Abstract
Angiostrongyliasis is a parasitic disease caused by nematodes of the genus Angiostrongylus. Distribution of this worm corresponds to the dispersal of its main intermediate host, the giant African land snail Achatina fulica. Genetic characterization can help identify parasitic pathogens and control the spreading of disease. The present study describes infection of A. fulica by Angiostrongylus, and provides a genetic outlook based on sequencing of specific regions. We collected 343 land snails from 22 provinces across six regions of Thailand between May 2017 and July 2018. Artificial digestion and Baermann’s technique were employed to isolate Angiostrongylus larvae. The worm and its intermediate host were identified by sequencing with specific nucleotide regions. Phylogenetic tree was constructed to evaluate the relationship with other isolates. A. fulica from Chaiyaphum province was infected with A. cantonensis, whereas snails collected from Phrae and Chiang Rai provinces were infected with A. malaysiensis. The maximum likelihood tree based on 74 A. fulica COI sequences revealed monophyletic groups and identified two haplotypes: AF1 and AF2. Only AF1, which is distributed in all regions of Thailand, harbored the larvae of A. cantonensis and A. malaysiensis. Two mitochondrial genes (COI and cytb) and two nuclear regions (ITS2 and SSU rRNA) were sequenced in 41 Angiostrongylus specimens. The COI gene indicated that A. cantonensis was closely related to the AC10 haplotype; whereas the cytb gene revealed two new haplotypes: AC19 and AC20. SSU rRNA was useful for the identification of A. cantonensis; whereas ITS2 was a good genetic marker for differentiating between A. cantonensis and A. malaysiensis. This study provides genetic information about the parasite Angiostrongylus and its snail intermediate host. The data in this work may be useful for further study on the identification of Angiostrongylus spp., the genetic relationship between intermediate host and parasite, and control of parasites.
Introduction
Angiostrongylus is a parasitic nematode from the superfamily Metastrongyloidea [1]. To date, 21 species of this genus have been reported around the world, with A. cantonensis and A. costaricensis being the most notable [2]. A. cantonensis is the causative agent of human angiostrongyliasis associated with eosinophilic meningitis or meningoencephalitis. Ocular and neuro-angiostrongyliasis are reported as sporadic in Asian counties [3–5]. A. costaricensis causes abdominal angiostrongyliasis and most cases are reported in South America [6,7]. Other veterinary-relevant species include A. malaysiensis, A. mackerrasae, and A. vasorum, which act as animal pathogens [2,8]; although, A. malaysiensis, which is epidemic in Asian countries, may cause also human angiostrongyliasis [9]. In Thailand, A. cantonensis, A. siamensis, A. malaysiensis, and Thaistrongylus harinasuti have been recorded in several hosts [8,10]. A. cantonensis is the main causative agent of human angiostrongyliasis in Thailand, whereas A. malaysiensis is reported with increasing extent in the Greater Mekong area. To complete the life cycle, A. cantonensis and A. malaysiensis use snails and terrestrial slugs as intermediate hosts, and rodents as their final hosts [11]. Humans are an accidental host, and become infected by ingesting Angiostrongylus larvae present in snails, slugs, paratenic hosts or on contaminated vegetables [12–14]. Clinical manifestations of human angiostrongyliasis include severe headache, and neck stiffness with eosinophilic meningitis or meningoencephalitis. Most cases of the disease are reported in Thailand, Taiwan, and southern China [1].
The giant African land snail Achatina fulica is an important intermediate host for A. cantonensis [15]. In the 19th Century, this land snail was dispersed by humans across the Indian Ocean from Africa to India, Sri Lanka, and Southeast Asia [16]. In Thailand, most snails were accidentally moved across geographic locations on agricultural products or transportation containers. A. fulica is abundant in tropical climates with warm, mild year-round temperatures and high humidity [17]. The snail causes damage to vegetables and other food crops [18,19]. The spreading of A. fulica was affected also by dispersal of the rat lungworm, particularly in the Pacific [20]. Not surprisingly, the giant African land snail has been listed among the 100 worst invasive species and is considered the most damaging land snail in the world [17]. However, only a few genetic studies of A. fulica have been reported to date and none of them in Thailand.
Genetic characterization is important for the identification of parasitic pathogens, as well as to control the spreading of disease. Sequencing and phylogenetic studies of A. cantonensis based on mitochondrial or ribosomal genes have been used to identify and study the evolution and distribution of this species. Several genes or nucleotide regions from A. cantonensis have been used in genetic studies so far: 66-kDa protein [21], ribosomal transcribed spacer (ITS) regions [22,23], small subunit (SSU) ribosomal RNA (18S rRNA) [22,24,25], cytochrome c oxidase subunit I (COI) [9,26], and cytochrome b (cytb) [27,28]. In comparison, only a few studies have tried to characterize A. malaysiensis in Thailand [9]. Therefore, to gain further knowledge about the genetic make-up of Angiostrongylus spp. and its natural intermediate host, A. fulica, in Thailand, the present study sought to observe larval infection by Angiostrongylus in A. fulica. Analysis of A. cantonensis COI, SSU rRNA, ITS2, and cytb gene sequences and the ITS2 region of A. malaysiensis enabled construction of a phylogenetic tree. Finally, sequencing of the A. fulica COI gene, allowed for haplotype analysis and genetic structure characterization of this snail species.
Methods
Ethics statement
The experimental protocol for the use of animals (snail intermediate host) in this study was approved by the Center for Animal Research of Naresuan University (Project Ethics Approval No: NU-AQ610711). The biosafety protocol was approved by the Naresuan University Institutional Biosafety Committee (Project Approval No: NUIBC MI 61-08-50).
Collection of Achatina fulica
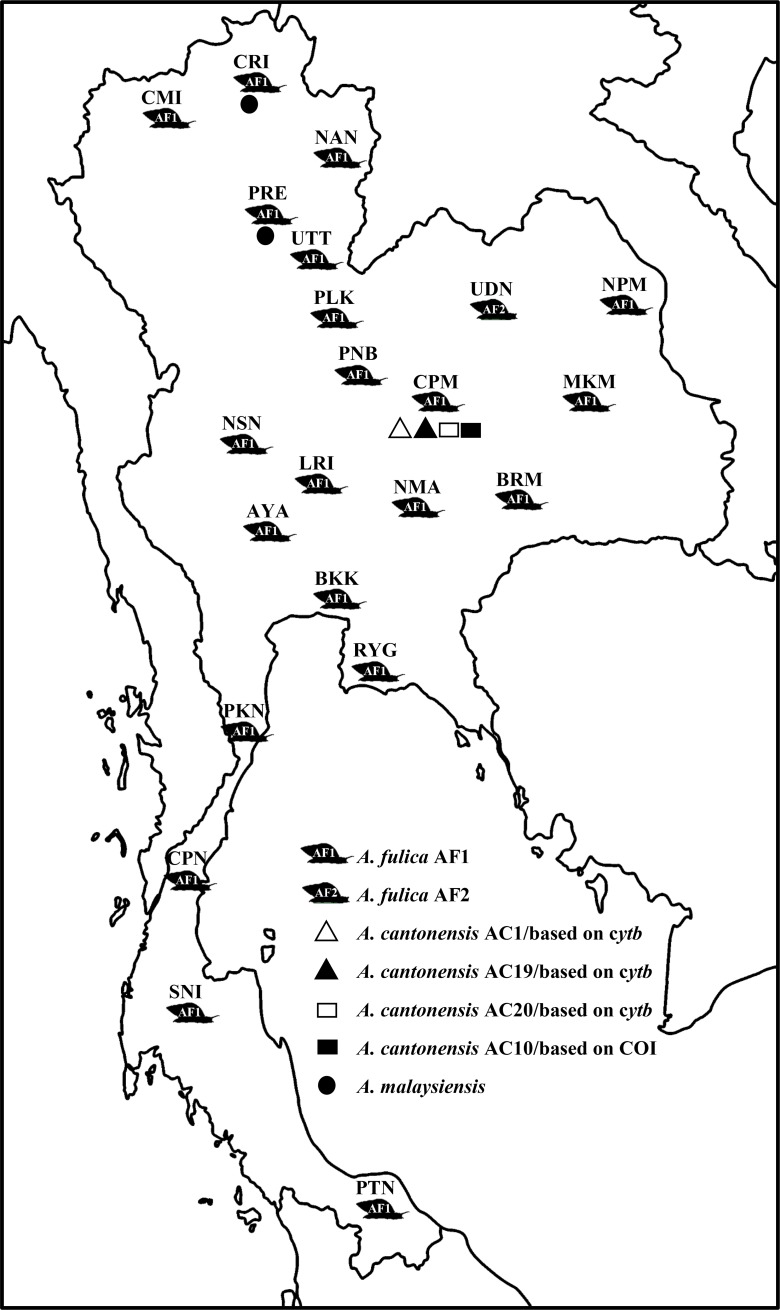
Achatina fulica was randomly collected between May 2017 and July 2018 from 22 provinces across Thailand (Uttaradit, Chiang Rai, Chiang Mai, Nan, Phrae, Phitsanulok, Phetchabun, Bangkok, Lop Buri, Phra Nakhon Si Ayutthaya, Nakhon Sawan, Rayong, Nakhon Ratchasima, Buri Ram, Maha Sarakham, Chaiyaphum, Udon Thani, Nakhon Phanom, Prachuap Khiri Khan, Pattani, Chumphon, and Surat Thani) (Fig 1and Table 1). No specific permission was required for sampling snail in public locations. The snails were collected from several habitats (e.g., under leaf litter and under or above dried trees) by hand picking and were placed in a plastic box with air ventilation. The snails were then transported at ambient temperature to the Department of Microbiology and Parasitology, Faculty of Medical Science, Naresuan University, Phitsanulok, Thailand. All specimens were identified through comparison of shell morphology according to previous studies [29,30] Schotman (1989) and Jena et al. (2017); the conical shell of A. fulica was identified as wider at its operculum and tapering at its apex. The size of the shell was approximately 9.0–12.0 cm in length and 4.0–5.0 cm in width. The coloration and vertical stripes were dark brownish, and alternated by a cream tinge.
Fig 1. Map of sampling location and distribution of Achatina fulica, Angiostrongylus cantonensis, and A. malaysiensis in Thailand.
Details of collection sites are given in Table 1.
Table 1. Locations in Thailand where Achatina fulica was collected and number of corresponding COI haplotypes.
| Regions | Location | Code | Latitude | Longitude | Number of samples | Haplotype name | Number of haplotypes |
|---|---|---|---|---|---|---|---|
| North | Uttaradit | UTT | 17.620088 | 100.099294 | 2 | AF1 | 2 |
| Chiang Rai | CRI | 19.907165 | 99.830955 | 4 | AF1 | 4 | |
| Chiang Mai | CMI | 18.706064 | 98.981716 | 2 | AF1 | 2 | |
| Nan | NAN | 18.775631 | 100.773041 | 4 | AF1 | 4 | |
| Phrae | PRE | 18.144577 | 100.140283 | 4 | AF1 | 4 | |
| Central | Phitsanulok | PLK | 17.036385 | 100.583513 | 3 | AF1 | 3 |
| Phetchabun | PNB | 16.301669 | 101.119280 | 4 | AF1 | 4 | |
| Bangkok | BKK | 13.756330 | 100.501765 | 4 | AF1 | 4 | |
| Lop Buri | LRI | 14.799508 | 100.653370 | 1 | AF1 | 1 | |
| Phra Nakhon Si Ayutthaya | AYA | 14.353212 | 100.568959 | 3 | AF1 | 3 | |
| Nakhon Sawan | NSN | 15.693007 | 100.122559 | 4 | AF1 | 4 | |
| East | Rayong | RYG | 12.707434 | 101.147351 | 3 | AF1 | 3 |
| Northeast | Nakhon Ratchasima | NMA | 14.979899 | 102.097769 | 2 | AF1 | 2 |
| Buri Ram | BRM | 14.993001 | 103.102919 | 3 | AF1 | 3 | |
| Maha Sarakham | MKM | 16.013201 | 103.161516 |
4 | AF1 | 4 | |
| Chaiyaphum | CPM | 15.806817 | 102.031502 | 4 | AF1 | 4 | |
| Udon Thani | UDN | 17.413841 | 102.787232 | 4 | AF2 | 4 | |
| Nakhon Phanom | NPM | 17.392039 | 104.769550 | 3 | AF1 | 3 | |
| South | Pattani | PTN | 6.761830 | 101.323254 | 4 | AF1 | 4 |
| Chumphon | CPN | 10.493049 | 99.180019 | 4 | AF1 | 4 | |
| Surat Thani | SNI | 9.138238 | 99.321748 | 4 | AF1 | 4 | |
| West | Prachuap Khiri Khan | PKN | 11.812367 | 99.797327 | 4 | AF1 | 4 |
Isolation of Angiostrongylus larvae from Achatina fulica
The body of A. fulica was removed from the shell. The mantle and foot of the land snail (approximately 25 mm3) were cut by a razor blade and kept at -20°C for further molecular analysis. To isolate Angiostrongylus larvae, most of the remaining snail’s body was mixed with 50–100 mL of 0.7% pepsin solution (Acros Organics, Geel, Belgium) and minced in a blender. The mixed solution was transferred to a beaker and incubated in a water bath at 37°C until the majority of the tissue was dissolved (1–2 h). The digested tissue solution was then placed in a Baermann apparatus, which consisted of a grass funnel connected to a short piece of rubber tubing at the outlet. The funnel, supported by a wire mesh, was covered with a layer of gauze and was let stand for 30–60 min to allow larval migration to the funnel’s neck. The filtered liquid containing mainly larvae was transferred to a Petri dish. The larvae were identified as described previously [11]. Angiostrongylus larvae were collected using a sterile Pasteur pipette under a stereomicroscope, transferred to a 1.5-mL microcentrifuge tube, and stored at -20°C for molecular analysis.
Extraction of genomic DNA
Genomic DNA from individual land snails and from third-stage larvae of Angiostrongylus was extracted using the NucleoSpin® Tissue kit (Macherey-Nagel, Duren, Germany) following the manufacturer’s instructions. The genomic DNA solution was checked by running it on a 0.8% agarose gel in 1× TBE buffer at 100 V. The gel was stained with ethidium bromide, followed by destaining with distilled water and photographed under u.v. light. The DNA solution was kept at -20°C for further use.
PCR and sequencing
PCR was used to amplify a partial region of the COI gene of A. fulica. A set of primers, AfCOI_F (5′-TGTGGGTTAGTTGGCACAGG-3′) and AfCOI_R (5′-TTAAGGGCGGGTACACAGTC-3′), was designed based on the deposited GenBank sequence (accession no. KT290318) using the Primer-BLAST program. The primers were used to amplify a 319-bp fragment. The reaction mixture was prepared in a total volume of 30 μL containing 3 μL of 10× buffer (1×), 2.1 μL of 25 mM MgCl2 (1.75 mM), 0.6 μL of 200 mM dNTPs (4 mM), 1.2 μL of 5 μM of each primer (0.2 μM), 0.3 μL of 5 U/mL Taq DNA polymerase (0.05 U/mL), 18.6 μL of distilled water, and 3 μL of DNA template (20–200 ng). The PCR conditions included initial denaturation at 95°C for 1 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 50°C for 40 s, extension at 72°C for 1 min, and final extension at 72°C for 5 min. The PCR was performed in a Biometra TOne Thermal cycler (Analytik Jena AG, Jena, Germany). The amplified products were analyzed by 1.2% agarose gel-electrophoresis, stained with ethidium bromide, destained with distilled water, and visualized and photographed under u.v. light. The amplified PCR products were then purified using a NucleoSpin® Gel and PCR Clean-up kit (Macherey-Nagel). Two volumes (56 μL) of buffer NTI were added to the tube containing the PCR product solution (28 μL). The mixture was then transferred onto a NucleoSpin® Gel and PCR Clean-up Column and centrifuged at 11,000 × g for 30 s. Buffer NT3 (700 μL) was added to the tube and this was centrifuged again at 11,000 × g for 30 s. The column was transferred to a new 1.5-mL microcentrifuge tube, followed by addition of 30 μL of elution buffer and incubation at room temperature (25°C) for 1 min. The tube was centrifuged at 11,000 × g for 1 min. The purified PCR product was checked on a 1.2% agarose gel in 1× TBE buffer at 100 V. The gel was stained with ethidium bromide, followed by destaining with distilled water and photographed under u.v. light. The purified PCR products were shipped to Macrogen Inc., Seoul, Korea, for sequencing in both forward and reverse directions.
For Angiostrongylus, PCR was used to amplify the selected nucleotide regions (COI, ITS2, SSU rRNA, and cytb for A. cantonensis and ITS2 for A. malaysiensis). Specific primers are listed in Table 2. The reaction mixture was prepared in a total volume of 30 μL, containing 15 μL of EconoTaq® PLUS 2× Master mix (1×; Lucigen Corporation, Middleton, WI, USA), 1.5 μL of 5 μM of each primer (0.25 μM), 9 μL of distilled water, and 3 μL of DNA template (20–200 ng). PCR conditions for amplifying COI, ITS, and SSU rRNA were as described by Rodpai et al. (2016); whereas those for amplifying cytb were as described by Dusitsittipon et al. (2015). All PCR amplifications were conducted in a Biometra TOne Thermal cycler. The amplified products were analyzed by 1.2% agarose gel-electrophoresis as mentioned above, and then purified using a NucleoSpin® Gel and PCR Clean-up kit as mentioned above. The PCR products were sequenced in both the forward and reverse direction by Macrogen Inc.
Table 2. Primers used for amplifying nucleotide regions in Angiostrongylus.
| Gene or region | Primer/(Reference) | Approximate amplicon size (bp) |
Target organism |
|---|---|---|---|
| SSU rRNA | Angi18S-1_forward 5′- AAAGTTAAGCCATGCATG -3′ Angi18S-2_reverse 5′- CATTCTTGGCAAATGCTTTCG -3′ [31] |
885 | A. cantonensis |
| cytb | Cytb-F 5′-TGAATAGACAGAATTTTAAGAG-3′ Cytb-R 5′-ATCAACTTAACATTACAGAAAC-3′ [27] |
853 | A. cantonensis |
| COI | AngiCOI_forward 5′- TTTTTTGGGCATCCTGAGGTTTAT -3′ AngiCOI_reverse 5′- CGAGGATAACCATGTAAACCAGC -3′ [9] |
605 | A. cantonensis |
| ITS2 | AngiITS2_forward 5'—ACATCTGGTTCAGGGTTGTT—3' AngiITS2_ reverse 5'—AGCATACAAGCACATGATCAC—3' [9] |
395 |
A. cantonensis A. malaysiensis |
Sequence and phylogenetic analysis
All sequences were edited by viewing the peak of the chromatogram in SeqMan II software (DNASTAR, Madison, WI, USA). Species identification of Angiostrongylus and Achatina was confirmed by a BLASTN search, whereby similarity with known sequences in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was determined. The maximum likelihood (ML), neighbour joining (NJ) and maximum parsimony (MP) phylogenetic trees were constructed based on a Kimura 2-parameter model for SSU rRNA and cytb alignment, Tamura 3-parameter model for ITS2 alignment, and the General Time Reversible model for COI alignment with 1,000 bootstrap replicates using MEGA 7 software [32]. Bayesian analysis were performed based on Markov chain Monte Carlo method in MrBayes v3.2 [33–35]
Haplotype diversity (h) and nucleotide diversity (𝜋) for A. fulica were calculated in ARLEQUIN, version 3.5.1.2 [36]. Population pairwise FST (p < 0.05) calculated in ARLEQUIN was used to infer the genetic structure of A. fulica from the six different sampled regions.
Results
Infection of Angiostrongylus in Achatina fulica
A total of 1,595 Angiostrongylus larvae were isolated from 343 A. fulica collected in 22 provinces across Thailand. These included 13 and 1,269 A. malaysiensis larvae isolated from A. fulica collected in Phrae and Chiang Rai provinces in northern Thailand, respectively; as well as 313 A. cantonensis larvae from A. fulica collected in Chaiyaphum province, northeast Thailand (S1 Table). A total of 41 Angiostrongylus spp. specimens and 74 individual land snails were randomly selected for further genetic studies.
Genetic characterization of Achatina fulica
A partial region of the COI gene from 74 individual snails collected in different locations was amplified by PCR and sequenced. PCR amplicons were 319 bp in length. Based on an edited stretch of 291 bp, all sequences showed high similarity (99–100%) with the known COI sequence of A. fulica (GenBank accession no. MF415341). All 74 sequences of A. fulica COI in the present study were deposited in the NCBI database (GenBank accession nos. MK858335-MK858408). The phylogenetic tree of the 74 COI sequences of A. fulica collected across Thailand revealed a monophyletic cluster with bootstrap and Bayesian posterior probability values (61/62/-/85%). It could be grouped together with A. fulica from the United Kingdom and the United States (Fig 2).
Fig 2. Maximum likelihood phylogenetic tree generated from 74 sequences of a partial COI sequence (291 bp) of A. fulica collected across Thailand.
Support values (ML bootstrap/NJ bootstrap/MP bootstrap/Bayesian posterior probabilities) show above the branches of the phylogenetic tree. At the branches of the tree, a dash (-) instead of a numerical support value indicates that a certain grouping was not seen by that method of analysis. Bold letters indicate sequences obtained in the present study. Archachatina marginata was used as the out-group. TH, Thailand; UK, United Kingdom; USA, United States of America; NG, Nigeria.
Two haplotypes were identified and named as AF1 and AF2 (Table 1and S1 Fig). All nucleotides in haplotypes AF1 and AF2 were the same except for the nucleotide at position 161, which corresponded to a “T” in AF1 and a “C” in AF2. Haplotype AF1 was found in the northern, central, eastern, northeastern, southern, and western regions of Thailand; whereas haplotype AF2 was found in Udon Thani province, northeast Thailand. Table 3shows the haplotype and nucleotide diversity of A. fulica COI sequences. In northeastern Thailand, haplotype and nucleotide diversity were found among 20 specimens. Population pairwise FST analysis revealed no genetic differentiation among populations from northern, central, eastern, northeastern, southern, and western Thailand (Table 4).
Table 3. Haplotype diversity (h) and nucleotide diversity (π) for six populations of A. fulica in Thailand based on mitochondrial COI gene sequences.
| Location | Number of samples | Haplotype diversity (h), mean±SD | Nucleotide diversity (π), mean±SD |
|---|---|---|---|
| Northern | 16 | 0 | 0 |
| Central | 19 | 0 | 0 |
| Eastern | 3 | 0 | 0 |
| Northeastern | 20 | 0.3368±0.1098 | 0.0011±0.0013 |
| Southern | 12 | 0 | 0 |
| Western | 4 | 0 | 0 |
| Total | 74 | 0.1037±0.0469 | 0.0003±0.0006 |
Table 4. Population pairwise FST from six populations of A. fulica based on mitochondrial COI gene sequences.
| Populations | Northern | Central | Eastern | Northeastern | Southern | Western |
|---|---|---|---|---|---|---|
| Northern | 0.000 | |||||
| Central | 0.000 | 0.000 | ||||
| Eastern | 0.000 | 0.000 | 0.000 | |||
| Northeastern | 0.135 | 0.152 | -0.064 | 0.000 | ||
| Southern | 0.000 | 0.000 | 0.000 | 0.107 | 0.000 | |
| Western | 0.000 | 0.000 | 0.000 | -0.012 | 0.000 | 0.000 |
Genetic characterization of Angiostrongylus
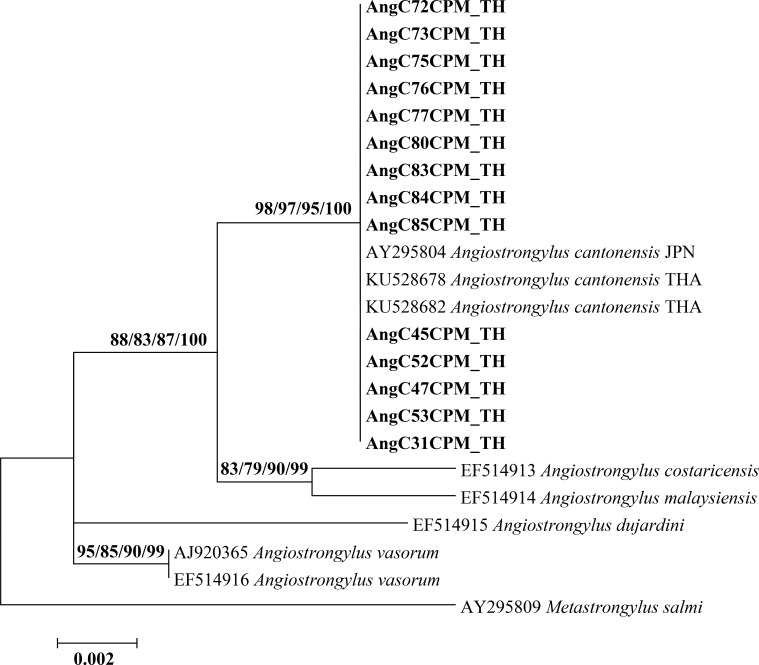
Partial sequences of SSU rRNA, COI, cytb, and ITS2 regions from 41 Angiostrongylus specimens were determined by PCR and sequencing. BLASTN search results relative to these four nucleotide regions are shown in Table 5. Based on 839 bp of the SSU rRNA gene, 14 specimens (GenBank accession nos. MK858285-MK858298) of Angiostrongylus showed 100% identity with A. cantonensis (GenBank accession no. KU528682). The maximum likelihood tree derived from all sequences of SSU rRNA from the present study was grouped together with A. cantonensis from Thailand (GenBank accession nos. KU528687 and KU528682) and Japan (GenBank accession no. AY295804) (Fig 3). Phylogenetic tree showed well support values (98/97/95/100%). There was no difference in p-distance for intraspecific divergence within A. cantonensis (S2 Table).
Table 5. BLASTN search based on SSU rRNA, ITS2, COI, and cytb regions of Angiostrongylus spp. (41 specimens) in Thailand.
| Code | Host/Location | No. of larvae | BLASTN identity (%) | Species identification | |||
|---|---|---|---|---|---|---|---|
| COI | SSU rRNA | cytb | ITS2 | ||||
| AngC30CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | ND | ND | ND | A. cantonensis |
| AngC31CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | 100 | 100 | 100 | A. cantonensis |
| AngC32CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | ND | ND | ND | A. cantonensis |
| AngC33CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | ND | ND | ND | A. cantonensis |
| AngC35CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | ND | 100 | 99 | A. cantonensis |
| AngC45CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | 100 | ND | 100 | A. cantonensis |
| AngC46CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | ND | ND | 100 | A. cantonensis |
| AngC47CPM_TH | A. fulica/Chaiyaphum | 1 | 100 | 100 | ND | 100 | A. cantonensis |
| AngC50CPM_TH | A. fulica/Chaiyaphum | 5 | 100 | ND | ND | 100 | A. cantonensis |
| AngC51CPM_TH | A. fulica/Chaiyaphum | 10 | ND | ND | 99 | 100 | A. cantonensis |
| AngC52CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | 99 | ND | A. cantonensis |
| AngC53CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | 100 | 100 | A. cantonensis |
| AngC72CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | 99 | 100 | A. cantonensis |
| AngC73CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | 100 | 100 | A. cantonensis |
| AngC74CPM_TH | A. fulica/Chaiyaphum | 10 | ND | ND | ND | 100 | A. cantonensis |
| AngC75CPM_TH | A. fulica/Chaiyaphum | 10 | ND | 100 | ND | 100 | A. cantonensis |
| AngC76CPM_TH | A. fulica/Chaiyaphum | 9 | 100 | 100 | 100 | 100 | A. cantonensis |
| AngC77CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | ND | 100 | A. cantonensis |
| AngC78CPM_TH | A. fulica/Chaiyaphum | 10 | ND | ND | ND | 100 | A. cantonensis |
| AngC79CPM_TH | A. fulica/Chaiyaphum | 10 | ND | ND | ND | 100 | A. cantonensis |
| AngC80CPM_TH | A. fulica/Chaiyaphum | 8 | ND | 100 | 100 | 100 | A. cantonensis |
| AngC81CPM_TH | A. fulica/Chaiyaphum | 6 | ND | ND | ND | 100 | A. cantonensis |
| AngC83CPM_TH | A. fulica/Chaiyaphum | 10 | ND | 100 | ND | 100 | A. cantonensis |
| AngC84CPM_TH | A. fulica/Chaiyaphum | 10 | 100 | 100 | 99 | 100 | A. cantonensis |
| AngC85CPM_TH | A. fulica/Chaiyaphum | 10 | ND | 100 | ND | ND | A. cantonensis |
| AngM107CRI_TH | A. fulica/Chiang Rai | 10 | ND | ND | ND | 100 | A. malaysiensis |
| AngM15CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM4CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM14CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM5CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM6CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM7CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM21CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM105CRI_TH | A. fulica/Chiang Rai | 10 | ND | ND | ND | 100 | A. malaysiensis |
| AngM70CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM108CRI_TH | A. fulica/Chiang Rai | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM109CRI_TH | A. fulica/Chiang Rai | 10 | ND | ND | ND | 100 | A. malaysiensis |
| AngM110CRI_TH | A. fulica/Chiang Rai | 10 | ND | ND | ND | 100 | A. malaysiensis |
| AngM40PRE_TH | A. fulica/Phrae | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM26PRE_TH | A. fulica/Phrae | 1 | ND | ND | ND | 100 | A. malaysiensis |
| AngM39PRE_TH | A. fulica/Phrae | 1 | ND | ND | ND | 100 | A. malaysiensis |
ND, not determined
Fig 3. Maximum likelihood phylogenetic tree of A. cantonensis based on a partial SSU rRNA sequence (839 bp).
Support values (ML bootstrap/NJ bootstrap/MP bootstrap/Bayesian posterior probabilities) show above the branches of the phylogenetic tree. Bold letters indicate sequences obtained in the present study. Metastrongylus salmi was used as the out-group. TH, THA, Thailand; JPN, Japan.
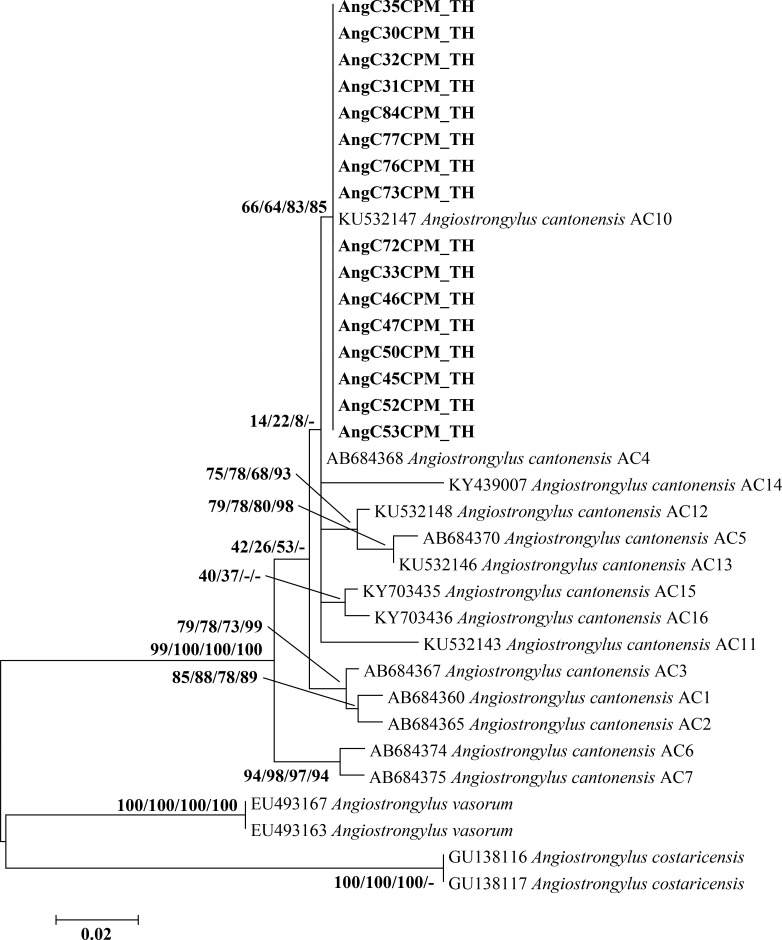
Based on a partial COI sequence (478 bp) of Angiostrongylus, 16 sequences (GenBank accession nos. MK734431-MK734446) from this study showed 100% similarity with the known sequence of A. cantonensis (GenBank accession no. KU532147). The maximum likelihood tree of 16 Angiostrongylus COI sequences was grouped together with the identified haplotypes of A. cantonensis AC10 (GenBank accession no. KU532147) (Fig 4). All sequences were formed a monophyletic group with support values of 66/64/83/85%. There was no difference in p-distance for intraspecific divergence within A. cantonensis (S3 Table).
Fig 4. Maximum likelihood tree based on a partial COI sequence (478 bp) of 16 samples of A. cantonensis from Thailand, together with A. cantonensis haplotypes AC1-AC16.
Support values (ML bootstrap/NJ bootstrap/MP bootstrap/Bayesian posterior probabilities) show above the branches of the phylogenetic tree. At the branches of the tree, a dash (-) instead of a numerical support value indicates that a certain grouping was not seen by that method of analysis. Bold letters indicate sequences obtained in the present study. A. vasorum and A. costaricensis were used as out-groups. TH, Thailand.
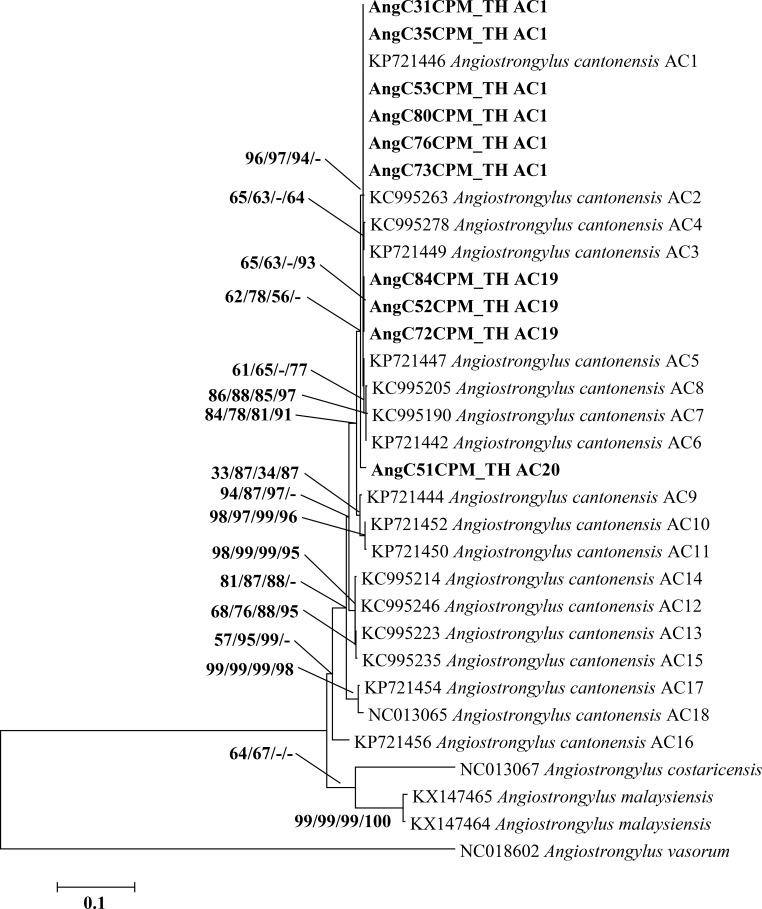
All ten cytb sequences (853 bp) from the present study (GenBank accession nos. MK858275-MK858284) showed 99–100% identity to the known sequence of A. cantonensis (GenBank accession no. KP721446). The phylogenetic tree of the cytb sequence revealed that ten sequences from the present study were closely related to A. cantonensis AC1 (GenBank accession no. KP721446), AC2 (GenBank accession no. KC995263), AC3 (GenBank accession no. KP721449), AC4 (GenBank accession no. KC995278), AC5 (GenBank accession no. KP721447), AC6 (GenBank accession no. KP721442), AC7 (GenBank accession no. KC995190), and AC8 (GenBank accession no. KC995205) (Fig 5). Six sequences from this study were similar to A. cantonensis haplotype AC1. In addition, three sequences were identified as belonging to the new haplotype AC19 with support values of 65/63/-/93% and one to the haplotype AC20 with support values of 62/78/56/-%. Comparison of nucleotide sequences between these two new haplotypes and 18 previously reported haplotypes is presented in S4 Table. Intraspecific distances within A. cantonensis ranged from <0.1% to 0.9% (S5 Table).
Fig 5. Maximum likelihood phylogenetic tree of A. cantonensis based on a partial cytb sequence (853 bp).
Support values (ML bootstrap/NJ bootstrap/MP bootstrap/Bayesian posterior probabilities) show above the branches of the phylogenetic tree. At the branches of the tree, a dash (-) instead of a numerical support value indicates that a certain grouping was not seen by that method of analysis. Bold letters indicate sequences obtained in the present study. A. costaricensis, A. malaysiensis, and A. vasorum are included in the tree. TH, Thailand.
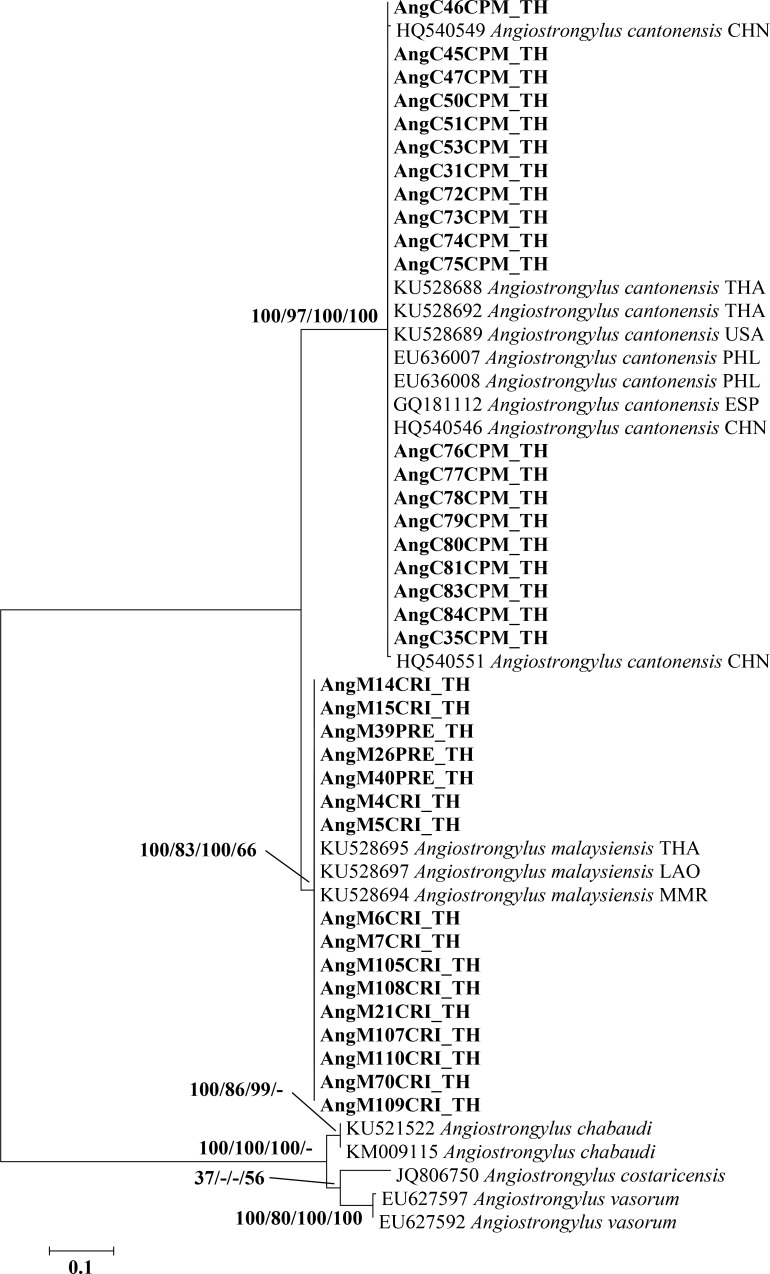
The ITS2 sequences (278 bp) from 20 specimens in the present study (GenBank accession nos. MK858299-MK858318) displayed 100% similarity to A. cantonensis (GenBank accession no. KU528692). A maximum likelihood tree showed that all 20 sequences fell in the A. cantonensis groups from Thailand (GenBank accession nos. KU528688 and KU528692), the Philippines (GenBank accession nos. EU636007 and EU636008), China (GenBank accession nos. HQ540546, HQ540549, and HQ540551), United States (GenBank accession no. KU528689), and Spain (GenBank accession no. GQ181112) (Fig 6). In addition, sixteen ITS2 sequences (268 bp) of Angiostrongylus larvae from this study (GenBank accession nos. MK858319-MK858334) showed 100% identity to A. malaysiensis (GenBank accession no. KU528697). Based on the maximum likelihood tree, all A. malaysiensis sequences fell in the A. malaysiensis groups from Thailand (GenBank accession no. KU528695), Laos (GenBank accession no. KU528697), and Myanmar (GenBank accession no. KU528694) (Fig 6). Two major clades of phylogenetic tree based on ITS2 sequences showed clearly distinguished between A. cantonensis and A. malaysiensis with support values of 100/97/100/100% and 100/83/100/66%, respectively (Fig 6). Interspecific distances between A. cantonensis and A. malaysiensis sequences ranged from 14.8% to 15.5% (S6 Table). Intraspecific distances among A. cantonensis samples were <0.4–0.7%. There was no difference in p-distance for intraspecific divergence within A. malaysiensis (S7 Table and S8 Table).
Fig 6. Maximum likelihood phylogenetic tree of A. cantonensis and A. malaysiensis based on a partial ITS2 sequence (278 bp).
Support values (ML bootstrap/NJ bootstrap/MP bootstrap/Bayesian posterior probabilities) show above the branches of the phylogenetic tree. At the branches of the tree, a dash (-) instead of a numerical support value indicates that a certain grouping was not seen by that method of analysis. Bold letters indicate sequences obtained in the present study. A. costaricensis, A. vasorum, and A. chabaudi are included in the tree. TH, THA, Thailand; CHN, China; PHL, Philippines; LAO, Laos; MMR, Myanmar; USA, United States of America; ESP, España.
Discussion
The present study describes infection of A. fulica with A. cantonensis and A. malaysiensis in Thailand. Earlier studies on the prevalence of A. cantonensis reported infection rates of up to 90% [37,38]; these estimates were lowered in later studies to 36.4% [39]. Recently, several surveys of A. cantonensis larvae in A. fulica were reported from various provinces in Thailand, their infection rates ranging between 1.1% and 7.6% [26,27,40]. Such low infection rates may be ascribed to variability in the presence and abundance of A. cantonensis in different environments, as well as to abiotic factors, such as humidity and temperature. The distribution of infected rats, the species of rats, and the interactions between gastropods and rats may determine the prevalence of A. cantonensis in snails [41]. In addition, we found A. fulica infected with A. malaysiensis in the north (Phrae and Chiang Rai provinces) of the country; in agreement with a previous report that detected A. malaysiensis in the northern Mae Hong Sorn and Tak provinces. Because this nematode is found also in the south (Phatthalung and Phang Nga provinces) of the country, close to Malaysia [9,28], A. fulica is a possible intermediate host for A. cantonensis and A. malaysiensis throughout Thailand. Moreover, A. fulica is implicated in an increased distribution of A. cantonensis in China and Japan [29,42]. At present, most human angiostrongyliasis cases are reported in the northeast of Thailand, but transmission of Angiostrongylus species reflects the dispersal of intermediate and definitive hosts. Accordingly, spreading of the African giant land snail can potentially augment the dispersion of angiostrongyliasis cases.
In the present study, genetic characterization of A. fulica collected from across Thailand was studied based on sequencing of the COI gene. The phylogenetic tree showed a monophyletic group for A. fulica in Thailand, suggesting that a single lineage of this snail had been introduced in the country. Interestingly, this lineage was closely related to A. fulica from the United Kingdom and the United States. Accordingly, these giant African land snail populations may share a common ancestor that was brought to each country by human intervention. In a previous study, A. fulica from Odisha state in India was closely related to A. fulica from Bangalore, Kerala, Africa, Cameroon, and China [30]. That result differs from the present one relating the Thai, UK, and USA isolates to a common origin, and demonstrates the existence of multiple lineages of this snail around the world, all originating from the African continent. The population genetic structure of A. fulica in Thailand revealed no difference between the six sampled regions. This uniformity may be due to gene flow within the A. fulica population in Thailand. Indeed, A. fulica was first introduced into Thailand from Malaysia in 1937 [43]. Five years after the first presumed entry, A. fulica population increased dramatically and expanded to several other parts of the country [44]. By analyzing the COI gene sequence, we identified two haplotypes (AF1 and AF2) of A. fulica. However, diversity between the two haplotypes was detected only in the northeast of Thailand, possibly as a result of the founder effect [45]. Importantly, in the present study, A. fulica haplotype AF1 from the northeast and north of Thailand was naturally infected with A. cantonensis and A. malaysiensis; whereas haplotype AF2, which is restricted to the northeast of the country, was not infected with any of the two Angiostrongylus species. At present, it is difficult to explain why A. fulica haplotype AF1 seems to be more susceptible to Angiostrongylus infection than haplotype AF2.
Genetic characterization of A. cantonensis in the present study was achieved through sequencing of SSU rRNA, ITS2, COI, and cytb nucleotide regions. Based on the COI maximum likelihood tree, A. cantonensis (16 specimens) collected from Chaiyaphum province was closely related to A. cantonensis AC10, which was collected from the closely located Khon Kaen province. Sixteen COI-based haplotypes (AC1-AC16) of A. cantonensis have been reported from several parts of the world [9,31,46]. Haplotypes AC1, AC2, AC3, AC5, and AC7 were reported in Japan; haplotypes AC8 and AC9 were reported in Brazil; and haplotype AC6 was found in China (31, 46). In Thailand, the different haplotypes of A. cantonensis appeared confined to specific localities: haplotype AC4 to Bangkok in the central part of the country, AC10 and AC11 to Khon Kaen province in the northeast, AC13 to Surat Thani province in the south, AC14 to Kanchanaburi province in the west, AC15 to Trat province in the east, and AC16 to Chanthaburi province in the east. This distribution corresponds to our finding of the AC10 haplotype in Chaiyaphum province in the northeast of Thailand. Therefore, the COI region represents a good marker for studying the genetic evolution and differentiation of Angiostrongylus spp. [47], as well as to distinguish geographical isolates of A. cantonensis [46] and to identify its haplotypes [48].
In the present study, SSU rRNA sequences of A. cantonensis isolated from Chaiyaphum province shared a single phylogenetic group. All 14 sequence samples were closely related to the Thai and Japanese isolates. This was confirmed by the lack of difference between intraspecific distances within A. cantonensis isolates. Previous studies have reported little variation of the nuclear small subunit (SSU) rRNA sequences within a nematode species but substantial divergence among species, allowing for species differentiation [22,24,31]. Therefore, the SSU rRNA gene has been used to identify A. cantonensis and for the discrimination of Angiostrongylus species [24,31].
In this study, A. cantonensis cytb sequences (10 specimens) from Chaiyaphum province were closely related to AC1-AC8 cytb haplotypes found across several provinces in Thailand. However, most sequences (six specimens) were similar to cytb haplotype AC1, suggesting that this may be the dominant haplotype in Thailand. In addition, we identified two new cytb haplotypes: AC19 (three sequences) and AC20 (one sequence). Previous studies reported 15 haplotypes (AC1-AC15) based on the cytb sequence in Thailand; two haplotypes (AC16 and AC18) were reported in China; and one haplotype (AC17) was reported in Hawaii [27,28]. In Thailand, cytb haplotypes were distributed at random throughout the country; AC1 in Phitsanulok and Prachuap Khiri Khan provinces; AC2 in Prachuap Khiri Khan province; AC3 in Chiang Rai province; AC4 in Phitsanulok province; AC5 in Chiang Mai province; AC6 in Samut Prakan province and Bangkok; AC7 in Bangkok; AC8 in Kanchanaburi province; AC9 in Bangkok; AC10 in Nan, Surat Thani, and Nakhon Si Thammarat provinces; AC11 in Khon Kaen province; AC12 in Nan and Lop Buri provinces; AC13 in Maha Sarakham province; AC14 in Lop Buri province; and AC15 in Maha Sarakham province [28]. A larger sample size may reveal a clearer relationship between the cytb haplotype of this worm and localization in Thailand.
ITS2 sequences revealed differences between A. cantonensis and A. malaysiensis. The genetic distance between A. cantonensis and A. malaysiensis was 14.8–15.5%, whereas intraspecific distances among A. cantonensis were <0.4–0.7% and there was no intraspecific divergence within A. malaysiensis. Our findings are similar to those reported previously [9,23,49] and suggest that the ITS2 sequence might be useful for the identification of Angiostrongylus species [23,49].
Conclusions
In summary, we describe here the genetic characterization of A. cantonensis and A. malaysiensis isolated from the giant African snail A. fulica in Thailand. Two haplotypes (AF1 and AF2) of A. fulica were identified for the first time based on sequencing of the COI gene. Only haplotype AF1 of A. fulica was infected with A. cantonensis and A. malaysiensis. This confirmed that A. cantonensis and A. malaysiensis were found across the country. A. fulica is the main intermediate host for transmission of Angiostrongylus spp. in nature. The COI and cytb genes of A. cantonensis are suitable for phylogenetic studies, whereas the SSU rRNA gene is appropriate for identification. The ITS2 nucleotide region represents a good genetic marker for distinguishing between A. cantonensis and A. malaysiensis. Two new additional cytb haplotypes of A. cantonensis (AC19 and AC20) were identified in this study. A larger sample will help future studies on the genetics of this nematode species. This study provides basic genetic information about the parasite Angiostrongylus and its snail intermediate host, A. fulica.
Supporting information
Each haplotype is represented by a circle. Sizes of circles are relative to number of individuals sharing specific haplotype.
(TIF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
This work was supported by Naresuan University (Grant number R2562B079). We would like to thank Professor Dr. Pairot Pramual, Faculty of Science, Maha Sarakham University for his guidance in analyzing the population genetics of Achatina fulica.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Naresuan University (Grant number R2562B079) to AV.
References
- 1.Wang QP, Lai DH, Zhu XQ, Chen XG, Lun ZR. Human angiostrongyliasis. Lancet Infect Dis. 2008; 8: 621–630. 10.1016/S1473-3099(08)70229-9 [DOI] [PubMed] [Google Scholar]
- 2.Spratt DM. Species of Angiostrongylus (Nematoda: Metastrongyloidea) in wildlife: a review. Int J Parasitol Parasites Wildl. 2015; 4: 178–189. 10.1016/j.ijppaw.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prociv P, Spratt DM, Carlisle M.S. Neuro-angiostrongyliasis: unresolved issue. Int J Parasitol. 2000; 30: 1295–1303. 10.1016/s0020-7519(00)00133-8 [DOI] [PubMed] [Google Scholar]
- 4.Diao Z, Wang J, Qi H, Li X, Zheng X, Yin C. Human ocular angiostrongyliasis: a literature review. Trop Doct. 2011; 41: 76–78. 10.1258/td.2010.100294 [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Nawa Y, Sawanyavisuth K, Lv Z, Wu ZD. Comprehensive review of ocular angiostrongyliasis with special reference to optic neuritis. Korean J Parasitol. 2013; 51: 613–619. 10.3347/kjp.2013.51.6.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez R, Dequi RM, Peruzzo L, Mesquita PM, Garcia E, Fornari F. Abdominal angiostrongyliasis: report of two cases with different clinical presentations. Rev Inst Med Trop Sao Paulo. 2008; 50: 339–341. 10.1590/s0036-46652008000600005 [DOI] [PubMed] [Google Scholar]
- 7.Quirós JL, Jiménez E, Bonilla R, Arce I, Hernández C, Jiménez Y. Abdominal angiostrongyliasis with involvement of liver histopathologically confirmed: a case report. Rev Inst Med Trop Sao Paulo. 2011; 53: 219–222. 10.1590/s0036-46652011000400008 [DOI] [PubMed] [Google Scholar]
- 8.Bhaibulaya M, Cross JH. Angiostrongylus malaysiensis (Nematoda: Metastrongylidae), a new species of rat lung-worm from Malaysia. Southeast Asian J Trop Med Public Health. 1971; 2: 527–533. [PubMed] [Google Scholar]
- 9.Rodpai R, Intapan PM, Thanchomnang T, Sanpool O, Sadaow L, Laymanivong S, et al. Angiostrongylus cantonensis and A. malaysiensis broadly overlap in Thailand, Lao PDR, Cambodia and Myanmar: a molecular survey of larvae in land snails. PLoS One. 2016; 11(8): e0161128 10.1371/journal.pone.0161128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohbayashi M, Kamiya M, Bhaibulaya M. Studies on parasites fauna of Thailand. I two new metastrongylid nematodes, Angiostrongylus siamensis sp. n. and Thaistrongylus harinasutai gen. et. sp. n. (Metastrongyloidae): Angiostrongylidae from wild rats. Jpn J Vet Res. 1979; 27: 5–10. [PubMed] [Google Scholar]
- 11.Eamsohana P. The rat lungworm Parastrongylus (= Angiostrongylus) cantonensis: parasitology, immunology, eosinophilic meningitis, epidemiology and laboratory diagnosis. Bangkok: Wankaew (IQ) Book Center; 2006. [Google Scholar]
- 12.Wan KS, Weng WC. 2004. Eosinophilic meningitis in a child raising snails as pets. Acta Trop. 2004; 90: 51–53. [DOI] [PubMed] [Google Scholar]
- 13.Thiengo SC, Simões Rde O, Fernande MA, Maldonado AJr. Angiostrongylus cantonensis and rat lungworm disease in Brazil. Hawaii J Med Public Health. 2013; 72(Suppl 2): 18–22. [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai HC, Chen YS, Yen CM. Human parasitic meningitis caused by Angiostrongylus cantonensis infection in Taiwan. Hawaii J Med Public Health. 2013; 72(Suppl 2): 26–27. [PMC free article] [PubMed] [Google Scholar]
- 15.Marquardt WC, Demaree RS, Grieve RB. Parasitology and vector biology. 2nd edn California: Academic Press; 2000. p. 702. [Google Scholar]
- 16.Mead AR. The giant African snail: A problem in economic malacology. Chicago: The University of Chicago Press; 1961. [Google Scholar]
- 17.Venette RC, Larson M. Mini Risk Assessment Giant African Snail, Achatina fulica Bowdich [Gastropoda: Achatinidae]. 2004. Available from: http://www.aphis.usda.gov/plant_health/plant_pest_info/pest_detection/downloads/pra/afulicapra.pdf. [Google Scholar]
- 18.Mead AR. Pulmonates, Vol. 2B: Economic malacology with particular reference to Achatina fulica. London: Academic Press; 1979. p. 150. [Google Scholar]
- 19.Raut SK, Barker GM. Achatina fulica Bowdich and other Achatinidae as pests in tropical agriculture In: Barker G.M. (Ed.). Molluscs as Crop Pests. Hamilton, New Zealand: CABI Publishing; 2002. pp. 55–114. [Google Scholar]
- 20.Alicata J. The presence of Angiostrongylus cantonensis in the islands of the Indian Ocean and probable role of the giant African snail, Achatina fulica, in the dispersal of the parasite to the Pacific islands. Can J Zool. 1966; 44: 1041–1049. 10.1139/z66-111 [DOI] [PubMed] [Google Scholar]
- 21.Eamsobhana P, Yong HS, Song SL, Gan XX, Prasartvit A, Tungtrongchitr A. Molecular phylogeography and genetic diversity of Angiostrongylus cantonensis and A. malaysiensis (Nematoda: Angiostrongylidae) based on 66-kDa protein gene. Parasitol Int. 2019; 68: 24–30. 10.1016/j.parint.2018.09.006 [DOI] [PubMed] [Google Scholar]
- 22.Foronda P, Lopez-Gonzalez M, Miquel J, Torres J, Segovia M, Abreu-Acosta N, et al. Finding of Parastrongylus cantonensis (Chen, 1935) in Rattus rattus in Tenerife, Canary Islands (Spain). Acta Trop. 2010; 114: 123–127. 10.1016/j.actatropica.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 23.Liu CY, Zhang RL, Chen MX, Li J, Ai L, Wu CY, et al. Characterization of Angiostrongylus cantonensis isolates from China by sequences of internal transcribed spacers of nuclear ribosomal DNA. J Anim Vet Adv. 2011; 10: 593–596. [Google Scholar]
- 24.Fontanilla IK, Wade CM. The small subunit (SSU) ribosomal (r) RNA gene as a genetic marker for identifying infective 3rd juvenile stage Angiostrongylus cantonensis. Acta Trop. 2008; 105: 181–186. 10.1016/j.actatropica.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 25.Eamsobhana P, Lim PE, Yong HS. Phylogenetics and systematics of Angiostrongylus lungworms and related taxa (Nematoda: Metastrongyloidea) inferred from the nuclear small subunit (SSU) ribosomal DNA sequences. J Helminthol. 2015; 89: 317–325. 10.1017/S0022149X14000108 [DOI] [PubMed] [Google Scholar]
- 26.Vitta A, Srisongcram N, Thiproaj J, Wongma A, Polsut W, Fukruksa C, et al. Phylogeny of Angiostrongylus cantonensis in Thailand based on cytochrome c oxidase subunit I gene sequence. Southeast Asian J Trop Med Public Health. 2016; 47: 377–386. [PubMed] [Google Scholar]
- 27.Dusitsittipon S, Thaenkham U, Watthanakulpanich D, Adisakwattana P, Komalamisra C. Genetic differences in the rat lungworm, Angiostrongylus cantonensis (Nematoda: Angiostrongylidae), in Thailand. J Helminthol. 2015; 89: 545–551. 10.1017/S0022149X14000388 [DOI] [PubMed] [Google Scholar]
- 28.Yong HS, Eamsobhana P, Song SL, Prasartvit A, Lim PE. Molecular phylogeography of Angiostrongylus cantonensis (Nematoda: Angiostrongylidae) and genetic relationships with congeners using cytochrome b gene marker. Acta Trop. 2015; 148: 66–71. 10.1016/j.actatropica.2015.04.020 [DOI] [PubMed] [Google Scholar]
- 29.Schotman CYL. Data sheet on the Giant African Land Snail Achatina fulica Bowdich (Mollusca: Achatinidae). In: PROVEG No. 19. FAO Regional Office of Latin America and the Caribbean Plant Quarantine Action Program; 1989. pp. 16–21. [Google Scholar]
- 30.Jena C, Sarkar S, Jalaja N. Krupanidhi S. Molecular phylogenetic relations of Achatina fulica based on partial sequence of COI gene. Natl Acad Sci Lett 2017; 40(2). 10.1007/s40009-017-0538-5 [DOI] [Google Scholar]
- 31.Tokiwa T, Harunari T, Tanikawa T, Komatsu N, Koizumi N, Tung KC, et al. Phylogenetic relationships of rat lungworm, Angiostrongylus cantonensis, isolated from different geographical regions revealed widespread multiple lineages. Parasitol Int. 2012; 61: 431–436. 10.1016/j.parint.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016; 33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yong HS, Song SL, Eamsobhana P, Goh SY, Lim PE. Complete mitochondrial genome reveals genetic diversity of Angiostrongylus cantonensis (Nematoda: Angiostrongylidae). Acta Trop. 2015; 152: 157–164. 10.1016/j.actatropica.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 35.Ayyagari VS, Sreerama K. Evaluation of haplotype diversity of Achatina fulica (Lissachatina) [Bowdich] from Indian sub-continent by means of 16S rDNA sequence and its phylogenetic relationships with other global populations. 3 Biotech. 2017; 7(4): 252 10.1007/s13205-017-0877-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10: 564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 37.Harinasuta C, Setasuban P, Radomyos P. Observations on Angiostrongylus cantonensis in rats and mollusks in Thailand. J Med Assoc Thai. 1965; 48: 158–172. [Google Scholar]
- 38.Setasuban P, Vajrasthira S, Harinasuta C. The preliminary observations on natural infection of rat lungworm (Angiostrongylus cantonensis) in rodents and intermediate host in Thailand. J Med Assoc Thai. 1968; 51: 156–157. [Google Scholar]
- 39.Pipitgool V, Sithithaworn P, Pongmuttasaya P, Hinz E. Angiostrongylus infections in rats and snails in northeast Thailand. Southeast Asian J Trop Med Public Health. 1997; 28: 190–193. [PubMed] [Google Scholar]
- 40.Tesana S, Srisawangwong T, Sithithaworn P, Laha T, Andrews R. Prevalence and intensity of infection with third stage larvae of Angiostrongylus cantonensis in mollusks from northeast Thailand. Am J Trop Med. Hyg. 2009; 80: 983–987. [PubMed] [Google Scholar]
- 41.Kim JR, Hayes KA, Yeung NW, Cowie RH. Correction: Diverse gastropod hosts of Angiostrongylus cantonensis, the rat lungworm, globally and with a focus on the Hawaiian Islands. PLOS One. 2018; 13(2): e0193556 10.1371/journal.pone.0193556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv S, Zhang Y, Liu H-X, Hu L, Yang K, Steinmann P, et al. Invasive snails and an emerging infectious disease: eesults from the first national survey on Angiostrongylus cantonensis in China. PLoS Negl Trop Dis. 2009; 3(2): e368 10.1371/journal.pntd.0000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kerdpreedee S. The giant African snail. Kasikorn. 1956; 29: 511–516. [Google Scholar]
- 44.Upatham S, Kruatrachue M, Baidikul V. Cultivation of the giant African snail, Achatina fulica. J Sci Soc. Thailand. 1988; 14: 25–40. [Google Scholar]
- 45.Fontanilla IKC, Sta. Maria IMP, Garcia JRM, Ghate H, Naggs F, Wade CM. Restricted genetic variation in populations of Achatina (Lissachatina) fulica outside of East Africa and the Indian Ocean Islands points to the Indian Ocean Islands as the earliest known common source. PLoS One. 2014; 9(9): e105151 10.1371/journal.pone.0105151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monte TC, Simões RO, Oliveira AP, Novaes CF, Thiengo SC, Silva AJ, et al. Phylogenetic relationship of the Brazilian isolates of the rat lungworm Angiostrongylus cantonensis (Nematoda: Metastrongylidae) employing mitochondrial COI gene sequence data. Parasit Vectors. 2012; 5: 248 10.1186/1756-3305-5-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eamsobhana P, Lim PE, Solano G, Zhang H, Gan X, Yong HS. Molecular differentiation of Angiostrongylus taxa (Nematoda: Angiostrongylidae) by cytochrome c oxidase subunit I (COI) gene sequences. Acta Trop. 2010; 116: 152–156. 10.1016/j.actatropica.2010.07.005 [DOI] [PubMed] [Google Scholar]
- 48.Červená B, Modrý D, Fecková B, Hrazdilová K, Foronda P, Alonso AM, et al. Low diversity of Angiostrongylus cantonensis complete mitochondrial DNA sequences from Australia, Hawaii, French Polynesia and the Canary Islands revealed using whole genome next-generation sequencing. Parasit Vectors. 2019; 12(1): 241 10.1186/s13071-019-3491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jefferies R, Shaw SE, Viney ME, Morgan ER. Angiostrongylus vasorum from South America and Europe represent distinct lineages. Parasitology. 2009; 136: 107–115. 10.1017/S0031182008005258 [DOI] [PubMed] [Google Scholar]