cocaine self-administration. The heatmaps show results from RNA-seq analyses of NAc after short- (24 h) or long-term (30 d) withdrawal from chronic self-administration of cocaine. Animals subjected to long-term withdrawal were given an intraperitoneal injection of saline or cocaine and placed back in their original chambers to determine if withdrawal plus context or context/drug re-exposure resulted in transcriptional priming or desensitization of genes. For both re-exposure paradigms, gene expression changes were mostly observed in magnitude and not direction. By applying the analytical technique of pattern analysis, unique gene lists were generated of transcripts that are regulated significantly and uniquely under specific conditions. For example, in the figure, only genes altered by cocaine/context re-exposure to a significantly greater extent than all other conditions are presented. Given that this is a unique list of genes that are up- and down-regulated by cocaine/context re-exposure but not context re-exposure alone, we hypothesize that these genes are primed or desensitized during withdrawal and activated or suppressed by re-exposure to the drug. Upstream regulator analysis predicted CREB and several nuclear receptors as the most prominent transcriptional regulators of these genes, suggesting an important role for these transcription factor families in priming/desensitizing the transcriptional response to cocaine in the NAc. (24 h – Coc) 24 hours after chronic cocaine self-administration, (30 d – Sal-Coc) 30 days after saline self-administration with an acute cocaine challenge (reflects the effects of acute cocaine exposure), (30 d – Coc-Sal) 30 days after cocaine self-administration with an acute saline challenge (reflects the effects of prolonged withdrawal plus context re-exposure), (30 d – Coc-Coc) 30 days after cocaine self-administration with an acute cocaine challenge (reflects the effects of prolonged withdrawal plus context and drug re-exposure). All conditions were compared to the same control group: 24 h after chronic saline self-administration. (Adapted from Walker et al. 2018, with permission from Elsevier, © 2018 Society of Biological Psychiatry.)
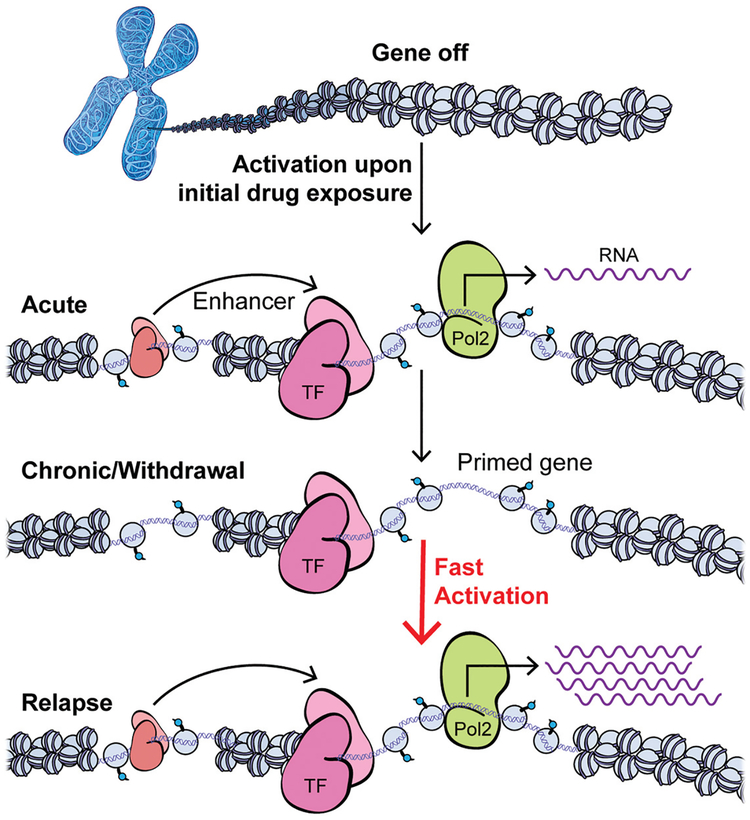
Figure 3.
Time-dependent control of gene regulation by drug exposure via epigenetic mechanisms. Chromatin mechanisms not only regulate the acute transcriptional response to drug exposure but further mediate latent effects at many genes that alter their inducibility to future stimuli. Transcription initiates within core promoters at the transcriptional start sites (TSSs) at 5′ ends of genes, which recruit RNA polymerase II (Pol2) and determine the accurate initiation position and direction of transcription. Efficient transcription is supported by enhancer elements (located at some distance from their target genes) that contain transcription factor (TF; “salmon”) binding sites and recruit a combination of TFs with a variety of cofactors to exert their overall regulatory function to control transcription from the targeted core promoter. Neuronal activation upon the initial drug exposure triggers intracellular signaling cascades that activate TFs and many other nuclear targets, including chromatin-regulatory proteins that modify histones and other proteins to regulate DNA accessibility, as well as transcription initiation and elongation. Early evidence suggests that chronic drug exposure causes extremely stable changes at the chromatin level that underlie transcriptional priming (shown) and desensitization (not shown) linked to drug addiction. Such gene priming/desensitization may remain latent during periods of withdrawal, when certain TFs and cofactors are bound to accessible chromatin without changing the steady state mRNA levels. However, future context and/or drug re-exposure can up-regulate a primed gene much faster and to a greater extent, based on the epigenetic changes induced by previous chronic drug exposure at regulatory promoter and enhancer regions.
We also used a data reduction technique, called exploratory factor analysis, to identify key components of self-administration behavior that are associated with specific patterns of gene regulation (Walker et al. 2018). We then combined the factor values within individual animals that reflected specific aspects of “addiction” to produce an “addiction index” for each animal in the study. The power of this technique was twofold: (1) animals were separated based on their performance during self-administration including the animals who self-administered saline, and (2) for the first time, genes were identified that were associated with addiction-related behaviors in an unbiased manner. Within the NAc, we found that genes associated with the addiction index were induced or suppressed to a greater extent after long-term withdrawal, suggesting once again that transcriptional correlates of reward-associated behaviors are primed or desensitized by volitional cocaine exposure. Several of the same transcription factors were predicted as upstream regulators of genes associated with the addiction index.
We observed significant overlap of those genes associated with behavior and those that were uniquely regulated by re-exposure to cocaine after prolonged withdrawal, within the NAc and other brain regions studied. In silico analysis revealed that overlapping genes were enriched for binding motifs for many of the same transcription factors (noted above) within their promoters. This finding led us to hypothesize that transcriptional reprogramming takes place during long-term withdrawal, which is associated with the degree of the prior addicted phenotype. Of interest is the observation that two of the most strongly implicated transcription factors in this analysis, CREB and nuclear receptors, have recently been identified as critical and co-dependent regulators of long-term potentiation (LTP) in the hippocampus (Bridi et al. 2017), thus supporting the hypothesis that these two transcription factor families are particularly important for transcriptional priming/desensitization during long-term abstinence from drug exposure.
CHROMATIN REGULATION IN DRUG ADDICTION
Investigations into how cocaine and other drugs of abuse dysregulate gene expression over long periods of time are critical to develop a mechanistic understanding of the molecular processes leading to drug addiction. As highlighted above, cocaine and other drugs of abuse lead to altered transcriptional states through the activation of several transcription factors. As one example, CREB is activated by several protein kinases, including PKA, Ca2+/calmodulin-dependent protein kinase IV (CaMKIV), and ribosomal S6 kinase (RSK). It is well established that activated CREB recruits other transcriptional coactivators that remodel the chromatin structure of gene-regulatory promoter and enhancer elements. For instance, following cocaine self-administration, rapid increases in CREB activity are linked to dynamic increases in promoter-proximal histone acetylation in the NAc (Kumar et al. 2005) at the Cdk5, Bdnf, and Fosb genes, all of which have key functions in neural plasticity and are induced in this region by several drugs of abuse (Hope et al. 1994; Bibb et al. 2001; Graham et al. 2007). Similar histone acetylation has been implicated during learning and memory formation and associated with induction of genes with key roles in hippocampal plasticity and LTP (Gräff and Tsai 2013).
Over the past decade, there has been a surge in research examining the complex epigenetic regulation that occurs in drug addiction, most especially in rodent models of cocaine addiction. Most evidence on the role of epigenetic processes in drug addiction derives from studies focused on histone acetylation, often by modifying the opposing activities of two families of enzymes—histone acetyltransferases (HATs) that acetylate histones and histone deacetylases (HDACs) that remove acetyl groups. For example, several studies using HDAC inhibitors (HDACis) showed that increased histone acetylation in NAc or other brain reward regions affects several addiction-related behaviors, including place conditioning, drug consumption, and withdrawal-related anxiety (Kumar et al. 2005; Rogge and Wood 2013; Walker et al. 2015). Generally, HAT enzymes acetylate and, therefore, neutralize the positive charge of specific histone lysine residues to “open” the chromatin structure at enhancers and gene promoters, leading to a permissive transcriptional state of chromatin and to active gene transcription (Berndsen and Denu 2008; Mews et al. 2017). Additionally, histone acetylation marks can be bound by protein subunits containing bromodomains, which are conserved within many chromatin-associated factors that have been found to regulate transcription-mediated processes in health and disease (Bannister and Kouzarides 2011; Filippakopoulos and Knapp 2014).
Although the effects of drugs of abuse are widespread, a picture of how histone acetylation changes throughout the brain after exposure to different drugs of abuse such as cocaine (Rogge and Wood 2013), morphine (Wang et al. 2014), and nicotine (Huang et al. 2013) is beginning to emerge. In general, acute or repeated exposure to drugs of abuse increases total tissue levels of acetylation of histones H3 and H4 in several brain reward regions, particularly in the NAc (Kumar et al. 2005; Schroeder et al. 2008; Chi et al. 2010; Levine et al. 2011; Sheng et al. 2011; Kennedy et al. 2013). Chromatin immunoprecipitation (ChIP) has been used to detect histone acetylation at selected candidate genes throughout the brain’s reward circuitry and a common set of genes that are regulated by drug-induced acetylation have been identified. These genes include, among others, immediate early genes such as cFos, Fosb, Bdnf, Cdk5, and Cpb, all of which are controlled by the transcription factor CREB (Nestler 2014). Intriguingly, as CREB is activated through Ca2+-dependent signaling, increasing levels of Ca2+-permeable AMPA receptors that result from chronic cocaine exposure (Dong and Nestler 2014) may provide a potential mechanism by which histone acetylation controls plasticity in a feed-forward loop to potentiate transcriptional responses to drug-associated stimuli. In this way, increases in Ca2+ per-meability and potentiated signaling through ΔFosB, CREB, and other key transcription factors that control the activity of chromatin-modifying enzymes could have lasting effects on the epigenetic landscape, causing the chromatin structure at key neuronal genes to be remodeled for long periods of time. However, although numerous studies suggest that cocaine and other drugs of abuse modulate several histone modifications to elicit transcriptionally active states and persistent changes in neuronal plasticity, most evidence linking epigenetic regulation to drug addiction remains indirect and fragmentary. Only recently has it become possible to experimentally target a specific type of histone or other chromatin modification to a single gene locus in a single type of neuron in a given brain region in vivo and show directly that such regulation is indeed sufficient to alter the transcriptional state of the targeted genes (Hamilton et al. 2018).
To improve our understanding of how epigenome modification dysregulates gene expression and promotes drug addiction, it is necessary to survey these changes genome-wide. An earlier study using the ChIP-chip method with antibodies against pan-acetylated H3 or H4 followed by promoter microarrays provided an initial, more complete map of genes in NAc that display altered histone H3 and H4 acetylation after chronic cocaine exposure (Renthal et al. 2009). Interestingly, whereas a large number of genes were found to be hyperacetylated following chronic cocaine administration, there was little overlap of genes displaying alterations of H3ac and H4ac, and most genes did not follow the expected expression pattern predicted by the altered histone modifications. These data indicate that additional gene-regulatory mechanisms work in concert with histone acetylation to control drug-induced changes in gene expression and alternative splicing, with histone acetylation contributing just a fraction of all epigenetic information controlling gene activity. Histone methylation is another well-characterized histone modification that can be associated with either activation or repression of gene expression depending on which lysine residues are methylated and the number of methyl groups added. Translating the role of histone methylation and its responses to drugs of abuse is therefore much more complex than histone acetylation, but several studies have associated the histone methylation state at gene promoters with controlling both transcription and splicing of genes involved in neural plasticity.
Whereas most of the work on drugs of abuse has focused on the role of repressive histone methylation in addiction, a prior study from our group used ChIP-seq coupled with RNA-seq to investigate how combinations of several methylation marks—H3K4me1, H3K4me3, and H3K36me3 (activating marks) and H3K9me2, H3k9me3, and H3K27me3 (repressive marks)—correlate with altered transcript levels after exposure to repeated cocaine (Feng et al. 2014). Although global changes across the genome were not observed, cocaine was found not only to induce modifications in each of these marks at numerous genes but also at putative enhancer elements. Enhancers function as integrated transcription factor binding platforms that deliver accessory factors to gene promoters to potentiate or repress transcription initiation or the transition to elongation. Other studies have found that repeated exposure to cocaine decreases total tissue levels of H3K9me2 and H3K9me3 in NAc (Maze et al. 2010, 2011), and investigations focused on specific genes suggest that reduced binding of the histone methyltransferase G9a and consequent reduced enrichment of H3K9me2 are linked to transcriptional priming (Maze et al. 2010).
Overall, these data indicate that the transcriptional response to drugs of abuse is defined by a complex combinatory pattern of modifications, and that distinct epigenetic signatures at promoters and enhancers are linked to gene priming or desensitization. However, all existing data sets were derived from whole tissue composed of highly heterogeneous populations of cells, all of which might be expected to respond to cocaine in a different way. Therefore, to develop a more precise mechanistic understanding of drug-induced epigenetic priming, it is essential to achieve the selective analysis of chromatin alterations in distinct neuronal subtypes, which has only recently become feasible technically for brain.
DISTINCT ROLES OF D1 AND D2 NAc MSNs IN DRUG ADDICTION
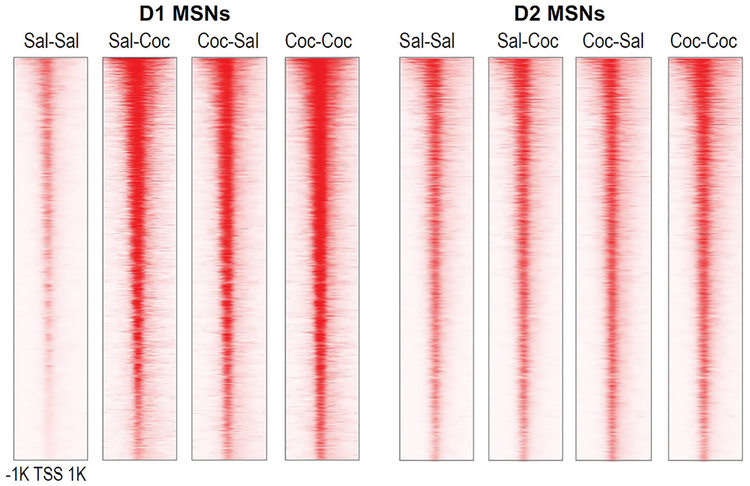
Ongoing studies of chromatin regulation in addiction models support the view that latent epigenetic changes alter the inducibility of many genes in the NAc, suggesting that cocaine-induced alterations in chromatin architecture “scar” gene-regulatory regions to permanently prime or desensitize genes for more rapid or greater degrees of induction or suppression in response to future stimuli (see Fig. 3). An important challenge is to determine which MSN subtypes are responsible. D1 and D2 MSNs in the NAc receive overlapping projections from cortical and sensory areas, and there is ample evidence that the two neuronal subtypes respond differently to both acute and chronic cocaine (Lobo et al. 2010; Calipari et al. 2016). For example, repeated cocaine exposure induces ΔFosB solely in D1 MSNs, and not in D2 MSNs, and ultimately appears to change the balance of inputs onto each of these striatal populations, leading to increased activity of D1 MSNs and perhaps decreased activity of D2 MSNs (Grueter et al. 2013). Therefore, it is imperative to investigate cocaine-induced alterations in chromatin structure in D1 and D2 MSNs separately. In recent years, the assay for transposase-accessible chromatin using sequencing (ATAC-seq) has become a fundamental tool of epigenomic research and is used to assess chromatin structure genome-wide to detect “open” chromatin regions, which can be indicative of active gene transcription or priming (Buenrostro et al. 2015). We defined chromatin accessibility genome-wide in D1 and D2 MSNs of the NAc using fluorescence-activated cell sorting (FACS) coupled to ATAC-seq after acute or chronic cocaine exposure and after prolonged withdrawal (Fig. 4; Mews et al. 2018). These data show that chromatin is overall less accessible in D1 MSNs than in D2 MSNs at baseline; however, a genome-wide “opening” of chromatin occurs selectively in D1 MSNs upon acute exposure to cocaine. Intriguingly, this D1-specific chromatin opening is sustained during prolonged withdrawal from chronic cocaine exposure and linked to even greater increases in chromatin accessibility upon an acute challenge injection of the drug. These data, although preliminary, indicate that remodeling of the D1 MSN epigenome may be particularly important for the persisting effects of cocaine-induced changes in gene regulation. We hypothesize that chronic cocaine exposure alters the gene-regulatory chromatin landscape especially in D1 MSNs in a way that primes cocaine-induced gene expression programs, which we posit are coupled to enduring changes in synaptic connectivity and behavior.
Figure 4.
Cell type–specific control of chromatin accessibility in NAc by acute and chronic cocaine exposure. ATAC-seq was performed on D1 and D2 MSNs isolated from NAc using FACS in two transgenic mouse lines that express EGFP-RPL10a in either subtype (Drd1a/Drd2a::EGFP-L10a). Each horizontal row reflects a single gene locus centered around its TSS and 1 kb up- and downstream. This ongoing study investigates immediate and long-term changes in chromatin architecture, following acute cocaine (Sal-Coc, 1 h after 20 mg/kg cocaine by intraperitoneal injection) and prolonged withdrawal after chronic cocaine exposure (Coc-Sal, 30 d following 10 d of cocaine injections) as well as after drug challenge in withdrawal animals (Coc-Coc, 1 h after cocaine challenge). Drug-induced changes in chromatin accessibility discriminate D1 from D2 MSNs, and D1-specific chromatin “opening” following chronic cocaine exposure is sustained even during prolonged periods of withdrawal.
Another important open question is: How do drug-induced patterns of epigenetic priming persist and escape the actions of normal chromatin-regulatory mechanisms? A wealth of evidence supports a strong (~50%) genetic component of susceptibility to addiction, thus advocating a more complex model of gene–environment interaction. Accordingly, increased disease susceptibility may be linked to detrimental genetic factors that remain harmless until chronic drug exposure or additional environmental insults (e.g., chronic stress). In other cases, environmental stressors during early development and adolescence may amplify the vulnerability of adults to drug addiction through epigenetic priming leading to unfavorable gene expression upon initial and repeated drug exposure. As new tools are developed to survey dynamic changes in chromatin structure within specific neural networks and engrams over time, future studies will reveal a more comprehensive view of how different epigenetic marks and circuit stability are persistently altered by drug exposure across the brain. As epigenetic aberrations may be reversible, a better mechanistic understanding of such chromatin scarring by drugs of abuse is anticipated to provide novel therapeutic avenues and pave the way to epigenetic interventions to treat drug addiction.
REFERENCES
- Baker DA, McFarland K, Lake RW, Shen H, Tang XC, Toda S, Kalivas PW. 2003. Neuroadaptations in cystine–glutamate exchange underlie cocaine relapse. Nat Neurosci 6: 743–749. doi: 10.1038/nn1069 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 1–15. doi: 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastle RM, Maze IS. 2019. Chromatin regulation in complex brain disorders. Curr Opin Behav Sci 25: 57–65. doi: 10.1016/j.cobeha.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Denu JM. 2008. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr Opin Struct Biol 18: 682–689. doi: 10.1016/j.sbi.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb JA, Chen J, Taylor JR, Svenningsson P, Nishi A, Snyder GL, Yan Z, Sagawa ZK, Ouimet CC, Nairn AC, et al. 2001. Effects of chronic exposure to cocaine are regulated by the neuronal protein Cdk5. Nature 410: 376–380. doi: 10.1038/35066591 [DOI] [PubMed] [Google Scholar]
- Bose J, Hedden SL, Lipari RN, Park-Lee E, Tice P. 2018. Use and mental health indicators in the United States: results from the 2017 National Survey on Drug Use and Health. Substance Abuse and Mental Health Services Administration (HHS Publication No. SMA 18–5068, NSDUH Series H-53), Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration, Rockville, MD: Retrieved from https://www.samhsa.gov/data/. [Google Scholar]
- Bridi MS, Hawk JD, Chatterjee S, Safe S, Abel T. 2017. Pharmacological activators of the NR4A nuclear receptors enhance LTP in a CREB/CBP-dependent manner. Neuropsychophar-macology 42: 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Prot Mol Biol 109: 21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calipari ES, Bagot RC, Purushothaman I, Davidson TJ, Yorgason JT, Peña CJ, Walker DM, Pirpinias ST, Guise KG, Rama-krishnan C, et al. 2016. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc Natl Acad Sci 113: 2726–2731. doi: 10.1073/pnas.1521238113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates HM, Heller EA, Lardner CK, Purushothaman I, Peña CJ, Walker DM, Cahill ME, Neve RL, Shen L, Bagot RC, et al. 2018. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol Psychiatry 84: 167–179. doi: 10.1016/j.biopsych.2017.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. 2010. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10: 457–469. doi: 10.1038/nrc2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad KL, Tseng KY, Uejima JL, Reimers JM, Heng LJ, Shaham Y, Marinelli M, Wolf ME. 2008. Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature 454: 118–121. doi: 10.1038/nature06995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. 2011. Epigenetic mechanisms in cognition. Neuron 70: 813–829. doi: 10.1016/j.neuron.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Nestler EJ. 2014. The neural rejuvenation hypothesis of cocaine addiction. Trends Pharmacol Sci 35: 374–383. doi: 10.1016/j.tips.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, et al. 2014. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol 15: R65. doi: 10.1186/gb-2014-15-4-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. 2014. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13: 337–356. doi: 10.1038/nrd4286 [DOI] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. 2013. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci 14: 97–111. doi: 10.1038/nrn3427 [DOI] [PubMed] [Google Scholar]
- Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW. 2007. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci 10: 1029–1037. doi: 10.1038/nn1929 [DOI] [PubMed] [Google Scholar]
- Grueter BA, Robison AJ, Neve RL, Nestler EJ, Malenka RC. 2013. ΔFosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc Natl Acad Sci 110: 1923–1928. doi: 10.1073/pnas.1221742110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton PJ, Lim CJ, Nestler EJ, Heller EA. 2018. Neuroepigenetic editing. Methods Mol Biol 1767: 113–136. doi: 10.1007/978-1-4939-7774-1_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Nye HE, Kelz MB, Self DW, Iadarola MJ, Nakabeppu Y, Duman RS, Nestler EJ. 1994. Induction of a long-lasting AP-1 complex composed of altered Fos-like proteins in brain by chronic cocaine and other chronic treatments. Neuron 13: 1235–1244. doi: 10.1016/0896-6273(94)90061-2 [DOI] [PubMed] [Google Scholar]
- Huang YY, Kandel DB, Kandel ER, Levine A. 2013. Nicotine primes the effect of cocaine on the induction of LTP in the amygdala. Neuropharmacology 74: 126–134. doi: 10.1016/j.neuropharm.2013.03.031 [DOI] [PubMed] [Google Scholar]
- Kau KS, Madayag A, Mantsch JR, Grier MD, Abdulhameed O, Baker DA. 2008. Blunted cystine–glutamate antiporter function in the nucleus accumbens promotes cocaine-induced drug seeking. Neuroscience 155: 530–537. doi: 10.1016/j.neuroscience.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han MH, et al. 2013. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci 16: 434–440. doi: 10.1038/nn.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. 2016. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry 3: 760–773. doi: 10.1016/S2215-0366(16)00104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourrich S, Rothwell PE, Klug JR, Thomas MJ. 2007. Cocaine experience controls bidirectional synaptic plasticity in the nucleus accumbens. J Neurosci 27: 7921–7928. doi: 10.1523/JNEUROSCI.1859-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. 2005. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci 8: 1450–1457. doi: 10.1038/nn1583 [DOI] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DEH, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, et al. 2005. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48: 303–314. doi: 10.1016/j.neuron.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Leshner AI. 1997. Addiction is a brain disease, and it matters. Science 278: 45–47. doi: 10.1126/science.278.5335.45 [DOI] [PubMed] [Google Scholar]
- Levine A, Huang YY, Drisaldi B, Griffin EA, Pollak DD, Xu S, Yin D, Schaffran C, Kandel DB, Kandel ER. 2011. Molecular mechanism for a gateway drug: epigenetic changes initiated by nicotine prime gene expression by cocaine. Sci Transl Med 3:107ra109. doi: 10.1126/scitranslmed.3003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, Dietz DM, Zaman S, Koo JW, Kennedy PJ, et al. 2010. Cell type–specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330: 385–390. doi: 10.1126/science.1188472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, et al. 2010. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327: 213–216. doi: 10.1126/science.1179438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. 2011. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci 108: 3035–3040. doi: 10.1073/pnas.1015483108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Calipari ES. 2017. Cross-talk between the epigenome and neural circuits in drug addiction. Prog Brain Res 235: 19–63. doi: 10.1016/bs.pbr.2017.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL. 2017. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546: 381–386. doi: 10.1038/nature22405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Kronman H, Ramakrishnan A, Sidoli S, Garcia B, Shen L, Nestler E. 2018. Mechanisms of epigenetic priming in cocaine addiction. Soc Neurosci Abs 161.04. [Google Scholar]
- Nestler EJ. 2014. Epigenetic mechanisms of drug addiction. Neuropharmacology 76: 259–268. doi: 10.1016/j.neuropharm.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce RC, Bell K, Duffy P, Kalivas PW. 1996. Repeated cocaine augments excitatory amino acid transmission in the nucleus accumbens only in rats having developed behavioral sensitization. J Neurosci 16: 1550–1560. doi: 10.1523/JNEUROSCI.16-04-01550.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid MS, Berger SP. 1996. Evidence for sensitization of cocaine-induced nucleus accumbens glutamate release. Neuroreport 7: 1325–1329. doi: 10.1097/00001756-199605170-00022 [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, et al. 2009. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron 62: 335–348. doi: 10.1016/j.neuron.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CM, Ren B. 2013. Mapping human epigenomes. Cell 155: 39–55. doi: 10.1016/j.cell.2013.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. 2011. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci 12: 623–637. doi: 10.1038/nrn3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Wood MA. 2013. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology 38: 94–110. doi: 10.1038/npp.2012.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder FA, Penta KL, Matevossian A, Jones SR, Konradi C, Tapper AR, Akbarian S. 2008. Drug-induced activation of dopamine D(1) receptor signaling and inhibition of class I/II histone deacetylase induce chromatin remodeling in reward circuitry and modulate cocaine-related behaviors. Neuropsychopharmacology 33: 2981–2992. doi: 10.1038/npp.2008.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Lv ZG, Wang L, Zhou Y, Hui B. 2011. Histone H3 phosphoacetylation is critical for heroin-induced place preference. Neuroreport 22: 575–580. doi: 10.1097/WNR.0b013e328348e6aa [DOI] [PubMed] [Google Scholar]
- Surmeier DJ, Ding J, Day M, Wang Z, Shen W. 2007. D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci 30: 228–235. doi: 10.1016/j.tins.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Heller EA, Nestler EJ. 2015. Regulation of chromatin states by drugs of abuse. Curr Opin Neurobiol 30: 112–121. doi: 10.1016/j.conb.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Loh YHE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, et al. 2018. Cocaine self-administration alters transcriptome-wide responses in the brain’s reward circuitry. Biol Psychiatry 84: 867–880. doi: 10.1016/j.biopsych.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lai J, Cui H, Zhu Y, Zhao B, Wang W, Wei S. 2014. Inhibition of histone deacetylase in the basolateral amygdala facilitates morphine context-associated memory formation in rats. J Mol Neurosci 55: 269–278. doi: 10.1007/s12031-014-0317-4 [DOI] [PubMed] [Google Scholar]
- Whitaker LR, Hope BT. 2018. Chasing the addicted en-gram: identifying functional alterations in Fos-expressing neuronal ensembles that mediate drug-related learned behavior. Learn Mem 25: 455–460. doi: 10.1101/lm.046698.117 [DOI] [PMC free article] [PubMed] [Google Scholar]