Abstract
Introduction
EGFR exon 20 insertions (EGFRex20ins) comprise an uncommon subset of EGFR activating alterations relatively insensitive to first and second-generation EGFR tyrosine kinase inhibitors (TKIs). However, recent early clinical data suggests these patients may benefit from newer generation EGFR-TKIs. Comprehensive genomic profiling (CGP) identifies a broad spectrum of EGFRex20ins and associated co-occurring genomic alterations (GA) present in non-small cell lung cancer (NSCLC)
Methods
Hybrid capture-based CGP was performed prospectively on 14,483 clinically annotated consecutive NSCLC specimens to a mean coverage depth of >650X for 236 or 315 cancer-related genes.
Results
Of 14,483 NSCLC cases, CGP identified 263 (1.8%) cases with EGFRex20ins, representing 12% (263/2,251) of cases with EGFR mutations. 64 unique EGFRex20ins were identified, most commonly D770_N771>ASVDN (21%) and N771_P772>SVDNP (20%). EGFR amplification occurred in 22% (57/263). The most common co-occurring GA effected TP53 (56%), CDKN2A (22%), CDKN2B (16%), NKX2-1 (14%) and RB1 (11%); co-occurring GA in other known lung cancer drivers were rare (5%). Average tumor mutational burden (TMB) was low (mean 4.3, range 0-40.3 mutations/Mb). Clinical outcomes to first- and second-generation EGFR TKIs were obtained for 5 patients and none responded.
Conclusion
In the largest series of EGFRex20ins NSCLC, diverse EGFRex20ins were detected in 12% of EGFR-mutant NSCLC, a higher frequency than previously reported in smaller single-institution studies. Clinical outcomes demonstrated lack of response to EGFR TKIs. TMB was low, consistent with non-smoking associated NSCLC. Comprehensive sequencing revealed increased proportion and wide variety of EGFRex20ins, representing a population of patients significant enough for focused efforts on effective interventions.
Keywords: EGFR Exon 20 Insertion, Genomics, Tumor Mutational Burden
Introduction
EGFR exon 19 deletions and EGFR exon 21 L858R represent the vast majority of EGFR activating mutations, and are exquisitely sensitive to approved EGFR TKIs such as erlotinib, gefitinib, and afatinib [1–3]. Less common EGFR mutations have variable sensitivity to EGFR inhibitors, though many still demonstrate clinical sensitivity to EGFR TKIs (e.g. EGFR G719X, L861Q, S768I)[4]. EGFR exon 20 insertions (EGFRex20ins) are a collection of EGFR driver mutations characterized by in-frame insertions that typically serve to constitutively upregulate EGFR kinase activity similar to sensitizing mutations (but are insensitive to 1st and 2nd gen EGFR-TKIs). These alterations have previously been reported to comprise approximately 4-10% of all EGFR mutant lung cancers [5–8]. The diverse array of exon 20 insertions and the challenges associated with identifying them may lead to underestimation of their true frequency. Although EGFRex20ins usually have the same transforming ability as more common EGFR activating mutations and are thus considered driver mutations, they are typically unresponsive to first and second-generation EGFR TKIs due to the modified structures of their kinase domains [5–8]. Very few of these EGFRex20ins such as A763_Y764insFQEA have demonstrated sensitivity to 1st and 2nd generation EGFR TKIs [9].
Several next-generation EGFR TKIs (some with pan-HER activity) have demonstrated pre-clinical activity against EGFRex20ins and are in clinical development (EGF816, AP32788, osimertinib, poziotinib)[5, 10, 11]. In particular, a strong signal of clinical activity was demonstrated with poziotinib with an overall response rate of 64% in the first 11 patients in an early phase clinical trial[12]. However, no EGFR-TKIs are currently approved for EGFRex20ins and their diversity of structures suggests that different insertion events may have divergent responsiveness to various EGFR TKIs. Thus, identifying the full array and scope of EGFRex20ins is of paramount importance.
Studies examining EGFRex20ins have generally been limited to cases from single institutions. Certain next-generation sequencing approaches such as comprehensive genomic profiling (CGP) permit sequencing of the entire EGFR gene to broadly assess for diverse EGFRex20ins and co-existing genomic alterations that may be relevant to the pathogenesis of EGFRex20ins-positive NSCLC. In addition to characterizing the clinical and pathologic characteristics of EGFRex20ins-positive NSCLC from a large dataset with molecular testing performed in the course of clinical care from multiple institutions, the purpose of this study was to assess the frequency and diversity of EGFRex20ins in NSCLC by their pattern of sequence alterations and co-occurring genomic alterations, which may impact the development of targeted treatments for these patients.
Methods
DNA was extracted from 40-um formalin-fixed paraffin-embedded sections. EGFRex20ins and co-occurring genomic alterations were identified by hybrid capture-based comprehensive genomic profiling performed during the course of clinical care on 14,483 consecutive NSCLC specimens to a mean coverage depth of >650X for 236 (version 1, July 2012 to August 2014) or 315 (version 2, August 2014 to June 2016) cancer related genes plus selected introns from 19 or 28 genes frequently rearranged in cancer. EGFR amplification was defined as estimated copy number greater than 6 copies[13]. Clinical data such as age, gender, stage, and histologic subtype were abstracted from the accompanying pathology report submitted by the ordering physician. Treatment outcomes from these patients were included where available. Testing was performed in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited reference laboratory (Foundation Medicine, Inc., Cambridge, MA). Patient samples were evaluated for genomic alterations (GAs), including base pair substitutions, insertions/deletions (indels), copy number alterations, and rearrangements, as described previously. Tumor mutational burden (TMB) was characterized as the number of somatic base substitution or indel alterations per megabase (MB) per previously described methods[14]. Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
Results
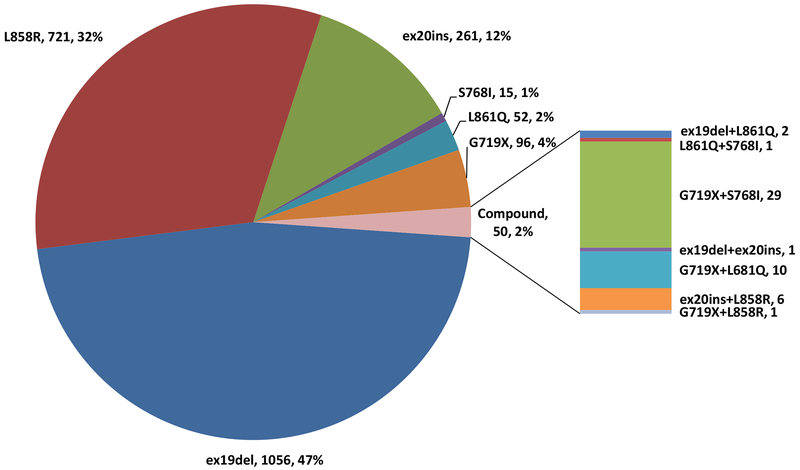
Comprehensive genomic profiling (CGP) performed on 14,483 NSCLC cases in the course of clinical care identified 2,251 cases with EGFR mutations, 263 of these cases were EGFRex20ins, representing 12% of all EGFR-mutant NSCLC and 1.8% of all NSCLC cases tested. EGFRex20ins were the third most common type of EGFR mutation detected following EGFR exon 19 deletions (47%) and EGFR L858R (32%). Other less common EGFR mutations detected in this large case series include G719X (4%), L861Q (2%), S768I (1%), and cases with compound EGFR-activating mutations (2%) (Figure 1).
Figure 1:
Frequency and Distribution of 2,251 EGFR mutations in NSCLC Detected by Comprehensive Genomic Profiling. Each alteration is shown as: mutation, number of cases, frequency.
Clinical and pathologic characteristics were available for all of the 263 patients with EGFRex20ins as summarized in Table 1. The large majority of cases EGFRex20ins were identified in lung adenocarcinoma (90%, 237/263), followed by NSCLC not otherwise specified (9.1%, 24/263), and sarcomatoid histology (0.7%, 2/263). The median age for EGFRex20-positive cases was 63 years, and 62% of patients were female. Of the 263 patients with EGFRex20ins-positive NSCLC, treatment outcomes were available for 11 patients. Five of those patients received either first or second-generation EGFR TKIs, with none of the five patients demonstrating a response to treatment and median time to progression of only 3.5 months (Supplementary Table 1). The remaining 6 patients did not receive EGFR targeted therapy.
Table 1:
Histologic and Clinical Characteristics of Non-Small Cell Lung Cancer Patients with Tumors Harboring EGFR Exon 20 Insertions
| Histologic Subtype | All | Adenocarcinoma | NSCLC NOS | Squamous/Adenosquamous | Sarcomatoid |
|---|---|---|---|---|---|
| Total Cases | 14483 | 10272 (71%) | 2197 (15%) | 1942 (13%) | 122 (0.8%) |
| N of patients with EGFR exon 20 Insertions | 263 | 237 (90.1%) | 24 (9.2%) | 0 | 2 (0.7%) |
| Median Age (range) | 63 (14-90) | 63 (14-90) | 71 (44-87) | 50.5 (45-56) | |
| Sex | |||||
| M | 100 | 89 | 10 | 1 | |
| F | 163 | 148 | 14 | 1 | |
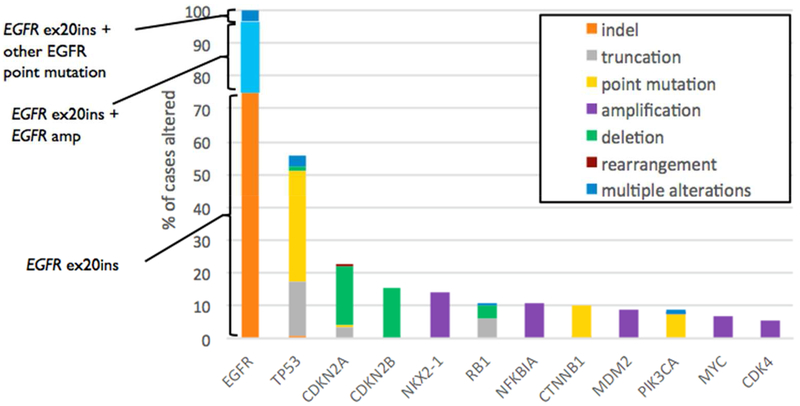
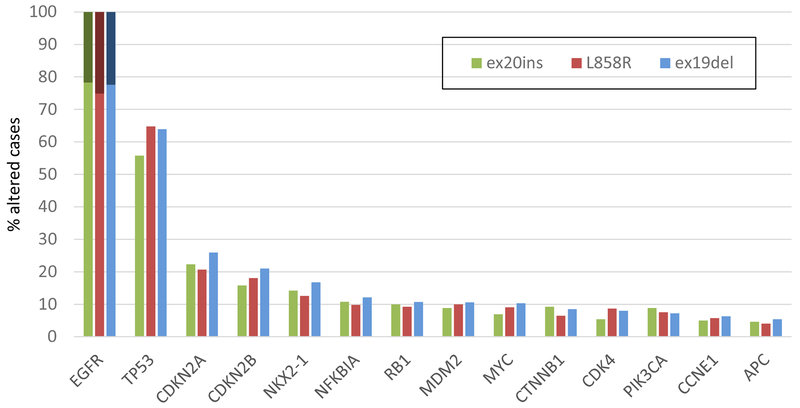
Sixty-four unique EGFRex20ins were identified, most commonly D770_N771>ASVDN (21%), N771_P772>SVDNP (20%) and N771_H773dupNPH (8%); 6% (15/263) harbored EGFR A763_Y764insFQEA, an EGFRex20ins with preclinical and clinical evidence demonstrating sensitivity to first- and second-generation EGFR-TKI (Figures 2 and 3). Putative co-occurring driver alterations in genes including EGFR (ex19del and L858R), HER2, MET and KRAS mutations tended to be mutually exclusive from EGFRex20ins, occurring only in 5% (12/263) of cases, and no co-occurring ALK, ROS1, or RET fusions or BRAF mutations were identified. Among EGFRex20ins cases, EGFR amplification occurred in 22% (57/263). Three cases with EGFRex20ins harbored co-occurring T790M, including 2 cases with A763_Y764insFQEA, which is sensitive to 1st and 2nd generation EGFR TKIs. The most common co-occurring alterations affected TP53 (56%), CDKN2A (22%), CDKN2B (16%), NKX2-1 (14%) and RB1 (11%) (Figure 4). Co-occurring alterations did not substantially differ between EGFRex20ins and canonical EGFR mutations (EGFR exon 19 deletion and EGFR L858R) that were negative for the most common acquired EGFR TKI resistance mutation, EGFR T790M (Table 2, Figure 4). Average tumor mutational burden (TMB) was low (mean 4.3, median 3.6, range 0-40.3 mutations/Mb) in EGFRex20ins cases -comparable with TMB in EGFR exon 19del and EGFR L858R and lower than EGFR-WT NSCLC (Table 2).
Figure 2:
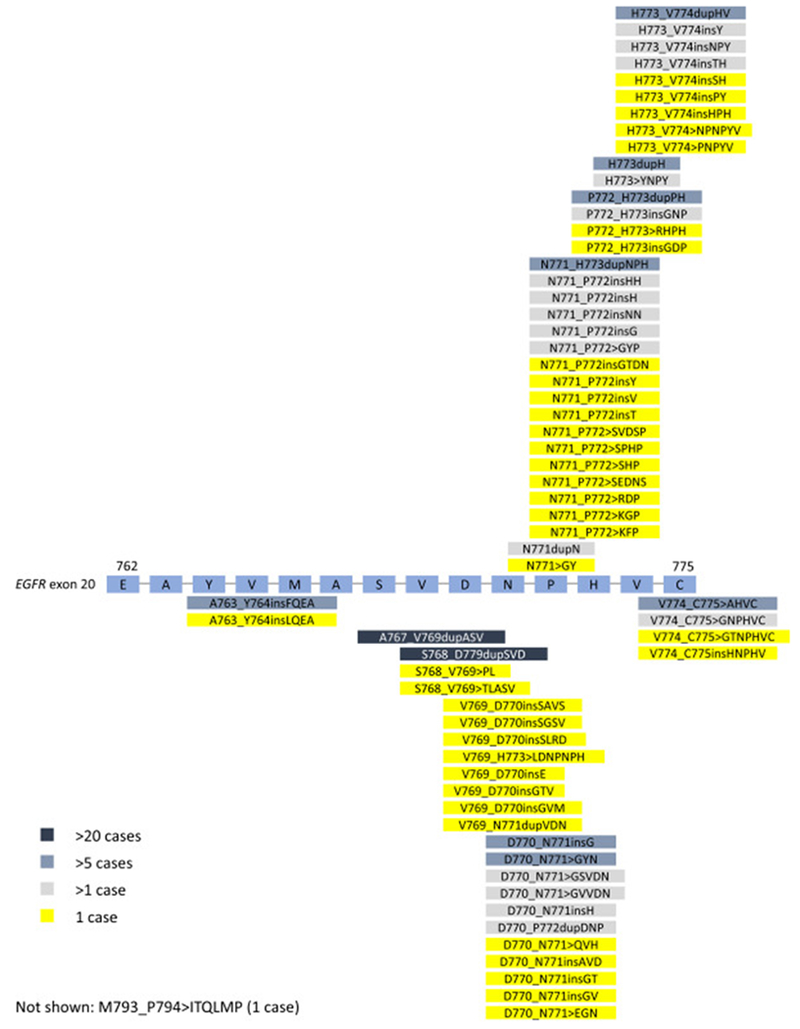
Schematic of genomic positions of EGFR Exon 20 Insertions Detected by Comprehensive Genomic Profiling. EGFR amino acids positions are indicated.
Figure 3:
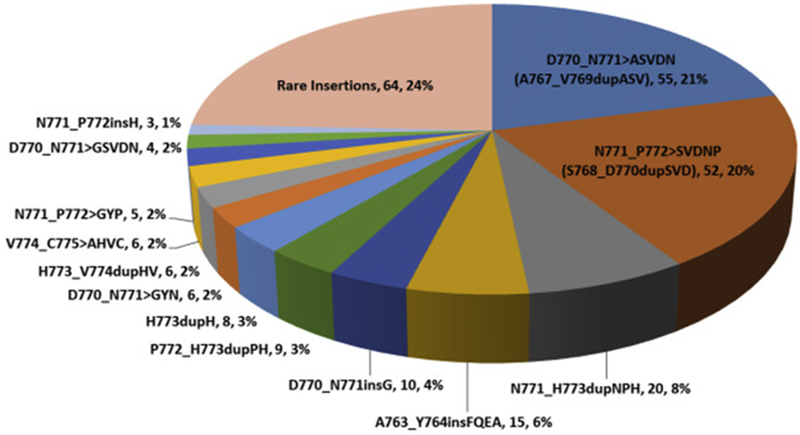
Frequency of Unique EGFR Exon 20 Insertions Detected by Comprehensive Genomic Profiling. Each alteration is shown as: Insertion (Alternative nomenclature), number of cases, frequency.
Figure 4.
A: Co-Occurring Genomic Alterations With a Frequency > 5% in NSCLC Harboring EGFR Exon 20 Insertions.
B: Longtail of frequently co-altered genes in NSCLC with EGFRex20ins L858R, ex19del. Only cases without co-occurring T790M mutation are shown: EGFR ex20ins (n=260), L858R (n=542), ex19del (n=776). Dark shading on the EGFR bars indicate co-occurring EGFR amplification.
Table 2:
Comparison of Molecular, Pathologic and Clinical Characteristics of EGFR-WT, EGFR-mutant (E19del/L858R positive / T790M negative) and EGFR ex20ins NSCLC. Mann Whitney test for age. Chi-square tests (2-sided, alpha <0.01) used for sex, mutation data, histology (Adenocarcinoma vs all others). TMB (tumor mutational burden) by category: 2-way ANOVA, Bonferroni posttest for significance.
| EGFR-WT NSCLC | EGFR-mutant NSCLC (E19del and L858R – T790M neg) | EGFR ex20ins NSCLC | p-value (EGFR-WT vs. EGFR ex20ins) | p-value (EGFR-mutant E19del and L858R vs. ex20ins) | p-value (EGFR-WT vs. EGFR-mutant E19del and L858R – T790M neg) | |
|---|---|---|---|---|---|---|
| Total Cases (N) | 12,551 | 1,318 | 260 | |||
| Median Age, years (range) | 65 (6-99) | 65 (25-95) | 63 (14-90) | 0.0007 | 0.02 | 0.07 |
| Sex, F/M (%F) | 6,246/6,30 4 (50%) |
888/430 (67%) | 161/99 (62%) | <0.0001 | 0.089 | <0.0001 |
| Histologic Subtype | ||||||
| Adenocarcinoma | 8,572 (68%) | 1,149 (87%) | 235 (90%) | <0.0001 | 0.15 | <0.0001 |
| Squamous/Adeno squamous | 1,835 (15%) | 50 (3.8%) | 0 | |||
| NSCLC-NOS | 2,027 (16%) | 115 (8.7%) | 23 (8.8%) | |||
| Sarcomatoid | 117 (0.9%) | 4 (0.3%) | 2 (0.8%) | |||
| Frequency of Co-Occurring Genomic Alterations | ||||||
| Concurrent EGFR Copy Number Gain | 355 (2.8%) | 311 (24%) | 57 (22%) | <0.0001 | 0.63 | <0.0001 |
| Concurrent TP53 alteration | 7,748 (62%) | 844 (64%) | 146 (56%) | 0.0773 | 0.0163 | 0.107 |
| Concurrent RB1 alteration | 771 (6.1%) | 133 (10%) | 28 (11%) | 0.0035 | 0.7413 | <0.0001 |
| Concurrent CDKN2A/2B alteration | 3,222 (26%) | 317 (24%) | 57 (22%) | 0.1939 | 0.5232 | 0.2113 |
| TMB (mutations per MB) Median | 8.1 | 3.6 | 3.6 | <0.0001 | 0.31 | <0.0001 |
| TMB Low (<5) | 3,785 (30%) | 832 (63%) | 179 (69%) | <0.01 | NS | <0.01 |
| TMB Intermediate Low (5-10) | 3,470 (28%) | 400 (30%) | 69 (27%) | <0.01 | NS | <0.01 |
| TMB Intermediate High (10-20) | 3,325 (26%) | 82 (6.2%) | 10 (3.8%) | <0.01 | NS | <0.01 |
| TMB High (>20) | 1,971 (16%) | 4 (0.3%) | 2 (0.8%) | NS | NS | NS |
Discussion
Herein we present the largest known dataset of EGFRex20ins NSCLC, where 64 unique EGFRex20ins sequence alterations are identified, reflecting the diversity of these EGFR driver mutations, which comprise 12% of EGFR-mutant NSCLC and 1.8% of all NSCLC in our series. With approximately 222,000 newly diagnosed lung cancer cases annually in the United States alone, this corresponds to about 3,000 newly diagnosed EGFRex20ins cases per year in the US [16].
EGFRex20ins were detected in 12% of EGFR-mut NSCLC cases, which is higher than previously reported in smaller single institution series (~4-10% frequency). Prior record of molecular testing was not available for the majority of cases. Several reasons may explain the higher frequency of EGFRex20ins detected. This may reflect referral bias with enrichment for lung cancers from never smoking patients. However, for other common EGFR alterations, frequencies detected were comparable to those reported in the literature (15.5% of all NSCLC cases). Thus, the higher frequency of EGFRex20ins is likely rather due to advancement of sequencing technology and improved detection of these alterations at lower mutant allele frequency by next-generation sequencing. Mutation specific assays in some previous studies also likely underestimate the number of EGFRex20ins given the diversity of EGFRex20ins we detected by next-generation sequencing.
A previous study of 27 cases at a single institution described 13 unique EGFRex20ins variants[6]. This dataset herein reports 263 cases including 64 unique EGFRex20ins variants with concomitant molecular alterations and tumor mutational burden (TMB), with a comparison of these co-occurring alterations to a large dataset of EGFR-wild-type and canonical EGFR-mutant NSCLC.
Sensitivity of the EGFR L858R and exon 19 deletions to EGFR TKIs is mediated by enhanced intrinsic affinity for EGFR TKI versus substrate ATP. Structural modeling and binding affinity studies of representative EGFR exon 20 insertions showed that 1st generation EGFR TKI binding affinity is similar to that of wild-type EGFR[9]. Identifying the spectrum of EGFRex20ins where next-generation EGFR TKI can fit into a sterically hindered EGFR exon 20 binding pocket is critical to demonstrating anti-tumor activity and improving clinical outcomes in patients NSCLC harboring diverse EGFRex20ins[9, 15, 16]
The majority of EGFRex20ins were detected in lung adenocarcinoma, though they were also found in a limited number of lung cancers with sarcomatoid histology. In this retrospective series, these insertions were detected at a higher frequency than reported in smaller series suggesting that EGFRex20ins are more prevalent than previously reported. Though this dataset represents patients that were selected specifically by their physician to be tested for analysis, which may not represent a random cross-section of NSCLC, the higher frequency of EGFRex20ins reported could be due in part to comprehensive sequencing of EGFR compared to limitations of non-comprehensive assays performed in prior studies. Nevertheless, EGFRex20ins were the third most common EGFR activating mutation following exon19del and L858R, and occur at approximately the same frequency as ROS1 gene fusions (~1% in our dataset; data not shown) and BRAF V600E mutations in NSCLC (~2% in our dataset; data not shown) where targeted therapies are approved with high rates of response and lengthy progression-free survival [17, 18]. Available treatment outcomes demonstrated lack of response to first- and second-generation EGFR TKIs such as erlotinib, gefitinib and afatinib, consistent with other series, and highlighting the need for new strategies to target EGFRex20ins [19]. The EGFR A763_Y764insFQEA insertion, for which sensitivity to first- and second-generation EGFR TKIs has been described, only comprised 6% of all EGFRex20ins in our series (thus representing less than 1% of all EGFR mutations). Although we detected over 60 EGFRex20ins, the most frequently occurring, D770_N771>ASVDN, N771_P772>SVDNP, and N771_H773dupNPH, represent about 50% of all detected EGFRex20ins in this series (Figure 3). Studies of newer generation EGFR TKIs demonstrating pre-clinical and clinical efficacy against these particular EGFRex20ins would thus impact the greatest number of patients. More recently, poziotinib has demonstrated robust pre-clinical and clinical activity; a confirmed response rate of 64% in EGFRex20ins NSCLC was noted in a phase 2 trial, albeit in a limited number of patients[12]. Poziotinib, due to it’s small size, can circumvent steric changes resulting from the exon 20 insertion to more effectively bind to mutant EGFR. Further understanding the spectrum of EGFRex20ins is important, as it is likely that among the 64 unique insertions we detected, there will be differential sensitivity to poziotinib and similar drugs due to the size and configuration of the drug binding pocket based on the specific EGFRex20ins variant.
The most common genomic alterations co-occurring with EGFRex20ins were tumor suppressor (e.g. TP53 and RB1) and cell cycle alterations (e.g. CDKN2A/2B). These occurred at comparable frequency in NSCLCs samples with EGFR exon 19del and EGFR L585R without EGFR T790M (Table 2, Figure 4). Since EGFR T790M is the most common acquired resistance mutation to first and second generation EGFR TKIs, these were excluded for this comparison to control as best as possible for additional genomic aberrations resulting from acquired EGFR TKI resistance. As is typical of other oncogenic drivers, the vast majority of EGFRex20ins were mutually exclusive of other putative oncogenic drivers in NSCLC such as canonical EGFR, KRAS, BRAF, MET, and HER2 mutations, MET amplification, and ALK, ROS1 and RET fusions.
EGFR amplification was detected in 22% of EGFRex20ins-positive NSCLCs, which is comparable to other EGFR mutations in our dataset when cases with co-occurring T790M are excluded (EGFR ex19del 23%, L858R 25%) and higher than EGFR-WT NSCLC (Figure 4, Table 2), as noted in other studies in EGFR-mutant NSCLC[20]. Treatment with EGFR monoclonal antibodies based on EGFR amplification and overexpression has modestly improved clinical outcomes of squamous NSCLC, and cetuximab added to afatinib appears to increase response rates compared to afatinib alone in patients with EGFR exon 19 deletion or L858R[21, 22]. Small series have described activity of the addition of cetuximab to EGFR TKIs in EGFRex20ins NSCLC[23, 24].
Another important aspect of study is analysis of tumor mutational burden (TMB), which has not been previously reported for EGFRex20ins. Immune checkpoint inhibition with PD-1 and PD-L1 antibodies has revolutionized the treatment of NSCLC and improved survival outcomes for many lung cancer patients[25]. However, patients with EGFR-mutant NSCLC appear to derive less clinical benefit from PD-1/PD-L1 blockade and in a recent meta-analysis do not have an overall survival benefit compared to chemotherapy[26]. In addition to PD-L1 expression, other features such as TMB appear to also be associated with clinical benefit to PD-1 / PD-L1 antibodies, potentially relating to an increase in neoantigen specific T-cell activity [27, 28]. TMB measured by comprehensive genomic profiling is comparable to measurements by whole exome sequencing and is typically higher in smoking associated lung cancer due to tobacco carcinogenesis[14, 29]. Our study shows comparable TMB in EGFRex20ins cases to classic (exon 19 del or L858R) EGFR-mutant NSCLC that is lower than EGFR-WT NSCLC, likely reflecting non-tobacco associated carcinogenesis (Table 2). Less than 4% (10/263) of EGFRex20ins-positive cases analyzed had intermediate-high (10-20 mutations/Mb) TMB, and only 0.7% (2/263) cases had high (>20 mutations/Mb) TMB (Table 2). Overall, low TMB suggests comparable lack of benefit to single agent PD-1 / PD-L1 antibodies in EGFRex20ins NSCLC as in NSCLC harboring more common EGFR mutations.
This study represents the largest series of EGFRex20ins with over 60 unique EGFR exon 20 insertions detected. With next-generation EGFR TKIs demonstrating potential activity in EGFRex20ins-positive NSCLC identifying patients with these molecular aberrations is paramount [5, 8, 11, 30, 31]. In light of newly available combination trials and clinical development of fourth-generation EGFR TKIs targeting EGFRex20ins, comprehensive genomic profiling to detect diverse EGFRex20ins in NSCLC is warranted.
Supplementary Material
Supplementary Table 1: Clinical Outcomes of EGFR Exon 20 Insertion NSCLC Treated with EGFR TKI.
Acknowledgements
This work was supported by a Paul Calabresi Career Development Award in Clinical Oncology National Institutes of Health grant 5 K12 CA 138464 (JWR).
Conflict of Interest: GMF, RM, PJS, JSR, VAM, SMA, and ABS are employees and have equity interest in Foundation Medicine, Inc. SIO received personal fees from AstraZeneca, Pfizer, Roche, Takeda, Novartis, Foundation Medicine outside the submitted work. NP received personal fees from AstraZeneca. NP received personal fees from Advisor & Honorarium from AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Eli Lilly , Foundation Medicine, Guardant360, MSD, Novartis, NovellusDx, Pfizer, Roche, and Takeda outside the submitted work. DRG received grants and other from AstraZeneca, other from Ariad, other from Bayer, grants from Bristol-Myers Squibb, other from Boehringer-Ingelheim, other from Celgene, grants and other from Clovis, grants and other from Genentech, other from Guardant Health, grants from Johnson&Johnson, grants and other from Lilly, other from Liquid Genomics, grants and other from Merck, other from Mirati, grants and other from Novartis, other from Peregrine, other from Pfizer, other from Roche Diagnostics, other from Synta, other from Trovogene outside the submitted work. JWR received personal fees from Takeda, personal fees from Celgene, personal fees from Clovis, personal fees from AbbVie, personal fees from Medtronic, grants from Merck, grants from AstraZeneca, grants from Novartis, grants from Millenium outside the submitted work.
Funding: This work was supported by the National Institutes of Health grant 5 K12 CA 138464 (JWR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mok TS, et al. , Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine, 2009. 361(10): p. 947–57. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, et al. , Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The lancet oncology, 2012. 13(3): p. 239–246. [DOI] [PubMed] [Google Scholar]
- 3.Sequist LV, et al. , Phase III Study of Afatinib or Cisplatin Plus Pemetrexed in Patients With Metastatic Lung Adenocarcinoma With EGFR Mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2013. 31(27): p. 3327–3334. [DOI] [PubMed] [Google Scholar]
- 4.Yang JC, et al. , Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol, 2015. 16(7): p. 830–8. [DOI] [PubMed] [Google Scholar]
- 5.Jia Y, et al. , EGF816 Exerts Anticancer Effects in Non-Small Cell Lung Cancer by Irreversibly and Selectively Targeting Primary and Acquired Activating Mutations in the EGFReceptor. Cancer Res, 2016. 76(6): p. 1591–602. [DOI] [PubMed] [Google Scholar]
- 6.Oxnard GR, et al. , Natural history and molecular characteristics of lung cancers harboring EGFR exon 20 insertions. J Thorac Oncol, 2013. 8(2): p. 179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arcila ME, et al. , EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther, 2013. 12(2): p. 220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riess JW, F. N, Martin M, Orme J, Staniszewska A, Menard L, Cuomo ME, O’Neill D, Ward R, Finlay R, McKerrecher D, Cheng M, Vang D, Tsai R, Ye C, Keck JG, Gandara DR, Mack PC, Cross D. , Antitumor activity of osimertinib in NSCLC harboring EGFR exon 20 insertions. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 2017. 35(15): p. 9030. [Google Scholar]
- 9.Yasuda H, et al. , Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med, 2013. 5(216): p. 216ra177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Floc’h N, et al. , Antitumor Activity of Osimertinib, an Irreversible Mutant-Selective EGFR Tyrosine Kinase Inhibitor, in NSCLC Harboring EGFR Exon 20 Insertions. Mol Cancer Ther, 2018. 17(5): p. 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francois Gonzalvez XZ, Huang Wei-Sheng, Baker Theresa E., Ning Yaoyu, Wardwell Scott D., Nadworny Sara, Zhang Sen, Das Biplab, Gong Yongjin, Greenfield Matthew T., Jang Hyun G., Kohlmann Anna, Li Feng, Taslimi Paul M., Tugnait Meera, Xu Yongjin, Ye Emily Y., Youngsaye Willmen W., Zech Stephan G., Zhang Yun, Zhou Tianjun, Narasimhan Narayana I., Dalgarno David C., Shakespeare William C. and Rivera Victor M., AP32788, a potent, selective inhibitor of EGFR and HER2 oncogenic mutants, including exon 20 insertions, in preclinical models. Cancer Research, 2016. Abstract 2644. [Google Scholar]
- 12.Robichaux JP, et al. , Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med, 2018. 24(5): p. 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frampton GM, et al. , Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol, 2013. 31(11): p. 1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalmers ZR, et al. , Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med, 2017. 9(1): p. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun CH, et al. , Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell, 2007. 11(3): p. 217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey KD, et al. , Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res, 2006. 66(16): p. 8163–71. [DOI] [PubMed] [Google Scholar]
- 17.Shaw AT, et al. , Crizotinib in ROS1-rearranged non-small-cell lung cancer. The New England journal of medicine, 2014. 371(21): p. 1963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Planchard D, et al. , Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol, 2016. 17(7): p. 984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naidoo J, et al. , Epidermal growth factor receptor exon 20 insertions in advanced lung adenocarcinomas: Clinical outcomes and response to erlotinib. Cancer, 2015. 121(18): p. 3212–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soh J, et al. , Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One, 2009. 4(10): p. e7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thatcher N, et al. , Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): an open-label, randomised, controlled phase 3 trial. Lancet Oncol, 2015. 16(7): p. 763–74. [DOI] [PubMed] [Google Scholar]
- 22.Janjigian YY, et al. , Dual Inhibition of EGFR with Afatinib and Cetuximab in Kinase Inhibitor-Resistant EGFR-Mutant Lung Cancer with and without T790M Mutations. Cancer discovery, 2014. 4(9): p. 1036–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsigelny IF, et al. , Molecular determinants of drug-specific sensitivity for epidermal growth factor receptor (EGFR) exon 19 and 20 mutants in non-small cell lung cancer. Oncotarget, 2015. 6(8): p. 6029–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheler J, et al. , Revisiting clinical trials using EGFR inhibitor-based regimens in patients with advanced non-small cell lung cancer: a retrospective analysis of an MD Anderson Cancer Center phase I population. Oncotarget, 2013. 4(5): p. 772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reck M, et al. , Pembrolizumab versus Chemotherapy for PD-Ll-Positive Non-Small-Cell Lung Cancer. N Engl J Med, 2016. 375(19): p. 1823–1833. [DOI] [PubMed] [Google Scholar]
- 26.Lee CK, et al. , Checkpoint Inhibitors in Metastatic EGFR-Mutated Non-Small Cell Lung Cancer-A Meta-Analysis. J Thorac Oncol, 2017. 12(2): p. 403–407. [DOI] [PubMed] [Google Scholar]
- 27.Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbone DP, et al. , First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med, 2017. 376(25): p. 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alexandrov LB, et al. , Mutational signatures associated with tobacco smoking in human cancer. Science, 2016. 354(6312): p. 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elamin YY, R. J, Lam VK, Tsao A, Lu C, Blumenschein G, Kurie J, Brahmer JR, Li S, Chen T, Estrada-Bernal A, Truini A, Nilsson M, Le AT, Tan Z, Zhang S, Doebele R, Politi K, Yang Z, Liu S, Wong KK, Heymach JV., The Preclinical and Clinical Activity of Poziotinib, a Potent, Selective Inhibitor of EGFR Exon 20 Mutant NSCLC. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer, 2017: p. OA12.01. [Google Scholar]
- 31.Horn L, et al. , Continued use of afatinib with the addition of cetuximab after progression on afatinib in patients with EGFR mutation-positive non-small-cell lung cancer and acquired resistance to gefitinib or erlotinib. Lung Cancer, 2017. 113: p. 51–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: Clinical Outcomes of EGFR Exon 20 Insertion NSCLC Treated with EGFR TKI.